Abstract
Dark-field microscope (DFM) analysis of nanoparticle binding signal is highly useful for a variety of research and biomedical applications, but current applications for nanoparticle quantification rely on expensive DFM systems. The cost, size, limited robustness of these DFMs limits their utility for non-laboratory settings. Most nanoparticle analyses use high-magnification DFM images, which are labor intensive to acquire and subject to operator bias. Low-magnification DFM image capture is faster, but is subject to background from surface artifacts and debris, although image processing can partially compensate for background signal. We thus mated an LED light source, a dark-field condenser and a 20× objective lens with a mobile phone camera to create an inexpensive, portable and robust DFM system suitable for use in non-laboratory conditions. This proof-of-concept mobile DFM device weighs less than 400g and costs less than $2000, but analysis of images captured with this device reveal similar nanoparticle quantitation results to those acquired with a much larger and more expensive desktop DFMM system. Our results suggest that similar devices may be useful for quantification of stable, nanoparticle-based activity and quantitation assays in resource-limited areas where conventional assay approaches are not practical.
Keywords: Dark field microscope, mobile phone, nanoparticle, ImageJ, Immunoassay
1. Introduction
Nanoparticles probes are useful in point-of-care assays, including lateral-flow chromatographic immunoassays, since antibody-labeled nanoparticles can be dried and stored under ambient conditions until use, unlike enzyme-linked antibodies employed by conventional immunoassays. These point-of-care devices are not quantitative, however, and require extensive development and validation (Chao et al., 2012; Sajid et al., 2015). Gold nanoparticles are used to quantify different targets (Hu et al., 2003; Rajendran, 2013; Wagner et al., 2014) in cell imaging, (Hu et al., 2009; X.-L. Li et al., 2016) biomolecular quantification, (T. Li et al., 2016; Yuan et al., 2016) and interaction studies (Jin et al., 2015; McFarland and Van Duyne, 2003), but these assays rely on complex and relatively non-robust equipment that limits their utility outside controlled laboratory settings. Portable spectrometers can quantify nanoparticles, but suffer from low throughput (Heider et al., 2012; Verma et al., 2016; Zuber et al., 2016) or have low sensitivity and require complex setup prior to their use for quantitation (Wang et al., 2017). DFM image analysis used to detect and quantify nanoparticle assays predominantly utilize high-magnification, since low-magnification (far-field) DFM images are sensitive to surface artifacts and debris that can mask nanoparticle signal. The size, cost and delicacy of these DFM systems limit their utility in field hospitals and other settings where these factors represent barriers. Attempts to develop more portable DFM approaches date back to at least 1958, (Goldman and Sawyer, 1958) when dermatologists still relied on DFM analysis to identify pathogens, including syphilis, responsible for certain skin lesions, (Brown and Frank, 2003) but these devices fell out of use upon development of more specific assays, effectively ending portable DFM development. Recent technology advances have spurred the use of mobile phone cameras for medical applications, including portable microscopy for numerous point-of-care diagnostics (Table S5), but none of these devices use DFM to quantify nanoparticle-based assays.
We now report the development of a mobile phone-based DFM (MDFM) system that can quantify nanoparticle signal for a variety of research and medical applications. We analyzed the potential of this approach to quantify methods that form the basis of most clinical assays, binding kinetics and biomarker quantification, as well as a novel nanoparticle-based serum diagnostic assay for tuberculosis. MDFM analysis of these assays yielded robust results that were similar to, albeit less sensitive than, those obtained with a much more expensive and cumbersome desktop DFM system. These results suggest that a lightweight, portable MDFM device allows simple and rapid assay quantitation, and may thus serve as a valuable platform for biomarker quantitation in resource-limited settings, where simplicity and robustness are more important than absolute assay sensitivity.
2. Material and methods
2.1 Image capture and processing
Solidworks 2013 CAD software (Dassault Systemes SolidWorks Corporation) was used to design the MDFM case, which was fabricated with black acrylonitrile-butadiene-styrene (ABS) using a 3D printing service (3D hubs). DDFM images were acquired under consistent lighting and magnification using an Olympus IX81 microscope equipped with a dark-field condenser, a 4× or 10× objective lens, and an Olympus DP71 digital camera, using a 1/45s exposure time. MDFM images were acquired using the Motorola Moto G2 camera to capture images from a slide holder case containing a dark-field condenser and a 10× or 20× objective lens and illuminated with a constant triple-LED white light source (Modgy, Inc.). Table S1 lists the components of these systems. All images were processed and quantified using our previously reported “DarkScatterMaster” (DSM) DFM algorithm(Sun et al., 2016) using the following software input parameters: contour threshold (Ct) = 253.020, center scale (S) = 0.8, type = Red, Low (Lt)/High (Ht) quantification limit: 0/62. Motorola Moto G2 (XT1068) images were captured with a 1/15 s exposure time with Open Camera (Version 1.32.1)(Harman, 2017) using an ISO 5000 configuration and allowing autofocus and 4× digital zoom. Magnification (M) was defined as the sample image height (hi) divided by the height of the sample object (ho), where ho was the target well diameter (1.5 mm) and hi was the diameter of this image in pixels multiplied by the resolution of the sensor chip (72 vs. 432 pixels/inch for MDFM and DDFM, respectively).
2.2 Binding affinity assay
Carboxyl-functionalized AuNRs (C12-25-650-TC-50, Nanopartz) were activated to covalently bond amine groups by mixing 40 μL of AuNR (4.22×1012/mL) with 20 μL of EDC/NHS-sulfo PBS (2 mg/mL of 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride and 1 mg/mL of N-hydroxysulfosuccinimide, Sigma-Aldrich) for 10 minutes at 25°C. These amine-reactive AuNRs were then PBS-washed and 1 μL of indicated AuNR concentrations were applied to replicate wells on 192-well amine-functionalized slides (2×1012 group/mm2, Arrayit), which were sonicated (Q500 Sonicator, Qsonica) for 8 minutes at 80% amplitude using a 5 second on/off cycle to accelerate hybridization. Slides were then washed for 10 min at 25°C with 0.01% Tween-20 in PBS (PBST, pH 7.0), and deionized water, and then air-dried for DFM imagery. Binding affinity was calculated by nonlinear curve fitting with Origin 2015 software (OrginLab Corporation).
2.3 Protein quantification assay
Protein A/G-modified 192-well slides (Arrayit) were blocked with 1 μL/well Pierce Protein-Free Blocking Buffer (Thermo Scientific) for 1 hour at 25°C, then incubated with the indicated amounts of biotinylated CD9 antibody (NB110-81616, Novus) for 1 hour at 25°C, and PBS-washed for 10 min at 25°C before hybridization with AuNR. Neutravidin-functionalized AuNR (Nanopartz C12-25-650-TN-50, 7×10−9 M) were PBS-diluted (40 μL AuNR to 200 μL PBS) after which 1 μL/well of AuNR was applied to replicate wells, which were sonicated (Q500 Sonicator, Qsonica) for 8 minutes at 80% amplitude using a 5 second on/off cycle to accelerate hybridization. After hybridization, slides were washed for 10 min at 25°C with 0.01% Tween-20 in PBS (PBST, pH 7.0), and deionized water, and then air-dried for DFM imagery.
2.4 Data analysis
Limits of detection (LOD) and quantification (LOQ) were defined as 3× and 10× the standard deviation of the assay blank, respectively. Assay precision was determined with five replicates of three samples analyzed in a single assay (intra-assay) or in three assays analyzed on three different days (inter-assay). Graphs were generated with Origin 2015 and Microsoft Excel.
3. Results and discussion
3.1 Optical design and characterization
To generate an inexpensive, lightweight, and portable device capable of sensitive far-field DFM image analysis for nanoparticle quantitation, we wrapped a triple-LED light source, a standard dark-field condenser and a 20× or a 10× objective lens in a 3D-printed case that mates these components to a mobile phone camera (Figure 1). We then compared the nanoparticle quantitation properties of this MDFM device with those of a standard desktop DFM (DDFM) system. Both systems used the same dark-field condenser, but differed in their light sources, objective lenses, cameras, and total system weight and cost (Table 1). It is difficult to compare the cost of our prototype MDFM system to all generic DDFM systems. However, given that the MDFM phone and its camera is much less expensive (< $200) than that any camera-equipped DDFM camera system (several thousand dollars), a MDFM system will always the less expensive option.
Figure 1.
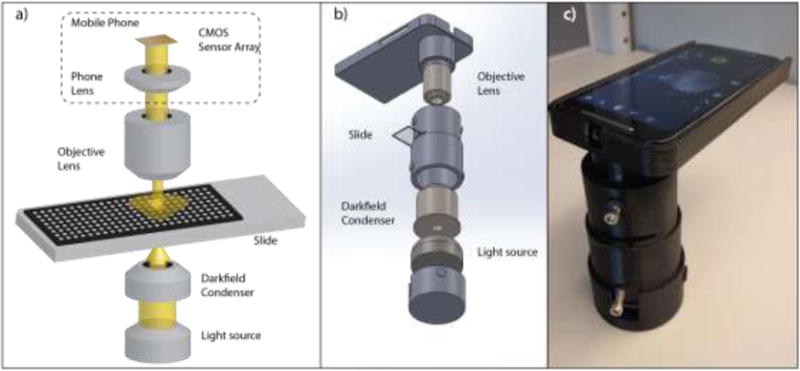
MDFM system a) layout schematic, b) assembly and c) working prototype.
Table 1.
Differences of the mobile and desktop DFM systems.
| Mobile DFM | Desktop DFM | |
|---|---|---|
| Light source | 3 LEDs (1,000 lux) | Halogen lamp (> 10,000 lux) |
| Camera model | Motorola XT1064 | Olympus DP71 |
| Maximum resolution | 3264 × 2448 JPEG (8.0 megapixels) |
4080 × 3072 (12.5 megapixels) |
| Camera sensor | CMOS | CCD |
| Weight | 0.38 kg | 26 kg |
| System cost with lens | $1,360 (10×)/$1,560 (20×) Motorola Moto G2* |
>$50,000 Olympus IX81 |
Cost details in Table S3
Comparison of the image quality produced by these systems found that MFDM images exhibited weaker and more uneven signal than similar DDFM images (Figure 2), most likely due to differences in their illumination systems and condenser alignment. The MDFM prototype employs a relatively weak (1,000-lux) triple-LED light source run off a 3V lithium battery, whereas the DDFM system uses a single 100 W (>10,000-lux) halogen lamp powered by an AC electrical outlet. We chose the LED system used in our MDFM for its low power draw and relatively high light output, but its three point LED arrangement is consistent with lighting artifacts observed in MDFM images. Both the MDFM condenser and case may exacerbate these artifacts, since the MDFM condenser is not optimized for this multi-source light setup and the case does not permit precise alignment of the condenser light path. MDFM image quality also suffers from the lower resolution of the MDFM CMOS vs. DDFM CCD sensor (72 vs. 432 pixels/inch). To compensate for this resolution difference, MDFM and DDFM employ different lenses to capture low-magnification (10× vs. 4×) and high-magnification (20× vs. 10×) images. The low fixed aperture of the MDFM camera (f/2) also limits depth of focus, but this should not negatively influence surface-based imagery for nanoparticle quantification.
Figure 2.

Comparison of MDFM, DDFM and desktop bright-field microscope images. MDFM images were captured with 20× and 10× objective lenses, and DDFM and desktop bright field microscope images were captured with 10× and 4× objective lenses.
Both MDFM and DDFM captured similar DFM signal intensity and image features from the same wells. DDFM images had more consistent lighting and better resolution (Figure 2), so that MDFM were less sensitive to DFM artifacts arising from sample well debris and surface defects. Improved MDFM cases milling could improve misalignment effects, which appeared to play a significant role in MDFM image variability.
The DDFM system required precise manual focus to capture images at a set magnification and working distance for each objective lens, while the MDFM system captured focused images over a wide range of working distances (3 to 10 mm) using a 10× lens, due to the autofocus function of its camera (Figure 3). MDFM did not exhibit such dynamic working ranges when using a 20× lens, however, due to the reduced working distance available for the autofocus function. The ability of the MDFM system to autofocus low magnification DFM images ensures that captured images are useful for image analysis, and reduces the time required to obtain this data regardless of the technical proficiency of the operator.
Figure 3.
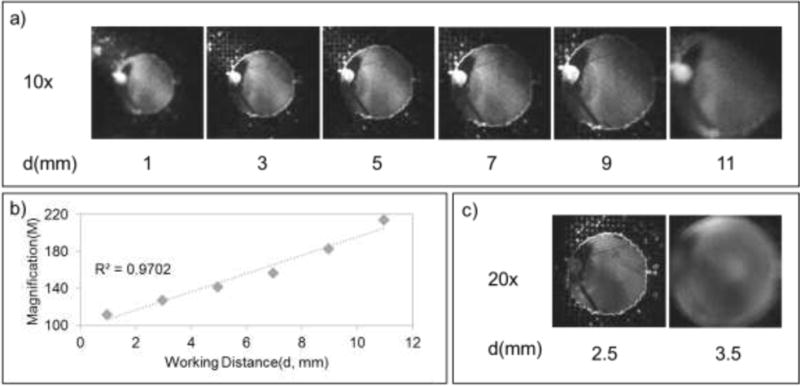
MDFM magnification. a) MDFM images captured with a 10× objective lens at different working distances, indicating the autofocus limits of the mobile phone’s camera. b) Correlation between working distance and MDFM magnification with a 10× objective lens. c) MDFM images captured with a 20× objective lens at different working distances. All images were converted to grayscale to highlight image focus.
The MDFM system magnified samples 370-fold and 110- to 210-fold when using 20× and 10× lenses, while the DDFM system magnified samples 375-fold and 150-fold using 10× and 4× lenses. MDFM images thus exhibited magnifications corresponding to 98.7% (high-power) and 58.6% to 112% (low-power) of the matching DDFM images, after adjusting for lens and sensor differences. Such differences should not greatly influence nanoparticle detection by MFDM vs DDFM systems, since low versus high magnification DDFM images produce similar signal, albeit with increased background and a correspondingly reduced dynamic range (Figure S1). MDFM and DDFM therefore achieve similar magnification for focused images, but MDFM has greater working distance flexibility, allowing rapid capture of focused images by individuals without DFM experience, which may be advantageous in resource-limited settings. Since both systems achieved similar magnification and altering the resolution of the DDFM system had no effect on image quantitation (Figure S4), it appears that illumination, light path alignment, and/or camera sensitivity differences account for the performance differences we observe between the DDFM and MDFM system.
3.2 Nanoparticle quantification assays
To evaluate MDFM capacity to analyze two common biological assay types (binding kinetics and biomarker concentration), we compared MDFM and DDFM results from binding affinity and protein quantitation assays we adapted for DFM nanoparticle quantification. We first examined a nanoparticle binding assay by measuring the interaction between carboxylic acid-functionalized gold nanorods (AuNR−) and an amine-modified slide (Figure 4a), expressing this interaction as a function of AuNR− concentration and the electric field potential at the liquid-solid interface, which obeys Boltzmann statistics.(Sun et al., 2017, 2015a, 2015b)
| (1) |
where the amount of AuNR available at the slide surface ([AuNR−]surface) was a function of [AuNR−]solution, the elementary charge q (1.60218×10−19 C), the surface potential ψD, the Boltzmann constant kB (1.38066×10−23 J/K) and temperature T. This equation simplified to
| (2) |
when ψD and T were held constant. Based on the Michaelis-Menten model at steady-state, (Johnson and Goody, 2011; Koshland et al., 1966; LÓPEZ-FIDALGO and WONG, 2002; McPherson, 1983) the surface binding rate was described as
| (3) |
determined by the equilibrium binding constant Kd, the maximum number of surface binding sites , and the input nanoparticle concentration constant [AuNR−]solution, so that Kd can be solved for by curve fitting. We applied this information and concentration-dependent DFM scatter responses from both the MDFM and DDFM systems to calculate the equilibrium binding constant (Kd) of this interaction. MDFM and DDFM response curves produced in this analysis yielded similar Kd values (0.0128 versus 0.0104), despite consistently lower MDFM signal throughout the entire range of the AuNR concentration curve (Figure 4b).
Figure 4.
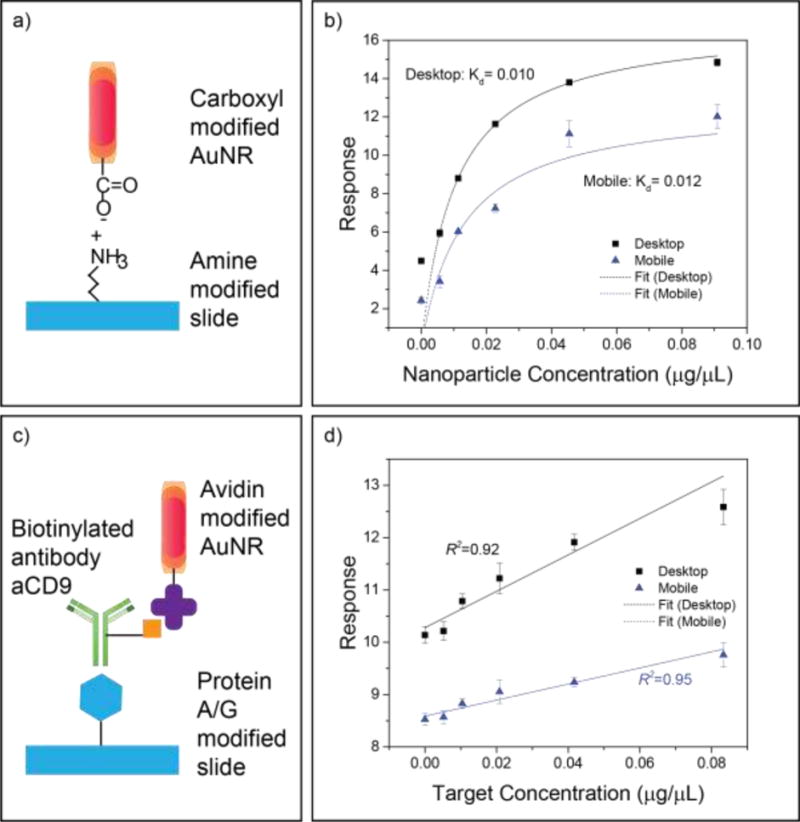
Comparison of MDFM and DDFM for binding affinity and protein quantification assays. (a and b) Assay scheme and binding affinity result. (c and d) Assay scheme and target (anti-CD9) quantification result. Data points represent the mean ± SEM of 5 sample replicates.
We next analyzed MDFM and DDFM performance to quantify the results of a protein binding assay that used protein A/G-modified slides to capture AuNR-conjugated antibodies (Figure 4c). Both the MDFM and DDFM responses strongly correlated with the target protein concentration; however, the MDFM response exhibited reduced background, less overall variability and greater linearity than the DDFM response, although it also exhibited a smaller dynamic response over the assay concentration range (Figure 4d).
The reduced dynamic range and/or shallower curves of MDFM versus DDFM signal in these assays resulted in higher limits of detection and quantification for the MDFM analyses (Table 2), reflected by 4.5-fold to 7-fold sensitivity decreases for these measurements in binding kinetics and biomarker quantification assays. MDFM results still revealed reasonable coefficients of variation for both intra-assay (3.1% to 7.8%) and inter-assay (8.1% to 13.7%) replicates (Table S2), although direct comparison of MDFM and DDFM results revealed an increase in MDFM versus DDFM intra-assay (0.9- to 3.2-fold) and inter-assay (1.3- to 3.0-fold) coefficient of variation. DFM image analysis can thus quantitate the nanoparticle-based versions of two assay types that constitute the majority of quantitative biological assays, with good sensitivity and precision, even when using an inexpensive MDFM system for image analysis.
Table 2.
Sensitivity of the quantitative assays
| Assay | (ng/μL) | MDFM | DDFM |
|---|---|---|---|
| Binding | LOD | 4.6 | 0.6 |
| affinity | LOQ | 15.3 | 2.1 |
|
| |||
| Protein | LOD | 135.2 | 30.1 |
| quantification | LOQ | 811.3 | 180.9 |
LOD: Limit of detection; LOQ: limit of quantitation
Nanoparticle-based quantitative bioassays are not in common usage, but exhibit certain advantages over conventional enzyme-based assays, since the nanoparticle probes are less labile and produce stable endpoint results. We therefore also analyzed MDFM performance with a variant of a novel assay that employs nanoparticle-based DFM signal to achieve results that are more rapid, sensitive and specific than can be achieved by conventional assays(Liang et al., 2017; Sun et al., 2016). We have previously used this approach to detect and monitor pancreatic cancer-derived exosomes in patient blood samples, while our current study applies a modification of this assay to distinguish patients with pediatric tuberculosis (TB) from their healthy controls (Figure S3). We chose TB for this analysis since it has high prevalence in developing countries, where the lack of resources and trained healthcare providers can inhibit effective diagnosis and disease control, highlighting the need for inexpensive and easy-to-use diagnostic methods. Our novel assay quantifies circulating levels of exosomes produced by cells infected with the TB bacillus Mycobacterium tuberculosis by detecting all such vesicles that contain lipoarabinomannan (LAM), the only marker suggested for TB screening by the WHO (World Health Organization, 2015). A proof-of-concept study, which used this assay to analyze patient serum samples without a separate exosome enrichment step, easily differentiated a small group of pediatric TB cases from their healthy controls, suggesting that MDFM analysis of this and other pathogens may have significant potential for rapid disease diagnosis, particularly in resource-limited settings. For example, these assays require reagents that can be stored dry at ambient temperature and reconstituted as needed (antibody-conjugated slides and probes) or that are inherently stable at ambient temperature (PBS, Tween and distilled water). Technicians can easily perform sample incubations at ambient temperature and analyze assay results on-site or transmit them to a central site for remote analysis by mobile. The ability to perform such assays at remote sites that may lack electricity is of significant advantage for the clinicians attempting to identify and immediately treat infected individuals in order to control disease outbreaks.
Lighting induced artifacts observed with the current MFDM prototype prevent its use for high-quality DFM imagery, but do not decrease its utility for nanoparticle-based quantitation assays once images are processed to correct for artifacts commonly associated with low-magnification far-field DFM images, including uneven lighting and other signal artifacts using our previously published DFM image processing approach.(Sun et al., 2016)
4. Conclusion
The current MDFM system rapidly captures focused DFM images, without requiring significant user intervention, and analysis of these images yields quantitative results that closely parallel those generated with matching DDFM images, albeit with slightly reduced sensitivity at low protein concentrations. MDFM thus exhibits strong potential as a hand-held DFM imaging system for bioassay quantitation in resource-limited areas that prevent the use of DDFM or other quantification methods. Reduced MDFM versus DDFM optical performance appears to derive primarily from reduced DFM signal quality due to weak and uneven sample illumination from a multi-LED light source and non-optimized optics in this proof-of-concept device. Both issues should be easily addressable through selection of a larger, single-source LED and case refinements, or other changes, to improve optical focusing to increase DFM image signal quality.
Supplementary Material
Highlights.
A mobile phone based dark-field far-field microscope was developed for nanoparticle quantification.
The microscope weighted 380g and cost less than 2000 with easy assemble and operation.
Quantification of bioassay with the microscope shown comparable result to standard dark-field microscope.
Potentiate a TB diagnosis assay for resource-limited area.
Acknowledgments
This research was supported in part by US National Institute of Allergy and Infectious Diseases grant R01Al113725-01A1 and R01AI122932-01A1 to YH. Authors would like to thank Drs. Christopher J. Lyon, Chang Liu, and Jia Fan for their invaluable advice. D.S. designed the study and conducted the experiments.
Abbreviations
- DFM
Dark field microscope
- DSM
DarkScatterMaster
- MDFM
Mobile dark field microscope
- DDFM
Desktop dark field microscope
- AuNR
gold nanorod
- FFLM
Far-field low magnification
- CV
coefficient of variation
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supporting Information
Supplementary data associated with this article can be found in the online version.
The authors declare no competing financial interest.
References
- Brown DL, Frank JE. Diagnosis and management of syphilis. Am Fam Physician. 2003;68:283–290. [PubMed] [Google Scholar]
- Chao CH, Wu CS, Huang CC, Liang JC, Wang HT, Tang PT, Lin LY, Ko FH. A rapid and portable sensor based on protein-modified gold nanoparticle probes and lateral flow assay for naked eye detection of mercury ion. Microelectron Eng. 2012;97:294–296. doi: 10.1016/j.mee.2012.03.015. [DOI] [Google Scholar]
- Goldman L, Sawyer F. Simple Microscope Hula-Hoop Skin. AMA Arch Dermatology. 1958;190:137. [Google Scholar]
- Harman M. Open Camera [WWW Document] 2017 sourceforge. URL http://opencamera.sourceforge.net/
- Heider EC, Trieu K, Moore AFT, Campiglia AD. Portable mercury sensor for tap water using surface plasmon resonance of immobilized gold nanorods. Talanta. 2012;99:180–185. doi: 10.1016/j.talanta.2012.05.037. [DOI] [PubMed] [Google Scholar]
- Hu M, Novo C, Funston A, Wang H, Staleva H, Zou S, Mulvaney P, Xia Y, Hartland GV, Raschke G, Feldmann J, Nichtl A, Kurzinger K, Franzl T, Kowarik S, Sonnichsen C, Klar TA, Hu R, Yong KT, Roy I, Ding H, He S, Prasad PN. Dark-field microscopy studies of single metal nanoparticles: understanding the factors that influence the linewidth of the localized surface plasmon resonance. J Mater Chem. 2003;113:2676–2684. doi: 10.1039/b714759g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu R, Yong KT, Roy I, Ding H, He S, Prasad PN. Metallic Nanostructures as Localized Plasmon Resonance Enhanced Scattering Probes for Multiplex Dark Field Targeted Imaging of Cancer Cells. J Phys Chem C Nanomater Interfaces. 2009;113:2676–2684. doi: 10.1021/jp8076672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin HY, Li DW, Zhang N, Gu Z, Long YT. Analyzing Carbohydrate-Protein Interaction Based on Single Plasmonic Nanoparticle by Conventional Dark Field Microscopy. ACS Appl Mater Interfaces. 2015;7:12249–12253. doi: 10.1021/acsami.5b02744. [DOI] [PubMed] [Google Scholar]
- Johnson KA, Goody RS. The original Michaelis constant: Translation of the 1913 Michaelis-Menten Paper. Biochemistry. 2011;50:8264–8269. doi: 10.1021/bi201284u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshland DE, Némethy G, Filmer D. Comparison of experimental binding data and theoretical models in proteins containing subunits. Biochemistry. 1966;5:365–385. doi: 10.1021/bi00865a047. [DOI] [PubMed] [Google Scholar]
- Li T, Xu X, Zhang G, Lin R, Chen Y, Li C, Liu F, Li N. Nonamplification Sandwich Assay Platform for Sensitive Nucleic Acid Detection Based on AuNPs Enumeration with the Dark-Field Microscope. Anal Chem. 2016;88:4188–4191. doi: 10.1021/acs.analchem.6b00535. [DOI] [PubMed] [Google Scholar]
- Li XL, Zhang ZL, Zhao W, Xia XH, Xu JJ, Chen HY. Oriented assembly of invisible probes: towards single mRNA imaging in living cells. Chem Sci. 2016;0:1–8. doi: 10.1039/C5SC04369G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang K, Liu F, Fan J, Sun D, Liu C, Lyon CJ, Bernard DW, Li Y, Yokoi K, Katz MH, Koay EJ, Zhao Z, Hu Y, Brinton LT, Sloane HS, Kester M, Kelly KA, Gyorgy B, Raposo G, Stoorvogel W, Anderson HC, Mulhall D, Garimella R, van der Pol E, Boing AN, Harrison P, Sturk A, Nieuwland R, Muralidharan-Chari V, Clancy JW, Sedgwick A, D’Souza-Schorey C, De Candia P, Tanaka Y, Rabinowits G, Gercel-Taylor C, Day JM, Taylor DD, Kloecker GH, Boukouris S, Mathivanan S, O’Driscoll L, Khan S, Théry C, Clayton A, Amigorena S, Raposo G, He M, Crow J, Roth M, Zeng Y, Godwin AK, Lane RE, Korbie D, Anderson W, Vaidyanathan R, Trau M, Kanwar SS, Dunlay CJ, Simeone DM, Nagrath S, Logozzi M, Shao H, Shao H, Thery C, Zitvogel L, Amigorena S, Im H, Melo SA, Peinado H, Yoshioka Y, Miyazaki T, Kato H, Fukuchi M, Nakajima M, Kuwano H, Duxbury MS, Ito H, Zinner MJ, Ashley SW, Whang EE, Mudali SV, Duxbury MS, Ito H, Zinner MJ, Ashley SW, Whang EE, Anker JN, Rodriguez-Lorenzo L, de la Rica R, Alvarez-Puebla RA, Liz-Marzan LM, Stevens MM, Choi Y, Park Y, Kang T, Lee LP, Nusz GJ, Ghosh SK, Pa T, Lee K, Cui Y, Lee LP, Irudayaraj J, Link S, El-Sayed MA, Shi L, Kalra H, Vaidyanathan R, Li D, Xie K, Wolff R, Abbruzzese JL, Freelove R, Walling AD, Ballehaninna UK, Chamberlain RS, Hidalgo M, Ballehaninna UK, Chamberlain RS, Del Villano BC, Jazieh KA, Foote MB, Diaz LA, Locker GY, Lowenfels AB, Logsdon CD, Pei H, Kinch MS, Carles-Kinch K, Chang Q, Jorgensen C, Pawson T, Hedley DW, Dobrzanski P, Ansuini H, Archin NM, Kowal J, Good DM, Lescuyer P, Hochstrasser D, Rabilloud T, Hori SS, Gambhir SS, Joshi GK, Brantley DM, Kinch MS, Carles-Kinch K, Evans DB, Yokoi K, Jensen MM, Jorgensen JT, Binderup T, Kjaer A. Nanoplasmonic quantification of tumour-derived extracellular vesicles in plasma microsamples for diagnosis and treatment monitoring. Nat Biomed Eng. 2017;1:21. doi: 10.1038/s41551-016-0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LÓPEZ-FIDALGO J, WONG WK. Design Issues for the Michaelis–Menten Model. J Theor Biol. 2002;215:1–11. doi: 10.1006/jtbi.2001.2497. [DOI] [PubMed] [Google Scholar]
- McFarland AD, Van Duyne RP. Single silver nanoparticles as real-time optical sensors with zeptomole sensitivity. Nano Lett. 2003;3:1057–1062. doi: 10.1021/nl034372s. [DOI] [Google Scholar]
- McPherson GA. A practical computer based approach to the analysis of radiologand binding experiments. Comput Prog Biomed. 1983;17:107–114. doi: 10.1016/0010-468x(83)90031-4. [DOI] [PubMed] [Google Scholar]
- Rajendran P. S Ynthesis and C Haracterization of M Etallic N Ano. J Opt Res. 2013;81:13–16. [Google Scholar]
- Sajid M, Kawde AN, Daud M. Designs, formats and applications of lateral flow assay: A literature review. J Saudi Chem Soc. 2015;19:689–705. doi: 10.1016/j.jscs.2014.09.001. [DOI] [Google Scholar]
- Sun D, Fan J, Liu C, Liu Y, Bu Y, Lyon CJ, Hu Y. A noise reduction method for quantifying nanoparticle light scattering in low magnification dark-field microscope far-field images. Anal Chem. 2016;88:12001–12005. doi: 10.1021/acs.analchem.6b03661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun D, Matsui H, Wu CN, Tabata H. Surface treatment on amorphous InGaZnO4 thin film for single-stranded DNA biosensing. Appl Surf Sci. 2015a;324:310–318. doi: 10.1016/j.apsusc.2014.10.115. [DOI] [Google Scholar]
- Sun D, Matsui H, Yamahara H, Liu C, Wu L. Sensors and Actuators B : Chemical A low-cost portable electrical sensor for hydroxyl ions based on amorphous InGaZnO4 thin film at room temperature. Sensors Actuators B Chem. 2017;239:679–687. doi: 10.1016/j.snb.2016.08.060. [DOI] [Google Scholar]
- Sun D, Yamahara H, Nakane R, Matsui H, Tabata H. Hydroxyl radical and thermal annealing on amorphous InGaZnO<inf>4</inf> films for DNA immobilizations. Colloids Surfaces B Biointerfaces. 2015b;130:119–125. doi: 10.1016/j.colsurfb.2015.04.024. [DOI] [PubMed] [Google Scholar]
- Verma MS, Tsuji JM, Hall B, Chen PZ, Forrest J, Jones L, Gu FX. Towards point-of-care detection of polymicrobial infections: Rapid colorimetric response using a portable spectrophotometer. Sens Bio-Sensing Res. 2016;10:15–19. doi: 10.1016/j.sbsr.2016.05.004. [DOI] [Google Scholar]
- Wagner T, Lipinski HG, Wiemann M. Dark field nanoparticle tracking analysis for size characterization of plasmonic and non-plasmonic particles. J Nanoparticle Res. 2014;16 doi: 10.1007/s11051-014-2419-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang LJ, Chang YC, Sun R, Li L. A multichannel smartphone optical biosensor for high-throughput point-of-care diagnostics. Biosens Bioelectron. 2017;87:686–692. doi: 10.1016/j.bios.2016.09.021. [DOI] [PubMed] [Google Scholar]
- World Health Organization. The use of lateral flow urine lipoarabinomannan assay (LF-LAM) for the diagnosis and screening of active tuberculosis in people living with HIV: policy 1–74. 2015 doi:978 92 4 150963 3. [Google Scholar]
- Yuan L, Wang X, Fang Y, Liu C, Jiang D, Wo X, Wang W, Chen HY. Digitizing Gold Nanoparticle-Based Colorimetric Assay by Imaging and Counting Single Nanoparticles. Anal Chem acs.analchem. 2016:5b04244. doi: 10.1021/acs.analchem.5b04244. [DOI] [PubMed] [Google Scholar]
- Zuber A, Purdey M, Schartner E, Forbes C, Van Der Hoek B, Giles D, Abell A, Monro T, Ebendorff-Heidepriem H. Detection of gold nanoparticles with different sizes using absorption and fluorescence based method. Sensors Actuators, B Chem. 2016;227:117–127. doi: 10.1016/j.snb.2015.12.044. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


