Abstract
Aberrantly expressed microRNAs have been implicated in lots of cancers. Reduced amounts of let-7g have been found in breast cancer tissues. The function of let-7g in bone metastasis of breast cancer remains poorly understood. This study is to explore the significance of let-7g and its novel target gene in bone metastasis of breast cancer.
The expression of let-7g or forkhead box C2 (FOXC2) was measured in human clinical breast cancer tissues with bone metastasis by using quantitative real-time Polymerase Chain Reaction (qRT-PCR). After transfection with let-7g or anti-let-7g in breast cancer cell linesMDA-MB-231or SK-BR3, qRT-PCR and Western blot were done to test the levels of let-7g and FOXC2. The effect of anti-let-7g and/ or FOXC2 RNA interference (RNAi) on cell migration in breast cancer cells was evaluated by using wound healing assay.
Clinically, qRT-PCR showed that FOXC2 levels were higher in breast cancer tissues with bone metastasis than those in their noncancerous counterparts. Let-7g was showed to be negatively correlated with FOXC2 in human breast cancer samples with bone metastasis. We found that enforced expression of let-7g reduced levels of FOXC2 protein by using Western blot in MDA-MB-231 cells. Conversely, anti-let-7g enhanced levels of FOXC2 in SK-BR3 cells. In terms of function, anti-let-7g accelerated migration of SK-BR3 cells. Interestingly, FOXC2 RNAi abrogated anti-let-7g-mediated migration in breast cancer cells. Thus, we conclude that let-7g suppresses cell migration through targeting FOXC2 in breast cancer. Our finding provides a new perspective for understanding the mechanism of bone metastasis in breast cancer.
Keywords: let-7g, FOXC2, Migration, Bone metastasis, Breast cancer
1. Introduction
Breast cancer is the most common cancer among women [1]. MicroRNAs (miRNAs) usually modulate the expression of target genes at the post-transcription level [2-4]. As one of the first tumor suppressor miRNAs to be identified, the let-7 family is composed of 13 members with overlapping and distinct functions in humans [5-7]. A report shows that let-7 expression is decreased and RAS protein is significantly higher in lung tumors [8], which is consistent with clinical observations in lung cancer [9]. A number of groups found inhibitory functions of the let-7 family in various types of tumors [6, 7, 10]. Reduced let-7 expression is reported to be associated with shortened postoperative survival in patients with cancer [11]. Let-7 is capable of targeting many oncogenic proteins, such as KRAS/HRAS [8, 12, 13], HMGA2 [13-16], and cyclin genes [17, 18]. Let-7g, one member of let-7 family, is involved in the development of hepatocellular carcinoma and breast cancer [19, 20]. Let-7g plays important roles in liver cancer through negatively regulating Bcl-xL or collagen type I α2 [19, 21]. However, the underlying mechanism by which let-7g functions in breast cancer metastasis remains unclear.
The transcription factor superfamily Forkhead-box (FOX) plays a role in differentiation, proliferation, migration, apoptosis, and metabolism [22, 23]. The change in FOX expression is involved in progression of various cancers through affecting epithelial–mesenchymal transition (EMT) or EMT regulatory pathways [24]. One member of FOX, FOXA1, is shown to be highly expressed in breast cancer [25]. FOXC2, another member, is able to promote metastasis and paclitaxel drug resistance [26, 27].
In the present study, we investigated the role of let-7g and its novel target gene in bone metastasis of breast cancer. We show that let-7g is negatively related to FOXC2 in breast tissues with bone metastasis and suppresses cell migration through regulating FOXC2 in breast cancer. Our finding contributes to understanding the mechanism of breast cancer metastasis mediated by let-7g.
2. Materials and methods
2.1. Patient samples
Twenty-five cases of clinical breast tumor tissues with bone metastasis and their corresponding peritumor tissues were obtained from the First Affiliated Hospital of Xi’an Medical University (Xi’an, China) after surgical resection. The patients consented that samples could be used for research. Patient information is listed in Table S1. Research Ethics Committee at the First Affiliated Hospital of Xi’an Medical University approved the study protocol.
Table S1.
Clinical characteristics of breast tumor samples
| No. | Age | Sex | Organ | Pathology diagnosis | Grade |
|---|---|---|---|---|---|
| 01 | 56 | F | Breast | Nonspecific infiltrating ductal carcinoma with bone metastasis | I |
| 02 | 47 | F | Breast | Little nonspecific infiltrating ductal carcinoma with bone metastasis | I |
| 03 | 34 | F | Breast | Nonspecific infiltrating ductal carcinoma with bone metastasis | I |
| 04 | 33 | F | Breast | Nonspecific infiltrating ductal carcinoma with bone metastasis | I |
| 05 | 38 | F | Breast | A little nonspecific infiltrating ductal carcinoma with bone metastasis | I |
| 06 | 46 | F | Breast | Little nonspecific infiltrating ductal carcinoma with bone metastasis | II |
| 07 | 43 | F | Breast | Nonspecific infiltrating ductal carcinoma with bone metastasis | II |
| 08 | 48 | F | Breast | Little nonspecific infiltrating ductal carcinoma with bone metastasis | II |
| 09 | 53 | F | Breast | Nonspecific infiltrating ductal carcinoma with bone metastasis | II |
| 10 | 62 | F | Breast | Nonspecific infiltrating ductal carcinoma with bone metastasis | II |
| 11 | 34 | F | Breast | Little nonspecific infiltrating ductal carcinoma with bone metastasis | II |
| 12 | 57 | F | Breast | Nonspecific infiltrating ductal carcinoma with bone metastasis | II |
| 13 | 36 | F | Breast | Nonspecific infiltrating ductal carcinoma with bone metastasis | II |
| 14 | 52 | F | Breast | Nonspecific infiltrating ductal carcinoma with bone metastasis | II |
| 15 | 51 | F | Breast | Nonspecific infiltrating ductal carcinoma with bone metastasis | III |
| 16 | 49 | F | Breast | Nonspecific infiltrating ductal carcinoma with bone metastasis | II |
| 17 | 40 | F | Breast | Nonspecific infiltrating ductal carcinoma with bone metastasis | II |
| 18 | 52 | F | Breast | Nonspecific infiltrating ductal carcinoma with bone metastasis | II |
| 19 | 48 | F | Breast | Nonspecific infiltrating ductal carcinoma with bone metastasis | II |
| 20 | 35 | F | Breast | Nonspecific infiltrating ductal carcinoma with bone metastasis | II |
| 21 | 39 | F | Breast | Nonspecific infiltrating ductal carcinoma with bone metastasis | II |
| 22 | 40 | F | Breast | Nonspecific infiltrating ductal carcinoma with bone metastasis | II |
| 23 | 45 | F | Breast | Little nonspecific infiltrating ductal carcinoma with bone metastasis | II |
| 24 | 33 | F | Breast | Nonspecific infiltrating ductal carcinoma with bone metastasis | II |
| 25 | 53 | F | Breast | Nonspecific infiltrating ductal carcinoma with bone metastasis | I-II |
2.2. Cell lines
Breast cancer cell lines MDA-MB-231 with high metastatic ability and SK-BR3 were grown in Dulbecco’s modified Eagle’s medium with 10% fetal bovine serum (Gibco, CA, USA). Cells were incubated at 37°C., 5% CO2.
2.3. Quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA was purified by Trizol (Invitrogen, USA). To test let-7g poly (A) polymerase (Ambion, USA) was added to polyadenylate total RNA. SuperScript™ IV Reverse Transcriptase (ThermoFisher Scientific, USA) was used to do reverse transcription. The primers used were as follows: FOXC2 forward, 5′-CCTACC TGAGCGAGCAGAAT-3′, reverse, 5′-ACCTTGACGA AGCACTCGTT-3′; GAPDH forward, 5′-AACGGATTTGGTCGTATTG-3′, reverse, 5′-GGAAGATGGTGATGGGATT-3′; let-7g forward, 5′- CTGTACAGGCCACTGCCTTGC-3′, reverse, 5′-GCGAGCACAGAATTAATACGAC-3′; U6 forward, 5′-CTCGCTTCGGCAGCACA-3′, reverse, 5′-AACTCTTCACTAATTTGCTG-3′.
2.4. Cell transfection
Lipofectamine 2000 reagent (Invitrogen, USA) was used to transfect miRNAs or siRNAs. Let-7g, anti-let-7g, and siFOXC2 (5′-GCAGTCTTATCTAACTATGATGCAA-3′) were synthesized by Sangon Biotech (Shanghai, China).
2.5. Western blot
Cells were lysed and proteins were extracted in RIPA buffer (Biomed, China). Equal amounts of protein were separated by SDS-PAGE and transferred to membranes. Membranes were blotted with anti-β-actin (NeoMarkers, USA) and anti-FOXC2 (Abcam, USA). Membranes were finally visualized by ImmobilonTM Western Chemiluminescent HRP Substrate (Millipore Corporation, USA).
Wound healing assay Breast
cancer cells were seeded into 6-well plates. Sterile plastic pipette tip was utilized to make a wound. Pictures of the wound were taken at 0h and 48h post scraping. At each time point the wound gaps were measured.
2.6. Statistical analysis
Each experiment was performed in triplicate. By comparing mean values (±SD) using a Student’s t test, statistical significance was estimated for independent groups (⋆⋆P < 0.01, ⋆⋆⋆P < 0.001). The correlation of let-7g with FOXC2 in clinical breast cancer tissues with bone metastasis was determined by Pearson’s correlation coefficient.
3. Results
3.1. Let-7g is negatively related to FOXC2 in breast cancer patients with bone metastasis
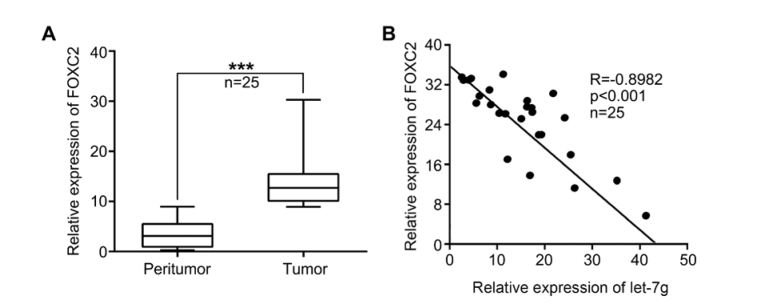
Our data showed that FOXC2 was highly expressed in 25 breast tumor samples with bone metastasis compared with peritumor samples (Figure. 1A). Furthermore, qRT-PCR analysis showed a negative correlation of let-7g with FOXC2 in clinical breast cancer tissues with bone metastasis (Pearson’s correlation coefficient r=-0.8982, p<0.001) (Figure. 1B), indicating that FOXC2 might be one target gene of let-7g in breast cancer. These results suggest that let-7g negatively correlates with FOXC2 in breast cancer tissues with bone metastasis.
Figure 1.
Let-7g negatively correlates with FOXC2 in human breast cancer samples with bone metastasis. (A) FOXC2 expression was tested by qRT-PCR in 25 breast tumor tissues with bone metastasis and peritumor tissues. (B) The association of let-7g with FOXC2 was analyzed by qRT-PCR in 25 breast cancer tissues with bone metastasis (Pearson’s correlation coefficient, r=-0.8982). (Statistical differences: *** P < 0.001; Student’s t test)
3.2. Let-7g is able to inhibit FOXC2 in breast cancer cells
To explore the modulation of FOXC2 by let-7g we transfected let-7g into MDA-MB-231 cells. We observed that let-7g could decrease the level of FOXC2 protein in MDA-MB-231 cells (Figure. 2A). Nevertheless, FOXC2 was rescued when anti-let-7g transfection depressed endogenous let-7g in SK-BR3 cells (Figure. 2B), further supporting that let-7g is critical for FOXC2 regulation in the cells. Transfection validation of let-7g and anti-let-7g by qRT-PCR analysis was performed in the cells (Figure. 2A and 2B). Our data illustrate that let-7g is likely to repress FOXC2 in breast cancer cells.
Figure 2.
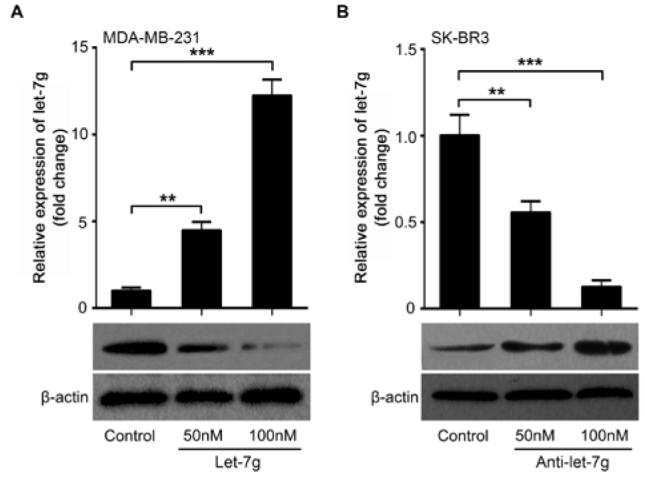
Let-7g inhibits FOXC2 expression in breast cancer cells. (A) The levels of FOXC2 protein were examined in MDA-MB-231 cells with the treatment of let-7g by Western blot analysis. The successful transfection of let-7g was confirmed by qRT-PCR analysis. (B) The levels of FOXC2 protein were determined in SK-BR3 cells with anti-let-7g treatment using Western blot. The successful transfection of anti-let-7g was accessed by qRT-PCR. (Statistical differences: ** P < 0.01, *** P < 0.001; Student’s t test)
3.3. Let-7g reduces migration of breast cancer cells via FOXC2
Next, we tried to clarify the function of let-7g in breast cancer. Wound healing assays revealed that cell migration was increased post-transfection with anti-let-7g in SK-BR3 cells, while FOXC2 knockdown abolished the induced-migration mediated by anti-let-7g (Figure. 3A and 3B). In general, our data indicate that let-7g is capable of inhibiting cell migration through FOXC2 in breast cancer cells.
Figure 3.
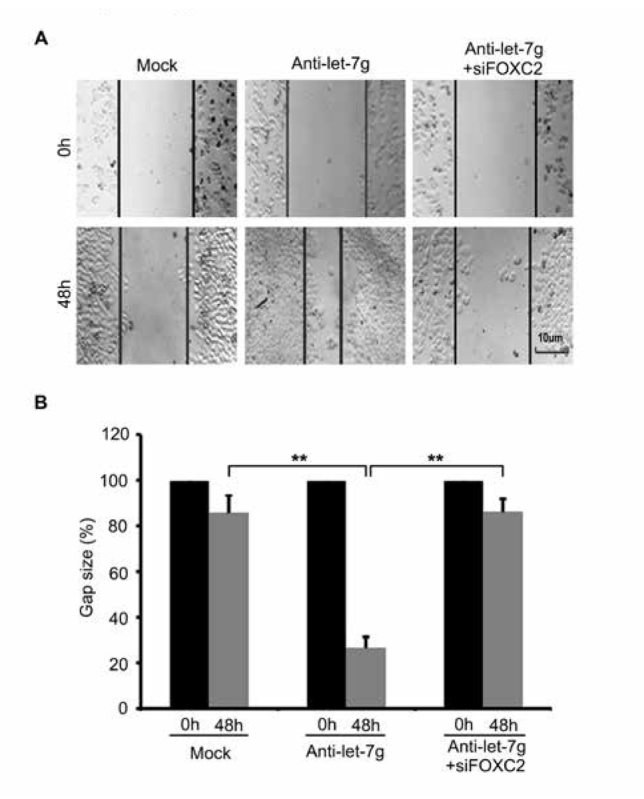
Let-7g reduces cell migration via FOXC2 in breast cancer. (A, B) Wound healing assays for anti-let-7g (or anti-let-7g/siFOXC2) effect on cell migration was performed in SK-BR3 cells. Scale bar = 10 μm. (Statistical differences: ** P < 0.01; Student’s t test)
4. Discussion
MiRNAs play important roles in multiple biological processes. MiRNAs take the roles of oncogenes or tumor suppressor genes through different mechanisms. Many studies show that let-7 is the most frequently found miRNA and is significantly related to clinical outcomes in cancers [11]. Some nanoparticle-based let-7 replacement therapy has been successfully tested in preclinical animal models of cancer [12, 28-31]. The functional characterization of the let-7 family in cancer represents a great opportunity to develop robust biomarkers and novel therapeutic strategies for this disease. Therefore, we are interested in the role of let-7g and its novel target gene in bone metastasis of breast cancer.
In our present study, we found that as one member of the FOX transcription factor family, FOXC2 was obviously up-regulated in clinical breast cancer tissues with bone metastasis. Moreover, a significant correlation between let-7g and FOXC2 was observed in clinical breast cancer samples. Ectopicly expressed let-7g significantly inhibited the expression of FOXC2 in breast cancer cells. Functionally, anti-let-7g promoted the migration of breast cancer cells in vitro. However, silencing of FOXC2 could abrogate the increase of cell migration induced by anti-let-7g in breast cancer.
Accumulating evidence shows that the expression of let-7 is usually lost, reduced, or deregulated in the majority of human cancers [2]. A report shows that restoration of let-7 expression inhibits proliferation of cancer cells of different kinds [8]. LIN28 induced-suppression of let-7 maturation mediated by TUTase is a potential target for pharmacologic inhibition by small chemical compounds [32]. Some let-7 family members have been reported to be negatively correlated with metastasis in cancers [19, 20, 33-35]. Notably, let-7g was specifically linked with breast cancer metastasis and poor patient survival [20]. Our results suggest that let-7g might be an ideal therapeutic target in breast cancer. Given every miRNA is capable of targeting many transcripts, the potential regulation in cancer afforded by the let-7 family might be different. Let-7 could reduce tumor cell migration via ITGB3 and MYH9 [33, 36]. The chemotactic response during tumor metastasis can be blocked by let-7 through CCR7 and CXCR4 [35, 37]. FOXC2 is one member of FOX transcription factor superfamily. FOXC2 could sensitize stem cell-rich breast cancer cells to PLK1 inhibition and induce the G2/M transition [38]. Recently, a report has showed that FOXC2 regulates EMT and metastasis in a p38-dependent manner in breast cancer [39]. We found that FOXC2 was a novel target gene of let-7g in breast cancer metastasis.
We presented a novel target gene of let-7g in driving bone metastasis of breast cancer. We found the negative correlation of FOXC2 and let-7g in clinical breast cancer tissues with bone metastasis. Let-7g is able to negatively regulate FOXC2 to inhibit cell migration in breast cancer. Therapeutically, the let-7g/FOXC2 axis might be a potential target for breast cancer treatment.
Footnotes
Conflicts of interest: There are no conflicts of interest.
References
- [1].Jemal A., Bray F., Center M.M., Ferlay J., Ward E., Forman D.. Global cancer statistics. CA Cancer J Clin. 2011;61(2):69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- [2].Yates L.A., Norbury C.J., Gilbet R.J.. The long and short of microRNA. Cell. 2013;153(3):516–519. doi: 10.1016/j.cell.2013.04.003. [DOI] [PubMed] [Google Scholar]
- [3].Wei Y., Schober A., Weber C.. Pathogenic arterial remodeling: the good and bad of microRNAs. Am J Physiol Heart Circ Physiol. 2013;304(8):H1050–1059. doi: 10.1152/ajpheart.00267.2012. [DOI] [PubMed] [Google Scholar]
- [4].Wang Z., Yao H., Lin S., Zhu X., Shen Z., Lu G.. et al. Transcriptional and epigenetic regulation of human microRNAs. Cancer Lett. 2013;331(1):1–10. doi: 10.1016/j.canlet.2012.12.006. [DOI] [PubMed] [Google Scholar]
- [5].Pasquinelli A.E., Reinhart B.J., Slack F., Martindale M.Q., Kuroda M.I., Maller B.. et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408(6808):86–89. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- [6].Bussing I., Slack F.J., Grosshans H.. let-7 microRNAs in development, stem cells and cancer. Trends Mol Med. 2008;14(9):400–409. doi: 10.1016/j.molmed.2008.07.001. [DOI] [PubMed] [Google Scholar]
- [7].Roush S., Slack F.J.. The let-7 family of microRNAs. Trends Cell Biol. 2008;18(10):505–516. doi: 10.1016/j.tcb.2008.07.007. [DOI] [PubMed] [Google Scholar]
- [8].Johnson S.M., Grosshans H., Shingara J., Byrom M., Jarvis R., Cheng A.. et al. RAS is regulated by the let-7 microRNA family. Cell. 2005;120(5):635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- [9].Takamizawa J., Konishi H., Yanagisawa K., Tomida S., Osada H., Endoh H.. et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64(11):3753–3756. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- [10].Boyerinas B., Park S.M., Hau A., Murmann A.E., Peter M.E.. The role of let-7 in cell differentiation and cancer. Endocr Relat Cancer. 2010;17(1):F19–36. doi: 10.1677/ERC-09-0184. [DOI] [PubMed] [Google Scholar]
- [11].Nair V.S., Maeda L.S., loannidis J.P.. Clinical outcome prediction by microRNAs in human cancer: a systematic review. J Natl Cancer Inst. 2012;104(7):528–540. doi: 10.1093/jnci/djs027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Kumar M,S, Erkeland S,J, Pester R,E, Chen C,Y, Ebert M,S, Sharp P,A. et al. Suppression of non-small cell lung tumor development by the let-7 microRNA family. Proc Natl Acad Sci U S A. 2008;105(10):3903–3908. doi: 10.1073/pnas.0712321105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Yu F., Yao H., Zhu P., Zhang X., Pan Q., Gong C.. et al. let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell. 2007;131(6):1109–1123. doi: 10.1016/j.cell.2007.10.054. [DOI] [PubMed] [Google Scholar]
- [14].Mayr C., Hemann MT, Bartel D.P.. Disrupting the pairing between let-7 and Hmga2 enhances oncogenic transformation. Science. 2007;315(5818):1576–1579. doi: 10.1126/science.1137999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Lee Y.S., Dutta A.. The tumor suppressor microRNA let-7 represses the HMGA2 oncogene. Genes Dev. 2007;21(9):1025–1030. doi: 10.1101/gad.1540407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Peng Y., Laser J., Shi G., Mittal K., Melamed J., Lee P.. et al. Antiproliferative effects by Let-7 repression of high-mobility group A2 in uterine leiomyoma. Mol Cancer Res. 2008;6(4):663–673. doi: 10.1158/1541-7786.MCR-07-0370. [DOI] [PubMed] [Google Scholar]
- [17].Schultz J., Lorenz P., Gross G., Ibrahim S., Kunz M.. MicroRNA let-7b targets important cell cycle molecules in malignant melanoma cells and interferes with anchorage-independent growth. Cell Res. 2008;18(5):549–557. doi: 10.1038/cr.2008.45. [DOI] [PubMed] [Google Scholar]
- [18].Johnson C.D., Esquela-Kerscher A., Stefani G., Byrom M., Kelnar K., Ovcharenko D.. et al. The let-7 microRNA represses cell proliferation pathways in human cells. Cancer Res. 2007;67(16):7713–7722. doi: 10.1158/0008-5472.CAN-07-1083. [DOI] [PubMed] [Google Scholar]
- [19].Ji J., Zhao L., Budhu A., Forgues M., Jia H.L., Qin L.X.. et al. Let-7g targets collagen type I alpha2 and inhibits cell migration in hepatocellular carcinoma. J Hepatol. 2010;52(5):690–697. doi: 10.1016/j.jhep.2009.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Qian P., Zuo Z., Wu Z., Meng X., Li G., Wu Z.. et al. Pivotal role of reduced let-7g expression in breast cancer invasion and metastasis. Cancer Res. 2011;71(20):6463–6474. doi: 10.1158/0008-5472.CAN-11-1322. [DOI] [PubMed] [Google Scholar]
- [21].Shimizu S., Takehara T., Hikita H., Kodama T., Miyagi T., Hosui A.. et al. The let-7 family of microRNAs inhibits Bcl-xL expression and potentiates sorafenib-induced apoptosis in human hepatocellular carcinoma. J Hepatol. 2010;52(5):698–704. doi: 10.1016/j.jhep.2009.12.024. [DOI] [PubMed] [Google Scholar]
- [22].Myatt S.S., Lam E.W.. The emerging roles of forkhead box (Fox) proteins in cancer. Nat Rev Cancer. 2007;7(11):847–859. doi: 10.1038/nrc2223. [DOI] [PubMed] [Google Scholar]
- [23].Wijchers P.J., Burbach J.P., Smidt M.P.. In control of biology: of mice, men and Foxes. Biochem J. 2006;397(2):233–246. doi: 10.1042/BJ20060387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Katoh M., Igarashi M., Fukuda H., Nakagama H., Katoh M.. Cancer genetics and genomics of human FOX family genes. Cancer Lett. 2013;328(2):198–206. doi: 10.1016/j.canlet.2012.09.017. [DOI] [PubMed] [Google Scholar]
- [25].Bernardo G.M., Bebek G., Ginther C.L., Sizemore S.T., Lozada K.L., Miedler J.D.. et al. FOXA1 represses the molecular phenotype of basal breast cancer cells. Oncogene. 2013;32(5):554–563. doi: 10.1038/onc.2012.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Mani S.A., Yang J., Brooks M., Schwaninger G., Zhou A., Miura N.. et al. Mesenchyme Forkhead 1 (FOXC2) plays a key role in metastasis and is associated with aggressive basal-like breast cancers. Proc Natl Acad Sci U S A. 2007;104(24):10069–10074. doi: 10.1073/pnas.0703900104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Hollier B.G., Tinnirello A.A., Werden S.J., Evans K.W., Taube J.H., Sarkar T.R.. et al. FOXC2 expression links epithelial-mesenchymal transition and stem cell properties in breast cancer. Cancer Res. 2013;73(6):1981–1992. doi: 10.1158/0008-5472.CAN-12-2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Esquela-Kerscher A. Trang., Wiggins J.F., Patrawala L., Cheng A., Ford L.. et al. The let-7 microRNA reduces tumor growth in mouse models of lung cancer. Cell Cycle. 2008;7(6):759–764. doi: 10.4161/cc.7.6.5834. [DOI] [PubMed] [Google Scholar]
- [29].Trang P., Medina P.P., Wiggins J.F., Ruffino L., Kelnar K., Omotola M.. et al. Regression of murine lung tumors by the let-7 microRNA. Oncogene. 2010;29(11):1580–1587. doi: 10.1038/onc.2009.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Trang P., Wiggins J.F., Daige C.L., Cho C., Omotola M., Brown D.. et al. Systemic delivery of tumor suppressor microRNA mimics using a neutral lipid emulsion inhibits lung tumors in mice. Mol Ther. 2011;19(6):1116–1122. doi: 10.1038/mt.2011.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Wang Y., Hu X., Greshock J., Shen L., Yang X., Shao Z.. et al. Genomic DNA copy-number alterations of the let-7 family in human cancers. PLoS One. 2012;7(9):e44399. doi: 10.1371/journal.pone.0044399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Viswanathan S.R., Daley G.Q.. Lin28: A microRNA regulator with a macro role. Cell. 2010;140(4):445–449. doi: 10.1016/j.cell.2010.02.007. [DOI] [PubMed] [Google Scholar]
- [33].Muller D.W., Bosserhoff A.K.. Integrin beta 3 expression is regulated by let-7a miRNA in malignant melanoma. Oncogene. 2008;27(52):6698–6706. doi: 10.1038/onc.2008.282. [DOI] [PubMed] [Google Scholar]
- [34].Han H.B., Gu J., Zuo H.J., Chen Z.G., Zhao W., Li M.. et al. Let-7c functions as a metastasis suppressor by targeting MMP11 and PBX3 in colorectal cancer. J Pathol. 2012;226(3):544–555. doi: 10.1002/path.3014. [DOI] [PubMed] [Google Scholar]
- [35].Yun J., Frankenberger C.A., Kuo W.L., Boelens M.C., Eves E.M., Cheng N.. et al. Signalling pathway for RKIP and Let-7 regulates and predicts metastatic breast cancer. EMBO J. 2011;30(21):4500–4514. doi: 10.1038/emboj.2011.312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Liang S., He L., Zhao X., Miao Y., Gu Y., Guo C.. et al. MicroRNA let-7f inhibits tumor invasion and metastasis by targeting MYH9 in human gastric cancer. PloS one. 2011;6(4):e18409. doi: 10.1371/journal.pone.0018409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Kim S.J., Shin J.Y., Lee K.D., Bae Y.K., Sung K.W., Nam S.J.. et al. MicroRNA let-7a suppresses breast cancer cell migration and invasion through downregulation of C-C chemokine receptor type 7. Breast Cancer Res. 2012;14(1):R14. doi: 10.1186/bcr3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Pietila M., Vijay G.V., Soundararajan R., Yu X., Symmans W.F., Sphyris N.. et al. FOXC2 regulates the G2/M transition of stem cell-rich breast cancer cells and sensitizes them to PLK1 inhibition. Sci Rep. 2016;(6):23070. doi: 10.1038/srep23070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Werden S.J., Sphyris N., Sarkar T.R., Paranjape A.N., LaBaff A.M., Taube J.H.. et al. Phosphorylation of serine 367 of FOXC2 by p38 regulates ZEB1 and breast cancer metastasis, without impacting primary tumor growth. Oncogene. 2016;35(46):5977–5988. doi: 10.1038/onc.2016.203. [DOI] [PMC free article] [PubMed] [Google Scholar]