The isolation and crystal structures of the title compounds from Hibiscus sabdariffa (Malvaceae) are described. Hibiscus acid dimethyl sulfoxide monosolvate forms a two-dimensional hydrogen-bonded motif, while hibiscus acid dimethyl ester (Z′ = 2) forms a one-dimensional hydrogen-bonded motif.
Keywords: crystal structure, natural products, hibiscus, lactone acids, hydrogen bonding
Abstract
The biologically active title compounds have been isolated from Hibiscus sabdariffa plants, hibiscus acid as a dimethyl sulfoxide monosolvate [systematic name: (2S,3R)-3-hydroxy-5-oxo-2,3,4,5-tetrahydrofuran-2,3-dicarboxylic acid dimethyl sulfoxide monosolvate], C6H6O7·C2H6OS, (I), and hibiscus acid dimethyl ester [systematic name: dimethyl (2S,3R)-3-hydroxy-5-oxo-2,3,4,5-tetrahydrofuran-2,3-dicarboxylate], C8H10O7, (II). Compound (I) forms a layered structure with alternating layers of lactone and solvent molecules, that include a two-dimensional hydrogen-bonding construct. Compound (II) has two crystallographically independent and conformationally similar molecules per asymmetric unit and forms a one-dimensional hydrogen-bonding construct. The known absolute configuration for both compounds has been confirmed.
Chemical context
Lactone acid producing plants, including Hibiscus sabdariffa (Malvaceae), have been documented to have significant potential in the traditional treatment of various diseases. H. sabdariffa Linn is a species of hibiscus from the Malvaceae family, commonly known as ‘Karkade’ or ‘red sorrel’. It is used in traditional medicine in the form of herbal teas or cold drinks for its hypotensive and diuretic effects and to lower body temperature and blood viscosity (Ali et al., 2005 ▸; Da-Costa-Rocha et al., 2014 ▸). Little attention has been paid to organic acids from H. sabdariffa, specifically hibiscus acid. However, studies have documented the activity of hibiscus acid and hibiscus acid methyl ester. These report an inhibitory effect against enzymes, such as α-amylase and α-glucosidase (Hansawasdi et al., 2000 ▸, 2001 ▸). As these compounds are not available commercially and to enable a study of their biological activities, we report on the extraction of hibiscus acid and hibiscus acid dimethyl ester from H. sabdariffa (Malvaceae), and on their purification and characterization. The crystal structures of the acid, as the dimethyl sulfoxide monosolvate, (I), and the diester, (II), are reported herein.
Structural commentary
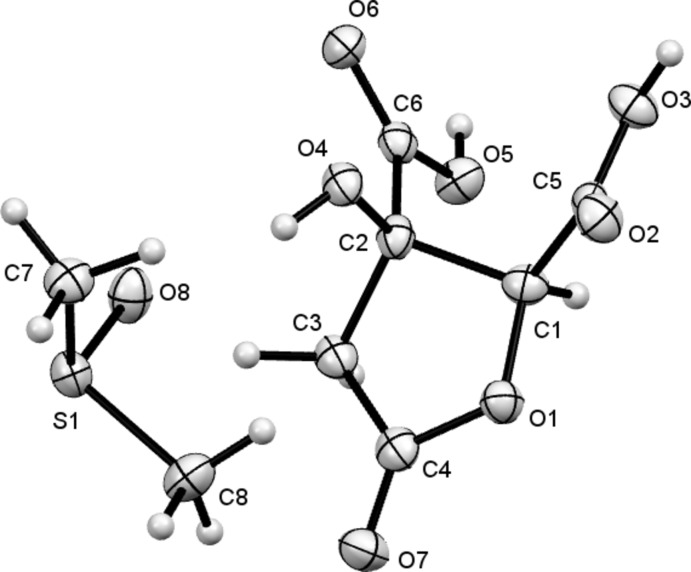
The crystal structures of the 1:1 dimethyl sulfoxide (DMSO) solvate of hibiscus acid, (I), and of hibiscus acid dimethyl ester, (II), are shown in Figs. 1 ▸ and 2 ▸. The COOR (R = H or Me) groups lie in equatorial positions on their rings and the absolute configuration of both species is confirmed by the Flack parameter values (Parsons et al., 2013 ▸), for arbitrarily named atoms in (I) [C2(R),C1(S), 0.00 (4)] and both arbitrarily named equivalent atoms in (II) [C3(R),C4(S) and C11(R),C12(S), 0.08 (17)] (Table 1 ▸). The absolute configuration found thus agrees with that originally proposed by Boll et al. (1969 ▸) for hibiscus acid. The structure of garcinia lactone, an epimer of hibiscus acid, has been reported (Mahapatra et al., 2007 ▸). The comparable molecular geometries of (I) and its epimer are similar. The five-membered ring of (I) adopts an envelope conformation, with the OH-bearing C2 atom 0.582 (6) Å out of the plane defined by the other four atoms.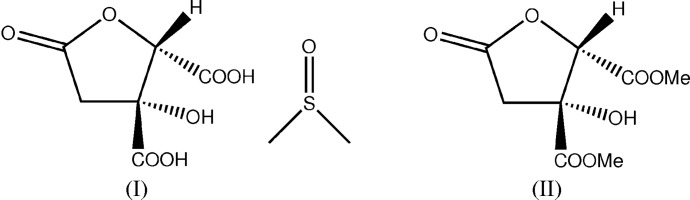
Figure 1.
The molecular structure of compound (I), with the atom labelling and 50% probability displacement ellipsoids.
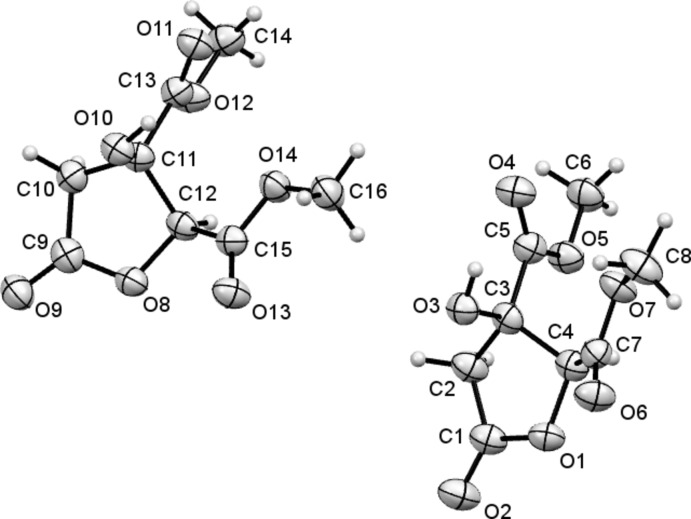
Figure 2.
The molecular structures of the two independent molecules comprising the asymmetric unit of (II), with the atom labelling and 50% probability displacement ellipsoids.
Table 1. Experimental details.
| (I) | (II) | |
|---|---|---|
| Crystal data | ||
| Chemical formula | C6H6O7·C2H6OS | C8H10O7 |
| M r | 268.24 | 218.16 |
| Crystal system, space group | Monoclinic, P21 | Monoclinic, P21 |
| Temperature (K) | 123 | 123 |
| a, b, c (Å) | 5.4258 (2), 8.9491 (3), 11.4365 (3) | 9.3057 (6), 7.6934 (6), 13.4012 (11) |
| β (°) | 94.092 (3) | 96.243 (7) |
| V (Å3) | 553.90 (3) | 953.74 (12) |
| Z | 2 | 4 |
| Radiation type | Cu Kα | Cu Kα |
| μ (mm−1) | 2.94 | 1.20 |
| Crystal size (mm) | 0.30 × 0.15 × 0.05 | 0.30 × 0.20 × 0.04 |
| Data collection | ||
| Diffractometer | Oxford Diffraction Gemini S CCD | Oxford Diffraction Gemini S CCD |
| Absorption correction | Multi-scan (CrysAlis PRO; Oxford Diffraction, 2010 ▸) | Multi-scan (CrysAlis PRO; Oxford Diffraction, 2010 ▸) |
| T min, T max | 0.554, 1.000 | 0.747, 1.000 |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 4397, 1854, 1640 | 8046, 3506, 2976 |
| R int | 0.054 | 0.036 |
| (sin θ/λ)max (Å−1) | 0.619 | 0.622 |
| Refinement | ||
| R[F 2 > 2σ(F 2)], wR(F 2), S | 0.047, 0.113, 1.05 | 0.044, 0.121, 1.10 |
| No. of reflections | 1854 | 3506 |
| No. of parameters | 169 | 281 |
| No. of restraints | 4 | 3 |
| H-atom treatment | H atoms treated by a mixture of independent and constrained refinement | H atoms treated by a mixture of independent and constrained refinement |
| Δρmax, Δρmin (e Å−3) | 0.44, −0.25 | 0.23, −0.22 |
| Absolute structure | Flack x determined using 698 quotients [(I +) − (I −)]/[(I +) + (I −)] (Parsons et al., 2013 ▸) | Flack x determined using 1098 quotients [(I +) − (I −)]/[(I +) + (I −)] (Parsons et al., 2013 ▸) |
| Absolute structure parameter | 0.00 (4) | 0.08 (17) |
The structure of (II) contains two crystallographically independent molecules (A and B) (Z′ = 2), whose molecular geometries differ only by small deviations in torsion angles, for example, C3—C5—O5—C6 in A is 175.1 (4)°, whilst the equivalent angle in B (C11—C13—O12—C—14) is 180.0 (4)°. As with structure (I), the five-membered rings adopt envelope conformations, with the OH-bearing C atoms lying out of the plane of the other four atoms, here by 0.505 (5) and 0.530 (5) Å for molecules A and B, respectively.
Supramolecular features
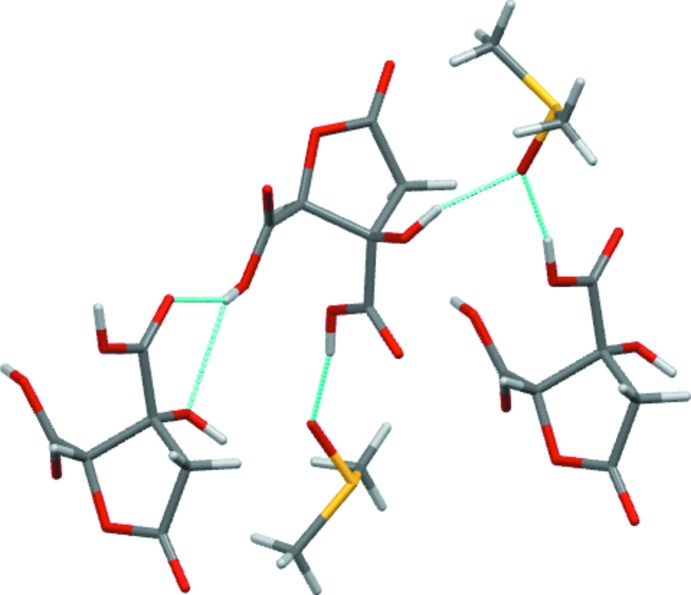
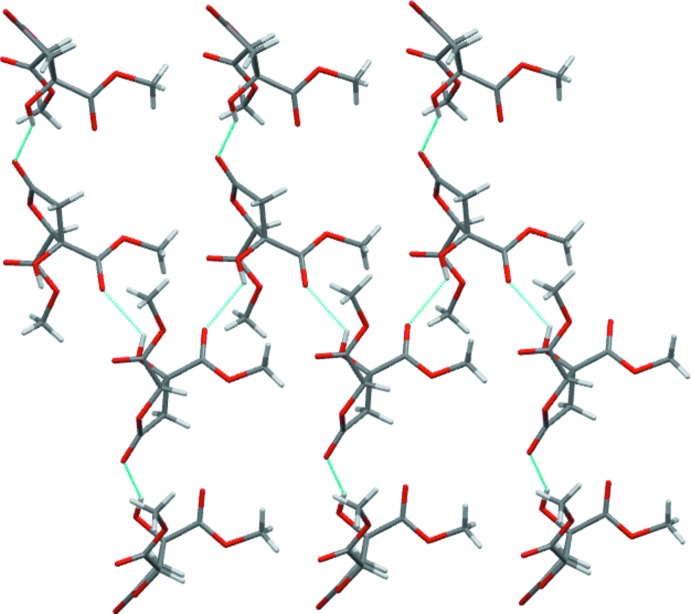
Despite containing two carboxylic acid functionalities, the structure of (I) does not feature the classic 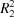 (8) carboxylic acid dimer motif. Instead, each of the three potential hydrogen-bond donors of the acid molecule form interactions with a total of three separate neighbouring molecules (Fig. 3 ▸). The H atom of the carboxylic acid group (O3—H) adjacent to the ether forms a bifurcated hydrogen bond that is accepted by the ROH and C=O functions (i.e. O4i and O6i) of one neighbour, whilst the other two donors, the second carboxylic acid (O5—H) and the hydroxy group (O4—H), form hydrogen bonds with atoms O8ii and O8 of DMSO solvent molecules, respectively (Table 2 ▸). These interactions combine to give a two-dimensional hydrogen-bonded layered structure, with DMSO and acid layers alternating along the c-cell direction (Fig. 4 ▸).
(8) carboxylic acid dimer motif. Instead, each of the three potential hydrogen-bond donors of the acid molecule form interactions with a total of three separate neighbouring molecules (Fig. 3 ▸). The H atom of the carboxylic acid group (O3—H) adjacent to the ether forms a bifurcated hydrogen bond that is accepted by the ROH and C=O functions (i.e. O4i and O6i) of one neighbour, whilst the other two donors, the second carboxylic acid (O5—H) and the hydroxy group (O4—H), form hydrogen bonds with atoms O8ii and O8 of DMSO solvent molecules, respectively (Table 2 ▸). These interactions combine to give a two-dimensional hydrogen-bonded layered structure, with DMSO and acid layers alternating along the c-cell direction (Fig. 4 ▸).
Figure 3.
Hydrogen-bonding contacts in (I).
Table 2. Hydrogen-bond geometry (Å, °) for (I) .
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O3—H1H⋯O4i | 0.87 (2) | 2.42 (4) | 2.996 (4) | 124 (3) |
| O3—H1H⋯O6i | 0.87 (2) | 1.98 (3) | 2.805 (4) | 158 (4) |
| O4—H3H⋯O8 | 0.87 (2) | 1.87 (3) | 2.714 (5) | 160 (7) |
| O5—H2H⋯O8ii | 0.89 (2) | 1.73 (2) | 2.603 (4) | 167 (5) |
Symmetry codes: (i) 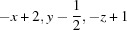 ; (ii)
; (ii) 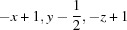 .
.
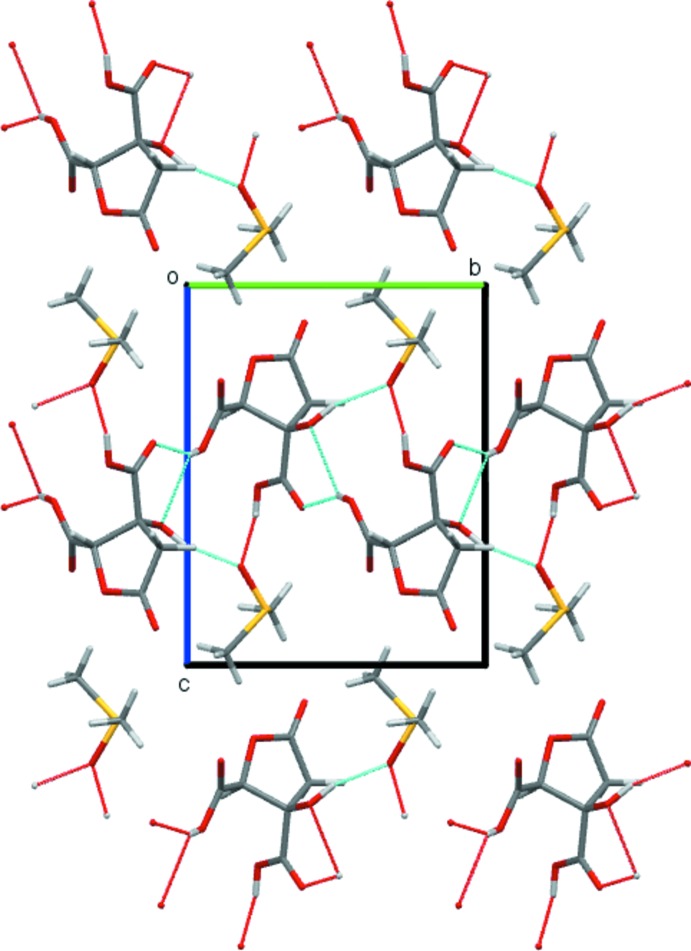
Figure 4.
The crystal packing of compound (I), viewed along the a axis.
Both independent molecules in the structure of (II) donate single hydrogen bonds through their OH groups, but only one molecule (A) acts as a hydrogen-bond acceptor (O3—H⋯O4i and O10—H⋯O2ii; Table 3 ▸). That a total of four carbonyl O atoms do not act as acceptors is probably related to the low ratio of classic hydrogen-bond donors to acceptors in this compound. In (II), the hydrogen bonding combines to give a four-molecule-wide one-dimensional ribbon of linked molecules that propagates parallel to the a axis (Fig. 5 ▸).
Table 3. Hydrogen-bond geometry (Å, °) for (II) .
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O3—H1H⋯O4i | 0.88 (1) | 2.36 (5) | 2.951 (4) | 125 (4) |
| O10—H2H⋯O2ii | 0.88 (1) | 2.03 (3) | 2.802 (4) | 147 (5) |
Symmetry codes: (i) 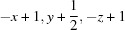 ; (ii)
; (ii) 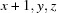 .
.
Figure 5.
A section of the extended structure of (II), with the hydrogen-bonded polymer extending left and right parallel to the a axis.
Database survey
A search of the Cambridge Structural Database (Version 5.37, searched June 2017; Groom et al., 2016 ▸) yielded few relevant structures. For hibiscus acid, only the structures of a Ca salt form (Glusker et al., 1972 ▸) and of the diastereomer mentioned previously (Mahapatra et al., 2007 ▸) have been reported. The closest relative of (II) to have been structurally described is a derivative with additional OH and Me substituents on the five-membered ring (Evans et al., 1997 ▸).
Synthesis and crystallization
Dried H. sabdariffa calyces were crushed to a powder (500 g) and extracted in a Soxhlet apparatus using 2500 ml each of hexane, ethyl acetate and methanol. The methanol extract was dried and concentrated at 313 K by rotatory evaporation, yielding about 125 g (25%) of crude extract. The methanol extract (2 g) was dissolved in about 2 ml of methanol and subjected to gel filtration chromatography (GFC) using a glass column packed with a wet slurry of 30 g of Sephadex LH20 in methanol. Vials were collected (5 ml each) after elution with 100% methanol, which led to isolation of pure hibiscus acid (0.5%). Crystals of (I) were obtained by recrystallisation from DMSO. For nonsolvated material, 1H NMR [OC(CD3)2]: 5.31 (1H, s), 3.23 (1H, d, J = 17.19 Hz), 2.77 (1H, d, J = 17.18 Hz). HRMS: found 189.0000; calculated 189.0035.
Hibiscus acid dimethyl ester, (II), was obtained from the methanol extract (20 g) using vacuum liquid chromatography (VLC) eluted with solvent systems in different ratios to increase the polarity. The ethyl acetate portion was evaporated and a thick paste was obtained. A pure precipitate of the compound (5%) was obtained by addition of propan-2-ol to the dried ethyl acetate fraction. 1H NMR [OC(CD3)2]: 5.35 (1H, s), 3.23 (1H, d, J = 17.28 Hz), 2.77 (1H, d, J = 17.31 Hz), 3.87 (3H, s), 3.76 (3H, s). HRMS: found 218.000; calculated 218.035.
Refinement
Crystal data, data collection and structure refinement details are summarized in Table 1 ▸. For all structures, C-bound H atoms were placed in their expected geometrical positions and treated as riding, with C—H = 0.95–0.99 Å and U iso(H) = 1.5U eq(C) for methyl C atoms and 1.2U eq(C) for the other H atoms. The absolute configuraion was determined for the molecules in both acid (I) for arbitrarily named atoms [C2(R),C1(S), Flack parameter 0.00 (4)] and both arbitrarily named equivalent atoms in (II) [C3(R),C4(S) (molecule A) and C11(R),C12(S) (molecule B), Flack parameter 0.08 (17)] (Parsons et al., 2013 ▸).
Supplementary Material
Crystal structure: contains datablock(s) I, II, global. DOI: 10.1107/S2056989017011902/zs2386sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989017011902/zs2386Isup2.hkl
Supporting information file. DOI: 10.1107/S2056989017011902/zs2386Isup4.cml
Structure factors: contains datablock(s) II. DOI: 10.1107/S2056989017011902/zs2386IIsup3.hkl
Supporting information file. DOI: 10.1107/S2056989017011902/zs2386IIsup5.cml
Additional supporting information: crystallographic information; 3D view; checkCIF report
Acknowledgments
We thank the College of Pharmacy, University of Misan, and the Ministry of Higher Education, Iraq, for funding AZ.
supplementary crystallographic information
(2S,3R)-3-Hydroxy-5-oxo-2,3,4,5-tetrahydrofuran-2,3-dicarboxylic acid dimethyl sulfoxide monosolvate (I) . Crystal data
| C6H6O7·C2H6OS | F(000) = 280 |
| Mr = 268.24 | Dx = 1.608 Mg m−3 |
| Monoclinic, P21 | Cu Kα radiation, λ = 1.5418 Å |
| a = 5.4258 (2) Å | Cell parameters from 2057 reflections |
| b = 8.9491 (3) Å | θ = 6.3–72.8° |
| c = 11.4365 (3) Å | µ = 2.94 mm−1 |
| β = 94.092 (3)° | T = 123 K |
| V = 553.90 (3) Å3 | Fragment from a square plate, colourless |
| Z = 2 | 0.30 × 0.15 × 0.05 mm |
(2S,3R)-3-Hydroxy-5-oxo-2,3,4,5-tetrahydrofuran-2,3-dicarboxylic acid dimethyl sulfoxide monosolvate (I) . Data collection
| Oxford Diffraction Gemini S CCD diffractometer | 1640 reflections with I > 2σ(I) |
| Radiation source: sealed tube | Rint = 0.054 |
| ω scans | θmax = 72.8°, θmin = 3.9° |
| Absorption correction: multi-scan (CrysAlis PRO; Oxford Diffraction, 2010) | h = −6→6 |
| Tmin = 0.554, Tmax = 1.000 | k = −10→8 |
| 4397 measured reflections | l = −14→14 |
| 1854 independent reflections |
(2S,3R)-3-Hydroxy-5-oxo-2,3,4,5-tetrahydrofuran-2,3-dicarboxylic acid dimethyl sulfoxide monosolvate (I) . Refinement
| Refinement on F2 | Hydrogen site location: mixed |
| Least-squares matrix: full | H atoms treated by a mixture of independent and constrained refinement |
| R[F2 > 2σ(F2)] = 0.047 | w = 1/[σ2(Fo2) + (0.0678P)2] where P = (Fo2 + 2Fc2)/3 |
| wR(F2) = 0.113 | (Δ/σ)max < 0.001 |
| S = 1.05 | Δρmax = 0.44 e Å−3 |
| 1854 reflections | Δρmin = −0.25 e Å−3 |
| 169 parameters | Absolute structure: Flack x determined using 698 quotients [(I+)-(I-)]/[(I+)+(I-)] (Parsons et al., 2013) |
| 4 restraints | Absolute structure parameter: 0.00 (4) |
(2S,3R)-3-Hydroxy-5-oxo-2,3,4,5-tetrahydrofuran-2,3-dicarboxylic acid dimethyl sulfoxide monosolvate (I) . Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
| Refinement. Refined as a 2-component inversion twin |
(2S,3R)-3-Hydroxy-5-oxo-2,3,4,5-tetrahydrofuran-2,3-dicarboxylic acid dimethyl sulfoxide monosolvate (I) . Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| S1 | 0.90564 (19) | 1.26444 (16) | 0.85370 (9) | 0.0235 (3) | |
| O1 | 0.6239 (6) | 0.7220 (4) | 0.8066 (3) | 0.0233 (9) | |
| O2 | 1.0711 (6) | 0.6125 (5) | 0.7467 (3) | 0.0285 (9) | |
| O3 | 0.9028 (6) | 0.5501 (5) | 0.5682 (3) | 0.0276 (8) | |
| O4 | 0.8575 (6) | 0.9112 (5) | 0.6333 (3) | 0.0232 (8) | |
| O5 | 0.3551 (6) | 0.7366 (5) | 0.4714 (3) | 0.0265 (9) | |
| O6 | 0.6572 (6) | 0.8844 (5) | 0.4153 (3) | 0.0254 (8) | |
| O7 | 0.4156 (6) | 0.8927 (5) | 0.9015 (3) | 0.0300 (9) | |
| O8 | 0.8239 (7) | 1.1798 (5) | 0.7411 (3) | 0.0295 (9) | |
| C1 | 0.6534 (8) | 0.6840 (7) | 0.6856 (4) | 0.0231 (11) | |
| H1 | 0.5185 | 0.6149 | 0.6557 | 0.028* | |
| C2 | 0.6288 (8) | 0.8370 (6) | 0.6206 (4) | 0.0219 (11) | |
| C3 | 0.4303 (8) | 0.9097 (7) | 0.6897 (4) | 0.0236 (11) | |
| H3A | 0.4466 | 1.0198 | 0.6901 | 0.028* | |
| H3B | 0.2627 | 0.8823 | 0.6567 | 0.028* | |
| C4 | 0.4814 (8) | 0.8461 (7) | 0.8109 (4) | 0.0244 (11) | |
| C5 | 0.9026 (8) | 0.6125 (7) | 0.6737 (4) | 0.0221 (11) | |
| C6 | 0.5504 (8) | 0.8209 (6) | 0.4898 (4) | 0.0216 (10) | |
| C7 | 1.2345 (8) | 1.2784 (8) | 0.8549 (4) | 0.0273 (12) | |
| H7A | 1.2788 | 1.3417 | 0.7897 | 0.041* | |
| H7B | 1.3053 | 1.1786 | 0.8465 | 0.041* | |
| H7C | 1.2997 | 1.3227 | 0.9292 | 0.041* | |
| C8 | 0.8906 (10) | 1.1285 (8) | 0.9679 (4) | 0.0309 (13) | |
| H8A | 0.9898 | 1.0411 | 0.9498 | 0.046* | |
| H8B | 0.7184 | 1.0981 | 0.9739 | 0.046* | |
| H8C | 0.9552 | 1.1720 | 1.0425 | 0.046* | |
| H2H | 0.308 (11) | 0.729 (8) | 0.396 (3) | 0.032 (17)* | |
| H1H | 1.043 (7) | 0.509 (7) | 0.555 (5) | 0.026 (15)* | |
| H3H | 0.865 (17) | 0.987 (7) | 0.682 (6) | 0.07 (3)* |
(2S,3R)-3-Hydroxy-5-oxo-2,3,4,5-tetrahydrofuran-2,3-dicarboxylic acid dimethyl sulfoxide monosolvate (I) . Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| S1 | 0.0241 (5) | 0.0207 (7) | 0.0254 (5) | 0.0013 (5) | 0.0002 (4) | −0.0021 (5) |
| O1 | 0.0228 (14) | 0.024 (3) | 0.0232 (15) | 0.0022 (13) | 0.0016 (11) | 0.0007 (13) |
| O2 | 0.0237 (16) | 0.030 (3) | 0.0320 (17) | 0.0050 (15) | 0.0000 (13) | 0.0007 (16) |
| O3 | 0.0213 (14) | 0.032 (3) | 0.0299 (16) | 0.0032 (15) | 0.0023 (12) | −0.0049 (16) |
| O4 | 0.0187 (14) | 0.021 (2) | 0.0299 (17) | −0.0025 (14) | 0.0029 (12) | −0.0026 (15) |
| O5 | 0.0248 (14) | 0.030 (3) | 0.0243 (14) | −0.0046 (15) | −0.0009 (11) | 0.0008 (14) |
| O6 | 0.0258 (15) | 0.024 (2) | 0.0267 (15) | −0.0024 (15) | 0.0046 (12) | 0.0010 (15) |
| O7 | 0.0308 (16) | 0.033 (3) | 0.0274 (17) | 0.0032 (16) | 0.0073 (13) | −0.0037 (17) |
| O8 | 0.0329 (18) | 0.025 (3) | 0.0296 (17) | 0.0006 (17) | −0.0058 (14) | 0.0030 (17) |
| C1 | 0.019 (2) | 0.027 (3) | 0.024 (2) | 0.000 (2) | 0.0022 (16) | −0.001 (2) |
| C2 | 0.0178 (19) | 0.018 (3) | 0.030 (2) | −0.0008 (19) | 0.0023 (16) | 0.001 (2) |
| C3 | 0.0184 (19) | 0.023 (3) | 0.030 (2) | −0.0014 (19) | 0.0027 (16) | −0.003 (2) |
| C4 | 0.019 (2) | 0.024 (3) | 0.030 (2) | −0.0040 (19) | 0.0025 (16) | −0.002 (2) |
| C5 | 0.023 (2) | 0.017 (3) | 0.028 (2) | 0.0014 (19) | 0.0073 (18) | 0.005 (2) |
| C6 | 0.019 (2) | 0.019 (3) | 0.027 (2) | 0.0031 (18) | 0.0028 (16) | −0.0004 (19) |
| C7 | 0.0204 (18) | 0.031 (4) | 0.030 (2) | −0.001 (2) | 0.0012 (16) | 0.001 (2) |
| C8 | 0.032 (2) | 0.033 (4) | 0.027 (2) | −0.004 (2) | 0.0027 (18) | 0.006 (2) |
(2S,3R)-3-Hydroxy-5-oxo-2,3,4,5-tetrahydrofuran-2,3-dicarboxylic acid dimethyl sulfoxide monosolvate (I) . Geometric parameters (Å, º)
| S1—O8 | 1.532 (4) | C1—C5 | 1.511 (6) |
| S1—C7 | 1.788 (5) | C1—C2 | 1.559 (8) |
| S1—C8 | 1.791 (6) | C1—H1 | 1.0000 |
| O1—C4 | 1.356 (7) | C2—C3 | 1.525 (6) |
| O1—C1 | 1.445 (6) | C2—C6 | 1.532 (6) |
| O2—C5 | 1.194 (6) | C3—C4 | 1.505 (7) |
| O3—C5 | 1.329 (6) | C3—H3A | 0.9900 |
| O3—H1H | 0.87 (3) | C3—H3B | 0.9900 |
| O4—C2 | 1.406 (6) | C7—H7A | 0.9800 |
| O4—H3H | 0.87 (3) | C7—H7B | 0.9800 |
| O5—C6 | 1.306 (6) | C7—H7C | 0.9800 |
| O5—H2H | 0.89 (3) | C8—H8A | 0.9800 |
| O6—C6 | 1.206 (6) | C8—H8B | 0.9800 |
| O7—C4 | 1.195 (6) | C8—H8C | 0.9800 |
| O8—S1—C7 | 105.7 (2) | C2—C3—H3B | 111.2 |
| O8—S1—C8 | 104.6 (3) | H3A—C3—H3B | 109.1 |
| C7—S1—C8 | 98.0 (3) | O7—C4—O1 | 121.5 (5) |
| C4—O1—C1 | 109.3 (4) | O7—C4—C3 | 128.3 (5) |
| C5—O3—H1H | 113 (4) | O1—C4—C3 | 110.1 (4) |
| C2—O4—H3H | 116 (6) | O2—C5—O3 | 125.8 (5) |
| C6—O5—H2H | 112 (4) | O2—C5—C1 | 125.6 (5) |
| O1—C1—C5 | 110.3 (4) | O3—C5—C1 | 108.6 (4) |
| O1—C1—C2 | 103.8 (4) | O6—C6—O5 | 125.7 (4) |
| C5—C1—C2 | 112.1 (4) | O6—C6—C2 | 122.1 (5) |
| O1—C1—H1 | 110.2 | O5—C6—C2 | 112.1 (4) |
| C5—C1—H1 | 110.2 | S1—C7—H7A | 109.5 |
| C2—C1—H1 | 110.2 | S1—C7—H7B | 109.5 |
| O4—C2—C3 | 113.3 (4) | H7A—C7—H7B | 109.5 |
| O4—C2—C6 | 109.0 (4) | S1—C7—H7C | 109.5 |
| C3—C2—C6 | 112.9 (4) | H7A—C7—H7C | 109.5 |
| O4—C2—C1 | 108.7 (4) | H7B—C7—H7C | 109.5 |
| C3—C2—C1 | 99.6 (4) | S1—C8—H8A | 109.5 |
| C6—C2—C1 | 113.0 (5) | S1—C8—H8B | 109.5 |
| C4—C3—C2 | 103.0 (4) | H8A—C8—H8B | 109.5 |
| C4—C3—H3A | 111.2 | S1—C8—H8C | 109.5 |
| C2—C3—H3A | 111.2 | H8A—C8—H8C | 109.5 |
| C4—C3—H3B | 111.2 | H8B—C8—H8C | 109.5 |
| C4—O1—C1—C5 | 148.2 (4) | C2—C3—C4—O7 | 161.1 (5) |
| C4—O1—C1—C2 | 27.9 (5) | C2—C3—C4—O1 | −17.9 (5) |
| O1—C1—C2—O4 | 82.0 (4) | O1—C1—C5—O2 | −13.6 (8) |
| C5—C1—C2—O4 | −37.1 (5) | C2—C1—C5—O2 | 101.5 (6) |
| O1—C1—C2—C3 | −36.8 (4) | O1—C1—C5—O3 | 166.6 (4) |
| C5—C1—C2—C3 | −155.9 (4) | C2—C1—C5—O3 | −78.3 (6) |
| O1—C1—C2—C6 | −156.8 (3) | O4—C2—C6—O6 | −10.2 (7) |
| C5—C1—C2—C6 | 84.1 (5) | C3—C2—C6—O6 | 116.7 (5) |
| O4—C2—C3—C4 | −83.0 (5) | C1—C2—C6—O6 | −131.2 (5) |
| C6—C2—C3—C4 | 152.4 (5) | O4—C2—C6—O5 | 172.0 (4) |
| C1—C2—C3—C4 | 32.3 (5) | C3—C2—C6—O5 | −61.2 (6) |
| C1—O1—C4—O7 | 174.3 (5) | C1—C2—C6—O5 | 50.9 (5) |
| C1—O1—C4—C3 | −6.6 (5) |
(2S,3R)-3-Hydroxy-5-oxo-2,3,4,5-tetrahydrofuran-2,3-dicarboxylic acid dimethyl sulfoxide monosolvate (I) . Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| O3—H1H···O4i | 0.87 (2) | 2.42 (4) | 2.996 (4) | 124 (3) |
| O3—H1H···O6i | 0.87 (2) | 1.98 (3) | 2.805 (4) | 158 (4) |
| O4—H3H···O8 | 0.87 (2) | 1.87 (3) | 2.714 (5) | 160 (7) |
| O5—H2H···O8ii | 0.89 (2) | 1.73 (2) | 2.603 (4) | 167 (5) |
Symmetry codes: (i) −x+2, y−1/2, −z+1; (ii) −x+1, y−1/2, −z+1.
Dimethyl (2S,3R)-3-Hydroxy-5-oxo-2,3,4,5-tetrahydrofuran-2,3-dicarboxylate (II) . Crystal data
| C8H10O7 | F(000) = 456 |
| Mr = 218.16 | Dx = 1.519 Mg m−3 |
| Monoclinic, P21 | Cu Kα radiation, λ = 1.5418 Å |
| a = 9.3057 (6) Å | Cell parameters from 3289 reflections |
| b = 7.6934 (6) Å | θ = 3.4–72.8° |
| c = 13.4012 (11) Å | µ = 1.20 mm−1 |
| β = 96.243 (7)° | T = 123 K |
| V = 953.74 (12) Å3 | Platey fragment, colourless |
| Z = 4 | 0.30 × 0.20 × 0.04 mm |
Dimethyl (2S,3R)-3-Hydroxy-5-oxo-2,3,4,5-tetrahydrofuran-2,3-dicarboxylate (II) . Data collection
| Oxford Diffraction Gemini S CCD diffractometer | 2976 reflections with I > 2σ(I) |
| Radiation source: sealed tube | Rint = 0.036 |
| ω scans | θmax = 73.4°, θmin = 3.3° |
| Absorption correction: multi-scan (CrysAlis PRO; Oxford Diffraction, 2010) | h = −11→11 |
| Tmin = 0.747, Tmax = 1.000 | k = −8→9 |
| 8046 measured reflections | l = −16→14 |
| 3506 independent reflections |
Dimethyl (2S,3R)-3-Hydroxy-5-oxo-2,3,4,5-tetrahydrofuran-2,3-dicarboxylate (II) . Refinement
| Refinement on F2 | Hydrogen site location: mixed |
| Least-squares matrix: full | H atoms treated by a mixture of independent and constrained refinement |
| R[F2 > 2σ(F2)] = 0.044 | w = 1/[σ2(Fo2) + (0.0568P)2 + 0.1462P] where P = (Fo2 + 2Fc2)/3 |
| wR(F2) = 0.121 | (Δ/σ)max < 0.001 |
| S = 1.10 | Δρmax = 0.23 e Å−3 |
| 3506 reflections | Δρmin = −0.22 e Å−3 |
| 281 parameters | Absolute structure: Flack x determined using 1098 quotients [(I+)-(I-)]/[(I+)+(I-)] (Parsons et al., 2013) |
| 3 restraints | Absolute structure parameter: 0.08 (17) |
Dimethyl (2S,3R)-3-Hydroxy-5-oxo-2,3,4,5-tetrahydrofuran-2,3-dicarboxylate (II) . Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
Dimethyl (2S,3R)-3-Hydroxy-5-oxo-2,3,4,5-tetrahydrofuran-2,3-dicarboxylate (II) . Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| O1 | 0.0127 (3) | 0.3894 (4) | 0.4143 (2) | 0.0393 (7) | |
| O2 | −0.0438 (4) | 0.4647 (5) | 0.2547 (2) | 0.0502 (8) | |
| O3 | 0.3364 (3) | 0.3776 (4) | 0.4119 (2) | 0.0399 (7) | |
| H1H | 0.407 (4) | 0.349 (7) | 0.458 (3) | 0.048* | |
| O4 | 0.4351 (3) | 0.0487 (4) | 0.4527 (2) | 0.0460 (8) | |
| O5 | 0.2148 (3) | −0.0633 (4) | 0.4061 (2) | 0.0432 (7) | |
| O6 | 0.1333 (3) | 0.5084 (4) | 0.5929 (2) | 0.0460 (7) | |
| O7 | 0.2626 (3) | 0.2625 (4) | 0.6205 (2) | 0.0431 (7) | |
| O8 | 0.5725 (3) | 0.4161 (4) | −0.0594 (2) | 0.0391 (7) | |
| O9 | 0.6406 (3) | 0.5063 (5) | −0.2045 (2) | 0.0484 (8) | |
| O10 | 0.8715 (3) | 0.4023 (4) | 0.0508 (2) | 0.0388 (7) | |
| H2H | 0.904 (5) | 0.376 (7) | 0.1128 (16) | 0.047* | |
| O11 | 0.9307 (3) | 0.0730 (4) | 0.1173 (2) | 0.0437 (7) | |
| O12 | 0.7678 (3) | −0.0374 (4) | −0.0028 (3) | 0.0467 (8) | |
| O13 | 0.5379 (3) | 0.5087 (4) | 0.1312 (2) | 0.0443 (7) | |
| O14 | 0.6548 (3) | 0.2682 (4) | 0.1911 (2) | 0.0414 (7) | |
| C1 | 0.0312 (5) | 0.3802 (6) | 0.3161 (3) | 0.0408 (9) | |
| C2 | 0.1501 (5) | 0.2555 (7) | 0.3003 (3) | 0.0424 (10) | |
| H2A | 0.2116 | 0.3012 | 0.2505 | 0.051* | |
| H2B | 0.1106 | 0.1412 | 0.2771 | 0.051* | |
| C3 | 0.2350 (4) | 0.2408 (6) | 0.4034 (3) | 0.0366 (9) | |
| C4 | 0.1125 (4) | 0.2751 (6) | 0.4721 (3) | 0.0377 (9) | |
| H4 | 0.0637 | 0.1638 | 0.4870 | 0.045* | |
| C5 | 0.3083 (5) | 0.0651 (6) | 0.4238 (3) | 0.0389 (9) | |
| C6 | 0.2722 (5) | −0.2380 (6) | 0.4145 (4) | 0.0477 (11) | |
| H6A | 0.3464 | −0.2521 | 0.3685 | 0.072* | |
| H6B | 0.1940 | −0.3216 | 0.3971 | 0.072* | |
| H6C | 0.3151 | −0.2587 | 0.4835 | 0.072* | |
| C7 | 0.1678 (4) | 0.3672 (6) | 0.5685 (3) | 0.0378 (9) | |
| C8 | 0.3289 (6) | 0.3351 (8) | 0.7144 (4) | 0.0564 (13) | |
| H8A | 0.3960 | 0.4279 | 0.7003 | 0.085* | |
| H8B | 0.3819 | 0.2437 | 0.7538 | 0.085* | |
| H8C | 0.2537 | 0.3826 | 0.7524 | 0.085* | |
| C9 | 0.6617 (5) | 0.4150 (6) | −0.1329 (3) | 0.0393 (9) | |
| C10 | 0.7821 (4) | 0.2878 (6) | −0.1067 (3) | 0.0396 (9) | |
| H10A | 0.8750 | 0.3340 | −0.1252 | 0.048* | |
| H10B | 0.7616 | 0.1755 | −0.1414 | 0.048* | |
| C11 | 0.7864 (4) | 0.2667 (6) | 0.0067 (3) | 0.0352 (9) | |
| C12 | 0.6230 (4) | 0.2963 (6) | 0.0191 (3) | 0.0358 (9) | |
| H12 | 0.5694 | 0.1840 | 0.0090 | 0.043* | |
| C13 | 0.8388 (4) | 0.0905 (6) | 0.0475 (3) | 0.0370 (9) | |
| C14 | 0.8023 (5) | −0.2154 (7) | 0.0250 (4) | 0.0478 (11) | |
| H14A | 0.8702 | −0.2623 | −0.0192 | 0.072* | |
| H14B | 0.7137 | −0.2851 | 0.0182 | 0.072* | |
| H14C | 0.8465 | −0.2193 | 0.0947 | 0.072* | |
| C15 | 0.5978 (4) | 0.3732 (6) | 0.1191 (3) | 0.0365 (9) | |
| C16 | 0.6380 (6) | 0.3211 (7) | 0.2932 (4) | 0.0504 (12) | |
| H16A | 0.6857 | 0.4334 | 0.3071 | 0.076* | |
| H16B | 0.6820 | 0.2338 | 0.3402 | 0.076* | |
| H16C | 0.5349 | 0.3319 | 0.3012 | 0.076* |
Dimethyl (2S,3R)-3-Hydroxy-5-oxo-2,3,4,5-tetrahydrofuran-2,3-dicarboxylate (II) . Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| O1 | 0.0391 (13) | 0.0337 (18) | 0.0436 (16) | 0.0050 (13) | −0.0023 (11) | −0.0018 (13) |
| O2 | 0.0564 (18) | 0.046 (2) | 0.0456 (17) | 0.0136 (15) | −0.0086 (14) | −0.0029 (14) |
| O3 | 0.0377 (14) | 0.0332 (18) | 0.0481 (16) | −0.0031 (13) | 0.0012 (12) | 0.0019 (13) |
| O4 | 0.0439 (16) | 0.040 (2) | 0.0519 (17) | 0.0044 (14) | −0.0055 (13) | −0.0016 (13) |
| O5 | 0.0441 (16) | 0.0288 (17) | 0.0561 (18) | −0.0004 (13) | 0.0023 (13) | 0.0014 (13) |
| O6 | 0.0513 (16) | 0.0371 (19) | 0.0477 (17) | 0.0081 (14) | −0.0031 (14) | −0.0090 (14) |
| O7 | 0.0528 (16) | 0.0314 (18) | 0.0429 (16) | 0.0050 (14) | −0.0048 (13) | 0.0029 (13) |
| O8 | 0.0386 (14) | 0.0344 (18) | 0.0431 (15) | 0.0009 (12) | −0.0014 (11) | 0.0040 (12) |
| O9 | 0.0510 (17) | 0.049 (2) | 0.0441 (17) | −0.0017 (15) | 0.0000 (14) | 0.0089 (14) |
| O10 | 0.0405 (14) | 0.0341 (19) | 0.0404 (15) | −0.0032 (13) | −0.0021 (11) | −0.0002 (12) |
| O11 | 0.0484 (16) | 0.0373 (18) | 0.0439 (16) | 0.0042 (14) | −0.0015 (13) | −0.0001 (13) |
| O12 | 0.0433 (16) | 0.0281 (19) | 0.066 (2) | 0.0007 (13) | −0.0051 (14) | −0.0063 (14) |
| O13 | 0.0493 (16) | 0.039 (2) | 0.0428 (16) | 0.0079 (14) | −0.0027 (13) | −0.0032 (14) |
| O14 | 0.0492 (15) | 0.0353 (18) | 0.0400 (15) | 0.0056 (14) | 0.0063 (12) | 0.0045 (13) |
| C1 | 0.044 (2) | 0.033 (3) | 0.043 (2) | 0.0020 (19) | −0.0042 (17) | −0.0038 (18) |
| C2 | 0.049 (2) | 0.036 (3) | 0.040 (2) | 0.0044 (19) | −0.0029 (17) | −0.0028 (18) |
| C3 | 0.041 (2) | 0.027 (2) | 0.041 (2) | −0.0035 (17) | 0.0022 (16) | −0.0005 (16) |
| C4 | 0.0380 (19) | 0.031 (2) | 0.043 (2) | −0.0024 (17) | −0.0014 (16) | 0.0003 (18) |
| C5 | 0.043 (2) | 0.037 (3) | 0.036 (2) | 0.0028 (18) | 0.0024 (16) | −0.0034 (17) |
| C6 | 0.055 (3) | 0.032 (3) | 0.056 (3) | 0.002 (2) | 0.005 (2) | 0.002 (2) |
| C7 | 0.0351 (18) | 0.039 (3) | 0.039 (2) | −0.0013 (17) | 0.0045 (15) | 0.0006 (18) |
| C8 | 0.069 (3) | 0.050 (3) | 0.046 (3) | 0.004 (2) | −0.015 (2) | 0.002 (2) |
| C9 | 0.045 (2) | 0.036 (3) | 0.037 (2) | −0.0065 (18) | −0.0001 (16) | −0.0026 (17) |
| C10 | 0.041 (2) | 0.037 (3) | 0.040 (2) | −0.0018 (18) | 0.0036 (16) | −0.0003 (17) |
| C11 | 0.0377 (19) | 0.029 (2) | 0.039 (2) | 0.0002 (16) | 0.0020 (15) | −0.0029 (17) |
| C12 | 0.039 (2) | 0.026 (2) | 0.042 (2) | −0.0024 (17) | −0.0012 (16) | 0.0026 (17) |
| C13 | 0.0368 (19) | 0.031 (2) | 0.043 (2) | −0.0014 (17) | 0.0050 (17) | −0.0033 (17) |
| C14 | 0.042 (2) | 0.036 (3) | 0.065 (3) | 0.000 (2) | 0.007 (2) | −0.004 (2) |
| C15 | 0.0339 (17) | 0.031 (2) | 0.044 (2) | −0.0025 (17) | 0.0017 (15) | 0.0001 (17) |
| C16 | 0.060 (3) | 0.048 (3) | 0.043 (2) | 0.005 (2) | 0.006 (2) | 0.008 (2) |
Dimethyl (2S,3R)-3-Hydroxy-5-oxo-2,3,4,5-tetrahydrofuran-2,3-dicarboxylate (II) . Geometric parameters (Å, º)
| O1—C1 | 1.347 (5) | C2—H2B | 0.9900 |
| O1—C4 | 1.442 (5) | C3—C5 | 1.526 (6) |
| O2—C1 | 1.210 (5) | C3—C4 | 1.562 (6) |
| O3—C3 | 1.410 (5) | C4—C7 | 1.514 (6) |
| O3—H1H | 0.880 (14) | C4—H4 | 1.0000 |
| O4—C5 | 1.208 (5) | C6—H6A | 0.9800 |
| O5—C5 | 1.320 (5) | C6—H6B | 0.9800 |
| O5—C6 | 1.446 (6) | C6—H6C | 0.9800 |
| O6—C7 | 1.189 (6) | C8—H8A | 0.9800 |
| O7—C7 | 1.334 (5) | C8—H8B | 0.9800 |
| O7—C8 | 1.451 (6) | C8—H8C | 0.9800 |
| O8—C9 | 1.355 (5) | C9—C10 | 1.501 (6) |
| O8—C12 | 1.439 (5) | C10—C11 | 1.524 (6) |
| O9—C9 | 1.188 (5) | C10—H10A | 0.9900 |
| O10—C11 | 1.401 (5) | C10—H10B | 0.9900 |
| O10—H2H | 0.876 (14) | C11—C13 | 1.522 (6) |
| O11—C13 | 1.204 (5) | C11—C12 | 1.565 (5) |
| O12—C13 | 1.328 (5) | C12—C15 | 1.506 (6) |
| O12—C14 | 1.446 (6) | C12—H12 | 1.0000 |
| O13—C15 | 1.202 (5) | C14—H14A | 0.9800 |
| O14—C15 | 1.324 (5) | C14—H14B | 0.9800 |
| O14—C16 | 1.452 (6) | C14—H14C | 0.9800 |
| C1—C2 | 1.497 (6) | C16—H16A | 0.9800 |
| C2—C3 | 1.519 (6) | C16—H16B | 0.9800 |
| C2—H2A | 0.9900 | C16—H16C | 0.9800 |
| C1—O1—C4 | 110.4 (3) | H8A—C8—H8B | 109.5 |
| C3—O3—H1H | 108 (4) | O7—C8—H8C | 109.5 |
| C5—O5—C6 | 116.8 (3) | H8A—C8—H8C | 109.5 |
| C7—O7—C8 | 114.6 (4) | H8B—C8—H8C | 109.5 |
| C9—O8—C12 | 110.5 (3) | O9—C9—O8 | 121.5 (4) |
| C11—O10—H2H | 110 (4) | O9—C9—C10 | 128.9 (4) |
| C13—O12—C14 | 119.2 (4) | O8—C9—C10 | 109.6 (4) |
| C15—O14—C16 | 116.1 (4) | C9—C10—C11 | 103.9 (3) |
| O2—C1—O1 | 120.7 (4) | C9—C10—H10A | 111.0 |
| O2—C1—C2 | 129.0 (4) | C11—C10—H10A | 111.0 |
| O1—C1—C2 | 110.3 (4) | C9—C10—H10B | 111.0 |
| C1—C2—C3 | 103.7 (3) | C11—C10—H10B | 111.0 |
| C1—C2—H2A | 111.0 | H10A—C10—H10B | 109.0 |
| C3—C2—H2A | 111.0 | O10—C11—C13 | 111.5 (3) |
| C1—C2—H2B | 111.0 | O10—C11—C10 | 107.1 (3) |
| C3—C2—H2B | 111.0 | C13—C11—C10 | 115.2 (4) |
| H2A—C2—H2B | 109.0 | O10—C11—C12 | 111.0 (3) |
| O3—C3—C2 | 107.1 (3) | C13—C11—C12 | 111.6 (3) |
| O3—C3—C5 | 111.4 (3) | C10—C11—C12 | 99.8 (3) |
| C2—C3—C5 | 114.0 (4) | O8—C12—C15 | 109.2 (3) |
| O3—C3—C4 | 110.6 (4) | O8—C12—C11 | 105.0 (3) |
| C2—C3—C4 | 100.6 (3) | C15—C12—C11 | 113.5 (3) |
| C5—C3—C4 | 112.6 (4) | O8—C12—H12 | 109.6 |
| O1—C4—C7 | 108.2 (3) | C15—C12—H12 | 109.6 |
| O1—C4—C3 | 104.8 (3) | C11—C12—H12 | 109.6 |
| C7—C4—C3 | 112.3 (3) | O11—C13—O12 | 125.7 (4) |
| O1—C4—H4 | 110.5 | O11—C13—C11 | 123.5 (4) |
| C7—C4—H4 | 110.5 | O12—C13—C11 | 110.8 (3) |
| C3—C4—H4 | 110.5 | O12—C14—H14A | 109.5 |
| O4—C5—O5 | 125.5 (4) | O12—C14—H14B | 109.5 |
| O4—C5—C3 | 123.5 (4) | H14A—C14—H14B | 109.5 |
| O5—C5—C3 | 111.0 (3) | O12—C14—H14C | 109.5 |
| O5—C6—H6A | 109.5 | H14A—C14—H14C | 109.5 |
| O5—C6—H6B | 109.5 | H14B—C14—H14C | 109.5 |
| H6A—C6—H6B | 109.5 | O13—C15—O14 | 125.9 (4) |
| O5—C6—H6C | 109.5 | O13—C15—C12 | 125.4 (4) |
| H6A—C6—H6C | 109.5 | O14—C15—C12 | 108.6 (4) |
| H6B—C6—H6C | 109.5 | O14—C16—H16A | 109.5 |
| O6—C7—O7 | 126.2 (4) | O14—C16—H16B | 109.5 |
| O6—C7—C4 | 125.9 (4) | H16A—C16—H16B | 109.5 |
| O7—C7—C4 | 107.9 (4) | O14—C16—H16C | 109.5 |
| O7—C8—H8A | 109.5 | H16A—C16—H16C | 109.5 |
| O7—C8—H8B | 109.5 | H16B—C16—H16C | 109.5 |
| C4—O1—C1—O2 | 179.0 (4) | C12—O8—C9—O9 | −179.1 (4) |
| C4—O1—C1—C2 | −0.5 (5) | C12—O8—C9—C10 | 0.3 (5) |
| O2—C1—C2—C3 | 160.9 (5) | O9—C9—C10—C11 | 158.1 (5) |
| O1—C1—C2—C3 | −19.7 (5) | O8—C9—C10—C11 | −21.2 (5) |
| C1—C2—C3—O3 | −86.2 (4) | C9—C10—C11—O10 | −84.8 (4) |
| C1—C2—C3—C5 | 150.1 (4) | C9—C10—C11—C13 | 150.5 (4) |
| C1—C2—C3—C4 | 29.3 (5) | C9—C10—C11—C12 | 30.9 (4) |
| C1—O1—C4—C7 | 139.9 (4) | C9—O8—C12—C15 | 142.4 (3) |
| C1—O1—C4—C3 | 19.9 (4) | C9—O8—C12—C11 | 20.3 (4) |
| O3—C3—C4—O1 | 82.7 (4) | O10—C11—C12—O8 | 81.3 (4) |
| C2—C3—C4—O1 | −30.2 (4) | C13—C11—C12—O8 | −153.6 (3) |
| C5—C3—C4—O1 | −152.0 (3) | C10—C11—C12—O8 | −31.4 (4) |
| O3—C3—C4—C7 | −34.5 (5) | O10—C11—C12—C15 | −37.9 (5) |
| C2—C3—C4—C7 | −147.5 (4) | C13—C11—C12—C15 | 87.1 (4) |
| C5—C3—C4—C7 | 90.8 (4) | C10—C11—C12—C15 | −150.6 (4) |
| C6—O5—C5—O4 | −5.2 (6) | C14—O12—C13—O11 | −1.2 (7) |
| C6—O5—C5—C3 | 175.1 (4) | C14—O12—C13—C11 | 180.0 (4) |
| O3—C3—C5—O4 | 6.5 (6) | O10—C11—C13—O11 | 8.2 (6) |
| C2—C3—C5—O4 | 127.9 (5) | C10—C11—C13—O11 | 130.5 (4) |
| C4—C3—C5—O4 | −118.4 (5) | C12—C11—C13—O11 | −116.6 (4) |
| O3—C3—C5—O5 | −173.8 (3) | O10—C11—C13—O12 | −173.0 (3) |
| C2—C3—C5—O5 | −52.4 (5) | C10—C11—C13—O12 | −50.6 (5) |
| C4—C3—C5—O5 | 61.3 (5) | C12—C11—C13—O12 | 62.3 (4) |
| C8—O7—C7—O6 | −1.3 (6) | C16—O14—C15—O13 | 2.1 (6) |
| C8—O7—C7—C4 | 178.3 (4) | C16—O14—C15—C12 | −178.9 (4) |
| O1—C4—C7—O6 | 1.0 (6) | O8—C12—C15—O13 | 3.7 (5) |
| C3—C4—C7—O6 | 116.1 (5) | C11—C12—C15—O13 | 120.6 (5) |
| O1—C4—C7—O7 | −178.6 (3) | O8—C12—C15—O14 | −175.2 (3) |
| C3—C4—C7—O7 | −63.4 (4) | C11—C12—C15—O14 | −58.4 (4) |
Dimethyl (2S,3R)-3-Hydroxy-5-oxo-2,3,4,5-tetrahydrofuran-2,3-dicarboxylate (II) . Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| O3—H1H···O4i | 0.88 (1) | 2.36 (5) | 2.951 (4) | 125 (4) |
| O10—H2H···O2ii | 0.88 (1) | 2.03 (3) | 2.802 (4) | 147 (5) |
Symmetry codes: (i) −x+1, y+1/2, −z+1; (ii) x+1, y, z.
References
- Ali, B. H., Al Wabel, N. & Blunden, G. (2005). Phytother. Res. 19, 369–375. [DOI] [PubMed]
- Altomare, A., Cascarano, G., Giacovazzo, C. & Guagliardi, A. (1993). J. Appl. Cryst. 26, 343–350.
- Boll, P. M., Sorensen, E. & Balieu, E. (1969). Acta Chem. Scand. 23, 286–293.
- Da-Costa-Rocha, I., Bonnlaender, B., Sievers, H., Pischel, I. & Heinrich, M. (2014). Food Chem. 165, 424–443. [DOI] [PubMed]
- Evans, D. A., Trotter, B. W. & Barrow, J. C. (1997). Tetrahedron, 53, 8779–8794.
- Glusker, J. P., Minkin, J. A. & Soule, F. B. (1972). Acta Cryst. B28, 2499–2505.
- Groom, C. R., Bruno, I. J., Lightfoot, M. P. & Ward, S. C. (2016). Acta Cryst. B72, 171–179. [DOI] [PMC free article] [PubMed]
- Hansawasdi, C., Kawabata, J. & Kasai, T. (2000). Biosci. Biotechnol. Biochem. 64, 1041–1043. [DOI] [PubMed]
- Hansawasdi, C., Kawabata, J. & Kasai, T. (2001). Biosci. Biotechnol. Biochem. 65, 2087–2089. [DOI] [PubMed]
- Macrae, C. F., Bruno, I. J., Chisholm, J. A., Edgington, P. R., McCabe, P., Pidcock, E., Rodriguez-Monge, L., Taylor, R., van de Streek, J. & Wood, P. A. (2008). J. Appl. Cryst. 41, 466–470.
- Mahapatra, S., Mallik, S. B., Rao, G. V., Reddy, G. C. & Guru Row, T. N. (2007). Acta Cryst. E63, o3869.
- Oxford Diffraction (2010). CrysAlis PRO. Oxford Diffraction Ltd, Yarnton, Oxfordshire, England.
- Parsons, S., Flack, H. D. & Wagner, T. (2013). Acta Cryst. B69, 249–259. [DOI] [PMC free article] [PubMed]
- Sheldrick, G. M. (2015). Acta Cryst. C71, 3–8.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I, II, global. DOI: 10.1107/S2056989017011902/zs2386sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989017011902/zs2386Isup2.hkl
Supporting information file. DOI: 10.1107/S2056989017011902/zs2386Isup4.cml
Structure factors: contains datablock(s) II. DOI: 10.1107/S2056989017011902/zs2386IIsup3.hkl
Supporting information file. DOI: 10.1107/S2056989017011902/zs2386IIsup5.cml
Additional supporting information: crystallographic information; 3D view; checkCIF report