Summary
Because of the high variability of seasonal influenza viruses and the eminent threat of influenza viruses with pandemic potential, there is great interest in the development of vaccines that induce broadly protective immunity. Most probably, broadly protective influenza vaccines are based on conserved proteins, such as nucleoprotein (NP). NP is a vaccine target of interest as it has been shown to induce cross‐reactive antibody and T cell responses. Here we tested and compared various NP‐based vaccine preparations for their capacity to induce humoral and cellular immune responses to influenza virus NP. The immunogenicity of protein‐based vaccine preparations with Matrix‐M™ adjuvant as well as recombinant viral vaccine vector modified Vaccinia virus Ankara (MVA) expressing the influenza virus NP gene, with or without modifications that aim at optimization of CD8+ T cell responses, was addressed in BALB/c mice. Addition of Matrix‐M™ adjuvant to NP wild‐type protein‐based vaccines significantly improved T cell responses. Furthermore, recombinant MVA expressing the influenza virus NP induced strong antibody and CD8+ T cell responses, which could not be improved further by modifications of NP to increase antigen processing and presentation.
Keywords: adjuvant, immunogenicity, influenza virus, Matrix‐M™, MVA, nucleoprotein, vaccine
Introduction
Influenza A (H1N1 and H3N2) and B viruses are responsible for seasonal epidemics in the human population and cause substantial morbidity and mortality in high‐risk groups such as elderly people. The antigenic properties of these viruses change regularly due to accumulation of mutations in the two major surface proteins, haemagglutinin (HA) and neuraminidase (NA), resulting in escape from pre‐existing virus‐neutralizing antibodies induced by previous infections or vaccinations (antigenic drift) 1, 2, 3. In addition to seasonal influenza viruses, avian and swine influenza A viruses – for example, viruses of the A(H5N1) 4, A(H5N6) 5 and A(H7N9) 6 subtype – cause occasional human infections. These zoonotic influenza viruses have the potential to cause pandemic outbreaks, as the human population is immunologically virtually naive.
Currently used inactivated vaccines against seasonal influenza viruses predominantly induce neutralizing antibodies against the globular head‐domain of HA that neutralize homologous influenza viruses efficiently 3. However, the breadth of reactivity of these antibodies is limited due to the high degree of variability in the head‐domain of the HA glycoprotein between different influenza viruses, resulting in reduced vaccine efficacy in case of a vaccine mismatch with epidemic strains 3, 7, 8, 9. Due to the antigenic drift of seasonal influenza viruses, vaccines need to be updated almost annually 10. Furthermore, in case of a pandemic outbreak it is essential that a tailor‐made vaccine can be produced rapidly, which proved to be difficult during the pandemic of 2009. Thus, alternative cross‐reactive correlates of protection induced by a ‘universal’ influenza vaccine with the capacity to provide intra‐ or intersubtypic immunity, such as virus‐specific T cells, have gained renewed interest.
Virus‐specific CD8+ cytotoxic T cells predominantly recognize internal viral antigens, such as matrix (M1) and nucleoprotein (NP), that are relatively conserved and contain epitopes shared by various subtypes of influenza virus. These cells are induced by natural infection but are induced inefficiently by inactivated influenza vaccines (reviewed in 11) 12, 13, 14. It has been shown that cross‐reactive virus‐specific CD8+ cytotoxic T cells contribute to reduction of disease duration and severity after infection with a heterologous influenza virus 12, 15. In addition, antibodies directed against the conserved stalk‐domain of HA have been identified, which display cross‐protective potential against infection with heterologous influenza viruses 16, 17, 18, 19, 20. Thus, the development of vaccines that induce broadly protective HA stalk‐specific antibodies and/or cellular immune responses against conserved proteins such as NP or M1 is highly desirable and is listed high on the research agenda (reviewed in 21, 22, 23).
Viral vaccine vectors, such as modified vaccinia virus Ankara (MVA), have been shown to efficiently induce antigen‐specific humoral and cellular responses (reviewed in 24, 25). Recombinant (r)MVA vaccines have been tested extensively in various animal models and multiple clinical trials against different pathogens, including influenza virus, and has proved that the use of rMVA‐based vaccines is safe 25, 26, 27, 28. As it is relatively easy to insert genes encoding antigens of interest into the genome of MVA, recombinant (r)MVA‐based vaccines can be rapidly produced 24, 25. rMVA drives de‐novo synthesis of one or multiple antigens of interest, leading to endogenous antigen processing and major histocompatibility complex (MHC) class I antigen presentation, which is important for the efficient induction of CD8+ T cells (reviewed in 29).
Humoral and cellular immunity could also be improved by the use of adjuvants in combination with for example virosomal, trivalent, split virion and inactivated influenza vaccines 30, 31, 32, 33, 34, 35, 36, 37. Addition of Matrix‐M™ adjuvant, made of saponins extracted from the tree Quillaja saponaria Molina 38, to influenza vaccines enhances significantly humoral responses and the induction of virus‐specific T cells in mice, ferrets and humans 31, 32, 33, 34, 35, 39, 40, 41. Furthermore, Matrix‐M™ adjuvant has been evaluated in three clinical Phase I studies using seasonal and pandemic (H5N1 and H7N9) influenza vaccines 32, 40, 42, 43. All studies showed promising results with increased humoral and cellular responses along with good safety data.
Most adjuvanted influenza vaccine studies have focused upon the immune response towards HA (reviewed in 44). Additionally, a couple of studies have shown that adjuvanted recombinant influenza virus NP protein vaccines are able to induce T helper type 1 (Th1)‐skewed CD4+ T cell response and protect C57BL/6 mice from influenza A virus challenge 45, 46. Here, we explore the effect of Matrix‐M™ adjuvant on a recombinant influenza virus NP protein‐based vaccine and compare it to rMVA‐NP vaccines in BALB/c mice. Accordingly, the immunogenicity of unadjuvanted or Matrix‐M™ adjuvanted wild‐type NP (NPwt) protein‐based vaccines and rMVA‐based vaccines expressing NPwt or modified NP, optimized for proteasomal processing 47, were investigated. With rMVA expressing modified NP, enhanced activation of a human CD8+ T cells in vitro was observed previously, but the use of these constructs did not improve NP‐specific CD8+ T cell responses compared to NPwt in the C57BL/6 mouse model 47. As we speculated that the immune response in C57BL/6 mice was directed mainly against the immunodominant NP366–374 epitope 47, 48, we next addressed the immunogenicity of rMVA vaccines expressing either NPwt or modified NP in BALB/c mice, which do not mount a response to this immunodominant NP epitope.
In this study, we show that Matrix‐M™ adjuvant can improve the immunogenicity of protein‐based NPwt vaccines substantially. Comparison of the two types of recombinant NP vaccines showed that while Matrix‐M™ adjuvant improved the immunogenicity of NPwt vaccine by enhancing the antibody titre and T cell responses compared to unadjuvanted NPwt vaccine, rMVA‐NPwt vaccines also induced antigen‐specific interferon (IFN)‐γ‐positive CD8+ T cells and higher IgG2a NP‐specific antibody titres. However, rMVA‐driven expression of NP modified to increase antigen processing did not improve the NP‐specific CD8+ T cell response in BALB/c mice.
Materials and methods
NPwt protein preparation
The NP was expressed in Escherichia coli BL21 as a His‐tagged maltose binding protein (MBP) fusion protein containing a DEVD sequence as cleavage site for murine (m)caspase3. Expression of the tagged MBP–NP fusion protein was induced by addition of 1 mM isopropyl β‐D‐1‐thiogalactopyranoside (IPTG). After 4h induction at 28°C the cells were harvested, lysed by sonication at 4°C followed by centrifugation. The supernatant was loaded on Ni‐Sepharose 6 FF (GE Healthcare, Amersham, UK) and proteins were eluted by applying an imidazole gradient. The 50 mM imidazole fractions containing the MBP–NP fusion protein were pooled and after incubation for 1 h at 37°C in the presence of 0·06 mg mcaspase3/mg fusion protein reloaded on Ni‐IMAC resin. Analysis by sodium dodecyl sulphate‐polyacrylamide gel electrophoresis/Coomassie Brilliant Blue (SDS‐PAGE/CBB) staining and Western blot showed that more than 90% pure NP was recovered in the flow‐through. NP was concentrated to 1 mg/ml and dialyzed against 25 mM HEPES, 300 mM NaCl pH 7·5. The lipopolysaccharide (LPS) content (< 0·25 EU/µg NP) was determined with EndoSafe‐PTS test (Charles River, Franklin, MA, USA).
Matrix‐M™ adjuvant
Novavax proprietary Matrix‐M™ adjuvant is a saponin‐based adjuvant consisting of two individually formed 40 nm‐sized particles, each with a different and well‐characterized saponin fraction (fraction‐A and fraction‐C, respectively). The Matrix‐A and C particles are formed by formulating purified saponin from the tree Quillaja saponaria Molina with cholesterol and phospholipid 49.
Generation of recombinant MVA
The rMVA constructs expressing NP under control of the early/late PsynII promotor were prepared as described recently 47. In short, the respective (modified) NP nucleotide sequences (PR8‐based, Accession number NC002019) were synthesized by Baseclear BV (Leiden, the Netherlands) and clonal rMVA viruses were prepared through transient mCherry‐dependent plaque selection in chicken embryo fibroblasts (CEF) 47, 50. To generate a final vaccine preparation, virus was propagated in CEF, purified by ultracentrifugation through 36% sucrose and reconstituted in 120 mM NaCl, 10 mM Tris‐HCl pH 7·4. rMVA‐NP constructs were previously extensively characterized in vitro by sequencing, Western blot, confocal microscopy and radiolabelling experiments 47.
Vaccination of BALB/c mice
Specified pathogen‐free female BALB/c mice were purchased from Charles River Laboratories and were 8–10 weeks of age at the start of the experiment. Animals were housed in Makrolon type 3 cages, had access to food and water ad libitum and animal welfare was observed on a daily basis. All experiments were conducted in strict compliance with European guidelines (EU directive on animal testing 2010/63/EU) and the protocol was approved by an independent animal experimentation ethical review committee (Uppsala djurförsöksetiska nämnd). Experiments were performed in two sets, the first set focused on the comparison of protein vaccines with MVA‐based vaccines (Table 1) and the second set focused on the comparison of NPwt with modified NP expressed by MVA (Table 2). BALB/c mice received two vaccinations (time interval of 4 weeks) with 108 plaque‐forming units (PFU) rMVA, 10 μg NPwt protein or 1 or 10 µg NPwt protein adjuvanted with 5 or 10 µg Matrix‐M™ adjuvant. All vaccines were administered subcutaneously in 100 μl at the base of the tail. Blood was sampled 21 days after the first vaccination and before mice were euthanized; 14 or 10 days after the second vaccination for the first and second set, respectively. Spleens were collected in phosphate‐buffered saline (PBS) during necropsy and subsequently single‐cells suspensions were prepared as described previously 34.
Table 1.
Vaccines used to assess the effect of Matrix‐M™ adjuvant on protein‐based wild‐type nucleoprotein (NPwt) vaccine immunogenicity
| Vaccine | n | Formulation | Adjuvant |
|---|---|---|---|
| Protein | 5 | 10 μg NPwt | – |
| 8 | 1 μg NPwt | 5 μg Matrix‐M™ adjuvant | |
| 8 | 10 μg NPwt | 5 μg Matrix‐M™ adjuvant | |
| 8 | 1 μg NPwt | 10 μg Matrix‐M™ adjuvant | |
| rMVA | 8 | 108 PFU rMVA–NPwt | – |
BALB/c mice were vaccinated twice at a 4‐week interval with the respective vaccines. Mice were euthanized 14 days after the second vaccination; n indicates the number of animals per group. PFU = plaque‐forming units; rMVA = recombinant Modified Vaccinia virus Ankara.
Table 2.
Recombinant modified Vaccinia virus Ankara (rMVA)‐based vaccines expressing (modified) nucleoprotein (NP) used to address optimization of the NP‐specific CD8+ T cell response
| Vaccine | n | Formulation |
|---|---|---|
| Protein | 10 | 10 μg NPwt + 5 μg Matrix‐M™ adjuvant |
| rMVA | 5 | 108 PFU wtMVA |
| 10 | 108 PFU rMVA–NPwt | |
| 10 | 108 PFU rMVA–NPmut | |
| 10 | 108 PFU rMVA–NPΔNLS | |
| 10 | 108 PFU rMVA–UbqNP |
BALB/c mice were vaccinated twice at a 4‐week interval with the respective vaccines. Mice were euthanized 10 days after the booster vaccination; n indicates the number of animals per group. PFU = plaque‐forming units.
Detection of NP‐specific antibodies
Maxisorp microplates (Nunc, Rochester, NY, USA) were coated O/N at 4°C with 50 ng/well NPwt protein (AmatsiQ‐Biologicals, Ghent, Belgium) in 0·05 M carbonate/bicarbonate (Sigma Aldrich, Hamburg, Germany) buffer pH 9·6. Plates were blocked for 1 h at room temperature using PBS with 0·05% Tween‐20 (PBST; Thermo Fisher Scientific, Fremont, CA, USA) pH 7·2–7·6 supplemented with 2% (W/V) milk powder (Semper, Sweden). A three‐ or fivefold dilution series of the sera was prepared in blocking buffer starting at 1 : 30 and incubated on the coated plates for 1 h. Blocking buffer and anti‐NP positive serum were used as a negative or positive control, respectively. Plates were washed with PBST and incubated with anti‐IgG1 (Star 132P; AbD Serotec, Raleigh, NC, USA) or anti‐IgG2a (Star 133P; AbD Serotec) horseradish peroxidase (HRP)‐conjugated antibodies – indicative for a Th2 or Th1 response, respectively 51 – for 1 h. Subsequently, plates were washed with PBST and 100 μl 3,3′,5,5′‐tetramethylbenzidine (TMB) peroxidase substrate (Svanova Biotech, Uppsala, Sweden) was added. Reactions were stopped by adding 1·8 M H2SO4 and absorbance was measured subsequently at 450 nm using a SpectraMax Plus 384 Microplate Reader (Molecular Devices Corporation, Sunnyvale, CA, USA). Immunoglobulin (Ig)G1 and IgG2a anti‐NP titres were calculated using a four‐parameter logistic equation (Softmax software; Molecular Devices). The inflection point of the titration curve, i.e. half maximal effective concentration (EC50) titre, was used for analysis.
FluoroSpot analysis of protein‐ or peptide‐stimulated splenocytes
The number of interleukin (IL‐)2 and IFN‐γ‐producing cells in single‐cell suspensions of the spleen was analysed using FluoroSpot assay (Mabtech, Nacka Strand, Sweden), according to the manufacturer's instructions. In brief, single‐cell suspensions were seeded in triplicate on filter plates at 0·25 × 106 cells/well in Roswell Park Memorial Institute (RPMI) medium (Sigma Aldrich) supplemented with 10% heat‐inactivated fetal bovine serum (FBS; Sigma Aldrich), 100 U/ml penicillin/100 µg/ml streptomycin/2 mM L‐glutamine (P/S/G; Sigma Aldrich) and stimulated with 1 µg NP (AmatsiQ‐Biologicals) or 5 µM synthetic peptide (epitope NP147–155, TYQRTRALV 52) per well. Concanavalin A (Sigma Aldrich) and RPMI supplemented with 10% FBS were used as positive and negative controls, respectively. Samples were incubated for 20 h at 37°C after which spots were developed and analysed using an AID ELR02 reader (Autoimmune Diagnostika GmbH, Strasburg, Germany).
Intracellular cytokine staining (ICS) of peptide‐stimulated splenocytes
Single‐cell suspensions of splenocytes were mock‐treated, stimulated with 5 µM synthetic peptide (epitope NP147–155) or stimulated 20 ng/ml phorbol myristate acetate (PMA) and 1 µg/ml ionomycin as positive control in Iscove's modified Dulbecco's medium (IMDM; Gibco, Carlsbad, CA, USA) supplemented with 5% FBS (Sigma Aldrich), P/S/G and GolgiStop. Stimulation lasted 12 h at 37°C, which is longer than the optimal 6 h incubation with GolgiStop. However, enough viable events were measured for reliable analysis. Cells were stained with fluorochrome‐labelled antibodies CD3eAPC–Cy7 (BD Pharmingen, USA), CD8bFITC (BD Pharmingen, San Diego, CA, USA), CD4PerCP (BD Pharmingen) and Aqua LIVE/DEAD (Invitrogen, Carlsbad, CA, USA). Subsequently, cells were fixed and permeabilized using BD Cytofix/Cytoperm™ Plus (BD Biosciences) and stained with IFN‐γPacific Blue (Biolegend, San Diego, CA, USA). Samples were analysed by flow cytometry using a FACS Celesta flow cytometer and FACS DIVA software (BD Biosciences).
Statistical analysis
NP‐specific antibody responses were analysed by a one‐way analysis of variance (anova) to establish statistically significant differences. FluoroSpot and ICS data were first assessed with a D'Agostino & Pearson omnibus normality test. Non‐normally distributed data (FluoroSpot set 1 and ICS set 2) were analysed using a Kruskal–Wallis rank test. If a normal distribution was assumed, a one‐way anova was performed to determine statistical significance (FluoroSpot set 2).
Results
Matrix‐M™ adjuvant enhanced immunogenicity of NPwt protein‐based vaccine
As Matrix‐M™ adjuvant has been shown to induce high levels of biologically active antibodies, balanced type Th1 and Th2 immune responses, multi‐functional T cells and cytotoxic CD8+ T lymphocytes in combination with various subunit vaccines 41, 53, 54, 55, we addressed the effect of addition of Matrix‐M™ adjuvant to NPwt protein‐based vaccines and compared this to unadjuvanted NPwt protein and rMVA‐NPwt vaccination. To this end, mice were vaccinated twice with one of three different NPwt protein + Matrix‐M™ adjuvant combinations, NPwt protein alone or rMVA‐NPwt (Table 1).
Vaccine immunogenicity was assessed by determining NP‐specific antibody titres after one or two vaccinations (Fig. 1). NP‐specific antibody responses induced by the protein‐based vaccines, with or without Matrix‐M™ adjuvant, could not be detected after a single vaccination (Fig. 1a and b). In contrast, 21 days after the primary vaccination an NP‐specific IgG2a response was detected after vaccination with rMVA‐NPwt (Fig. 1b). At 14 days after either protein or rMVA‐NPwt booster vaccination NP‐specific antibody responses were detected. Protein‐based NPwt vaccines predominantly induced an IgG1 antibody response, whereas rMVA‐NPwt induced both IgG1 and IgG2a NP‐specific antibodies with a bias towards IgG2a (Fig. 1c,d). The NP‐specific IgG1 responses induced by protein‐based vaccines and rMVA‐NPwt were comparable (Fig. 1c). Although not statistically significant, the immunogenicity of the protein‐based NPwt vaccines was enhanced by the addition of Matrix‐M™ adjuvant to the vaccine formulation, particularly for 10 μg NPwt + 5 μg Matrix‐M™ adjuvant (Fig. 1c,d).
Figure 1.
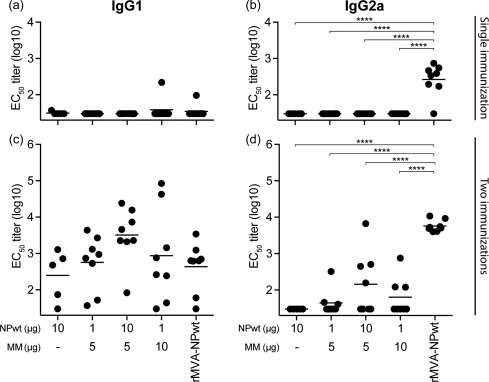
Nucleoprotein (NP)‐specific antibody responses after NP wild‐type (NPwt) protein (with or without Matrix‐M™ adjuvant) or recombinant Modified Vaccinia virus Ankara (rMVA)–NPwt vaccination. The immunoglobulin (Ig)G1 and IgG2a NP‐specific antibody responses 21 days after the primary vaccination (a,b) or 14 days after the booster vaccination (c,d). IgG1 (a,c) or IgG2a (b,d) serum antibodies were detected using purified NPwt protein and anti‐IgG1 or anti‐IgG2a horseradish peroxidase (HRP)‐conjugated antibodies. Mean of each group is indicated. MM = Matrix‐M™ adjuvant. ****P < 0·0001.
Next, the vaccine‐induced NP‐specific T cell responses were evaluated. T cell activation was measured by stimulation of splenocytes obtained 14 days after the booster vaccination and subsequent detection of IL‐2‐, IFN‐γ‐ or IL‐2/IFN‐γ‐producing NP‐specific splenocytes. NP‐specific T cell responses induced by protein‐based vaccine formulations were enhanced significantly by Matrix‐M™ adjuvant (Fig. 2). Furthermore, Matrix‐M™ adjuvanted NPwt protein vaccines induced more IL‐2‐producing T cells compared to rMVA‐NPwt vaccination (Fig. 2a). In contrast, no differences in IFN‐γ‐ or IL‐2/IFN‐γ‐producing cells between the Matrix‐M™ adjuvanted NPwt protein vaccines and the rMVA‐NPwt vaccine were observed (Fig. 2b,c). Collectively, NPwt protein vaccines, especially with the Matrix‐M™ adjuvant, and rMVA‐NPwt were immunogenic in BALB/c mice. In general, rMVA‐NPwt induced stronger IgG2a and similar IgG1 NP‐specific antibody responses compared to protein‐based vaccines. Addition of Matrix‐M™ adjuvant to protein‐based NP vaccine improved T cell responses significantly, but not antibody responses.
Figure 2.
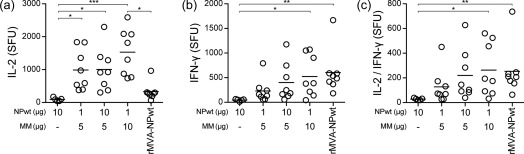
Vaccination with wild‐type nucleoprotein (NPwt) protein with Matrix‐M™ adjuvant or recombinant Modified Vaccinia virus Ankara (rMVA)–NPwt induced NP‐specific splenocyte responses. Spleens obtained 14 days after the booster vaccination were stimulated with purified NPwt protein. The number of interleukin (IL‐)2 (a), interferon (IFN‐)γ (b) and IL‐2/IFN‐γ (c)‐producing T cells was used to determine NP‐specific splenocyte activation in spot‐forming units (SFU)/106 cells. Samples were tested in triplicate. Mean of each group is indicated. MM = Matrix‐M™ adjuvant. *P < 0·0306; **P < 0·0082; ***P = 0·0005.
Modifications of NP do not enhance NP‐specific CD8+ T cell responses in BALB/c mice
In order to optimize the NP‐specific CD8+ T cell response, modifications were made to the NP protein that aimed at increasing protein processing and subsequent antigen presentation. Various NP mutant constructs were generated. First, the nuclear localization signal (NLS) of NP was mutated (NPmut) or deleted (NPΔNLS) to retain the protein in the cytosol and increase the amount of protein available for proteasomal processing 47, 56, 57. Secondly, NP was fused to ubiquitin (UbqNP), a small molecule marking proteins for proteasomal degradation 47, 58. Previously, we have shown that these modified proteins could be expressed by rMVA and were immunogenic in C57BL/6 mice 47. To determine whether the modified NP constructs are capable of inducing stronger NP‐specific T cell responses than NPwt in BALB/c mice, animals were vaccinated twice with rMVA‐NPwt, rMVA‐NPmut, rMVA‐NPΔNLS or rMVA‐UbqNP. Wild‐type (wt)MVA was used as a negative empty‐vector control. The rMVA‐based vaccines were compared to vaccination with 10 μg NPwt protein formulated with 5 μg Matrix‐M™ adjuvant (Table 2).
NP‐specific antibody responses were determined using serum collected 21 and 10 days after the first and booster vaccination, respectively. Similar to the results obtained in the initial experiment, vaccination with NPwt protein + Matrix‐M™ adjuvant did not induce an NP‐specific antibody response after a single vaccination. However, after two vaccinations both IgG1 and IgG2a NP‐specific antibodies were detected with a bias towards IgG1 (Fig. 3a,c). The IgG1 response of the adjuvanted protein vaccine was significantly higher than the response induced by the rMVA‐NP constructs (Fig. 3c). rMVA‐NPwt induced NP‐specific IgG2a antibodies after a single vaccination. Similar results were obtained for vaccination with rMVA‐NPmut. In contrast, rMVA expressing NPΔNLS (not statistically significant) or UbqNP seemed less immunogenic (Fig. 3b). The differences in the induction of NP‐specific IgG2a antibodies by the various rMVA‐NP constructs was not detected after two vaccinations (Fig. 3d). NP‐specific antibody responses induced by the empty vector (wt)MVA were never observed (Fig. 3).
Figure 3.
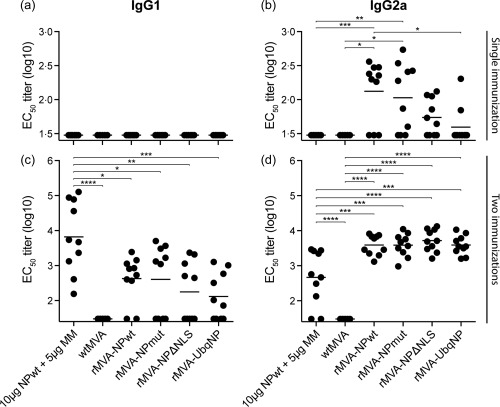
Nucleoprotein (NP)‐specific antibody responses induced by vaccination with NP wild‐type (NPwt) protein + Matrix‐MTM adjuvant or the respective recombinant Modified Vaccinia virus Ankara (rMVA)–NP constructs. Immunoglobulin (Ig)G1 and IgG2a NP‐specific antibody responses in mice 21 days after the primary vaccination (a,b) or 14 days after booster vaccination (c,d). IgG1 (a,c) or IgG2a (b,d) serum antibodies were detected using purified NPwt protein and anti‐IgG1 or anti‐IgG2a horseradish peroxidase (HRP)‐conjugated antibodies. Mean for each group is indicated. MM = Matrix‐M™ adjuvant. *P < 0·0432; **P < 0·0068; ***P < 0·0009; ****P < 0·0001.
Subsequently, NP‐specific T cell responses induced by vaccination with 10 μg NPwt protein with 5 μg Matrix‐M™ adjuvant or the respective rMVA‐NP vaccines were addressed using two different assays. Using the FluoroSpot assay, the induction of specific T cells was demonstrated by detection of IL‐2‐, IFN‐γ‐ or IL‐2/IFN‐γ‐producing cells after splenocyte stimulation with an NP147–155 peptide, representing a CD8+ T cell epitope. wtMVA vaccination was used as a negative control and did not induce any NP147–155‐specific T cell responses (Fig. 4). NP147–155‐specific IL‐2‐, IFN‐γ‐ or IL‐2/IFN‐γ‐producing T cells were detected for all rMVA‐NP vaccine groups (Fig. 4a–c). In general, the number of IL‐2‐, IFN‐γ‐ or IL‐2/IFN‐γ‐producing cells induced by the respective rMVA‐NP vaccines were similar, although the number of virus‐specific T cells induced by rMVA‐NPΔNLS seemed slightly elevated and by rMVA‐UbqNP seemed slightly reduced compared to rMVA‐NPwt (Fig. 4a–c). Similar to the first experiment, NPwt protein + Matrix‐M™ adjuvant vaccination induced similar numbers of IL‐2‐ or IL‐2 + IFN‐γ‐producing cells compared to rMVA‐NPwt. In contrast, the numbers of IFN‐γ‐producing cells after protein‐based vaccination was significantly lower than after rMVA‐NP vaccination (Fig. 4b).
Figure 4.
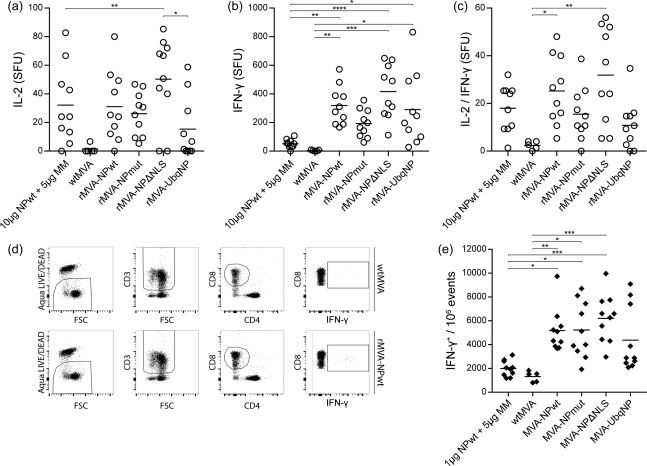
Vaccine‐induced nucleoprotein (NP)‐specific T cell responses. (a–c) Splenocytes were stimulated with the NP147–155 peptide after which the number of interleukin (IL‐)2 (a), interferon (IFN‐)γ (b) or IL‐2/IFN‐γ (c) spot‐forming units (SFU)/106 splenocytes were determined. Samples were measured in triplicate. The mean for each group is indicated. (d) Splenocytes were stimulated with the NP147–155 peptide for 12 h. Using flow cytometry, live splenocytes were selected and subsequently CD3+, CD8+ and IFN‐γ+ cells were gated. Representative graphs of mice vaccinated with wild‐type Modified Vaccinia virus Ankara (wtMVA) as a control or recombinant MVA (rMVA)–NPwt are shown. (e) IFN‐γ production of CD3+CD8+ splenocytes per 106 events measured using flow cytometry. Mean for each group is indicated. MM = Matrix‐MTM adjuvant. *P < 0·0227; **P = 0·0073; ***P = 0·0005; ****P < 0·0001.
The vaccine‐induced NP‐specific T cell responses were also assessed using flow cytometry. IFN‐γ production after stimulation of splenocytes with NP147–155 peptide was used to measure induction of NP‐specific CD3+CD8+ T cells (Fig. 4d). A similar pattern to the FluoroSpot data was observed; all rMVA‐NP vaccine groups induced NP‐specific CD3+CD8+ IFN‐γ‐producing cells and, although not statistically significant, rMVA‐NPΔNLS induced slightly elevated and rMVA‐UbqNP slightly reduced numbers of IFN‐γ‐producing T cells compared to rMVA‐NPwt (Fig. 4e). The use of NPwt protein, in combination with Matrix‐M™ adjuvant, induced an NP147–155‐specific CD8+ T cell response inefficiently (Fig. 4e). Thus, vaccination with rMVA‐NP induced stronger NP‐specific cytotoxic CD8+ T cells than with adjuvanted recombinant NP preparation. However, MVA‐driven expression of NP modified in order to increase its processing and presentation did not result in increased T cell responses in BALB/c mice.
Discussion
In this study, we investigated the immunogenicity of various influenza virus NP‐based vaccines. Influenza virus NP is relatively conserved and an interesting target for the induction of (cross‐)protective immune responses to influenza virus. Both NP‐specific antibodies 59, 60, 61 and T cells 62 have been shown to contribute to protective immunity to influenza virus infections. Here, we compared the immunogenicity of unadjuvanted and Matrix‐M™ adjuvanted protein and rMVA expressing NPwt or modified NP for their capacity to induce influenza virus NP‐specific antibody and T cell responses in BALB/c mice.
Differential induction of NP‐specific antibodies was observed after protein or MVA‐based NP vaccination (Fig. 1). As rMVA expresses full‐length NP similar to the way it is expressed in influenza virus infection, it is likely that NP is exposed on the surface of rMVA‐NP infected cells, leading potentially to more efficient induction of antibody responses after a single vaccination compared to vaccination with NPwt protein. The effect of this differential induction of antibody responses on a challenge infection was not addressed in this study.
As MVA is a replication‐deficient virus capable of infecting cells it has intrinsic adjuvant capacities, such as activation of the innate immune system 63, 64 – including activation of Toll‐like receptors (TLRs) 65. Matrix‐M™ adjuvant also recruits and activates innate immune cells, but does not activate TLRs. The Matrix‐M™ adjuvant increases antigen uptake and processing 49, 66, 67, resulting in a significant increase in the cellular immune response to NP, as demonstrated by the detection of IL‐2‐ and/or IFN‐γ‐producing splenocytes that were stimulated with NP in vitro. Also, after vaccination with rMVA‐NPwt, potent cellular immune responses were observed dominated by IFN‐γ+‐ or IL‐2+/IFN‐γ+‐producing cells, although the number of cells that produced IL‐2 only seemed lower compared to vaccination with Matrix‐M™ adjuvanted NP (Fig. 2). Of note, the concentration of protein or adjuvant used for vaccination did not make a significant difference in vaccine immunogenicity.
In an attempt to optimize the NP‐specific CD8+ T cell response induced by vaccination, we previously generated rMVA constructs expressing modified NP. We have shown that deletion of the NLS or fusion of NP to ubiquitin increased degradation of NP and enhanced activation of NP‐specific T cells in vitro. However, these modifications did not improve CD8+ T cell responses in vivo or protection from a lethal challenge with influenza A virus in C57BL/6 mice 47. We hypothesized that MVA‐NPwt already induced an optimal T cell response in C57BL/6 (H‐2Kb/Db) mice, because these mice mount a highly dominant CD8+ T cells response to a single epitope located in NP (NP366–374), which might mask any potential positive effects of the modifications. As this epitope is not recognized in in BALB/c mice (H‐2Kd/Db), we addressed if differences in immunogenicity of the various rMVA‐NP constructs could be detected in this mouse model and compared it to NPwt + Matrix‐M™ protein‐based vaccination.
The NP‐specific IgG1 response induced by NPwt protein with Matrix‐M™ adjuvant was significantly higher than the response induced by the rMVA‐NP constructs (Fig. 3c), similar to the trend in the first experiment (Fig. 1c). Furthermore, NPΔNLS and particularly UbqNP modifications resulted in lower IgG2a responses after a single vaccination, and lower IgG1 responses after two vaccinations compared to NPwt (Fig. 3b,c). As these vaccines were designed to enhance NP degradation, the amount of antigen and the time window that these antigens are available for antibody recognition and induction of B cell responses is limited. Therefore, induction of lower antibody responses by these constructs is not surprising. However, after two immunizations comparable IgG2a responses were observed.
The induction of NP‐specific CD8+ T cells was detected after stimulation of splenocytes with the H‐2Kd restricted CD8+ T cell epitope NP147–155 using the FluoroSpot assay and flow cytometry after intracellular IFN‐γ staining. IFN‐γ responses measured by the FluoroSpot assay were significantly higher after vaccination with rMVA‐NP constructs compared to NPwt + Matrix‐M™ vaccination, whereas comparable low numbers of IL‐2‐ and IFN‐γ/IL‐2‐producing cells were detected. Using flow cytometry, no NP147–155‐specific CD8+ T cells were detected after vaccination with NPwt + Matrix‐M™ adjuvant, indicating that the protein vaccine generates a mainly antigen‐specific CD4+ T cell response. This can be concluded from the experiment shown in Fig. 2, in which splenocytes were stimulated with NP protein which predominantly activates CD4+ T cells.
Both in the FluoroSpot assay and using flow cytometry, rMVA‐NPwt induced a NP‐specific CD8+ T cell response that could not be improved by modifying the NP protein (Fig. 4). Notably, rMVA‐UbqNP even induced slightly lower levels of NP‐specific T cells compared to rMVA expressing NPwt, NPmut or NPΔNLS (Fig. 4), possibly in contrast to our hypothesis, the result of reduced availability of NP for direct antigen processing and presentation or cross‐presentation 68, 69, 70. We have shown previously in pulse‐chase experiments that the degradation kinetics vary for the different NP constructs; NPΔNLS and particularly UbqNP had a higher degradation rate than NPwt and NPmut 47. Therefore, the kinetics of the induction of the T cell response could differ between the different rMVA‐NP constructs. In general, experiments in C57BL/6 and BALB/c mice show that NPwt is already optimally processed and presented, and that this cannot be improved by increasing the number of peptides available for presentation to T cells.
In conclusion, addition of the Matrix‐M™ adjuvant to NPwt protein‐based vaccines enhanced immunogenicity significantly, resulting in the induction of IgG2a as well as IgG1 antibody responses and increased T cell responses. Furthermore, rMVA‐based NP vaccines seem to be capable of inducing a more diverse antibody (IgG1 and IgG2a) and cellular (IL‐2, IFN‐γ, IL‐2/IFN‐γ) response compared to protein‐based NP vaccines, and only a single vaccination is enough to induce IgG2a antibody responses. The humoral and cellular immune response induced by rMVA expressing NPwt in BALB/c mice could not be enhanced further by increasing NP protein degradation. These results show that NP does not need any modifications to induce an optimal immune response.
Disclosure
S. E. M. and L. S. are employees of Novavax and hold stock and/or stock options in the company. Other authors report no conflict of interest.
Acknowledgements
The authors would like to thank Heidi De Gruyter and Stella van Trierum for their help in generating the rMVA constructs. Jurgen Haustraete is acknowledged for providing the BL21codon + pICA2 E. coli strain and the mCaspase enzyme and Dr Cecilia Carnrot, Eva Spennare and Dr. Carolina Lunderius Andersson for excellent technical assistance. This work was supported financially by the European Research Council FP7 project FLUNIVAC (project number 602604).
References
- 1. Koel BF, Burke DF, Bestebroer TM et al Substitutions near the receptor binding site determine major antigenic change during influenza virus evolution. Science 2013; 342:976–9. [DOI] [PubMed] [Google Scholar]
- 2. Westgeest KB, de Graaf M, Fourment M et al Genetic evolution of the neuraminidase of influenza A (H3N2) viruses from 1968 to 2009 and its correspondence to haemagglutinin evolution. J Gen Virol 2012; 93:1996–2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wright PF, Neumann G, Kawaoka Y. Orthomyxoviruses In: Knipe D, Howley P, eds. Field's virology, 6th edn. Philadelphia, PA: Lippincott Williams & Wilkins, 2013:1186–243. [Google Scholar]
- 4. World Health Organization (WHO) . Cumulative number of confirmed human cases for avian influenza A(H5N1) reported to WHO, 2003–2017. Available on: http://www.who.int/influenza/human_animal_interface/2017_03_16_tableH5N1.pdf?ua=1 Accessed 10/04/2017.
- 5. World Health Organization (WHO) . Human infections with avian influenza A(H5N6) virus. Available at: http://www.who.int/csr/don/07-december-2016-ah5n6-china/en/ Accessed 10/04/2017.
- 6. World Health Organization (WHO) . Human infection with avian influenza A(H7N9) virus in China. Available at: http://www.wpro.who.int/outbreaks_emergencies/H7N9/en/ Accessed on 10/04/2017.
- 7. Skowronski DM, Chambers C, Sabaiduc S et al Interim estimates of 2014/15 vaccine effectiveness against influenza A(H3N2) from Canada's Sentinel Physician Surveillance Network, January 2015. Euro Surveill 2015; 20:1–18. [DOI] [PubMed] [Google Scholar]
- 8. Pebody RG, Warburton F, Ellis J et al Low effectiveness of seasonal influenza vaccine in preventing laboratory‐confirmed influenza in primary care in the United Kingdom: 2014/15 mid‐season results. Euro Surveill 2015; 20:21025. [PubMed] [Google Scholar]
- 9. Flannery B, Clippard J, Zimmerman RK et al Early estimates of seasonal influenza vaccine effectiveness– United States, January 2015. Morb Mortal Wkly Rep 2015; 64:10–5. [PMC free article] [PubMed] [Google Scholar]
- 10. World Health Organization (WHO) . Recommended composition of influenza virus vaccines for the use in the 2017–2018 northern hemisphere influenza season. Available at: http://www.who.int/influenza/vaccines/virus/recommendations/201703_recommendation.pdf?ua=1 Accessed on 10/04/2017.
- 11. Altenburg AF, Rimmelzwaan GF, de Vries RD. Virus‐specific T cells as correlate of (cross‐) protective immunity against influenza. Vaccine 2015; 33:500–6. [DOI] [PubMed] [Google Scholar]
- 12. Sridhar S, Begom S, Bermingham A et al Cellular immune correlates of protection against symptomatic pandemic influenza. Nat Med 2013; 19:1305–12. [DOI] [PubMed] [Google Scholar]
- 13. He XS, Holmes TH, Zhang C et al Cellular immune responses in children and adults receiving inactivated or live attenuated influenza vaccines. J Virol 2006; 80:11756–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Forrest BD, Pride MW, Dunning AJ et al Correlation of cellular immune responses with protection against culture‐confirmed influenza virus in young children. Clin Vaccine Immunol 2008; 15:1042–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bender BS, Croghan T, Zhang L, Small PA Jr. Transgenic mice lacking class I major histocompatibility complex‐restricted T cells have delayed viral clearance and increased mortality after influenza virus challenge. J Exp Med 1992; 175:1143–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Corti D, Suguitan AL Jr, Pinna D et al Heterosubtypic neutralizing antibodies are produced by individuals immunized with a seasonal influenza vaccine. J Clin Invest 2010; 120:1663–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Throsby M, van den Brink E, Jongeneelen M et al Heterosubtypic neutralizing monoclonal antibodies cross‐protective against H5N1 and H1N1 recovered from human IgM+ memory B cells. PLOS ONE 2008; 3:e3942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ekiert DC, Bhabha G, Elsliger MA et al Antibody recognition of a highly conserved influenza virus epitope. Science 2009; 324:246–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Margine I, Krammer F, Hai R et al Hemagglutinin stalk‐based universal vaccine constructs protect against group 2 influenza A viruses. J Virol 2013; 87:10435–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ellebedy AH, Krammer F, Li GM et al Induction of broadly cross‐reactive antibody responses to the influenza HA stem region following H5N1 vaccination in humans. Proc Natl Acad Sci USA 2014; 111:13133–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. de Vries RD, Altenburg AF, Rimmelzwaan GF. Universal influenza vaccines, science fiction or soon reality? Expert Rev Vaccines 2015; 14:1299–301. [DOI] [PubMed] [Google Scholar]
- 22. Krammer F, Palese P, Steel J. Advances in universal influenza virus vaccine design and antibody mediated therapies based on conserved regions of the hemagglutinin. Curr Top Microbiol Immunol 2015; 386:301–21. [DOI] [PubMed] [Google Scholar]
- 23. de Vries RD, Altenburg AF, Rimmelzwaan GF. Universal influenza vaccines: a realistic option? Clin Microbiol Infect 2016; 22 Suppl 5:S120–S4. [DOI] [PubMed] [Google Scholar]
- 24. Altenburg AF, Kreijtz JH, de Vries RD et al Modified vaccinia virus Ankara (MVA) as production platform for vaccines against influenza and other viral respiratory diseases. Viruses 2014; 6:2735–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. de Vries RD, Rimmelzwaan GF. Viral vector‐based influenza vaccines. Hum Vaccin Immunother 2016; 12:2881–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Stickl H, Hochstein‐Mintzel V, Mayr A, Huber HC, Schafer H, Holzner A. [MVA vaccination against smallpox: clinical tests with an attenuated live vaccinia virus strain (MVA) (author's translation)]. Dtsch Med Wochenschr 1974; 99:2386–92. [DOI] [PubMed] [Google Scholar]
- 27. Verheust C, Goossens M, Pauwels K, Breyer D. Biosafety aspects of modified vaccinia virus Ankara (MVA)‐based vectors used for gene therapy or vaccination. Vaccine 2012; 30:2623–32. [DOI] [PubMed] [Google Scholar]
- 28. Ramezanpour B, Haan I, Osterhaus A, Claassen E. Vector‐based genetically modified vaccines: exploiting Jenner's legacy. Vaccine 2016; 34:6436–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hewitt EW. The MHC class I antigen presentation pathway: strategies for viral immune evasion. Immunology 2003; 110:163–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ott G, Barchfeld GL, Van Nest G. Enhancement of humoral response against human influenza vaccine with the simple submicron oil/water emulsion adjuvant MF59. Vaccine 1995; 13:1557–62. [DOI] [PubMed] [Google Scholar]
- 31. Madhun AS, Haaheim LR, Nilsen MV, Cox RJ. Intramuscular Matrix‐M‐adjuvanted virosomal H5N1 vaccine induces high frequencies of multifunctional Th1 CD4+ cells and strong antibody responses in mice. Vaccine 2009; 27:7367–76. [DOI] [PubMed] [Google Scholar]
- 32. Cox RJ, Pedersen G, Madhun AS et al Evaluation of a virosomal H5N1 vaccine formulated with Matrix M adjuvant in a phase I clinical trial. Vaccine 2011; 29:8049–59. [DOI] [PubMed] [Google Scholar]
- 33. Pedersen G, Major D, Roseby S, Wood J, Madhun AS, Cox RJ. Matrix‐M adjuvanted virosomal H5N1 vaccine confers protection against lethal viral challenge in a murine model. Influenza Other Respir Viruses 2011; 5:426–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Magnusson SE, Reimer JM, Karlsson KH, Lilja L, Bengtsson KL, Stertman L. Immune enhancing properties of the novel Matrix‐M adjuvant leads to potentiated immune responses to an influenza vaccine in mice. Vaccine 2013; 31:1725–33. [DOI] [PubMed] [Google Scholar]
- 35. Cox F, Baart M, Huizingh J et al Protection against H5N1 influenza virus induced by Matrix‐M adjuvanted seasonal virosomal vaccine in mice requires both antibodies and T cells. PLOS ONE 2015; 10:e0145243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wee JL, Scheerlinck JP, Snibson KJ et al Pulmonary delivery of ISCOMATRIX influenza vaccine induces both systemic and mucosal immunity with antigen dose sparing. Mucosal Immunol 2008; 1:489–96. [DOI] [PubMed] [Google Scholar]
- 37. Mastelic Gavillet B, Eberhardt CS, Auderset F et al MF59 mediates its B cell adjuvanticity by promoting T follicular helper cells and thus germinal center responses in adult and early life. J Immunol 2015; 194:4836–45. [DOI] [PubMed] [Google Scholar]
- 38. Lovgren K, Morein B. The requirement of lipids for the formation of immunostimulating complexes (iscoms). Biotechnol Appl Biochem 1988; 10:161–72. [PubMed] [Google Scholar]
- 39. Radosevic K, Rodriguez A, Mintardjo R et al Antibody and T‐cell responses to a virosomal adjuvanted H9N2 avian influenza vaccine: impact of distinct additional adjuvants. Vaccine 2008; 26:3640–6. [DOI] [PubMed] [Google Scholar]
- 40. Pedersen GK, Sjursen H, Nostbakken JK, Jul‐Larsen A, Hoschler K, Cox RJ. Matrix M(TM) adjuvanted virosomal H5N1 vaccine induces balanced Th1/Th2 CD4(+) T cell responses in man. Hum Vaccin Immunother 2014; 10:2408–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Liu YV, Massare MJ, Pearce MB et al Recombinant virus‐like particles elicit protective immunity against avian influenza A(H7N9) virus infection in ferrets. Vaccine 2015; 33:2152–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Study of Parenterally Administrated Adjuvanted Seasonal Influenza Vaccine in Healthy Elderly Volunteers . Webaddress: https://clinicaltrials.gov/ct2/show/NCT01444482. Identifier: NCT01444482.
- 43. A(H7N9) VLP Antigen Dose‐Ranging Study With Matrix‐M1™ Adjuvant . Webaddress: https://clinicaltrials.gov/ct2/show/NCT02078674. Identifier: NCT02078674.
- 44. Trombetta CM, Montomoli E. Influenza immunology evaluation and correlates of protection: a focus on vaccines. Expert Rev Vaccines 2016; 15:967–76. [DOI] [PubMed] [Google Scholar]
- 45. Cargnelutti DE, Sanchez MA, Alvarez P, Boado L, Mattion N, Scodeller EA. Enhancement of Th1 immune responses to recombinant influenza nucleoprotein by Ribi adjuvant. New Microbiol 2013; 36:145–51. [PubMed] [Google Scholar]
- 46. Macleod MK, David A, Jin N et al Influenza nucleoprotein delivered with aluminium salts protects mice from an influenza A virus that expresses an altered nucleoprotein sequence. PLOS ONE 2013; 8:e61775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Altenburg AF, van de Sandt CE, van Trierum SE et al Increased protein degradation improves influenza virus nucleoprotein‐specific CD8+ T cell activation in vitro but not in C57BL/6 mice. J Virol 2016; 90:10209–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Flynn KJ, Belz GT, Altman JD, Ahmed R, Woodland DL, Doherty PC. Virus‐specific CD8+ T cells in primary and secondary influenza pneumonia. Immunity 1998; 8:683–91. [DOI] [PubMed] [Google Scholar]
- 49. Lovgren Bengtsson K, Morein B, Osterhaus AD. ISCOM technology‐based Matrix M adjuvant: success in future vaccines relies on formulation. Expert Rev Vaccines 2011; 10:401–3. [DOI] [PubMed] [Google Scholar]
- 50. Kreijtz JH, Suezer Y, van Amerongen G et al Recombinant modified vaccinia virus Ankara‐based vaccine induces protective immunity in mice against infection with influenza virus H5N1. J Infect Dis 2007; 195:1598–606. [DOI] [PubMed] [Google Scholar]
- 51. Luckheeram RV, Zhou R, Verma AD, Xia B. CD4(+)T cells: differentiation and functions. Clin Dev Immunol 2012; 2012:925135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lambe T, Carey JB, Li Y et al Immunity against heterosubtypic influenza virus induced by adenovirus and MVA expressing nucleoprotein and matrix protein‐1. Sci Rep 2013; 3:1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Magnusson SE, Karlsson KH, Reimer JM et al Matrix‐M adjuvanted envelope protein vaccine protects against lethal lineage 1 and 2 West Nile virus infection in mice. Vaccine 2014; 32:800–8. [DOI] [PubMed] [Google Scholar]
- 54. Verstrepen BE, Oostermeijer H, Fagrouch Z et al Vaccine‐induced protection of rhesus macaques against plasma viremia after intradermal infection with a European lineage 1 strain of West Nile virus. PLOS ONE 2014; 9:e112568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Bengtsson KL, Song H, Stertman L et al Matrix‐M adjuvant enhances antibody, cellular and protective immune responses of a Zaire Ebola/Makona virus glycoprotein (GP) nanoparticle vaccine in mice. Vaccine 2016; 34:1927–35. [DOI] [PubMed] [Google Scholar]
- 56. Cros JF, Garcia‐Sastre A, Palese P. An unconventional NLS is critical for the nuclear import of the influenza A virus nucleoprotein and ribonucleoprotein. Traffic 2005; 6:205–13. [DOI] [PubMed] [Google Scholar]
- 57. Ozawa M, Fujii K, Muramoto Y et al Contributions of two nuclear localization signals of influenza A virus nucleoprotein to viral replication. J Virol 2007; 81:30–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Qian SB, Ott DE, Schubert U, Bennink JR, Yewdell JW. Fusion proteins with COOH‐terminal ubiquitin are stable and maintain dual functionality in vivo . J Biol Chem 2002; 277:38818–26. [DOI] [PubMed] [Google Scholar]
- 59. Carragher DM, Kaminski DA, Moquin A, Hartson L, Randall TD. A novel role for non‐neutralizing antibodies against nucleoprotein in facilitating resistance to influenza virus. J Immunol 2008; 181:4168–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. LaMere MW, Lam HT, Moquin A et al Contributions of antinucleoprotein IgG to heterosubtypic immunity against influenza virus. J Immunol 2011; 186:4331–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Lamere MW, Moquin A, Lee FE et al Regulation of antinucleoprotein IgG by systemic vaccination and its effect on influenza virus clearance. J Virol 2011; 85:5027–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Taylor PM, Askonas BA. Influenza nucleoprotein‐specific cytotoxic T‐cell clones are protective in vivo. Immunology 1986; 58:417–20. [PMC free article] [PubMed] [Google Scholar]
- 63. Price GE, Soboleski MR, Lo CY et al Vaccination focusing immunity on conserved antigens protects mice and ferrets against virulent H1N1 and H5N1 influenza A viruses. Vaccine 2009; 27:6512–21. [DOI] [PubMed] [Google Scholar]
- 64. Norder M, Becker PD, Drexler I, Link C, Erfle V, Guzman CA. Modified vaccinia virus Ankara exerts potent immune modulatory activities in a murine model. PLOS ONE 2010; 5:e11400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Delaloye J, Roger T, Steiner‐Tardivel QG et al Innate immune sensing of modified vaccinia virus Ankara (MVA) is mediated by TLR2–TLR6, MDA‐5 and the NALP3 inflammasome. PLOS Pathog 2009; 5:e1000480. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 66. Watson DL, Lovgren K, Watson NA, Fossum C, Morein B, Hoglund S. Inflammatory response and antigen localization following immunization with influenza virus ISCOMs. Inflammation 1989; 13:641–9. [DOI] [PubMed] [Google Scholar]
- 67. Sjolander A, Bengtsson KL, Morein B. Kinetics, localization and cytokine profile of T cell responses to immune stimulating complexes (iscoms) containing human influenza virus envelope glycoproteins. Vaccine 1997; 15:1030–8. [DOI] [PubMed] [Google Scholar]
- 68. Gasteiger G, Kastenmuller W, Ljapoci R, Sutter G, Drexler I. Cross‐priming of cytotoxic T cells dictates antigen requisites for modified vaccinia virus Ankara vector vaccines. J Virol 2007; 81:11925–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Yewdell JW. Designing CD8+ T cell vaccines: it's not rocket science (yet). Curr Opin Immunol 2010; 22:402–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Wei J, Zanker DD, Di Carluccio AR et al Varied role of ubiquitylation in generating MHC class I peptide ligands. J Immunol 2017; 198:3835–45. [DOI] [PMC free article] [PubMed] [Google Scholar]


