Abstract
The molecular signature and functional properties of astroglial subtypes in the adult CNS remain largely undefined. By using translational ribosome affinity purification followed by RNA-Seq, we profiled astroglial ribosome-associated (presumably translating) mRNAs in major cortical and subcortical brain regions (cortex, hippocampus, caudate-putamen, nucleus accumbens, thalamus, and hypothalamus) of BAC aldh1l1-translational ribosome affinity purification (TRAP) mice (both sexes). We found that the expression of astroglial translating mRNAs closely follows the dorsoventral axis, especially from cortex/hippocampus to thalamus/hypothalamus posteriorly. This region-specific expression pattern of genes, such as synaptogenic modulator sparc and transcriptional factors (emx2, lhx2, and hopx), was validated by qRT-PCR and immunostaining in brain sections. Interestingly, cortical or subcortical astrocytes selectively promote neurite growth and synaptic activity of neurons only from the same region in mismatched cocultures, exhibiting region-matched astrocyte to neuron communication. Overall, these results generated new molecular signature of astrocyte types in the adult CNS, providing insights into their origin and functional diversity.
SIGNIFICANCE STATEMENT We investigated the in vivo molecular and functional heterogeneity of astrocytes inter-regionally from adult brain. Our results showed that the expression pattern of ribosome-associated mRNA profiles in astrocytes closely follows the dorsoventral axis, especially posteriorly from cortex/hippocampus to thalamus/hypothalamus. In line with this, our functional results further demonstrated region-selective roles of cortical and subcortical astrocytes in regulating cortical or subcortical neuronal synaptogenesis and maturation. These in vivo studies provide a previously uncharacterized and important molecular atlas for exploring region-specific astroglial functions.
Keywords: astrocyte, dorsoventral axis, heterogeneity, RNA-seq, TRAP
Introduction
Although the diverse morphology of mammalian astrocytes has been long characterized by early Golgi staining (Somjen, 1988), astrocytes are historically regarded as homogeneous cell populations, largely due to the observation that they are typically nonelectrically excitable cells in the mammalian CNS (Kettenmann, 2005). It has become increasingly evident that astrocytes exhibit significant functional and molecular heterogeneity (Chaboub and Deneen, 2012), especially in a region-dependent manner (Zhang and Barres, 2010; Bayraktar et al., 2014). Mature cortical or hippocampal astrocytes typically occupy significantly larger domain size than hypothalamic astrocytes with a higher number of fine processes (Morel et al., 2014). Expression of characteristic astroglial glutamate transporters GLAST and GLT1 that mediate glutamate uptake function is region-dependent (Kerkerian et al., 1982; Danbolt, 2001; Regan et al., 2007). Other important astroglial functional features or phenotypes, such as gap-junction coupling (Batter et al., 1992) and intracellular Ca2+ responses (Oberheim et al., 2012), are also manifested differently across CNS regions (Chai et al., 2017). Astrocyte fate-mapping results showed that astrocytes are organized into spatial domains, especially in the spinal cord, which exhibit domain-restricted interaction with motor neurons (Hochstim et al., 2008; Tsai et al., 2012; Molofsky et al., 2014). Astrocytes have also been considered active regulators of synaptogenesis, neural circuit formation, and synaptic transmission (Haydon and Carmignoto, 2006; Eroglu and Barres, 2010). However, whether and how these regulatory functions are region and circuit specific remains largely unknown. In addition to the inter-regional heterogeneity, intraregional astroglial subpopulations and their functional diversity in the brain remain essentially unexplored.
Subcortical brain regions have a distinct glia to neuron ratio (∼11:1) compared with the cortex (∼4:1) (Azevedo et al., 2009), perform specific physiological functions, and are often associated with psychiatric and neurodegenerative diseases. Several studies have previously characterized gene expression profiles in subcortical astrocytes (Bachoo et al., 2004; Yeh et al., 2009). However, these studies were all performed from cultured astrocytes, limiting their in vivo relevance. In addition, previous attempts to characterize in vivo astrocyte gene profile have largely focused on the cortex (and cerebellum) (Doyle et al., 2008) or at developing (<2 weeks) ages (Cahoy et al., 2008; Zhang et al., 2014). Moreover, although region-specific gene expression studies have been performed in rodent and human brains (Lein et al., 2007; Ng et al., 2009; Ramasamy et al., 2014), these gene expression datasets represent all cell types, not astrocyte-specific or enriched gene expression. By performing a cell-surface marker screening, a recent study begins to characterize astrocyte subpopulations from multiple brain regions of BAC aldh1l1-eGFP reporter mice, including the subcortical thalamus region (John Lin et al., 2017).
Here we used a translational ribosome affinity purification (TRAP)/RNA-Seq approach to investigate the molecular heterogeneity of adult astrocytes in various brain regions. We also investigated the heterogeneous modulation of representative dorsal and ventral brain region's astrocytes on neurite growth and synaptic activity of neurons using in vitro mismatched cocultures.
Materials and Methods
Animals.
BAC aldh1l1-TRAP (Doyle et al., 2008) and BAC aldh1l1-eGFP (Cahoy et al., 2008) transgenic mice (C57BL/6FVB mixed background) were obtained from the GENSAT project through The Jackson Laboratory. All mice were maintained on a 12 h light/dark cycle with food and water ad libitum. Both male and female mice were used for all experiments. All procedures were in strict accordance with the National Institutes of Health Guide for the care and use of laboratory animals and were approved by the Tufts University Institutional Animal Care and Use Committee.
Immunohistochemistry in mouse brain and confocal imaging.
Mice were deeply anesthetized with ketamine (100 mg/kg) + xylazine (10 mg/kg) in saline by intraperitoneal injection and perfused intracardially with 4% PFA in PBS. The brains were dissected and kept in 4% PFA overnight at 4°C, then cryoprotected by immersion in 30% sucrose for 24–48 h. Brains were embedded and frozen in OCT-Compound Tissue-Tek (Sakura). Coronal sections (10–20 μm) were prepared with a cryostat (Leica HM525). For immunostaining of ALDH1L1, Sparc, and Emx2, sections were mounted on slides, then incubated with blocking buffer (1% BSA, 5% goat or donkey serum, 0.2% Triton X-100 in PBS) for 30 min at room temperature. Primary antibodies for Sparc (R&D Systems, #AF-942, 1:200), ALDH1L1 (Neuromab, #73-140, 1:5), and Emx2 (Assay Biotech, 1:50) were incubated overnight at 4°C in blocking buffer. After washing slides three times in PBS, corresponding secondary antibody (1:2000) was added for 120 min at room temperature. The sections were rinsed three times in PBS before mounting. All images were obtained with a confocal laser scanning microscope (15–20 μm Z stack with 1 μm step, A1R Nikon) or fluorescent microscope (Zeiss AXIO Imager with ApoTome). The βIII-tubulin (R&D Systems, 1:1000) immunostaining was performed with standard procedures in cocultures.
FIJI open source plug-ins were used to quantify Sparc and Emx2, immunoreactivity from colocalized astrocytes (indicated by the eGFP reporter). In FIJI, maximum projections were generated from confocal images for each channel and the merged images. The number of eGFP-labeled astrocytes in the field was manually counted using the region of interest manager. The freehand selection tool was then used to outline labeling in immunostaining images that were colocalized with eGFP-labeled astrocytes in the merged images. Measurements of average intensity within the freehand region were calculated through FIJI's region of interest manager. For neurite quantification, images were imported to Fiji-ImageJ and the Simple Neurite Tracer plugin was used to trace neurons, which will generate the length of the neurite. The measurements were then uploaded into GraphPad Prism, and a cumulative frequency curve was generated.
Primary astrocyte and neuron cocultures.
Cortices of postnatal day 0–3 (P0–P3) mouse pups were used for cortical astrocyte cultures. Cells were plated at a density of 1.0 × 106 cells/ml in 6-well plate or in 100 mm dish. Astrocyte medium was composed of DMEM supplemented with 10% FBS (Sigma) and 1% penicillin/streptomycin. Astrocytes become 90% confluent at 6–7 d in culture and are ready for experiment. For astrocyte and neuron cocultures, cortical neurons were isolated from embryonic 14- to 16-d-old mouse brains. Embryonic (E14–E16) or P1–P3 pups brains with cortex, hippocampus, olfactory bulb, and cerebellum removed were used for subcortical neuron and astrocyte cultures, respectively. The coculture medium was composed of neurobasal medium, 2% B27 neurobasal supplement, 5% FBS, 2 mm glutamine, 1% of 100× GlutaMAX, and 1% penicillin/streptomycin. Freshly prepared neurons (1–2 × 106 cells) were plated on top of astrocytes that had grown on 24-well or 6-well cell culture plates for 6–7 d.
Electrophysiology.
Whole-cell patch-clamp recordings of cortical neurons in neuron-astrocyte cocultures were obtained by Multiclamp 700B amplifier, filtered at 2 kHz, and sampled at 10 kHz with Digidata 1322A (Molecular Devices). Cocultured cells were continuously perfused with aCSF with a flow rate of 1–2 ml/min bubbled with 95% O2 and 5% CO2. Neurons from coculture were identified by bright-field Nikon Eclipse e600FN microscopy with a 40× water-immersion lens (numerical aperture 0.8) and infrared illumination. Input resistance and capacitance were measured in voltage-clamp mode with 500 ms 5 mV step pulse from −70 mV holding potential. Cell capacitance was calculated by first obtaining the decay time constant of the transient current by the 5 mV step pulse and dividing this number by the series resistance. To measure the miniature EPSCs (mEPSCs), cells were recorded in voltage-clamp mode at a holding potential of −70 mV in the presence of 1 μm TTX and 50 μm picrotoxin. A pClamp 9.2 (Molecular Devices) was used for data acquisition and storage. Analysis of mEPSC was performed using Minianalysis 6.0 software (Synaptosoft). At least 5 min of spontaneous activity was recorded from each cell with access resistance measured before and after the recording. Access resistance (Ra) was monitored following membrane rupture and dialysis, and recordings were abandoned if Ra >30 mΩ. Recordings with an unstable or high noise (>4 pA root mean square) were excluded. An event amplitude threshold was set at 5 pA above the baseline, and at least 300 events were analyzed per cell. A minimum recording length of 2 min without apparent mEPSCs was defined as a silent cell.
Affinity purification of translating RNA, amplification, and sequencing library construction.
Thick coronal sections (2 mm) of the adult (P70) BAC aldh1l1 TRAP mouse brain were prepared on a mouse brain matrix based on the mouse brain atlas. Somatosensory cortex (ctx), hippocampus (hippo), thalamus (thal), hypothalamus (hypo), nucleus accumbens (nacc), and caudate-putamen (cpu) were then carefully dissected out from corresponding coronal sections (see Fig. 1A). Ribosomal bound translating mRNAs from astrocytes in these regions were extracted using immunoprecipitation procedures, as previously described (Doyle et al., 2008). Briefly, the anti-eGFP antibody (mouse clone HtzGFP-19C8, Memorial Sloan Kettering Cancer Center) was coupled to magnetic beads (Dynabeads, Invitrogen) as described in the kit instructions. Tissue lysates were prepared and mixed with eGFP antibody-coupled beads for immunoprecipitation. Following immunoprecipitation, RNA was isolated using the Trizol reagent and precipitated with isopropanol. For each region, 3–5 independent TRAP isolations were performed from BAC aldh1l1 TRAP mice. For hypothalamus, caudate-putamen, and nucleus accumbens, 2 or 3 mice were pooled together for a single sample. The TRAP isolation typically yields 10–200 ng RNA, depending on the region.
Figure 1.
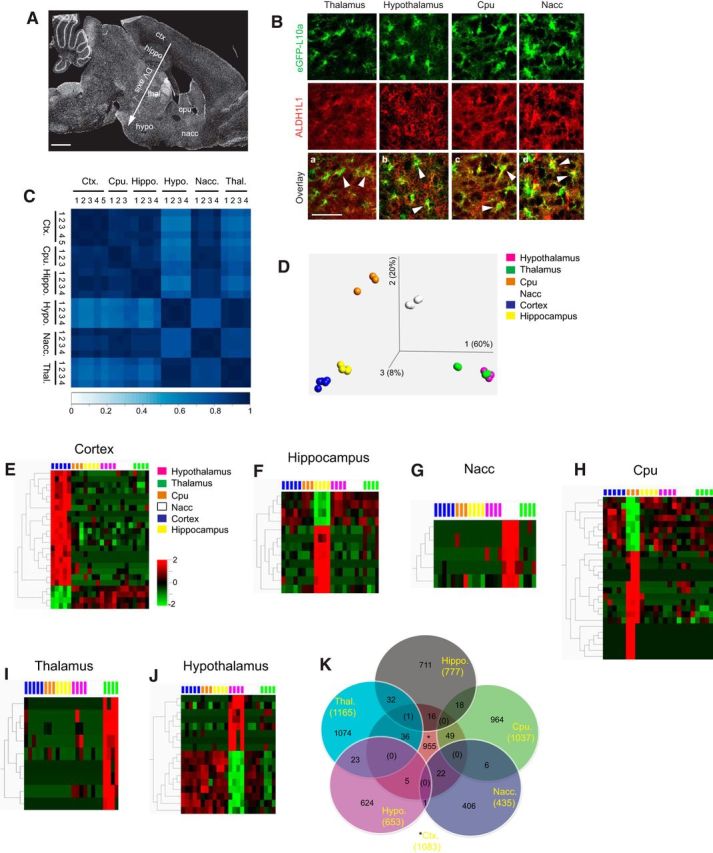
RNA-Seq analysis of TRAP-isolated translating mRNAs from adult astrocytes in various brain regions of BAC aldh1l1 TRAP mice. A, Illustration of brain regions analyzed on a representative sagittal brain section. Ctx, Somatosensory cortex; hippo, hippocampus; thal, thalamus; hypo, hypothalamus; nacc, nucleus accumbens; cpu, caudate-putamen; DV axis, dorsoventral axis. Scale bar, 500 μm. B, Representative ALDH1L1 immunostaining in subcortical brain regions of BAC aldh1l1-TRAP mice (P70). Scale bar, 50 μm. White arrows indicate colocalization between ALDH1L1 immunostaining and eGFP fluorescence. C, Spearman rank correlation of sample replicates for each region. D, Principal component analysis plot of RNA-Seq results of all samples. Unique mRNA expression signature of cortex (E), hippocampus (F), caudate-putamen (Cpu, G), thalamus (H), nucleus accumbens (Nacc, I), and hypothalamus (J). Each region's astrocyte RNA-Seq dataset was compared with all other regions to identify the unique mRNA signature. q value, FDR-adjusted p value. q < 0.001 for Ctx and Hippo; q < 0.01 for all other regions. K, A partial Venn diagram with relatively enriched gene expression from each region's astrocytes. Values in yellow in parentheses are the number of genes that are enriched in each region's astrocytes. Values in italics and in parentheses are the number of genes that are shared from multiple (3) regions' astrocytes. Only intersection up to 3 regions is displayed, due to the large number of total intersections from 6 regions. FDR q value <0.01 was used to identify the genes that are relatively enriched in each region's astrocytes.
All RNA samples were amplified, size selected, and RNA-sequencing libraries were constructed by closely following manufacturer's kit protocols unless otherwise stated. Briefly, the Ovation RNA-Seq system version 2 (Nugen) was used for sample amplification (Nugen). We used 20 ng RNA as input for amplification. After amplification, samples were processed with a MinElute DNA reaction clean up kit (QIAGEN). To eliminate double-stranded RNA or DNA, samples were treated with single-strand nuclease (S1, Promega). Following nuclease treatment, samples were purified with QIAGEN Minelute kit again. Approximately 1500 ng of amplified sample was sonicated to select for 200 bp fragments (M220, Covaris). After sonication, samples were purified with QIAGEN Minelute kit again. Finally, samples (600 ng) were constructed into a sequencing library with the Trueseq RNA sample prep kit (Illumina).
RNA-Seq analysis.
The 100 bp paired-end sequencing for all prepared sequencing libraries was performed at the Tufts Core Facility using Illumina HiSeq 2500 Sequencer. RNA-sequencing libraries were assigned randomly to sequencing lanes to avoid lane bias. FastQC program was used to examine the quality of generated reads. For mouse samples, we usually obtained 25–50 million reads for each library with a Phred quality score >28. The total number of mapped reads for each region was at least 50 million, sufficient for calculation of reliable Fragments Per Kilobase of Mapped Reads (FPKM) values and subsequent differential gene expression analysis. The Tuxedo suite of tools (Tophat/Cufflinks/Cuffdiff) was used to analyze RNA-Seq reads (Trapnell et al., 2012). Reads were aligned to the reference mouse genome (July 2007 NCBI37/mm9) using the Tophat (version 2.0.8) software; >50% of both read pairs were mapped for all samples. Cufflinks (version 2.1.1) was used to calculate FPKM values with −G option for all mapped transcripts.
Cuffdiff2 was used to identify significant changes in mRNA expression among different samples using the biological replicates with a default q value (FDR-adjusted p value) cutoff of 0.05 (q < 0.05). Custom R-scripts were used for calculating the correlation coefficient. The principal component analysis was performed, and heatmaps were generated from the Qlucore software with variable q values (q < 0.01 or q < 0.001). We also performed qRT-PCR on a group of genes with a wide range of FPKM values and found a reverse linear correlation between Ct and FPKM values for examined genes across all region samples (data not shown), indicating that FPKM values indeed correlate well with the relative abundance of individual mRNAs.
qRT-PCR.
For qRT-PCR of mouse genes, RNA isolated with the TRAP procedure or isolated from FAC sorted cells was converted to cDNA using a high archive cDNA synthesis kit (Applied Biosystems). The relative expression levels of selective genes were measured using SYBR Green (Invitrogen) reagent, and specific primers for analyzed genes were chosen from the PrimerBank (https://pga.mgh.harvard.edu/primerbank/).
Experimental design and statistical analysis.
The sample size and statistical approach used for each experiment were described in each method section and in figure legends. Briefly, for TRAP/RNA-Seq, 3–5 independent TRAP isolations per region were performed from BAC aldh1l1 TRAP mice; for qRT-PCR, 2–6 independent TRAP samples from 4–6 mice (both genders) per region were used. For immunostaining, 90–200 astrocytes per region from 2 mice were used; for coculture neurite quantification, 141–174 neurons were quantified and 36–48 neurons were patched from three independent cultures. The G*Power (version 3.1) was used to estimate the reasonable sample size. All analyses were performed using GraphPad Prism6. All values were plotted using the mean ± SEM, except the neurite length, which was converted to cumulative frequency. The Kolmogorov–Smirnov test was used to analyze significance for the cumulative frequency curves. For multiple groups (>2), one-way ANOVA was used to analyze the variance, followed by a Tukey post hoc test to compare multiple groups. For two-group comparison, Student's t test was used. Statistical significance was tested at a 95% (p < 0.05) confidence level.
Results
Astroglial translating mRNA expression pattern along the dorsal to ventral axis of the adult brain
Previous in vitro (Bachoo et al., 2004; Yeh et al., 2009) and in vivo (Doyle et al., 2008) studies to characterize region-specific astroglial transcriptomes were intrinsically limited due to significant influences of culture conditions or a focus only on the cortex and cerebellum. We decide to focus on 6 major brain regions, cortex (ctx), hippocampus (hippo), thalamus (thal), hypothalamus (hypo), caudate-putamen (cpu), and nucleus accumbens (nacc), as shown in Figure 1A. These regions allow us to examine the molecular profile pattern of astrocytes along the dorsoventral axis. The aldh1l1 promoter has been previously shown to be selectively active in adult astrocytes (Cahoy et al., 2008; Yang et al., 2011), and the ALDH1L1 immunostaining selectively labels cortical and hippocampal astrocytes (Cahoy et al., 2008; Clasadonte et al., 2016). However, it remains unexplored whether the aldh1l1 promoter-driven eGFP-L10a expression selectively overlaps with endogenous ALDH1L1 protein expression in astrocytes from subcortical brain regions, especially thalamus and hypothalamus. We first performed ALDH1L1 immunostaining on brain sections of BAC aldh1l1 TRAP mice. We found widespread colocalization (Fig. 1Ba–Bd, white arrows) of ALDH1L1 immunoreactivity with eGFP fluorescence, an indication of L10a subunit expression, in all subcortical regions examined. In contrast, no colocalization of eGFP fluorescence with NeuN immunoreactivity was observed (data not shown).
To profile astroglial mRNAs (presumably translating mRNAs) from selected regions, we isolated ribosome-bound mRNAs using the TRAP procedure (Doyle et al., 2008) from each region of BAC aldh1l1-TRAP mice (P70) and prepared sequencing libraries for RNA-Seq. We obtained a total number of 50–100 million reads from 3–5 sequencing libraries per region that is sufficient for subsequent mapping and bioinformatic analysis. Our biological replicates produced highly similar sequencing reads with the correlation coefficient generally >0.8 among replicates (Fig. 1C). To determine the overall variations of RNA-Seq datasets of all samples, we performed principal component analysis (q < 0.001) and found that hypothalamus and thalamus RNA-Seq datasets are closely grouped with a 60% of overall variation along the principal component 1 (PC1, Fig. 1D), suggesting that hypothalamus and thalamus astrocyte RNA-Seq datasets are mostly different from astrocyte RNA-Seq datasets of other regions. Second, the analysis showed that Cpu and Nacc astrocyte RNA-Seq datasets have a 20% of variation along the PC2 (Fig. 1D), indicating a more similar astroglial-translating mRNA profile between these two regions. The principal component analysis suggests that astrocyte-translating mRNA profile is closely associated with their anatomical location. Notably, a large number of astrocyte mRNAs were similarly expressed across brain regions. The highly expressed genes (log2FPKM > 10) in adult astrocytes (e.g., cst3, aldoc, ndrg2, mt1, and mt2) all have very similar FPKM values across different brain regions (Table 1).
Table 1.
Top expressed genes in adult astrocytes from individual mouse brain regions (shown in FPKM)
| Gene symbol | Ctx | Gene symbol | Hippo | Gene symbol | Nacc | Gene symbol | Cpu | Gene symbol | Thal | Gene symbol | Hypo |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Cst3 | 6598.62 | Cst3 | 8616.47 | Cst3 | 6186.01 | Cst3 | 7439.16 | Ttr | 5482.97 | Cst3 | 5994.07 |
| Aldoc | 3900.21 | Aldoc | 4760.01 | Ndrg2 | 6142.8 | Ttr | 7279.24 | Cst3 | 5266.85 | Ndrg2 | 5761.92 |
| Ndrg2 | 3197.53 | Mt2 | 4752.79 | Mt2 | 4405.63 | Mt2 | 4271.83 | Ndrg2 | 5174.98 | Aldoc | 5261.3 |
| Mt2 | 3008.21 | Dbi | 3927.37 | Dbi | 4050.09 | Mt1 | 4180.62 | Dbi | 3844.69 | Dbi | 3352.61 |
| Mt1 | 2574.61 | Mt1 | 3877.14 | Aldoc | 3661.01 | Dbi | 3596.49 | Mt2 | 2930.55 | Mt2 | 2687.04 |
| Glul | 2321.23 | Ndrg2 | 3861.61 | Mt1 | 3092.03 | Lars2 | 3565.6 | Aldoc | 2561.19 | Mt1 | 2590.29 |
| Dbi | 2051.17 | Cpe | 2590.3 | Apoe | 2759.57 | Apoe | 2576.81 | Mt1 | 2426.17 | Cpe | 2515.64 |
| Sparcl1 | 1758.45 | Glul | 2319.83 | Glul | 2488.46 | Rpl41 | 2574.89 | Apoe | 2229.18 | Apoe | 2032.85 |
| Snap25 | 1737.69 | Apoe | 2016.91 | Ckb | 2400.8 | Ndrg2 | 2548.03 | Ptgds | 2192.33 | Ckb | 2004.46 |
| Apoe | 1687.66 | Rpl41 | 1779.62 | Cpe | 1707.21 | Aldoc | 2453.2 | Cpe | 1986.47 | Sparcl1 | 1856.57 |
| Cpe | 1583.4 | Sparcl1 | 1658.48 | Sparcl1 | 1630.02 | Ckb | 2367.09 | Sparc | 1937.7 | Sparc | 1815.51 |
| Ptgds | 1580.17 | Lars2 | 1486.69 | Rpl41 | 1611.68 | Glul | 1765.95 | Ckb | 1704.31 | Glul | 1553.78 |
| Ckb | 1539.06 | Gstm1 | 1469.18 | Gstm1 | 1566.73 | Hist1h4d | 1707.83 | Sparcl1 | 1524.72 | Atp1b2 | 1502.49 |
| Gstm1 | 1394.81 | Ckb | 1391.11 | Lars2 | 1469.83 | Cox8a | 1453.67 | Glul | 1516.55 | Lars2 | 1469.55 |
| Rpl41 | 1246.43 | Slc1a2 | 1369.93 | Ywhae | 1394.46 | Gstm1 | 1187.64 | Rpl41 | 1320.16 | Rpl41 | 1380.42 |
| Tspan7 | 1118.53 | Tspan7 | 1238.74 | Sparc | 1276.94 | Atp5k | 1172.14 | Snap25 | 1223.16 | Ywhae | 1203.27 |
| Gstm5 | 1107.49 | Hist1h4d | 1110.87 | Nnat | 1057.93 | Gstm5 | 1171.29 | Nnat | 1198.21 | Ttr | 1137.24 |
| Ywhae | 1092.91 | Ywhae | 1108.02 | Cox8a | 1046.81 | Cox4i1 | 1150.81 | Lars2 | 1191.24 | Gstm1 | 1064.01 |
| Slc1a2 | 1042.71 | Gstm5 | 1093.61 | S100a1 | 1006.17 | Cpe | 1121.74 | Atp1b2 | 1157.9 | ||
| Clu | 1056.85 | Uqcr10 | 1046.62 | Ywhae | 1138.03 | ||||||
| Gstm1 | 1098.92 |
We further analyzed the unique regional astroglial mRNA signature from each region sample by comparing its RNA-Seq datasets with those of all other regions. For cortex and hippocampus, the enriched mRNA signature is identified with a more stringent q value (q < 0.001; Fig. 1E,F), whereas enriched mRNA signatures were identified from the rest of regions with a q < 0.01 (Fig. 1G–J). It is worthwhile to point out that subcortical region (Nacc, Cpu, and thalamus) astrocytes tend to have very uniquely expressed mRNAs that are not expressed in astrocytes from any other region examined. The representative top uniquely expressed genes in astrocytes of each region are listed in Table 2. Because these stringent q values only identified a small number of highly enriched (or uniquely expressed) genes, we also used less stringent q value (q < 0.1) to obtain larger number of genes that are relatively enriched in each region's astrocytes. The number of these genes in each region's astrocytes is shown in the Venn diagram (Fig. 1K, yellow, inside parentheses). The Venn diagram also showed the shared or enriched genes in each region's astrocytes. Because of the large total number of potential intersections between regions and the limited dimensions of the Venn diagram, we only displayed a subset of intersections (up to 3 regions) on the Venn diagram.
Table 2.
Representative top uniquely expressed genes (FPKM values) in astrocytes of different brain regions
| Gene symbol | Ctx | Hippo | Nacc | Cpu | Thal | Hypo |
|---|---|---|---|---|---|---|
| Kcnh7 | 10.9926 | 2.93249 | 2.0687425 | 2.421673333 | 3.007585 | 2.5854425 |
| Dkkl1 | 2.276714 | 0.2032365 | 0.1910775 | 0.528168333 | 0 | 0 |
| Tmem215 | 1.4593886 | 0.339576425 | 0.27376925 | 0.026210033 | 0.173311775 | 0.09126255 |
| Tmem132d | 1.4099348 | 0.30782775 | 0.388022 | 0.448074 | 0.44882775 | 0.418008 |
| Wnt10a | 0.8273358 | 0.187030275 | 0.1529765 | 0.030291 | 0.213710775 | 0.09525035 |
| Gene symbol | Hippo | Ctx | Nacc | Cpu | Thal | Hypo |
| Epcam | 1.8520275 | 0.10431866 | 0.07008275 | 0.057363333 | 0.17043325 | 0.105862 |
| Klk8 | 1.590955 | 0.2160908 | 0.04458025 | 0.048059 | 0.3311085 | 0.06985675 |
| Slc9a4 | 1.051725 | 0.05654138 | 0.0121237 | 0 | 0.1087185 | 0.005753575 |
| Krt2 | 0.79760175 | 0.0518049 | 0.0361653 | 0 | 0.010108 | 0.03271405 |
| Gene symbol | Nacc | Ctx | Hippo | Cpu | Thal | Hypo |
| Gnrh1 | 7.64711 | 0 | 0 | 0 | 0.103087 | 0 |
| Ucn3 | 3.342701 | 0.00957572 | 0 | 0 | 0.07933075 | 0.1961695 |
| Sash3 | 0.902787 | 0.05357386 | 0.04546785 | 0 | 0.07857125 | 0 |
| Slc25a34 | 185.7559 | 22.03586 | 17.823475 | 88.4137 | 19.389425 | 22.9692 |
| Gene symbol | Cpu | Ctx | Hippo | Nacc | Thal | Hypo |
| Prss56 | 1.961656667 | 0.01256876 | 0 | 0.41571525 | 0.013522075 | 0.087599475 |
| Acvrl1 | 1.78357 | 0.1244332 | 0 | 0.3183685 | 0.078967425 | 0.034625325 |
| Gm9833 | 0.777352667 | 0.09345392 | 0.0688435 | 0.017100425 | 0.150218275 | 0.1458375 |
| Lpar3 | 0.654218333 | 0.07840754 | 0.04563125 | 0.12758975 | 0.04096125 | 0.0561428 |
| Sprr1a | 0.615105 | 0 | 0 | 0 | 0.0403725 | 0.033528 |
| Gene symbol | Thal | Ctx | Hippo | Nacc | Cpu | Hypo |
| Defb25 | 1.53967 | 0.088143 | 0 | 0 | 0 | 0.3512075 |
| Eci3 | 0.77785825 | 0.09008554 | 0.070807 | 0.059224675 | 0.140395 | 0.1260191 |
| Mc3r | 0.40689525 | 0.01790572 | 0.020278 | 0.0930006 | 0.0445781 | 0.17774925 |
| Igsf5 | 0.3334155 | 0.03858668 | 0 | 0 | 0.077578333 | 0.0600255 |
| Pou2af1 | 0.2483465 | 0.0087492 | 0.0207151 | 0.057930875 | 0.045642233 | 0.016692175 |
| Gene symbol | Hypo | Ctx | Hippo | Nacc | Cpu | Thal |
| Gm13546 | 0.70789275 | 0.0247092 | 0.05235275 | 0 | 0 | 0.273812 |
| Serpinb1c | 0.24681725 | 0 | 0 | 0.066517 | 0.038012 | 0.101944 |
| Gucy2c | 0.21579575 | 0.00566476 | 0 | 0.024330425 | 0.0292756 | 0.298065 |
| AI317395 | 0.16049125 | 0.01412448 | 0.01195745 | 0.0280599 | 0 | 0.1552747 |
| Slc19a3 | 0.133734025 | 0.06637662 | 0.036552825 | 0.017952025 | 0.038626667 | 0.3096302 |
Uniquely expressed genes in astrocytes of cortex region. These genes were identified by comparing RNA-seq datasets of cortical astrocytes to RNA-seq datasets of all other regional astrocytes with a FDR adjusted P value (q value < 0.001). Download Table 2-1, XLSX file (13.9KB, xlsx)
Uniquely expressed genes in astrocytes of hippocampus region (q < 0.001). These genes were identified by comparing RNA-seq datasets of hippocampal astrocytes to RNA-seq datasets of all other regional astrocytes with a FDR adjusted P value (q value < 0.001). Download Table 2-2, XLSX file (12.8KB, xlsx)
Uniquely expressed genes in astrocytes of Nacc region (q < 0.01). These genes were identified by comparing RNA-seq datasets of nucleus accubens astrocytes to RNA-seq datasets of all other regional astrocytes with a FDR adjusted P value (q value < 0.01). Download Table 2-3, XLSX file (9.9KB, xlsx)
Uniquely expressed genes in astrocytes of Cpu region (q < 0.01). These genes were identified by comparing RNA-seq datasets of caudate putamen astrocytes to RNA-seq datasets of all other regional astrocytes with a FDR adjusted P value (q value < 0.01). Download Table 2-4, XLSX file (15.9KB, xlsx)
Uniquely expressed genes in astrocytes of thalamus region (q < 0.01). These genes were identified by comparing RNA-seq datasets of thalamic astrocytes to RNA-seq datasets of all other regional astrocytes with a FDR adjusted P value (q value < 0.01). Download Table 2-5, XLSX file (10.3KB, xlsx)
Uniquely expressed genes in astrocytes of hypothalamus region (q < 0.01). These genes were identified by comparing RNA-seq datasets of hypothalamic astrocytes to RNA-seq datasets of all other regional astrocytes with a FDR adjusted P value (q value < 0.01). Download Table 2-6, XLSX file (11KB, xlsx)
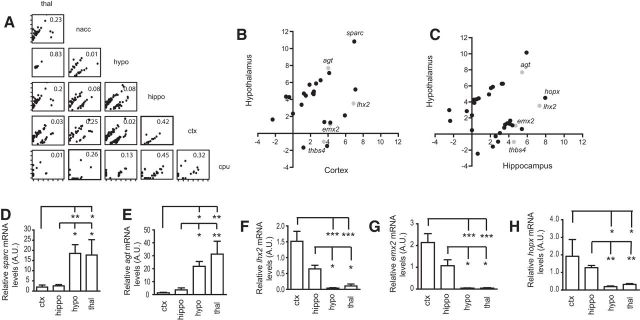
To further determine how the astroglial translating mRNA profiles are correlated among brain regions, we performed cuffdiff analysis (Trapnell et al., 2012) with region-specific TRAP/RNA-Seq datasets between any given two regions and then generated pairwise correlation plots using FPKM values of differentially expressed genes identified by cuffdiff. Again, we found that the similarity of adult astroglial translating mRNA profiles is highly dependent upon the anatomical location of compared regions. As shown in Figure 2A, thalamic and hypothalamic astrocytes share a highly similar profile of translating mRNAs (r = 0.83). Molecular profiles of translating mRNAs in cortical and hippocampal astrocytes are also similar with r = 0.42. In contrast, comparisons of anatomically distant regions, such as ctx/hippo versus hypo (r = 0.02 or 0.08) or nacc versus hypo (r = 0.01), yield extremely low r values (Fig. 2A), suggesting that the profile of translating mRNAs in astrocytes from these regions are highly different. Direct comparisons of astroglial gene profiles between dorsal (ctx/hippo) and ventral (thal/hypo) regions further identified a number of genes that were significantly and differentially expressed between ctx and hypo (Fig. 2B; for full list, see Fig. 2-1), or between hippo and hypo (Fig. 2C; for full list, see Fig. 2-2). Interestingly, many of these identified genes overlap between ctx and hippo (Fig. 2B; see italicized genes in Fig. 2-1; and Fig. 2-2). In particular, sparc mRNA, which encodes a synaptogenesis-modulating molecule (Sparc), has highly contrasting FPKMs between hypothalamus and hippocampus/cortex, and its expression pattern is consistent with its in situ hybridization results from the Allen brain atlas. Interestingly, the FPKM values of the sparcl1 mRNA that encodes SPARC-like 1 (also known as Hevin), a functional binding partner of Sparc, are similar between dorsal (FPKM: 1758 in cortex and 1658 in hippocampus) and ventral (FPKM: 1524 in thalamus and 1856 in hypothalamus) regions. Several transcription factors (hopx, lhx2, and emx2) and a representative enzyme (agt) also showed opposite expression patterns in astrocytes between dorsal forebrain (ctx/hippo) and ventral subcortical (thal/hypo) regions. These comparisons, together with Figure 1D, strongly suggest that the profile of astroglial translating mRNAs in brain regions follows a dorsal (cortex/hippocampus) to ventral (thalamus/hypothalamus) pattern.
Figure 2.
Differentially expressed translating mRNAs in astrocytes along the dorsoventral axis of the adult brain. A, Pairwise plots of differentially translating mRNAs in astrocytes between regions examined. These genes were identified using the pairwise cuffdiff analysis from region-specific TRAP/RNA-Seq datasets. Genes with FDR-adjusted p value (q value) <0.05 were considered significantly different. Correlation coefficient (r) value: the correlation of FPKM values between any two regions. Scatter plot of genes that were significantly differentially expressed between hypothalamus and cortex (B) or between hypothalamus and hippocampus (C). These genes are also shown in extended data (Figure 2-1; and Figure 2-2). B, C, Gray represents overlapped genes. qRT-PCR of differentially expressed genes sparc (D), F(3,11) = 6.01, p = 0.011; agt (E), F(3,11) = 10.16, p = 0.002; lhx2 (F), F(3,12) = 4.9, p = 0.02; emx2 (G), F(3,10) = 8.6, p = 0.004; hopx (H), F(3,10) = 3.84, p = 0.04, as determined by one-way ANOVA test, in TRAP-isolated RNA samples from dorsal ctx/hippo and ventral hypo/thal regions. *p < 0.05 (one-way ANOVA with a Tukey post hoc test). **p < 0.01 (one-way ANOVA with a Tukey post hoc test). ***p < 0.001 (one-way ANOVA with a Tukey post hoc test). n = 2–6 independent TRAP samples from 4–6 mice per region. GFAP mRNA was used as the endogenous control to normalize loading mRNA quantity.
Genes that are differentially expressed in astrocytes between cortex and hypothalamus. These genes were identified using the cuffdiff pairwise analysis between RNA-seq datasets of cortical and hypothalamic astrocytes (q < 0.05). Download Figure 2-1, XLSX file (17.1KB, xlsx)
Genes that are differentially expressed in astrocytes between hippocampus and hypothalamus. These genes were identified using the cuffdiff pairwise analysis between RNA-seq datasets of hippocampal and hypothalamic astrocytes (q < 0.05). Download Figure 2-2, XLSX file (12.3KB, xlsx)
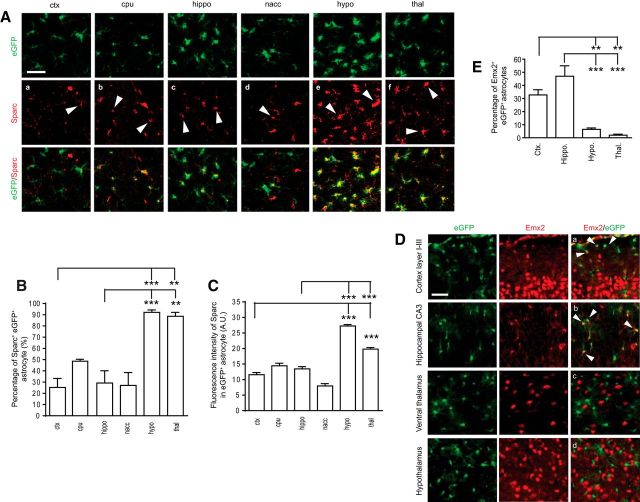
We next validated the mRNA levels of these genes in new TRAP isolated samples from representative ctx/hippo and hypo/thal regions using qRT-PCR. As shown in Figure 2D, E, sparc and agt mRNA levels were significantly higher in hypo/thal regions than in ctx/hippo regions. In contrast, lhx2, emx2, and hopx mRNA levels were significantly higher in ctx/hippo than in hypo/thal (Fig. 2F–H), consistent with the TRAP/RNA-Seq results. We also performed immunostaining of representative differentially expressed genes to determine whether their protein expression also follows dorsal to ventral patterns. For sparc, we found clear Sparc immunoreactivity in all brain regions, with hypothalamus being the highest (Fig. 3A). In addition, Sparc immunoreactivity almost completely colocalizes (∼90%) with eGFP-L10a fluorescence in hypothalamus and thalamus but is much less colocalized (20%–30%) in other brain regions examined (Fig. 3B). Subsequent quantification of Sparc immunoreactivity in eGFP+ astrocytes found that the Sparc intensity in astrocytes gradually increased along the dorsoventral axis, particularly from cortex to hypothalamus, which has >2-fold higher Sparc immunoreactivity in astrocytes (Fig. 3C). These results suggest a highly selective expression of Sparc protein in hypothalamic/thalamic astrocytes. Emx2 is known to be expressed dorsally in the forebrain, playing important roles for cortex and hippocampus development (Cecchi and Boncinelli, 2000). We found that Emx2 immunoreactivity was also frequently colocalized (30%–50%; Fig. 3D) with eGFP+ astrocytes in BAC aldh1l1-TRAP mice in cortex and hippocampus (Fig. 3Da,Db, white arrows), in addition to neuronal colocalization. The intensity of Emx2 in cortical and hippocampal astrocytes was comparable (quantification not shown). However, we observed only rare (<5%) colocalization of Emx2 with eGFP-L10a in thalamus and hypothalamus (Fig. 3Dc,Dd,E). These region-specific expression patterns likely imply a selective function of these proteins in astrocytes of different brain regions.
Figure 3.
Differential immunostaining patterns of the representative synaptogenic modulator and the transcriptional factor in astrocytes of the adult brain. A, Immunostaining of Sparc in brain regions of BAC aldh1l1-TRAP mice (P60). Scale bar, 50 μm. White arrows indicate Sparc immunoreactivity in eGFP+ astrocytes. B, Percentage of Sparc+eGFP+ astrocytes in each brain region examined. n = 90–215 astrocytes/region from 2 mice (F(5,913) = 17.2, p < 0.0001). C, Intensity of Sparc immunofluorescence in eGFP+ astrocytes in brain regions examined. n = 50–118 astrocytes/region from 2 mice (F(5,410) = 108, p < 0.0001). D, Immunostaining of Emx2 in brain regions of BAC aldh1l1-TRAP mice (P60). Scale bar, 50 μm. White arrows indicate Emx2 immunoreactivity in eGFP+ astrocytes. E, Percentage of Emx2+eGFP+ astrocytes in each brain region examined. n = 90–110 astrocytes/region from 2 mice (F(3,224) = 22.6, p < 0.0001). **p < 0.01 (one-way ANOVA and a Tukey post hoc test). ***p < 0.001 (one-way ANOVA and a Tukey post hoc test).
Dorsoventral region-selective astroglial modulation on neurite outgrowth and synaptic activity of neurons
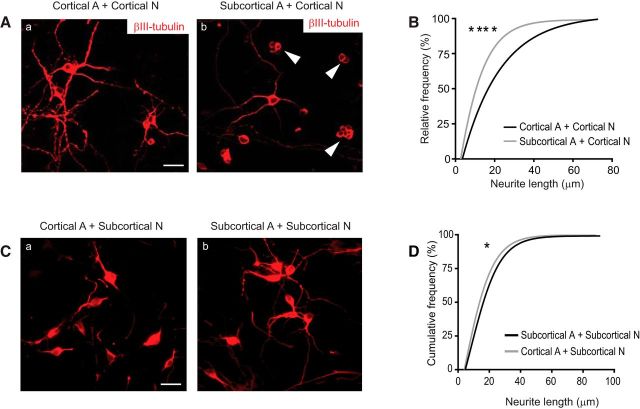
Astrocytes have been recently suggested to actively modulate neuronal synaptogenesis and maturation through secretion of several extracellular matrix proteins (Eroglu and Barres, 2010). The spatial specificity of astrocyte-mediated modulation of synaptogenesis and maturation, however, remains largely unknown. To assess whether astrocytes derived from different regions differentially modulate neuronal growth and maturation, we first prepared cortical neurons with either cortical or subcortical (no hippocampus) astrocyte cocultures. While we observed robust neurite growth of cortical neurons when cocultured with cortical astrocytes, revealed by the abundant βIII-tubulin staining signals (Fig. 4Aa), we found that many cortical neurons bear no or minimal neurites (<2 μm) in the presence of subcortical astrocytes (Fig. 4Ab, white arrows). In addition, quantification of neurite length on neurite-bearing cortical neurons found that a greater number of cortical neurons appeared to have overall significantly longer neurites when cocultured with cortical than subcortical astrocytes (Fig. 4B).
Figure 4.
Dorsoventral region-selective astroglial modulation on neurite outgrowth of neurons. A, Representative neuronal morphology of cortical neurons cocultured with cortical or subcortical astrocytes. White arrows indicate neurons bearing no processes. Scale bar, 20 μm. B, Cumulative frequency of the neurite length of cortical neurons cocultured with cortical or subcortical astrocytes. Only neurite-bearing neurons (>2 μm) were quantified. n = 141–162 neurons from three independent cultures. C, Representative neuronal morphology of subcortical neurons cocultured with cortical or subcortical astrocytes. Scale bar, 20 μm. D, Cumulative frequency of the neurite length of subcortical neurons cocultured with cortical or subcortical astrocytes. n = 150–174 neurons from three independent cultures. ****p < 0.0001 (Kolmogorov–Smirnov test). *p < 0.05 (Kolmogorov–Smirnov test).
To determine whether subcortical astrocytes preferentially promote subcortical neurons during early development, we next prepared subcortical neuron with subcortical or cortical astrocyte cocultures. Interestingly, subcortical neurons develop significantly longer neurites when they were cocultured with subcortical astrocytes (Fig. 4Cb) than with cortical astrocytes (Fig. 4Ca). Especially, individual subcortical neurons appear to bear multiple neurites with the presence of subcortical but not cortical astrocytes (Fig. 4C), although the overall neurite length per neuron in subcortical neurons cocultured with cortical astrocytes is not as drastically different as in the subcortical neurons cocultured with subcortical astrocytes (Fig. 4D). Subcortical neurons without any neurites when cocultured with cortical astrocytes were also rarely observed. We also performed the live/dead cell viability assay (Thermo Fisher Scientific) in our cultures and rarely found dead neurons in all cocultures (data not shown), suggesting that mismatched astrocytes do not lead to abnormal neuronal death in cocultures. Overall, these results suggest an interesting region-matched effect of astrocytes in promoting neurite growth, of which cortical astrocytes are less selective compared with subcortical astrocytes.
We also examined the basic electrophysiological properties and synaptic activity of cortical and subcortical neurons in these cocultures. We noticed that subcortical neurons are quite heterogeneous compared with more homogeneous glutamatergic cortical neurons. We generally are able to identify 61% (27 of 44 patched neurons) glutamatergic-like neurons in subcortical neuron and subcortical astrocyte cocultures. This percentage decreases to 54% (26 of 48 patched neurons) when subcortical neurons are cocultured with cortical astrocytes. In contrast, we routinely identify 80% (29 of 36 patched) glutamatergic-like neurons in cortical neuron and cortical astrocyte cocultures, although it decreases to 70% (28 of 39) in cortical neuron and subcortical astrocyte cocultures. The glutamatergic-like neurons are determined by their typical firing patterns, as shown in Figure 5A. Because of the heterogeneous neuronal populations in cocultures, to make relevant and meaningful comparisons, we only focused on neurons that exhibited typical glutamatergic-like neuron firing properties in all cocultures before continuing with all other recordings.
Figure 5.
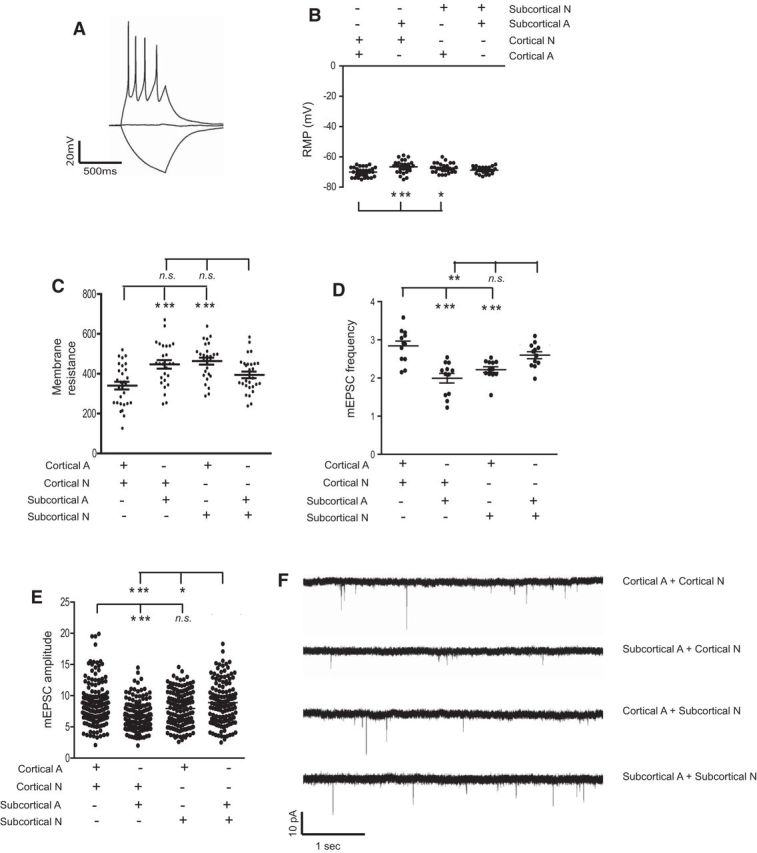
Dorsoventral region-selective astroglial modulation on synaptic activity of neurons. A, Representative firing pattern of the glutamatergic-like neurons patched in both cortical and subcortical cocultures; Resting membrane potential (RMP, B) and membrane resistance (C) of cortical or subcortical neurons cocultured with cortical or subcortical astrocytes (F(3,106) = 6.1, p = 0.0007 for the RMP and F(3,106) = 9.0, p < 0.0001 for the membrane resistance), as determined by one-way ANOVA test; n = 28 or 29 neurons from three independent cultures. Quantitative summary of frequency (D) and amplitude (E) of mEPSC recordings (F(3,44) = 12.5, p < 0.0001 for the frequency and F(3,576) = 15.4, p < 0.0001 for the amplitude), as determined by one-way ANOVA test; n = 26–29 neurons from three independent cultures. *p < 0.05 (one-way ANOVA test with a Tukey post hoc test). **p < 0.01 (one-way ANOVA test with a Tukey post hoc test). ***p < 0.001 (one-way ANOVA test with a Tukey post hoc test). F, Representative mEPSC recording traces of cortical or subcortical neurons cocultured with cortical or subcortical astrocytes, respectively.
We found that the resting membrane potential of cortical or subcortical neurons cocultured with cortical or subcortical astrocytes is generally similar, except with cortical neurons cocultured with subcortical astrocytes, which is −5 mV more depolarized (Fig. 5B). Similarly, membrane resistance of cortical neurons cocultured with subcortical astrocytes is 106 mΩ increased compared with cortical neurons cocultured with cortical astrocytes (Fig. 5C). This increase in membrane resistance was also observed in subcortical neurons cocultured with cortical astrocytes (Fig. 5C), suggesting that cortical neurons cocultured with subcortical astrocytes or subcortical neurons cocultured with cortical astrocytes may not have ion channels developed similarly as neurons cocultured with astrocytes from the same region. We next recorded the mEPSC of cortical or subcortical neurons in mismatched cocultures to examine the functional properties of neuronal synapses in these cocultures. Cortical neurons cocultured with cortical astrocytes formed the most active synapses with the highest frequency (2.84 Hz) and amplitude (8.8 pA) of mEPSCs (Fig. 5D,E). Similarly, subcortical neurons cocultured with subcortical astrocytes also formed active synapses with comparable frequency (2.6 Hz) and amplitude (8.9 pA) of mEPSCs. However, cortical neurons cocultured with subcortical astrocytes or subcortical neurons cocultured with cortical astrocytes have much less active synapses with significantly reduced frequency (2.0 and 2.2 Hz, respectively) of mEPSCs (Fig. 4D,E). Interestingly, subcortical neurons cocultured with cortical astrocytes have significantly higher mEPSC amplitude (7.9 pA) and slightly higher mEPSC frequency (2.2 Hz) compared with that (6.8 pA and 2.0 Hz) of cortical neurons cocultured with subcortical astrocytes. Representative mEPSC traces from each coculture are shown in Figure 5F. These results demonstrate that astroglial modulation of synaptic maturation is region-selective, especially to the subcortical astrocytes. This is consistent to the observation above that subcortical astrocytes are particularly less competent to promote neurite growth of cortical neurons.
Discussion
Although astrocytes are considered heterogeneous (Chaboub and Deneen, 2012; Bayraktar et al., 2014), in vivo molecular evidence to support this notion has not been well established. By TRAP isolation of translating astroglial mRNAs from representative brain regions and subsequent RNA-Seq analysis, we provide in vivo evidence for the molecular heterogeneity of brain astrocytes. Interestingly, our results showed that the expression pattern of ribosome-associated mRNA profiles in astrocytes closely follows the dorsoventral axis, especially posteriorly from cortex/hippocampus to thalamus/hypothalamus. In line with this, our functional results further demonstrated that astrocyte-mediated modulation on neuronal neurite growth and synaptic activity is region-selective. These results represent renewed effort to define the in vivo molecular heterogeneity of adult astrocytes.
Our current study is in contrast to a recent in vivo study (John Lin et al., 2017) to define astrocyte diversity based on cell surface antigen and aldh1l1-eGFP reporter in several aspects, including rationale (regional vs cell surface antigen), approach (TRAP vs immune/FACS), read-out (mRNA vs cell surface protein antigen), and brain region's astrocyte sequenced (cortical/subcortical regions vs cortex/olfactory bulb/brainstem). In our study, we found no significant difference in the mRNA levels of several cell surface antigens (CD51, CD71, and CD63) that were used to define astrocyte subpopulations in the previous study (John Lin et al., 2017). In addition to the difference of approach and read-out described above, this could also be due to the coexistence of multiple astrocyte subpopulations (with different percentage) in different brain regions, from which the overall astrocyte gene signatures are mixed. Consequently, we also noted that unique mRNA expression signature in astrocytes from different brain regions is not commonly found in our RNA-Seq datasets (Fig. 1; Table 2), even with stringent q values. These uniquely expressed mRNAs also tend to have small FPKM values, indicating their low abundance in the overall transcriptome. In contrast, astroglial mRNAs with differential FPKM values in different brain regions are more commonly identified (Fig. 2), becoming the major form of molecular heterogeneity of astrocytes.
Neural circuitry that controls our behavior and cognition often involves multiple brain regions. Astrocytes are suggested to participate in the assembly of neural circuitry by actively regulating generation (Christopherson et al., 2005), maturation (Allen et al., 2012), and elimination (Stevens et al., 2007) of synapses through the secretion of an array of extracellular matrix molecules. How these molecules specifically regulate local neural circuitry in different brain regions (and cortical layers) remains essentially unknown. Our results found that cortical or subcortical astrocytes preferentially promote neurite growth and synaptic activity of neurons from the same region, especially the subcortical astrocytes. These results suggest that astroglial modulation of synaptogenesis and synaptic activity is not only neuronal type-dependent, as previously described (Christopherson et al., 2005), but also is region-matched, even for the similar type of glutamatergic-like neurons.
It is likely that cortical astrocytes either secrete or express unique signals/ligands on the surface, which can be selectively recognized by cortical neurons. However, these signals/ligands are likely not sufficiently produced from subcortical astrocytes. On the other hand, subcortical astrocytes may secrete or express different sets of signals or ligands on the surface for their recognition by subcortical glutamatergic-like neurons for their neurite growth and synaptic activity. Thus, there are likely to have two distinct sets of pathways that are separately involved in cortical or subcortical astrocyte-mediated modulation on cortical or subcortical neuronal properties, respectively. These signals/ligands involved in these distinct pathways are likely to be differentially expressed in cortical or subcortical regions. It is also possible that astrocytes from different regions may differentially express molecules that selectively antagonize synaptogenesis of neurons that are not from the same region. Indeed, we found that Sparc expression is selectively high in the hypothalamus/thalamus, but very low in the cortex/hippocampus, with its immunoreactivity overlapping primarily with eGFP+ astrocytes only in hypothalamus/thalamus regions of BAC aldh1l1-TRAP mice. It is known that Sparc antagonizes the synaptogenic functions of Hevin (Kucukdereli et al., 2011) (encoded by the sparcl1 gene), also secreted from astrocytes, in glutamatergic synaptogenesis. Because we observed no significant changes of Hevin expression between dorsal and ventral brain regions, the selectively high expression of Sparc in subcortical astrocytes may selectively suppress Hevin's synaptogenic function to cortical neurons. Because subcortical astrocytes may use different sets of signals/ligands to promote neurite growth and synaptic maturation of subcortical neurons, the high expression of Sparc may only affect neurite growth and synaptic functions of cortical but not subcortical glutamatergic-like neurons. These results highlight the need for the region/nuclei-specific investigation of astroglial functions in the adult brain. Our current profile of the ribosome-associated mRNA (presumably actively translating mRNA) provides a useful in vivo molecular atlas for exploring region-specific astroglial functions.
Region-specific molecular properties of astrocytes have important implications in CNS pathology initiation and progression. The selective susceptibility of certain neurons in brain regions/nuclei is one of the characteristic but unsolved features of major neurodegenerative diseases. By characterizing specific molecular functions of astrocytes in different brain regions, our profiling data enable investigation of whether these specific functions are altered, which may selectively contribute to neuronal dysfunction and even cell death in these pathologic regions of neurodegenerative diseases. Similarly, it also provides new insights about how astrocytes are involved in psychiatric disorders, which typically manifest a region-specific neuronal or circuitry dysfunction.
How astroglial heterogeneity arises is not clearly understood. Astrocyte fate-mapping experiments (Hochstim et al., 2008; Tsai et al., 2012) in spinal cord and forebrain suggest that astrocytes are derived in a region-restricted manner from radial glia's domains of origin. As a result, it is conceivable that astrocyte heterogeneity is intrinsically defined by the origin of radial glial cells, which follows early embryonic dorsoventral (and posteroanterior) axis patterning. Indeed, dorsal Bergmann glia and ventral velate astrocytes in the cerebellum show opposite expression patterns for a number of synaptic proteins, such as GluA1, GluA4, GLAST, Kir4.1, and GLT1 (Farmer et al., 2016). Our astrocyte TRAP profiling found that dorsoventral regional astroglial gene expression is the most distinct among all regions examined, supporting the notion that molecular heterogeneity in adult astrocytes is formed in early development. How intrinsic mechanisms and early patterning signals interact to shape molecular heterogeneity remains to be determined. Meanwhile, localized neuronal signals, together with other extrinsic signals (e.g., growth factors), are also likely to influence specific molecular and functional properties of astrocytes in each brain region or nucleus during the postnatal astrocyte maturation phase. We previously showed that selective loss of VGluT1 neuronal glutamatergic signaling selectively affects the morphological and molecular maturation of cortical, but not hypothalamic astrocytes (Morel et al., 2014). Interestingly, we found differential expression levels for a number of transcriptional regulators in astrocytes (emx2, lhx2, and hopx). Whether and how these transcriptional factors mediate either localized neuronal or growth factor signals in directing astrocyte heterogeneity during postnatal development will be investigated in the future.
Footnotes
This work was supported by National Institutes of Health Grants R01MH099554, R01MH106490, and R21NS087391 to Y.Y. We thank Anna Nathanson for optimizing Sparc staining; and Tufts Center for Neuroscience Research and Tufts core facility genomics for imaging and sequencing support.
The authors declare no competing financial interests.
References
- Allen NJ, Bennett ML, Foo LC, Wang GX, Chakraborty C, Smith SJ, Barres BA (2012) Astrocyte glypicans 4 and 6 promote formation of excitatory synapses via GluA1 AMPA receptors. Nature 486:410–414. 10.1038/nature11059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azevedo FA, Carvalho LR, Grinberg LT, Farfel JM, Ferretti RE, Leite RE, Jacob Filho W, Lent R, Herculano-Houzel S (2009) Equal numbers of neuronal and nonneuronal cells make the human brain an isometrically scaled-up primate brain. J Comp Neurol 513:532–541. 10.1002/cne.21974 [DOI] [PubMed] [Google Scholar]
- Bachoo RM, Kim RS, Ligon KL, Maher EA, Brennan C, Billings N, Chan S, Li C, Rowitch DH, Wong WH, DePinho RA (2004) Molecular diversity of astrocytes with implications for neurological disorders. Proc Natl Acad Sci U S A 101:8384–8389. 10.1073/pnas.0402140101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batter DK, Corpina RA, Roy C, Spray DC, Hertzberg EL, Kessler JA (1992) Heterogeneity in gap junction expression in astrocytes cultured from different brain regions. Glia 6:213–221. 10.1002/glia.440060309 [DOI] [PubMed] [Google Scholar]
- Bayraktar OA, Fuentealba LC, Alvarez-Buylla A, Rowitch DH (2015) Astrocyte development and heterogeneity. Cold Spring Harb Perspect Biol 7:a020362. 10.1101/cshperspect.a020362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahoy JD, Emery B, Kaushal A, Foo LC, Zamanian JL, Christopherson KS, Xing Y, Lubischer JL, Krieg PA, Krupenko SA, Thompson WJ, Barres BA (2008) A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J Neurosci 28:264–278. 10.1523/JNEUROSCI.4178-07.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cecchi C, Boncinelli E (2000) Emx homeogenes and mouse brain development. Trends Neurosci 23:347–352. 10.1016/S0166-2236(00)01608-8 [DOI] [PubMed] [Google Scholar]
- Chaboub LS, Deneen B (2012) Developmental origins of astrocyte heterogeneity: the final frontier of CNS development. Dev Neurosci 34:379–388. 10.1159/000343723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai H, Diaz-Castro B, Shigetomi E, Monte E, Octeau JC, Yu X, Cohn W, Rajendran PS, Vondriska TM, Whitelegge JP, Coppola G, Khakh BS (2017) Neural circuit-specialized astrocytes: transcriptomic, proteomic, morphological, and functional evidence. Neuron 95:531–549. 10.1016/j.neuron.2017.06.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christopherson KS, Ullian EM, Stokes CC, Mullowney CE, Hell JW, Agah A, Lawler J, Mosher DF, Bornstein P, Barres BA (2005) Thrombospondins are astrocyte-secreted proteins that promote CNS synaptogenesis. Cell 120:421–433. 10.1016/j.cell.2004.12.020 [DOI] [PubMed] [Google Scholar]
- Clasadonte J, Morel L, Barrios-Camacho CM, Chiang MS, Zhang J, Iyer L, Haydon PG, Yang Y (2016) Molecular analysis of acute and chronic reactive astrocytes in the pilocarpine model of temporal lobe epilepsy. Neurobiol Dis 91:315–325. 10.1016/j.nbd.2016.03.024 [DOI] [PubMed] [Google Scholar]
- Danbolt NC. (2001) Glutamate uptake. Prog Neurobiol 65:1–105. 10.1016/S0301-0082(00)00067-8 [DOI] [PubMed] [Google Scholar]
- Clasadonte J, Morel L, Barrios-Camacho CM, Chiang MS, Zhang J, Iyer L, Haydon PG, Yang Y (2008) Application of a translational profiling approach for the comparative analysis of CNS cell types. Cell 135:749–762. 10.1016/j.cell.2008.10.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eroglu C, Barres BA (2010) Regulation of synaptic connectivity by glia. Nature 468:223–231. 10.1038/nature09612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer WT, Abrahamsson T, Chierzi S, Lui C, Zaelzer C, Jones EV, Bally BP, Chen GG, Théroux JF, Peng J, Bourque CW, Charron F, Ernst C, Sjöström PJ, Murai KK (2016) Neurons diversify astrocytes in the adult brain through sonic hedgehog signaling. Science 351:849–854. 10.1126/science.aab3103 [DOI] [PubMed] [Google Scholar]
- Haydon PG, Carmignoto G (2006) Astrocyte control of synaptic transmission and neurovascular coupling. Physiol Rev 86:1009–1031. 10.1152/physrev.00049.2005 [DOI] [PubMed] [Google Scholar]
- Hochstim C, Deneen B, Lukaszewicz A, Zhou Q, Anderson DJ (2008) Identification of positionally distinct astrocyte subtypes whose identities are specified by a homeodomain code. Cell 133:510–522. 10.1016/j.cell.2008.02.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- John Lin CC, Yu K, Hatcher A, Huang TW, Lee HK, Carlson J, Weston MC, Chen F, Zhang Y, Zhu W, Mohila CA, Ahmed N, Patel AJ, Arenkiel BR, Noebels JL, Creighton CJ, Deneen B (2017) Identification of diverse astrocyte populations and their malignant analogs. Nat Neurosci 20:396–405. 10.1038/nn.4493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerkerian L, Nieoullon A, Dusticier N (1982) Brain glutamate uptake: regional distribution study from sensorimotor areas in the cat. Neurochem Int 4:275–281. 10.1016/0197-0186(82)90064-X [DOI] [PubMed] [Google Scholar]
- Kettenmann H. (2015) Neuroglia, Ed 2 New York: Oxford UP. [Google Scholar]
- Kucukdereli H, Allen NJ, Lee AT, Feng A, Ozlu MI, Conatser LM, Chakraborty C, Workman G, Weaver M, Sage EH, Barres BA, Eroglu C (2011) Control of excitatory CNS synaptogenesis by astrocyte-secreted proteins Hevin and SPARC. Proc Natl Acad Sci U S A 108:E440–E449. 10.1073/pnas.1104977108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, Boe AF, Boguski MS, Brockway KS, Byrnes EJ, Chen L, Chen L, Chen TM, Chin MC, Chong J, Crook BE, Czaplinska A, Dang CN, Datta S, Dee NR, et al. (2007) Genome-wide atlas of gene expression in the adult mouse brain. Nature 445:168–176. 10.1038/nature05453 [DOI] [PubMed] [Google Scholar]
- Molofsky AV, Kelley KW, Tsai HH, Redmond SA, Chang SM, Madireddy L, Chan JR, Baranzini SE, Ullian EM, Rowitch DH (2014) Astrocyte-encoded positional cues maintain sensorimotor circuit integrity. Nature 509:189–194. 10.1038/nature13161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel L, Higashimori H, Tolman M, Yang Y (2014) VGluT1+ neuronal glutamatergic signaling regulates postnatal developmental maturation of cortical protoplasmic astroglia. J Neurosci 34:10950–10962. 10.1523/JNEUROSCI.1167-14.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng L, Bernard A, Lau C, Overly CC, Dong HW, Kuan C, Pathak S, Sunkin SM, Dang C, Bohland JW, Bokil H, Mitra PP, Puelles L, Hohmann J, Anderson DJ, Lein ES, Jones AR, Hawrylycz M (2009) An anatomic gene expression atlas of the adult mouse brain. Nat Neurosci 12:356–362. 10.1038/nn.2281 [DOI] [PubMed] [Google Scholar]
- Oberheim NA, Goldman SA, Nedergaard M (2012) Heterogeneity of astrocytic form and function. Methods Mol Biol 814:23–45. 10.1007/978-1-61779-452-0_3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramasamy A, Trabzuni D, Guelfi S, Varghese V, Smith C, Walker R, De T, De T, De T, Coin L, de Silva R, Cookson MR, Singleton AB, Hardy J, Ryten M, Weale ME (2014) Genetic variability in the regulation of gene expression in ten regions of the human brain. Nat Neurosci 17:1418–1428. 10.1038/nn.3801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regan MR, Huang YH, Kim YS, Dykes-Hoberg MI, Jin L, Watkins AM, Bergles DE, Rothstein JD (2007) Variations in promoter activity reveal a differential expression and physiology of glutamate transporters by glia in the developing and mature CNS. J Neurosci 27:6607–6619. 10.1523/JNEUROSCI.0790-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somjen GG. (1988) Nervenkitt: notes on the history of the concept of neuroglia. Glia 1:2–9. 10.1002/glia.440010103 [DOI] [PubMed] [Google Scholar]
- Stevens B, Allen NJ, Vazquez LE, Howell GR, Christopherson KS, Nouri N, Micheva KD, Mehalow AK, Huberman AD, Stafford B, Sher A, Litke AM, Lambris JD, Smith SJ, John SW, Barres BA (2007) The classical complement cascade mediates CNS synapse elimination. Cell 131:1164–1178. 10.1016/j.cell.2007.10.036 [DOI] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L (2012) Differential gene and transcript expression analysis of RNA-Seq experiments with TopHat and Cufflinks. Nat Protoc 7:562–578. 10.1038/nprot.2012.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai HH, Li H, Fuentealba LC, Molofsky AV, Taveira-Marques R, Zhuang H, Tenney A, Murnen AT, Fancy SP, Merkle F, Kessaris N, Alvarez-Buylla A, Richardson WD, Rowitch DH (2012) Regional astrocyte allocation regulates CNS synaptogenesis and repair. Science 337:358–362. 10.1126/science.1222381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Vidensky S, Jin L, Jie C, Lorenzini I, Frankl M, Rothstein JD (2011) Molecular comparison of GLT1+ and ALDH1L1+ astrocytes in vivo in astroglial reporter mice. Glia 59:200–207. 10.1002/glia.21089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh TH, Lee DY, Gianino SM, Gutmann DH (2009) Microarray analyses reveal regional astrocyte heterogeneity with implications for neurofibromatosis type 1 (NF1)-regulated glial proliferation. Glia 57:1239–1249. 10.1002/glia.20845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Barres BA (2010) Astrocyte heterogeneity: an underappreciated topic in neurobiology. Curr Opin Neurobiol 20:588–594. 10.1016/j.conb.2010.06.005 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Chen K, Sloan SA, Bennett ML, Scholze AR, O'Keeffe S, Phatnani HP, Guarnieri P, Caneda C, Ruderisch N, Deng S, Liddelow SA, Zhang C, Daneman R, Maniatis T, Barres BA, Wu JQ (2014) An RNA-Sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J Neurosci 34:11929–11947. 10.1523/JNEUROSCI.1860-14.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Uniquely expressed genes in astrocytes of cortex region. These genes were identified by comparing RNA-seq datasets of cortical astrocytes to RNA-seq datasets of all other regional astrocytes with a FDR adjusted P value (q value < 0.001). Download Table 2-1, XLSX file (13.9KB, xlsx)
Uniquely expressed genes in astrocytes of hippocampus region (q < 0.001). These genes were identified by comparing RNA-seq datasets of hippocampal astrocytes to RNA-seq datasets of all other regional astrocytes with a FDR adjusted P value (q value < 0.001). Download Table 2-2, XLSX file (12.8KB, xlsx)
Uniquely expressed genes in astrocytes of Nacc region (q < 0.01). These genes were identified by comparing RNA-seq datasets of nucleus accubens astrocytes to RNA-seq datasets of all other regional astrocytes with a FDR adjusted P value (q value < 0.01). Download Table 2-3, XLSX file (9.9KB, xlsx)
Uniquely expressed genes in astrocytes of Cpu region (q < 0.01). These genes were identified by comparing RNA-seq datasets of caudate putamen astrocytes to RNA-seq datasets of all other regional astrocytes with a FDR adjusted P value (q value < 0.01). Download Table 2-4, XLSX file (15.9KB, xlsx)
Uniquely expressed genes in astrocytes of thalamus region (q < 0.01). These genes were identified by comparing RNA-seq datasets of thalamic astrocytes to RNA-seq datasets of all other regional astrocytes with a FDR adjusted P value (q value < 0.01). Download Table 2-5, XLSX file (10.3KB, xlsx)
Uniquely expressed genes in astrocytes of hypothalamus region (q < 0.01). These genes were identified by comparing RNA-seq datasets of hypothalamic astrocytes to RNA-seq datasets of all other regional astrocytes with a FDR adjusted P value (q value < 0.01). Download Table 2-6, XLSX file (11KB, xlsx)
Genes that are differentially expressed in astrocytes between cortex and hypothalamus. These genes were identified using the cuffdiff pairwise analysis between RNA-seq datasets of cortical and hypothalamic astrocytes (q < 0.05). Download Figure 2-1, XLSX file (17.1KB, xlsx)
Genes that are differentially expressed in astrocytes between hippocampus and hypothalamus. These genes were identified using the cuffdiff pairwise analysis between RNA-seq datasets of hippocampal and hypothalamic astrocytes (q < 0.05). Download Figure 2-2, XLSX file (12.3KB, xlsx)