Abstract
We have recently shown that the mammalian nucleolar protein Bop1 is involved in synthesis of the 28S and 5.8S ribosomal RNAs (rRNAs) and large ribosome subunits in mouse cells. Here we have investigated the functions of the Saccharomyces cerevisiae homolog of Bop1, Erb1p, encoded by the previously uncharacterized open reading frame YMR049C. Gene disruption showed that ERB1 is essential for viability. Depletion of Erb1p resulted in a loss of 25S and 5.8S rRNAs synthesis, while causing only a moderate reduction and not a complete block in 18S rRNA formation. Processing analysis showed that Erb1p is required for synthesis of 7S pre-rRNA and mature 25S rRNA from 27SB pre-rRNA. In Erb1p-depleted cells these products of 27SB processing are largely absent and 27SB pre-rRNA is under-accumulated, apparently due to degradation. In addition, depletion of Erb1p caused delayed processing of the 35S pre-rRNA. These findings demonstrate that Erb1p, like its mammalian counterpart Bop1, is required for formation of rRNA components of the large ribosome particles. The similarities in processing defects caused by functional disruption of Erb1p and Bop1 suggest that late steps in maturation of the large ribosome subunit rRNAs employ mechanisms that are evolutionarily conserved throughout eukaryotes.
INTRODUCTION
In eukaryotes, ribosome biogenesis is a highly complex process that involves the assembly of four ribosomal RNAs (rRNAs) and about 80 ribosomal proteins into 60S and 40S ribosome subunits within a specialized nuclear compartment, the nucleolus. Our current understanding of ribosome synthesis is largely organized around observed rRNA processing steps. In yeast, three rRNAs (18S, 5.8S and 25S) are derived from a single 35S precursor rRNA (pre-rRNA) transcribed by RNA polymerase I. The primary transcript contains, in addition to sequences present in the mature ribosome, flanking regions designated 5′ and 3′ external transcribed sequences (5′-ETS and 3′-ETS) and intervening sequences termed internal transcribed spacers 1 and 2 (ITS1 and ITS2). The primary transcript is extensively modified by methylation and pseudouridylation and processed via a series of endo- and exonucleolytic reactions. This process ultimately generates the mature 18S, 5.8S and 25S rRNAs and appears to occur in parallel with assembly of ribosomal particles. A large number of trans-acting factors required for ribosome biogenesis have been identified to date, including more than 150 small nucleolar RNAs (snoRNAs) and multiple non-ribosomal proteins (for reviews see 1,2).
Much of our current knowledge of rRNA processing comes from studies in Saccharomyces cerevisiae. Ribosome synthesis in other organisms is not well characterized largely due to the lack of comparably amenable systems for genetic manipulation. However, insights obtained from studies of yeast ribosome biogenesis should have relevance in other eukaryotes. rRNA structure and processing are generally similar among a wide range of eukaryotes (3) and recent sequencing efforts have revealed that many known factors in yeast ribosome biogenesis have counterparts throughout the eukaryotic kingdom.
In order to characterize rRNA processing in mammalian cells, we have employed a strategy of inducible expression of dominant negative mutants to inactivate the function of critical proteins involved in ribosome biogenesis. Using this strategy, we have recently identified and characterized a novel mouse nucleolar protein, Bop1, that is involved in rRNA processing and ribosome assembly (4). Bop1Δ, an N-terminally truncated form of Bop1, acts as a dominant negative inhibitor of Bop1 function in mouse cells, causing a block of synthesis of mature 28S and 5.8S rRNAs and a deficiency in 60S ribosomal subunit assembly (4). Analysis of pre-rRNA processing revealed that in Bop1Δ-expressing cells there is reduced conversion of the 36S precursor to 32S pre-rRNA, which in turn is blocked from being processed to 28S and 12S/5.8S rRNAs, but instead is degraded. Expression of Bop1Δ thus completely blocks formation of mature 28S and 5.8S rRNAs, although it has only a minor effect on synthesis of 18S rRNA and production of 40S subunits (4). Bop1 contains WD repeats, a structural motif involved in intra- and intermolecular protein interactions (5,6).
To understand the function of Bop1 further, we sought to establish the role of its homolog in yeast. In this work we identify YMR049C, the S.cerevisiae homolog of mammalian Bop1, as an essential gene required for synthesis of 25S and 5.8S rRNAs. Together with our previous results, these findings demonstrate that Bop1 is an essential factor in large ribosomal subunit biogenesis, conserved from yeast to mammals both structurally and functionally. In reference to the important role of this protein in eukaryotic ribosome biogenesis, we named the yeast gene ERB1 (yeast BOP1 is an unrelated gene). Bop1/Erb1p is the first protein for which a detailed analysis of pre-rRNA processing effects has been performed in both yeast and mammalian cells. The functional disruption of ERB1 in yeast and Bop1 in mouse cells provides a unique opportunity for parallel analysis of pre-rRNA processing in these two species, thereby contributing to our understanding of mechanisms of ribosome formation in higher eukaryotes.
MATERIALS AND METHODS
Strains, media and genetic methods
Saccharomyces cerevisiae strains used in this work are derivatives of the diploid strain resulting from crossing BY4741 with BY4742 (7). YMS0086 and YDP0135 are isogenic strains (MATa ura3Δ0 his3Δ1 leu2Δ0 lys2Δ0 met15Δ0 erb1::URA3) carrying plasmids pGAL1yBOPfl-HIS/CEN (CEN, HIS3, pGAL1::ERB1) and pDP943-33 (CEN, LEU2, pGAL1::HA-ERB1), respectively. YMS0092 has the endogenous wild-type ERB1 allele and carries plasmid pRS413 (CEN, HIS3), but is otherwise isogenic with YMS0086. Growth and genetic manipulation of yeast cells were performed by established methods (8).
Cloning, disruption and rescue of ERB1
The high fidelity Platinum Pfx DNA polymerase (Gibco BRL) was used for all PCR cloning. A fragment of the ERB1 locus was cloned by PCR from yeast genomic DNA into the PCR-Script vector (Stratagene), using primers GTCTCTAGCAAAGGGAG and GGTGTGCTAGCCAAATCC (–408 and +2126 relative to the start codon of ERB1, respectively). A fragment containing 197 bp of 5′-untranslated sequence and 1167 bp of ERB1 coding sequence was removed with NruI and BglII and replaced with a SmaI–BamHI fragment from YCp50 (9) containing URA3. The resulting plasmid was cut with XmnI to generate a fragment for disrupting ERB1 in a diploid strain (BY4741 crossed with BY4742). Gene disruption was carried out using a standard method (10). Correct integration was confirmed by Southern blot analysis.
A galactose-inducible ERB1 expression plasmid was constructed to allow rescue of haploids inheriting the erb1::URA3 allele. Primers CGGGATCCGGTCAATGATGGCTAAG (BamHI site underlined, start codon of ERB1 in bold) and GGTGTGCTAGCCAAATCC (+2126 relative to the start codon of ERB1) were used to amplify ERB1 from the cosmid SC9796 (ATCC). A 625 bp BamHI–EcoRI fragment of this PCR product containing the 5′-portion of the ERB1 open reading frame was ligated with an EcoRI–SstII fragment from SC9796 that contained the remaining 1797 bp of the ERB1 coding sequence and 617 bp of 3′ flanking sequence. The complete ERB1 coding fragment was cloned into BamHI- and SstII-cut pRS316GAL1 (11; a gift of Dr S. Liebman). The inducible expression cassette from the resulting construct was excised with ApaI and FspI and ligated with an ApaI–SmaI fragment of pRS413 (12) to make pGAL1yBOPfl-HIS/CEN. The diploid strains containing the erb1::URA3 allele were transformed with this plasmid, sporulated and tetrads were dissected on YPGal plates. From several resulting erb1::URA3[pGAL1::ERB1] haploids, which showed identical phenotypes, one strain, YMS0086, was selected for subsequent experiments.
To obtain HA-tagged Erb1p, a synthetic oligonucleotide encoding the HA tag was inserted upstream of the ERB1 start codon in pGAL1yBOPfl-HIS/CEN and the Bsp120I–EagI fragment containing the pGAL1::HA-ERB1 cassette was cloned into Bsp120I-cut pRS319a (13). The resulting expression plasmid pDP943-33 was used to replace pGAL1yBOPfl-HIS/CEN in YMS0086 by plasmid shuffling and one of the His–Leu+ clones, YDP0135, was used for further experiments. The phenotypes of erb1::URA3 complementation with wild-type ERB1 and HA–ERB1 were similar, indicating that HA-tagged Erb1p and wild-type Erb1p are functionally equivalent.
Depletion of Erb1p and protein analysis
Cells grown in YPGal to mid log phase were depleted of Erb1p by shifting to YPD medium. Protein lysates were prepared as described (8), except that the lysis buffer was additionally supplemented with 10% (v/v) protease inhibitor cocktail (Sigma) and 1 mM EDTA. Protein concentration in lysates was determined using the RC DC assay (Bio-Rad) and samples normalized for total protein were separated on a 10% SDS–polyacrylamide gel. Proteins were transferred to a nitrocellulose membrane, stained with Ponceau S to verify equal transfer and probed with HA.11 antibody (Babco), followed by detection with ECL reagents (NEN).
Pulse–chase labeling of RNA
For pulse–chase analysis, 25–30 ml of culture grown to an OD600 of 0.6–0.8 were precipitated, washed with medium lacking methionine or uracil and resuspended in 1 ml of the same medium at 30°C. l-[Methyl-3H]methionine or [5,6-3H]uracil (NEN) was added to 100 µCi/ml and after 2 min labeling the mixture was chased by 4-fold dilution with medium containing 5 mM methionine or 2 mM uracil. Cells were collected from aliquots removed at 0, 2, 8 and 16 min of chase by brief centrifugation and immediately frozen on a dry ice/ethanol mixture. RNA was isolated from frozen cells by acid phenol extraction (14).
Northern and primer extension analyses of steady-state RNA levels
The sequence of oligonucleotide yU3-1 used for detection of U3 snoRNA is ACCAAGTTGGATTCAGTGGC; other oligonucleotides used in northern hybridizations are identical to those previously described (15). Primer extensions were done with 1 µg total RNA and 2 pmol of labeled primer in a 20 µl reaction mixture containing 200 U MMLV reverse transcriptase (Gibco BRL), 1× first strand buffer supplied with the enzyme, 10 mM DTT, 1 mM dNTPs, 10 U RNasin (Promega) and 10% formamide for 40 min at 42°C. Aliquots of 1/15 of the reaction products were separated on a 7.5 M urea–6% polyacrylamide gel alongside a dideoxy sequencing reaction to determine the positions of primer extension stops. All hybridizations and primer extensions were analyzed on a Storm 860 phosphorimager and quantitated using ImageQuant 5.2 software (Molecular Dynamics).
RESULTS
Highly conserved homologs of Bop1 exist throughout the Eukaryota
A database search revealed genes homologous to mouse Bop1 in various eukaryotic species that display high levels of sequence similarity with a well-conserved domain structure (Fig. 1). All these proteins contain WD repeats that are similarly positioned in the C-terminal portion of the protein sequence. Based on an analysis of multiple aligned sequences, we now recognize five distinct WD repeats (Fig. 1), whereas only four WD repeats were initially described in the mouse Bop1 sequence (16). Individual repeats show various levels of divergence from the canonical structure (5). Two additional WD-like segments are identified in some organisms within the gap between WD repeats 1 and 2, but their scores fall below significance level. The absence of critical amino acid residues and lack of evolutionary conservation preclude their assignment as true WD repeats. However, this leads to an interesting speculation that in the ‘primordial’ Bop1 the entire C-terminus (starting with WD1, as shown in Fig. 1) may have consisted entirely of WD repeats. In addition to a well-conserved core, all these proteins contain an N-terminal domain of 50–150 amino acid residues that is not conserved at the sequence level, but has a similar composition predominated by polar residues such as serine, threonine, aspartate and glutamate.
Figure 1.
Homology of the Erb1/Bop1 primary structure in eukaryotes. Organisms and sequence accession nos are: Sce, S.cerevisiae (NP_013764); Spo, Schizosaccharomyces pombe (S54549); Ath, Arabidopsis thaliana (AAD25679); Cel, Caenorhabditis elegans (T26995); Dme, Drosophila melanogaster (T13579); Mmu, Mus musculus (NP_038509); Hsa, Homo sapiens (BAA09473, incomplete sequence). Initial alignment was obtained with ClustalX (34) and then manually edited using GeneDoc (35). Black shading indicates identical residues and conservative substitutions in all sequences; grey shading indicates such residues in >50% sequences. WD repeats 1–5 were identified by a search against the Pfam database (36) in all seven sequences. The WD1 repeat is the closest to the canonical WD40 structure (5). Numbers within parentheses in each sequence indicate lengths of small unique amino acid insertions and the N-terminal domains, which are not conserved at the primary structure level (see text).
These data show that Bop1 is structurally conserved throughout the Eukaryota. All eukaryotic organisms with sequenced genomes appear to contain a single copy of this gene. No homologs of Bop1 were found in Archaea or Eubacteria.
ERB1, the yeast homolog of mammalian Bop1, is an essential gene
The mouse Bop1 protein has been previously shown to participate in the regulation of ribosome biogenesis (4) and cell cycle control (16,17), based on the analysis of its dominant-negative mutant protein, Bop1Δ. To investigate the yeast homolog of Bop1, we replaced the ERB1 allele (the previously uncharacterized open reading frame YMR049C) with a URA3 marker cassette (see Materials and Methods). Tetrad analysis of the resulting diploid strain showed 2:2 segregation of viable to non-viable spores (Fig. 2). All viable spores were Ura–, indicating that disruption of ERB1 is incompatible with vegetative growth. We constructed a centromeric plasmid for conditional expression of ERB1 from the glucose-repressible GAL1 promoter (see Materials and Methods). Expression of Erb1p from this plasmid supported growth of erb1::URA3 strains on YPGal plates, whereas repression of Erb1p with glucose on YPD plates prevented growth (Fig. 2).
Figure 2.
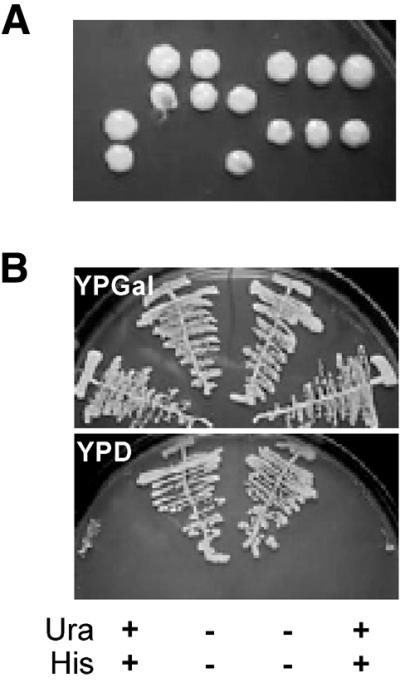
Disruption and rescue of ERB1. (A) Tetrad analysis of an ERB1/erb1::URA3 diploid strain shows 2:2 segregation of viable spores, indicating that ERB1 is essential for viability. (B) Four haploid products from one tetrad obtained by sporulation of an ERB1/erb1::URA3 strain carrying a CEN, HIS3, pGAL1::ERB1 plasmid were streaked on YPGal (top) or YPD (bottom) plates. The Ura and His genotypes of the four clones are indicated at the bottom of the figure, showing that growth of erb1::URA3 haploids is restored by expression of the plasmid-borne, galactose-inducible ERB1.
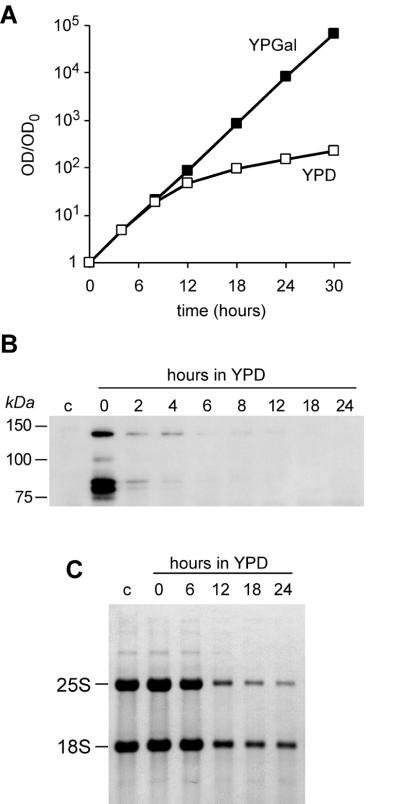
Effects of ERB1 disruption on growth were next studied in strain YDP0135, in which the GAL1 promoter drove expression of HA-tagged Erb1p. The growth rate of this strain on galactose-containing medium was exponential and similar to that of the wild-type strain (Fig. 3A and data not shown). Repression of the promoter by shifting cells to glucose-containing YPD medium led to a significant impediment of growth at 8–12 h after medium shift (Fig. 3A). We analyzed expression of HA–Erb1p in this strain by immunoblotting using antibodies against the HA tag. In cells grown in galactose-containing medium we detected several bands resulting from expression of HA-tagged Erb1p that were absent in the control strain containing non-tagged Erb1p (Fig. 3B). The largest form of HA–Erb1p migrated in SDS–polyacrylamide gels at ∼140 kDa, which is larger than the calculated molecular mass of Erb1p (93 kDa). This anomalous mobility is also observed for mouse Bop1, which migrates at >100 kDa but has a calculated molecular mass of 83 kDa (4). Several other HA-tagged polypeptides that are also detected in the HA–Erb1p strain, including two prominent bands migrating at 80–85 kDa, may represent proteolytically processed forms of Erb1p (Fig. 3B). The amounts of all these proteins were significantly reduced ∼2 h after a shift from galactose- to glucose-containing medium, indicating that Erb1p is indeed depleted (Fig. 3B).
Figure 3.
Effects of Erb1p depletion on growth rate and rRNA. (A) Increase in OD600 was measured in parallel in two YDP0135 cultures, one of which was grown in YPGal medium and the other transferred to YPD at time 0. Cultures were periodically diluted to maintain OD600 < 1.0 throughout the experiment. (B) Protein lysates were prepared from YDP0135 cells (pGAL1::HA-ERB1) at the indicated times after transfer to YPD and from the control YMS0086 strain (pGAL1::ERB1) (lane c), normalized by protein content, and analyzed by immunoblotting with an anti-HA tag antibody. (C) RNA was extracted from the same cultures as used for protein analysis. Equal amounts of total RNA, as determined by absorbance at 260 nm, were separated on a formaldehyde-containing 1.2% agarose gel, transferred to a nylon membrane and stained with methylene blue.
Depletion of Erb1p impairs synthesis of 25S and 5.8S rRNA
Since Bop1 is involved in processing of pre-rRNA in mouse cells (4), we tested whether inhibition of cell growth caused by depletion of Erb1p may be related to defects in pre-rRNA synthesis. Analysis of total RNA prepared from YDP0135 cells after repression of ERB1 showed a decrease in rRNA, with a progressively declining ratio of 25S to 18S rRNA (Fig. 3C), indicating that 25S rRNA is preferentially affected. This result suggested that, like its mouse homolog, Erb1p plays a role in biogenesis of the large ribosome subunit rRNA.
Expression of Bop1Δ, a dominant negative form of Bop1, in mouse cells affects several processing steps in 28S rRNA formation (4). We therefore tested whether depletion of its yeast homolog may affect pre-rRNA processing (Fig. 4). First, we analyzed pre-rRNA processing by pulse–chase labeling of RNA with [methyl-3H]methionine (Fig. 5, upper panels). To show that the observed effects are not due to altered methylation, we performed a similar analysis in cells labeled with [3H]uracil (Fig. 5, lower panels).
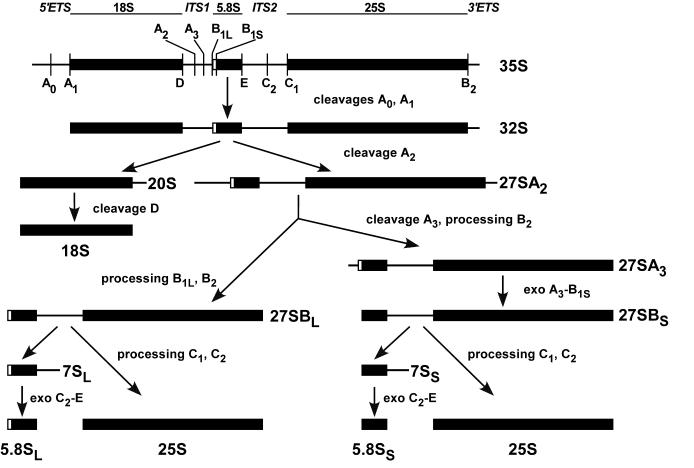
Figure 4.
Pre-rRNA processing in S.cerevisiae. The structure and major sites of processing of the primary 35S pre-rRNA are shown at the top. The external transcribed sequences 5′-ETS and 3′-ETS flank the ends of the mature 18S, 5.8S and 25S rRNA sequences, which are separated by internal transcribed spacers ITS1 and ITS2. Initially, 35S pre-rRNA is cleaved in succession at sites A0 and A1 to generate 32S pre-rRNA. The cleavage of 32S pre-rRNA at site A2 separates the two processing branches that lead to formation of mature rRNAs in the small and large ribosome subunits. The 20S precursor undergoes cleavage at site D after export to the cytoplasm, yielding mature 18S rRNA. The 27SA2 precursor is processed via two alternative pathways to form the long and short forms of 5.8S rRNA and 25S rRNA. The major pathway, which generates the 5.8SS form, proceeds through cleavage of 27SA2 at site A3 within ITS1 and processing at site B2, giving rise to 27SA3, followed by exonucleolytic processing from A3 to site B1S, which generates 27SBS pre-rRNA. In the minor pathway that leads to 5.8SL formation, 27SBL pre-rRNA is generated by processing at site B1L, which occurs by an as yet unidentified mechanism, and processing at B2. The subsequent processing of both 27SB precursors at sites C1 and C2 is probably identical and gives rise to the mature 25S rRNA and 7S pre-rRNAs. The latter are trimmed by exonucleases from the 3′-end to site E, yielding mature 5.8S rRNA species.
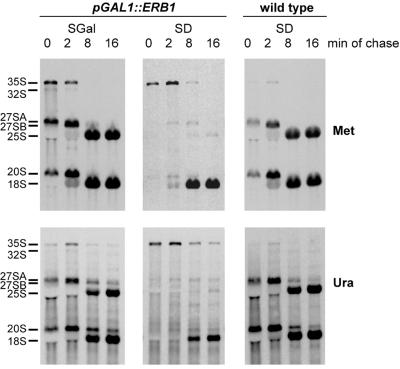
Figure 5.
Pulse–chase analysis of pre-rRNA processing. YMS0086 cells were grown in galactose-containing synthetic SGal medium (left) or transferred to glucose-containing SD medium for 20 h to deplete Erb1p (center). RNA from the control strain YMS0092 containing wild-type ERB1 is shown on the right. Cells were labeled with l-[methyl-3H]methionine (upper) or [5,6-3H]uracil (lower) for 2 min and chased with non-radioactive methionine or uracil, respectively, for 2, 8 and 16 min as described in Materials and Methods. The same total amount of RNA was loaded on each lane for methyl methionine-labeled samples and the same c.p.m. per lane were loaded for uracil-labeled samples. RNA was separated by formaldehyde–agarose gel electrophoresis, transferred to a nylon membrane and visualized by fluorography.
The kinetics of pre-rRNA processing in erb1::URA3 cells grown on galactose-containing medium were essentially similar to those in the wild-type strain (Fig. 5, compare left and right panels). Depletion of Erb1p in glucose-containing medium produced strong defects in pre-rRNA processing. In pulse–chase labeling of non-depleted cells, 27SA and 20S pre-rRNAs, the products of processing of the primary 35S pre-rRNA at sites A0, A1 and A2 (see Fig. 4), are already visible after 2 min labeling (Fig. 5, left, 0 min chase). In contrast, highly labeled 27SA and 20S pre-rRNAs are not apparent at any point in Erb1p-depleted cells, while 35S pre-rRNA takes longer to process and remains detectable up to 8 min of chase (Fig. 5; the center top panel demonstrates this point better because of a more efficient chase with methionine labeling). Although processing of 35S pre-rRNA is delayed after Erb1p depletion, it is not completely blocked: by 8 min of chase most of the material from the labeled 35S pre-rRNA in these cells is processed beyond the A2 step, judging from accumulation of labeled 18S rRNA.
Whereas the transfer of label to 18S rRNA still occurs in Erb1p-depleted cells, virtually no labeled 25S rRNA can be detected after 16 min of chase in these cells (Fig. 5, center panels). In control cells expressing Erb1p on galactose medium formation of 25S and 18S rRNAs is essentially complete at this time (Fig. 5, left), as it is in wild-type cells (Fig. 5, right). The disappearance of label from the 35S precursor and the absence of label in processing intermediates to 25S rRNA in Erb1p-depleted cells indicates that these precursors are degraded, rather than being extremely slowly converted to 25S rRNA. Analysis of low molecular weight RNAs labeled with [3H]uracil also showed loss of synthesis of 5.8S rRNA after Erb1p depletion (data not shown). Thus, Erb1p depletion causes a virtually complete processing block in the branch that leads to formation of 25S and 5.8S rRNAs (Fig. 4) but does not prevent 18S rRNA synthesis, consistent with the decreasing 25S/18S rRNA ratio in these cells (Fig. 3C).
Pre-rRNA processing anomalies in Erb1p-depleted cells
To determine which processing steps are affected by Erb1p depletion, we isolated RNA from depleted, non-depleted and wild-type cells, hybridized them with oligonucleotide probes designed to detect various pre-rRNA intermediates (Fig. 6) and quantified the results on a phosphorimager. Consistent with the results of the pulse–chase experiments, all hybridization probes complementary to pre-rRNA revealed an increase in steady-state levels of 35S pre-rRNA, indicating that the efficiency of cleavage at site A0 in the 5′-ETS is diminished. The product of A0 processing, 33S pre-rRNA, is rapidly converted by cleavage at site A1 to 32S pre-rRNA, whose subsequent processing at site A2 generates the 20S precursor to 18S rRNA and the 27SA2 precursor to 25S and 5.8S rRNAs (Fig. 4). The decrease in levels of 32S pre-rRNA (Figs 5 and 6) suggests that depletion of Erb1p does not significantly affect processing at site A2. As noted above, pulse–chase analysis shows accumulation of significant amounts of labeled 18S rRNA after 8–16 min of chase (Fig. 5), implying that the A1 and A2 processing steps must occur relatively efficiently. However, the steady-state level of the 20S precursor to 18S rRNA, revealed by hybridization with probe y002, undergoes an 8–10-fold decrease after 18–24 h of Erb1p depletion (Fig. 6). Although these two results may at first seem contradictory, they can be explained by the lack of inhibition of further processing of 20S pre-rRNA in Erb1p-depleted cells. Thus, the combination of lowered influx due to delayed processing of 35S pre-rRNA with rapid subsequent conversion to 18S RNA may account for the low steady-state levels of the 20S precursor observed in northern hybridizations (Fig. 6) and the absence of a strongly labeled 20S band in pulse–chase labeling (Fig. 5).
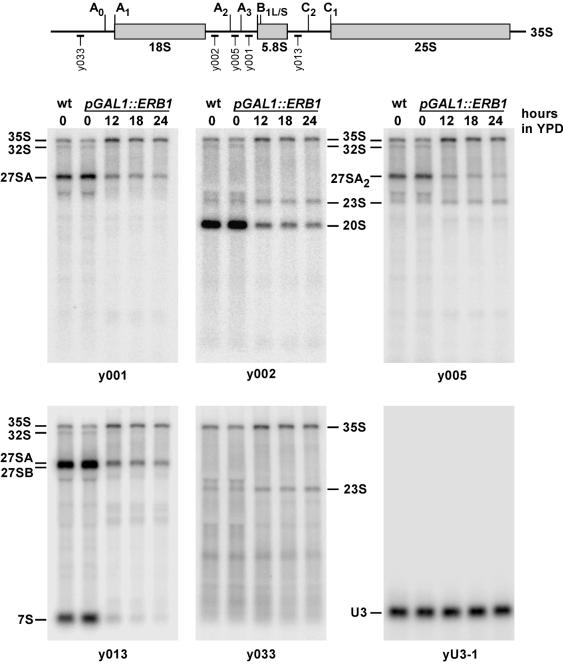
Figure 6.
Northern analysis of pre-rRNA processing effects produced by Erb1p depletion. YDP0135 cells grown in YPGal were transferred to YPD for the time indicated. Equal amounts of total RNA were separated by agarose gel electrophoresis, blotted and hybridized with labeled oligonucleotide probes complementary to different regions of the pre-rRNA transcript (shown at the top). Hybridization of the same membrane with a probe detecting U3 snoRNA, which is not affected by Erb1p depletion, was performed as a loading control. Lane wt, RNA from an isogenic wild-type ERB1 strain YMS0092.
Hybridizations with probes y002, y005 and y033 detected an ∼2-fold accumulation of 23S RNA after Erb1p depletion (Fig. 6). This RNA is considered an aberrant product formed by cleavage of 35S pre-rRNA at site A3, bypassing processing at sites A0–A2, often observed when pre-rRNA processing is perturbed (1). The increase in 23S RNA thus also indicates that cleavage at site A0 is delayed. However, we detect no accumulation of other aberrant RNA species, such as 21S or 22S RNAs (1,15), arguing that once A0 processing is completed, subsequent processing at sites A1 and A2 is not significantly inhibited.
The steady-state levels of the 27SA2 pre-rRNA detected with probe y005 show a 4–7-fold decrease and the combined levels of the 27SA2 and 27SA3 pre-rRNAs detected with probe y001 undergo a 3–5-fold decrease at 18–24 h after shifting pGAL1::ERB1 cells to glucose-containing medium, as compared with non-depleted cells (Fig. 6). A slowed synthesis due to delayed processing at A0 likely contributes to the decrease in 27SA pre-rRNA steady-state levels. It is not clear whether 27SA pre-rRNA may also be destabilized. Interestingly, the ratio between the levels of the two products of A2 cleavage, 27SA2 and 20S pre-rRNAs (Fig. 4), is slightly increased in Erb1p-depleted cells (1.3–2-fold), suggesting that further processing of 27SA2 pre-rRNA may in fact occur with reduced efficiency. The end products of the processing branch that starts with 27SA pre-rRNA (see Fig. 4) are not synthesized in depleted cells: pulse–chase analysis shows a virtually complete absence of 25S and 5.8S rRNAs (see above) and hybridization with probe y013 reveals the disappearance of 7S pre-rRNA, a precursor to 5.8S rRNA (Fig. 6).
We next examined the levels of processing intermediates formed from 27SA pre-rRNA. 27SA2 pre-rRNA is predominantly processed at site A3 to give rise to 27SA3 pre-rRNA and also processed at site B1L to generate 27SBL pre-rRNA. The subsequent processing of 27SA3 RNA involves exonucleolytic trimming from site A3 to site B1S, generating 27SBS pre-rRNA (Fig. 4). To determine how steady-state levels of the 27SA and 27SB pre-rRNAs are affected by Erb1p depletion, we performed primer extension with oligo y013 (Fig. 7). In good agreement with the above hybridization results, the intensity of the primer extension stop at site A2 was reduced (5-fold at 24 h), while the signal at A3 modestly (1.4-fold) increased, indicating an increase in steady-state levels of 27SA3 pre-rRNA. The altered A3/A2 ratio in Erb1p-depleted cells may be, at least in part, due to preferential cleavage of the slowly processed 35S pre-rRNA at site A3, which also generates 23S RNA (see above).
Figure 7.
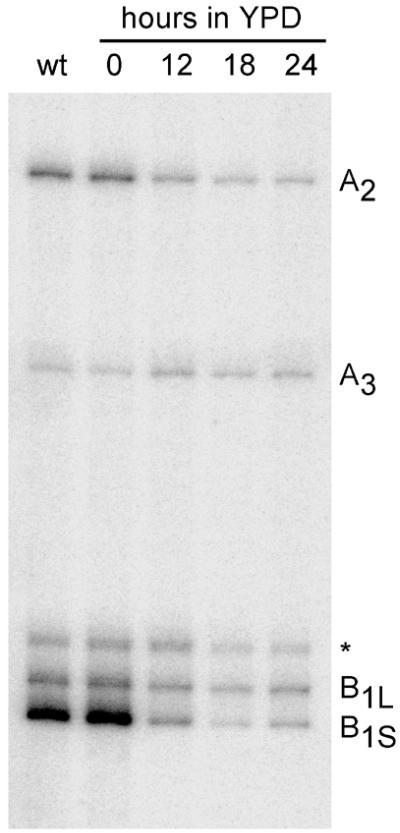
Primer extension analysis of processing in ITS1. Total RNA was isolated from the wild-type ERB1 strain YMS0092 (lane wt) and the pGAL1::HA-ERB1 strain YDP0135 grown on YPGal medium or transferred to YPD medium for the indicated times to deplete Erb1p. Primer extensions were performed on equal amounts of RNA using oligonucleotide y013 (see Fig. 6) as described in Materials and Methods. Positions of primer extension stops (indicated on the right) match the previously determined major processing sites within ITS1 (31); the asterisk indicates an additional stop of unidentified nature observed in this strain.
The intensity of the primer extension stop corresponding to processing sites B1S and B1L decreased after Erb1p depletion (Fig. 7). The effect was especially striking for the B1S stop, which was greatly diminished (13–33-fold reduction), while the stop corresponding to site B1L decreased to a lesser extent (2–3-fold). One possible interpretation of the decrease in the B1S signal relative to B1L is that processing of 27SA3 to 27SBS pre-rRNA may be more inhibited than that of 27SA2 to 27SBL pre-rRNA. However, the strong decrease in the B1S signal is disproportional to the relatively small increase in A3, arguing that this effect is unlikely due to blocked 27SA3 to 27SBS processing. Since no precursors are formed in Erb1p-depleted cells beyond 27SB pre-rRNAs and 27SB pre-rRNAs themselves do not accumulate, the bulk of RNA in these precursors appears to be destined for degradation. This raises the possibility that the stronger decrease in the B1S signal may also be affected by a more rapid degradation of the newly formed 27SBS pre-rRNA as compared with 27SBL.
The pulse–chase data (Fig. 5) are consistent with the interpretation that 27SB pre-rRNA does not accumulate but is degraded in Erb1p-depleted cells. With both types of labeling (Fig. 5, center) only a minute amount of 27S bands can be detected, whereas in control samples of non-depleted cells the 27SB products are clearly visible starting at 2 min of chase and are later converted to 25S rRNAs (Fig. 5, left). The precise pathway of pre-rRNA degradation in Erb1p-depleted cells, however, remains to be determined.
In summary, the most significant effect of Erb1p depletion in yeast cells is the absence of synthesis of 25S rRNA and 7S pre-rRNA from 27SB pre-rRNAs. This is accompanied by under-accumulation of 27SB pre-rRNAs, suggesting that either these pre-rRNAs or their immediate products are destabilized as a result of Erb1p deficiency. No other steps in pre-rRNA processing are completely blocked, although processing of 35S pre-rRNA, and possibly 27SA pre-rRNA, is delayed.
DISCUSSION
In this study we have examined the role of Erb1p, the S.cerevisiae homolog of mammalian Bop1, in processing of rRNA. Gene disruption of ERB1 in yeast showed that this gene is essential for cell viability. In a conditional expression system, depletion of Erb1p caused a virtually complete inhibition of synthesis of 25S and 5.8S rRNA. Synthesis of 18S rRNA in these cells was not blocked, although this rRNA was formed at a reduced rate, in agreement with the observation that defects in 25S/5.8S rRNA synthesis always inhibit to some extent 18S rRNA synthesis (1). The levels of 5S rRNA were not significantly affected by Erb1p depletion (data not shown). These findings are consistent with our previous observations showing that interfering with mouse Bop1 function results in inhibition of 28S and 5.8S rRNA maturation (4) and suggest that Erb1p and Bop1 serve parallel functions in yeast and mammalian cells.
Analysis of pre-rRNA processing by hybridization and primer extension demonstrates that depletion of Erb1p in yeast affects multiple steps in the synthesis of 25S and 5.8S rRNAs. The first affected step appears to be cleavage of 35S pre-rRNA at site A0. Both 20S and 27SA pre-rRNA levels are decreased at least partially due to the delayed A0 processing. The subsequent fate of these precursors is different: while 18S rRNA is synthesized, albeit with a reduced efficiency, no mature rRNAs are formed from 27SA pre-rRNA. The steady-state levels of different 27S pre-rRNA species decrease to varying degrees, reflecting a complex balance between their synthesis, processing and degradation rates. The most striking is depletion of 27SB pre-rRNA, which occurs largely due to under-accumulation of 27SBS. Further products of 27SB pre-rRNA processing are virtually absent in Erb1p-depleted cells, apparently due to accelerated degradation of either 27SB pre-rRNAs or their derivatives.
A number of yeast proteins have been previously shown to be involved in 25S/5.8S rRNA synthesis (for reviews see 1,2). Notably, depletion of some of these proteins leads to accumulation of 27SB pre-rRNA, apparently because of a delay in its further processing. Examples of such factors include Nip7p (18), Nop56p (19), putative methyltransferases Nop2p (20) and Spb1p (21) and helicases Spb4p (22), Dbp3p (23) and Dbp10p (24). Deficiencies in several other factors appear to increase 27S pre-rRNA turnover. For example, reduced accumulation of 27S pre-rRNA, presumably through increased degradation, was reported for depletion of Nop8p (25); it was not, however, shown whether 27SB pre-rRNA was preferentially affected, as in Erb1p-depleted cells. Under-accumulation of 27SB pre-rRNA was observed in cells depleted for Dbp6p (26), Dbp7p (27) and Nop4p/Nop77p (28,29). In these cells, however, both the B1S and B1L pathways were similarly affected and 27SA3 pre-rRNA appeared to be depleted as well. The pre-rRNA processing defects produced by Erb1p depletion most closely resemble defects observed after depletion of Rlp7p (30). Deficiency of Rlp7p similarly causes an increase in steady-state levels of 27SA3 and a strong depletion of 27SBS, with a little or no effect on 27SBL. Rlp7p was proposed to function by recruiting exonucleases to the 27SA3 precursor. Initiation of exonucleolytic processing at site A3 might also be negatively affected by depletion of Erb1p, although the relatively small increase in 27SA3 in these cells is difficult to interpret unequivocally because the A3/A2 ratio is also altered by preferential cleavage at site A3 in slowly processed 35S pre-rRNA.
While Erb1p depletion could potentially affect processing of 27SA3 to 27SBS, Erb1p is not absolutely required for this step. The intensity of the primer extension stop at site B1S is reduced, but this stop is still detectable after depletion of Erb1p (Fig. 7). Moreover, strains carrying mutations in Rat1p and Xrn1p that are unable to perform normal processing from site A3 to site B1S display a ladder of aberrant RNA forms with 5′-ends extending from B1S to A3 (31). Similar RNAs also accumulate in a temperature-sensitive mutant of the nucleolar protein Ebp2p, which may be involved in 27SA3 processing (32). No such extended forms are observed in primer extension reactions with RNA from Erb1p-depleted cells (Fig. 7). We therefore favor a model in which the strong decrease in the B1S signal occurs primarily through destabilization of 27SBS pre-rRNA. One possible scenario is that Erb1p depletion may lead to misassembly of pre-ribosomal particles, whereby the 5′→3′ exonucleases that enter at site A3 proceed to degrade pre-rRNA beyond their normal stop at site B1S. Exonucleolytic digestion initiating from the ITS1 region has been previously suggested to function as a quality control mechanism that degrades pre-rRNA in incorrectly assembled ribosomes (31). The different mechanism of 5′-end formation in B1L processing (1) could render 27SBL pre-rRNAs less succeptible to exonucleolytic degradation, thus making their levels less affected in Erb1p-depleted cells.
The exact biochemical function of Erb1p is unknown at present. The mammalian homolog Bop1 co-fractionates with 32S pre-rRNA-containing complexes (4) and forms tight, salt-resistant bonds with nucleolar RNPs (D.G.Pestov, unpublished data). Erb1p was recently identified as a component of the 66S RNP complexes isolated by purification of tagged Nop7p, which contain more than 40 proteins and appear to represent intermediates of 60S ribosome subunit assembly in yeast (P.Harnpicharnchai, J.Jakovljevic, E.Horsey, T.Miles, J.Roman, M.Rout and J.Woolford, personal communication). Remarkably, depletion of several other components of these complexes, including Nop7p, Drs1p (33) and Rlp7p (30), also inhibits processing of 27S precursors, suggesting that cooperation between these proteins is critical for proper maturation of 27S pre-rRNA. Erb1p/Bop1 does not display any known RNA-binding motifs, although the presence of WD motifs suggests its participation in protein–protein interactions. Erb1p/Bop1 may therefore serve as an assembly factor that coordinates protein interactions within RNPs in which processing of 27S pre-rRNA (36S/32S pre-rRNA in mammalian cells) takes place. Detailed analysis of interactions between Erb1p and other proteins in the Nop7p-containing complexes may provide important insights into the molecular mechanisms of formation of the 60S ribosome particle.
There is substantial similarity between effects on pre-rRNA processing caused by depletion of Erb1p in yeast cells and transdominant mutants of Bop1 in mouse cells (4). Interference with function of both genes similarly blocks formation of two components of large ribosome subunits, 25S and 5.8S rRNAs in yeast and 28S and 5.8S rRNAs in mammalian cells. The functional, as well as structural (Fig. 1), conservation between mouse Bop1 and yeast Erb1p strongly suggests that these proteins perform similar functions in pre-rRNA processing and thus are most likely orthologous. Unfortunately, our efforts to complement Erb1p deficiency with mammalian Bop1 have been unsuccessful thus far. Despite the use of different vectors, we could not detect any expression of full-length mouse Bop1 in yeast cells, suggesting that either some presently unidentified feature of the Bop1 cDNA prevents its efficient expression or the mammalian protein is rapidly degraded in yeast cells.
Disruption of Bop1 and Erb1p functions presents a unique opportunity for comparative analysis of pre-rRNA processing mechanisms in yeast and mammalian cells. Like its yeast counterpart, Bop1 has been previously implicated in processing within the ITS1 of mouse cells (4). In mammalian cells Bop1 functions were analyzed by using inducible expression of transdominant mutant Bop1Δ (4). This analysis showed that inhibition of Bop1 function slows early steps in pre-rRNA processing, but most severely affects processing of 36S and 32S pre-rRNAs, which are structurally homologous to 27SA and 27SB pre-rRNAs in yeast. In a striking similarity to the effects of Erb1p depletion in yeast, Bop1Δ causes accumulation of 36S pre-rRNA and the absence of products of processing of 32S pre-rRNA (4). These studies thus demonstrate a key role of Erb1p/Bop1 in several processing events that lead to synthesis of 25S/28S and 5.8S rRNA in diverse eukaryotic organisms.
We initially identified the N-terminally truncated form of Bop1, Bop1Δ, in a genetic selection for reversible inhibitors of the cell cycle in mammalian cells (16). Remarkably, induction of Bop1Δ causes cell cycle arrest in asynchronously growing cells within one cell cycle, but it does not decrease cell viability (17). In contrast, yeast cells depleted of Erb1p continue growing at an unaltered rate for several generations, gradually slowing down only after ∼8 h of depletion (Fig. 3). These kinetics of growth inhibition are very similar to those observed for a number of other yeast proteins whose depletion blocks rRNA synthesis.
Does Bop1 have an additional function in mammalian cells related to cell cycle control that is lacking in Erb1p? While we cannot rule out this possibility, it is notable that the Bop1 sequence does not exhibit any obvious additional domains that could account for such a function (Fig. 1). It is possible that the difference may rather be in the fundamentally different ways that mammalian and yeast cells respond to defects in ribosome biosynthesis. The slow decline in proliferation rate of yeast cells most likely results from gradual ribosome depletion caused by distribution of pre-existing ribosomes between dividing cells and their naturally occuring degradation. In contrast, mammalian cells may have evolved a special system that monitors defects in ribosome biogenesis and blocks cell cycle progression when they are detected. We have recently proposed that this response to ‘nucleolar stress’ may be a built-in protective mechanism that prevents DNA synthesis under sub-optimal conditions, thereby reducing the chances of acquiring mutations that could be deleterious in the context of a multicellular organism (17). This hypothesis is supported by the findings that inactivation of p53 abolishes the stringent cell cycle block caused by Bop1Δ expression and allows for slow proliferation of cells despite a pre-rRNA processing block (17). These data suggest that mammalian cells possess a special checkpoint mechanism linked with ribosome biosynthesis which at least partially depends on p53. It will be extremely interesting to determine whether functional disruption of other proteins that participate in ribosome biogenesis may lead to similar cell cycle effects in mammalian cells. These studies will require broad application of the knowledge of ribosome biogenesis gained in the yeast system. The conservation of function of the proteins involved, as this study of Erb1p/Bop1 illustrates, should facilitate this exploration.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Dr Sue Liebman for invaluable advice and help and Dr John Woolford for his permission to mention data prior to publication. This work was supported by NIH grant CA52220 to L.F.L.
References
- 1.Venema J. and Tollervey,D. (1999) Ribosome synthesis in Saccharomyces cerevisiae. Annu. Rev. Genet., 33, 261–311. [DOI] [PubMed] [Google Scholar]
- 2.Kressler D., Linder,P. and de la Cruz,J. (1999) Protein trans-acting factors involved in ribosome biogenesis in Saccharomyces cerevisiae. Mol. Cell. Biol., 19, 7897–7912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eichler D.C. and Craig,N. (1994) Processing of eukaryotic ribosomal RNA. Prog. Nucleic Acid Res. Mol. Biol., 49, 197–239. [DOI] [PubMed] [Google Scholar]
- 4.Strezoska Z., Pestov,D.G. and Lau,L.F. (2000) Bop1 is a mouse WD40 repeat nucleolar protein involved in 28S and 5.8S rRNA processing and 60S ribosome biogenesis. Mol. Cell. Biol., 20, 5516–5528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Neer E.J., Schmidt,C.J., Nambudripad,R. and Smith,T.F. (1994) The ancient regulatory-protein family of WD-repeat proteins. Nature, 371, 297–300. [DOI] [PubMed] [Google Scholar]
- 6.Smith T.F., Gaitatzes,C., Saxena,K. and Neer,E.J. (1999) The WD repeat: a common architecture for diverse functions. Trends Biochem. Sci., 24, 181–185. [DOI] [PubMed] [Google Scholar]
- 7.Brachmann C.B., Davies,A., Cost,G.J., Caputo,E., Li,J., Hieter,P. and Boeke,J.D. (1998) Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast, 14, 115–132. [DOI] [PubMed] [Google Scholar]
- 8.Adams A., Gottschling,D.E., Kaiser,C.A. and Stearns,T. (1998) Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 9.Rose M.D., Novick,P., Thomas,J.H., Botstein,D. and Fink,G.R. (1987) A Saccharomyces cerevisiae genomic plasmid bank based on a centromere-containing shuttle vector. Gene, 60, 237–243. [DOI] [PubMed] [Google Scholar]
- 10.Rothstein R.J. (1983) One-step gene disruption in yeast. Methods Enzymol., 101, 202–211. [DOI] [PubMed] [Google Scholar]
- 11.Liu H., Krizek,J. and Bretscher,A. (1992) Construction of a GAL1-regulated yeast cDNA expression library and its application to the identification of genes whose overexpression causes lethality in yeast. Genetics, 132, 665–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sikorski R.S. and Hieter,P. (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics, 122, 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sikorski R.S. and Boeke,J.D. (1991) In vitro mutagenesis and plasmid shuffling: from cloned gene to mutant yeast. Methods Enzymol., 194, 302–318. [DOI] [PubMed] [Google Scholar]
- 14.Schmitt M.E., Brown,T.A. and Trumpower,B.L. (1990) A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res., 18, 3091–3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Allmang C., Mitchell,P., Petfalski,E. and Tollervey,D. (2000) Degradation of ribosomal RNA precursors by the exosome. Nucleic Acids Res., 28, 1684–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pestov D.G., Grzeszkiewicz,T.M. and Lau,L.F. (1998) Isolation of growth suppressors from a cDNA expression library. Oncogene, 17, 3187–3197. [DOI] [PubMed] [Google Scholar]
- 17.Pestov D.G., Strezoska,Z. and Lau,L.F. (2001) Evidence of p53-dependent cross-talk between ribosome biogenesis and the cell cycle: effects of nucleolar protein Bop1 on G1/S transition. Mol. Cell. Biol., 21, 4246–4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zanchin N.I., Roberts,P., DeSilva,A., Sherman,F. and Goldfarb,D.S. (1997) Saccharomyces cerevisiae Nip7p is required for efficient 60S ribosome subunit biogenesis. Mol. Cell. Biol., 17, 5001–5015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gautier T., Berges,T., Tollervey,D. and Hurt,E. (1997) Nucleolar KKE/D repeat proteins Nop56p and Nop58p interact with Nop1p and are required for ribosome biogenesis. Mol. Cell. Biol., 17, 7088–7098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hong B., Brockenbrough,J.S., Wu,P. and Aris,J.P. (1997) Nop2p is required for pre-rRNA processing and 60S ribosome subunit synthesis in yeast. Mol. Cell. Biol., 17, 378–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kressler D., Rojo,M., Linder,P. and de la Cruz,J. (1999) Spb1p is a putative methyltransferase required for 60S ribosomal subunit biogenesis in Saccharomyces cerevisiae. Nucleic Acids Res., 27, 4598–4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.de la Cruz J., Kressler,D., Rojo,M., Tollervey,D. and Linder,P. (1998) Spb4p, an essential putative RNA helicase, is required for a late step in the assembly of 60S ribosomal subunits in Saccharomyces cerevisiae. RNA, 4, 1268–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weaver P.L., Sun,C. and Chang,T.H. (1997) Dbp3p, a putative RNA helicase in Saccharomyces cerevisiae, is required for efficient pre-rRNA processing predominantly at site A3. Mol. Cell. Biol., 17, 1354–1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Burger F., Daugeron,M.C. and Linder,P. (2000) Dbp10p, a putative RNA helicase from Saccharomyces cerevisiae, is required for ribosome biogenesis. Nucleic Acids Res., 28, 2315–2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zanchin N.I. and Goldfarb,D.S. (1999) Nip7p interacts with Nop8p, an essential nucleolar protein required for 60S ribosome biogenesis and the exosome subunit Rrp43p. Mol. Cell. Biol., 19, 1518–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kressler D., de la Cruz,J., Rojo,M. and Linder,P. (1998) Dbp6p is an essential putative ATP-dependent RNA helicase required for 60S-ribosomal-subunit assembly in Saccharomyces cerevisiae. Mol. Cell. Biol., 18, 1855–1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Daugeron M.C. and Linder,P. (1998) Dbp7p, a putative ATP-dependent RNA helicase from Saccharomyces cerevisiae, is required for 60S ribosomal subunit assembly. RNA, 4, 566–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun C. and Woolford,J.L. (1994) The yeast NOP4 gene product is an essential nucleolar protein required for pre-rRNA processing and accumulation of 60S ribosomal subunits. EMBO J., 13, 3127–3135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Berges T., Petfalski,E., Tollervey,D. and Hurt,E.C. (1994) Synthetic lethality with fibrillarin identifies NOP77p, a nucleolar protein required for pre-rRNA processing and modification. EMBO J., 13, 3136–3148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dunbar D.A., Dragon,F., Lee,S.J. and Baserga,S.J. (2000) A nucleolar protein related to ribosomal protein L7 is required for an early step in large ribosomal subunit biogenesis. Proc. Natl Acad. Sci. USA, 97, 13027–13032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Henry Y., Wood,H., Morrissey,J.P., Petfalski,E., Kearsey,S. and Tollervey,D. (1994) The 5′ end of yeast 5.8S rRNA is generated by exonucleases from an upstream cleavage site. EMBO J., 13, 2452–2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huber M.D., Dworet,J.H., Shire,K., Frappier,L. and McAlear,M.A. (2000) The budding yeast homolog of the human EBNA1-binding protein 2 (Ebp2p) is an essential nucleolar protein required for pre-rRNA processing. J. Biol. Chem., 275, 28764–28773. [DOI] [PubMed] [Google Scholar]
- 33.Ripmaster T.L., Vaughn,G.P. and Woolford,J.L. (1992) A putative ATP-dependent RNA helicase involved in Saccharomyces cerevisiae ribosome assembly. Proc. Natl Acad. Sci. USA, 89, 11131–11135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thompson J.D., Gibson,T.J., Plewniak,F., Jeanmougin,F. and Higgins,D.G. (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res., 25, 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nicholas K.B., Nicholas,H.B.,Jr and Deerfield,D.W. (1997) GeneDoc: analysis and visualization of genetic variation. EMBnet News, 4(2). www.hgmp.mrc.ac.uk/embnet.news/vol4_2/genedoc.html [Google Scholar]
- 36.Bateman A., Birney,E., Durbin,R., Eddy,S.R., Howe,K.L. and Sonnhammer,E.L. (2000) The Pfam protein families database. Nucleic Acids Res., 28, 263–266. [DOI] [PMC free article] [PubMed] [Google Scholar]