This review by Deschamps and Duboule discusses recent advances in collinear regulation of Hox genes in space and time as well as novel concepts associated with large-scale regulation and chromatin structure during the development of both axial and limb patterns.
Keywords: chromatin, collinearity, embryos, Hox genes, TAD, transcription, stem cells
Abstract
Collinear regulation of Hox genes in space and time has been an outstanding question ever since the initial work of Ed Lewis in 1978. Here we discuss recent advances in our understanding of this phenomenon in relation to novel concepts associated with large-scale regulation and chromatin structure during the development of both axial and limb patterns. We further discuss how this sequential transcriptional activation marks embryonic stem cell-like axial progenitors in mammals and, consequently, how a temporal genetic system is further translated into spatial coordinates via the fate of these progenitors. In this context, we argue the benefit and necessity of implementing this unique mechanism as well as the difficulty in evolving an alternative strategy to deliver this critical positional information.
“In order to stop at any determined point in time, there is a lever to lock the triple brake”
How to construct a time machine (Alfred Jarry 1899)
During the development of vertebrate animals, Hox genes provide positional information to the emerging embryonic axial tissues, thereby instructing them how to undergo appropriate morphogenesis (Krumlauf 1994; Wellik 2009). While this local patterning of embryonic tissues by various combinations of Hox proteins likely emerged early during evolution, the underlying molecular mechanisms are diverse, depending on the ontogenetic and phylogenetic contexts. The strong adaptive value of this system ultimately relied on distinct distributions of proteins rather than the genetic mechanisms used to deliver this information. Consequently, various mechanisms seem to have evolved in bilaterian animals, depending on their developmental strategies to secure an appropriate spatial coordination in Hox gene expression along the emerging embryonic axes. However, in many classes of bilateria, this coordination is associated with the process of “collinearity,” whereby clustered Hox genes are functional in a series of spatial domains that recapitulates the order of the respective genes in their genomic cluster (Lewis 1978; Gaunt et al. 1988). Nevertheless, this rule has many exceptions, and animals as diverse as the urochordate Oikopleura or the fruit fly Drosophila display collinear Hox gene expression either without any Hox clusters or with a split series of genes, respectively (Harding et al. 1985; Seo et al. 2004; see Duboule 2007), thus illustrating that various mechanisms can eventually deliver similar information.
Vertebrates organize their bodies through progressive growth from anterior to posterior structures. Hox genes are activated in a timed sequence, which follows their 3′-to-5′ genomic order—a process referred to as “temporal collinearity” (Dolle et al. 1989; Izpisua-Belmonte et al. 1991). This time-controlled transcriptional activation (the Hox clock) has been observed thus far only in animals containing an intact cluster of genes and occurs in a growth zone, a progenitor region located at the posterior aspect of the extending body axis. Therefore, temporal collinearity is a property displayed by animals with an anterior-to-posterior developmental progression in time, whereas strategies relying mostly on other developmental principles that do not involve a progressive anterior-to-posterior determination do not call for this process. As a consequence, they may not display an intact Hox cluster (Duboule 1994). In Drosophila, for instance, transcriptional activation of Hox genes is not sequential in time but instead depends on the regional activities of both gap and segmentation genes (Ingham and Martinez-Arias 1986; Casares and Sanchez-Herrero 1995).
Mammals achieve the appropriate Hox-mediated spatial patterning through the initial timed-sequenced activation of their Hox clusters in response to early embryonic signals (Forlani et al. 2003; Neijts et al. 2016). During axial elongation, they use this early timing mechanism to synchronize Hox gene expression with the progressive generation of the trunk and tail from the posterior embryonic growth zone (Neijts et al. 2016, 2017). This Hox clock is critical for the spatial distribution of the patterning information both in various axial tissues and along the appendicular axes (Fig. 1). Here we discuss recent progress in understanding the molecular mechanisms at work in mouse embryos, which orchestrate their Hox expression program during embryogenesis. In the context of a permissive genome topology, initial Hox gene activation responds to time-dependent embryonic signals acting on enhancers located on the early side of the cluster, and collinear Hox gene expression is relayed to the regional tissue anlagen that generates axial structures. The cis-acting mechanisms underlying this time-dependent developmental sequence may constitute an important constraint in the organization of an evolutionarily conserved Bauplan in vertebrates.
Figure 1.
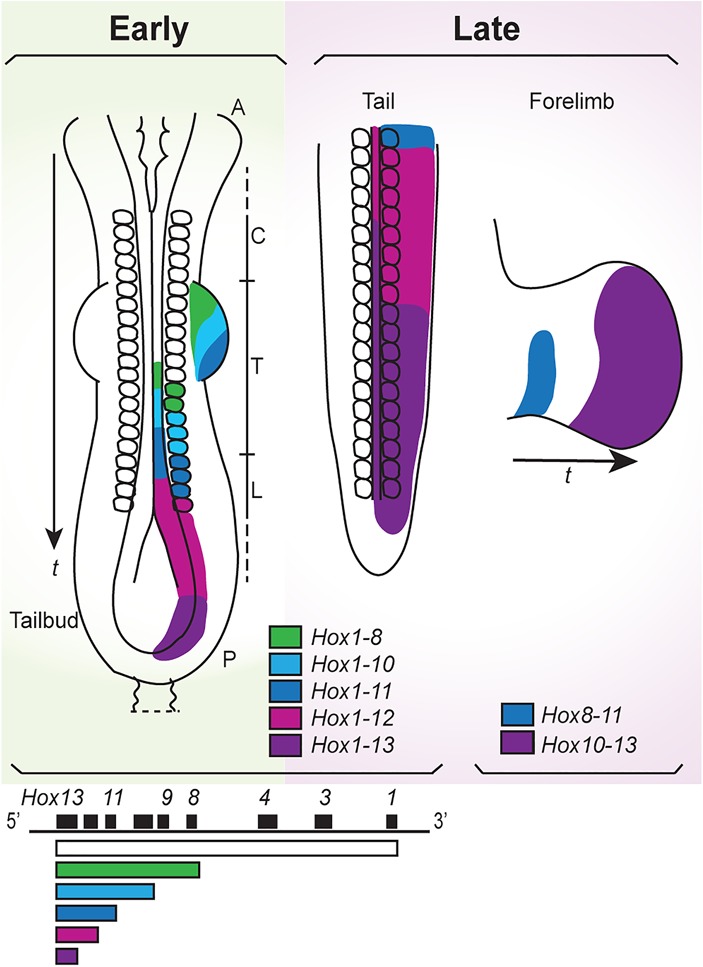
Collinear expression of Hox genes during the development of trunk axial tissues and limbs. (Left) Early: Schematic drawings of posteriorly overlapping transcript domains of HoxD genes in developing trunk axial tissues and early limb buds of an embryonic day 9.5 (E9.5) mouse embryo. Hox-expressing tissues are neural tube (midline); somites (blocks along the neural tube) labeled as cervical (C), thoracic (T) and lumbar (L); forelimb mesoderm (lateral bulges at the level of somites 7–12); and nascent mesoderm and neurectoderm in the tailbud. (t) Time of anterior to posterior development. Colinearity between the positions of the genes in the cluster and the anterior extension of their expression domains along the antero–posterior embryonic axis is illustrated both in the schematic embryo and by the bars under the cluster drawn below. Hox1 is expressed from an anterior limit that is the most rostral of all Hox genes in the embryo (expression not shown in the embryo; no color in the bar), and Hox8 is expressed from an anterior limit that is less rostral than that of Hox1 in the embryo. Posterior to this Hox8 anterior expression boundary, all genes between 1 and 8 are expressed (green color in the schematic embryo and in the bar corresponding to Hox8). Similarly, posterior to the Hox10 expression boundary in the embryo, all genes between Hox1 and Hox10 are expressed (lighter blue in the schematic embryo and in the bar corresponding to Hox10), and similar representations illustrate the expression of Hox11, Hox12, and Hox13. These domains thus tend to overlap posteriorly in the embryo, like Russian dolls. Expression of other Hox genes is not shown. (Right) Late: Hox transcript distribution in the E10.5 developing tailbud and forelimb bud. The two domains in late forelimb buds mark the future proximal (arm and forearm) and distal (digits) parts of the adult limbs, respectively. (t) Time of development of proximal to distal limb structures. Color codes indicate the cumulative amounts of combinations of Hox transcripts. Anterior is to the top in all schemes.
Embryonic timing and Hox gene expression
In murine embryos, the positional identities of post-occipital tissues (i.e., located from below the skull to more caudal areas) are acquired in progenitor cells located in the posterior embryonic growth zone. This zone includes the rostral third of the primitive streak as well as its flanking epiblast. Its posterior-most portion produces the lateral and paraxial mesoderm from mesoderm progenitors (MPs) (Lawson et al. 1991; Kinder et al. 1999), whereas the rostral-most aspect of the streak abutting the node (the node–streak border) contains stem cell-like progenitors for both the mesoderm and neurectoderm (Cambray and Wilson 2002, 2007). These latter bipotent progenitors, called neuromesodermal progenitors (NMPs), appear in the embryo at around embryonic day 7.5 (E7.5), just before the first somite is formed (Wymeersch et al. 2016). They constitute a tightly regulated and likely changing population of self-renewing cells, feeding the elongating axis with new mesoderm and neurectoderm tissues. As they differentiate, NMPs progressively populate more posterior embryonic axial tissues (Tzouanacou et al. 2009; Wilson et al. 2009). These progenitors, which are initially present in the anterior streak, are subsequently found in the tailbud between E10.5 and E13.5, where they contribute to the formation of the most caudal part of the axis.
Hox genes are initially switched on in the posterior streak area as a response to Wnt3 signals (Neijts et al. 2016). Their expression domains subsequently spread anteriorly through a process that does not involve cell migration (Deschamps and Wijgerde 1993; Gaunt and Strachan 1994). The rostral expansion of the Wnt3 (and later Wnt3a) domain at gastrulation is followed by a similar expansion of 3′-located Hox transcripts toward the progenitor region (Fig. 2). Successively more 5′-located Hox genes are induced thereafter and follow the same dynamics. As a result, the anterior streak region, which contains the MPs and NMPs, sequentially transcribes an increasing number of more “posterior” Hox genes. Thus, an early Wnt-dependent wave of collinear Hox gene transcription along the streak (phase 1 expression) instructs MPs and NMPs, the descendants of which assume embryonic spatial patterning. In this context, early to later axial progenitors acquire group 1–13 combinatorial Hox addresses at the time that they release descendants that generate tissues at various future anterior to posterior body levels. This process provides synchronicity between the acquisition of genetic positional information over time (temporal collinearity) and the anterior to posterior levels where progenitors carrying this information contribute during axial elongation (spatial collinearity)—a translation from temporal information to a set of spatial coordinates.
Figure 2.
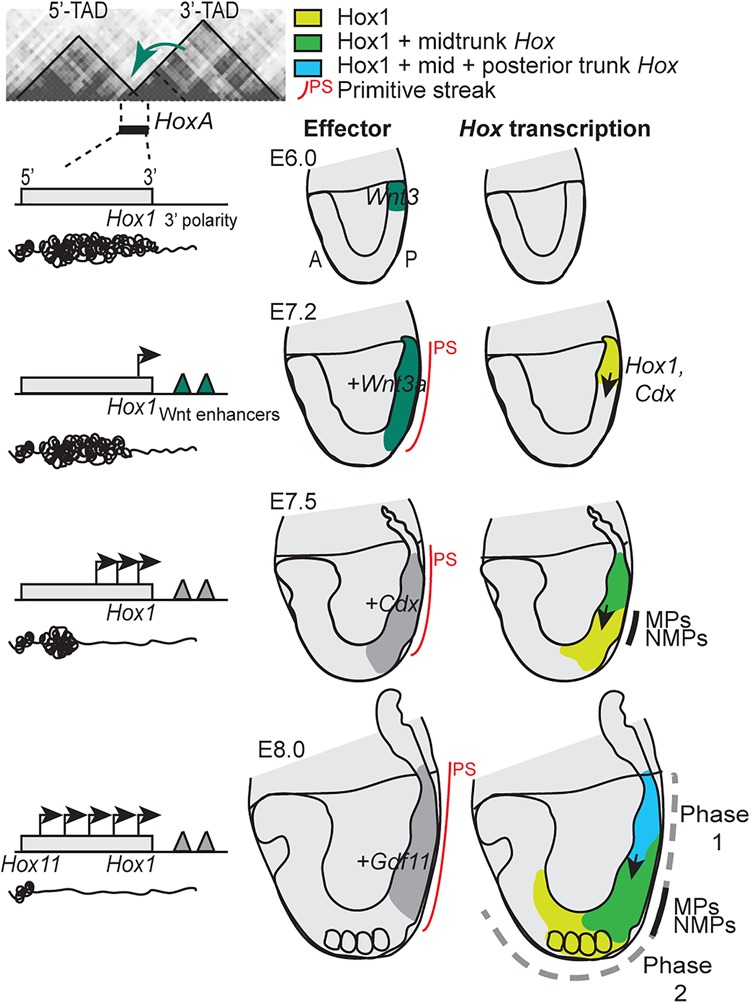
Temporal collinearity of Hox gene transcription is relayed by axial progenitors into spatially collinear expression domains. Schematics showing the early expression of HoxA genes. The HoxA cluster is represented in reversed genomic orientation in order to appear comparable with HoxD (Fig. 3). Out of the Hox1 to Hox13 genes, only the 3′-most-located Hox1 and 5′-most-located gene expressed at the stages considered (Hox11) are indicated below the cluster (light-gray box). (From top to bottom) Developmental stages and associated genomic events at the Hox locus (left) and transcription domains of Hox genes (embryos at the right) and of their inducers (embryos on the left). (E6.0) Preferential interactions between 3′-located Hox genes and the 3′ topologically associating domain (TAD). No Hox expression was detected, but priming of the 3′ flanking region (open chromatin) is shown in the Wnt3-expressing region. (E7.2) Hox (and Cdx) transcriptional initiation by Wnt3 and Wnt3a via Wnt-dependent enhancers in the proximal 3′ flanking region. (Light green) Expression domain of Hox1 and Cdx. (E7.5) Mid-trunk Hox gene activation (dark-green domain) through the binding of Cdx products in the cluster and further chromatin opening. Mid-trunk Hox domains reach the axial progenitor region (or the posterior growth zone) in the anterior part of the streak. (E8.0; early somite) Gdf11-induced activation of posterior trunk Hox genes (blue). Expansion of trunk Hox gene transcripts (Hox1 [light green] and mid-trunk Hox [dark green]) into the axial progenitor region (end of phase 1). The color code indicates cumulative amounts of Hox transcript combinations. The expression domains overlap posteriorly, as explained in the legend for Figure 1. For all embryos, anterior is to the left. (PS) Primitive streak. (Arrows) transcriptional activation. Genome topology at the top left is from Dixon et al. (2012).
Directionality of the Hox clock
In vertebrates, all Hox genes of a cluster are transcribed from the same DNA strand, and the first to be expressed are members of groups 1 and 2 (i.e., those genes located at “3′ ends” of the gene clusters) followed by more 5′-located genes. However, while an inversion in this transcriptional directionality (starting from group 13) led to early lethality (Zakany et al. 2004), the factors determining and initiating this obligatory time sequence are not yet fully understood. They may involve a pre-existing favorable genomic topology, which would skew gene activation toward one side of the gene clusters. The asymmetric distribution of Wnt-responsive enhancer sequences on the early side of the cluster (Fig. 2; Neijts et al. 2016) also likely constitutes such a bias. A few hours after, the more centrally located Hox genes belonging, for example, to groups 4–10 are activated, and their transcription is further enhanced by a feed-forward effect of Cdx gene products, which are Wnt-dependent transcription factors initially expressed like 3′-located Hox genes. This stimulating effect of Cdx proteins over these central Hox gene transcriptions (Neijts et al. 2017; see also Bel-Vialar et al. 2002; van den Akker et al. 2002) seems to occur through binding sites within the Hox clusters, as shown in the case of the HoxA locus (Mazzoni et al. 2013; Amin et al. 2016; Neijts et al. 2017).
On the other hand, enhancement of transcription of the more 5′-located “posterior” Hox genes (e.g., group 11) responds to the presence of Gdf11, a TGFβ signal released by posterior embryonic tissues after E8.0 (Liu et al. 2001; Liu 2006; Jurberg et al. 2013). Therefore, beyond a pre-existing chromatin architecture, the precise timing of distinct signals during the progression of embryogenesis appears to determine a global collinear activation of Hox gene transcription: Wnt3 and Wnt3a signaling induces anterior genes (Neijts et al. 2016), and Cdx proteins stimulate trunk/central genes (Neijts et al. 2017), whereas Gdf11 further activates group 11 Hox genes (Gaunt et al. 2013) as well as groups 12 and 13 (our unpublished data). While the temporal progression within these various groups of genes responding to the same factor remains to be understood, it may rely on a progressive and directional opening of a chromatin structure (see below).
Axial progenitors translate the Hox clock into spatial coordinates
The onset of temporal collinearity is determined by Wnt signaling, a parameter dependent on the developmental timing of the embryo itself (Neijts et al. 2016). In contrast, the end of sequential Hox gene activation is fixed by the transcription of the last genes of the clusters, members of group 13 (Neijts et al. 2017). These genes play a crucial role in retroinhibiting the functions of more anterior HOX proteins and arrest axial elongation (Young et al. 2009), thus explaining the early lethality of mice carrying an inverted Hox cluster (see above). This negative feedback may involve a dominant-negative effect, referred to as posterior prevalence; i.e., the prevalent function of posterior HOX proteins over their more anterior neighbors. This effect was observed in many instances and may rely on various mechanisms (e.g., see Gonzalez-Reyes et al. 1990; Duboule and Morata 1994; Yekta et al. 2008; Durston and Zhu 2015). It coincides with both the slowing down of axial tissue production and a reduction in the size of the presomitic mesoderm (PSM) (Gomez and Pourquie 2009; Denans et al. 2015). In this view, the Hox clock must be tightly synchronized with the intrinsic timing of the embryo, the developmental clock.
Because these initial collinear transcript domains expand anteriorly into the region of axial progenitors, early MPs and NMPs express 3′-located Hox genes exclusively and thus have an anterior identity, whereas progenitors present at a later time express more 5′-located Hox genes in addition to anterior genes and thus have a more posterior identity (Figs. 2, 3). Subsequently, progenitors transmit these identities to their differentiated cell progenies, which contribute to the elongating tissues along the growing axis. The transmission of these identities marks the end of the early phase of Hox gene transcription (phase 1) (Figs. 2, 3). A second phase of Hox expression (phase 2) then takes place as the axial progenitors generate the elongating neurectoderm and mesoderm, with different regulatory processes depending on both the germ layer and the axial level.
Figure 3.
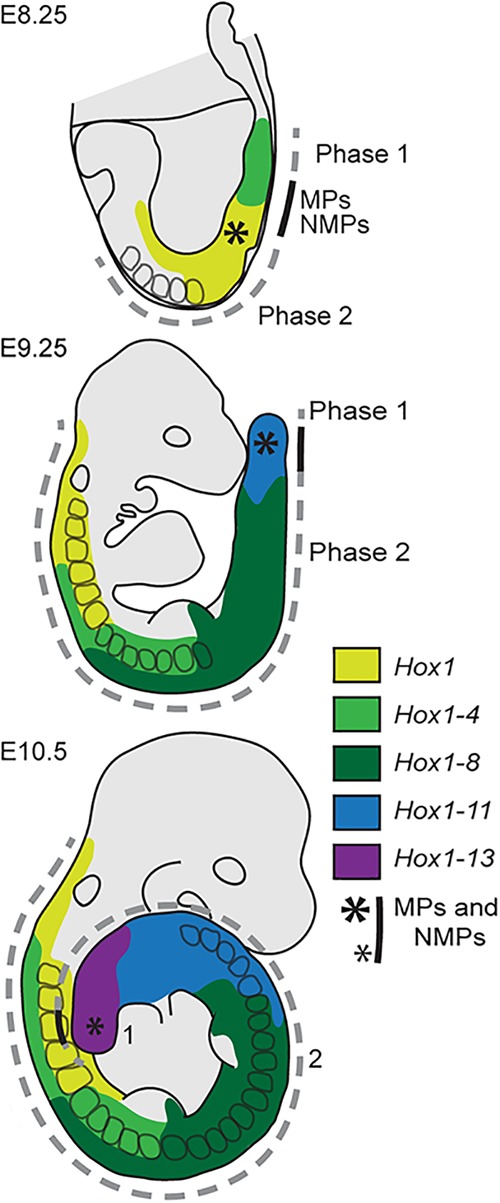
Early temporal collinearity of Hox gene transcription is relayed by the axial progenitors into spatially collinear expression domains in the emerging differentiated tissues. (E8.25) The transcript domains of Hox1 followed by that of Hox2 to Hox4 (phase 1, temporal collinearity) expand anteriorly and reach the axial progenitor region ([*] MPs and NMPs) before expanding further anteriorly during phase 2. (E9.25) Transcriptional initiation of the next Hox genes has occurred (mid-trunk Hox4 to Hox8), and activation of more posterior Hox genes takes place (as shown with Hox11). The expression domains expand anteriorly toward the axial progenitor area in those tissues generated by the descendants of the MPs and NMPs (phase 2, spatial collinearity). (E10.5) Hox13 is now transcriptionally activated and expressed strongly posteriorly, where it counteracts further axial extension. The Wnt and Fgf pathways are weakened, and the progenitor niche becomes deficient, with fewer and fewer MPs and NMPs (smaller asterisk). The color code identifies the combinations of Hox genes expressed along the axis at the different stages. MPs and NMPs are found around the anterior part of the primitive streak, just posterior to the node (indentation) at E8.25 and in the tailbud at E9.25 and E10.5. For the E8.25 embryo, anterior is to the left; for the E9.25 and E10.5 embryos, anterior is to the top.
Therefore, Hox patterns are established in time in a collinear manner during phase 1 and instruct early to late axial progenitors about their anterior to posterior identity, respectively. These identities are further transmitted to their differentiated progenies, which expand and contribute to axial extension. These Hox patterns are maintained during phase 2, which occurs at early somite stages for 3′-located (or “anterior”) Hox genes (Figs. 2, 3) and continues until after the embryo has turned, when progenitors for the posterior trunk are located in the tailbud (Fig. 3, E9.25 embryo). This lasts until E10.5, when the tailbud contains only a decreasing population of progenitors for tail tissues. Therefore, from an early to a late developmental stage, axial progenitors for anterior to more posterior axial tissues first acquire their Hox addresses and subsequently relay this information to their progeny in the emerging differentiated neurectoderm and mesoderm.
On top of these global regulations, additional gene- and/or tissue-specific regulatory events may fine-tune the Hox expression domains in axial tissues. While the rostral expression boundary of any given Hox gene will eventually be more anterior in the neurectoderm than in the mesoderm, which may reflect distinct properties of these cellular subsets produced by the progenitor pools, expression might either be reinforced by autoregulatory loops (Popperl et al. 1995) or undergo a late retinoic acid-dependent anterior shift in other cases (Oosterveen et al. 2003).
Relay from early temporal to spatial Hox expression: a mechanism conserved in vertebrates
Recent findings in zebrafish (e.g., Steventon et al. 2016) in addition to previous literature suggest that the translation of an initial Hox expression timing into successive anterior-to-posterior instructions to axial tissues is a feature conserved throughout vertebrates despite the apparent differences in how these organisms gastrulate. In amphibian and murine embryos, for example, Wnt signals precede the onset of Hox gene activation in a ring around the marginal zone in Xenopus laevis (Holland 2002) or along the corresponding structure in mice—the primitive streak, respectively. In both species, sequential Hox gene transcription further expands into presumptive trunk tissues as gastrulation proceeds, as schematized for mice (Figs. 2, 3; see also Deschamps and van Nes 2005) and described for Xenopus (Wacker et al. 2004). However, slight differences exist; Hox transcripts are found in a large marginal zone in amphibians that converges toward the blastopore lip, whereas the tiny initial murine Hox domains expand anteriorly along the streak toward the regions containing MPs and NMPs. The situation in bird embryos is somewhat similar to that in mice (Iimura et al. 2007; McGrew et al. 2008), while the situation in fish is closer to that of amphibians (Alexandre et al. 1996). The embryonic domain fated to contribute to trunk tissues is thus larger in early fish and amphibian embryos than in mice, where a small population of axial progenitors ensures most of the axial growth in the trunk. In zebrafish, NMPs contribute significantly to axial growth only in the tailbud (Martin and Kimelman 2008; Steventon et al. 2016), and a low number of multipotent progenitors are found at relatively late stages in the Xenopus tailbud (Davis and Kirschner 2000).
Despite these differences, the translation of sequential Hox gene activation into spatial cues in axial tissue anlagen occurs in all vertebrates thus far examined, including amphibians (Durston et al. 2012). Since this feature is crucial for patterning along the main axis and because it is dependent on a timing device that is itself intrinsically linked to the physical arrangement of genes along a genomic cluster, we conclude that the meta-cis arrangement of Hox genes is constrained by the necessity to properly and safely implement the Hox clock. The existence of this particular constraint in all vertebrates was proposed earlier to coincide with the narrow passage of the phylotypic hourglass; i.e., a short time window during which various developmental strategies converge toward completion of axial growth (Duboule 1994; Raff 1996). As we discussed here, recent data indicate that the core of this obligatory process includes the relay of temporally acquired Hox codes to the precursors of anterior to posterior axial tissues.
A link between the Hox clock and the somitic clock?
Once the successive waves of Hox gene expression have progressively labeled the axial progenitors for trunk tissues in the node-abutting posterior growth zone (Figs. 2, 3), the progenitors transmit their Hox addresses to their daughter cells in both the mesoderm and neurectoderm of the emerging axial tissues. Early progenitors relay an anterior code, while later progenitors transmit a more posterior genetic address until the progenitor population is exhausted by the end of axial elongation. In nascent paraxial mesoderm, the proliferation of Hox-instructed cells takes place concomitantly with the segmentation of the PSM into somites, a series of discrete compartments that foreshadows the future vertebrae. The sequential production of somites results from an oscillatory mechanism (the “somitic clock”) (Palmeirim et al. 1997), and potential connections between the two clocks were documented in both mice (Zakany et al. 2001) and chicks (Dubrulle et al. 2001), where early expression of several anterior Hox genes was observed to cycle and follow a particular phase of the somitogenesis cycle (Pourquie 2003 and references therein).
Nevertheless, this connection between both clocks has not yet been functionally validated, and thus a mechanism synchronizing these clocks is still elusive. The necessary genetic approaches are complicated due to the high level of redundancy found in the developing PSM, where all four Hox gene clusters are at work. Alternatively, the somitic clock may operate independently from the Hox clock; i.e., by segmenting a tissue where the set of Hox addresses would already be properly distributed due to its earlier activation.
Hox genes and the control of body length
The length of the trunk depends on the activity of axial progenitors. In addition to niche factors, which are essential for maintaining these progenitors (Wilson et al. 2009; Neijts et al. 2014), the pluripotency factor Oct4 is a crucial player in determining progenitor activity. A sufficiently high level of Oct4 expression in the posterior aspect of the early mouse embryo is essential to maintain the pluripotency network active in the epiblast, and Oct4 levels normally decrease at the three- to five-somite stage (Osorno et al. 2012). Experimental stimulation of early Oct4 expression produced a longer trunk in mice, as shown by using the Cdx2 promoter driving Oct4 in early mouse embryos. Aires et al. (2016) increased both the level and the time of expression of Oct4 in posterior epiblasts, including in the caudal lateral epiblast and the NMP region. This presumably overruled the reduction in Oct4 expression and associated decline in pluripotency, which normally occurs in posterior epiblasts after the three- to five-somite stage, and allowed for an extended activity of the pluripotency network, leading to a longer trunk.
Hox genes, together with the related Cdx genes, also play a role in trunk extension. Therefore, they do more than merely confer axial identity to the emerging tissues. Cdx genes are required in a dosage-dependent manner for the generation of post-occipital embryonic axial tissues (van Rooijen et al. 2012). Cdx2 transcriptionally activates Wnt and Fgf signaling pathway components in posterior embryonic tissues (Amin et al. 2016) and thus maintains the proficiency of the axial progenitor niche. Accordingly, experimental stimulation of both the Wnt and Fgf pathways rescues the axial truncation of Cdx mutants at least in part (Young et al. 2009; van Rooijen et al. 2012). Middle or trunk Hox genes can substitute for Cdx genes in axial extension, demonstrating that these Hox gene products stimulate trunk growth, presumably by maintaining a proficient niche of axial progenitors. The trunk growth-stimulating action of Cdx and Hox genes is amplified by a feed-forward activation of Hox genes by Cdx2 (Neijts et al. 2017).
After the trunk-to-tail transition, posterior Hox genes become highly expressed in most caudal embryonic tissues. These genes—in particular Hox13—retroinhibit more anterior Hox genes (Denans et al. 2015; Beccari et al. 2016; Sheth et al. 2016). In addition, they antagonize Cdx2 and more centrally located Hox genes in their task of axial stimulation by directly interfering with activation of the Wnt and Fgf pathways (Amin et al. 2016). Posterior Hox genes thus intervene to slow down and interrupt the axial elongation process by preventing Cdx and middle Hox genes from further activating axial progenitors, MPs, and NMPs at caudal axial levels.
Early temporal collinearity is required for subsequent expression during development
The time-sequenced activation of murine Hox genes in the posterior primitive streak (Fig. 2, phase 1) inherently leads to their sequential transcription in the precursors of axial tissues. This corollary is also observed in birds (Iimura and Pourquie 2006), amphibians (Wacker et al. 2004), and fish (Alexandre et al. 1996), indicating that this feature is an evolutionarily conserved mechanism, fundamental for the organization of the vertebrate body plan. Here again, the underlying molecular mechanism is not yet fully understood. It nevertheless involves the concomitant and directional release of H3K27me3 (Soshnikova and Duboule 2009; Neijts et al. 2016), a histone H3 modification introduced by the Polycomb-repressive complex 2 (PRC2) and specific for inactive chromatin regions (see Margueron and Reinberg 2011). Through the recruitment of PRC1, these negative domains form compact structures from which transcribed Hox genes selectively escape, depending on the developmental context (Noordermeer et al. 2011; Mazzoni et al. 2013; Kundu et al. 2017).
At developmental time points earlier than the gastrulation stage epiblast, as in inner cell mass (ICM)-derived embryonic stem cells (ESCs), Hox genes are not transcribed. In these cells, the PRC2 coverage is not as dense as in Hox-negative differentiated cells (Noordermeer et al. 2011), likely due to the presence of bivalent positive and negative chromatin modifications of histone H3 (Bernstein et al. 2006). This moderately compacted poised state can also be observed through high-resolution stochastic optical reconstruction microscopy (STORM) (Fabre et al. 2015). These observations suggest that Hox gene expression and function at later developmental stages and in adult tissues are contingent on their initial phase of activation, which thus fixes the part of the gene cluster that remains at work at a given body level (the “open for business” model) (Duboule 1992 and references therein; Gaunt 2015 and references therein; Maeda and Karch 2015 and references therein).
Open for business or open by business?
While the mechanism underlying the “open for business” model comprises similar components in flies and mammals (i.e., the establishment of dynamic “open” and “closed” chromatin domains early on) (Soshnikova and Duboule 2009; Bowman et al. 2014), the mammalian version implies that the extent of such domains is regulated initially as a function of time, unlike in Drosophila. In fact, the highly versatile removal of PRC-dependent histone modifications on chromatin along with the progression of transcription in time and the concurrent change in the three-dimensional (3D) structure (Noordermeer et al. 2014) raise the question of causality: Do chromatin domains set or maintain the expression of subsets of Hox genes or, alternatively, only reflect the transcriptional mechanisms at work?
While recent work in Drosophila suggests a causal role for Polycomb-dependent chromatin modifications in the epigenetic memory of a given transcriptional state (Coleman and Struhl 2017), the situation in vertebrates, which lack clear Polycomb response elements (PREs), is unclear. The cellular memory of various combinations of transcribed Hox genes and hence their transmission to daughter cells may indeed rely on a reinforcement of PRC-based compaction of the silent parts of Hox clusters after an initial temporal activation in early embryos. However, this activation may not depend on the removal of “negative” chromatin marks but instead may trigger this transition in chromatin structure. Hox genes might thus remain differentially active in the progenies of axial stem cells (Fig. 2, phase 2) either permanently or transiently. Interestingly, silent genes may be reactivated at later time points in various cells and tissues where their function was subsequently co-opted during evolution. In such cases, subgroups of Hox genes are often found (re)activated together, as in the cases of spinal motoneurons (Dasen and Jessell 2009; Mazzoni et al. 2013), the intestinal caecum (Delpretti et al. 2013), or the developing metanephros (Di-Poi et al. 2007), in response to potent remote enhancer sequences.
The various subgroups of contiguous Hox genes showing either sustained transcription or subsequent reactivation could be defined by the distribution of bound CTCF proteins, as the deletion of specific CTCF-binding sites within the HoxA and HoxC clusters was found recently to locally disturb the organization of chromatin domains and cause Hox gene misregulation, leading to patterning defects and motoneuron misspecification in mice (Narendra et al. 2015, 2016). As active versus inactive domains of Hox clusters are labeled by the absence or presence of Polycomb complexes, an interplay between these complexes and CTCF to establish these domains may occur (Narendra et al. 2015). In some normal or pathological cases, however, particular Hox genes seem to be transcribed in isolation from their immediate neighbors (e.g., see Seifert et al. 2015), thus escaping any type of collinear regulation, a situation difficult to reconcile with the above-mentioned hypothesis.
Genomic topologies and dynamic architectures
The collinear regulation observed during trunk extension (described above) has long been thought to rely mostly on mechanisms involving regulatory sequences internal to the gene clusters (Spitz et al. 2005), such as enhancer sharing (Gould et al. 1997), based on results from transgenic animals where isolated (groups of) Hox genes were activated at about the right anterior–posterior positions. However, recent work indicates that the surrounding 3D chromatin architecture plays a critical role. Indeed, both the correct implementation of the collinear mechanism and the selection of which subgroups of Hox genes ought to be transcribed at any given time in various embryonic and adult contexts as well as in stem cells depends at least in part on regulatory sequences located outside the gene clusters.
Both HoxA and HoxD (i.e., the two mammalian clusters showing the most compact and complete structure) are flanked by two topologically associating domains (TADs), which are constitutive chromatin domains (Dixon et al. 2012; Nora et al. 2012) containing distinct sets of enhancers (Andrey et al. 2013; Berlivet et al. 2013; Neijts et al. 2016). At the HoxA locus, the 3′-located TAD (referred to here as “3′ TAD”) appears to contain enhancers necessary for activation of the earliest Hox gene (Fig. 2; Neijts et al. 2016; Cao et al. 2017), as if interactions between the gene cluster and the flanking regulatory landscape were required to properly control the timing of the system. Deletion of two discrete 3′ enhancers was shown to impair HoxA activation and strongly alter the profile of 3D interactions between Hoxa1 and its 3′ and 5′ surroundings in differentiating ESCs (Cao et al. 2017). This shows that 3D chromatin architecture at the 3′ side of the Hox locus is dynamic and intimately linked to enhancer-driven gene activation. This may be one of the reasons why global interactions between Hox genes and this particular 3′-located landscape seem to be evolutionarily more conserved than interaction occurring at the 5′ side of the gene cluster, as shown by the presence of a single large TAD in amphioxus (Acemel et al. 2016), whereas the presence of two distinct TADs was observed from fish to mammals (Woltering et al. 2014). Subsequently, these two TADs help to partition regulatory inputs to those subsets of Hox gene targets that are transcribed in a variety of contexts. This segregation of productive interactions between either 3′-located Hox genes and their associated 3′ TAD or 5′-located genes and the other TADs have been well characterized in the context of appendicular development.
Limb axes (temporal collinearity)2
Once progressive Hox gene activation has occurred, driven by sequences within the 3′ TAD, different parts of the Hox cluster are recruited along with the development of specific structures but always in relation to either one of the two flanking TADs. For example, the subset of Hoxd genes transcribed in the developing caecum or in mammary buds interacts only with the 3′ TAD, whereas another subset of genes expressed in the developing genitalia is regulated by sequences located within the 5′ TAD (Delpretti et al. 2013; Lonfat et al. 2014; Schep et al. 2016). This selectivity and exclusiveness in TAD functions (the 5′ TAD and 3′ TAD are never active at the same time) underlie collinear regulation during the development of our appendicular axes. There again, indeed, gene alignment is used in various ways to elicit a global collinear response in both time and space.
Limb buds arise from the lateral plate mesoderm at defined axial levels. At early stages of limb budding, Hoxd genes are transcribed in the same spatio–temporal order as during trunk extension. During this early phase (“limb phase 1”) (Fig. 4), the Hox clock is implemented, and 3′-located genes are activated sequentially at the posterior aspect of the limb buds, leading to nested transcript domains, as found with Russian dolls (Fig. 3; Dolle et al. 1989; Nelson et al. 1996; Tarchini and Duboule 2006). During this initial phase of transcription, the underlying mechanism is likely identical to that at work during trunk extension and specifically involves interactions with the 3′ TAD, as shown by targeted chromosome engineering (Andrey et al. 2013). The outcome of this first wave of transcription is to help grow and establish proximal patterns (the arm and the forearm) (Zakany and Duboule 2007 and references therein).
Figure 4.
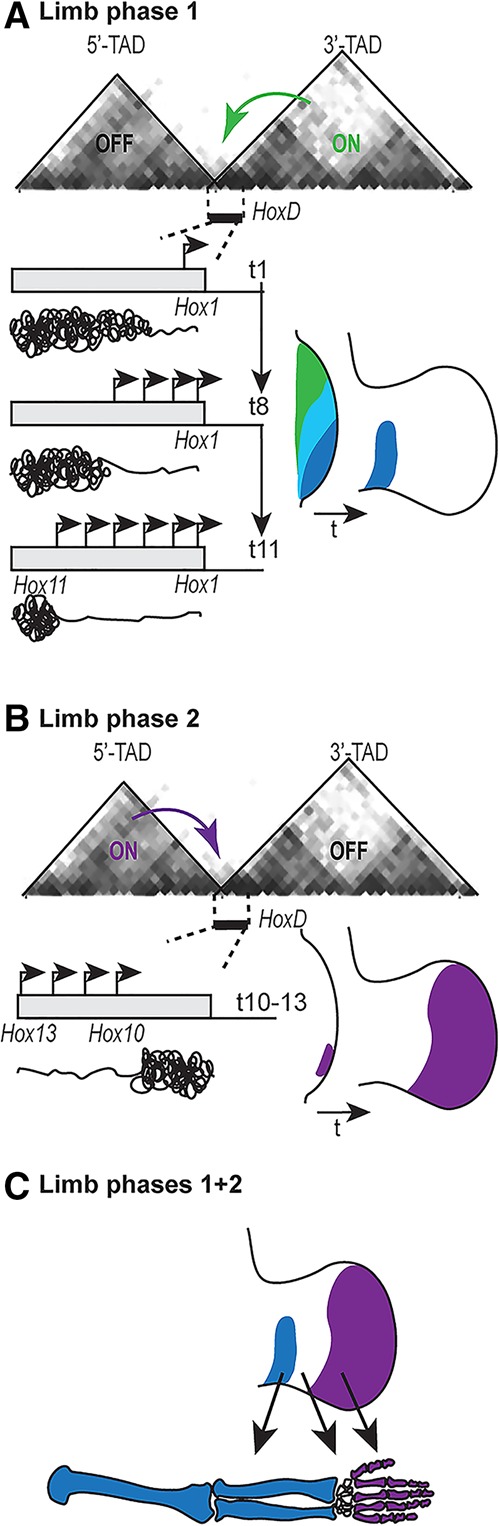
Two temporal sequences in Hoxd gene activation during limb bud development. (A) Limb phase 1. HoxD in the early bud is controlled (green arrow) exclusively by enhancers located in the 3′ TAD. Time sequence of Hoxd activation (t1 to t11) is the same as in the trunk (Fig. 1), and the nested expression domains generated occupy the proximal part of the E12.5 limb bud (right limb bud) and pattern the long bones along with genes from the HoxA cluster. Arrows are transcriptionally active genes, corresponding to open chromatin. (B) Limb phase 2. Between 24 and 48 h after the start of phase 1, enhancers in the 5′ TAD are switched on and activate Hoxd10 to Hoxd13 (purple arrow). HOX13 products then switch off the 3′ TAD enhancers in cells located at the posterior-distal margin. These cells expand and generate the autopods (hands and feet). (C) Combination of limb phases 1 and 2. The 3′ and 5′ TADs operate sequentially, leaving a zone of low Hox expression as the limb bud grows distally, which generates the future mesopods (wrist and ankle). Anterior is to the top, and distal to the right. The color code for Hox gene expression is the same as in Figure 1.
Transcription timing vs. evolutionary timing
About 24 h after budding, however, cells located at the posterior-distal aspect of the bud start expressing simultaneously a series of “terminal” Hoxd genes, including Hoxd13. This is the start of limb expression phase 2, which helps pattern the distal part of the limbs (the hands and feet). In these posterior-distal cells, Hoxd genes are no longer controlled by the 3′ TAD but instead are controlled by the opposite 5′ TAD (Fig. 3; Montavon et al. 2011; Andrey et al. 2013). At the time the 5′ TAD is switched on, the 3′ TAD enhancers terminate operating such that a second type of temporal collinearity is established, whereby enhancers located 3′ to the HoxD cluster work earlier than a large series of 5′-located enhancers. This topological switch in regulations is due in part to a retroactive negative effect of HOX13 proteins upon the activity of the 3′ TAD. This was shown by the genetic removal of the appropriate HOX13 proteins, which prevented the 3′ TAD from switching off; its enhancers continued to operate in most distal limb bud cells, where 5′ TAD enhancers should have normally operated (Beccari et al. 2016; Sheth et al. 2016).
In contrast, the mechanism involved in activating the 5′ TAD remains unknown. Using dissociated limb bud cells cultured in vitro, it was shown recently that the activation of the full set of Hoxd genes requires the presence of Wnt, Fgf8, and Shh signaling. In such a system, however, the switch from 3′ to 5′ TAD regulation was not clearly detected (Rodrigues et al. 2017). Notably, this 5′ TAD does not seem to exist in amphioxus (see above), raising the possibility of an evolutionary sequence in the appearance of these global regulatory landscapes, with the 5′ TAD emerging subsequently, with the evolution of vertebrates and their appendages (Acemel et al. 2016).
Therefore, during limb development, temporal collinearity comes under two different flavors, which likely reflect distinct key evolutionary steps. Initially, at the time that appendicular skeletons appeared, particular Hox combinations were recruited in cells from the lateral plate mesoderm, corresponding to the places of limb bud emergence. These cells had inherited their Hox code from their MPs, according to the temporal collinearity rule in the primitive streak. In these cells, the Hox clock further processed the state of “cluster opening” that had been memorized in space at these body levels during trunk extension. In this respect, patterning of proximal limb structures shares a genetic “history” with that of the major body axis (Woltering and Duboule 2010).
In a subsequent mechanistic phase, which probably appeared along with the emergence of the autopods (hands and feet), a distinct regulatory topology was recruited to control the last Hoxd genes into the newly appearing most distal pieces of our limbs. This second type of temporal collinearity involves 5′ TAD enhancers being switched on after 3′ TAD proximal enhancers and just before the latter regulatory sequences are switched off, thus somewhat recapitulating the evolutionary sequence of events leading to the actual morphology of the tetrapod limb. Interestingly, unlike in the trunk as mentioned previously, the relationship between both phases of Hox gene activation and the presence of limb stem cells remains elusive. The existence of such cells throughout limb bud development is controversial, and hence the restricted cell population expressing Hoxd13 and Hoxa13 at the start of limb phase 2 may either already have a transitory proximal fate or be kept at an early step of mesoderm commitment (for discussion, see Tabin and Wolpert 2007). In any case, however, these future distal cells went through the activation of the full repertoire of Hoxd genes before expanding distally.
The rationale of temporal collinearity
While other Hox-unrelated gene clusters are regulated coordinately (e.g., see Kim and Dean 2012), this precision in meta-cis regulation associated with temporal collinearity is thus far unique to Hox gene clusters. This time process requires a complete series of genes to be clustered, and hence the physical neighborhood of Hox genes at their genomic loci is a fundamental parameter to ensure a faithful anterior-to-posterior patterning in vertebrates. It is likely that this particular process was selected and conserved in vertebrates as well as in all animals developing their body through an anterior-to-posterior time progression (Duboule 2007) because of the double security that it offers in the processing of the patterning information. First, the progressive opening of Hox clusters from Hox1 to Hox13 following a set of signals and chromatin reorganization allows, in principle, the use of the same few activators for series of genes, the main parameter being promoter accessibility. This may greatly simplify the task, as a precise and sequential activation of series of up to 13 genes within a short time window based solely on a “classical” gene regulatory network in trans would have been prone to errors and variations, which cannot be tolerated in the developmental contexts concerned. Likewise, this meta-cis organization secures the proper order of the patterning information emitted, since, for example, group 7 Hox genes cannot be activated before group 5.
Second, this structural parameter also ensures that the Hox genes terminating the process, mostly the members of group 13, are not activated before the whole repertoire of information has been dispatched. Group 13 proteins exert a dominant-negative effect over other HOX proteins through posterior prevalence, leading to the abrupt termination of growth and patterning wherever such proteins are produced, as observed by either spontaneous mutations or induced gain-of-function experiments (e.g., see Herault et al. 1997; Young et al. 2009). Therefore, an essential prerequisite of the system is to prevent Hox13 genes from being activated before all other genes have achieved their tasks. However, it is equally important that Hox13 genes are activated as soon as the series has reached its end. In this context, it is likely that the most parsimonious way to properly solve this conundrum was to select such a meta-cis regulation, which may explain the evolutionary success of temporal collinearity.
Acknowledgments
We are grateful to Carina van Rooijen for her help with graphic work, and Roel Neijts for critical reading of the manuscript. We also thank all past and present members of the two laboratories for their collaboration and for participating in the emergence and development of the ideas discussed above. We apologize for not citing all of the relevant literature due to space limitations and aging memories.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.303123.117.
Freely available online through the Genes & Development Open Access option.
References
- Acemel RD, Tena JJ, Irastorza-Azcarate I, Marletaz F, Gomez-Marin C, de la Calle-Mustienes E, Bertrand S, Diaz SG, Aldea D, Aury JM, et al. 2016. A single three-dimensional chromatin compartment in amphioxus indicates a stepwise evolution of vertebrate Hox bimodal regulation. Nat Genet 48: 336–341. [DOI] [PubMed] [Google Scholar]
- Aires R, Jurberg AD, Leal F, Novoa A, Cohn MJ, Mallo M. 2016. Oct4 is a key regulator of vertebrate trunk length diversity. Dev Cell 38: 262–274. [DOI] [PubMed] [Google Scholar]
- Alexandre D, Clarke JD, Oxtoby E, Yan YL, Jowett T, Holder N. 1996. Ectopic expression of Hoxa-1 in the zebrafish alters the fate of the mandibular arch neural crest and phenocopies a retinoic acid-induced phenotype. Development 122: 735–746. [DOI] [PubMed] [Google Scholar]
- Amin S, Neijts R, Simmini S, van Rooijen C, Tan SC, Kester L, van Oudenaarden A, Creyghton MP, Deschamps J. 2016. Cdx and T Brachyury co-activate growth signaling in the embryonic axial progenitor niche. Cell Rep 17: 3165–3177. [DOI] [PubMed] [Google Scholar]
- Andrey G, Montavon T, Mascrez B, Gonzalez F, Noordermeer D, Leleu M, Trono D, Spitz F, Duboule D. 2013. A switch between topological domains underlies HoxD genes collinearity in mouse limbs. Science 340: 1234167. [DOI] [PubMed] [Google Scholar]
- Beccari L, Yakushiji-Kaminatsui N, Woltering JM, Necsulea A, Lonfat N, Rodriguez-Carballo E, Mascrez B, Yamamoto S, Kuroiwa A, Duboule D. 2016. A role for HOX13 proteins in the regulatory switch between TADs at the HoxD locus. Genes Dev 30: 1172–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bel-Vialar S, Itasaki N, Krumlauf R. 2002. Initiating Hox gene expression: in the early chick neural tube differential sensitivity to FGF and RA signaling subdivides the HoxB genes in two distinct groups. Development 129: 5103–5115. [DOI] [PubMed] [Google Scholar]
- Berlivet S, Paquette D, Dumouchel A, Langlais D, Dostie J, Kmita M. 2013. Clustering of tissue-specific sub-TADs accompanies the regulation of HoxA genes in developing limbs. PLoS Genet 9: e1004018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. 2006. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125: 315–326. [DOI] [PubMed] [Google Scholar]
- Bowman SK, Deaton AM, Domingues H, Wang PI, Sadreyev RI, Kingston RE, Bender W. 2014. H3K27 modifications define segmental regulatory domains in the Drosophila bithorax complex. eLife 3: e02833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cambray N, Wilson V. 2002. Axial progenitors with extensive potency are localised to the mouse chordoneural hinge. Development 129: 4855–4866. [DOI] [PubMed] [Google Scholar]
- Cambray N, Wilson V. 2007. Two distinct sources for a population of maturing axial progenitors. Development 134: 2829–2840. [DOI] [PubMed] [Google Scholar]
- Cao K, Collings CK, Marshall SA, Morgan MA, Rendleman EJ, Wang L, Sze CC, Sun T, Bartom ET, Shilatifard A. 2017. SET1A/COMPASS and shadow enhancers in the regulation of homeotic gene expression. Genes Dev 31: 787–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casares F, Sanchez-Herrero E. 1995. Regulation of the infraabdominal regions of the bithorax complex of Drosophila by gap genes. Development 121: 1855–1866. [DOI] [PubMed] [Google Scholar]
- Coleman RT, Struhl G. 2017. Causal role for inheritance of H3K27me3 in maintaining the OFF state of a Drosophila HOX gene. Science 356: eaai8236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasen JS, Jessell TM. 2009. Hox networks and the origins of motor neuron diversity. Curr Top Dev Biol 88: 169–200. [DOI] [PubMed] [Google Scholar]
- Davis RL, Kirschner MW. 2000. The fate of cells in the tailbud of Xenopus laevis. Development 127: 255–267. [DOI] [PubMed] [Google Scholar]
- Delpretti S, Montavon T, Leleu M, Joye E, Tzika A, Milinkovitch M, Duboule D. 2013. Multiple enhancers regulate Hoxd genes and the Hotdog LncRNA during cecum budding. Cell Rep 5: 137–150. [DOI] [PubMed] [Google Scholar]
- Denans N, Iimura T, Pourquie O. 2015. Hox genes control vertebrate body elongation by collinear Wnt repression. eLife 4: e04379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deschamps J, van Nes J. 2005. Developmental regulation of the Hox genes during axial morphogenesis in the mouse. Development 132: 2931–2942. [DOI] [PubMed] [Google Scholar]
- Deschamps J, Wijgerde M. 1993. Two phases in the establishment of HOX expression domains. Dev Biol 156: 473–480. [DOI] [PubMed] [Google Scholar]
- Di-Poi N, Zakany J, Duboule D. 2007. Distinct roles and regulations for HoxD genes in metanephric kidney development. PLoS Genet 3: e232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. 2012. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485: 376–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolle P, Izpisua-Belmonte JC, Falkenstein H, Renucci A, Duboule D. 1989. Coordinate expression of the murine Hox-5 complex homoeobox-containing genes during limb pattern formation. Nature 342: 767–772. [DOI] [PubMed] [Google Scholar]
- Duboule D. 1992. The vertebrate limb: a model system to study the Hox/HOM gene network during development and evolution. Bioessays 14: 375–384. [DOI] [PubMed] [Google Scholar]
- Duboule D. 1994. Temporal colinearity and the phylotypic progression: a basis for the stability of a vertebrate Bauplan and the evolution of morphologies through heterochrony. Development 120: 135–142. [PubMed] [Google Scholar]
- Duboule D. 2007. The rise and fall of Hox gene clusters. Development 134: 2549–2560. [DOI] [PubMed] [Google Scholar]
- Duboule D, Morata G. 1994. Colinearity and functional hierarchy among genes of the homeotic complexes. Trends Genet 10: 358–364. [DOI] [PubMed] [Google Scholar]
- Dubrulle J, McGrew MJ, Pourquie O. 2001. FGF signaling controls somite boundary position and regulates segmentation clock control of spatiotemporal Hox gene activation. Cell 106: 219–232. [DOI] [PubMed] [Google Scholar]
- Durston AJ, Zhu K. 2015. A time space translation hypothesis for vertebrate axial patterning. Semin Cell Dev Biol 42: 86–93. [DOI] [PubMed] [Google Scholar]
- Durston A, Wacker S, Bardine N, Jansen H. 2012. Time space translation: a Hox mechanism for vertebrate A–P patterning. Curr Genomics 13: 300–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabre PJ, Benke A, Joye E, Nguyen Huynh TH, Manley S, Duboule D. 2015. Nanoscale spatial organization of the HoxD gene cluster in distinct transcriptional states. Proc Natl Acad Sci 112: 13964–13969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forlani S, Lawson KA, Deschamps J. 2003. Acquisition of Hox codes during gastrulation and axial elongation in the mouse embryo. Development 130: 3807–3819. [DOI] [PubMed] [Google Scholar]
- Gaunt SJ. 2015. The significance of Hox gene collinearity. Int J Dev Biol 59: 159–170. [DOI] [PubMed] [Google Scholar]
- Gaunt SJ, Strachan L. 1994. Forward spreading in the establishment of a vertebrate Hox expression boundary: the expression domain separates into anterior and posterior zones, and the spread occurs across implanted glass barriers. Dev Dyn 199: 229–240. [DOI] [PubMed] [Google Scholar]
- Gaunt S, Sharpe PT, Duboule D. 1988. Spatially restricted domains of homeo-gene transcripts in mouse embryos: relation to a segmented body plan. Development 104: 169–179. [Google Scholar]
- Gaunt SJ, George M, Paul YL. 2013. Direct activation of a mouse Hoxd11 axial expression enhancer by Gdf11/Smad signalling. Dev Biol 383: 52–60. [DOI] [PubMed] [Google Scholar]
- Gomez C, Pourquie O. 2009. Developmental control of segment numbers in vertebrates. J Exp Zool B Mol Dev Evol 312: 533–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Reyes A, Urquia N, Gehring WJ, Struhl G, Morata G. 1990. Are cross-regulatory interactions between homoeotic genes functionally significant? Nature 344: 78–80. [DOI] [PubMed] [Google Scholar]
- Gould A, Morrison A, Sproat G, White RA, Krumlauf R. 1997. Positive cross-regulation and enhancer sharing: two mechanisms for specifying overlapping Hox expression patterns. Genes Dev 11: 900–913. [DOI] [PubMed] [Google Scholar]
- Harding K, Wedeen C, McGinnis W, Levine M. 1985. Spatially regulated expression of homeotic genes in Drosophila. Science 229: 1236–1242. [DOI] [PubMed] [Google Scholar]
- Herault Y, Fraudeau N, Zakany J, Duboule D. 1997. Ulnaless (Ul), a regulatory mutation inducing both loss-of-function and gain-of-function of posterior Hoxd genes. Development 124: 3493–3500. [DOI] [PubMed] [Google Scholar]
- Holland LZ. 2002. Heads or tails? Amphioxus and the evolution of anterior–posterior patterning in deuterostomes. Dev Biol 241: 209–228. [DOI] [PubMed] [Google Scholar]
- Iimura T, Pourquie O. 2006. Collinear activation of Hoxb genes during gastrulation is linked to mesoderm cell ingression. Nature 442: 568–571. [DOI] [PubMed] [Google Scholar]
- Iimura T, Yang X, Weijer CJ, Pourquie O. 2007. Dual mode of paraxial mesoderm formation during chick gastrulation. Proc Natl Acad Sci 104: 2744–2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingham PW, Martinez-Arias A. 1986. The correct activation of Antennapedia and bithorax complex genes requires the fushi tarazu gene. Nature 324: 592–597. [DOI] [PubMed] [Google Scholar]
- Izpisua-Belmonte JC, Falkenstein H, Dolle P, Renucci A, Duboule D. 1991. Murine genes related to the Drosophila AbdB homeotic genes are sequentially expressed during development of the posterior part of the body. EMBO J 10: 2279–2289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurberg AD, Aires R, Varela-Lasheras I, Novoa A, Mallo M. 2013. Switching axial progenitors from producing trunk to tail tissues in vertebrate embryos. Dev Cell 25: 451–462. [DOI] [PubMed] [Google Scholar]
- Kim A, Dean A. 2012. Chromatin loop formation in the β-globin locus and its role in globin gene transcription. Mol Cells 34: 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinder SJ, Tsang TE, Quinlan GA, Hadjantonakis AK, Nagy A, Tam PP. 1999. The orderly allocation of mesodermal cells to the extraembryonic structures and the anteroposterior axis during gastrulation of the mouse embryo. Development 126: 4691–4701. [DOI] [PubMed] [Google Scholar]
- Krumlauf R. 1994. Hox genes in vertebrate development. Cell 78: 191–201. [DOI] [PubMed] [Google Scholar]
- Kundu S, Ji F, Sunwoo H, Jain G, Lee JT, Sadreyev RI, Dekker J, Kingston RE. 2017. Polycomb repressive complex 1 generates discrete compacted domains that change during differentiation. Mol Cell 65: 432–446 e435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson KA, Meneses JJ, Pedersen RA. 1991. Clonal analysis of epiblast fate during germ layer formation in the mouse embryo. Development 113: 891–911. [DOI] [PubMed] [Google Scholar]
- Lewis EB. 1978. A gene complex controlling segmentation in Drosophila. Nature 276: 565–570. [DOI] [PubMed] [Google Scholar]
- Liu JP. 2006. The function of growth/differentiation factor 11 (Gdf11) in rostrocaudal patterning of the developing spinal cord. Development 133: 2865–2874. [DOI] [PubMed] [Google Scholar]
- Liu JP, Laufer E, Jessell TM. 2001. Assigning the positional identity of spinal motor neurons: rostrocaudal patterning of Hox-c expression by FGFs, Gdf11, and retinoids. Neuron 32: 997–1012. [DOI] [PubMed] [Google Scholar]
- Lonfat N, Montavon T, Darbellay F, Gitto S, Duboule D. 2014. Convergent evolution of complex regulatory landscapes and pleiotropy at Hox loci. Science 346: 1004–1006. [DOI] [PubMed] [Google Scholar]
- Maeda RK, Karch F. 2015. The open for business model of the bithorax complex in Drosophila. Chromosoma 124: 293–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R, Reinberg D. 2011. The Polycomb complex PRC2 and its mark in life. Nature 469: 343–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin BL, Kimelman D. 2008. Regulation of canonical Wnt signaling by Brachyury is essential for posterior mesoderm formation. Dev Cell 15: 121–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzoni EO, Mahony S, Peljto M, Patel T, Thornton SR, McCuine S, Reeder C, Boyer LA, Young RA, Gifford DK, et al. 2013. Saltatory remodeling of Hox chromatin in response to rostrocaudal patterning signals. Nat Neurosci 16: 1191–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrew MJ, Sherman A, Lillico SG, Ellard FM, Radcliffe PA, Gilhooley HJ, Mitrophanous KA, Cambray N, Wilson V, Sang H. 2008. Localised axial progenitor cell populations in the avian tail bud are not committed to a posterior Hox identity. Development 135: 2289–2299. [DOI] [PubMed] [Google Scholar]
- Montavon T, Soshnikova N, Mascrez B, Joye E, Thevenet L, Splinter E, de Laat W, Spitz F, Duboule D. 2011. A regulatory archipelago controls Hox genes transcription in digits. Cell 147: 1132–1145. [DOI] [PubMed] [Google Scholar]
- Narendra V, Rocha PP, An D, Raviram R, Skok JA, Mazzoni EO, Reinberg D. 2015. CTCF establishes discrete functional chromatin domains at the Hox clusters during differentiation. Science 347: 1017–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra V, Bulajic M, Dekker J, Mazzoni EO, Reinberg D. 2016. CTCF-mediated topological boundaries during development foster appropriate gene regulation. Genes Dev 30: 2657–2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neijts R, Simmini S, Giuliani F, van Rooijen C, Deschamps J. 2014. Region-specific regulation of posterior axial elongation during vertebrate embryogenesis. Dev Dyn 243: 88–98. [DOI] [PubMed] [Google Scholar]
- Neijts R, Amin S, van Rooijen C, Tan S, Creyghton MP, de Laat W, Deschamps J. 2016. Polarized regulatory landscape and Wnt responsiveness underlie Hox activation in embryos. Genes Dev 30: 1937–1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neijts R, Amin S, van Rooijen C, Deschamps J. 2017. Cdx is crucial for the timing mechanism driving colinear Hox activation and defines a trunk segment in the Hox cluster topology. Dev Biol 422: 146–154. [DOI] [PubMed] [Google Scholar]
- Nelson CE, Morgan BA, Burke AC, Laufer E, DiMambro E, Murtaugh LC, Gonzales E, Tessarollo L, Parada LF, Tabin C. 1996. Analysis of Hox gene expression in the chick limb bud. Development 122: 1449–1466. [DOI] [PubMed] [Google Scholar]
- Noordermeer D, Leleu M, Splinter E, Rougemont J, De Laat W, Duboule D. 2011. The dynamic architecture of Hox gene clusters. Science 334: 222–225. [DOI] [PubMed] [Google Scholar]
- Noordermeer D, Leleu M, Schorderet P, Joye E, Chabaud F, Duboule D. 2014. Temporal dynamics and developmental memory of 3D chromatin architecture at Hox gene loci. eLife 3: e02557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nora EP, Lajoie BR, Schulz EG, Giorgetti L, Okamoto I, Servant N, Piolot T, van Berkum NL, Meisig J, Sedat J, et al. 2012. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature 485: 381–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oosterveen T, Niederreither K, Dolle P, Chambon P, Meijlink F, Deschamps J. 2003. Retinoids regulate the anterior expression boundaries of 5′ Hoxb genes in posterior hindbrain. EMBO J 22: 262–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osorno R, Tsakiridis A, Wong F, Cambray N, Economou C, Wilkie R, Blin G, Scotting PJ, Chambers I, Wilson V. 2012. The developmental dismantling of pluripotency is reversed by ectopic Oct4 expression. Development 139: 2288–2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmeirim I, Henrique D, Ish-Horowicz D, Pourquie O. 1997. Avian hairy gene expression identifies a molecular clock linked to vertebrate segmentation and somitogenesis. Cell 91: 639–648. [DOI] [PubMed] [Google Scholar]
- Popperl H, Bienz M, Studer M, Chan SK, Aparicio S, Brenner S, Mann RS, Krumlauf R. 1995. Segmental expression of Hoxb-1 is controlled by a highly conserved autoregulatory loop dependent upon exd/pbx. Cell 81: 1031–1042. [DOI] [PubMed] [Google Scholar]
- Pourquie O. 2003. The segmentation clock: converting embryonic time into spatial pattern. Science 301: 328–330. [DOI] [PubMed] [Google Scholar]
- Raff RA. 1996. The shape of life: genes, development and the evolution of animal form. University of Chicago Press, Chicago. [Google Scholar]
- Rodrigues AR, Yakushiji-Kaminatsui N, Atsuta Y, Andrey G, Schorderet P, Duboule D, Tabin CJ. 2017. Integration of Shh and Fgf signaling in controlling Hox gene expression in cultured limb cells. Proc Natl Acad Sci 114: 3139–3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schep R, Necsulea A, Rodriguez-Carballo E, Guerreiro I, Andrey G, Nguyen Huynh TH, Marcet V, Zakany J, Duboule D, Beccari L. 2016. Control of Hoxd gene transcription in the mammary bud by hijacking a preexisting regulatory landscape. Proc Natl Acad Sci 113: E7720–E7729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert A, Werheid DF, Knapp SM, Tobiasch E. 2015. Role of Hox genes in stem cell differentiation. World J Stem Cells 7: 583–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo HC, Edvardsen RB, Maeland AD, Bjordal M, Jensen MF, Hansen A, Flaat M, Weissenbach J, Lehrach H, Wincker P, et al. 2004. Hox cluster disintegration with persistent anteroposterior order of expression in Oikopleura dioica. Nature 431: 67–71. [DOI] [PubMed] [Google Scholar]
- Sheth R, Barozzi I, Langlais D, Osterwalder M, Nemec S, Carlson HL, Stadler HS, Visel A, Drouin J, Kmita M. 2016. Distal limb patterning requires modulation of cis-regulatory activities by HOX13. Cell Rep 17: 2913–2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soshnikova N, Duboule D. 2009. Epigenetic temporal control of mouse Hox genes in vivo. Science 324: 1320–1323. [DOI] [PubMed] [Google Scholar]
- Spitz F, Herkenne C, Morris MA, Duboule D. 2005. Inversion-induced disruption of the Hoxd cluster leads to the partition of regulatory landscapes. Nat Genet 37: 889–893. [DOI] [PubMed] [Google Scholar]
- Steventon B, Duarte F, Lagadec R, Mazan S, Nicolas JF, Hirsinger E. 2016. Species-specific contribution of volumetric growth and tissue convergence to posterior body elongation in vertebrates. Development 143: 1732–1741. [DOI] [PubMed] [Google Scholar]
- Tabin C, Wolpert L. 2007. Rethinking the proximodistal axis of the vertebrate limb in the molecular era. Genes Dev 21: 1433–1442. [DOI] [PubMed] [Google Scholar]
- Tarchini B, Duboule D. 2006. Control of Hoxd genes’ collinearity during early limb development. Dev Cell 10: 93–103. [DOI] [PubMed] [Google Scholar]
- Tzouanacou E, Wegener A, Wymeersch FJ, Wilson V, Nicolas JF. 2009. Redefining the progression of lineage segregations during mammalian embryogenesis by clonal analysis. Dev Cell 17: 365–376. [DOI] [PubMed] [Google Scholar]
- van den Akker E, Forlani S, Chawengsaksophak K, de Graaff W, Beck F, Meyer BI, Deschamps J. 2002. Cdx1 and Cdx2 have overlapping functions in anteroposterior patterning and posterior axis elongation. Development 129: 2181–2193. [DOI] [PubMed] [Google Scholar]
- van Rooijen C, Simmini S, Bialecka M, Neijts R, van de Ven C, Beck F, Deschamps J. 2012. Evolutionarily conserved requirement of Cdx for post-occipital tissue emergence. Development 139: 2576–2583. [DOI] [PubMed] [Google Scholar]
- Wacker SA, Jansen HJ, McNulty CL, Houtzager E, Durston AJ. 2004. Timed interactions between the Hox expressing non-organiser mesoderm and the Spemann organiser generate positional information during vertebrate gastrulation. Dev Biol 268: 207–219. [DOI] [PubMed] [Google Scholar]
- Wellik DM. 2009. Hox genes and vertebrate axial pattern. Curr Top Dev Biol 88: 257–278. [DOI] [PubMed] [Google Scholar]
- Wilson V, Olivera-Martinez I, Storey KG. 2009. Stem cells, signals and vertebrate body axis extension. Development 136: 1591–1604. [DOI] [PubMed] [Google Scholar]
- Woltering JM, Duboule D. 2010. The origin of digits: expression patterns versus regulatory mechanisms. Dev Cell 18: 526–532. [DOI] [PubMed] [Google Scholar]
- Woltering JM, Noordermeer D, Leleu M, Duboule D. 2014. Conservation and divergence of regulatory strategies at Hox loci and the origin of tetrapod digits. PLoS Biol 12: e1001773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wymeersch FJ, Huang Y, Blin G, Cambray N, Wilkie R, Wong FC, Wilson V. 2016. Position-dependent plasticity of distinct progenitor types in the primitive streak. eLife 5: e10042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yekta S, Tabin CJ, Bartel DP. 2008. MicroRNAs in the Hox network: an apparent link to posterior prevalence. Nat Rev Genet 9: 789–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young T, Rowland JE, van de Ven C, Bialecka M, Novoa A, Carapuco M, van Nes J, de Graaff W, Duluc I, Freund JN, et al. 2009. Cdx and Hox genes differentially regulate posterior axial growth in mammalian embryos. Dev Cell 17: 516–526. [DOI] [PubMed] [Google Scholar]
- Zakany J, Duboule D. 2007. The role of Hox genes during vertebrate limb development. Curr Opin Genet Dev 17: 359–366. [DOI] [PubMed] [Google Scholar]
- Zakany J, Kmita M, Alarcon P, de la Pompa JL, Duboule D. 2001. Localized and transient transcription of Hox genes suggests a link between patterning and the segmentation clock. Cell 106: 207–217. [DOI] [PubMed] [Google Scholar]
- Zakany J, Kmita M, Duboule D. 2004. A dual role for Hox genes in limb anterior-posterior asymmetry. Science 304: 1669–1672. [DOI] [PubMed] [Google Scholar]


