Abstract
We report a method for serial X-ray crystallography at X-ray free electron lasers (XFELs), which allows for full use of the current 120 Hz repetition rate of the Linear Coherent Light Source (LCLS). Using a micro-patterned silicon chip in combination with the high-speed Roadrunner goniometer for sample delivery we were able to determine the crystal structures of a picornavirus, bovine enterovirus 2 (BEV2), and the cytoplasmic polyhedrosis virus type 18 polyhedrin. Total data collection times were less than 14 and 10 minutes, respectively. Our method requires only micrograms of sample and will therefore broaden the applicability of serial femtosecond crystallography to challenging projects for which only limited sample amounts are available. By synchronizing the sample exchange to the XFEL repetition rate, our method allows for the most efficient use of the limited beamtime available at XFELs and should enable a substantial increase in sample throughput at these facilities.
Introduction
X-ray crystallography has been the dominant method for the determination of high-resolution virus structures in the last 30 years. For non-enveloped viruses, numerous X-ray crystallographic structures have been solved, at resolutions of up to 1.4 Å1. Due to the large unit cell dimensions and limited size of the crystals, Bragg reflections from virus crystals are typically weak2,3. Thus, X-ray structure determination of virus crystals are ideally carried out at highly brilliant X-ray sources, allowing a large number of photons to be focused into a small spot, ideally matching the size of the virus crystals4. A further challenge for virus crystallography is radiation damage. Structure determination from biological macromolecules are usually carried out at cryogenic temperatures in order to reduce the detrimental effect of ionizing radiation on crystal diffraction5–7.While finding appropriate conditions for cryo-protection is often straightforward for many protein crystals, this has remained a challenge for virus crystals since they only possess weak crystal contacts and a small increase in crystal mosaicity often results in overlapping reflections and a reduction of the measured resolution2,3. Thus so far relatively few virus structures have been determined at cryogenic temperatures1 and most work is performed at room temperature4.
A promising method for biological structure determination from virus crystals is Serial Femtosecond X-ray Crystallography (SFX)8–11 at X-ray Free Electron Lasers (XFELs), a technique well-suited to room temperature experiments, overcoming classical radiation dose limits by several orders of magnitude12,13. By taking up to hundreds of thousands of snapshots of nano- to micrometer-sized crystals, more than 80 biomolecular structures have been solved using SFX to date. The method has also been successfully extended to the time domain to investigate protein kinetics and enzyme reactions14,15. SFX therefore has not only the potential to yield high-resolution information about the virus structure itself, it also allows investigation of dynamics, since many viral proteins undergo structural changes during their life cycles, for example due to protein interactions with cellular receptors and pH change during entry through the endosome16–18.
A continuing limiting factor for virus SFX in particular is efficient sample delivery, as typically only microgram amounts of virus crystals are available. Most SFX experiments today are carried out using gas dynamic virtual nozzles (GDVNs) and high-viscosity gel matrices typically requiring more than 500 µg (~370 nL) of sample for structure determination19–26.
In a first attempt of virus crystallography at XFELs,27 104 micro-crystals of the Sindbis virus of ~700 Å unit cell dimension were suspended in a slow-moving stream of agarose and exposed to XFEL pulses. Using this approach, diffraction patterns were obtained with up to ~40 Å resolution and a 0.8% hit rate, defined by the percentage of images containing a certain number of Bragg spots. Due to the technological challenges, especially regarding the amount of sample required, no virus structure has yet been determined at an XFEL.
A promising sample delivery approach for SFX is the use of solid sample supports, also referred to as fixed targets28–37, especially if sample is limited. Here, several tens to thousands of crystals are loaded onto a structured solid support and automatically raster-scanned during X-ray exposure. A major challenge for fixed-target experiments is fast and precise scanning of these supports and synchronization of predefined sample positions with the arrival of XFEL pulses.
So far, such experiments have been performed in step-scanning mode, rendering them less efficient than other sample delivery methods due to the relatively long data collection times34,38. Another obstacle for fixed-target experiments is the relatively high background scattering level, mainly caused by air scattering from the direct beam. Further contributions originate from scattering by non-sample material such as the surrounding mother liquor, or Kapton or Mylar foil typically used as sealing materials to prevent crystal dehydration.
Here, we report a method for fixed-target serial crystallography at low background levels and a sample exchange rate of 120 Hz, requiring only micrograms of sample. We demonstrate the applicability of our method for virus structure determination, shown for an intact virus, BEV2, and cytoplasmic polyhedrosis virus type 18 polyhedrin (CPV18).
BEV2 belongs to the virus family Picornaviridae, genus Enterovirus, and is a non-enveloped, positive-stranded RNA virus, 30 nm in diameter, which is endemic in cattle environments39–41. Unlike some other picornaviruses, it is not a serious economic or animal health threat and is therefore a suitable model system for the investigation of biological processes such as virus uncoating, universal to all enteroviruses. Enteroviruses (including BEV2) are stabilized by lipid cofactors such as sphingosine, which bind to a hydrophobic pocket of the VP1 capsid protein42. Potent binders to this pocket have potential as antivirals (by preventing uncoating) and so we have investigated the specificity of binding natural and synthetic moieties in this pocket. BEV2 crystals represent a challenging system for current SFX experiments, possessing a large unit cell constant of 436.6 Å, and typically only microgram quantities of small crystals are available.
Cytoplasmic polyhedrosis viruses (CPVs) are found as parasites in many insects and cause significant losses in silkworm cocoon harvests43. CPV strains have polyhedra crystals varying in size from hundreds of nanometers to several micrometers and contain several thousands of CPV particles43–46, protecting them against environmental insults. CPV polyhedra were chosen as a well-established and robust model system for SFX data collection at cryogenic temperatures47.
Results
High-speed fixed-target structure determination approach
Micro-crystals of BEV2 and CPV18 were measured by fixed-target SFX using the Roadrunner goniometer installed at the XPP instrument at the Linac Coherent Light Source (Supplementary Fig. 1). The Roadrunner setup consists of high-precision x and y piezo-motor driven scanning stages mounted on a horizontal translation stage and a vertical rotation axis (Supplementary Fig. 2, also see Methods section for details). A high-magnification inline microscope allows visualization of samples and their support structure (Supplementary Fig. 3). The Roadrunner setup is capable of data collection at both room temperature and cryogenic temperatures.
To reduce air scattering of X-rays most of the path of the direct beam in air is enclosed in capillary shields, both upstream and downstream of the sample, reducing the free path of the direct beam in air to 20 mm (Supplementary Fig. 4). By streaming helium gas across the remaining unenclosed direct beam behind the sample, the number of photons scattered by air is further reduced (Fig. 1). Combining these two approaches, air scattering can be reduced by a factor of about 8.
Figure 1. Low background experimental setup for fast fixed-target SFX experiments using the Roadrunner goniometer.
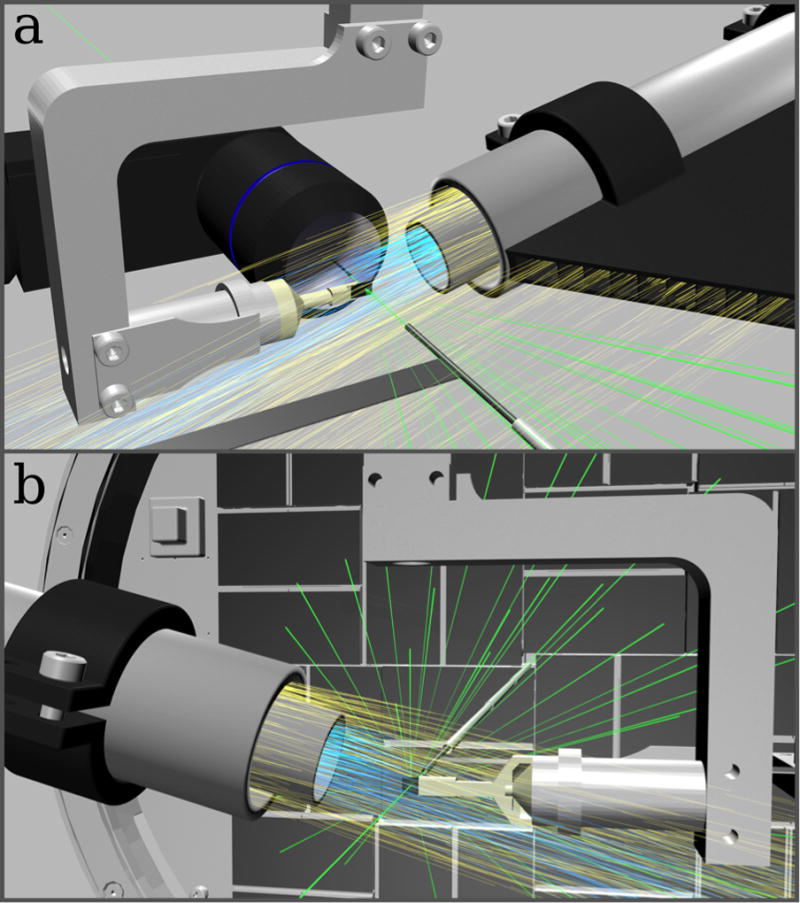
(a) Front view: The silicon chip is raster scanned through the X-ray beam (green) while maintained in a continuous stream of humidified air (blue). A helium sheath flow (yellow) is used to confine the humidity stream and to reduce air scattering. Air scattering is further reduced by helium injection along the beam path. An inline microscope is used for proper chip alignment and definition of the scanning grid. (b) Back view: X-ray diffraction caused by the sample crystals is recorded with a Cornell-SLAC hybrid pixel array detector (CSPAD). After hitting the sample, the primary beam is enclosed by a molybdenum tubule and additional steel tubules, which further absorb air-scattered photons. In b, the inline microscope is not shown for clarity.
By using micro-patterned chips made of single-crystalline silicon as substrate material, the background scattering signal caused by the support can be further reduced31. Dehydration of the crystals is prevented either by keeping them constantly in a stream of humidified gas32,48 or by flash freezing and collecting data at cryogenic temperatures31 (Supplementary Fig. 5). With this approach, sealing of the sample holders e.g. with Mylar foil, which would result in an increased background scattering level, is not required.
Our silicon chip (Fig. 2a) provides 22,500 pores for crystals31. For sample loading, 2–3 µL of sample suspension is pipetted onto the chip so that the amount of sample material used is typically in the range of a few micrograms, depending on the crystal sizes, desired coverage of the chip membrane and the amount of sample available31,32. The resulting arrangement of the crystals is due to the pore pattern and allows for a highly efficient measurement strategy by shooting through all pores with the FEL pulses in a fully automated procedure (Fig. 2b,c). With the Roadrunner control software the scan points are defined by drawing a grid, which is graphically overlaid on the inline-microscope image (Supplementary Fig. 6). For data collection the coordinates of scan points are downloaded to the motion controller and the entire chip is scanned in a meander-scan manner (Fig. 2d,e, for details see Methods section). With this approach most of the crystalline material is used for the diffraction experiment and not wasted.
Figure 2. Design of the micro-patterned silicon chip and data collection strategy.
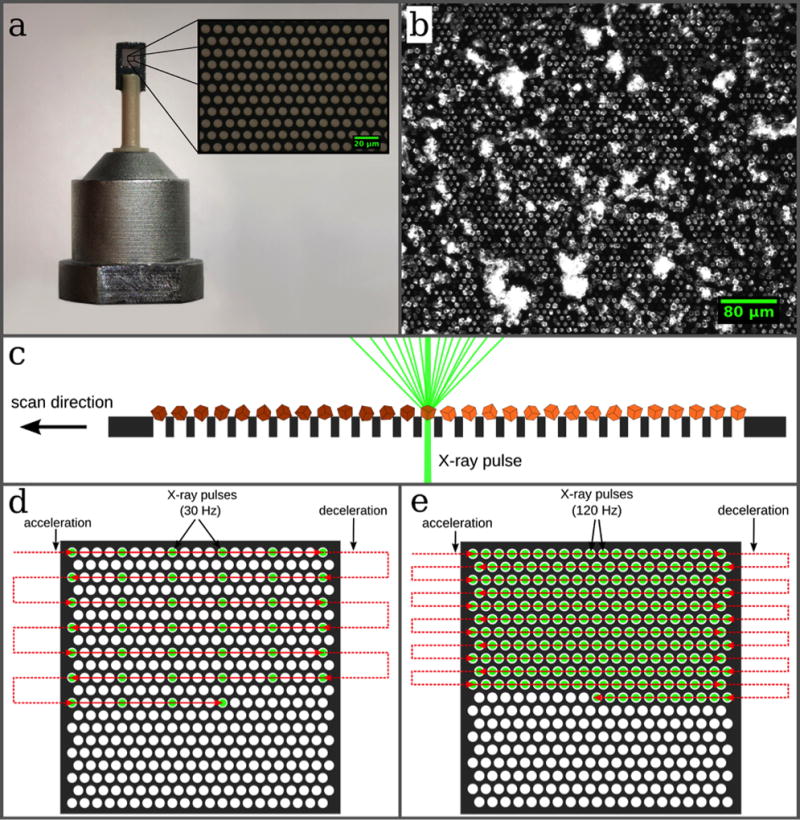
(a) The chip is attached to a plastic rod for the purpose of thermal isolation. The membrane part within the outer frame consists of micropores with diameters of typically 4 µm–8 µm, which are arranged in a triangular grid (a, inset). (b) The chip acts as a sample holder for more than 20,000 microcrystals, which largely organize themselves according to the pore pattern. (c) After loading, the microcrystals are scanned through the X-ray beam. By shooting through the micropores in the chips the interaction of the X-rays with any support material is further minimized. (d, e) Scanning strategies for measurements performed at room temperature and cryogenic temperatures, respectively (see text for details).
To achieve fixed-target data collection rates of 120 Hz we have developed an improved version of the so-called fly-scan. In a conventional fly-scan the sample is accelerated and then moved along a predefined trajectory at constant velocity. This approach would in principle allow data collection at 120 Hz, but in most cases results in lower hit rates than obtained with our method since the X-ray pulses would not necessarily always hit through the pores where the crystals are located. To achieve higher hit rates – and thereby requiring significantly less sample – more precise motion control is required. Such an approach demands, in addition to velocity control, also phase control of the movement of the stages with respect to the arrival of the X-ray pulses. The synchronized movement assures that every X-ray pulse hits through a pore (Fig. 2c–e, also see Methods section, and Supplementary Figure 7).
BEV2 and CPV18 data collection
In the case of BEV2 samples, which were measured at room temperature, diffraction images were collected at an FEL pulse rate of 30 Hz per line, resulting in an effective frame rate of up to 12.2 images/s per chip, when taking the time for line switching into account (Table 1). A maximum hit rate of more than 9% for one chip and on average of about 5% was achieved for BEV2. The relatively low hit rates in this case are due to the fact that the density of the crystals on the chip was low. As only limited amounts of sample were available we aimed at making most efficient use of the available sample instead of optimizing hit rates. For room-temperature data collection, it was not possible to run at the full LCLS frame rate of 120 Hz since crystals in the neighboring compartments were already pre-damaged by the wings of the X-ray beam. By shooting every 4th hole (40 µm separation) in the horizontal direction and every 2nd row in the vertical direction (20 µm separation) no effects of pre-damage were observed (Fig. 2d). With a maximum speed of the horizontal scanning stage of 2.5 mm/s data collection at 60 Hz with 40 µm separation or at 120 Hz with 20 µm separation would have also been possible.
Table 1.
Data collection parameters and hit rates for individual runs. For spot-finding parameters see Supplementary Table 1.
| Run number |
Sample | Temperature Conditions |
Number of images |
Acquisition time [s] |
Effective acquisition rate [images/s] |
Number of hits* |
Hit rate [%] |
Indexed |
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| 294 | BEV2 | 293 K | 2280 | 230.7 | 9.9 | 159 | 7.0 | 124 |
| 296 | BEV2 | 293 K | 180 | 14.8 | 12.2 | 17 | 9.4 | 16 |
| 298 | BEV2 | 293 K | 1950 | 178.0 | 11.0 | 76 | 3.9 | 69 |
| 301 | BEV2 | 293 K | 2015 | 180.7 | 11.2 | 150 | 7.4 | 113 |
| 303 | BEV2 | 293 K | 2387 | 214.4 | 11.1 | 44 | 1.8 | 30 |
| 47 | CPV18 | 100 K | 19028 | 566.1 | 33.6 | 13424 | 70.5 | 16739 |
Images containing equal to or more than 50 (BEV2) and 20 (CPV18) strong spots were considered as a hit, respectively.
For CPV18, data collection was performed at cryogenic temperatures with the full LCLS repetition rate of 120 Hz, resulting in an average data collection rate of 33.6 images/s (Fig. 2e). From these images more than 70% were classified as a hit. In other runs, we were able to achieve hit rates of more than 90%. No pre-damage of the neighboring crystals by the wings of the X-ray beam could be observed for CPV18, which is probably due to reduced diffusion rates of free radicals and the resulting higher radiation tolerance of macromolecular crystals at cryogenic temperatures5–7.
Image quality and background analysis
The diffraction patterns extended to a resolution of 2.3 Å for BEV2 and 2.4 Å for CPV18. An example diffraction image (Fig. 3a) obtained from a BEV2 crystal illustrates the high quality of the diffraction patterns obtained using the Roadrunner goniometer. The low background contribution of the chip results in high-resolution diffraction data with high signal-to-noise ratios. We analyzed the averaged background signal of the measured diffraction images (Fig. 3b) and compared it to that of an SFX experiment where a liquid jet was used for sample delivery (Fig. 3c). The azimuthally averaged background signal as a function of resolution clearly shows that in the diffraction images measured with our fixed-target setup, background is dominated by air scattering, which is most prominent at resolutions lower than 10 Å (Fig. 3d). The chip itself consists of single-crystalline silicon and therefore does not contribute to any background signal. The absence of a water ring for room-temperature data collection reveals the efficient removal of mother liquor during sample loading. In typical liquid jet experiments, the averaged background signal shows a strong water ring around 3.1 Å.
Figure 3. Exemplary BEV2 diffraction pattern and comparison of background scattering levels achievable with different sample delivery methods.
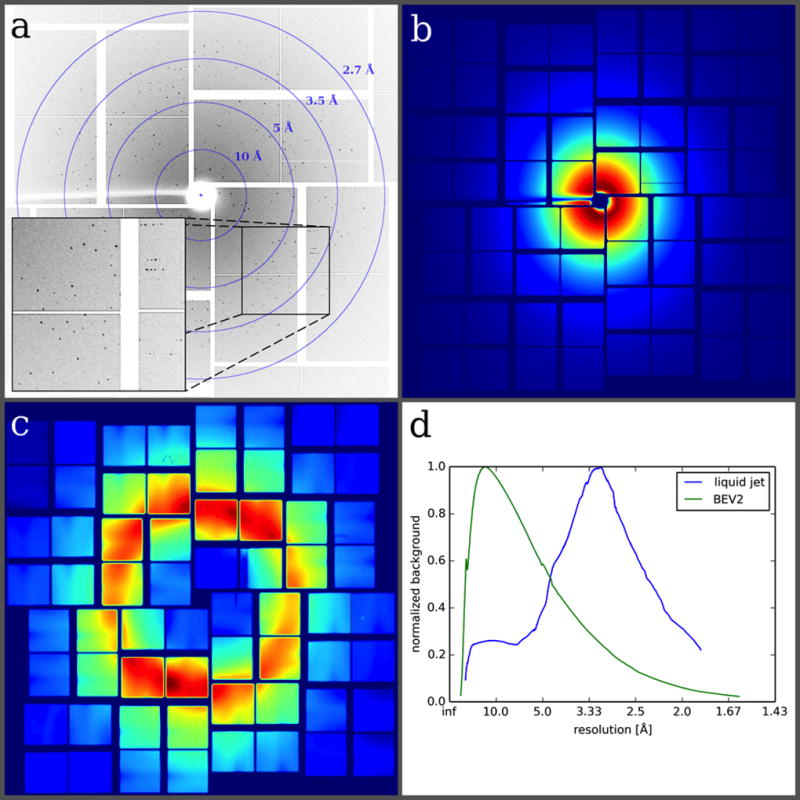
(a) Diffraction image of BEV2 microcrystals obtained at the XPP instrument at LCLS using the micro-patterned silicon chip as a sample holder. (b) Due to the efficient removal of successive mother liquor during sample loading, no water ring is observed in the averaged background image of the chip. (c) For comparison, an averaged background image from a typical SFX liquid jet experiment with CPV 17 crystals47 is shown. (d) The azimuthally averaged radial distribution of both images is plotted as a function of resolution. Both curves are normalized since measurements were performed under different experimental conditions and therefore a direct comparison was not possible.
Structure determination
Summary information regarding data collection and structure refinement for both samples are given in Supplementary Table 2 (PDB codes: 5MQU for BEV2 and 5MQW for CPV18). The structure of BEV2 was solved using diffraction data obtained from 5 chips, from a total of about 446 crystal hits in less than 14 minutes of scanning time. For CPV18 the complete structure could be solved with the data obtained from only one single chip, collected in less than 10 minutes. Unfortunately, the resolution was limited by the dimensions of the detector for the given detector distance in this case. Structure refinement (based on prior models) yielded high-quality electron density maps (Fig. 4) with model R-values of Rwork/Rfree = 23.3/25.7 % for BEV2 (to 2.3 Å, data were measureable to 2.0 Å resolution but the resulting map was only marginally improved and the statistics were significantly worse) and 11.3/14.5 % for CPV18 (to 2.4 Å), respectively.
Figure 4. Overall structure of BEV2 and corresponding high-resolution electron density maps.
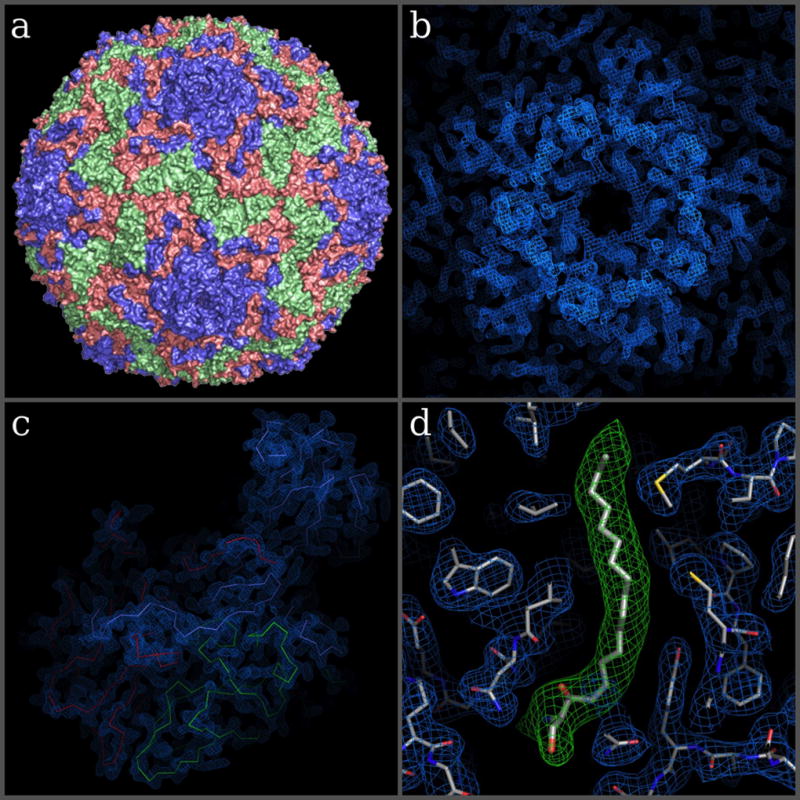
(a) Surface representation of BEV2 particle as viewed towards an icosahedral 2-fold axis. VP1, VP2 and VP3 are shown in blue, green and red, respectively. (b–c) Electron density maps after one cycle of 5-fold real space averaging using the phases calculated from the current refined model showing the electron density around the 5-fold in b and for a biological protomer in c. (c) C-alpha traces of VP1-3, colored as in a. (d) A close-up view of the electron density for protein residues around the pocket factor binding site of VP1 (blue mesh and thinner sticks) and density for the pocket factor (thicker sticks show a sphingosine fitted to the density, while the green density is for a simulated annealing omit map).
The CPV18 data were measured with high redundancy (>100 fold) and were of very high quality (Rsplit 9.2%, CC1/2 0.993), supporting the excellent refinement. In contrast the BEV2 data were derived from only 324 crystals and had a multiplicity of only 2 and the merging statistics were correspondingly poor (Rsplit 0.486%, CC1/2 0.746). We therefore performed two tests to determine if the amplitudes contained enough information to determine the high-resolution structure in the absence of accurate phase information. Firstly, we extended the phases using the 5-fold NCS averaging and solvent flattening from 5 Å to 2.5 Å resolution. The result was an excellent map, with largely successful phase recovery (for details see Methods section and Supplementary Fig. 8). Secondly, we performed molecular replacement starting from a distantly related virus (FMDV type A22, sequence identity 19.5%). As expected the initial map showed significant bias, however this was eliminated by cyclic averaging, again resulting in a high quality electron density map (see Methods and Supplementary Figures 9 and 10).
The BEV2 structure was determined as part of an investigation of the specificity of the hydrophobic pocket in VP1 for different fatty acids. The structure revealed that despite co-crystallising the virus with lauric acid, the pocket factor present in the particles was indistinguishable from that observed in native particles, which is well modelled as sphingosine (Fig. 4d), explaining biophysical observations that lauric acid has essentially no effect on the stability of the virus particles.
The CPV18 structure is similar to recently published structures of isolated crystals of CPV1831,49. A part of the electron density map is shown in Supplementary Figures 11 and 12.
Discussion
To our knowledge, we report the first structure of a virus particle determined at an X-ray Free Electron Laser by means of serial crystallography. The electron density maps obtained for BEV2 provide a high level of detail, sufficient to demonstrate that lauric acid cannot displace sphingosine from the VP1 pocket, which is the major target for the design of anti-enterovirus compounds. We further showed that the method is also applicable to data collection at cryogenic temperatures, where hit rates of more than 70% were achieved and the structure of CPV18 crystals was solved from the measurement of one chip loaded with about 4 µg of protein.
The periodic arrangement of the crystals on our chip in combination with the Roadrunner goniometer allows for very effective use of beam time. With data collection rates of 120 Hz during a line scan, combined with hit rates of more than 70%, we were able to obtain up to 29.6 indexable diffraction patterns per second. Sample loading onto the chip is very efficient and no precious crystalline material is lost. The method is likely more reliable than liquid jet experiments, which often suffer from clogging of the nozzles and settling of the crystals, leading to substantial downtimes during the experiments.
A further benefit of our method is the ultra-low sample consumption, which requires orders of magnitude less sample compared to current liquid jet methods at XFELs and also significantly less compared to room temperature experiments at synchrotrons. A synchrotron structure4 of the apo form of the BEV2 capsid was previously determined at 2.1 Å resolution based on the measurement of 28 crystals of a cubic edge length of about 50 µm, which amounts to a total crystal volume of 3.5 nL. Our work is based on data collected from 446 much smaller crystals with a cubic edge length of only about 8 µm corresponding to a total volume of 228 pL. Co-crystallization of BEV2 with lauric acid limits the achievable crystal size and renders these crystals far too small for a conventional synchrotron structure determination. The obligatory use of the XFEL therefore not only provided the high intensity X-ray pulses required to generate strong enough diffraction to solve the ligand-bound virus structure, but also reduced the total sample amount used for structure factor calculation by 15-fold. It is notable that high-quality phases and hence electron density map could be derived from amplitudes assembled from so little material, and such a low multiplicity XFEL data set assembled from only 324 crystals. We attribute this to the high quality of the data obtained from this experimental setup, to the advances in data processing methods, and in part to the 5-fold non-crystallographic symmetry.
In the current setup the X-ray scattering background is dominated by air scattering from the short remaining beam path in humidified air or cold nitrogen gas, respectively. By further reducing the path of the primary beam in air and by replacing air or nitrogen with helium we aim to significantly reduce the background level in future experiments to achieve higher resolution data from even smaller crystals. A larger chip design with up to 200,000 micro-pores in combination with faster scanning stages will allow longer data collection runs at frame rates of up to 1 kHz, which will result in even more efficient use of beamtime both at XFELs and synchrotron facilities.
Online Methods
A detailed step-by-step protocol of our Method is provided as Supplementary Protocol and also available on the Protocol Exchange from Nature Protocols50.
Sample preparation
Bovine enterovirus 2 light fraction (BEV2) was prepared at the Department of Microbiology, University of Leeds, UK by Prof. D. Rowlands’ group. Particles were purified by ultracentrifugation over a 30% sucrose cushion followed by a 15 – 45% sucrose gradient. Fractions containing BEV2 particles were pooled, incubated with lauric acid and pelleted by ultracentrifugation by James Kelly. To remove excess lauric acid, particles were solubilized in 20 mM HEPES pH 8.0, 200 mM NaCl and 0.5% NP40 at 4°C, and pelleted again by ultracentrifugation using a 30% sucrose cushion. Pellet was solubilized in 20 mM HEPES pH 8.0, 200 mM NaCl. Crystals were grown from the purified concentrated particles by vapor diffusion in Greiner ‘CrystalQuick X’ plates at 293 K. Drops of 100 nL of BEV2 particles (A280 = 5) were mixed with 100 nL drops51 of the precipitant (1.5 M ammonium sulfate, 0.1 M Tris pH 8) and equilibrated against 35 µL of precipitant. More than 100 small crystals, with typical dimensions of 8 × 8 × 8 µm3, appeared in each drop within hours. An image of the BEV2 crystals is shown in Supplementary Figure 13. CPV18 polyhedrin crystals were prepared as described in reference 31.
Chip design and fabrication
The chip design is illustrated in Figure 2. The chips are made from single-crystalline silicon by UV lithography and have overall dimensions of 2.5 × 4 mm2 with a thickness of 0.1 mm. The inner membrane part with an area of 1.5 × 1.5 mm2 is thinned down to a thickness of 10 µm and provides a hexagonal dense pattern of pores with diameters between 4 µm and 8 µm and a 10 µm periodicity (inset Figure 2a). The chips are glued to plastic pins, which can be mounted on conventional magnetic caps routinely used in macromolecular crystallography. More details about chip design, fabrication, loading and handling can be found in reference 31.
Pre-orientation of the chips
With the extremely high X-ray intensity per FEL pulse, Bragg reflections arising from the silicon chip material can easily damage the detector. Hence it is essential to know the exact angular orientation of the chips with respect to the incident X-ray beam in order to avoid these Bragg reflections. As a reference mark, the magnetic caps carrying the chips were modified by removing some material at the lower rim of the caps as shown in Figure 2a. All chips glued to the plastic pins were then oriented and fixed in such a way that the chip surface was always parallel to the face of the magnetic caps.
Sample loading
Sample loading is performed by applying 2–3 µL of crystal suspension to the upper side of the chip. Additional mother liquor is then removed by soaking with a wedge of filter paper attached to the lower side of the chip (see reference 31 for details). The chip allows for data collection at both room temperature (BEV2) and cryogenic temperatures (CPV18). For room-temperature data collection the samples are loaded onto the chips directly at the experimental setup. Similar to recently performed synchrotron experiments32, a humidified gas stream with adjustable relative humidity was used to prevent the crystals from drying out during loading and data collection48,52. For BEV2 data collection the relative humidity was set to 96%. The experimental setup can be also used for data collection at cryogenic temperatures. The major difference is simply the replacement of the humidity stream used for room temperature data collection by a cold nitrogen gas stream. In this case pre-frozen samples on the chips are directly mounted on the goniometer.
Roadrunner goniometer
For the experiment, we have designed a special goniometer, the main part of which is a fast piezo-motor driven x,y translation stage for fast raster scanning of the chips carrying the samples. A technical overview drawing of the Roadrunner goniometer is provided in Supplementary Figure 1. The setup consists of three major components, a high-resolution inline sample-viewing microscope, the high-precision goniometer itself, and a post sample beam pipe unit, all mounted on a common support frame structure. With outer dimensions of 250 mm along the beam direction, a width of 400 mm and a height of 515 mm the entire setup is compact and can be therefore easily installed at different experimental endstations such as XPP (as in the case presented here), the new MFX endstation at LCLS, or other X-ray sources.
The first element in the X-ray beam path, the inline sample-viewing microscope, is shown and described in more detail in Supplementary Figure 3 and the corresponding figure caption. It provides a high-resolution image of the samples mounted on the goniometer and is used for precise alignment of the chips with respect to the X-ray beam. The X-ray beam passes through a molybdenum collimator tube inserted into the hole of the objective lens with an inner diameter of 0.35 mm. The capillary is utilized to prevent X-ray damage to the microscope lenses. It extends to only 3 mm from the sample position to reduce air scattering along the beam path.
The micro-patterned silicon chip carrying the samples is mounted on a high-precision goniometer axis. A technical drawing of the goniometer and a detailed description of is functionality is provided in Supplementary Figure 2. Main element is the high x,y precision stage for scanning of the chips synchronized to the time structure of the X-ray pulses. The x,y scanning stage is controlled by a DMC-4080 motion controller from Galil. The motion controller is capable of synchronizing the two axes of the scanning stage to the repetition rate of the LCLS beam at 120 Hz. The synchronization was done on a line-by-line basis to insure that each X-ray pulse hits the center of the holes in the silicon chip. For each line the motion controller was provided with the starting point, the number of scan points (number of holes), the angular orientation of the line, and the repetition rate of the LCLS beam. The synchronization scheme is illustrated and described in more detail in Supplementary Figure 7. Upon start, the controller moves the scanning stages to a defined position before the first scan point and sends a trigger signal to the LCLS control system. This trigger signal induces a defined sequence of TTL signals to be sent from the LCLS control system to the motion controller to allow the scanning stage to reach a constant speed and the position of the chip pores to be in phase with the arrival of the X-ray pulses at the pre-defined beam position. Once the starting point of the grid is reached, the pulse picker opens and the X-rays are hitting the crystals located in the pores of the chip. At each scan point the current position is read out by the controller and any error is instantaneously injected into the control loop and compensated for in order to prevent accumulation of the errors. After a predefined number of pulses (equal to the number of pores selected) has been reached, the pulse picker closes and the scanner decelerates before switching to the next line of the chip. This sequence is repeated for all rows of the chip in a meander-scan like manner.
After interacting with the sample the direct undiffracted beam is guided into a beam pipe, which is shown and described in Supplementary Figure 4. By enclosing the beam in a beam pipe all X-rays scattered by air are absorbed in the walls of the tube and thereby do not contribute to background scattering on the detector.
Roadrunner control system and software
Alignment of the goniometer setup, control of individual motors, pre-alignment of the individual chips, definition of the scan grid, and data collection is controlled by a custom software written in the Python programming language with the underlying control system TANGO. The software provides a Graphical User Interface (GUI) for easy and efficient operation. A screenshot of the Roadrunner GUI together with a more detailed functionality of the software is provided in Supplementary Figure 6. Style and functionality of the GUI are adapted from GUIs typically used at protein crystallography beamlines. The software is available for free download (http://doi.org/10.5281/zenodo.571598).
Data collection
Measurements were conducted on the XPP instrument at the Linac Coherent Light Source (LCLS) at SLAC under experiment number xpph9015. An X-ray energy of 9.5 keV was chosen for the experiment as it provides a good compromise between detector efficiency and pulse intensity on one hand and X-ray absorption by the silicon chips on the other. The X-ray beam size at the sample was 3 × 3 µm2. X-ray pulse energies were attenuated to 40% of the full flux. A photograph of the Roadrunner setup installed at the XPP instrument at LCLS is shown in Supplementary Figure 5.
For measurements performed at room temperature, it was observed that, with the aforementioned procedure, only the first shot of each line yielded useful diffraction data, probably due to pre-damage of the subsequent crystals by the wings of the X-ray beam. It was therefore necessary to increase the displacement of subsequent pulses in order to prevent damage of the crystals caused by the previous X-ray pulse. For this reason the beam shutter was used to chop the repetition rate of the laser to 30 Hz, shooting only every 4th hole in the chip (Figure 2d). In addition, during the scan only every 2nd line of the chip membrane was scanned. In this way an effective acquisition rate of up to 12.2 images/s was achieved for room-temperature measurements.
At cryogenic temperatures in each line the chip was translated with a speed of 1.2 mm/s so that the displacement of subsequent pulses matched the distance between two neighboring holes in the chip (Figure 2e). This way a maximum data acquisition rate of 120 Hz could be achieved within a line. After the end of a line was reached, the chip moved to the next line and scanned in reverse direction. This allowed scanning of the entire chip membrane with about 19,000 collected detector frames in less than 10 minutes, resulting in an effective data acquisition rate of 33.6 images/s (see Table 1).
Data evaluation/structure refinement
Diffraction images considered hits were isolated from the XTC streams using cctbx.xfel and passed into the data processing pipeline cppxfel53. Technical difficulties with indexing the BEV2 diffraction patterns drove the development of a new indexing algorithm called TakeTwo54. Due to the increased indexing rate from this algorithm, the TakeTwo algorithm was used to index both the BEV2 and CPV18 samples. Integration, initial orientation matrix refinement and post-refinement were carried out similarly to previous publications47,53. Geometry was refined initially with cctbx.xfel and then further refined using the geometry algorithm in cppxfel using the spot predictions from the indexing solutions and the nearest peak pixel value. For BEV2, a 2 × 2 foreground integration window was used to match the spot size, and care was taken to ensure the background subtraction region did not overlap with a neighboring spot. After geometry refinement, the accuracy of spot prediction allowed interpolation between pixels to be used. After post-refinement, the BEV2 data were reintegrated with the updated orientation matrix to more accurately predict the spot positions. For CPV18, the integration window was 5 × 5 due to the larger spot size.
For the BEV2 samples 446 detector frames out of 8,812 collected images from 5 different chips were classified as possible hits (see Table 1). Out of these, 352 indexed diffraction patterns could be obtained, of which 324 diffraction patterns were included in the final dataset and used for structure refinement. To test whether the amplitudes were sufficiently accurate and complete to support phase determination, an initial map was calculated from phase information using the known BEV coordinates truncated to 5 Å. Density modification, non-crystallographic symmetry (NCS) averaging and gradual phase extension was performed from 5 Å to 2.5 Å, providing a good interpretable map with clear side chain density (Supplementary Fig. 8, which also shows the relationship between the phases derived from phase extension and those derived from an averaged map obtained from the synchrotron data). The atomic coordinates were rebuilt into the map derived from phase extension to remove bias from the model reported by Axford et al.4, using CNS55 with strict NCS constraints. The resulting BEV2 electron densities are shown in Figure 4. In a second test phases derived from the suitably placed capsid of FMDV A22 were combined with amplitudes from BEV2 (to a resolution of 2.3 Å). The level of sequence identity between the capsid protein of these two viruses was only 19.5%. This map was then refined by cyclic density modification and NCS averaging (Supplementary Figures 9 and 10).
For the CPV18 sample, 13,424 diffraction images out of 19,028 collected images were regarded as hits, all from one single chip. Images were indexed using the multiple lattice version of the TakeTwo algorithm, producing 16,739 indexing solutions. Up to 5 diffraction patterns could be indexed on a single image due to multiple hits (Supplementary Fig. 14). Finally, 9,293 patterns were included in the final dataset. For structure refinement of CPV18, phases were introduced from PDB code 4OTS as a template file and the structure was refined using Phenix56. Further data evaluation details are summarized in Supplementary Table 2. Part of the electron density map for CPV18 is shown in Supplementary Figures 11 and 12.
Statistics
Statistical measures addressing the quality of the two refined structures are provided in Supplementary Table 2.
Supplementary Material
Acknowledgments
We thank LCLS staff for their support during experiment LH90. Use of the Linac Coherent Light Source (LCLS), SLAC National Accelerator Laboratory, is supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences under Contract No. DE-AC02-76SF00515. This work was further supported by the European Cluster of Advanced Laser Light Sources (EUCALL) and the Virtual Institute VH-VI-403 of the Helmholtz Association. H. M. Ginn was supported by the Wellcome Trust (studentship 075491/04). D. I. Stuart was supported by the Medical Research Council, grant MR/N00065X/1 and previously G1000099. N. K. Sauter was supported by US National Institutes of Health grants GM102520 and GM117126 for data processing methods. We further thank J. Bergtholdt from DESY for preparation of the technical drawings of the Roadrunner setup.
Footnotes
Author Contributions
P.R., T.P., P.F., J.M., A.W., and A.M. designed the experiment. P.R., G.S., K.H., T.S.W., R.D., I.V., M.W., A.K., J.K., and D.J.R. were involved in sample preparation. P.R., T.P., G.S., K.H., J.M., P.F., R.D., B.R., M.S., S.N., D.S.D., R.A.M., C.D., A.W., and A.M. participated in data collection. P.R., H.M.G., A.S.B., I.D.Y., T.M.C., N.K.S., J.R., E.E.F., and D.I.S. analyzed the data. P.R., H.M.G., D.I.S., A.W., and A.M. wrote the manuscript.
Competing Financial Interests Statement
A. Meents is one of the CEOs and shareholder of the DESY spin-off company Suna-Precision GmbH. T. Pakendorf and B. Reime are further shareholders. Suna-Precision sells technical equipment for experiments with X-rays including different micro-structured silicon chips for serial crystallography experiments. I. Vartiainen is shareholder and CTO of the company FinnLitho, which produced the silicon chips used for the experiment.
Accession Codes: Solved structures were deposited in the Protein Data Bank (PDB) under PDB IDs 5MQU and 5MQW for BEV2 and CPV18, respectively. Source data for Figure 3d is provided as a separate data file.
Code Availability: The Roadrunner control software is available under doi 10.5281/zenodo.571598 (direct link: http://doi.org/10.5281/zenodo.571598).
References
- 1.Zocher G, et al. A Sialic Acid Binding Site in a Human Picornavirus. PLoS Pathog. 2014;10:e1004401. doi: 10.1371/journal.ppat.1004401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fry EE, Abrescia NGA, Stuart DI. In: Macromolecular Crystallography: conventional and high-throughput methods. Sanderson MR, Skelly JV, editors. 2007. pp. 245–264. [Google Scholar]
- 3.Fry EE, Grimes J, Stuart DI. Virus crystallography. Mol. Biotechnol. 1999;12:13–23. doi: 10.1385/MB:12:1:13. [DOI] [PubMed] [Google Scholar]
- 4.Axford D, et al. In situ macromolecular crystallography using microbeams. Acta Crystallogr. Sect. D Biol. Crystallogr. 2012;68:592–600. doi: 10.1107/S0907444912006749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hope H. Cryocrystallography of biological macromolecules: a generally applicable method. Acta Crystallogr. Sect. B Struct. Sci. Cryst. Eng. Mater. 1988;44:22–26. doi: 10.1107/s0108768187008632. [DOI] [PubMed] [Google Scholar]
- 6.Owen RL, Rudiño-Piñera E, Garman EF. Experimental determination of the radiation dose limit for cryocooled protein crystals. Proc. Natl. Acad. Sci. U. S. A. 2006;103:4912–4917. doi: 10.1073/pnas.0600973103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meents A, Gutmann S, Wagner A, Schulze-Briese C. Origin and temperature dependence of radiation damage in biological samples at cryogenic temperatures. Proc. Natl. Acad. Sci. U. S. A. 2010;107:1094–1099. doi: 10.1073/pnas.0905481107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chapman HN, et al. Femtosecond X-ray protein nanocrystallography. Nature. 2011;470:73–77. doi: 10.1038/nature09750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boutet S, et al. High-resolution protein structure determination by serial femtosecond crystallography. Science. 2012;337:362–364. doi: 10.1126/science.1217737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Redecke L, et al. Natively inhibited Trypanosoma brucei cathepsin B structure determined by using an X-ray laser. Science. 2013;339:227–30. doi: 10.1126/science.1229663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ayyer K, et al. Macromolecular diffractive imaging using imperfect crystals. Nature. 2016;530:202–206. doi: 10.1038/nature16949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Neutze R, Wouts R, van der Spoel D, Weckert E, Hajdu J. Potential for biomolecular imaging with femtosecond X-ray pulses. Nature. 2000;406:752–757. doi: 10.1038/35021099. [DOI] [PubMed] [Google Scholar]
- 13.Barty A, et al. Self-terminating diffraction gates femtosecond X-ray nanocrystallography measurements. Nat. Photonics. 2011;6:35–40. doi: 10.1038/nphoton.2011.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kupitz C, et al. Serial time-resolved crystallography of photosystem II using a femtosecond X-ray laser. Nature. 2014;513:261–265. doi: 10.1038/nature13453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tenboer J, et al. Time-resolved serial crystallography captures high-resolution intermediates of photoactive yellow protein. Science. 2014;346:1242–1246. doi: 10.1126/science.1259357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Perera R, Khaliq M, Kuhn RJ. Closing the door on flaviviruses: Entry as a target for antiviral drug design. Antiviral Res. 2008;80:11–22. doi: 10.1016/j.antiviral.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Connolly SA, Jackson JO, Jardetzky TS, Longnecker R. Fusing structure and function: a structural view of the herpesvirus entry machinery. Nat. Rev. Microbiol. 2011;9:369–381. doi: 10.1038/nrmicro2548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harrison JS et al. Role of Electrostatic Repulsion in Controlling pH-Dependent Conformational Changes of Viral Fusion Proteins. Structure. 2013;21:1085–1096. doi: 10.1016/j.str.2013.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weierstall U, et al. Lipidic cubic phase injector facilitates membrane protein serial femtosecond crystallography. Nat. Commun. 2014;5:3309. doi: 10.1038/ncomms4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.DePonte DP. Gas dynamic virtual nozzle for generation of microscopic droplet streams. J. Phys. D. Appl. Phys. 2008;41:195505. [Google Scholar]
- 21.Caffrey M, Cherezov V, et al. Crystallizing membrane proteins using lipidic mesophases. Nat. Protoc. 2009;4:706–731. doi: 10.1038/nprot.2009.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu W, et al. Serial Femtosecond Crystallography of G Protein-Coupled Receptors. Science. 2013;342:1521–1524. doi: 10.1126/science.1244142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fromme R, et al. Serial femtosecond crystallography of soluble proteins in lipidic cubic phase. IUCrJ. 2015;2:545–551. doi: 10.1107/S2052252515013160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sugahara M, et al. Grease matrix as a versatile carrier of proteins for serial crystallography. Nat. Methods. 2015;12:61–3. doi: 10.1038/nmeth.3172. [DOI] [PubMed] [Google Scholar]
- 25.Sugahara M, et al. Oil-free hyaluronic acid matrix for serial femtosecond crystallography. Sci. Rep. 2016;6:24484. doi: 10.1038/srep24484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Conrad CE, et al. A novel inert crystal delivery medium for serial femtosecond crystallography. IUCrJ. 2015;2:421–30. doi: 10.1107/S2052252515009811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lawrence RM, et al. Serial femtosecond X-ray diffraction of enveloped virus microcrystals. Struct. Dyn. 2015;2:041720. doi: 10.1063/1.4929410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zarrine-Afsar A, et al. Crystallography on a chip. Acta Crystallogr. Sect. D Biol. Crystallogr. 2012;68:321–323. doi: 10.1107/S0907444911055296. [DOI] [PubMed] [Google Scholar]
- 29.Hunter MS, et al. Fixed-target protein serial microcrystallography with an x-ray free electron laser. Sci. Rep. 2014;4:6026. doi: 10.1038/srep06026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mueller C, et al. Fixed target matrix for femtosecond time-resolved and in situ serial micro-crystallography. Struct. Dyn. 2015;2:054302. doi: 10.1063/1.4928706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roedig P, et al. A micro-patterned silicon chip as sample holder for macromolecular crystallography experiments with minimal background scattering. Sci. Rep. 2015;5:10451. doi: 10.1038/srep10451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roedig P, et al. Room-temperature macromolecular crystallography using a micro-patterned silicon chip with minimal background scattering. J. Appl. Crystallogr. 2016;49:968–975. doi: 10.1107/S1600576716006348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oghbaey S, et al. Fixed target combined with spectral mapping: approaching 100% hit rates for serial crystallography. Acta Crystallogr. Sect. D Biol. Crystallogr. 2016;72:944–955. doi: 10.1107/S2059798316010834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cohen AE, et al. Goniometer-based femtosecond crystallography with X-ray free electron lasers. Proc. Natl. Acad. Sci. U. S. A. 2014;111:17122–7. doi: 10.1073/pnas.1418733111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murray TD, et al. A high-transparency, micro-patternable chip for X-ray diffraction analysis of microcrystals under native growth conditions. Acta Crystallogr. Sect. D Biol. Crystallogr. 2015;71:1987–97. doi: 10.1107/S1399004715015011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baxter EL, et al. High-density grids for efficient data collection from multiple crystals. Acta Crystallogr. Sect. D Biol. Crystallogr. 2016;72:2–11. doi: 10.1107/S2059798315020847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Owen RL, et al. Low-dose fixed-target serial synchrotron crystallography. Acta Crystallogr. Sect. D Struct. Biol. 2017;73:373–378. doi: 10.1107/S2059798317002996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sherrell DA, et al. A modular and compact portable mini-endstation for high-precision, high-speed fixed target serial crystallography at FEL and synchrotron sources. J. Synchrotron Radiat. 2015;22:1372–1378. doi: 10.1107/S1600577515016938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smyth M, et al. Preliminary crystallographic analysis of bovine enterovirus. J. Mol. Biol. 1993;231:930–932. doi: 10.1006/jmbi.1993.1339. [DOI] [PubMed] [Google Scholar]
- 40.Smyth M, et al. Implications for viral uncoating from the structure of bovine enterovirus. Nat. Struct. Biol. 1995;2:224–31. doi: 10.1038/nsb0395-224. [DOI] [PubMed] [Google Scholar]
- 41.Goens SD, Botero S, Zemla A, Zhou CE, Perdue ML. Bovine enterovirus 2: complete genomic sequence and molecular modelling of a reference strain and a wild-type isolate from endemically infected US cattle. J. Gen. Virol. 2004;85:3195–203. doi: 10.1099/vir.0.80159-0. [DOI] [PubMed] [Google Scholar]
- 42.Filman DJ, et al. Structural factors that control conformational transitions and serotype specificity in type 3 poliovirus. EMBO J. 1989;8:1567–79. doi: 10.1002/j.1460-2075.1989.tb03541.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Coulibaly F, et al. The molecular organization of cypovirus polyhedra. Nature. 2007;446:97–101. doi: 10.1038/nature05628. [DOI] [PubMed] [Google Scholar]
- 44.Ji X, et al. How baculovirus polyhedra fit square pegs into round holes to robustly package viruses. EMBO J. 2010;29:505–14. doi: 10.1038/emboj.2009.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Anduleit K, et al. Crystal lattice as biological phenotype for insect viruses. Protein Sci. 2005;14:2741–2743. doi: 10.1110/ps.051516405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ji X, et al. Polyhedra structures and the evolution of the insect viruses. J. Struct. Biol. 2015;192:88–99. doi: 10.1016/j.jsb.2015.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ginn HM, et al. Structure of CPV17 polyhedrin determined by the improved analysis of serial femtosecond crystallographic data. Nat. Commun. 2015;6:6435. doi: 10.1038/ncomms7435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sanchez-Weatherby J, et al. Improving diffraction by humidity control: A novel device compatible with X-ray beamlines. Acta Crystallogr. Sect. D Biol. Crystallogr. 2009;65:1237–1246. doi: 10.1107/S0907444909037822. [DOI] [PubMed] [Google Scholar]
- 49.Axford D, Ji X, Stuart DI, Sutton G. In cellulo structure determination of a novel cypovirus polyhedrin. Acta Crystallogr. Sect. D Biol. Crystallogr. 2014;70:1435–41. doi: 10.1107/S1399004714004714. [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods References
- 50.Roedig P, et al. Sample Preparation and Data Collection for High-Speed Fixed-Target Serial Femtosecond Crystallography. Protocol Exchange. 2017 doi: 10.1038/protex.2017.059. [DOI] [Google Scholar]
- 51.Walter TS, et al. A procedure for setting up high-throughput nanolitre crystallization experiments. Crystallization workflow for initial screening, automated storage, imaging and optimization. Acta Crystallogr. Sect. D Biol. Crystallogr. 2005;61:651–657. doi: 10.1107/S0907444905007808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wheeler MJ, Russi S, Bowler MG, Bowler MW. Measurement of the equilibrium relative humidity for common precipitant concentrations: Facilitating controlled dehydration experiments. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2012;68:111–114. doi: 10.1107/S1744309111054029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ginn HM, Evans G, Sauter NK, Stuart DI. On the release of cppxfel for processing X-ray free-electron laser images. J. Appl. Crystallogr. 2016;49:1065–1072. doi: 10.1107/S1600576716006981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ginn HM, et al. TakeTwo : an indexing algorithm suited to still images with known crystal parameters. Acta Crystallogr. Sect. D Biol. Crystallogr. 2016;72:956–965. doi: 10.1107/S2059798316010706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brunger AT. Version 1.2 of the Crystallography and NMR system. Nat. Protoc. 2007;2:2728–2733. doi: 10.1038/nprot.2007.406. [DOI] [PubMed] [Google Scholar]
- 56.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


