Abstract
Objective
The aim of this study was to investigate the possible migration of proliferating peripheral retinal pigment epithelial (RPE) cells and their association with differential gene expressions.
Materials and Methods
The RPE layer was obtained from the inner aspect of the eyeball of dark agouti rats (12-13 weeks old) and was mounted on glass slides. The peripheral RPE cell proliferation was evaluated using bromodeoxyuridine immunohistochemistry (n = 10). The cell migration was examined using the Dil tracer technique (n = 40) at the end of weeks 6, 10, 14 and 18. Affymetrix microarray analysis was used to investigate differential gene expressions in peripheral and central RPE cells, which was authenticated by RT-PCR using 4 RPE-specific genes (n = 10).
Results
In this study, peripheral RPE cells divided and appeared in clusters, but equatorial and central RPE cells rarely divided. The peripheral RPE cells migrated to the central RPE region in a time-dependent manner up to the end of week 14, but not later. The microarray analysis showed the expression of 9,645 out of a total of 35,220 genes studied. Among the 9,645 genes, 573 were differentially expressed (438 were upregulated and 135 were downregulated) in peripheral RPE cells as compared to central RPE cells. Of these 573 genes, 56 were associated with signaling pathways related to the regulation of cell proliferation, including Pax6, TGFβ, BMP and Wnt, and 404 were associated with pathways of cell migration.
Conclusions
In this study, peripheral RPE cells divided and migrated to the central region. This process was associated with differential gene expressions in these cells.
Key Words: Cell cycle, Gene expression, Microarray, Retina, Retinal cell proliferation
Introduction
The retinal pigment epithelium (RPE) forms a formidable blood-retina barrier that is necessary for normal and efficient photoreceptor functions [1]. The ability of peripheral RPE cells to divide in rodents has been proven using immunohistochemical labeling of cell proliferation markers. such as bromodeoxyuridine, Ki67 and proliferating cell nuclear antigen [2,3]. It has been widely reported that newly generated peripheral RPE cells replace dead ones due to aging at the central RPE [2,4]. In addition, RPE cell proliferation could be clinically important when choosing appropriate cells for transplantation as a treatment modality for diseases such as age-related macular degeneration [5]. In order to replace the dead cells at the central RPE, the peripheral RPE cells have to migrate, but evidence in favor of the migration is lacking, although previous studies have postulated a migration hypothesis [2,4].
Although peripheral RPE cells proliferate, both under normal and disease conditions [6], the molecular pathways responsible for this phenomenon are not clearly defined. RPE cell proliferation may be related to possible differences in the gene expressions in cells of the two RPE regions. Gene analysis studies of human RPE cells have shown differential expression of at least 11 genes in the two RPE regions, and these genes were associated with the execution of apoptosis, differentiation, metabolism, oxidative stress and matrix regulation [7]. Another study showed the differential expression of 1-5s% of genes related to extracellular matrix regulation [8]. However, a detailed gene analysis study in RPE cells, which may be useful to understand the underlying mechanisms that are responsible for peripheral RPE cells, has not been performed. Hence, the objectives of the present study were to investigate peripheral RPE cell proliferation and migration, and to investigate differential gene expressions in the RPE regions.
Materials and Methods
Animals
Adult male dark agouti (DA) rats (12-13 weeks old) were procured from the Animal Resources Center, Faculty of Medicine, Kuwait University, Kuwait. The animals were housed under standard laboratory conditions (23°C, 50s% humidity, 12:12 h light and dark cycle) in plastic cages with sawdust bedding. The animals were fed with standard laboratory chow and tap water ad libitum. This experiment was conducted in accordance with the guidelines stipulated by Kuwait University and according to the ARRIVE guidelines (Animal Research Reporting in vivo Experiments). It was also approved by the Institutional Animal Ethics Committee.
Evaluation of RPE Cell Proliferation
5-bromo-2′-deoxyuridine (bromodeoxyuridine) was obtained from Sigma-Aldrich (New York, N.Y., USA) and dissolved in NaOH/phosphate-buffered saline (PBS). The rats (n = 5) were intraperitoneally (i.p.) injected with 50 mg/kg body weight of bromodeoxyuridine once daily for 5 days a week for 4 months. At the end of the 4th month, the animals were killed the next day. The eyes were dissected and immunohistochemistry was carried out according to a previously described procedure [2] to evaluate the RPE cell proliferation. By using a fluorescence microscope, the number of bromodeoxyuridine-labeled cells in each RPE layer was counted under a 20× objective. The locations of the labeled cells were mapped on a floral diagram of the RPE layer.
Evaluation of RPE Cell Migration
The 1,1′-dioctadecyl-3,3,3′,3′-tetramethylindocarbocyanine perchlorate (Dil crystal tracer; Molecular Probes, Eugene, Oreg., USA) technique was used to investigate the hypothesis of RPE cell migration. The rats (n = 40) were anesthetized using a mixture of ketamine (Fort Dodge, London, UK) and dormitor (Pfizer, Tadworth, UK) diluted in sterile water (0.2 ml/100 g, i.p.). A small amount (2-3 µl) of very fine DiI crystals was injected into the peripheral retina at two locations. The rats were killed after 6, 10, 14 and 18 weeks from the day of injections and the eyeballs were removed and fixed in 4s% paraformaldehyde. The RPE was stained using Alexa Fluor 488, DAPI and phalloidin (1:50; Invitrogen) diluted in PBS for 1 h, washed and mounted in Vectashield®. The RPE layer was observed under a fluorescent microscope and Dil crystal fluorescing RPE cells were identified. The distances traveled by the fluorescing cells from the sites of the injections were measured.
Microarray Analysis
Ten DA rats were killed using CO2 asphyxiation. The eyes were removed, immersed in RNAlater for 5 min (Qiagen, Hilden, Germany) and the RPE layer was separated from the peripheral and central regions as described previously [2]. The RPE layers were pooled and collected in separate nuclease-free tubes containing RNAlater. The tissue samples were stored at −20°C for gene analysis.
Total RNA was extracted from 100 mg of tissue using the TRIzol method as previously described [9,10]. The amount of RNA was measured using a NanoDrop spectrophotometer 8000 and its quality was assessed with a Bioanalyzer 2100 (Agilent Technologies, Santa Clara, Calif., USA). The microarray was conducted using a GeneChip® 3′ IVT Express kit according to the procedure described in the assay manual (Affymetrix, Santa Clara, Calif., USA).
Affymetrix Rat 230_2 dataset quality control was performed using the Expression Console software (Affymetrix). Data set analysis and visualization were done using EASANA® (GenoSplice; www.genosplice.com), which is based on GenoSplice FAST DB® annotations. Normalization, background correction and the annotation of data were performed as previously described [11,12,13]. Genes were considered significantly regulated when the fold change between peripheral RPE and central RPE cells was ≥1.5 and the uncorrected p value from the unpaired Student t test was ≤0.05.
Hierarchical Clustering and Pathway and Gene Ontology
The distance from the gene signal in a given sample to the corresponding average in all the samples was calculated for each regulated gene. Corresponding values were displayed and clustered with MeV4.6.2 from the Institute of Genome Research using the Euclidean distance and average linkage clustering. Significant Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways [14,15] and Gene Ontology (GO) terms were retrieved using the Database for Annotation, Visualization and Integrated Discovery (DAVID) [16,17].
Reverse Transcription-Polymerase Chain Reaction
RT-PCR was performed for five genes that are known to be indisputably expressed in RPE: Lhx9 (LIM homeobox 9; assay ID Rn00710498_m1), Pax6 (paired box 6; assay ID Rn00689608_m1), Igfbp2 (insulin-like growth factor binding protein; assay ID Rn00565473_m1), Mitf (microphthalmia-associated transcription factor; assay ID Rn01425758_m1) and Otx2 (orthodenticle homeobox 2; assay ID Rn01414596_m1). The RNA (500 ng) from each sample was reverse transcribed into cDNA using the Quantitect reverse transcription kit (Qiagen) and used as templates for the RT-PCR reactions. The reactions were performed in an RT-PCR system (7500 Fast; Applied Biosystems) using commercially available TaqMan Gene Expression Assays (Applied Biosystems). Rat GAPDH (ID NM_017008.3) served as an endogenous control gene. The RPE-derived rhodopsin homolog (Rrh; ID Rn01521911_m1) was used as a control to evaluate neural retina contamination in RPE samples. The samples were tested in triplicate for each primer and fold changes in gene expressions were calculated [18] using 7500 Fast system software, version 1.4 (Applied Biosystems).
Results
Some Peripheral RPE Cells Proliferate and Migrate in the Normal Retina
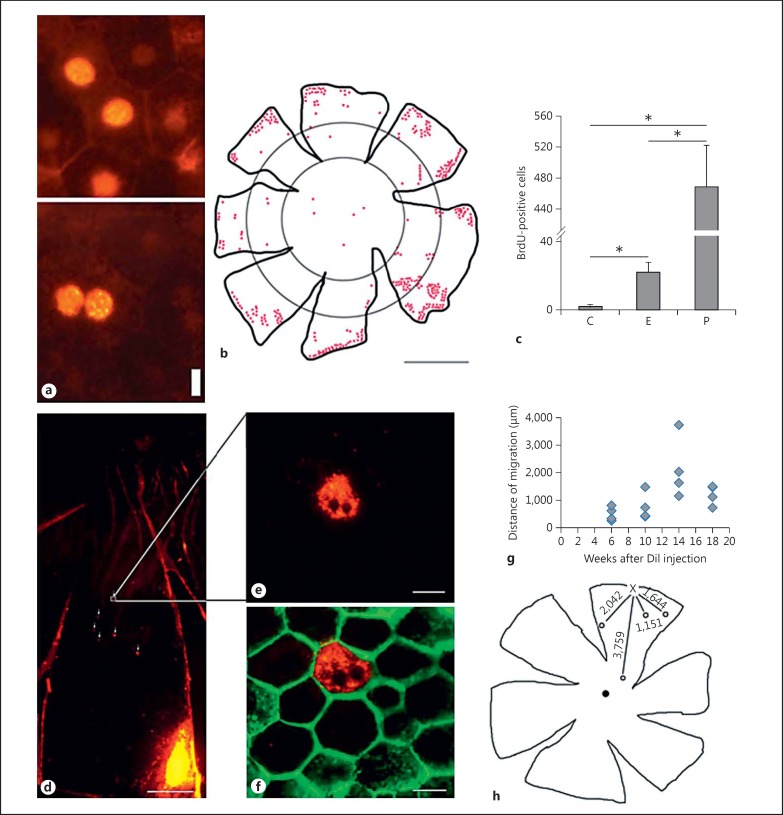
A total of 468 dividing cells appeared as clusters at the peripheral RPE region while 22 dividing cells were found at the equatorial and 3 cells at the central RPE regions (p < 0.001; fig. 1a-c). Autofluorescent Dil-labeled peripheral RPE cells were present farther away from the sites of the injections at or near the center of the RPE (fig. 1d). The migrating peripheral RPE cells traveled in a time-dependent manner up to the 14th week (fig. 1g, h). Interestingly, increasing the sample time to 18 weeks did not increase the distance of cell migration (fig. 1g).
Fig. 1.
Quantification of RPE cell proliferation and migration in adult DA rats. a Representative photomicrograph of binucleated and mononucleated RPE cells indicating BrdU-labeled cells. Scale bar = 10 µm. b Distribution of BrdU-labeled cells mostly in the peripheral RPE cells. Scale bar = 2 mm. c The number of BrdU-labeled cells at the three RPE regions: central (C), equatorial (E) and peripheral (P). d A low-magnification photomicrograph showing the DiI crystal injection site on the lower right; a few Dil-labeled RPE cells are indicated by arrows. e, f A fluorescent DiI crystal-incorporated cell from a is shown at a higher magnification. In f, the same cell shown in e is presented with phalloidin counterstain to mark the cell membrane. Scale bars = 500 µm (d) and 10 µm (e, f). g Distances (in µm) travelled by migrating RPE cells at the end of weeks 6, 10, 14 and 18, measured from the sites of Dil crystal injections at the peripheral retina. h The floral representation of distances travelled by migrating RPE cells by week 14. The letter X indicates the site of the injection and the numbers indicate the distances (in µm) travelled by the migrating cells.
Peripheral RPE Cells Show Differential Gene Expressions as Compared to Central RPE Cells
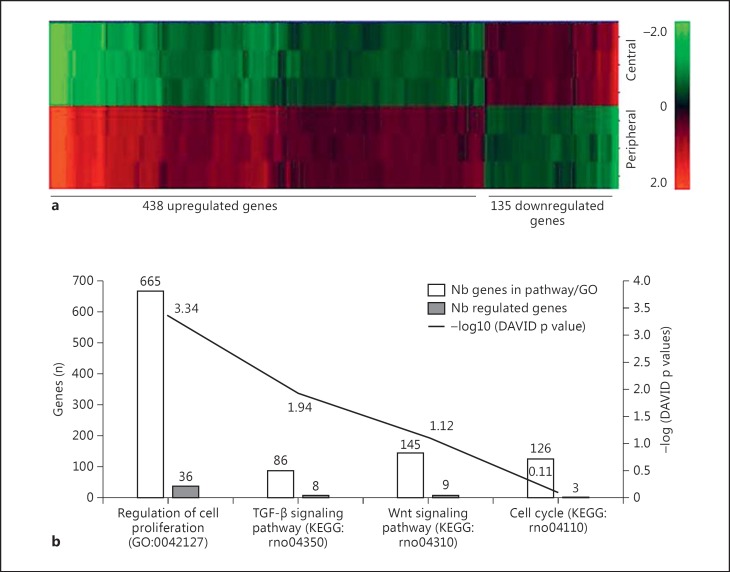
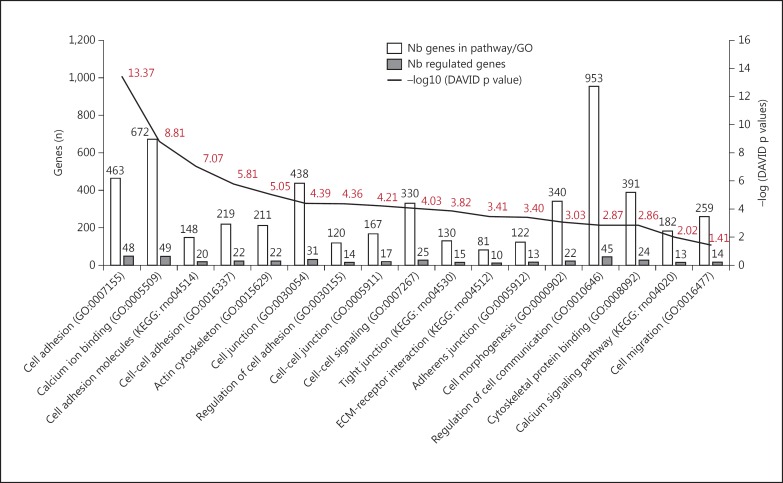
The microarray analysis for 35,220 transcripts in the RPE layer showed the expression of 9,645 genes, out of which 573 genes presented differential expressions in central and peripheral RPE regions. Of these 573 differentially expressed genes, 438 (76.44s%) were upregulated (mean fold change of 3.89; median fold change of 2.02) and 135 (23.56s%) were downregulated (mean fold change of −2.11; median fold change of 1.64) in the peripheral RPE as compared to that in the central RPE (438 downregulated and 135 upregulated genes; fig. 2a). Among the differentially expressed genes in the peripheral RPE, 56 (9.77s%) were associated with signaling pathways related to cell proliferation (see online suppl. tables 1-4; see www.karger.com/doi/10.1159/000446480 for all online suppl. material) while 404 (70.51s%) were associated with pathways of cell migration (fig. 3).
Fig. 2.
a Hierarchical clustering of differentially expressed genes in the two regions of the RPE layer. The color intensities show the extent of gene expressions in peripheral RPE cells either above (red) or below (green) the average gene expression (indicated by 0.0). b KEGG pathways and GO terms relating to proliferation. White bars correspond to the number of genes involved in KEGG pathways/GO terms and the gray bars correspond to the number of regulated genes involved in the 4 signaling pathways associated with cell proliferation. The dark curve corresponds to -log (DAVID p value) for each KEGG pathway/GO term (uncorrected p values are indicated above and below the curve, calculated using the EASE score (modified Fisher's exact p value) from DAVID.
Fig. 3.
KEGG pathways/GO terms relating to migration. White bars correspond to the number of genes involved in the KEGG pathways/GO terms and the gray bars correspond to the number of regulated genes for the 17 signaling pathways associated with cell migration. The black line corresponds to -log (DAVID p value) for each KEGG pathway/GO terms (uncorrected p values are indicated above and below the curve, calculated using the EASE score (modified Fisher's exact p value) from DAVID.
The genes associated with cell proliferation were related to: (i) the regulation of cell proliferation, (ii) the transforming growth factor-β (TGF-β) signaling pathway, (iii) the Wnt signaling pathway and (iv) cell cycle regulation (fig. 2b; online suppl. tables 1-4). The noteworthy upregulated genes responsible for cell proliferation and migration included Pax6 (18.82-fold), catenin (cadherin-associated protein)-delta 2 (Ctnnd2; 6.82-fold), Ctnna2 (1.81-fold), TGF-β2 (2.02-fold), BMP-2 (2.51-fold), BMP-4 (3.07-fold) and BMP-7 (3.9-fold).
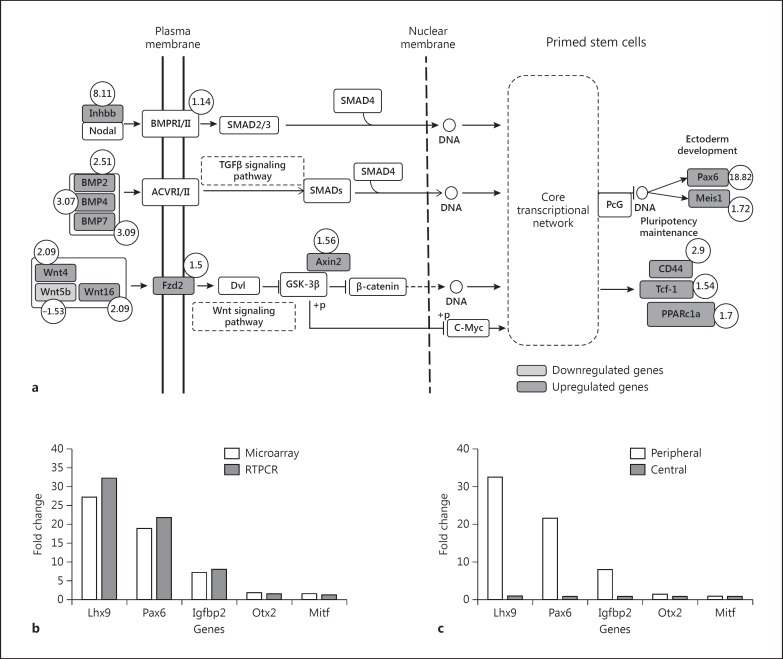
Peripheral RPE Cell Proliferation and Migration Are Associated with Upregulated BMP and Wnt Signaling Pathways
Although 573 genes showed up- or downregulations in the peripheral RPE cells as compared to the central RPE cells, two important pathways seemed to have an association with the peripheral RPE cell proliferation and migration. The BMP-2 and BMP-4 were upregulated, which appeared to have initiated the transcription of the Pax6 and Meis-1 genes (fig. 4a).
Fig. 4.
a BMP and Wnt signaling pathways in peripheral RPE cells. Fold changes for upregulated (dark gray) and downregulated (light gray) genes in peripheral RPE cells as compared to those in central RPE cells are indicated. The fold changes are indicated by numbers inside circles. b Comparison of RT-PCR results for the 5 genes with those of the microarray in peripheral RPE cells indicating the reproducibility of fold changes by the two methods. c RT-PCR results for the 5 genes indicating fold changes in the peripheral RPE as compared to those in the central RPE.
The analysis and pathway building for Wnt signaling showed upregulations of the Wnt4, Wnt16, Fzd1 and Axin2 genes. This pathway upregulated the transcription factor genes of cluster domain 44 (CD44), T cell factor 1 (Tcf-1) and peroxisome proliferator-activated receptor gamma, coactivator 1 alpha (PPARc1α). However, there was no change in β-catenin signaling, which is the key component of the canonical Wnt pathway (fig. 4a; online suppl. table 3). The analysis of the noncanonical Wnt pathway showed upregulations of the Tcf-1 and PPARc1α genes, but not the other genes of this pathway (fig. 4a). The Cas+s+ level also played a major role in the noncanonical pathway, although in this study the level of Cas+s+ in RPE cells was not estimated.
Microarray Results Are Comparable to RT-PCR Results
The RT-PCR analysis for the representative genes (Lhx9, Pax6, Igfbp2, Otx2 and Mitf) indicated that their expression levels were comparable to those seen in the microarray analysis. All these genes, except Mitf, were upregulated in peripheral RPE cells as compared to central RPE cells (p < 0.05; fig. 4b, c). The Rrh gene did not show any expression in the RPE sample, indicating a lack of neural retina contamination.
Discussion
The main findings of this study were peripheral RPE cell proliferation and migration towards the central region, and the upregulation of genes related to the cell proliferation and migration, including the BMP and Wnt pathways. These findings showed that a higher number of proliferating cells was seen at the peripheral RPE region than at the other two regions. The reasons for peripheral RPE cell proliferation and migration are not entirely clear, which could be due to their intrinsic nature to divide, as they develop later compared with the central RPE cells. Because the cells at the equatorial and central regions are very closely and tightly packed due to their stronger cell junctions, it might be difficult for the newly formed cells to migrate centripetally.
It is probable that not all dividing peripheral RPE cells migrate to the central region and that the cell migration could be an end result of RPE cellular remodeling and repositioning in the epithelium. In addition, whether or not any dividing central RPE cells migrate to the peripheral region is also unclear, but it is highly unlikely because rarely do any central RPE cells divide, and even if they do, the migration would not be possible because in the central region the cells are tightly packed [2].
The microarray analysis study showed that none of the signaling pathways that are usually upregulated in peripheral RPE cells during disease conditions had any changes, indicating that they did not regulate the proliferation of these cells [19,20,21]. These pathways include, but are not limited to, protein kinases C [19], mitogen-activated protein kinases [20] and PI3K/Akt pathways [21]. These proproliferative molecular pathways become active and stimulate RPE cell proliferation and migration due to disruptions to the blood-retina barrier in diabetes [22] and retinopathies [23]. In this study the rats were normal, hence the barrier was intact and therefore the lack of changes in the abovementioned pathways could be due to intact cellular functions. If the blood-retina barrier is disrupted, as happens in retinopathies, RPE cells are exposed to some blood-borne growth factors, which induce proproliferative signaling pathways and RPE cell proliferation [24]. In this study, considering that the blood-retina barrier was normal in the rats, what could have stimulated some peripheral RPE cells to proliferate in the normal retina? Our results showed no differential gene expressions for protein kinases, mitogen-activated protein kinases, cell cycle progression proteins and PI3k/Akt. Although their protein expressions were not estimated, which might or might not have been differentially expressed, based on the microarray study, it is clear that both normal retinal regions present comparable levels of gene expressions for the abovementioned proteins that are actively involved in cell proliferation. Another issue relates to the upregulation of so many genes controlling cell junctions (adherens junctions and tight junctions) in the peripheral RPE region as compared to the central RPE region. Logically, if peripheral RPE cells have to migrate centripetally, then the cell junctions between themselves and between the cells and the basement membrane should be loose and yielding; however, the upregulations of the cell junction-related genes at the peripheral RPE region in our study indicated otherwise. So, this structural feature of the peripheral RPE could be another impediment for cell proliferation and migration.
Although numerous genes were up- or downregulated in the peripheral as compared to central RPE cells, the most important finding was the upregulations of the BMP and Wnt signaling pathways. In this study, BMP-2, BMP-4 and BMP-7 showed increases in peripheral RPE cells, thereby indicating their role in cell proliferation. The BMPs, a group of proteins that belong to the TGF-β superfamily, play a major role in retinal regeneration from progenitor cells by upregulating the canonical BMP/Smad pathway [25,26]. However, the Smad genes did not show any upregulation in peripheral RPE cells, indicating that the canonical BMP pathway was not operational in these cells. Moreover, if the Smad genes were regulated, there would have been regulations of at least mitogen-activated protein kinases [27], which was not the case in our study, reflecting the fact that the Smad genes maintained a balance between the two RPE regions in normal retinas.
In this study, the finding that the gene expression of Pax6 - which is transcriptionally downstream of BMPs - showed more than an 18-fold increase in peripheral RPE cells indicated that peripheral RPE cells were more capable of dividing than the central RPE cells, and that this involved the BMP pathway. Previous studies indicated that the Pax6 gene along with Mitf inhibited the formation of the retina, but, conversely, by interacting with retinogenic genes it promoted retinogenesis [28]. The Pax6 gene is highly upregulated in newts during retinal regeneration, suggesting its involvement in RPE proliferation [29]. From our results, it appears that Wnt4 and Wnt16 are maintained at higher levels in normal peripheral RPE cells along with that of Frizled2 and Axin2. However, β-catenin levels did not show any significant changes between the two RPE regions. The latter finding indicates that transcriptional upregulation of β-catenin is not necessary for peripheral RPE cell proliferation, but its protein form stimulates RPE cell proliferation upon translocation to their nuclei, as reported previously [3]. Hence, the upregulated BMP and Wnt signaling showed an association with RPE cell proliferation.
We did not observe any changes in the expression of MKI67 or PCNA (proliferating cell nuclear antigen) genes in peripheral RPE cells in contrast to the expressions of their protein forms as markers for cell proliferation [2]. The lack of changes in MKI67 and PCNA gene expressions was unexpected, but this could be due to undetectable increases in these genes as the number of proliferating cells in peripheral RPE was abysmally small (∼5 cells/day), or their upregulation might be only at protein levels rather than at gene levels. In addition, the genes quantified in this microarray study did not show striking increases or decreases in peripheral RPE cells as compared to that in central RPE cells. The RNA was extracted from whole peripheral or central RPE regions, which included a small number of dividing cells (in peripheral RPE) and a remaining large number of quiescent cells; therefore, even if the dividing cells had upregulated genes, the effect would have been neutralized by the RNA of the quiescent cells, which lacked the gene expressions.
Conclusion
In this study, the proproliferative and promigratory genes were differentially expressed in peripheral RPE cells compared to central RPE cells. These differences in gene expressions, especially in the upregulation of the BMP and Wnt pathways, could underlie RPE cell migration and proliferation. Furthermore, peripheral RPE cells divided and migrated towards the central RPE region, which was reached by some of the migrating cells.
Acknowledgements
This study was funded by the Kuwait University grant No. MA02/09 and General Facility grant No. SRUL02/13. The technical assistance of Dr. Josely George, Sindhu Jacob, Ms. Fatemah Al-Ruwaieh and Ms. S. Varghese is gratefully acknowledged. We would also like to thank GenoSplice for the gene chip analysis. We would like to thank Prof. Glen Jeffery for his support.
References
- 1.Strauss O. The retinal pigment epithelium in visual function. Physiol Rev. 2005;85:845–881. doi: 10.1152/physrev.00021.2004. [DOI] [PubMed] [Google Scholar]
- 2.Al-Hussaini H, Kam JH, Vugler A, et al. Mature retinal pigment epithelium cells are retained in the cell cycle and proliferate in vivo. Mol Vis. 2008;14:1784–1791. [PMC free article] [PubMed] [Google Scholar]
- 3.Kokkinopoulos I, Shahabi G, Colman A, et al. Mature peripheral RPE cells have an intrinsic capacity to proliferate; a potential regulatory mechanism for age-related cell loss. PLoS One. 2011;6:e18921. doi: 10.1371/journal.pone.0018921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Del Priore LV, Kuo YH, Tezel TH. Age-related changes in human RPE cell density and apoptosis proportion in situ. Invest Ophthalmol Vis Sci. 2002;43:3312–3318. [PubMed] [Google Scholar]
- 5.Alexander P, Thomson HA, Luff AJ, et al. Retinal pigment epithelium transplantation: concepts, challenges, and future prospects. Eye (Lond) 2015;29:992–1002. doi: 10.1038/eye.2015.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gilbert C, Hiscott P, Unger W, et al. Inflammation and the formation of epiretinal membranes. Eye (Lond) 1988;2((suppl)):S140–S156. doi: 10.1038/eye.1988.140. [DOI] [PubMed] [Google Scholar]
- 7.Ishibashi K, Tian J, Handa JT. Similarity of mRNA phenotypes of morphologically normal macular and peripheral retinal pigment epithelial cells in older human eyes. Invest Ophthalmol Vis Sci. 2004;45:3291–3301. doi: 10.1167/iovs.04-0168. [DOI] [PubMed] [Google Scholar]
- 8.van Soest SS, de Wit GM, Essing AH, et al. Comparison of human retinal pigment epithelium gene expression in macula and periphery highlights potential topographic differences in Bruch's membrane. Mol Vis. 2007;13:1608–1617. [PubMed] [Google Scholar]
- 9.Al-Mulla F, Al-Tamimi R. Uses of microarray platforms in cancer: a correlative study between genomic copy number changes and their expression at mRNA and protein levels. Methods Mol Biol. 2007;382:77–95. doi: 10.1007/978-1-59745-304-2_6. [DOI] [PubMed] [Google Scholar]
- 10.Peirson SN, Butler JN. RNA extraction from mammalian tissues. Methods Mol Biol. 2007;362:315–327. doi: 10.1007/978-1-59745-257-1_22. [DOI] [PubMed] [Google Scholar]
- 11.Bignon A, Regent A, Klipfel L, et al. Dusp4-mediated accelerated T-cell senescence in idiopathic CD4 lymphopenia. Blood. 2015;125:2507–2518. doi: 10.1182/blood-2014-08-598565. [DOI] [PubMed] [Google Scholar]
- 12.Cottu P, Bieche I, Assayag F, et al. Acquired resistance to endocrine treatments is associated with tumor-specific molecular changes in patient-derived luminal breast cancer xenografts. Clin Cancer Res. 2014;20:4314–4325. doi: 10.1158/1078-0432.CCR-13-3230. [DOI] [PubMed] [Google Scholar]
- 13.Lustremant C, Habeler W, Plancheron A, et al. Human induced pluripotent stem cells as a tool to model a form of Leber congenital amaurosis. Cell Reprogram. 2013;15:233–246. doi: 10.1089/cell.2012.0076. [DOI] [PubMed] [Google Scholar]
- 14.Doganay S, Evereklioglu C, Er H, et al. Comparison of serum NO, TNF-alpha, IL-1beta, sIL-2R, IL-6 and IL-8 levels with grades of retinopathy in patients with diabetes mellitus. Eye (Lond) 2002;16:163–170. doi: 10.1038/sj.eye.6700095. [DOI] [PubMed] [Google Scholar]
- 15.Kanehisa M, Goto S, Sato Y, et al. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2012;40:D109–D114. doi: 10.1093/nar/gkr988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lorenz B, Gyurus P, Preising M, et al. Early-onset severe rod-cone dystrophy in young children with RPE65 mutations. Invest Ophthalmol Vis Sci. 2000;41:2735–2742. [PubMed] [Google Scholar]
- 17.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 18.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative pcr and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 19.Palma-Nicolas JP, Lopez E, Lopez-Colome AM. Thrombin stimulates RPE cell motility by PKC-ζ- and NF-κB-dependent gene expression of MCP-1 and CINC-1/GRO chemokines. J Cell Biochem. 2010;110:948–967. doi: 10.1002/jcb.22608. [DOI] [PubMed] [Google Scholar]
- 20.Parrales A, Lopez E, Lee-Rivera I, et al. ERK1/2-dependent activation of mTOR/mTORC1/p70S6K regulates thrombin-induced RPE cell proliferation. Cell Signal. 2013;25:829–838. doi: 10.1016/j.cellsig.2012.12.023. [DOI] [PubMed] [Google Scholar]
- 21.Sasore T, Kennedy B. Deciphering combinations of PI3k/AKT/mTOR pathway drugs augmenting anti-angiogenic efficacy in vivo. PLoS One. 2014;9:e105280. doi: 10.1371/journal.pone.0105280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gariano RF, Hu D, Helms J. Expression of angiogenesis-related genes during retinal development. Gene Expr Patterns. 2006;6:187–192. doi: 10.1016/j.modgep.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 23.Umazume K, Tsukahara R, Liu L, et al. Role of retinal pigment epithelial cell β-catenin signaling in experimental proliferative vitreoretinopathy. Am J Pathol. 2014;184:1419–1428. doi: 10.1016/j.ajpath.2014.01.022. [DOI] [PubMed] [Google Scholar]
- 24.Qin D, Zheng XX, Jiang YR. Apelin-13 induces proliferation, migration, and collagen I mRNA expression in human RPE cells via PI3K/Akt and MEK/Erk signaling pathways. Mol Vis. 2013;19:2227–2236. [PMC free article] [PubMed] [Google Scholar]
- 25.Du Y, Xiao Q, Yip HK. Regulation of retinal progenitor cell differentiation by bone morphogenetic protein 4 is mediated by the Smad/Id cascade. Invest Ophthalmol Vis Sci. 2010;51:3764–3773. doi: 10.1167/iovs.09-4906. [DOI] [PubMed] [Google Scholar]
- 26.Haynes T, Gutierrez C, Aycinena JC, et al. BMP signaling mediates stem/progenitor cell-induced retina regeneration. Proc Natl Acad Sci USA. 2007;104:20380–20385. doi: 10.1073/pnas.0707202104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Derynck R, Zhang YE. Smad-dependent and Smad-independent pathways in TGF-β family signalling. Nature. 2003;425:577–584. doi: 10.1038/nature02006. [DOI] [PubMed] [Google Scholar]
- 28.Bharti K, Gasper M, Ou J, et al. A regulatory loop involving PAX6, MITF, and WNT signaling controls retinal pigment epithelium development. PLoS Genet. 2012;8:e1002757. doi: 10.1371/journal.pgen.1002757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kaneko Y, Matsumoto G, Hanyu Y. Pax-6 expression during retinal regeneration in the adult newt. Dev Growth Differ. 1999;41:723–729. doi: 10.1046/j.1440-169x.1999.00476.x. [DOI] [PubMed] [Google Scholar]