Abstract
Recent advances in the molecular characterization of cancers has triggered interest in developing a new taxonomy of disease in oncology with the goal of using the molecular profile of a patient’s tumor to predict response to treatment. Image-guided needle biopsy is central to this “Precision Medicine” effort. In this review, we first discuss the current role of biopsy in relation to clinical examples of molecular medicine. We then outline important bottlenecks to the advancement of precision medicine and highlight the potential role of image guided biopsy to address these challenges.
Keywords: Molecular medicine, image-guided needle biopsy, “precision-medicine”
Introduction
Over the past decade a tremendous amount of effort has been invested into translational research programs geared toward the advancement of “molecular medicine” (or “personalized medicine” or “precision medicine”). Broadly speaking, the goal of this research is to define a new taxonomy of disease, in which patients receive tailored treatments based on specific molecular characteristics of their tumor(1). Whether meritorious or perilous (2), there is little doubt regarding plans for continued investment. In 2015 President Obama announced a research initiative “that aims to accelerate progress toward a new era of precision medicine…[driven by] extensive characterization of biologic specimens (cell populations, proteins, metabolites, RNA, and DNA)”(3). Accurate and informative tissue biopsies are essential in this new era.
In oncology, biopsies are used to establish diagnosis of malignancy, identify tumor histology, and confirm presence of metastases for staging(4). With the advent of advanced imaging and image-guidance technologies, there has been an increase in the number of biopsies, largely performed by radiologists (5). In the era of molecular medicine, the role of image-guided percutaneous biopsies is now expanding as a growing compendium of molecular assays and biomarkers are used to determine prognosis, to predict treatment response, and to detect disease progression or treatment resistance. Biopsies are integral to clinical trials, particularly adaptive clinical trials that attempt to study drug effects and identify relevant biomarkers (6). Recognizing this tremendous growth, the Society of Interventional Radiology (SIR) recently convened a research consensus panel comprised of a multidisciplinary group of experts to address how the field should evolve in the coming years(7). In this article we first review the current utility of image-guided biopsy in relation to molecular medicine and then discuss some unresolved issues and open questions facing the community.
The current role of image guided biopsy in molecular medicine
Biopsy in the era of molecular medicine: success stories
Advances in molecular medicine over the past 15 years represent a paradigm shift in oncology. One of the most striking examples of this is in lung cancer. In 2002, the Eastern Cooperative Oncology Group published results of a large randomized study comparing four different chemotherapy regimens in patients with advanced non-small-cell lung cancer.(8) The authors reported that none of the regimens provided an advantage over the others with abysmal response rates (19%) and median survival (7.9 months). Response rates of drugs that targeted epidermal growth factor receptor (EGFR) were known to produce dramatic responses, but were also limited to a small percentage of patients. However, it was soon recognized that these responses were seen in patients that carried activating EGFR mutations in their tumors(9, 10). Patients receiving EGFR inhibitors with biopsy-proven EGFR mutations demonstrated much higher response rates compared with traditional chemotherapy(11). Just three years later, investigators identified a subset of lung tumors with chromosomal aberrations in the anaplastic lymphoma kinase gene (ALK) (12). Targeted therapies with ALK inhibitors also improved response rates in this patient population.(13, 14) Over the course of the past decade, the classification of lung adenocarcinomas has moved from histology-based to clinically relevant molecular subsets (Figure 1). Biopsies for somatic mutation testing to guide patient management has become routine practice (15).
Figure 1.
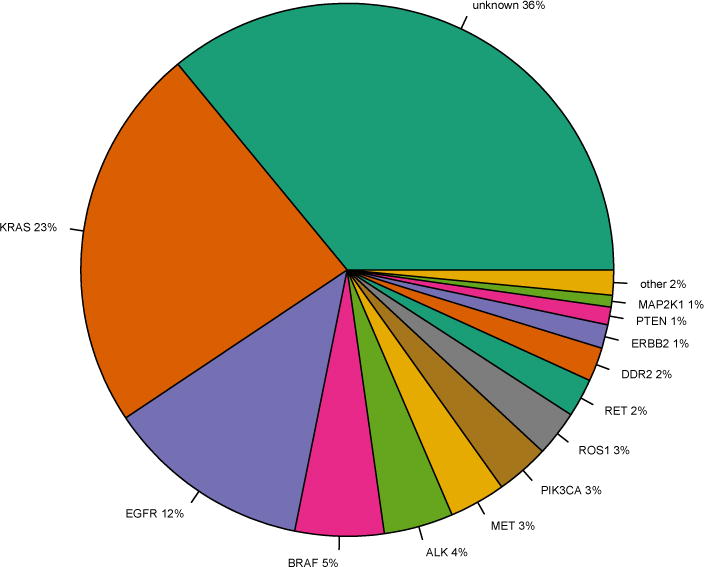
Driver mutation profile of lung adenocarcinomas. Generated using data from cbio-portal based on 5 studies of lung adenocarcinoma(65). Targeted small molecule inhibitors are either approved or in development for all of the above mutations (66). “Other” category includes NRAS, FGFR1, and AKT1 which represent <1% of driver mutations.
Examples of molecular prognostic and predictive biomarkers abound in other cancer types. In patients with breast cancer, erb-b2 receptor tyrosine kinase 2 (ERBB2) amplification (or protein overexpression) has long been known to be associated with poor prognosis (16). These patients frequently respond to trastuzumab, an antibody that blocks the ERBB2 receptor (17). Breast cancer tissue ERBB2 testing is now commonly performed alongside estrogen and progesterone receptor status assessment, and multiple diagnostic multi-gene assays are used to predict recurrence (18, 19). Patients with melanoma are tested for BRAF V600E mutations to identify potential responders to vemurafenib (20). Patients with colorectal cancers are examined for KRAS (and possibly other downstream effectors of the EGFR signaling pathway) to determine candidates for EGFR inhibitors (21–23). Metastatic bladder cancer patients with TSC1 mutations show durable remission when treated with everolimus (24). BRCA mutation carriers with various solid tumors show response to PARP inhibitors.(25) In some cases of rare and understudied tumors, treatment options may be derived from the presence of common mutations (26). Histologically unclassifiable cancers, or those without a known primary site, may be particularly amenable to such a strategy (27). Over-expression of programmed death ligand-1 (PD-L1) in tumor cells is associated with objective response in patients treated Anti-PD-1 antibody, suggesting a role for biopsy to select patients best suited for immunotherapy.(28)
Clinical trials and biopsies
Promising exemplars have spurred many centers to develop and institute multiplexed genomic profiling with the intent of expediting relevant clinical trial enrollment (29). Indeed, the role of biopsies has expanded with more frequent incorporation of biomarker studies into clinical trials.(30), (31, 32) Predictive biomarkers offer the opportunity to streamline drug development by (a) enabling smaller trial designs and (b) identifying biologically based targets (6). Serial biopsies of the same or different tumors may be obtained throughout the course of treatment for these purposes (33).
Interventional oncology procedures
In the field of interventional oncology, there is a growing interest in identifying both prognostic and predictive markers of treatment response. Predictors of loco-regional cancer treatment efficacy, such as after tumor embolization or ablation, have traditionally focused on technical variables such as ablation margin and tumor size (34, 35). However, paralleling a shift toward molecularly guided systemic treatments, several investigators have recently focused on molecular biomarkers of local tumor response. For example, KRAS status prior to lung adenocarcinoma ablation was an independent predictor of local recurrence (36); thus prior knowledge of mutation status could influence decision-making regarding thermal ablation size, or possibly result in triage to a different treatment modality. Similar advances in biomarker discovery are being pursued with colorectal liver metastases where KRAS mutation status was found to predict poor prognosis after radio-embolization(37). Gaba et al, recently reported hepatocellular carcinomas with complete response to trans-arterial chemoembolization demonstrated upregulation in genes associated with pre-treatment chemotherapy-sensitivity and mitosis (38), although they did not report any correction for multiple testing. Further studies such as these are certain to shed light on the role of biomarkers in interventional oncology therapies. In the near future, compendiums of tissue biopsy profiles linked to patient outcomes are likely to guide best practices in oncology.
Histopathology
Conventional histopathology remains central to tumor classification.(39) For example, some cancers share mutations but respond differently to the same targeted therapy. In melanoma patients, BRAF V600E mutations are common and demonstrate excellent response rates to BRAF inhibitors. Yet, colorectal cancer patients with the same mutation show poor response rates (40). Another example is seen in the lung cancer literature. In 2011, the International Association for the Study of Lung Cancer, the American Thoracic Society, and the European Respiratory Society proposed a new classification system for lung adenocarcinoma.(41) Invasive lung adenocarcinoma tumor was subdivided into lepidic, acinar, papillary, micropapillary, solid, colloid, and invasive mucinous adenocarcinoma. Multiple studies in the surgery literature have since validated the prognostic utility of this classification with respect to recurrence patterns and post-recurrence survival, confirming the role of micropapillary and solid histologic subtypes as predictors of poor prognosis and high local recurrence even in completely resected early-stage lung adenocarcinomas(42, 43). The molecular correlates of these subtypes remain poorly understood, underscoring the continued relevance of histology.
Open questions and challenges
Despite the advances in molecular characterization of tumors, there are several unresolved issues that represent critical bottlenecks to the advancement of molecular medicine (Table 1).
Table 1.
| Bottlenecks to the advancement of molecular medicine | Relationship to image-guided biopsy |
|---|---|
|
| |
| Sample Quality | Standardization |
| Heterogeneity | Spatial and temporal sampling |
| Tumor Resistance | Molecular imaging guidance |
| Assessing Response | Radio-genomics |
| Liquid Biopsy | Correlative studies |
Specimen quality
A large phase II precision medicine trial was recently paused after an interim analysis showed only 87% of cases submitted completed tumor testing, largely due to poor sample quality.(44) Biopsy specimens were inadequate for testing in 26% of cases in a multicenter study of driver mutations in lung adenocarcinomas.(45) Given the patient risk and financial cost of biopsy, specimen quality and sufficiency are essential to the oncology community.
Currently there are no standard guidelines for tissue acquisition and preparation. Variables in tissue acquisition include technique (fine needle aspiration, core biopsy), needle gauge, number of samples, spatial sampling method within a tumor, number of sites, site location, primary vs metastasis. Formalin-fixed paraffin-embedded is the most widely used tissue preparation method and multiple molecular methods for DNA, RNA, and protein extraction have been developed.(46) However, formalin induced degradation, especially for RNA and protein is problematic. Additionally, standard decalcification methods in bone effects the quantity and quality of nucleic acids in the specimen.(47) Cellular and genetic preservation may become even more critical with the emergence of single cell analytic platforms.(48) Biopsy samples vary in tumor quantity; DNA, RNA, and protein quantity vary by organ and tumor. These issues may in part be offset by manual microdissection(46), but potentially important information about the tumor stroma may be ignored. Rapid on-site evaluation (ROSE) of imprint cytology or core biopsy touch preparation has been shown to improve diagnostic adequacy rate, (49) although it remains unclear whether ROSE can improve adequacy with respect to molecular characterization.
Heterogeneity
Tumor heterogeneity represents an important challenge for personalized medicine. Heterogeneity may be spatial within a single tumor, or occur across metastatic sites and time points. Gerlinger et al demonstrated that single biopsy specimens revealed only a minority of genetic aberrations when compared to the entire tumor(50) in renal cancer. Multiple biopsy sampling improved mutation detection, depending upon mutation prevalence.(51) In a cohort of lung adenocarcinomas, single-region sampling was sufficient to identify the majority of known cancer mutations(52). The role of spatial heterogeneity may therefore be dependent on the temporal evolution of the individual tumor. Differences in tumor grade between primary sites and synchronous/metachronous metastases have been reported in neuroendocrine tumors.(53) Studies systematically examining tumor heterogeneity are likely to inform treatment approaches in the coming years.
Assessing Resistance
Excitement regarding targeted therapies has been tempered by the inevitable development of treatment resistance.(54) For example, most lung cancer patients with activating EGFR mutations have median responses of approximately one year before therapeutic efficacy wanes. Resistance is attributed to a T790M point mutation in exon 20 in approximately one-half of such cases.(55) Several newer EGFR inhibitors are in development to overcome this resistance, commonly prompting re-biopsy to identify the T790M mutation.(56) Since resistance may only be seen in spatially localized subclonal populations in a tumor, mutationally representative tissue sampling remains a formidable challenge. Techniques that facilitate increase needle biopsy targeting specificity, such as real-time or multi-modal PET/CT fusion imaging, may be beneficial in this regard (7).
Assessing tumor response
The effectiveness of targeted therapies is generally evaluated using the standardized imaging-based Response Evaluation Criteria in Solid Tumors (RECIST). Fernandes et al contend that tumor size based response criteria do not directly measure what they define as meaningful progression (local invasion and metastasis) and should therefore be replaced (57). The authors’ position is in part informed by the now well-established view of cancer as not a disease of abnormal cells, but a disease of abnormal cells within a tumor microenvironment (58). Quantitative and molecular imaging are offered as potential alternatives to assess tumor response, especially in the setting of targeted (non-cytotoxic) therapies (59–61). Correlative studies of imaging features with molecular changes via biopsies (e.g. radiogenomics (62)) will likely play an important role in this transition.
Liquid biopsies
Many investigators are seeking to develop biomarker analytics platforms based on blood samples rather than percutaneous tissue biopsies. So-called “liquid biopsies” denote the capture and extraction of circulating tumor cells (CTCs) or fragments of DNA (ctDNA). Since these methods are minimally invasive compared to other sampling alternatives, they have several potential advantages including lower patient risk and cost (7). Successful liquid biopsy technologies would simplify serial assessment of tumor response or acquired resistance. At the European Lung Cancer Conference (ELCC) 2016, several authors presented the feasibility of liquid biopsy to predict benefit in patients with lung adenocarcinoma. In one study, there was high concordance between T790M positive plasma and T790M tissue biopsies and in a second study there was high correlation between T790M-positive plasma and response rates to T790M-targeting EGFR inhibitor.(63, 64) However, unstable molecules such as RNA and protein are less likely to be reliably extracted and low volume disease may not be detectable. The fraction of ctDNA in a sample limits the sensitivity of such approaches. The previously discussed issue of tumor heterogeneity may also be relevant here, as it is unclear whether hematogenous tumor elements will be sufficiently representative to guide effective treatments. At least some of these issues may be addressed with systematic correlative studies between image guided biopsies and liquid biopsies.
Summary
We have reviewed the central role of image-guided biopsy in the era of molecular medicine. We have also highlighted several bottlenecks to the advancement of the field and which represent challenges and opportunities for interventional oncologists participating in image-guided biopsies.
References
- 1.National Research Council Committee on AFfDaNToD. Toward Precision Medicine: Building a Knowledge Network for Biomedical Research and a New Taxonomy of Disease. Washington (DC): National Academies Press (US) National Academy of Sciences; 2011. The National Academies Collection: Reports funded by National Institutes of Health. [PubMed] [Google Scholar]
- 2.Joyner MJ, Paneth N, Ioannidis JP. What Happens When Underperforming Big Ideas in Research Become Entrenched? Jama. 2016 doi: 10.1001/jama.2016.11076. [DOI] [PubMed] [Google Scholar]
- 3.Collins FS, Varmus H. A New Initiative on Precision Medicine. New England Journal of Medicine. 2015;372(9):793–5. doi: 10.1056/NEJMp1500523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marshall D, Laberge JM, Firetag B, Miller T, Kerlan RK. The changing face of percutaneous image-guided biopsy: molecular profiling and genomic analysis in current practice. J Vasc Interv Radiol. 2013;24(8):1094–103. doi: 10.1016/j.jvir.2013.04.027. [DOI] [PubMed] [Google Scholar]
- 5.Kwan SW, Bhargavan M, Kerlan RK, Jr, Sunshine JH. Effect of advanced imaging technology on how biopsies are done and who does them. Radiology. 2010;256(3):751–8. doi: 10.1148/radiol.10092130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bradley E. Incorporating biomarkers into clinical trial designs: points to consider. Nat Biotechnol. 2012;30(7):596–9. doi: 10.1038/nbt.2296. [DOI] [PubMed] [Google Scholar]
- 7.Tam AL, Lim HJ, Wistuba II, Tamrazi A, Kuo MD, Ziv E, et al. Image-Guided Biopsy in the Era of Personalized Cancer Care: Proceedings from the Society of Interventional Radiology Research Consensus Panel. J Vasc Interv Radiol. 2016;27(1):8–19. doi: 10.1016/j.jvir.2015.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schiller JH, Harrington D, Belani CP, Langer C, Sandler A, Krook J, et al. Comparison of Four Chemotherapy Regimens for Advanced Non–Small-Cell Lung Cancer. New England Journal of Medicine. 2002;346(2):92–8. doi: 10.1056/NEJMoa011954. [DOI] [PubMed] [Google Scholar]
- 9.Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350(21):2129–39. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 10.Pao W, Miller V, Zakowski M, Doherty J, Politi K, Sarkaria I, et al. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci U S A. 2004;101(36):13306–11. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mitsudomi T, Morita S, Yatabe Y, Negoro S, Okamoto I, Tsurutani J, et al. Gefitinib versus cisplatin plus docetaxel in patients with non-small-cell lung cancer harbouring mutations of the epidermal growth factor receptor (WJTOG3405): an open label, randomised phase 3 trial. The Lancet Oncology. 2010;11(2):121–8. doi: 10.1016/S1470-2045(09)70364-X. [DOI] [PubMed] [Google Scholar]
- 12.Soda M, Choi Yl, Fau - Enomoto M, Enomoto M, Fau - Takada S, Takada S, Fau - Yamashita Y, Yamashita Y, Fau - Ishikawa S, Ishikawa S, Fau - Fujiwara S-i, et al. Identification of the transforming EML4-ALK fusion gene. non-small-cell lung cancer. 1476–4687 (Electronic) [Google Scholar]
- 13.Kwak EL, Bang Yj, Fau - Camidge DR, Camidge Dr, Fau - Shaw AT, Shaw At, Fau - Solomon B, Solomon B, Fau - Maki RG, Maki Rg, Fau - Ou S-HI, et al. Anaplastic lymphoma kinase inhibition. non-small-cell lung cancer. 1533–4406 (Electronic) [Google Scholar]
- 14.Shaw AT, Kim Dw, Fau - Nakagawa K, Nakagawa K, Fau - Seto T, Seto T, Fau - Crino L, Crino L, Fau - Ahn M-J, Ahn Mj, Fau - De Pas T, et al. Crizotinib versus chemotherapy in advanced ALK-positive lung cancer. 1533–4406(Electronic) [Google Scholar]
- 15.Kris MG, Johnson BE, Berry LD, et al. USing multiplexed assays of oncogenic drivers in lung cancers to select targeted drugs. JAMA. 2014;311(19):1998–2006. doi: 10.1001/jama.2014.3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Slamon Dj, Fau - Clark GM, Clark Gm, Fau - Wong SG, Wong Sg, Fau - Levin WJ, Levin Wj, Fau - Ullrich A, Ullrich A, Fau - McGuire WL, McGuire WL. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. doi: 10.1126/science.3798106. (0036-8075 (Print) [DOI] [PubMed] [Google Scholar]
- 17.Vogel CL, Cobleigh Ma, Fau - Tripathy D, Tripathy D, Fau - Gutheil JC, Gutheil Jc, Fau - Harris LN, Harris Ln, Fau - Fehrenbacher L, Fehrenbacher L, Fau - Slamon DJ, et al. Efficacy and safety of trastuzumab as a single agent in first-line treatment of HER2-overexpressing metastatic breast cancer. (0732-183X (Print) [Google Scholar]
- 18.van ‘t Veer LJ, Dai H, Fau - van de Vijver MJ, van de Vijver Mj, Fau - He YD, He Yd, Fau - Hart AAM, Hart Aa, Fau - Mao M, Mao M, Fau - Peterse HL, et al. Gene expression profiling predicts clinical outcome of breast cancer. (0028-0836(Print) [Google Scholar]
- 19.Paik S, Shak S, Fau - Tang G, Tang G, Fau - Kim C, Kim C, Fau - Baker J, Baker J, Fau - Cronin M, Cronin M, Fau - Baehner FL, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. doi: 10.1056/NEJMoa041588. 1533–4406(Electronic) [DOI] [PubMed] [Google Scholar]
- 20.Flaherty KT, Puzanov I, Kim KB, Ribas A, McArthur GA, Sosman JA, et al. Inhibition of Mutated, Activated BRAF in Metastatic Melanoma. New England Journal of Medicine. 2010;363(9):809–19. doi: 10.1056/NEJMoa1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Amado RG, Wolf M, Fau - Peeters M, Peeters M, Fau - Van Cutsem E, Van Cutsem E, Fau - Siena S, Siena S, Fau - Freeman DJ, Freeman Dj, Fau - Juan T, et al. Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. 1527–7755(Electronic) [Google Scholar]
- 22.Barault L, Veyrie N, Jooste V, Lecorre D, Chapusot C, Ferraz JM, et al. Mutations in the RAS-MAPK, PI(3)K (phosphatidylinositol-3-OH kinase) signaling network correlate with poor survival in a population-based series of colon cancers. Int J Cancer. 2008;122(10):2255–9. doi: 10.1002/ijc.23388. [DOI] [PubMed] [Google Scholar]
- 23.De Roock W, Claes B, Bernasconi D, De Schutter J, Biesmans B, Fountzilas G, et al. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: a retrospective consortium analysis. Lancet Oncol. 2010;11(8):753–62. doi: 10.1016/S1470-2045(10)70130-3. [DOI] [PubMed] [Google Scholar]
- 24.Iyer G, Hanrahan Aj, Fau - Milowsky MI, Milowsky Mi, Fau - Al-Ahmadie H, Al-Ahmadie H, Fau - Scott SN, Scott Sn, Fau - Janakiraman M, Janakiraman M, Fau - Pirun M, et al. Genome sequencing identifies a basis for everolimus sensitivity. 1095–9203(Electronic) [Google Scholar]
- 25.Fong PC, Boss DS, Yap TA, Tutt A, Wu P, Mergui-Roelvink M, et al. Inhibition of Poly(ADP-Ribose) Polymerase in Tumors from BRCA Mutation Carriers. New England Journal of Medicine. 2009;361(2):123–34. doi: 10.1056/NEJMoa0900212. [DOI] [PubMed] [Google Scholar]
- 26.Collazo-Lorduy A, Castillo-Martin M, Wang L, Patel V, Iyer G, Jordan E, et al. Urachal Carcinoma Shares Genomic Alterations with Colorectal Carcinoma and May Respond to Epidermal Growth Factor. Inhibition. doi: 10.1016/j.eururo.2016.04.037. LID - S0302-2838(16)30147-6 [pii] LID - 10.1016/j.eururo.2016.04.037 [doi]. 1873–7560(Electronic) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ross JS, Wang K, Gay L, et al. Comprehensive genomic profiling of carcinoma of unknown primary site: New routes to targeted therapies. JAMA Oncology. 2015;1(1):40–9. doi: 10.1001/jamaoncol.2014.216. [DOI] [PubMed] [Google Scholar]
- 28.Topalian SL, Hodi FS, Brahmer JR, Gettinger SN, Smith DC, McDermott DF, et al. Safety, Activity, and Immune Correlates of Anti–PD-1 Antibody in Cancer. New England Journal of Medicine. 2012;366(26):2443–54. doi: 10.1056/NEJMoa1200690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hyman DM, Solit DB. Tumor Genetic Screening Programs: A Call to Action. doi: 10.1200/JCO.2015.61.9296. 1527–7755(Electronic) [DOI] [PubMed] [Google Scholar]
- 30.Basik M, Aguilar-Mahecha A, Rousseau C, Diaz Z, Tejpar S, Spatz A, et al. Biopsies: next-generation biospecimens for tailoring therapy. Nat Rev Clin Oncol. 2013;10(8):437–50. doi: 10.1038/nrclinonc.2013.101. [DOI] [PubMed] [Google Scholar]
- 31.Sikorski R, Yao B. Parallel Paths to Predictive Biomarkers in Oncology: Uncoupling of Emergent Biomarker Development and Phase III Trial Execution. Science Translational Medicine. 2009;1(10):10p. doi: 10.1126/scitranslmed.3000287. s1-ps1. [DOI] [PubMed] [Google Scholar]
- 32.Antoniou M, Jorgensen AL, Kolamunnage-Dona R. Biomarker-Guided Adaptive Trial Designs in Phase II and Phase III: A Methodological Review. PLoS ONE. 2016;11(2):e0149803. doi: 10.1371/journal.pone.0149803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barker AD, Sigman Cc, Fau - Kelloff GJ, Kelloff Gj, Fau - Hylton NM, Hylton Nm, Fau - Berry DA, Berry Da, Fau - Esserman LJ, Esserman LJ. I-SPY 2: an adaptive breast cancer trial design in the setting of neoadjuvant chemotherapy. doi: 10.1038/clpt.2009.68. 1532–6535(Electronic) [DOI] [PubMed] [Google Scholar]
- 34.de Baere T, Palussiere J, Auperin A, Hakime A, Abdel-Rehim M, Kind M, et al. Midterm local efficacy and survival after radiofrequency ablation of lung tumors with minimum follow-up of 1 year: prospective evaluation. Radiology. 2006;240(2):587–96. doi: 10.1148/radiol.2402050807. [DOI] [PubMed] [Google Scholar]
- 35.Okuma T, Matsuoka T, Yamamoto A, Oyama Y, Hamamoto S, Toyoshima M, et al. Determinants of local progression after computed tomography-guided percutaneous radiofrequency ablation for unresectable lung tumors: 9-year experience in a single institution. Cardiovasc Intervent Radiol. 2010;33(4):787–93. doi: 10.1007/s00270-009-9770-9. [DOI] [PubMed] [Google Scholar]
- 36.Ziv E, Erinjeri JP, Yarmohammadi H, Boas FE, Petre EN, Gao S, et al. Lung Adenocarcinoma: Predictive Value of KRAS Mutation Status in Assessing Local Recurrence in Patients Undergoing Image-guided Ablation. Radiology. 2016:160003. doi: 10.1148/radiol.2016160003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lahti SJ, Xing M, Zhang D, Lee JJ, Magnetta MJ, Kim HS. KRAS Status as an Independent Prognostic Factor for Survival after Yttrium-90 Radioembolization Therapy for Unresectable Colorectal Cancer Liver Metastases. J Vasc Interv Radiol. 2015;26(8):1102–11. doi: 10.1016/j.jvir.2015.05.032. [DOI] [PubMed] [Google Scholar]
- 38.Gaba RC, Groth JV, Parvinian A, Guzman G, Casadaban LC. Gene expression in hepatocellular carcinoma: pilot study of potential transarterial chemoembolization response biomarkers. J Vasc Interv Radiol. 2015;26(5):723–32. doi: 10.1016/j.jvir.2014.12.610. [DOI] [PubMed] [Google Scholar]
- 39.Dietel M, Johrens K, Laffert MV, Hummel M, Blaker H, Pfitzner BM, et al. A 2015 update on predictive molecular pathology and its role in targeted cancer therapy: a review focussing on clinical relevance. Cancer Gene Ther. 2015;22(9):417–30. doi: 10.1038/cgt.2015.39. [DOI] [PubMed] [Google Scholar]
- 40.Barras D. BRAF Mutation in Colorectal Cancer: An Update. Biomark Cancer. 2015;7(Suppl 1):9–12. doi: 10.4137/BIC.S25248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Travis WD, Brambilla E, Noguchi M, Nicholson AG, Geisinger KR, Yatabe Y, et al. International association for the study of lung cancer/american thoracic society/european respiratory society international multidisciplinary classification of lung adenocarcinoma. J Thorac Oncol. 2011;6(2):244–85. doi: 10.1097/JTO.0b013e318206a221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nitadori J, Bograd AJ, Kadota K, Sima CS, Rizk NP, Morales EA, et al. Impact of micropapillary histologic subtype in selecting limited resection vs lobectomy for lung adenocarcinoma of 2cm or smaller. J Natl Cancer Inst. 2013;105(16):1212–20. doi: 10.1093/jnci/djt166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ujiie H, Kadota K, Chaft JE, Buitrago D, Sima CS, Lee MC, et al. Solid Predominant Histologic Subtype in Resected Stage I Lung Adenocarcinoma Is an Independent Predictor of Early, Extrathoracic, Multisite Recurrence and of Poor Postrecurrence Survival. J Clin Oncol. 2015;33(26):2877–84. doi: 10.1200/JCO.2015.60.9818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Conley BA, Doroshow JH. Molecular analysis for therapy choice: NCI MATCH. doi: 10.1053/j.seminoncol.2014.05.002. 1532–8708(Electronic) [DOI] [PubMed] [Google Scholar]
- 45.Sholl LM, Aisner Dl, Fau - Varella-Garcia M, Varella-Garcia M, Fau - Berry LD, Berry Ld, Fau - Dias-Santagata D, Dias-Santagata D, Fau - Wistuba II, Wistuba Ii, Fau - Chen H, et al. Multi-institutional Oncogenic Driver Mutation Analysis in Lung Adenocarcinoma: The Lung Cancer Mutation Consortium Experience. doi: 10.1097/JTO.0000000000000516. 1556-1380(Electronic) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dietel M, Johrens K, Fau - Laffert M, Laffert M, Fau - Hummel M, Hummel M, Fau - Blaker H, Blaker H, Fau - Muller BM, Muller Bm, Fau - Lehmann A, et al. Predictive molecular pathology and its role in targeted cancer therapy: a review focussing on clinical relevance. doi: 10.1038/cgt.2013.13. 1476–5500(Electronic) [DOI] [PubMed] [Google Scholar]
- 47.Singh VM, Salunga Rc, Fau - Huang VJ, Huang Vj, Fau - Tran Y, Tran Y, Fau - Erlander M, Erlander M, Fau - Plumlee P, Plumlee P, Fau - Peterson MR, et al. Analysis of the effect of various decalcification agents on the quantity and quality of nucleic acid (DNA and RNA) recovered from bone biopsies. doi: 10.1016/j.anndiagpath.2013.02.001. 1532–8198(Electronic) [DOI] [PubMed] [Google Scholar]
- 48.Van Loo P, Voet T. Single cell analysis of cancer genomes. doi: 10.1016/j.gde.2013.12.004. 1879–0380(Electronic) [DOI] [PubMed] [Google Scholar]
- 49.Schmidt RL, Witt Bl, Fau - Lopez-Calderon LE, Lopez-Calderon Le, Fau - Layfield LJ, Layfield LJ. The influence of rapid onsite evaluation on the adequacy rate of fine-needle aspiration cytology: a systematic review and meta-analysis. doi: 10.1309/AJCPEGZMJKC42VUP. 1943–7722(Electronic) [DOI] [PubMed] [Google Scholar]
- 50.Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, et al. Intratumor Heterogeneity and Branched Evolution Revealed by Multiregion Sequencing. New England Journal of Medicine. 2012;366(10):883–92. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sankin A, Hakimi Aa, Fau - Mikkilineni N, Mikkilineni N, Fau - Ostrovnaya I, Ostrovnaya I, Fau - Silk MT, Silk Mt, Fau - Liang Y, Liang Y, Fau - Mano R, et al. The impact of genetic heterogeneity on biomarker development in kidney cancer assessed by multiregional sampling. doi: 10.1002/cam4.293. 2045–7634(Electronic) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang J, Fujimoto J, Zhang J, Wedge DC, Song X, Zhang J, et al. Intratumor heterogeneity in localized lung adenocarcinomas delineated by multiregion sequencing. doi: 10.1126/science.1256930. 1095–9203(Electronic) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Grillo F, Fau - Albertelli M, Albertelli M, Fau - Brisigotti MP, Brisigotti Mp, Fau - Borra T, Borra T, Fau - Boschetti M, Boschetti M, Fau - Fiocca R, Fiocca R, Fau - Ferone D, et al. Grade Increases in Gastroenteropancreatic Neuroendocrine Tumor Metastases Compared to the Primary Tumor. doi: 10.1159/000439434. 1423-0194(Electronic) [DOI] [PubMed] [Google Scholar]
- 54.Lovly CM, Shaw AT. Molecular pathways: resistance to kinase inhibitors and implications for therapeutic strategies. doi: 10.1158/1078-0432.CCR-13-1610. 1078-0432(Print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nguyen KS, Neal JW, Wakelee H. Review of the current targeted therapies for non-small-cell lung cancer. doi: 10.5306/wjco.v5.i4.576. 2218–4333(Electronic) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Arcila ME, Oxnard Gr, Fau - Nafa K, Nafa K, Fau - Riely GJ, Riely Gj, Fau - Solomon SB, Solomon Sb, Fau - Zakowski MF, Zakowski Mf, Fau - Kris MG, et al. Rebiopsy of lung cancer patients with acquired resistance to EGFR inhibitors and enhanced detection of the T790M mutation using a locked nucleic acid-based assay. doi: 10.1158/1078-0432.CCR-10-2277. 1078-0432(Print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fernandes M, Rosel D, Brabek J. Translation in solid cancer: are size-based response criteria an anachronism? Clin Transl Oncol. 2015;17(1):1–10. doi: 10.1007/s12094-014-1207-5. [DOI] [PubMed] [Google Scholar]
- 58.Allen M, Louise Jones J. Jekyll and Hyde: the role of the microenvironment on the progression of cancer. doi: 10.1002/path.2803. 1096–9896(Electronic) [DOI] [PubMed] [Google Scholar]
- 59.Serkova NJ. Translational imaging endpoints to predict treatment response to novel targeted anticancer agents. doi: 10.1016/j.drup.2011.04.004. 1532–2084(Electronic) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mountz JM, Yankeelov TE, Rubin DL, Buatti JM, Erikson BJ, Fennessy FM, et al. Letter to cancer center directors: Progress in quantitative imaging as a means to predict and/or measure tumor response in cancer therapy trials. J Clin Oncol. 2014;32(19):2115–6. doi: 10.1200/JCO.2014.55.8866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.James ML, Gambhir SS. A molecular imaging primer: modalities, imaging agents, and applications. Physiol Rev. 2012;92(2):897–965. doi: 10.1152/physrev.00049.2010. [DOI] [PubMed] [Google Scholar]
- 62.Kuo MD, Yamamoto S. Next generation radiologic-pathologic correlation in oncology: Rad-Path 2.0. AJR Am J Roentgenol. 2011;197:990–7. doi: 10.2214/AJR.11.7163. [DOI] [PubMed] [Google Scholar]
- 63.Jenkins S, Yang J, Ramalingam S, Yu K, Patel R, Weston S, et al., editors. European Lung Cancer Conference. Geneva, Switzerland: 2016. Plasma ctDNA analysis for detection of EGFR T790M mutation in patients (pts) with EGFR mutation-positive advanced non-small cell lung cancer (aNSCLC) [Google Scholar]
- 64.Oxnard G, Thress K, Alden R, Lawrance R, Paweletz C, Cantarini M, et al., editors. European Lung Cancer Conference. Geneva, Switzerland: 2016. Plasma genotyping for predicting benefit from osimertinib in patients (pts) with advanced NSCLC. [Google Scholar]
- 65.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):l1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lovly CM, Horn L, Pao W. Molecular Profiling of Lung Cancer. 2016 [updated March 28, 2016] Available from: https://www.mycancergenome.org/content/disease/lung-cancer/


