Abstract
Cardiac fibrosis remains an important health concern, but the study of fibroblast biology has been hindered by a lack of effective means for identifying and tracking fibroblasts. Recent advances in fibroblast-specific lineage tags and reporters have permitted a better understanding of these cells. After injury multiple cell types have been implicated as the source for extracellular matrix producing cells, but emerging studies suggest that resident cardiac fibroblasts contribute substantially to the remodeling process. In this review, we discuss recent findings regarding cardiac fibroblast origin and identity. Our understanding of cardiac fibroblast biology and fibrosis is still developing and will expand profoundly in the next few years, with many of the recent findings regarding fibroblast gene expression and behavior laying down the groundwork for interpreting the purpose and utility of these cells before and after injury.
Keywords: Cardiac fibroblast, fibrosis, myofibroblast, cardiovascular remodeling
Cardiac fibrosis occurs after heart injury, inflammation, or during aging. The accumulation of extracellular matrix (ECM) results in stiffening of the heart and decreased cardiac function1. Based on its known role in ECM production, the principle cell type implicated in the fibrotic remodeling process is the cardiac fibroblast. Although defining this cell and its behavior is essential for developing approaches to reduce the adverse effects of fibroblast activation, there is still much ambiguity regarding its origin, function, gene expression, and signaling pathways. In this review, we will focus on recent studies that shed light on the nature of these cells and provide data that fibroblast origins and gene expression may not be as diverse as previously thought. In addition, we outline new mechanisms for studying these cells. The overall goal is to establish a consensus for identifying and describing resident cardiac fibroblast behaviors in the hopes of discovering signaling pathways for controlling fibroblast activities in pathological situations. The majority of the described studies focus on the cardiac fibroblast population in the mouse, but conservation between human and murine heart biology suggests that findings in the mouse may pertain to human fibrosis and remodeling2,3.
Cardiac fibroblast identity
Definition by function
Typically, a cardiac fibroblast is defined as a cell that produces connective tissue. Unlike the connective tissue of bone and tendon, which is organized into regular patterns of collagen4, heart ECM is dense, irregular, and composed of collagens, proteoglycans, and glycoproteins5,6. Heart structural components that are produced by fibroblasts include periostin, vimentin, fibronectin, and collagen types I, III, V, and VI (reviewed by Snider7). Although fibroblasts are considered the predominant manufacturer of these proteins, several other cell types in the heart can also express these ECM components (Table 1). Basing cell categorization on dynamically and stress-induced genes is a primary difficulty in defining and studying the fibroblast.
Table 1.
Published markers for cardiac fibroblasts.
| Method of Detection | E12.5 | E14.5 – E18.5 | Adult | Injury Phase | Reference(s) | ||
|---|---|---|---|---|---|---|---|
| Nuclear | WT1 | GT, IHC | Ep, CM | Ep, EC, CM | Ep, ECLow | Ep, P/V, EC | 18, 31, 114, 115, 118 |
| Tcf21 | GT, IHC | Ep | F, Ep | F, Ep | F, Ep | 7, 19, 33, 53, 54, 116 | |
| Cytosollic | FSP1 | GT, IHC | NE | F*, P/V | F*, P/V, EC, IC | F*, P/V, IC, EC | 7, 117, 26, 44, 53, 118 |
| Prolyl-4-hydroxylase | IHC | F, EC | F, IC | 26, 107, 117, 119 | |||
| Cytoskeletal | Vimentin | IHC | En | F, P/V | F, P/V, EC | F, P/V | 7, 21, 33, 53, 117, 120, 121 |
| αSMA | GT, IHC | Ep, CM | P/V, CM | P/V | F*, P/V | 26, 33, 53, 118, 122 | |
| Cell membrane | PDGFRα | GT, IHC | Ep | F | F, CPC | F | 21, 26, 33, 40, 53, 105, 123, 124 |
| MEFSK4 | FC | F, P/VLow, IC | F, P/VLow, IC | 40 | |||
| DDR2 | FC, IHC | Ep | F*, P/V, En | F, P/V | F, P/V | 7, 41, 43, 53, 124–127 | |
| CD90 | FC, IHC | F*, P/V, IC, EC | F*, P/V, IC, EC | F*, P/V, IC, EC | 40, 42, 53, 118, 121, 128 | ||
| Sca1 | FC, GT, IHC | NE | F*, CPC | F*, CPC | 40, 45, 105, 129, 130 | ||
| Extracellular | Periostin | GT, IHC | Ep | F, P/V | NE | F* | 7, 33, 41, 44, 53, 70, 117 |
| Fibronectin | IHC | F, EC | F, EC | F* | 7, 33, 46, 118 | ||
| ED-A fibronectin | IHC | NE | NE | F*, EC | 117, 131, 132 | ||
| Collagen type I | GT, IHC | Ep | F, P/V | F, P/V | F, EC, CM | 7, 21, 33, 41, 53, 58, 70 | |
| Collagen type III | IHC | F, P/V | F, P/V | F, P/V | 7, 41, 133 | ||
| FAP | FC, IHC | F | 51, 117 | ||||
Adding to the confusion of understanding the cardiac fibroblast is the use of many different terms including: fibrocyte8,9, telocyte10, myofibroblast11, protomyofibroblast12, mesenchymal cell, and stromal cell. Each of these categories reflects a definition that varies depending on the author and demonstrates a lack of consensus regarding these cells. For the purpose of this review we will refer to fibroblasts in an uninjured heart as resting fibroblasts and in an injured heart as activated fibroblasts. We use these terms to be inclusive of the various fibroblast populations. Fortunately, recent studies have provided a refined view of the resident cardiac fibroblast and demonstrate that these cells are responsive to injury and are likely the dominant producer of ECM.
Definition by origin
Developmental biologists suggested many years ago that cardiac fibroblasts have a distinct embryonic origin13. Specifically, data in the avian system demonstrated that the epicardium undergoes the process of epithelial-to-mesenchymal transition (EMT) and contributes to cardiac fibroblasts and vascular smooth muscle cells (VSMC)13–17 (Figure 1). With the discovery of epicardium specific genes, such as WT118, Tcf2119 and Tbx1820, these initial observations have recently been reconfirmed using lineage-tracing methods in the mouse19–22. Lineage tracing is a heritable method of tagging cells that permits the later identification of the original cell and its progeny23. With the advent of new mouse lines that permit the genetic manipulation of fibroblasts24, investigators have elucidated several important findings.
Figure 1.
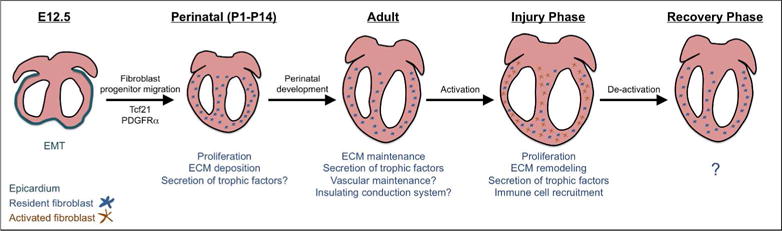
Murine cardiac fibroblast stages and function. Cardiac fibroblasts are derived from epicardial and endocardial progenitors after embryonic day 12.5 (E12.5). Tcf21 and PDGFRα direct fibroblast development at this stage. Fibroblast progenitors enter the ventricles and proliferate in the first week after birth. It is at this time that fibroblasts begin to deposit and degrade extracellular matrix. In the uninjured adult heart, proposed roles for fibroblasts include secretion of trophic factors, ECM surveillance, conduction system insulation, cardiomyocyte electrical coupling, and vascular maintenance. During the injury phase, fibroblasts proliferate, deposit ECM, and recruit inflammatory cells. Recent data has shown that after the proliferative phase of injury, previously activated cardiac fibroblasts can revert to a resting fibroblast gene profile. EMT; epithelial-to-mesenchymal transition.
First was the discovery that cardiac fibroblasts develop from two origins rather than one. Two independent groups found that populations of fibroblasts residing in the interventricular septum and right ventricle do not form from the epicardium but instead have an endothelial origin, constituting roughly 20% of the myocardial resident fibroblasts25,26. Second, in contrast to being a stochastically determined cell population, recent findings demonstrate that differentiation of the cardiac fibroblast requires specific signals. We found that two unrelated genes are essential for cardiac fibroblast formation. Disruption of expression of either Tcf21, a bHLH transcription factor, or PDGFRα, a receptor tyrosine kinase, results in loss of epicardial-derived ventricular fibroblasts19,21. In the absence of either of these two genes, not only is there a lack of fibroblasts, but also the expression of ECM components in the left ventricle is disrupted. These data suggest that there are no alternative sources for fibroblasts during developmental stages. It remains to be determined if these same genes impact the developmental program of endocardial-derived fibroblasts. Additionally, these data demonstrate that the cardiac fibroblast is not a default lineage and that EMT-derived fibroblast progenitors are the dominant cell type resulting in the resident cardiac fibroblast population.
The use of genetic marking systems to follow the resident cardiac fibroblast lineage after injury has also cast doubt on the likelihood that circulating27,28, endothelial29, hematopoietic9, or epicardial30–32 cells contribute significantly to fibrotic remodeling after injury25,26,33. Following pressure overload, the endocardial- and epicardial-derived fibroblasts appeared to respond similarly in regard to gene expression and proliferation. Additionally, the combination of these two fibroblast populations accounted for nearly 100% of the matrix producing cells26,34. In one instance, a Col1a1 transgenic reporter mouse34 was used to identify the ECM producing cell population26. Cell lineage tracing techniques, bone marrow chimeras, and parabiosis also failed to identify a significant contribution of other cell lineages to the expanding fibroblast population25,26. Although both of the aforementioned studies utilized pressure overload injury to induce fibrosis, similar results have also discounted the contribution of other cell types to the fibroblast pool after myocardial infarction and catecholamine induced fibrosis33. These studies used lineage tracing to investigate the contribution of endothelial, VSMCs, and hematopoietic cells to the expanding, activated fibroblast population and found little to no evidence for these cell types giving rise to fibroblasts. These experiments also demonstrated that the majority of the responding matrix producing cells were resident fibroblasts33. Taken together, these recent publications suggest that resident fibroblasts account for the majority of activated fibroblasts that respond to injury in the mouse heart.
Cardiac fibroblast gene expression
Heterogeneity
Historically, the fibroblast population has been considered heterogeneous based on protein expression, cell size, ability to proliferate after activation, and developmental origin35–39. Until recently, fibroblast studies have been hindered by the lack of means for identification in vivo, which required the use of in vitro culture. The markers initially available to study cardiac fibroblasts (CD90, Sca1, and αSMA) are indeed differentially expressed by fibroblasts26,40,41 leading to the suggestion that resting fibroblasts are an amalgam of cell populations. These ideas were reinforced by the notion that activated fibroblasts also derive from disparate cell types. The new tools available to study fibroblast biology have demonstrated that the fibroblast population may not be as diverse as previously thought. For example, even though cardiac fibroblasts come from two different developmental origins, gene expression analyses observed overlapping genetic profiles when comparing fibroblasts of the two origins either in uninjured or pressure overload conditions. In sham hearts, fibroblasts genes such as Col1a1, Col1a2, and PDGFRα were expressed at similar levels between the two populations of fibroblasts26. After injury, ECM and growth factor expression increases were observed when compared to sham, but there was no significant difference between the two fibroblast types25,26. Recent single cell analyses of fibroblasts have also demonstrated comparable profiles of gene expression (up-regulation of Postn, αSMA, Adam12, Lox, Wisp1, and DDR2) after activation33,41.
Markers for cardiac fibroblasts
In the past, the markers most often used to identify fibroblasts were CD90 (or Thy1)42, discoidin domain receptor 2 (DDR2)43, fibroblast specific protein 1 (FSP1)44, Sca145, fibronectin46, vimentin47, and collagen types I and III48–50 (Table 1). Even though FSP1, fibroblast activating protein (FAP)51, and the fibronection splice variant ED-A52 are upregulated during cardiac fibrosis, αSMA53 has been the most commonly used marker for activated fibroblasts.
As mentioned earlier, developmental studies have revealed that transcription factor Tcf21 and receptor tyrosine kinase PDGFRα are required during fibroblast formation and continue to be expressed in adult fibroblasts25,26,54. Possibly because Tcf21 is a transcription factor, it is often difficult to detect by IHC. PDGFRα expression, on the other hand, is readily detectable by IHC but recognizes rare stem cell populations in the heart55. Unfortunately, PDGFRα antibodies designed for flow cytometric applications are not robust for cardiac fibroblasts40. Recently, we have described a commercially available monoclonal antibody, MEFSK4, which identifies an antigen expressed by PDGFRα+, Col1a1+ murine, cardiac fibroblasts. One drawback to use of this antibody is that it also recognizes surface antigens on pericytes and granulocytes40. Periostin, an ECM protein expressed developmentally by cardiac fibroblasts but not adult resting fibroblasts, is highly upregulated after a variety of injuries and appears to be a distinguishing marker for activated fibroblasts7,33,41,44.
The use of markers such as collagen, fibronectin, and periostin stem from the functional definition of fibroblasts. As these proteins are secreted, IHC identification of cells expressing these markers can be technically difficult and subjective. Additionally, many ECM proteins are expressed in multiple cell types. For example, collagen can also be expressed by valve interstitial cells, VSMCs, and pericytes56–58.
Cytoskeletal and surface markers, such as vimentin, FSP1, Sca1, CD90 and DDR2, are not secreted and thus can be used to identify fibroblasts directly by IHC or flow cytometry. Unfortunately, these markers are not specific and require exclusion of non-fibroblast cell populations (Table 1). For example, FSP1, originally thought to be fibroblast specific, is found in a limited number of cardiac fibroblasts and is expressed by immune cells26. After both pressure overload and myocardial infarction, FSP1 expression broadens and is expressed by VMSCs and endothelial cells26,44.
Some previously used fibroblast markers, such as CD90 and Sca1, have been recently reevaluated with newly developed fibroblast tools, and these proteins appear to be expressed in a subset of cardiac fibroblasts40 (Table 1). Therefore, previous analyses with these markers may have underestimated the resident fibroblasts. The expression of αSMA, previously the gold standard for identifying activated fibroblasts, has also been reevaluated. Investigators found that αSMA staining identified about 15% of fibroblasts after TAC26 and 35% of fibroblasts after angiotensin II treatment in lesional areas41. Therefore, studies using αSMA expression as the sole readout for activated fibroblasts may have underrepresented the activated fibroblast population.
Genetic tools
Mouse lines such as PDGFRαGFP 59 and Collagen1a1-GFP 34 are one avenue for reliably observing the resident cardiac fibroblast population (Table 1). These lines have been used to further characterize and validate fibroblast markers26,33,40. Fibroblast specific inducible Cre mouse lines such as Tcf21mCrem 60, PeriostinmCrem 33, and PeriostinCreERT241 provide unique opportunities for tracing and genetically manipulating resident and activated fibroblasts. Given the described heterogeneity of fibroblasts, it is surprising that the PDGFRαGFP, Collagen1a1-GFP, and Tcf21mCrem lines were found to label the resident fibroblast population discretely, and this homogeneous cell population uniformly expressed the antigen recognized by the aforementioned MEFSK4 antibody41. The generation of reporter and Cre lines that specifically label both resting60 and activated33,41 fibroblasts in the heart will enable research to finally examine the role of the fibroblast, and fibroblast specific genes, during all stages of activation.
Cardiac fibroblast function
Development and resting
Although the above data demonstrate that resident cardiac fibroblasts respond to injury by producing components of the ECM, additional roles of the cardiac fibroblast in uninjured hearts remain a mystery. Without the ability to use genetic tools and well-defined markers, early studies often relied on cell morphology to identify these cells. A common notion was that cardiac fibroblasts comprised a majority of the non-cardiomyocytes of the heart61–63, but we have demonstrated that endothelial cells, not fibroblasts, are the most populous cell type in the human and murine heart40.
Although not the major constituent, it is likely that cardiac fibroblasts play an important part of normal heart physiology. In fact, many functions have been attributed to fibroblasts, but these proposed cellular activities are often deduced after in vitro culture and need to be verified in vivo (Figure 1). Matrix degradation, conduction system insulation, cardiomyocyte electrical coupling, vascular maintenance, and stress sensing are all potential aspects of fibroblast cell biology (reviewed in Baudino64, Souders65, and Snider7). Although cardiac fibroblasts are likely to perform these duties, it is unclear if they are the only cells capable of such feats. Certainly, the production of fibrillar collagens during development and disease is an accepted and documented fibroblast activity66, but recent data suggests that pericytes and/or mesenchymal progenitors can also produce ECM components in response to injury67,68.
Another example of a purported fibroblast role is insulation of the conduction system. Although a direct role for fibroblasts has not been proven, the best data supporting the idea that the annulus fibrosis buffers the myocardium from the atrioventricular node is the mechanical inhibition of epicardial migration in the avian heart69. An epicardial origin for the cells of the annulus fibrosis has been determined, but other than expression of ECM genes, an insulating role for these cells was not documented70,71.
Given that in vivo data designating the predominant roles of resting fibroblasts is lacking, more efforts should be focused on the activities of these cells in non-pathological conditions. A revised understanding of the developing and resting cardiac fibroblast population will further expand our knowledge of cellular processes assigned to fibroblasts.
Cardiac fibroblast activation (myofibroblast)
Because cardiac fibrosis contributes to many forms of heart disease, much attention has focused on behaviors of activated fibroblasts (Figure 1). The first step in such studies involves the ability to identify the cell of interest. In the field of wound healing and cardiac fibrosis, the terms protomyofibroblast and myofibroblast are often used to indicate the subpopulation of fibroblasts that are responsible for tissue remodeling. The term myofibroblast was originally coined to describe a cell that had morphological characteristics of both smooth muscle cells and fibroblasts during skin wound healing72.
The first mention of cardiac myofibroblasts was in the 1970s73,74. These cells could be distinguished from resting cells by morphology, including serrated nuclei, increased cytoplasm, microfilament bundles, and well defined endoplasmic reticulum and Golgi complex11,75. Later, skin myofibroblasts were documented to contract collagen in vitro and thus provide a unique and essential role in wound repair by providing tension75,76. With the advent of an α smooth muscle actin (αSMA) antibody permitting the identification of these microfilament bundles77, myofibroblasts were found in other injured organs78,79. Expression of the microfilament proteins, αSMA, transgelin, or caldesmon became the gold standard for identifying myofibroblasts80–82. Subsequent studies suggested that transforming growth factor β (TGFβ) stimulation induced αSMA76 and because TGFβ also induces collagen production, it was suggested that αSMA could be used to identify collagen producing cells after heart injury. As time passed, these changes in gene expression were considered a process of cell conversion or transdifferentiation into a new cell type.
Given the previous lack of markers and associated difficulty in identifying and studying the fibroblast in vivo, analyses were typically performed in vitro49,83–85. Notably, these in vitro studies may not have appreciated the added mechanical stress caused by substrate stiffness in culture49,86. Researchers observed that fibroblasts in culture fail to acquire quiescent features after stimulation removal, supporting the concept that myofibroblasts were a terminally differentiated cell type86. However, these studies did not take into account the mechanical stress from a non-physiological system on these fibroblasts. Thus, saying that fibroblast activation is an irreversible differentiation process may not accurately describe the reversible change in gene expression that occurs in vivo.
Recent studies have identified transcription factors that are involved in the functions of activated cardiac fibroblasts. Two of these proteins are scleraxis46,87,88, which is downstream of TGFβ signaling and involved in ECM synthesis, and myocardin-related transcription factors (MRTFs)89, which are involved in cytoskeletal changes and upregulation of αSMA expression during fibroblast activation. This information suggests that rather than a differentiation process, the changes in gene expression of fibroblasts after cardiac injury is more likely to be a response to changes in growth factor signaling and an increase in tissue stiffness (reviewed by van Putten90). Given recent findings, we would like to suggest a simplified nomenclature from myofibroblast to activated fibroblast. This would broaden the population of cells to investigate after injury and also reflect the other dynamic changes in gene expression, such as proliferation, ROS production and recruitment of inflammatory cells25,26,33,41,45,91.
Alternative cell sources after injury
Contrary to the accepted developmental origin of the resting fibroblast, the origin of the activated fibroblast is historically much less clear and is still debated. As the activated fibroblast was considered a newly differentiated cell type, it was feasible that the cells responding to the injury could come from a variety of sources. Using lineage tracing and the limited tools available to study fibroblast biology, activated fibroblasts were described to differentiate from multiple cell types. Studies suggested that activated fibroblasts differentiated from either endothelial cells via endothelial-to-mesenchymal transition29 or infiltrating immune cells from bone marrow9,27,28. However, these studies relied on lineage tracing using Tie1-Cre92 and the FSP1-GFP93 mouse lines. The recent realization that some populations of fibroblasts derive from an endothelial progenitor could provide an alternative explanation for the presence of endothelial lineages within the fibroblast population. Additionally, FSP1-GFP expression has also been reported in immune and endothelial cells26,44.
Recently, pericytes, mesenchymal cells associated with the microvasculature, have also been identified as a potential source of injury-induced matrix-producing cells. The ablation of Gli1-expressing pericytes resulted in a pronounced reduction in fibrosis, suggesting a role for pericytes in matrix production67. Other studies focusing on αV integrin signaling also point to a role for signaling through pericytes in promoting fibrosis after heart injury68. While these studies do implicate pericytes as an additional contributor to the fibrotic process, the mechanism of these actions remains unclear. For example, another study identified two populations of pericytes in the heart, type 1 and type 2. They found that type 1 pericytes expanded after myocardial infarction but did not express Collagen type I57. Intriguingly, Gli1 expressing pericytes mentioned above comprise only a small portion of the perivascular cell population67, suggesting that these cells may serve a role in regulating fibrosis rather than directly contributing to the act of ECM deposition94. These initial studies indicate that more data is required before the direct and indirect functions of pericytes during cardiac fibrosis can be elucidated.
Reversal of activation
Generally, it was believed that activated fibroblasts undergo apoptosis and disappear following the completion of tissue repair95. For example, fibroblast apoptosis occurs via a TNFα-mediated response in skeletal muscle96. In other organs, however, studies indicate that activated fibroblasts have the capacity to revert to a resting fibroblast as determined by reduction in αSMA expression97–99. To study the fate of activated cardiac fibroblasts after injury, a reversible model of cardiac fibrosis was investigated. Angiotensin II and phenylephrine (AngII-PE) infusion cause rapid fibroblast activation, but upon drug cessation fibrosis recedes. The activated fibroblast lineage was marked using a mouse line, PeriostinmCrem, and the cells were followed over time. After two weeks the marked fibroblasts were still present, but gene expression had reverted back to a resting fibroblast profile33. Interestingly, these reverted cells were more susceptible to re-activation, similar to a memory B or T cell response. This type of fibroblast reversion has also been observed in liver fibroblasts100,101 and supports the idea that activation is more a change in gene expression than a conversion of the fibroblast into another cell type.
Cardiac fibroblast plasticity
There is the current concept that fibroblasts are versatile and can interconvert readily into other cell types, but cellular reprogramming efforts have demonstrated that fibroblast reprogramming is often inefficient102,103, suggesting that these cells may not be as plastic as previously believed. Many past experiments demonstrating fibroblast transdifferentiation were performed in vitro on minimally characterized cell populations.
Nonetheless, recent studies have documented the ability of fibroblasts to convert to other cell types including adipocytes104, cardiomyocytes55,105, and endothelial cells106. One example is the description of the cardiac fibroblast colony-forming unit (cCFU-F). A Sca1+, PDGFRα+, CD31− population of cells from mouse heart was observed to have long term culture capabilities and could differentiate into cardiomyocytes, endothelial cells, and smooth muscle cells105. While a similar differentiation capacity of heart resident PDGFRα expressing cells was observed in a recent study, these cells were not identified as fibroblasts and were considered a resident stem cell population55. Both of these studies relied on in vitro culture with subsequent transplantation to generate nascient cardiomyocytes. Although evidence for spontaneous conversion of fibroblasts to cardiomyocytes in vivo has not been observed, there are several examples of fibroblast to cardiomyocyte conversion with cellular reprogramming after injury107–109. The efficiency of this conversion was less than 2%107 in the area of reprogramming and would need optimization for any practical application.
While the ability of fibroblasts to differentiate into adipocytes has been shown in skeletal muscle110,111, only recently has it been suggested that a cardiac fibroblast progenitor can differentiate into adipocytes104. It is unclear if fibroblasts themselves can form adipocytes in vivo, but recent data does suggest that a subset of epicardial derivatives contribute to adipocytes that are present in the atrio-ventricular groove and epicardial fat112,113. Finally, although the conversion of vascular endothelial cells into fibroblasts appears to be a minor contribution to fibrosis25,26,33, lineage analysis using a Col1a2CreERT mouse line suggests that some fibroblasts may adopt properties of endothelial cells after injury106.
Conclusion
Recent developments in tools to study fibroblast biology have enabled a more detailed and physiologic understanding of the fibroblast, as most original studies were limited in markers and to in vitro models. Even though cardiac fibroblasts have two developmental origins, these populations respond similarly to cardiac injury and are the predominant fibroblast source. The term “myofibroblast” was initially used to distinguish between the fibroblast and a new cell type that arose during the fibrotic response. However, recent advances in fibroblast tools have allowed us to gain a better understanding of fibroblast activation, gene expression, and behavior. These data suggest that an activated fibroblast arises from a tissue resident fibroblast and can revert back to a resting fibroblast. While progress is evident in the study of fibroblast biology and fibrosis, there remain key questions to be answered regarding the role of the fibroblast in physiology and disease.
Acknowledgments
Sources of Funding
This work was supported by National Heart, Lung, and Blood Institute grants HL074257 to M.D. Tallquist and F31HL126512 to M.J. Ivey.
Footnotes
Disclosures
None.
References
- 1.Brown RD, Ambler SK, Mitchell MD, Long CS. The cardiac fibroblast: therapeutic target in myocardial remodeling and failure. Annu Rev Pharmacol Toxicol. 2005;45:657–687. doi: 10.1146/annurev.pharmtox.45.120403.095802. [DOI] [PubMed] [Google Scholar]
- 2.Haidara MA, et al. Heart Failure Models: Traditional and Novel Therapy. Current vascular pharmacology. 2015;13:658–69. doi: 10.2174/1570161113666150212151506. [DOI] [PubMed] [Google Scholar]
- 3.Gittenberger-de Groot AC, Bartelings MM, Poelmann RE, Haak MC, Jongbloed MR. Embryology of the heart and its impact on understanding fetal and neonatal heart disease. Seminars in fetal & neonatal medicine. 2013;18:237–44. doi: 10.1016/j.siny.2013.04.008. [DOI] [PubMed] [Google Scholar]
- 4.Kjaer M. Role of extracellular matrix in adaptation of tendon and skeletal muscle to mechanical loading. Physiological reviews. 2004;84:649–698. doi: 10.1152/physrev.00031.2003. [DOI] [PubMed] [Google Scholar]
- 5.Van Den Borne SW, et al. Myocardial remodeling after infarction: the role of myofibroblasts. Nature Reviews Cardiology. 2010;7:30–37. doi: 10.1038/nrcardio.2009.199. [DOI] [PubMed] [Google Scholar]
- 6.Frangogiannis NG. Matricellular proteins in cardiac adaptation and disease. Physiological reviews. 2012;92:635–688. doi: 10.1152/physrev.00008.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Snider P, et al. Origin of cardiac fibroblasts and the role of periostin. Circulation research. 2009;105:934–947. doi: 10.1161/CIRCRESAHA.109.201400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hiatt JL, Gartner LP. Color Atlas and Text of Histology. Lippincott Williams & Wilkins; 2013. [Google Scholar]
- 9.Abe R, Donnelly SC, Peng T, Bucala R, Metz CN. Peripheral blood fibrocytes: differentiation pathway and migration to wound sites. The Journal of Immunology. 2001;166:7556–7562. doi: 10.4049/jimmunol.166.12.7556. [DOI] [PubMed] [Google Scholar]
- 10.Varga I, et al. The functional morphology and role of cardiac telocytes in myocardium regeneration. Canadian Journal of Physiology and Pharmacology. 2016 doi: 10.1139/cjpp-2016-0052. [DOI] [PubMed] [Google Scholar]
- 11.Gabbiani G, Ryan G, Majno G. Presence of modified fibroblasts in granulation tissue and their possible role in wound contraction. Experientia. 1971;27:549–550. doi: 10.1007/BF02147594. [DOI] [PubMed] [Google Scholar]
- 12.Tomasek JJ, Gabbiani G, Hinz B, Chaponnier C, Brown RA. Myofibroblasts and mechano-regulation of connective tissue remodelling. Nature reviews Molecular cell biology. 2002;3:349–363. doi: 10.1038/nrm809. [DOI] [PubMed] [Google Scholar]
- 13.Gittenberger-de Groot AC, Vrancken Peeters MP, Mentink MM, Gourdie RG, Poelmann RE. Epicardium-derived cells contribute a novel population to the myocardial wall and the atrioventricular cushions. Circulation research. 1998;82:1043–52. doi: 10.1161/01.res.82.10.1043. [DOI] [PubMed] [Google Scholar]
- 14.Mikawa T, Gourdie RG. Pericardial mesoderm generates a population of coronary smooth muscle cells migrating into the heart along with ingrowth of the epicardial organ. Developmental biology. 1996;174:221–32. doi: 10.1006/dbio.1996.0068. [DOI] [PubMed] [Google Scholar]
- 15.Dettman RW, Denetclaw W, Ordahl CP, Bristow J. Common epicardial origin of coronary vascular smooth muscle, perivascular fibroblasts, and intermyocardial fibroblasts in the avian heart. Developmental biology. 1998;193:169–181. doi: 10.1006/dbio.1997.8801. [DOI] [PubMed] [Google Scholar]
- 16.Manner J. Does the subepicardial mesenchyme contribute myocardioblasts to the myocardium of the chick embryo heart? A quail-chick chimera study tracing the fate of the epicardial primordium. The Anatomical record. 1999;255:212–26. doi: 10.1002/(sici)1097-0185(19990601)255:2<212::aid-ar11>3.3.co;2-o. [DOI] [PubMed] [Google Scholar]
- 17.Perez-Pomares J, Macias D, Garcia-Garrido L, Munoz-Chapuli R. Contribution of the primitive epicardium to the subepicardial mesenchyme in hamster and chick embryos. Developmental dynamics. 1997;210:96–105. doi: 10.1002/(SICI)1097-0177(199710)210:2<96::AID-AJA3>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 18.Wessels A, et al. Epicardially derived fibroblasts preferentially contribute to the parietal leaflets of the atrioventricular valves in the murine heart. Developmental biology. 2012;366:111–24. doi: 10.1016/j.ydbio.2012.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Acharya A, et al. The bHLH transcription factor Tcf21 is required for lineage-specific EMT of cardiac fibroblast progenitors. Development. 2012;139:2139–2149. doi: 10.1242/dev.079970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cai CL, et al. A myocardial lineage derives from Tbx18 epicardial cells. Nature. 2008;454:104–108. doi: 10.1038/nature06969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smith CL, Baek ST, Sung CY, Tallquist MD. Epicardial-derived cell epithelial-to-mesenchymal transition and fate specification require PDGF receptor signaling. Circulation research. 2011;108:e15–e26. doi: 10.1161/CIRCRESAHA.110.235531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou B, et al. Epicardial progenitors contribute to the cardiomyocyte lineage in the developing heart. Nature. 2008;454:109–113. doi: 10.1038/nature07060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dymecki S, Ray R, Kim J. Mapping cell fate and function using recombinase-based intersectional strategies. Methods in enzymology. 2010;477:183. doi: 10.1016/S0076-6879(10)77011-7. [DOI] [PubMed] [Google Scholar]
- 24.Swonger JM, Liu JS, Ivey MJ, Tallquist MD. Genetic tools for identifying and manipulating fibroblasts in the mouse. Differentiation. 2016 doi: 10.1016/j.diff.2016.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ali SR, et al. Developmental heterogeneity of cardiac fibroblasts does not predict pathological proliferation and activation. Circulation research. 2014;115:625–635. doi: 10.1161/CIRCRESAHA.115.303794. [DOI] [PubMed] [Google Scholar]
- 26.Moore-Morris T, et al. Resident fibroblast lineages mediate pressure overload-induced cardiac fibrosis. The Journal of clinical investigation. 2014;124:2921. doi: 10.1172/JCI74783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haudek SB, et al. Bone marrow-derived fibroblast precursors mediate ischemic cardiomyopathy in mice. Proceedings of the National Academy of Sciences. 2006;103:18284–18289. doi: 10.1073/pnas.0608799103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Van Amerongen M, et al. Bone marrow derived myofibroblasts contribute functionally to scar formation after myocardial infarction. The Journal of pathology. 2008;214:377–386. doi: 10.1002/path.2281. [DOI] [PubMed] [Google Scholar]
- 29.Zeisberg EM, et al. Endothelial-to-mesenchymal transition contributes to cardiac fibrosis. Nature medicine. 2007;13:952–961. doi: 10.1038/nm1613. [DOI] [PubMed] [Google Scholar]
- 30.Zhou B, Pu WT. Epicardial epithelial-to-mesenchymal transition in injured heart. Journal of cellular and molecular medicine. 2011;15:2781–2783. doi: 10.1111/j.1582-4934.2011.01450.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van Wijk B, Gunst QD, Moorman AF, van den Hoff MJ. Cardiac regeneration from activated epicardium. PloS one. 2012;7:e44692. doi: 10.1371/journal.pone.0044692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou B, et al. Adult mouse epicardium modulates myocardial injury by secreting paracrine factors. The Journal of clinical investigation. 2011;121:1894–904. doi: 10.1172/JCI45529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kanisicak O, et al. Genetic lineage tracing defines myofibroblast origin and function in the injured heart. Nature communications. 2016;7 doi: 10.1038/ncomms12260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yata Y, et al. DNase I–hypersensitive sites enhance α1 (I) collagen gene expression in hepatic stellate cells. Hepatology. 2003;37:267–276. doi: 10.1053/jhep.2003.50067. [DOI] [PubMed] [Google Scholar]
- 35.Lekic P, Pender N, McCulloch C. Is fibroblast heterogeneity relevant to the health, diseases, and treatments of periodontal tissues? Critical Reviews in Oral Biology & Medicine. 1997;8:253–268. doi: 10.1177/10454411970080030201. [DOI] [PubMed] [Google Scholar]
- 36.Ko SD, Page RC, Narayanan A. Fibroblast heterogeneity and prostaglandin regulation of subpopulations. Proceedings of the National Academy of Sciences. 1977;74:3429–3432. doi: 10.1073/pnas.74.8.3429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bordin S, Page R, Narayanan A. Heterogeneity of normal human diploid fibroblasts: isolation and characterization of one phenotype. Science. 1984;223:171–173. doi: 10.1126/science.6691142. [DOI] [PubMed] [Google Scholar]
- 38.Hassell T, Stanek E. Evidence that healthy human gingiva contains functionally heterogeneous fibroblast subpopulations. Archives of oral biology. 1983;28:617–625. doi: 10.1016/0003-9969(83)90010-9. [DOI] [PubMed] [Google Scholar]
- 39.Angello JC, Pendergrass WR, Norwood TH, Prothero J. Proliferative potential of human fibroblasts: an inverse dependence on cell size. Journal of cellular physiology. 1987;132:125–130. doi: 10.1002/jcp.1041320117. [DOI] [PubMed] [Google Scholar]
- 40.Pinto AR, et al. Revisiting cardiac cellular composition. Circulation research. 2016;118:400–409. doi: 10.1161/CIRCRESAHA.115.307778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kaur H, et al. Targeted Ablation of Periostin-Expressing Activated Fibroblasts Prevents Adverse Cardiac Remodeling in Mice. Circulation research. 2016;118:1906–1917. doi: 10.1161/CIRCRESAHA.116.308643. [DOI] [PubMed] [Google Scholar]
- 42.Hudon-David F, Bouzeghrane F, Couture P, Thibault G. Thy-1 expression by cardiac fibroblasts: lack of association with myofibroblast contractile markers. Journal of molecular and cellular cardiology. 2007;42:991–1000. doi: 10.1016/j.yjmcc.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 43.Goldsmith EC, et al. Organization of fibroblasts in the heart. Developmental dynamics. 2004;230:787–794. doi: 10.1002/dvdy.20095. [DOI] [PubMed] [Google Scholar]
- 44.Kong P, Christia P, Saxena A, Su Y, Frangogiannis NG. Lack of specificity of fibroblast-specific protein 1 in cardiac remodeling and fibrosis. American Journal of Physiology-Heart and Circulatory Physiology. 2013;305:H1363–H1372. doi: 10.1152/ajpheart.00395.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Furtado MB, et al. Cardiogenic genes expressed in cardiac fibroblasts contribute to heart development and repair. Circulation research. 2014;114:1422–34. doi: 10.1161/CIRCRESAHA.114.302530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bagchi RA, Lin J, Wang R, Czubryt MP. Regulation of fibronectin gene expression in cardiac fibroblasts by scleraxis. Cell and Tissue Research. 2016:1–11. doi: 10.1007/s00441-016-2439-1. [DOI] [PubMed] [Google Scholar]
- 47.Goodpaster T, et al. An immunohistochemical method for identifying fibroblasts in formalin-fixed, paraffin-embedded tissue. Journal of Histochemistry & Cytochemistry. 2008;56:347–358. doi: 10.1369/jhc.7A7287.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vasquez C, Benamer N, Morley GE. The cardiac fibroblast: functional and electrophysiological considerations in healthy and diseased hearts. Journal of cardiovascular pharmacology. 2011;57:380. doi: 10.1097/FJC.0b013e31820cda19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Santiago JJ, et al. Cardiac fibroblast to myofibroblast differentiation in vivo and in vitro: expression of focal adhesion components in neonatal and adult rat ventricular myofibroblasts. Developmental dynamics. 2010;239:1573–1584. doi: 10.1002/dvdy.22280. [DOI] [PubMed] [Google Scholar]
- 50.Chapman D, Weber KT, Eghbali M. Regulation of fibrillar collagen types I and III and basement membrane type IV collagen gene expression in pressure overloaded rat myocardium. Circulation research. 1990;67:787–794. doi: 10.1161/01.res.67.4.787. [DOI] [PubMed] [Google Scholar]
- 51.Tillmanns J, et al. Fibroblast activation protein alpha expression identifies activated fibroblasts after myocardial infarction. Journal of molecular and cellular cardiology. 2015;87:194–203. doi: 10.1016/j.yjmcc.2015.08.016. [DOI] [PubMed] [Google Scholar]
- 52.Ffrench-Constant C, Van de Water L, Dvorak HF, Hynes RO. Reappearance of an embryonic pattern of fibronectin splicing during wound healing in the adult rat. The Journal of cell biology. 1989;109:903–14. doi: 10.1083/jcb.109.2.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Travers JG, Kamal FA, Robbins J, Yutzey KE, Blaxall BC. Cardiac Fibrosis: The Fibroblast Awakens. Circulation research. 2016;118:1021–40. doi: 10.1161/CIRCRESAHA.115.306565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Braitsch CM, Kanisicak O, van Berlo JH, Molkentin JD, Yutzey KE. Differential expression of embryonic epicardial progenitor markers and localization of cardiac fibrosis in adult ischemic injury and hypertensive heart disease. Journal of molecular and cellular cardiology. 2013;65:108–119. doi: 10.1016/j.yjmcc.2013.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Noseda M, et al. PDGFR-alpha demarcates the cardiogenic clonogenic Sca1+ stem-progenitor cell in adult murine myocardium. Nature communications. 2015;6 doi: 10.1038/ncomms7930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lin SL, Kisseleva T, Brenner DA, Duffield JS. Pericytes and perivascular fibroblasts are the primary source of collagen-producing cells in obstructive fibrosis of the kidney. The American journal of pathology. 2008;173:1617–1627. doi: 10.2353/ajpath.2008.080433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Birbrair A, et al. Type-1 pericytes accumulate after tissue injury and produce collagen in an organ-dependent manner. Stem cell research & therapy. 2014;5:1. doi: 10.1186/scrt512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ponticos M, Partridge T, Black CM, Abraham DJ, Bou-Gharios G. Regulation of collagen type I in vascular smooth muscle cells by competition between Nkx2.5 and deltaEF1/ZEB1. Molecular and cellular biology. 2004;24:6151–61. doi: 10.1128/MCB.24.14.6151-6161.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hamilton TG, Klinghoffer RA, Corrin PD, Soriano P. Evolutionary divergence of platelet-derived growth factor alpha receptor signaling mechanisms. Molecular and cellular biology. 2003;23:4013–4025. doi: 10.1128/MCB.23.11.4013-4025.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Acharya A, Baek ST, Banfi S, Eskiocak B, Tallquist MD. Efficient inducible Cre-mediated recombination in Tcf21cell lineages in the heart and kidney. Genesis. 2011;49:870–877. doi: 10.1002/dvg.20750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nag A. Study of non-muscle cells of the adult mammalian heart: a fine structural analysis and distribution. Cytobios. 1979;28:41–61. [PubMed] [Google Scholar]
- 62.Bergmann O, et al. Dynamics of cell generation and turnover in the human heart. Cell. 2015;161:1566–1575. doi: 10.1016/j.cell.2015.05.026. [DOI] [PubMed] [Google Scholar]
- 63.Banerjee I, Fuseler JW, Price RL, Borg TK, Baudino TA. Determination of cell types and numbers during cardiac development in the neonatal and adult rat and mouse. American Journal of Physiology-Heart and Circulatory Physiology. 2007;293:H1883–H1891. doi: 10.1152/ajpheart.00514.2007. [DOI] [PubMed] [Google Scholar]
- 64.Baudino TA, Carver W, Giles W, Borg TK. Cardiac fibroblasts: friend or foe? American journal of physiology Heart and circulatory physiology. 2006;291:H1015–26. doi: 10.1152/ajpheart.00023.2006. [DOI] [PubMed] [Google Scholar]
- 65.Souders CA, Bowers SL, Baudino TA. Cardiac fibroblast: the renaissance cell. Circulation research. 2009;105:1164–76. doi: 10.1161/CIRCRESAHA.109.209809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Eghbali M. Cardiac Adaptation in Heart Failure. Springer; 1992. Cardiac fibroblasts: function, regulation of gene expression, and phenotypic modulation; pp. 183–189. [DOI] [PubMed] [Google Scholar]
- 67.Kramann R, et al. Perivascular Gli1+ progenitors are key contributors to injury-induced organ fibrosis. Cell Stem Cell. 2015;16:51–66. doi: 10.1016/j.stem.2014.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Henderson NC, et al. Targeting of [alpha] v integrin identifies a core molecular pathway that regulates fibrosis in several organs. Nature medicine. 2013;19:1617–1624. doi: 10.1038/nm.3282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kolditz DP, et al. Epicardium-derived cells in development of annulus fibrosis and persistence of accessory pathways. Circulation. 2008;117:1508–17. doi: 10.1161/CIRCULATIONAHA.107.726315. [DOI] [PubMed] [Google Scholar]
- 70.Zhou B, von Gise A, Ma Q, Hu YW, Pu WT. Genetic fate mapping demonstrates contribution of epicardium-derived cells to the annulus fibrosis of the mammalian heart. Developmental biology. 2010;338:251–261. doi: 10.1016/j.ydbio.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lockhart MM, Phelps AL, van den Hoff MJ, Wessels A. The Epicardium and the Development of the Atrioventricular Junction in the Murine Heart. Journal of developmental biology. 2014;2:1–17. doi: 10.3390/jdb2010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Majno G, Gabbiani G, Hirschel B, Ryan G, Statkov P. Contraction of granulation tissue in vitro: similarity to smooth muscle. Science. 1971;173:548–550. doi: 10.1126/science.173.3996.548. [DOI] [PubMed] [Google Scholar]
- 73.Lagace R, Delage C, Boutet M. Light and electron microscopic study of cellular proliferation in carcinoid heart disease. Recent advances in studies on cardiac structure and metabolism. 1974;10:605–616. [PubMed] [Google Scholar]
- 74.Kischer C, Shetlar M. Electron microscopic studies of connective tissue repair after myocardial injury. Texas reports on biology and medicine. 1978;39:357–369. [PubMed] [Google Scholar]
- 75.Gabbiani G, Hirschel B, Ryan G, Statkov P, Majno G. Granulation tissue as a contractile organ: A study of structure and function. The Journal of experimental medicine. 1972;135:719–734. doi: 10.1084/jem.135.4.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Desmoulière A, Geinoz A, Gabbiani F, Gabbiani G. Transforming growth factor-beta 1 induces alpha-smooth muscle actin expression in granulation tissue myofibroblasts and in quiescent and growing cultured fibroblasts. The Journal of cell biology. 1993;122:103–111. doi: 10.1083/jcb.122.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Skalli O, et al. A monoclonal antibody against alpha-smooth muscle actin: a new probe for smooth muscle differentiation. The Journal of cell biology. 1986;103:2787–2796. doi: 10.1083/jcb.103.6.2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Darby I, Skalli O, Gabbiani G. a-Smooth muscle actin is transiently expressed by myofibroblasts during experimental wound healing. Lab Invest. 1990;63:21–29. [PubMed] [Google Scholar]
- 79.Frangogiannis NG, Michael LH, Entman ML. Myofibroblasts in reperfused myocardial infarcts express the embryonic form of smooth muscle myosin heavy chain (SMemb) Cardiovascular research. 2000;48:89–100. doi: 10.1016/s0008-6363(00)00158-9. [DOI] [PubMed] [Google Scholar]
- 80.Qiu P, Feng XH, Li L. Interaction of Smad3 and SRF-associated complex mediates TGF-beta signals to regulate SM22 transcription during myofibroblast differentiation. Journal of molecular and cellular cardiology. 2003;35:1407–1420. doi: 10.1016/j.yjmcc.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 81.Lazard D, et al. Expression of smooth muscle-specific proteins in myoepithelium and stromal myofibroblasts of normal and malignant human breast tissue. Proceedings of the National Academy of Sciences. 1993;90:999–1003. doi: 10.1073/pnas.90.3.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sappino A, Schurch W, Gabbiani G. Differentiation repertoire of fibroblastic cells: expression of cytoskeletal proteins as marker of phenotypic modulations. Laboratory investigation; a journal of technical methods and pathology. 1990;63:144–161. [PubMed] [Google Scholar]
- 83.Arora PD, Narani N, McCulloch CA. The compliance of collagen gels regulates transforming growth factor-beta induction of alpha-smooth muscle actin in fibroblasts. The American journal of pathology. 1999;154:871–882. doi: 10.1016/s0002-9440(10)65334-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Carver W, Nagpal ML, Nachtigal M, Borg TK, Terracio L. Collagen expression in mechanically stimulated cardiac fibroblasts. Circulation research. 1991;69:116–22. doi: 10.1161/01.res.69.1.116. [DOI] [PubMed] [Google Scholar]
- 85.Wang J, Chen H, Seth A, McCulloch CA. Mechanical force regulation of myofibroblast differentiation in cardiac fibroblasts. American Journal of Physiology-Heart and Circulatory Physiology. 2003;285:H1871–H1881. doi: 10.1152/ajpheart.00387.2003. [DOI] [PubMed] [Google Scholar]
- 86.Petrov VV, Fagard RH, Lijnen PJ. Stimulation of collagen production by transforming growth factorβ-1 during differentiation of cardiac fibroblasts to myofibroblasts. Hypertension. 2002;39:258–263. doi: 10.1161/hy0202.103268. [DOI] [PubMed] [Google Scholar]
- 87.Espira L, et al. The basic helix-loop-helix transcription factor scleraxis regulates fibroblast collagen synthesis. Journal of molecular and cellular cardiology. 2009;47:188–195. doi: 10.1016/j.yjmcc.2009.03.024. [DOI] [PubMed] [Google Scholar]
- 88.Roche PL, et al. Role of scleraxis in mechanical stretch-mediated regulation of cardiac myofibroblast phenotype. American journal of physiology Cell physiology. 2016;311:C297–307. doi: 10.1152/ajpcell.00333.2015. [DOI] [PubMed] [Google Scholar]
- 89.Small EM. The actin-MRTF-SRF gene regulatory axis and myofibroblast differentiation. Journal of cardiovascular translational research. 2012;5:794–804. doi: 10.1007/s12265-012-9397-0. [DOI] [PubMed] [Google Scholar]
- 90.van Putten S, Shafieyan Y, Hinz B. Mechanical control of cardiac myofibroblasts. Journal of molecular and cellular cardiology. 2016;93:133–42. doi: 10.1016/j.yjmcc.2015.11.025. [DOI] [PubMed] [Google Scholar]
- 91.Chen W, Frangogiannis NG. Fibroblasts in post-infarction inflammation and cardiac repair. Biochimica et biophysica acta. 2013;1833:945–53. doi: 10.1016/j.bbamcr.2012.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gustafsson E, Brakebusch C, Hietanen K, Fassler R. Tie-1-directed expression of Cre recombinase in endothelial cells of embryoid bodies and transgenic mice. Journal of cell science. 2001;114:671–676. doi: 10.1242/jcs.114.4.671. [DOI] [PubMed] [Google Scholar]
- 93.Iwano M, et al. Evidence that fibroblasts derive from epithelium during tissue fibrosis. The Journal of clinical investigation. 2002;110:341–350. doi: 10.1172/JCI15518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Murray IR, et al. Skeletal and cardiac muscle pericytes: Functions and therapeutic potential. Pharmacology & therapeutics. 2016 doi: 10.1016/j.pharmthera.2016.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Desmouliere A, Redard M, Darby I, Gabbiani G. Apoptosis mediates the decrease in cellularity during the transition between granulation tissue and scar. The American journal of pathology. 1995;146:56. [PMC free article] [PubMed] [Google Scholar]
- 96.Lemos DR, et al. Nilotinib reduces muscle fibrosis in chronic muscle injury by promoting TNF-mediated apoptosis of fibro/adipogenic progenitors. Nature medicine. 2015;21:786–794. doi: 10.1038/nm.3869. [DOI] [PubMed] [Google Scholar]
- 97.Hecker L, Jagirdar R, Jin T, Thannickal VJ. Reversible differentiation of myofibroblasts by MyoD. Experimental cell research. 2011;317:1914–1921. doi: 10.1016/j.yexcr.2011.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Garrison G, et al. Reversal of myofibroblast differentiation by prostaglandin e2. American journal of respiratory cell and molecular biology. 2013;48:550–558. doi: 10.1165/rcmb.2012-0262OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Artaud-Macari E, et al. Nuclear factor erythroid 2-related factor 2 nuclear translocation induces myofibroblastic dedifferentiation in idiopathic pulmonary fibrosis. Antioxidants & redox signaling. 2013;18:66–79. doi: 10.1089/ars.2011.4240. [DOI] [PubMed] [Google Scholar]
- 100.Kisseleva T, et al. Myofibroblasts revert to an inactive phenotype during regression of liver fibrosis. Proceedings of the National Academy of Sciences. 2012;109:9448–9453. doi: 10.1073/pnas.1201840109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Troeger JS, et al. Deactivation of hepatic stellate cells during liver fibrosis resolution in mice. Gastroenterology. 2012;143:1073–1083.e22. doi: 10.1053/j.gastro.2012.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kim JB, et al. Pluripotent stem cells induced from adult neural stem cells by reprogramming with two factors. Nature. 2008;454:646–650. doi: 10.1038/nature07061. [DOI] [PubMed] [Google Scholar]
- 103.Aoi T, et al. Generation of pluripotent stem cells from adult mouse liver and stomach cells. Science. 2008;321:699–702. doi: 10.1126/science.1154884. [DOI] [PubMed] [Google Scholar]
- 104.Lombardi R, et al. Cardiac Fibro-Adipocyte Progenitors Express Desmosome Proteins and Preferentially Differentiate to Adipocytes Upon Deletion of the Desmoplakin Gene. Circulation research. 2016;119:41–54. doi: 10.1161/CIRCRESAHA.115.308136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Chong JJ, et al. Adult cardiac-resident MSC-like stem cells with a proepicardial origin. Cell Stem Cell. 2011;9:527–540. doi: 10.1016/j.stem.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ubil E, et al. Mesenchymal-endothelial transition contributes to cardiac neovascularization. Nature. 2014;514:585–590. doi: 10.1038/nature13839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Song K, et al. Heart repair by reprogramming non-myocytes with cardiac transcription factors. Nature. 2012;485:599–604. doi: 10.1038/nature11139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Chen JX, et al. Inefficient reprogramming of fibroblasts into cardiomyocytes using Gata4, Mef2c, and Tbx5. Circulation research. 2012;111:50–55. doi: 10.1161/CIRCRESAHA.112.270264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Qian L, et al. In vivo reprogramming of murine cardiac fibroblasts into induced cardiomyocytes. Nature. 2012;485:593–598. doi: 10.1038/nature11044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Joe AW, et al. Muscle injury activates resident fibro-adipogenic progenitors that facilitate myogenesis. Nature cell biology. 2010;12:153–163. doi: 10.1038/ncb2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Uezumi A, Fukada S-i, Yamamoto N, Takeda S-i, Tsuchida K. Mesenchymal progenitors distinct from satellite cells contribute to ectopic fat cell formation in skeletal muscle. Nature cell biology. 2010;12:143–152. doi: 10.1038/ncb2014. [DOI] [PubMed] [Google Scholar]
- 112.Yamaguchi Y, et al. Adipogenesis and epicardial adipose tissue: a novel fate of the epicardium induced by mesenchymal transformation and PPAR-gamma activation. Proceedings of the National Academy of Sciences. 2015;112:2070–2075. doi: 10.1073/pnas.1417232112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Liu Q, et al. Epicardium-to-fat transition in injured heart. Cell research. 2014;24:1367. doi: 10.1038/cr.2014.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zeng B, Ren X-f, Cao F, Zhou X-y, Zhang J. Developmental patterns and characteristics of epicardial cell markers Tbx18 and Wt1 in murine embryonic heart. Journal of biomedical science. 2011;18:1. doi: 10.1186/1423-0127-18-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Duim SN, Kurakula K, Goumans MJ, Kruithof BP. Cardiac endothelial cells express Wilms’ tumor-1: Wt1 expression in the developing, adult and infarcted heart. Journal of molecular and cellular cardiology. 2015;81:127–135. doi: 10.1016/j.yjmcc.2015.02.007. [DOI] [PubMed] [Google Scholar]
- 116.Robb L, et al. epicardin: A novel basic helix-loop-helix transcription factor gene expressed in epicardium, branchial arch myoblasts, and mesenchyme of developing lung, gut, kidney, and gonads. Developmental dynamics. 1998;213:105–113. doi: 10.1002/(SICI)1097-0177(199809)213:1<105::AID-AJA10>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 117.Matthijs Blankesteijn W. Has the search for a marker of activated fibroblasts finally come to an end? J Mol Cell Cardiol. 2015;88:120–3. doi: 10.1016/j.yjmcc.2015.10.005. [DOI] [PubMed] [Google Scholar]
- 118.Strutz F, et al. Identification and characterization of a fibroblast marker: FSP1. The Journal of cell biology. 1995;130:393–405. doi: 10.1083/jcb.130.2.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Pilling D, Fan T, Huang D, Kaul B, Gomer RH. Identification of markers that distinguish monocyte-derived fibrocytes from monocytes, macrophages, and fibroblasts. PloS one. 2009;4:e7475. doi: 10.1371/journal.pone.0007475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Lane EB, Hogan BL, Kurkinen M, Garrels JI. Co-expression of vimentin and cytokeratins in parietal endoderm cells of early mouse embryo. Nature. 1983;303:701–4. doi: 10.1038/303701a0. [DOI] [PubMed] [Google Scholar]
- 121.Franke WW, Schmid E, Osborn M, Weber K. Intermediate-sized filaments of human endothelial cells. The Journal of cell biology. 1979;81:570–80. doi: 10.1083/jcb.81.3.570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Yuan SM. alpha-Smooth Muscle Actin and ACTA2 Gene Expressions in Vasculopathies. Brazilian journal of cardiovascular surgery. 2015;30:644–9. doi: 10.5935/1678-9741.20150081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kang J, et al. PDGF-A as an epicardial mitogen during heart development. Developmental dynamics : an official publication of the American Association of Anatomists. 2008;237:692–701. doi: 10.1002/dvdy.21469. [DOI] [PubMed] [Google Scholar]
- 124.Moore-Morris T, Cattaneo P, Puceat M, Evans SM. Origins of cardiac fibroblasts. Journal of molecular and cellular cardiology. 2016;91:1–5. doi: 10.1016/j.yjmcc.2015.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Morales MO, Price RL, Goldsmith EC. Expression of Discoidin Domain Receptor 2 (DDR2) in the developing heart. Microscopy and microanalysis : the official journal of Microscopy Society of America, Microbeam Analysis Society, Microscopical Society of Canada. 2005;11:260–7. doi: 10.1017/S1431927605050518. [DOI] [PubMed] [Google Scholar]
- 126.Shyu KG, Chao YM, Wang BW, Kuan P. Regulation of discoidin domain receptor 2 by cyclic mechanical stretch in cultured rat vascular smooth muscle cells. Hypertension. 2005;46:614–21. doi: 10.1161/01.HYP.0000175811.79863.e2. [DOI] [PubMed] [Google Scholar]
- 127.DeLeon-Pennell KY. May the fibrosis be with you: Is discoidin domain receptor 2 the receptor we have been looking for? Journal of molecular and cellular cardiology. 2016;91:201–203. doi: 10.1016/j.yjmcc.2016.01.006. [DOI] [PubMed] [Google Scholar]
- 128.Vitetta ES, Boyse E, Uhr J. Isolation and characterization of a molecular complex containing thy-1 antigen from the surface of murine thymocytes and t cells. European journal of immunology. 1973;3:446–453. doi: 10.1002/eji.1830030714. [DOI] [PubMed] [Google Scholar]
- 129.Wang X, et al. The role of the sca-1+/CD31- cardiac progenitor cell population in postinfarction left ventricular remodeling. Stem cells. 2006;24:1779–88. doi: 10.1634/stemcells.2005-0386. [DOI] [PubMed] [Google Scholar]
- 130.Valente M, Nascimento DS, Cumano A, Pinto-do OP. Sca-1+ cardiac progenitor cells and heart-making: a critical synopsis. Stem cells and development. 2014;23:2263–73. doi: 10.1089/scd.2014.0197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Serini G, et al. The fibronectin domain ED-A is crucial for myofibroblastic phenotype induction by transforming growth factor-beta1. The Journal of cell biology. 1998;142:873–81. doi: 10.1083/jcb.142.3.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Arslan F, et al. Lack of fibronectin-EDA promotes survival and prevents adverse remodeling and heart function deterioration after myocardial infarction. Circulation research. 2011;108:582–92. doi: 10.1161/CIRCRESAHA.110.224428. [DOI] [PubMed] [Google Scholar]
- 133.Baek ST, Tallquist MD. Nf1 limits epicardial derivative expansion by regulating epithelial to mesenchymal transition and proliferation. Development. 2012;139:2040–9. doi: 10.1242/dev.074054. [DOI] [PMC free article] [PubMed] [Google Scholar]


