Abstract
Extension of the genetic code for the introduction of nonnatural amino acids into proteins was examined by using five-base codon–anticodon pairs. A streptavidin mRNA containing a CGGUA codon at the Tyr54 position and a tRNAUACCG chemically aminoacylated with a nonnatural amino acid were added to an Escherichia coli in vitro translation system. Western blot analysis indicated that the CGGUA codon is decoded by the aminoacyl-tRNA containing the UACCG anticodon. HPLC analysis of the tryptic fragment of the translation product revealed that the nonnatural amino acid was incorporated corresponding to the CGGUA codon without affecting the reading frame adjacent to the CGGUA codon. Another 15 five-base codons CGGN1N2, where N1 and N2 indicate one of four nucleotides, were also successfully decoded by aminoacyl-tRNAs containing the complementary five-base anticodons. These results provide a novel strategy for nonnatural mutagenesis as well as a novel insight into the mechanism of frameshift suppression.
INTRODUCTION
Extension of the genetic code is an important step to introduce nonnatural amino acids into proteins. Extension was first achieved using a nonsense UAG codon that could be decoded by a chemically aminoacylated nonsense suppressor tRNACUA (1,2). This methodology has been widely used to study structure and function of proteins (3–7) and the mechanism of translation (8–10). However, the nonsense suppression strategy is not highly efficient because of competition with a release factor, although some modifications have been reported (11). In addition, more than two different nonnatural amino acids cannot be introduced into a single protein, because of the limited types of stop codons. To make nonnatural mutagenesis more useful and more practical, we need to develop an alternative strategy for extension of the codon system that can achieve efficient and multiple incorporation of nonnatural amino acids. Bain et al. reported a nonnatural codon–anticodon pair containing isoC and isoG as a candidate for the extended codon–anticodon pair (12). However, the mRNA and tRNA containing the nonnatural bases must be synthesized chemically.
Recently we reported that four-base codons such as AGGU and CGGG are decoded by chemically aminoacylated tRNAs containing complementary four-base anticodons (13–15). When a tRNACCCG chemically aminoacylated with a nonnatural amino acid was added to an Escherichia coli or rabbit reticulocyte in vitro translation system together with a mRNA containing a CGGG codon, the nonnatural amino acid was successfully introduced into the directed position of the protein (15). Moreover, we have achieved incorporation of two different nonnatural amino acids into a single protein by using two independent four-base codons (16). In this article we report that even five-base codons can be decoded by aminoacylated tRNAs containing five-base anticodons. This five-base codon system has the advantage that up to 16 five-base codons may be derived from a single triplet codon. In addition, investigation of five-base decoding may provide a novel insight into the mechanism of frameshifting.
Five-base codon–anticodon pairing has not yet been proposed, but two unusual findings related to extended base pairing have been reported. First, decoding of five consecutive bases as a single codon was observed in an E.coli hopR mutant, in which GUGUG was translated into valine by a mutant tRNAVal containing a three-base anticodon (17). This phenomenon is explained in terms of tRNA hopping. Secondly, tRNAs having nine-base anticodon loops were synthesized by inserting one base on either the 5′- or 3′-side of the four-base anticodon and some of them could decode the four-base codon in vivo (18). This report shows the ability of the ribosome to accept a nine-base anticodon loop.
MATERIALS AND METHODS
Plasmid
A synthetic streptavidin gene was purchased from R&D Systems Europe. The streptavidin gene was cloned into a pGEMEX-1 (Promega)-derived vector, which was constructed for a previous study (15). The resulting plasmid contained the streptavidin gene fused with a T7 tag and a His tag at the N- and C-termini, respectively, for western blotting and metal chelation affinity purification. Substitution of the Tyr54 position of streptavidin by five-base codons CGGXX was carried out by PCR using oligonucleotides CAGTCAGTACYYCCGGCGGGA, where YY is the complementary sequence of XX. A synthetic yeast tRNAPhe gene containing a CCCG anticodon was previously constructed in pUC18 (15). Substitution of CCCG by five-base anticodons XXCCG was carried out in a similar manner using oligonucleotides GCCAGACTXXCCGAATC.
In vitro translation
The mRNA and nitrophenylalanyl-tRNA were prepared as previously reported (15). The reaction mixture for in vitro translation (10 µl) contained 2 µl of E.coli S30 extract (Promega for linear template), 12 mM magnesium acetate, 0.1 mM each of amino acids except arginine, 0.01 mM arginine, 16 µg of mRNA and 6.25 µg of aminoacyl-tRNA unless stated otherwise. The reaction mixture was incubated at 37°C for 1 h, then cooled to 0°C.
Western blot analysis
The reaction mixture for in vitro translation (2 µl) was mixed with gel loading buffer (38 µl) and 5 µl of the resulting solution was analyzed by 15% SDS–PAGE. Western blotting was carried out on a PVDF membrane (Bio-Rad) using an anti-T7 tag monoclonal antibody (Novagen) and ProtoBlot II AP system (Promega). The efficiency of five-base decoding was estimated by comparing the band intensity of the full-length protein with those of serial dilutions of in vitro translated wild-type streptavidin.
Dot blot analysis
The reaction mixture (0.5 µl) was diluted in 200 µl of TBS (20 mM Tris–HCl, 150 mM NaCl, pH 7.5) and spotted onto a nitrocellulose membrane using a vacuum blotter. After washing twice for 5 min with TBS, the membrane was incubated with 3% gelatin in TBS at 37°C for 30 min and then with 2 µg/ml biotinylated alkaline phosphatase (Zymed) in 1% gelatin in TBS for 30 min. After washing three times with TBST (TBS containing 0.02% Tween-20) for 5 min and once with TBS briefly, the membrane was incubated in NBT/BCIP solution at 37°C for 30 min, then washed with water and dried.
HPLC analysis of tryptic fragment
A tRNA aminoacylated with ɛ-nitrobenzoxadiazolyl-l-lysine [Lys(NBD)] (see Fig. 2B) was prepared as described (16). Translation of the CGGUA mutant in the presence of Lys(NBD)-tRNAUACCG was carried out as described above on a 20 µl scale. The full-length product was purified on a Ni–NTA affinity column (Qiagen) under denaturing conditions. The eluate (50 mM phosphate buffer, pH 5.0, containing 8 M urea) was diluted with 7 vol of 50 mM Tris–HCl pH 7.5, 1 mM CaCl2, then incubated with 1 µg/ml trypsin (Promega) at 37°C for 30 min. Reverse phase HPLC analysis was on an ODS column (µBondasphere 5µ 100 Å; Waters) with a linear gradient from 0.1% TFA to acetonitrile over 50 min and a flow rate of 1 ml/min. The eluate was monitored with a fluorescence detector (λex 470 nm, λem 530 nm) and the fluorescent fraction was collected. After evaporation of the solvent, the fraction was subjected to MALDI-TOF mass spectrometry (Voyager DE-Pro; PE Biosystems).
Figure 2.
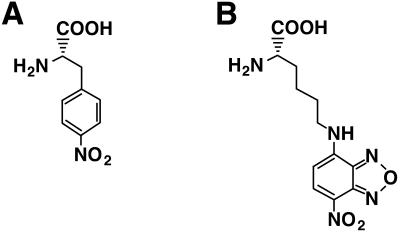
Structures of nonnatural amino acids. (A) Nitrophenylalanine (nitroPhe). (B) ɛ-Nitrobenzoxadiazolyl-l-lysine [Lys(NBD)].
The chemical synthesis of Lys(NBD)-Val-Leu-Thr-Gly-Arg was carried out as follows. NBD-Cl (0.4 mg) and α-Fmoc-Lys-Val-Leu-Thr-Gly-Arg (0.4 mg) synthesized on a peptide synthesizer (Pioneer, PE Biosystems) were added to a mixture of 50 µl of methanol and 10 µl of 4% NaHCO3. After incubation at 55°C for 1 h, Fmoc-Lys(NBD)-Val-Leu-Thr-Gly-Arg was isolated by reverse phase HPLC. Removal of the Fmoc group was by incubation in 20% piperidine/DMF at 37°C for 2 h, then the desired product was purified by reverse phase HPLC. Mass spectrum (FAB) calculated for C35H58N13O11 (MH+) 836.4379, found 836.4391.
RESULTS AND DISCUSSION
Design of the expression system
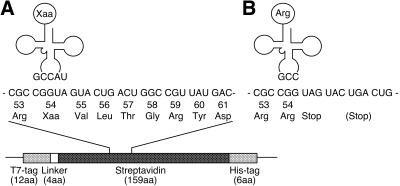
We employed a streptavidin expression system to evaluate five-base decoding, as previously reported for a four-base codon system (13,15). At first, a combination of a CGGUA codon and a UACCG anticodon was investigated. Since CGG is a minor codon in the E.coli translation system, the use of CGG-derived five-base codons will avoid serious competition with endogenous arginyl-tRNACCG. The CGGUA codon was introduced into the Tyr54 position of the streptavidin gene and the UACCG anticodon was introduced into the anticodon of a synthetic yeast tRNAPhe gene. As shown in Figure 1, when the CGGUA codon is recognized by the aminoacyl-tRNAUACCG, the five-base codon will be translated into the nonnatural amino acid and a correct reading frame will be maintained. However, when the first three bases of the CGGUA sequence are read as a triplet codon by endogenous arginyl-tRNACCG, translation will be terminated at a subsequent stop codon. As a result, five-base decoding will yield full-length streptavidin with the correct amino acid sequence except that the nonnatural amino acid corresponding to the CGGUA codon is incorporated, whereas failure of five-base decoding will yield a truncated protein. These two products are easily distinguishable by gel electrophoresis.
Figure 1.
Nucleotide and amino acid sequence of the 53–61 region of a mutated streptavidin. (A) A tRNAUACCG aminoacylated with a nonnatural amino acid (Xaa) decodes a CGGUA five-base codon and the correct reading frame is maintained. (B) When the aminoacyl-tRNAUACCG does not work, then the CGG triplet will be read by an endogenous arginyl-tRNACCG and translation will be stopped at a UAG codon. In the cases of other five-base codons, translation will be terminated at a UGA codon three triplets downstream of the CGG.
p-Nitrophenylalanine (nitroPhe, Fig. 2A) was used as the nonnatural amino acid, because it had been incorporated into streptavidin in good yield (14,15) and mutated streptavidin containing nitroPhe at the Tyr54 position was found to be active in biotin binding (14).
Incorporation of nonnatural amino acids using a CGGUA–UACCG codon–anticodon pair
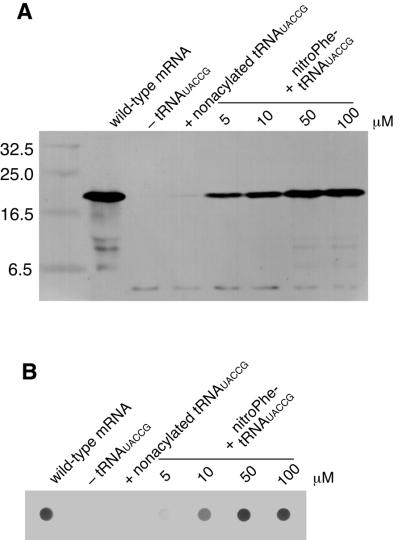
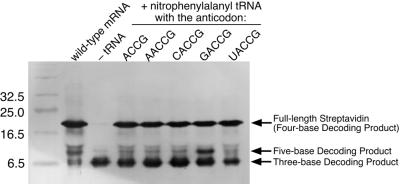
The mutated mRNA containing the CGGUA codon at the Tyr54 position and the tRNAUACCG chemically aminoacylated with nitroPhe were added to the E.coli in vitro translation system. Then, the reaction mixture was analyzed by SDS–PAGE, followed by western blot analysis using an anti-T7 tag antibody to detect translation products. As shown in Figure 3A, in the absence of tRNAUACCG and in the presence of non-aminoacylated tRNAUACCG, only a truncated product was observed instead of full-length streptavidin. This truncated product would result from three-base decoding at the Tyr54 position followed by termination of translation. In the presence of tRNAUACCG aminoacylated with nitroPhe, however, full-length streptavidin was successfully synthesized. The yield of full-length streptavidin increased with increasing concentration of aminoacyl-tRNAUACCG. These results suggest that the reading frame of the mutated mRNA is correctly maintained by addition of aminoacylated tRNAUACCG and, as a consequence, the nonnatural amino acid corresponding to the CGGUA codon is successfully incorporated. The ineffectiveness of non-aminoacylated tRNAUACCG supports the idea that the tRNA cannot be aminoacylated by any endogenous aminoacyl-tRNA synthetases. The biotin-binding activity of the translation product was evaluated by dot blot analysis using biotinylated alkaline phosphatase. As shown in Figure 3B, biotin-binding activity was observed only when nitroPhe-tRNAUACCG was added to the translation reaction. Because the binding activity of mutated streptavidin containing nitroPhe at the Tyr54 position was unaffected (14), the dot blotting results strongly suggest that nitroPhe is incorporated at the directed position without any alteration of the following amino acid sequence.
Figure 3.
Incorporation of a nonnatural amino acid into the Tyr54 position of streptavidin under direction of the CGGUA codon. (A) Western blot and (B) dot blot analysis of translation of the mutated streptavidin mRNA containing a CGGUA codon at the Tyr54 position in the presence of a tRNAUACCG aminoacylated with nitroPhe. Translation products were detected using an anti-T7 tag antibody and an alkaline phosphatase-labeled anti-mouse IgG for western blotting. Biotin-binding activity was evaluated with biotinylated alkaline phosphatase for dot blotting.
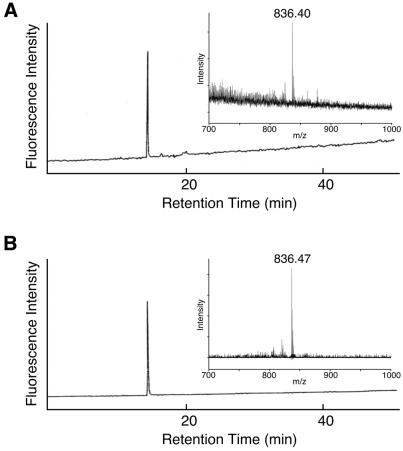
Furthermore, incorporation of the nonnatural amino acid was directly confirmed using a fluorescently labeled amino acid, Lys(NBD) (Fig. 2B; 16). Mutated mRNA containing the CGGUA codon at the Tyr54 position and tRNAUACCG aminoacylated with Lys(NBD) were added to the translation mixture. Full-length streptavidin was purified on a Ni–NTA affinity column under denaturing conditions, then digested with trypsin to generate a 54–59 amino acid fragment (Fig. 1A). The trypsin digestion reaction mixture was applied to a reverse phase HPLC column equipped with a fluorescence detector (λex 470 nm, λem 530 nm). As shown in Figure 4, a single peak was observed and its retention time was found to be identical to that of the chemically synthesized peptide Lys(NBD)-Val-Leu-Thr-Gly-Arg. In addition, the mass spectrum of the fluorescent peptide fragment was identical to that of the chemically synthesized peptide (Fig. 4). These results demonstrate that the nonnatural amino acid is incorporated corresponding to the CGGUA codon and exclude other possible mechanisms, such as a –1 frameshift and two consecutive +1 frameshifts.
Figure 4.
HPLC analysis of a tryptic fragment of a mutated streptavidin synthesized in the presence of Lys(NBD)-tRNAUACCG. The eluate was monitored with a fluorescence detector (λex 470 nm, λem 530 nm). (Inset) MALDI-TOF mass spectra of the fluorescent fraction. (A) Tryptic fragment of the mutated streptavidin. (B) Chemically synthesized peptide Lys(NBD)-Val-Leu-Thr-Gly-Arg.
Effect of a mismatch at the fifth letter of the five-base codon
Previous studies have shown that some tRNAs containing four-base anticodons are insensitive to mismatches at the fourth letter of four-base codons (19,20). Bossi and Smith proposed that the first letters of the four-base anticodons of such tRNAs sterically interfere with reading the adjacent in-frame codon by the next tRNA (21). Therefore, it is likely that the fifth letter of the five-base codon is not involved in codon–anticodon pairing. To test this possibility, single base substitutions at the fifth letter of the five-base codon were introduced (Fig. 5). When mRNA containing a CGGUG or CGGUC codon was added to the translation mixture together with nitroPhe-tRNAUACCG, no full-length streptavidin was synthesized. In the case of the CGGUU codon, however, a small amount of full-length streptavidin was produced. In a similar manner, single base substitutions at the first letter of the five-base anticodon were introduced. When mRNA containing the CGGUA codon was added to the translation mixture together with the nitroPhe-tRNA containing a CACCG or AACCG anticodon, a negligible amount of full-length streptavidin was synthesized. In the case of the GACCG anticodon, however, a small amount of full-length streptavidin was produced. These results indicate that the fifth letter of the five-base codons is recognized by the first letter of the five-base anticodons. U–U and A–G pairs, however, suggest that the recognition is partially loose.
Figure 5.
Effect of mismatch at the fifth letter of five-base codons in five-base decoding. Each pair of mRNA containing a CGGUX (X indicates one of four nucleotides) codon at the Tyr54 position and a nitroPhe-tRNA containing a YACCG (Y indicates one of four nucleotides) anticodon was added to the in vitro translation. Full-length streptavidin was visualized by western blotting.
Other five-base codon–anticodon pairs
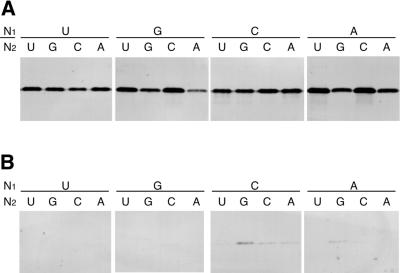
The five-base decoding system may derive up to 16 extended codons from a single three-base codon. Mutated mRNAs containing five-base codons CGGN1N2, where N1 and N2 indicate one of four bases, at the Tyr54 position were added to the translation mixture together with nitroPhe-tRNAs containing the complementary five-base anticodons. Western blot analysis showed that all combinations of complementary five-base codon–anticodon pairs gave full-length streptavidin (Fig. 6A). On the other hand, negligible amounts of full-length streptavidin were produced in the presence of non-aminoacylated tRNAs (Fig. 6B). These results suggest that the 16 five-base codons are successfully decoded by aminoacylated tRNAs containing the complementary five-base anticodons. The fact that any bases are allowed for N1 and N2 indicates that five-base decoding is effective even when the CGG triplet is followed by a stop codon (UAG), minor codons (AGG, CGG and GGG) or major codons.
Figure 6.
Western blot analysis of translation of a mutated streptavidin mRNA containing a CGGN1N2 codon (N1 and N2 indicate one of four nucleotides) at the Tyr54 position. (A) In the presence of a nitroPhe-tRNA with a complementary anticodon. (B) In the presence of a non-aminoacylated tRNA with a complementary anticodon.
Efficiency of five-base decoding was estimated by densitometric analysis of western blots and is summarized in Table 1. The highest yield of full-length streptavidin was observed in the case of the CGGAC codon (42% relative to wild-type streptavidin). This efficiency is much higher than that of amber suppression (12%; 1), but rather lower than that of the CGGG four-base codon (70%; 15).
Table 1. Efficiency (%) of five-base decoding by nitroPhe-tRNAs containing complementary five-base anticodons.
| Fourth letter | Fifth letter |
|
|
|
| |
U |
G |
C |
A |
| U | 18 | 14 | 8 | 16 |
| G | 19 | 9 | 24 | 5 |
| C | 21 | 15 | 29 | 34 |
| A | 30 | 12 | 42 | 13 |
The values are expressed as yields of full-length streptavidin relative to that of wild-type streptavidin.
Mechanism of five-base decoding
The above results suggest that the ribosome moves by five bases along the five-base codon–anticodon pair. This phenomenon might be interpreted in terms of a single-step quintuplet translocation. However, codon–anticodon pairing by five consecutive bases would cause severe deformation of the anticodon loop, resulting in failure of the translocation process. Farabaugh and co-workers proposed a model for +1 frameshift suppression (22,23), arguing against a classical quadruplet translocation model. They proposed that a +1 frameshift occurs in the P site after triplet translocation, even though frameshift suppressor tRNA contains a four-base anticodon. The idea that frameshifting occurs not in the A but in the P site is supported by the finding that binding of peptidyl-tRNA in the P site is weaker than in the A site (24).
Considering the Farabaugh and Björk model for +1 frameshift suppression, it is expected that five-base decoding is caused by an additional +1 frameshift in the P site after the initial +1 frameshift. To examine this possibility, four-base decoding by tRNAs containing five-base anticodons was investigated. mRNA containing a four-base CGGU codon at the Tyr83 position of streptavidin (15) and aminoacylated tRNAs containing XACCG anticodons were added to the translation mixture. As shown in Figure 7, full-length streptavidin was successfully synthesized when the four-base codon (CGGU) and five-base anticodon (NACCG) pairs were used, as well as the four-base codon (CGGU) and four-base anticodon (ACCG) pair. This result indicates that the CGGU codon can be decoded as a single four-base codon by tRNAs containing XACCG anticodons. Addition of nitroPhe-tRNAGACCG, however, yielded another truncated product of ∼10 kDa. This product could be generated through five-base decoding of the four-base codon CGGU plus the first letter C of the next CGU codon. Such five-base decoding gives a truncated product terminated at a UAA codon 27 triplets downstream of the CGGUC. It should be noted that the four-base decoding product in the cases of five-base codons at the Tyr54 position might be terminated at a UGA codon 7 triplets downstream of the CGGU (Fig. 1A), but it could not be distinguished from the three-base decoding product by western blotting.
Figure 7.
Western blot analysis of translation of a mutated streptavidin mRNA containing a CGGU codon at the Tyr83 position in the presence of nitroPhe-tRNAs containing four- or five-base anticodons. The partial sequence of the mutated mRNA is AAC-AAC-CGGU-CGU-AAU-GCG-CAC-AGC-GCC-ACU-ACG-UGG-UCU-GGC-CAA-UAC-GUU-GGC-GGU-GCU-GAG-GCU-CGU-AUC-AAC-ACU-CAG-UGG-CUG-UUA-ACA. The first underlined UAA appears when the CGG is decoded as a three-base codon by an endogenous arginyl-tRNACCG. The second underlined UAA appears when CGGUC is decoded as a five-base codon by the nitroPhe-tRNAGACCG.
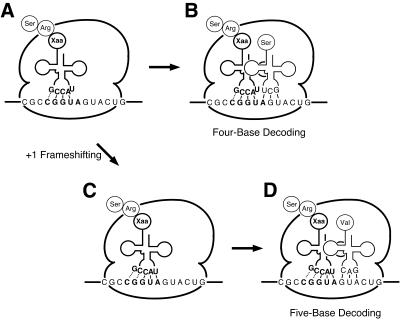
Here we propose a model for five-base decoding by tRNA containing a five-base anticodon (Fig. 8). At first, the peptidyl-tRNA containing the five-base anticodon binds to the P site with the reading frame shifted by one base (Fig. 8A). According to the Farabaugh and Björk model for the sufJ suppressor (22), this intermediate would be produced not by quadruplet translocation but by triplet translocation followed by a +1 frameshift through isomerization of the codon–anticodon complex. At this stage it is uncertain whether the first letter of the five-base codon interacts with the fifth letter of the five-base anticodon.
Figure 8.
A model for five-base decoding. A peptidyl-tRNA containing a five-base anticodon binds to the P site with the reading frame shifted by one base (A). Binding of Ser-tRNAGCU to the empty ribosomal A site results in four-base decoding (B). On the other hand, base pairing of the fifth letter of the five-base codon with the first letter of the five-base anticodon leads to an additional +1 frameshift (C). Binding of Val-tRNAGAC to the empty ribosomal A site results in five-base decoding (D).
If the next Ser-tRNAGCU binds to the empty A site of this intermediate ribosome complex, four-base decoding will be observed (Fig. 8B). On the other hand, if the fifth letter of the five-base codon pairs with the first letter of the five-base anticodon prior to binding of Ser-tRNAGCU, the additional +1 frameshift will occur in the P site (Fig. 8C). At this stage base pairing of the first and the second letters of the five-base codon with the fifth and the fourth letters of the five-base anticodon, respectively, is uncertain. Then, the next Val-tRNAGAC binds to the empty ribosomal A site to yield the five-base decoding product (Fig. 8D). The present model implies that five-base decoding is intrinsically inefficient because the additional +1 frameshift competes with binding of an aminoacyl-tRNA to the empty A site (Fig. 8B). The lower efficiency of five-base decoding than that of four-base decoding may be accounted for by this model. Moreover, this model may explain the accurate recognition of the first letter of the five-base anticodon observed in Figure 5. Although U as the first letter of triplet anticodons can pair with all four bases, U as the first letter of a five-base anticodon can only pair with A (or U to a lesser extent). In our model pairing of the first letter of the five-base anticodon occurs in the ribosomal P site in competition with binding of the next aminoacyl-tRNA, whereas that of triplet anticodons occurs in the A site without competition. These differences may explain the different specificities of the first letters of three-base and five-base anticodons.
CONCLUSIONS
For the first time we have demonstrated five-base decoding by using aminoacyl-tRNAs containing five-base anticodons in the E.coli in vitro translation system. This novel frameshift strategy will find wide application, such as multiple incorporation of nonnatural amino acids into proteins. The results in Figure 5 indicate that the CGGUA and CGGUG codons can be used independently, because the CGGUA codon cannot be recognized by tRNACACCG and the CGGUG codon cannot be recognized by tRNAUACCG. Such independent five-base codons will enable us to introduce various kinds of nonnatural amino acids into a single protein.
The present study also demonstrates the feasibility and utility of the application of chemically aminoacylated frameshift suppressor tRNA. In previous studies of frameshift suppression, suppressor tRNAs must be aminoacylated by any of aminoacyl-tRNA synthetases. Most aminoacyl-tRNA synthetases, however, recognize anticodons as identity determinants, which limits mutation of the anticodon loop of frameshift suppressor tRNA. Since the chemical aminoacylation method is free from enzymatic recognition, it will serve to probe structure–function relationships of frameshift suppressor tRNAs.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Prof. Shigeyuki Yokoyama (The University of Tokyo) for his important suggestions and Prof. Shinji Toyota (Okayama University of Science) for his measurement of the FAB mass spectrum. This research was supported by a Grant-in-Aid for Specially Promoted Research from the Ministry of Education, Culture, Sports, Science and Technology, Japan (no. 11102003).
References
- 1.Noren C.J., Anthony-Cahill,S.J., Griffith,M.C. and Schultz,P.G. (1989) A general method for site-specific incorporation of unnatural amino acids into proteins. Science, 244, 182–188. [DOI] [PubMed] [Google Scholar]
- 2.Bain J.D., Glabe,C.G., Dix,T.A., Chamberlin,A.R. and Diala,E.S. (1989) Biosynthetic site-specific incorporation of a non-natural amino acid into a polypeptide. J. Am. Chem. Soc., 111, 8013–8014. [Google Scholar]
- 3.Cornish V.W., Mendel,D. and Schultz,P.G. (1995) Probing protein structure and function with an expanded genetic code. Angew. Chem. Int. Ed. Engl., 34, 621–633. [Google Scholar]
- 4.Gillmore M.A., Steward,L.E. and Chamberlin,A.R. (1999) Incorporation of noncoded amino acids by in vitro protein biosynthesis. Top. Curr. Chem., 202, 77–99. [Google Scholar]
- 5.Short G.F.,III, Lodder,M., Laikhter,A.L., Arslan,T. and Hecht,S.M. (1999) Caged HIV-1 protease: dimerization is independent of the ionization state of the active site aspartates. J. Am. Chem. Soc., 121, 478–479. [Google Scholar]
- 6.Dougherty D.A. (2000) Unnatural amino acids as probes of protein structure and function. Curr. Opin. Chem. Biol., 4, 645–652. [DOI] [PubMed] [Google Scholar]
- 7.Kanamori T., Nishikawa,S., Nakai,M., Shin,I., Schultz,P.G. and Endo,T. (1999) Uncoupling of transfer of the presequence and unfolding of the mature domain in precursor translocation across the mitochondrial outer membrane. Proc. Natl Acad. Sci. USA, 96, 3634–3639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Karginov V.A., Mamaev,S.V. and Hecht,S.M. (1997) In vitro suppression as a tool for the investigation of translation initiation. Nucleic Acids Res., 25, 3912–3916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Killian J.A., Van Cleve,M.D., Shayo,Y.F. and Hecht,S.M. (1998) Ribosome-mediated incorporation of hydrazinophenylalanine into modified peptide and protein analogues. J. Am. Chem. Soc., 120, 3032–3042. [Google Scholar]
- 10.Karginov A.V., Lodder,M. and Hecht,S.M. (1999) Facile characterization of translation initiation via nonsense codon suppression. Nucleic Acids Res., 27, 3283–3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Short G.F.,III, Golovine,S.Y. and Hecht,S.M. (1999) Effects of release factor 1 on in vitro protein translation and the elaboration of proteins containing unnatural amino acids. Biochemistry, 38, 8808–8819. [DOI] [PubMed] [Google Scholar]
- 12.Bain J.D., Switzer,C., Chamberlin,A.R. and Benner,S.A. (1992) Ribosome-mediated incorporation of a non-standard amino acid into a peptide through expansion of the genetic code. Nature, 356, 537–539. [DOI] [PubMed] [Google Scholar]
- 13.Hohsaka T., Ashizuka,Y., Murakami,H. and Sisido,M. (1996) Incorporation of nonnatural amino acids into streptavidin through in vitro frame-shift suppression. J. Am. Chem. Soc., 118, 9778–9779. [Google Scholar]
- 14.Murakami H., Hohsaka,T., Ashizuka,Y. and Sisido,M. (1998) Site-directed incorporation of p-nitrophenylalanine into streptavidin and site-to-site photoinduced electron transfer from a pyrenyl group to a nitrophenyl group on the protein framework. J. Am. Chem. Soc., 120, 7520–7529. [Google Scholar]
- 15.Hohsaka T., Kajihara,D., Ashizuka,Y., Murakami,H. and Sisido,M. (1999) Efficient incorporation of nonnatural amino acids with large aromatic groups into streptavidin in in vitro protein synthesizing systems. J. Am. Chem. Soc., 121, 34–40. [Google Scholar]
- 16.Hohsaka T., Ashizuka,Y., Sasaki,H., Murakami,H. and Sisido,M. (1999) Incorporation of two different nonnatural amino acids independently into a single protein through extension of the genetic code. J. Am. Chem. Soc., 121, 12194–12195. [Google Scholar]
- 17.Falahee M.B., Weiss,R.B., O’Connor,M., Doonan,S., Gesteland,R.F. and Atkins,J.F. (1988) Mutants of translational components that alter reading frame by two steps forward or one step back. J. Biol. Chem., 263, 18099–18103. [PubMed] [Google Scholar]
- 18.Tuohy T.M., Thompson,S., Gesteland,R.F. and Atkins,J.F. (1992) Seven, eight and nine-membered anticodon loop mutants of tRNA2Arg which cause +1 frameshifting. Tolerance of DHU arm and other secondary mutations. J. Mol. Biol., 228, 1042–1054. [DOI] [PubMed] [Google Scholar]
- 19.Bossi L. and Roth,J. (1981) Four-base codons ACCA, ACCU and ACCC are recognized by frameshift suppressor sufJ. Cell, 25, 489–496. [DOI] [PubMed] [Google Scholar]
- 20.Gaber R.F. and Culbertson,M.R. (1984) Codon recognition during frameshift suppression in Saccharomyces cerevisiae. Mol. Cell. Biol., 4, 2052––2061.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bossi L. and Smith,D.M. (1984) Suppressor sufJ: a novel type of tRNA mutant that induces translational frameshifting. Proc. Natl Acad. Sci. USA, 81, 6105–6109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qian Q., Li,J.N., Zhao,H., Hagervall,T.G., Farabaugh,P.J. and Björk,G.R. (1998) A new model for phenotypic suppression of frameshift mutations by mutant tRNAs. Mol. Cell, 1, 471–482. [DOI] [PubMed] [Google Scholar]
- 23.Farabaugh P.J. and Björk,G.R. (1999) How translational accuracy influences reading frame maintenance. EMBO J., 18, 1427–1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dabrowski M., Spahn,C.M. and Nierhaus,K.H. (1995) Interaction of tRNAs with the ribosome at the A and P sites. EMBO J., 14, 4872–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]