Abstract
Precision medicine is based on accurate diagnosis and tailored intervention through the use of omics and clinical data together with epidemiology and environmental exposures. Precision medicine should be achieved with minimum adverse events and maximum efficacy in patients with chronic kidney disease (CKD). In this review, the breakthroughs of omics in CKD and the application of systems biology are reviewed. The potential role of transforming growth factor-β1 in the targeted intervention of renal fibrosis is discussed as an example of how to make precision medicine work for CKD.
Key Words: Precision medicine, Chronic kidney disease, Omics, Transforming growth factor-β1, Renal fibrosis
Significance of the Study
• With increasing knowledge of omics and the development of computational models, precision medicine is coming of age in nephrology in order to identify specific biomarkers and develop targeted therapy. The breakthroughs of omics in chronic kidney disease (CKD) and the application of systems biology in the potential role of transforming growth factor-β1 in the targeted intervention of renal fibrosis exemplify the use of precision medicine in CKD.
Introduction
With increasing knowledge of human omics and the development of computational tools for biomedical analysis, precision medicine is unveiled as an accurate diagnostic modality and targeted treatment (using the right dose and time), with minimum adverse events and maximum efficacy [1]. Precision medicine requires the following critical elements: clinical informatics and bioinformatics, computational science, mathematics, and systems biology [2]. In recent years, precision medicine has made great progress; for example, the novel classifications of cancers are based on molecular testing of genetic markers. This has led to targeted drugs such as imatinib for patients with chronic myeloid leukemia who have a BCR-ABL mutation and crizotinib for those with lung cancer who carry the EML4-ALK fusion gene [1]. In kidney transplant patients, urinary-cell messenger ribonucleic acid (mRNA) profile relevant to acute cellular rejection has been identified as a noninvasive diagnostic biomarker, leading to early antirejection therapy and monitoring response of drug treatment [3].
Chronic kidney disease (CKD) is a major health and socioeconomic burden [4]. Renal fibrosis is the common pathway for CKD of various origins, resulting in end-stage renal disease [4,5]. The injury results in local inflammation and the production of proinflammatory cytokines, which contribute to the recruitment of inflammatory cells that synthesize and release profibrotic cytokines. This induces the activation and recruitment of matrix-producing cells and epithelial-to-mesenchymal transition (EMT), which ultimately leads to renal fibrosis in CKD pathogenesis [6]. Transforming growth factor (TGF)-β1 is both a well-known EMT inducer and profibrotic molecule that participates in the pathogenesis of renal fibrosis [6].
In this report, the knowledge and understanding of omics (emerging technologies for analyzing big data) as well as novel diagnostic and therapeutic strategies for CKD are reviewed. The molecular pathways associated with TGF-β1 are discussed as potential therapeutic targets for progressive CKD in order to shed light on the role of precision medicine in CKD management.
Genomic, Epigenetic, and Transcriptional Studies of CKD
Series of studies have reported that specific genetic polymorphisms, epigenetic and transcriptional variations, could increase the risk of CKD [3,5,7]. There are two basic approaches for identifying candidate genes of CKD. First, the genotype-to-phenotype approach: perform unbiased screening to identify the genes which are associated with CKD, and then unveil the mechanistic relevance through experimental studies. Second, the phenotype-to-genotype approach: screen the specific genes which have been confirmed in renal pathogenesis to analyze the polymorphisms, variations on transcription or expression, in the susceptibility of CKD [3].
Genome-wide association studies (GWASs) use genomic variations, termed single nucleotide polymorphisms (SNPs), to identify regions of the genome which are associated with the disease status or a clinical phenotype [5,8]. Multiple GWASs have been conducted to expand the knowledge of genetic variants of CKD which reveal the pathways and mechanisms, and potentially represent the targets for therapeutic interventions [7,9]. For example, GWASs identified the variants in the promoter of the UMOD gene, SNP rs4293393, which increased UMOD expression. Uromodulin overexpression led to salt-sensitive hypertension by upregulating Na-K-Cl transporter (NKCC2) phosphorylation, and contributed to renal damage. This mechanism indicated that pharmacological inhibition of NKCC2 would be more effective in lowering blood pressure in hypertensive patients who were homozygous for UMOD promoter risk variants than in other hypertensive patients, and the uromodulin might be a therapeutic target for lowering blood pressure and preserving renal function [10].
The epigenetic state determines the expression and transcription of genes by DNA methylation of CpG dinucleotides, nucleosomal histone modifications, and other mechanisms [11,12]. Different from primary DNA sequences, epigenetic modifications are reversible and susceptible, depending on different time, locations, nutritional status, and environmental exposure; therefore these factors can be modified to suppress disease progression in clinical practice. The DNA methylation changes in CKD patients have shown both DNA hypomethylation and hypermethylation [13]. The hypermethylation downregulated the expression of the renoprotective gene KLOTHO and was associated with CKD progression; thus, KLOTHO might be a potential epigenetic drug target to suppress CKD progression [14].
The microRNAs (miRNAs) are short (20–23 nucleotides in length), noncoding, endogenous, single-stranded RNA molecules that regulate target mRNAs at the posttranscriptional level. miRNAs inhibit protein synthesis, either by inhibition of translation or mRNA degradation [15]. The miRNAs are more stable than mRNA and present in different body fluids, especially in urine, suggesting that miRNAs could serve as ideal biomarkers of CKD of various pathologies [12]. Previous studies have demonstrated that urinary miRNA abundance was variable in CKD (Table 1). miRNA-29 was reported to regulate the expression of collagen genes and extracellular matrix proteins which are involved in modulating renal fibrosis, negatively correlating with tubulointerstitial fibrosis in urine exosome [16]. Identifying the regulation and function of these miRNAs in renal pathogenesis might pinpoint miRNAs as new therapeutic targets for progressive CKD and renal fibrosis.
Table 1.
Main findings of omics associated with renal fibrosis: positive and negative correlations
| Field | Positive correlation | Negative correlation | Suppl. ref. No. |
|---|---|---|---|
| Genomics | IL-18 (+137 GG, −607CC); TGF-β1 (–509 TT); AS (int 2 CC); UMOD (rs12917707; rs12446492); ELMO1 gene; Nox2 gene | TGF- β1 (+869 TT); UMOD (rs13333226, rs12917707); Sirt1 gene | 1 – 10 |
| Epigenomics | DNA methylation (e.g., Klotho promoter); histone modifications (e.g., profibrotic and ER stress-related genes) | apelin-13, KLF4 | 11 – 16 |
| Transcriptomics | miR-192, miR-29, miR-21, miR-150, mRNA (e.g., APE1, AT1R, CXCR4, THBS1, TRIB1) | miR-93, miR-217, miR-200a, miR-26a, mRNA (e.g., BMP7, CD2AP) | 13, 17 – 25 |
| Proteomics | TGF-β1, α-SMA, NGAL, KIM-1, CD147, CXCL1, annexin A1, HE4, NGAL, MBL, MMP-7, MMP-9, CTGF, uVDBP, periostin, CKD273 peptides | HO-1, E-cadherin | 26 – 37 |
| Metabolomics | cystatin C, lipids (e.g., ectopic, oxidized), glycolysis, acetoacetate, phosphorylcholine/choline, H-1 NMR-based metabonomics | pyruvate, glycine, L-carnitine | 12, 38 – 43 |
Proteomics and Metabolomics for CKD
The biggest challenge of studies in proteomics and metabolomics in CKD is the identification of potential biomarkers. The term “biomarker” is defined as a quantitative biological molecule or pathological process that is typically used for the diagnosis of a disease or the assessment of disease activity. Biomarkers of CKD mainly indicate the proteins and metabolites from blood, urine, and kidney biopsy specimens; recently, the molecules from DNA and miRNA analyses can also be biomarkers. The ideal biomarkers of CKD should accurately predict an individual's risk of developing CKD, detect and classify the disease early based on the individual molecular mechanism, and then monitor the response to therapy for further adjustment.
Proteomics serve not only as a screening method to identify potential biomarkers but also as a diagnostic method itself [17]. The majority of urinary proteins are generated by the kidney and, hence, hold substantial information on renal pathogenesis. With ongoing studies on urinary proteome, the abundance of molecular information will replace kidney biopsy [18]. The CKD273 classifier, based on 273 urinary peptides, was well suited for the early detection of CKD and prognosis of progression [19]. The PRIORITY study validated the use of CKD273 to detect diabetic nephropathy (DN) from type 2 diabetic patients, who had no sign of DN with normal estimated glomerular filtration rate and normoalbuminuria. These patients will be randomized to the groups of low-dose spironolactone plus standard treatment and placebo plus standard treatment, aiming at demonstrating the benefit of the proteomics-guided intervention [20].
Podocyte phospholipase A2 receptor (PLA2R) is an antigenic target in autoimmune membranous nephropathy (MN). Anti-PLA2R antibodies in serum and glomerular deposits could serve as diagnostic biomarkers of idiopathic MN [21] and could also differentiate idiopathic MN from secondary MN [22], with high specificity and sensitivity. Moreover, in anti-PLA2R-associated idiopathic MN, serum titers of anti-PLA2R antibodies were positively correlated with proteinuria and disease activity and could predict the remission and relapse of idiopathic MN. Thus, anti-PLA2R antibody titers could have an important role in the selection of patients for immunosuppressive therapy and in monitoring the response to treatment [23].
The human metabolome is the product of multiple physiological and pathophysiological processes and might provide a novel insight into CKD etiology and progression. In contrast to traditional biochemical approaches that focus on a single metabolite, metabolomics involves a series of metabolites to gain an overall understanding of metabolism [24]. Compared with other omics technologies, there are fewer metabolites (3 × 103) than there are genes (2 × 104), transcripts (>106), proteins (>106), and posttranslational modified proteins (>107) [5]. Metabolomic biomarkers of diabetes were reported as sugar metabolites, ketone bodies, free fatty acids, and branched chain amino acids [25]. Distinct phospholipids, sphingolipids, and sphingomyelins in serum could identify DN from type 1 and type 2 diabetic patients. Urinary levels of acylcarnitines and hippuric acid were associated with early kidney damage and reflected alterations in β-oxidation and uremic toxin elimination, respectively [26]. Furthermore, increases in γ-butyrobetaine, citrulline, symmetric dimethylarginine, and kynurenine as well as a decrease in azelaic acid (β-oxidation) indicated the progression from mirco-DN to macro-DN [27].
Systems Biology Brings Forward Precision Medicine for CKD
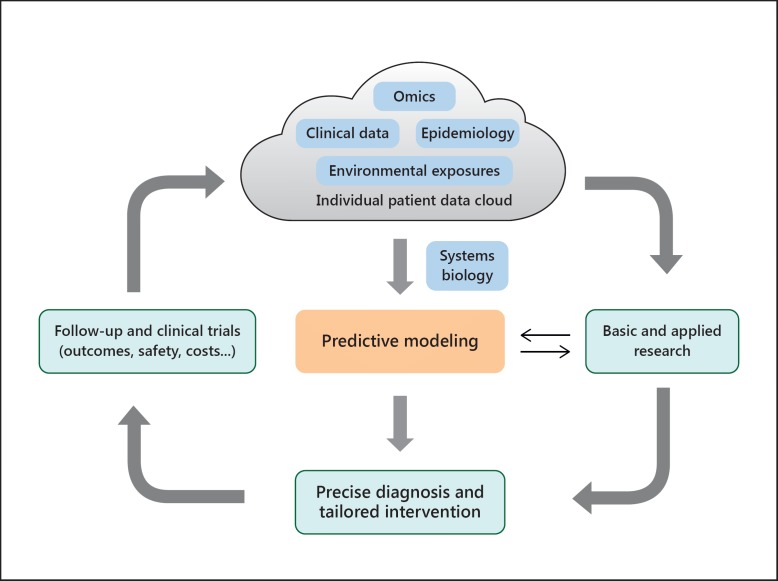
Systems biology is the computational and mathematical modeling of complex biological systems that can integrate the big data in omics as well as clinical phenotypes to gain a mechanistic understanding of disease [28]. Systems biology will help physicians to understand the function of the whole organism in order to expand the scope of precision medicine by establishing the predictive models and achieve the tailored intervention. For the nephrologist, each patient's disease should be analyzed according to data cloud and systems biology to establish diagnosis or classification at the molecular level and select appropriate treatments. Then, the patient's response should be monitored, and clinical trials should be conducted to adjust the predictive models and personalized treatments (Fig. 1) [29]. RNA microarrays have been used to analyze gene expression profiles in renal glomerular and tubular samples from diabetic kidney disease (DKD) patients. Then, weighted gene co-expression network analysis identified 10 modules of genes in tubuli and 12 modules in glomeruli, which indicated that dysregulation of cell proliferation might contribute to the development of DKD, and those genes could be therapeutic targets for DKD [30].
Fig. 1.
Schema of precision medicine for CKD. The figure visualizes the order and complex interconnections of the numerous interacting components which give rise to precision medicine.
The Role of TGF-β1 in Renal Fibrosis
Renal fibrosis is the hallmark of CKD, leading to irreversible loss of tissue and impaired kidney function. Understanding the mechanisms of renal fibrosis would bring new taxonomy of CKD and targeted therapy in order to prevent or delay the progression to end-stage renal disease (Table 1). TGF-β1 plays an important role in the pathogenesis of fibrosis in both the glomerular and interstitial compartments of the kidney via induction of EMT, accumulation of extracellular matrix, and activation of myofibroblasts [6].
TGF-β1 is encoded by the TGFB1 gene located at chromosome 19q13.1. The coding region contains 7 exons and 6 introns encoding for a 390 amino acid polypeptide, which consists of a signal peptide (amino acids 1–29), a latency-associated peptide (amino acids 30–278), and the TGF-β1 domain (amino acids 279–390). A homodimer of the C-terminal 112 amino acid TGF-β1 domain constitutes the biologically active TGF-β1. A previous study demonstrated that 8 SNPs were associated with increased CKD risk [3]. A subset of 179 co-regulated genes, which might present the TGF-β1-driven signature in renal fibrosis, were defined from human DN renal biopsies [31]. The targeted intervention of TGF-β1 and its co-regulated genes might offer novel therapeutic approaches in attenuating the initiation and progression of renal fibrosis.
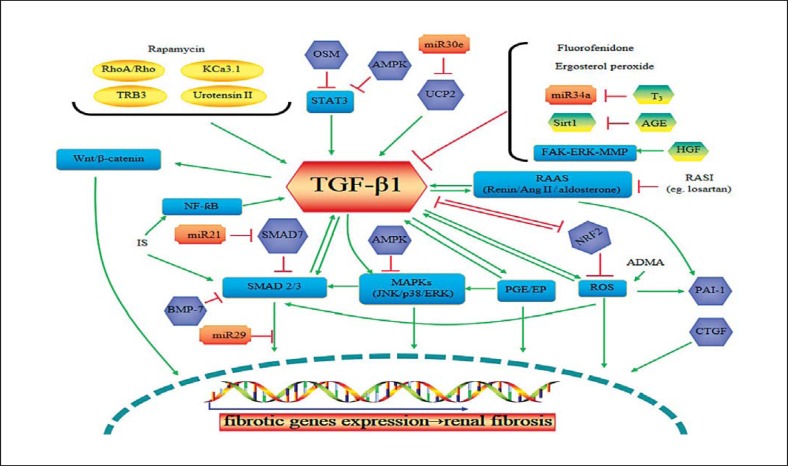
TGF-β1 is a well-known biomarker of CKD. Its elevated level in both the serum and urine of CKD patients turned out to be positively correlated with the level of estimated glomerular filtration rate [32], reflecting the activation of renal fibrotic progression. Molecules and drugs regulate, or are regulated by, TGF-β1 via different signal pathways which could influence the expression of fibrotic related genes that might result in renal fibrosis (Fig. 2) [33,34,35,36,37,38,39]. With the knowledge of TGF-β1-related pathways, CKD might be refined and treated via these underlying molecular mechanisms.
Fig. 2.
The potential targeted intervention of TGF-β1 in renal fibrosis. Green arrows represent upregulation or activation of the downstream molecules and pathways, while red lines represent downregulation or inhibition. TRB3, tribbles homolog; OSM, oncostatin M; AMPK, adenosine monophosphate-activated protein kinase; UCP2, uncoupling protein 2; STAT3, signal transducer and activator of transcription 3; PGE, prostaglandin E; ERK, extracellular signal-regulated kinase; ROS, reactive oxygen species; PAI-1, plasminogen activator inhibitor-1; MAPK, mitogen-activated protein kinase; JNK, c-Jun N-terminal kinase; AGE, advanced glycation end product; FAK, focal adhesion kinase; T3, thyroid hormone; HGF, hepatocyte growth factor; CTGF, connective tissue growth factor; PAI-1, plasminogen activator inhibitor type 1; BMP-7, bone morphogenetic protein-7; ADMA, asymmetric dimethylarginine; NRF2, nuclear factor-erythroid 2-related factor 2; RAAS, renin-angiotensin-aldosterone system.
TGF-β antagonists which reduce or block the activation of TGF-β1, including anti-TGF-β antibodies, the TGF-β receptor antagonist, and the targets of inhibiting TGF-β synthesis, might ameliorate renal fibrosis. Fresolimumab, a human monoclonal antibody that neutralizes all 3 isoforms of TGF-β, is being studied in a phase 2 clinical trial in patients with steroid-resistant primary focal segmental glomerulosclerosis. LY2382770, an antibody that neutralizes the bioactivity of TGF-β1, is also being investigated in a phase 2 clinical trial in DN patients. GW788388, a TGF-β receptor antagonist and small interfering RNA for the TGF-β receptor, could also inhibit TGF-β signaling activity. Pirfenidone, with antifibrotic and anti-inflammatory activities via the inhibition of TGF-β1 synthesis, has been demonstrated to be safe and effective in many disorders such as idiopathic pulmonary fibrosis, multiple sclerosis, and liver fibrosis. Clinical trials of pirfenidone in CKD have been performed in patients with focal segmental glomerulosclerosis and DN. The results suggested that pirfenidone might be an effective agent to slow renal function decline in CKD [40]. In addition, the inhibition of TGF-β signaling by Smad7, bone morphogenetic protein-7 agonists, connective tissue growth factor inhibitors, in vivo delivery of small interfering RNA, as well as all other molecules and pathways in Figure 2 could be potential therapeutic targets to inhibit or slow the progressive loss of kidney function in patients with CKD.
Conclusion
Precision medicine promises to combine omics data with clinical phenotypes to develop new clinical implementation that can predict CKD risks and therapeutic responses in a very precise way. For instance, by understanding the mechanisms of TGF-β1-related renal fibrosis, CKD would be refined by different pathogeneses of renal fibrosis depending on its involved pathway individually, and thus more precise biomarkers and therapeutic strategies would be achieved.
Disclosure Statement
The authors have no conflicts of interest to declare.
References
- 1.Suthanthiran M, Schwartz JE, Ding RC, et al. Urinary-cell mRNA profile and acute cellular rejection in kidney allografts. New Engl J Med. 2013;369:20–31. doi: 10.1056/NEJMoa1215555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jameson JL, Longo DL. Precision medicine – personalized, problematic, and promising. N Engl J Med. 2015;372:2229–2234. doi: 10.1056/NEJMsb1503104. [DOI] [PubMed] [Google Scholar]
- 3.Bjoern T, Michael Z. Contribution of genetics and epigenetics to progression of kidney fibrosis. Nephrol Dial Transplant. 2014;29:72–79. doi: 10.1093/ndt/gft025. [DOI] [PubMed] [Google Scholar]
- 4.Lindquist JA, Mertens PR. Myofibroblasts, regeneration or renal fibrosis – is there a decisive hint? Nephrol Dial Transplant. 2013;28:2678–2681. doi: 10.1093/ndt/gft247. [DOI] [PubMed] [Google Scholar]
- 5.Atzler D, Schwedhelm E, Zeller T. Integrated genomics and metabolomics in nephrology. Nephrol Dial Transplant. 2014;29:1467–1474. doi: 10.1093/ndt/gft492. [DOI] [PubMed] [Google Scholar]
- 6.Carew RM, Wang B, Kantharidis P. The role of EMT in renal fibrosis. Cell Tissue Res. 2012;347:103–116. doi: 10.1007/s00441-011-1227-1. [DOI] [PubMed] [Google Scholar]
- 7.O'Seaghdha CM, Fox CS. Genome-wide association studies of chronic kidney disease: what have we learned? Nat Rev Nephrol. 2012;8:89–99. doi: 10.1038/nrneph.2011.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Witasp A, Ekstrom TJ, Schalling M, et al. How can genetics and epigenetics help the nephrologist improve the diagnosis and treatment of chronic kidney disease patients? Nephrol Dial Transplant. 2014;29:972–980. doi: 10.1093/ndt/gfu021. [DOI] [PubMed] [Google Scholar]
- 9.Kottgen A. Genome-wide association studies in nephrology research. Am J Kidney Dis. 2010;56:743–758. doi: 10.1053/j.ajkd.2010.05.018. [DOI] [PubMed] [Google Scholar]
- 10.Trudu M, Janas S, Lanzani C, et al. Common noncoding UMOD gene variants induce salt-sensitive hypertension and kidney damage by increasing uromodulin expression. Nat Med. 2013;19:1655–1660. doi: 10.1038/nm.3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dwivedi RS, Herman JG, McCaffrey TA, et al. Beyond genetics: epigenetic code in chronic kidney disease. Kidney Int. 2011;79:23–32. doi: 10.1038/ki.2010.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Papadopoulos T, Belliere J, Bascands JL, et al. miRNAs in urine: a mirror image of kidney disease? Expert Rev Mol Diagn. 2015;15:361–374. doi: 10.1586/14737159.2015.1009449. [DOI] [PubMed] [Google Scholar]
- 13.Smyth LJ, McKay GJ, Maxwell AP, et al. DNA hypermethylation and DNA hypomethylation is present at different loci in chronic kidney disease. Epigenetics. 2014;9:366–376. doi: 10.4161/epi.27161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Young GH, Wu VC. Klotho methylation is linked to uremic toxins and chronic kidney disease. Kidney Int. 2012;81:611–612. doi: 10.1038/ki.2011.461. [DOI] [PubMed] [Google Scholar]
- 15.Lorenzen JM, Haller H, Thum T. MicroRNAs as mediators and therapeutic targets in chronic kidney disease. Nat Rev Nephrol. 2011;7:286–294. doi: 10.1038/nrneph.2011.26. [DOI] [PubMed] [Google Scholar]
- 16.Wang B, Komers R, Carew R, et al. Suppression of microRNA-29 expression by TGF-β1 promotes collagen expression and renal fibrosis. J Am Soc Nephrol. 2012;23:252–265. doi: 10.1681/ASN.2011010055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goncalves A, Bertucci F. Clinical application of proteomics in breast cancer: state of the art and perspectives. Med Princ Pract. 2011;20:4–18. doi: 10.1159/000319544. [DOI] [PubMed] [Google Scholar]
- 18.Mischak H. Pro: Urine proteomics as a liquid kidney biopsy: no more kidney punctures! Nephrol Dial Transplant. 2015;30:532–537. doi: 10.1093/ndt/gfv046. [DOI] [PubMed] [Google Scholar]
- 19.Argiles A, Siwy J, Duranton F, et al. CKD273, a new proteomics classifier assessing CKD and its prognosis. PLoS One. 2013;8:e62837. doi: 10.1371/journal.pone.0062837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Siwy J, Schanstra JP, Argiles A, et al. Multicentre prospective validation of a urinary peptidome-based classifier for the diagnosis of type 2 diabetic nephropathy. Nephrol Dial Transplant. 2014;29:1563–1570. doi: 10.1093/ndt/gfu039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Du Y, Li J, He F, et al. The diagnosis accuracy of PLA2R-AB in the diagnosis of idiopathic membranous nephropathy: a meta-analysis. PLoS One. 2014;9:e104936. doi: 10.1371/journal.pone.0104936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dai H, Zhang H, He Y. Diagnostic accuracy of PLA2R autoantibodies and glomerular staining for the differentiation of idiopathic and secondary membranous nephropathy: an updated meta-analysis. Sci Rep. 2015;5:8803. doi: 10.1038/srep08803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ronco P, Debiec H. Pathophysiological advances in membranous nephropathy: time for a shift in patient's care. Lancet. 2015;385:1983–1992. doi: 10.1016/S0140-6736(15)60731-0. [DOI] [PubMed] [Google Scholar]
- 24.Zhao YY, Cheng XL, Wei F, et al. Intrarenal metabolomic investigation of chronic kidney disease and its TGF-β1 mechanism in induced-adenine rats using UPLC Q-TOF/HSMS/MSE. J Proteome Res. 2013;12:692–703. doi: 10.1021/pr3007792. [DOI] [PubMed] [Google Scholar]
- 25.Suhre K, Meisinger C, Doring A, et al. Metabolic footprint of diabetes: a multiplatform metabolomics study in an epidemiological setting. PLoS One. 2010;5:e13953. doi: 10.1371/journal.pone.0013953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van der Kloet FM, Tempels FWA, Ismail N, et al. Discovery of early-stage biomarkers for diabetic kidney disease using ms-based metabolomics (FinnDiane study) Metabolomics. 2012;8:109–119. doi: 10.1007/s11306-011-0291-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hirayama A, Nakashima E, Sugimoto M, et al. Metabolic profiling reveals new serum biomarkers for differentiating diabetic nephropathy. Anal Bioanal Chem. 2012;404:3101–3109. doi: 10.1007/s00216-012-6412-x. [DOI] [PubMed] [Google Scholar]
- 28.Jain KK. Synthetic biology and personalized medicine. Med Princ Pract. 2013;22:209–219. doi: 10.1159/000341794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Duffy DJ. Problems, challenges and promises: perspectives on precision medicine. Brief Bioinform. 2016;17:494–504. doi: 10.1093/bib/bbv060. [DOI] [PubMed] [Google Scholar]
- 30.Tang W, Gao Y, Li Y, et al. Gene networks implicated in diabetic kidney disease. Eur Rev Med Pharmacol Sci. 2012;16:1967–1973. [PubMed] [Google Scholar]
- 31.Brennan EP, Morine MJ, Walsh DW, et al. Next-generation sequencing identifies TGF-β1-associated gene expression profiles in renal epithelial cells reiterated in human diabetic nephropathy. Biochim Biophys Acta. 2012;1822:589–599. doi: 10.1016/j.bbadis.2012.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Levin A, Rigatto C, Barrett B, et al. Biomarkers of inflammation, fibrosis, cardiac stretch and injury predict death but not renal replacement therapy at 1 year in a Canadian chronic kidney disease cohort. Nephrol Dial Transplant. 2014;29:1037–1047. doi: 10.1093/ndt/gft479. [DOI] [PubMed] [Google Scholar]
- 33.Hamzeh MT, Sridhara R, Alexander LD. Cyclic stretch-induced TGF-β1 and fibronectin expression is mediated by β1-integrin through c-Src- and STAT3-dependent pathways in renal epithelial cells. Am J Physiol Renal Physiol. 2015;308:F425–F436. doi: 10.1152/ajprenal.00589.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen KH, Hsu HH, Lee CC, et al. The AMPK agonist AICAR inhibits TGF-β1-induced activation of kidney myofibroblasts. PLoS One. 2014;9:e106554. doi: 10.1371/journal.pone.0106554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Overstreet JM, Samarakoon R, Meldrum KK, et al. Redox control of p53 in the transcriptional regulation of TGF-β1 target genes through SMAD cooperativity. Cell Signal. 2014;26:1427–1436. doi: 10.1016/j.cellsig.2014.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bondi CD, Manickam N, Lee DY, et al. NAD(P)H oxidase mediates TGF-β1-induced activation of kidney myofibroblasts. J Am Soc Nephrol. 2010;21:93–102. doi: 10.1681/ASN.2009020146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu C, Daishan J, Jing W, et al. Prostaglandin E2 EP1 receptor enhances TGF-β1-induced mesangial cell injury. Int J Mol Med. 2015;35:285–293. doi: 10.3892/ijmm.2014.1979. [DOI] [PubMed] [Google Scholar]
- 38.Samarakoon R, Dobberfuhl AD, Cooley C, et al. Induction of renal fibrotic genes by TGF-β1 requires eGFR activation, p53 and reactive oxygen species. Cell Signal. 2013;25:2198–2209. doi: 10.1016/j.cellsig.2013.07.007. [DOI] [PubMed] [Google Scholar]
- 39.Zeisberg M, Hanai J, Sugimoto H, et al. BMP-7 counteracts TGF-β1-induced epithelial-to-mesenchymal transition and reverses chronic renal injury. Nat Med. 2003;9:964–968. doi: 10.1038/nm888. [DOI] [PubMed] [Google Scholar]
- 40.Lee S-Y, Kim SI, Choi ME. Therapeutic targets for treating fibrotic kidney diseases. Transl Res. 2014;165:512–530. doi: 10.1016/j.trsl.2014.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]