Abstract
Lipid molecules are able to selectively interact with specific sites on integral membrane proteins, and modulate their structure and function. Identification and characterisation of these sites is of importance for our understanding of the molecular basis of membrane protein function and stability, and may facilitate the design of lipid-like drug molecules. Molecular dynamics simulations provide a powerful tool for the identification of these sites, complementing advances in membrane protein structural biology and biophysics. We describe recent notable biomolecular simulation studies which have identified lipid interaction sites on a range of different membrane proteins. The sites identified in these simulation studies agree well with those identified by complementary experimental techniques. This demonstrates the power of the molecular dynamics approach in the prediction and characterization of lipid interaction sites on integral membrane proteins.
Keywords: Lipid-protein interaction, Lipid binding site, MD simulation, Cholesterol, Cardiolipin, PIP2
1. Introduction
Cells are separated from their environment and compartmentalised by membranes. These barriers are composed of lipid bilayers (with the various lipid species distributed asymmetrically between the two leaflets of the bilayer), into which proteins are embedded. Parallel advances in lipidomics [1] and in the structural biology of membrane proteins [2, 3] over the past decade have revealed some of the complexities of the composition of cell membranes. Thus, the structures of ca. 1500 membrane proteins have been determined (http://blanco.biomol.uci.edu/mpstruc/) and the cellular lipidome is estimated to contain 40,000 lipid species (http://www.lipidmaps.org/data/structure/) [4]. The lipidome of membranes varies according to cell age, metabolic state, stage in cell cycle, organelle, and spatial location; resulting in a complex protein-lipid interactome. In addition to providing a bilayer environment, it is increasingly appreciated that the function of embedded proteins can be modulated by interactions with this complex lipid mixture [5–12]. Of particular interest, an emerging feature present within the protein-lipid interactome is that certain lipid molecules can selectively bind to specific sites on integral membrane proteins, and modulate both their structure and their function [13].
As recently reviewed [14], we now possess over 100 structures of membrane proteins containing electron density interpreted as bound lipid molecules. Structural identification of specific lipid binding sites aids our mechanistic understanding of lipid modulation of protein function, such as in the case of Kir2.2 and PIP2 [15]. Identification of sites of allosteric modulation on proteins is also of interest for the assessment of protein druggability [16]. The majority of membrane protein structures containing bound lipid molecules were solved using x-ray crystallography. In many cases such structures have been obtained from crystals grown in the presence of detergent. It is likely that the lipids observed represent a biased sample of tight binding lipids, and in some cases the molecular identity of the observed electron density corresponding to detergent and/or lipid may be uncertain. This may change as more membrane protein structures are determined using crystals obtained from lipidic cubic phases [17] which better approximate a native membrane environment.
Molecular dynamics simulations allow membrane protein structures to be computationally re-embedded into lipid bilayers, and their dynamic interactions with surrounding lipid molecules to be characterised [18]. A number of recent simulation studies probing lipid interactions have identified specific lipid binding sites. These sites show good agreement with those identified from a range of structural studies. A number of other, presumably weaker, binding sites can also be resolved. Whilst these weaker sites may not always be observed by x-ray crystallography, there are a number of other biophysical techniques which allow us to probe lipid interactions with membrane proteins, including e.g. fluorescence spectroscopy [19], EPR [20], NMR [21], and mass spectrometry [13]. These techniques provide further points of reference for simulation studies membrane protein/lipid interactions.
Within this review article we survey recent simulation studies which identify lipid interaction sites on membrane proteins. We focus on specific binding of lipids to defined sites on membrane proteins. We also focus on channels, receptors, and transporters, for which there are functional and structural data on the biological importance of lipid/protein interactions. Overall we find molecular dynamics simulations to have strong predictive power and to be well-suited for identification of these sites. Additionally the simulation approach provides a means for further characterisation of the identified sites, for instance by estimations of lipid binding affinities [22–24], as well as enabling functional insight into mechanisms of lipid modulation [25–27].
1.1. Lipid modulation of membrane protein function
The functions of a range of membrane proteins are known to be modulated by their lipid environment, including potassium channels [6, 7], receptor tyrosine kinases (RTKs) [8], G-protein coupled receptors (GPCRs) [9, 10], solute transporters such as BetP [5] and the ADP/ATP carrier [28], redox proteins such as cytochrome c oxidase [29], and certain P-type ATPases [30]. Such lipid modulation can influence several different aspects of protein function, including effects on the activity of a membrane protein, modulating protein-protein interactions, and altering cellular localisation by sequestering a protein to spatially defined regions of a membrane. In certain cases, a lipid may represent a native ligand for the protein rather than an allosteric modulator, as is the case for the sphingosine-1-phosphate (S1P) and lysophosphatidic acid (LPA) group of lipid-activated GPCRs [31].
In a number of cases structural, biophysical and functional assays have been combined to provide a detailed picture of lipid modulation. For example, this is the case for eukaryotic inward rectifying potassium ion (Kir) channels. Functional assays revealed Kir channels to be dependent on the presence of the anionic lipid phosphatidyl inositol-4,5-bisphosphate (PIP2) for activation. Subsequently, simulation studies [25, 26] and crystal structures [15] revealed four specific PIP2 binding sites and enabled the mechanism of PIP2 channel modulation to be structurally rationalised (See Section 2.1.2). In other cases careful biochemical analysis has revealed a functional dependence on certain lipid species, but the mechanism of modulation remains unclear. This is the case for the epidermal growth factor receptor (EGFR/ErbB1) which is a single-pass transmembrane receptor known to be modulated by an array of lipids, including PIP species and the glycolipid GM3 [8]. However exactly how these lipid species control receptor activity remains unclear, with proposals including an influence of receptor dimerization propensity, direct conformational stabilization and orientation effects, and larger scale lipid-induced clustering of the receptor.
1.2. Lipid interaction sites on integral membrane proteins
Lipids interact with membrane proteins via multiple modes. The presence of integral membrane proteins may induce formation of a lipid ‘annulus’ around the protein. Due to interactions with the protein, lipids within this annulus exhibit decreased motional freedom compared to their non-interacting bulk counterparts, and are detectable by EPR [32, 33]. This immobilizing effect of the protein may extend beyond the first shell of directly interacting annular lipids, leading to further outer shells with a lesser extent of lipid immobilisation, as suggested by MD simulations [34, 35]. In addition certain lipid species may bind to specific sites on the membrane protein surface – often described as ‘non-annular’ lipids. Binding may be driven by formation of physicochemical interactions between the lipid and protein surface, as well as by complementary geometry, for instance ‘slotting’ of lipid molecules into ‘grooves’ on the protein surface [36] or binding at the interface between subunits [37]. Binding sites may tightly coordinate the lipid [15], or act to cause weaker and more dynamic localisation [38]. Efforts have been made to describe general features of lipid binding sites and sequence interaction motifs, such as for cholesterol [39] and cardiolipin [40].
1.3. Biomolecular simulation approaches for lipid binding site identification
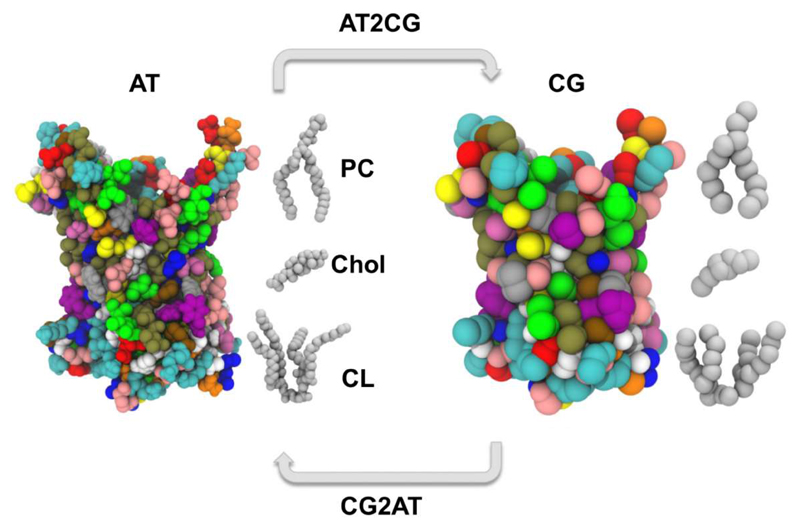
Molecular dynamics (MD) simulations provide a powerful tool to characterise the dynamics and interactions of membrane proteins with surrounding lipid molecules [18]. However, the computational cost of the simulations is such that length scales beyond microseconds are not currently readily accessible [41], especially for extended systems containing multiple membrane proteins. This has prompted the development of more approximate coarse-grained (CG) representations of membrane lipids and proteins in MD simulations [42, 43] in which groups of atoms are represented as single particles (Figure 1). Reducing the number of particles in the system reduces the computational demand involved in running the simulation and thus allows access to longer time and length scales, with the caveat that the level of approximation made in a given CG model has to be matched to the underlying biological interactions being probed. CG simulations can thus allow significantly enhanced lipid exploration of the protein surface and candidate binding sites, whilst sacrificing the finer detail of lipid-protein interactions. These approximations may be reconciled to some degree by conversion of the endpoint of a CG system back to atomistic detail [44, 45] and subsequently running an atomistic simulation to assess the validity of the CG system arrangement; so called (serial) multiscale modelling [46]. The MARTINI CG force field has been most widely applied in the field of protein-lipid interactions. We note that CG simulations may now extend to hundreds of microseconds [23], and contain many hundreds of protein molecules [47], whilst atomistic simulations of individual proteins may reach tens of microseconds duration using high performance computing resources [48].
Figure 1.
Schematic of a multiscale approach to modelling and simulation of lipid interactions with an integral membrane protein. The ADP/ATP carrier (ANT1/AAC1; PDB: 1OKC [36]) is depicted as spheres coloured by residue type, at both the atomistic (left) and CG (right) scale. Phosphatidylcholine (PC), cholesterol (Chol), and cardiolipin (CL) molecules are shown as grey spheres.
The structure of a membrane protein used as initial input for MD simulations may be from X-ray diffraction, cryoelectron microscopy, or NMR. If the 3D structure of the protein is not known experimentally, in some cases a model may be built by modelling [35]. The membrane protein is then embedded into a lipid bilayer. This may be achieved either by self-assembly simulations [49] in which short simulations are run to allow the spontaneous formation of a bilayer around an integral membrane protein, or by a number of methods which insert a membrane protein into a pre-assembled bilayer [50–52]. Advances in lipid parameterisation [53], along with a growing appreciation of the in vivo compositional complexity of lipid membranes [54] is leading to simulations of proteins in complex bilayers containing multiple lipid species. At the simplest level such mixed bilayers may contain two lipid species, while at their most complex they may provide approximations of in vivo plasma membrane composition [55, 56]. Such mixed lipid systems allow us to address competition between different lipid species for interaction with a given protein, in addition to providing a better approximation of lipid-lipid interactions which are linked to, and may influence, protein-lipid interactions. Such mixed lipid systems can now be routinely assembled in CG [55, 56]. Simulations of mixed lipid systems are also becoming more common at the atomistic level, utilising a number of recent tools (e.g. [57, 58] including as the Membrane Builder extension of the CHARMM-GUI [59] to facilitate automated construction of complex bilayers in all atom detail. Some of the equilibration and sampling problems arising from timescale limitations being combated by increases in processing power and algorithm efficiency, as well as the ability to reversibly convert between CG and atomistic levels (Figure 1) [44, 45]. Thus in the latter approach the membrane protein of interest may first be simulated in a mixed lipid bilayer using CG methods to equilibrate the system and make an assessment of how lipids interact on extended timescales, before converting the system to atomistic detail to further refine and characterise the observed lipid-protein interactions. This multiscale approach has been successfully used to identify lipid binding sites on a number of membrane proteins [23, 25, 26, 60].
Potential lipid binding sites may also be identified by docking calculations, using e.g. AutoDock [61]. However these methods do not generally take into account the membrane environment in which the interactions occur. Thus protein/lipid configurations identified by docking may require refinement by subsequent molecular dynamics simulation. Combined use of molecular docking and simulation has enabled identification of lipid binding sites on several types of membrane protein, including e.g. cholesterol interactions with Cys-loop receptors [62, 63] and with Kir channels [22].
2. Characterisation of lipid interaction sites: a digest of recent simulation studies
2.1. Channels
Lipids modulate the function of a number of channels. For example, the canonical bacterial potassium channel KcsA requires bound anionic lipid molecules for full activity, and both crystallographic [64] and simulation studies [65] have previously indicated the presence of a binding site for an anionic lipid molecule located between two arginine sidechains at the subunit/subunit interface of the trimeric channel protein. More recent studies have focussed on two species of lipid thought to regulate a number of channels in mammalian cell membranes, namely PIP2 and cholesterol.
2.1.2. Inward rectifying potassium (Kir) channels
Kir channels are tetrameric integral membrane proteins controlling the selective permeation of K+ ions across cell membranes. They have critical involvement in molecular processes ranging from control of the resting membrane potential to regulation of insulin secretion in pancreatic β cells [66]. Of particular interest Kir channels have a requirement for PIP2 for maximal activation [6, 67] which may be considered as lipid agonism of these channels [68].
The interaction of PIP2 with Kir channels has been explored using combined CG and atomistic simulations [25]. Three structures of Kir channels were simulated: KirBac1.1, a Kir3.1-KirBac1.3 chimera, and a homology model of Kir6.2. These structures were initially converted to CG representation and embedded into POPC bilayers, each of which contained 4 PIP2 lipid molecules within the inner leaflet of the bilayer. Analysis of time-averaged density maps for PIP2 revealed preferential localisation to a single site, present on all four subunits. The same site was observed for all three Kir channels. The binding site was formed by a cluster of basic and amphipathic aromatic residues at the interface between subunits. Once bound, PIP2 lipids did not dissociate from the identified sites even during extended (5 μs) CG simulations, suggesting tight binding. Conversion of CG snapshots of the channel with bound PIP2 lipids to atomistic models followed by short simulations enabled assessment of binding site interactions. Key residues seen to form contacts with the PIP2 lipid head group in the simulations had previously been implicated in PIP2 interactions with Kir2.1 [6, 69, 70]. This was not the case for residues seen in the simulations to form contacts with the lipid tail regions, which formed more transient interactions with the protein and were therefore not suggested to be major binding determinants.
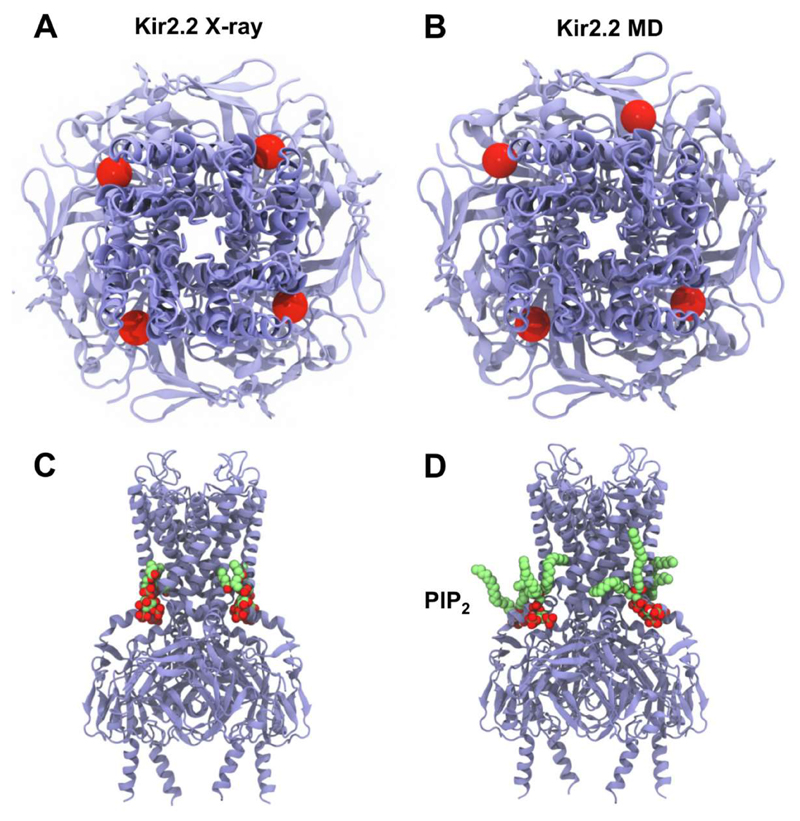
Subsequent to this study, a high resolution X-ray structure of Kir2.2 in complex with four short chain (dioctanoyl) PIP2 lipids was determined [15], revealing binding sites and interactions in good agreement with the simulation-based predictions [25]. This is despite the caveat that the structures used for simulations employed homology models and eukaryotic-prokaryotic chimeras. Further studies [26] extended the comparison by multiscale simulations of the PIP2 bound structure (PDB: 3SPI[15]), as well as other Kir2.2 X-ray structures including the apo state (PDB: 3JYC[71]), and a phosphatidic acid bound state (PDB: 3SPC[15]). This study provided a ‘like for like’ comparison of PIP2 binding to Kir2.2, supporting the sites initially identified via simulations of other Kir subtypes. These multiscale simulations showed unambiguously that PIP2 lipids bound at the same four sites identified in the crystal structure (Figure 2). The good agreement between multiple simulation and experimental approaches is suggestive of the accuracy of the binding site predictions, as well as the utility of multiscale simulations in their identification.
Figure 2.
Similarity in PIP2 binding sites identified by crystallography [15] (A,C) and by simulation [25, 26] (B,D). (A, B) View from the extracellular side down the pore axis of the tetrameric Kir2.2 channel with the four PIP2 α-phosphate groups (bridging the glycerol and inositol moieties) indicated as red spheres. (C, D) side view of the channel showing bound PIP2 molecules (lime green carbons and red oxygens).
Kir channels have also been shown to be regulated by cholesterol [72, 73], in a stereospecific fashion suggestive of the existence of a specific cholesterol binding site on the channel proteins [7] [74]. An integrated molecular docking and simulation approach has been used to investigate cholesterol binding to Kir2.1 [22], leading to the identification of two cholesterol binding sites. The validity of the sites was tested by mutagenesis and electrophysiology, and showed good agreement with the MD predictions. Docking and simulation studies were also used to compare predicted cholesterol interactions of bacterial (KirBac) and mammalian Kir channels [73]. Overall, these two studies suggest a consensus cholesterol binding region in the cytoplasmic half of the TM domains of Kir channels with the ligand also interacting with residues on the slide helix located at the membrane surface. For a recent comprehensive review of cholesterol interactions with membrane proteins as studied by molecular dynamics see [75].
2.1.2. Pentameric ligand gated ion-channels
The pentameric ligand gated ion-channels (pLGICs) are a major class of neurotransmitter receptor [76]. Both the nicotinic acetylcholine receptor (nAChR) and the y-aminobutyric acid receptor (GABAAR) are sensitive to membrane cholesterol [11, 12, 77], and the nAChR also shows functional sensitivity to anionic lipids [78] and to membrane hydrophobic thickness [79]. The interactions of cholesterol with the nAChR [62, 80] and the GABAAR [63] have both been explored using docking and simulations. In a study by Brannigan et al., [62] docking calculations were performed to screen the nAChR Torpedo cryo-EM structure (PDB: 2BG9) for possible cholesterol binding sites [62]. A total of three sites per subunit were identified (15 for the whole protein). These sites were buried both within grooves on the protein surface, and more deeply within non-membrane exposed pockets. A more recent study [81] identified the presence of cholesterol interaction motifs within the transmembrane domain of nAChR, which co-localised with a number of the sites suggested by Brannigan et al., as well as a number of additional solvent exposed sites. Interestingly the sites identified in these computational studies corresponded to gaps in the original cryo-EM structure (PDB: 2BG9). The possible role of cholesterol at the identified sites was addressed by 25 ns atomistic simulations both in the presence and the absence of bound cholesterol [62]. In the absence of cholesterol the binding sites were initially filled with water. During the simulation water was expelled and the structure was seen to collapse. In contrast in the presence of bound cholesterol the original conformation was more faithfully maintained, and contacts between the pore and agonist binding domain proposed to be critical for gating were seen to be maintained. The results overall suggested 15 cholesterol molecules bind to the pentameric nAChR structure and act to provide structural integrity to the receptor. Interactions of nAChR with phosphatidic acid (PA) have also been explored by simulations [80], which suggested potential differences in the interaction modes of anionic PA and zwitterionic lipids, although the relatively short duration (10 ns) of the simulations precludes more detailed comparisons.
In a second study, cholesterol interactions with the related GABAAR were explored [63]. In lieu of a high resolution structure of GABAAR, the authors built a homology model based on GluCl (PDB: 3RHW [82]) and used this structure for initial protein coordinates within simulations. Five equivalent cholesterol binding pockets positioned between subunits of the pentameric receptor were identified by docking (Figure 3). Atomistic simulations were performed starting from a configuration with cholesterol docked into these sites. Cholesterol was seen to undergo some reorientation within all five sites. During a 200 ns simulation the cholesterol molecules at two sites were seen to dissociate within 20 ns, but were subsequently seen to rebind at around 80-110 ns with similar but presumably slightly more favourable binding modes. The remaining three cholesterol molecules remained bound. The general stability of cholesterol and the rebinding events observed during the simulations are supportive of the validity of the sites suggested from docking. A second model of cholesterol bound to GABAAR was built based on the ivermectin binding sites in the GluCl (ivermectin is a relatively hydrophobic ligand) crystal structure [82]. The binding pockets in this model were identical to those seen from docking and spontaneous rebinding events during atomistic simulation, though some differences in cholesterol orientation were observed. Thus the different approaches seemed to converge in terms of cholesterol interactions. Comparison of the behaviour of the protein in the cholesterol-bound and the apo state simulations suggested that in the absence of cholesterol the pore radius of the channel decreased, whereas with bound cholesterol the channel showed an increased tendency to adopt a more open conformation. This suggests that cholesterol may in part exert its channel modulating properties via direct binding and stabilisation of an open conformation of the GABAAR.
Figure 3.
Location of cholesterol binding sites between the five subunits of the pentameric GABAAR [63]. Initial coordinates were predicted via docking, and the structure shown is post-simulation. (A) sideview, and (B) extracellular view of the five transmembrane domains, with cholesterol molecules represented as van der Waals spheres. We thank Dr. Jérôme Hénin and Dr. Grace Brannigan for providing coordinates used to generate the figure.
2.1.3. Mechanosensitive channels
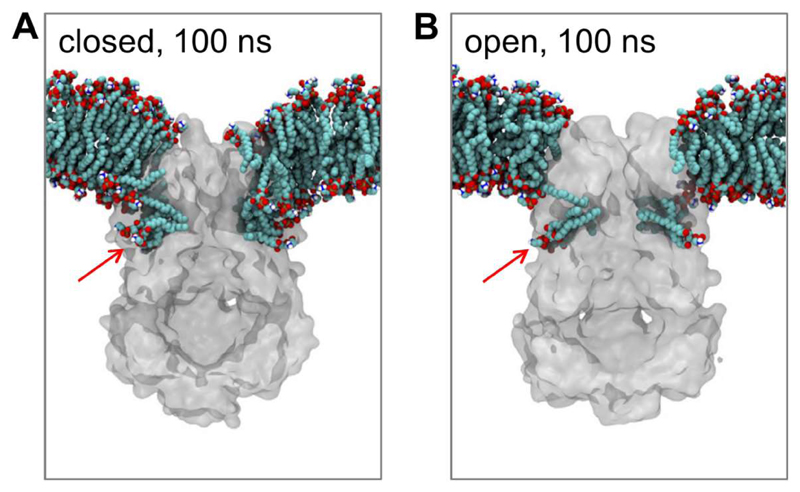
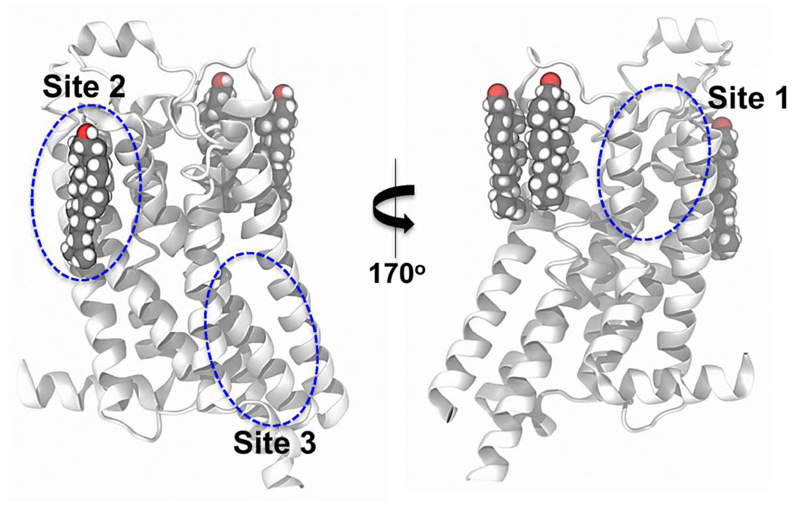
Mechanosensitive channels open in response to an increase in tension (i.e. stretching) of the membrane. A recent combined structural, biophysical and computational study has revealed the role of bound lipid molecules in mechanosensing by the E. coli MscS channel [27]. X-ray diffraction and simulation together identified lipid tails bound inside pockets formed by the TM helices. Significantly the number of lipid acyl chains occupying the pockets decreased upon channel opening (Figure 4). This suggested an activation mechanism whereby an increase in membrane tension perturbed lipid binding within the pocket, thus destabilizing the closed state and thus promoting channel opening.
Figure 4.
MscS/lipid interactions comparing closed (PDB: 2OAU) and open (PDB: 5AJI) conformations. Cut-away slices are shown of snapshots at 100 ns from atomistic simulations of the closed (A) and open (B) conformation of MscS in POPE/POPG (4:1) bilayers [27]. The proteins are shown in grey and the lipids in cyan (carbons) and red (oxygens). The red arrow shows the lipid binding cavity whose occupancy changes between the closed and the open conformation. Figure modified from [27].
2.2. G protein coupled receptors
G protein coupled receptors (GPCRs) are a large superfamily of integral membrane proteins composed of seven transmembrane helices [83]. The complexities of the interactions of GPCRs with cholesterol and other lipids are starting to be unravelled [84]. Cholesterol has been suggested to modulate various aspects of GPCR biology including stability [9, 10], oligomeric organisation [85], and ligand binding activity [86, 87]. Cholesterol molecules have been found co-crystallised in a number of GPCR X-ray structures [88–91], suggesting possible modulation by direct binding. Molecular simulation has been applied to identify lipid binding sites on a range of GPCRs including the β2-adrenergic receptor (β2AR) [92–94], the β1-adrenergic receptor (β1AR) [95], the Serotonin1A [96] and Serotonin2A receptors [97], the A2A adenosine receptor [24], Rhodopsin [98, 99], the Cannabinoid 2 (CB2) receptor [100], and the S1P1 receptor [47].
Cholesterol binding sites have been identified on the A2A adenosine receptor by long timescale all-atom MD simulations [24] and by X-ray crystallography [90]. In a study by Lee and Lyman [24], two independent 800 ns all-atom simulations of the adenosine-bound structure (PDB: 2YDO) were performed in a PC:cholesterol lipid bilayer. Over the course of the simulations cholesterol molecules bound to three distinct sites on the receptor. Once bound, a single cholesterol molecule occupied each site for the duration of the simulation. However despite these sites remaining occupied, the binding modes of cholesterol were seen to be dynamic. The simulation data were used to estimate cholesterol interaction energies at each site via an inverse Boltzmann approach, yielding energies of the order of kT, suggesting cholesterol is only weakly associated with the receptor. Concomitant to this study an X-ray structure of the A2A adenosine receptor was published with three co-crystallised cholesterol molecules (Figure 5) [90]. Both the simulation and crystallographic studies [24, 90] agreed on site II. The differences in sites I and III between the crystal structure and simulations may reflect the relatively weak and dynamic nature of the cholesterol interactions, as well as sampling constraints.
Figure 5.
Cholesterol interaction sites on the A2A Adenosine receptor. (A) The location of cholesterol molecules (van der Waals spheres) resolved in a high resolution crystal structure of A2A (PDB: 4EIY) are shown [90]. The approximate positions of cholesterol binding sites suggested by molecular simulation [24] are encircled in blue. Extracellular loop 2 is omitted for clarity.
CG simulations provide a means to significantly extend sampling of cholesterol/GPCR interactions. Thus, Sengupta et al. explored cholesterol binding to the Serotonin1A receptor [96] using CG simulations of up to 80 µs duration with the GPCR embedded in a PC:cholesterol bilayer. A number of sites were identified within both leaflets. On the basis of their lipid contact dynamics these sites were predicted to have a relatively weak propensity to bind cholesterol. Interestingly one site co-localised with a cholesterol consensus motif (CCM), and another with a cholesterol recognition amino acid consensus (CRAC) motif. However the significance of such motifs in general remains uncertain. These sites showed differences with those seen for the closely related Serotonin2A receptor [97], raising the question of how to best evaluate and validate simulation-based predictions.
A subsequent study utilized comparable CG simulations to investigate cholesterol interactions with the β2AR [93]. These simulations were performed in bilayers containing two β2AR molecules, allowing an exploration of cholesterol influence on GPCR dimerization. In the absence of cholesterol, protein dimerization was observed via an interface involving TM4 and TM5 from each monomer. Titration of increasing amounts of cholesterol into the lipid bilayer and subsequent simulation led to dissolution of this interface and emergence of a new interface involving helices TM1 and TM2 from each monomer. The TM4+TM5 and TM1+TM2 interfaces for β2AR identified by CG simulation are reminiscent of those seen in atomistic simulations of β1AR [95]. Computation of cholesterol density maps and cholesterol contact dynamics led to identification of three binding sites on the β2AR in the outer leaflet, and four within the inner leaflet [101]. One of these sites was formed on TM4 where it formed interactions with residues of a CCM motif. The binding modes at this site were dynamic, as seen for other GPCRs [24, 96]. Notably cholesterol was seen at this position in a crystal structure of β2AR [102]. In the cholesterol-containing bilayers this binding site became occupied by the sterol, leading to disruption of the involvement of TM4 in the TM4+TM5 interface. These results suggest that cholesterol may modulate dimerization of the β2AR via direct competition between lipid-protein and protein-protein interactions. This provides an example of the use of molecular simulations to explore effects of lipid binding on protein oligomerization and organization within membranes. These aspects have also been explored in large scale simulations of rhodopsin embedded in single lipid-species bilayer containing tens of protein molecules [103–105].
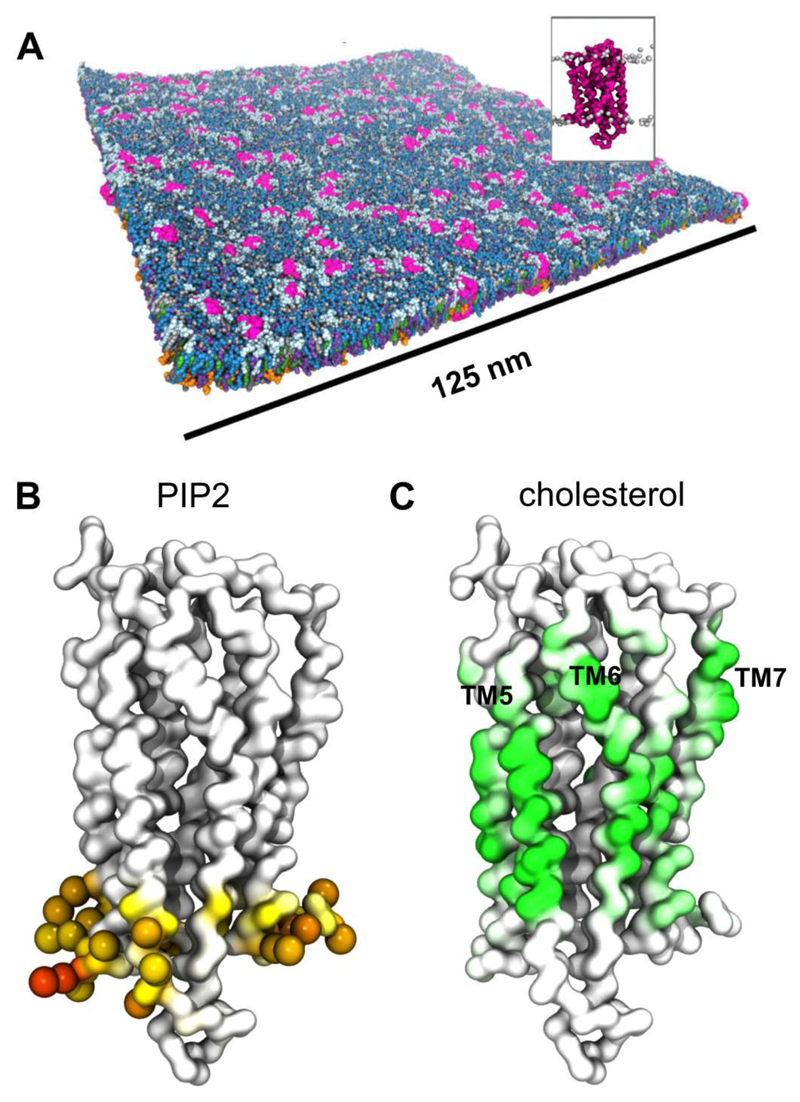
Recent large simulations of the S1P1 receptor in a plasma membrane model have provided further insights into the possible influence of lipid interactions on GPCR oligomerisation [47]. CG simulations of a 140 x 140 nm2 patch of membrane (Figure 6) containing 144 copies of the S1P1 receptor and with a lipid composition mimicking that of a mammalian plasma membrane (extracellular leaflet PC:PE:Sph:GM3:Chol = 40:10:15:10:25; intracellular PC:PE:PS:PIP2:Chol = 10:40:15:10:25) were run for 10 µs. Analysis of the simulations revealed transient formation of S1P1 dimers, trimers, and higher order oligomers. Interactions of cholesterol and of PIP2 with the GPCR were observed (Figure 6). Detailed examination of the S1P1 dimers observed in the simulation suggested that cholesterol may help to mediate the protein-protein interactions.
Figure 6.
Lipid interactions of the S1P1 receptor. (A) 144 S1P1 GPCR molecule (pink; see inset figure) embedded in a bilayer the lipid composition of which corresponds to that of a mammalian cell membrane. The image shown is from the end of a 10 µs CG-MD simulation [47]. (B) Model of the S1P1 receptor coloured according to the level of PIP2 phosphoryl head group interaction, from white (no interaction) to orange (high interaction). Residues with high levels of PIP2 interaction (>75% of simulation time) are shown as spheres. (C) The S1P1 receptor coloured according to the degree of cholesterol interaction from white (no interaction) to green (high interaction). We thank Dr. Heidi Koldsø for the figure. Figure (adapted) reprinted with permission from [47]. Copyright (2015) American Chemical Society.
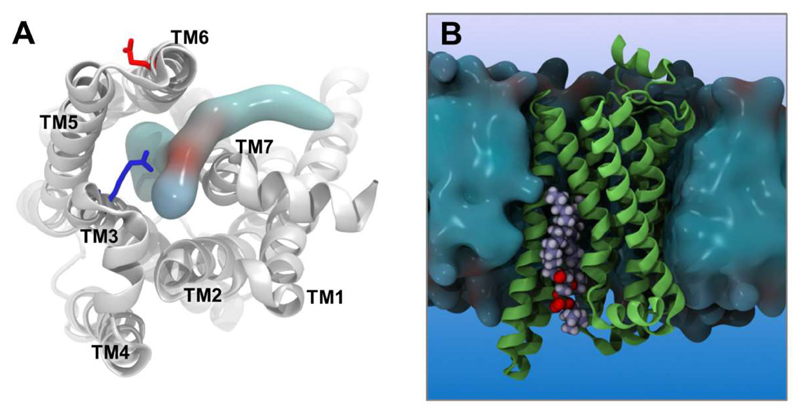
Simulations have also been used to explore how anionic phospholipids may modulate β2AR function [92]. From an extensive (0.25 ms) set of all-atom simulations it was possible to observe an anionic lipid, phosphatidylglycerol (PG), entering the core of the activated β2AR laterally via an opening between the cytoplasmic portions of helices TM6 and TM7 (Figure 7). Once bound the PG molecule formed electrostatic interactions with the protein which inhibited the formation of the ionic lock, a key interaction thought to stabilise the inactive state of the receptor. Entry of the PG lipid thus led to an increase in stability of the active state of the transmembrane domain, providing a testable mechanism which may explain the experimental observation that anionic lipids can enhance the activity of certain GPCRs [106] including the β2AR [107].
Figure 7.
PG/PC binding site within an activated state of the β2AR [92]. (A) Penetration of a PC lipid into the core of the β2AR. The residues R3.50 (blue) and E6.30 (red) of the ionic lock are indicated as sticks. (B) Side view of the receptor embedded in a lipid bilayer (blue surface) with a bound PC lipid (spheres, oxygens in red). We thank Dr. Chris Neale and Prof. Angel García for coordinates and simulation data used to produce the figure.
A number of other examples of identification of lipid binding sites on GPCRs are available [47, 94, 95, 98, 99]. These include long timescale atomistic simulations of the β1AR[95] and CB2 receptors [100], and multiscale simulations of rhodopsin [98, 99]; as well as a recent study on possible effects of omega-3 fatty acids on oligomerization of A2A adenosine and of dopamine D2 receptors in the context of neuropsychiatric disease [108]. For a recent detailed review of the application of molecular simulations to explore GPCR-cholesterol interactions see [38].
2.3. Other receptors: receptor tyrosine kinases
A second major superfamily of membrane receptors are those which have a single membrane spanning helix, and which form functional dimers in cell membranes. These include the receptor tyrosine kinases (RTKs) [109] and also a number of other families including e.g. integrins [110]. These receptors are characterised by extensive extracellular domains and single pass transmembrane helices. Many crystal structures are known for extracellular and intracellular domains, but a high resolution structure of an intact RTK remains elusive. A number of NMR structures of TM helix dimers from RTKs are known [111–114].
Experimental studies have shown that lipid interactions can modulate RTK activation, especially of the EGFR [8, 115]. There have been a number of simulation studies of: (i) models of the intact EGFR receptor in mixed lipid bilayers [116, 117]; and of (ii) the transmembrane helix of the EGFR and its dimers [35, 118, 119]. There has also been a recent study combining experiments and simulations to explore lipid interactions of the EphA2 receptor transmembrane region and ectodomain, and their influence on the conformation and orientation of the ectodomain relative to the membranes [120].
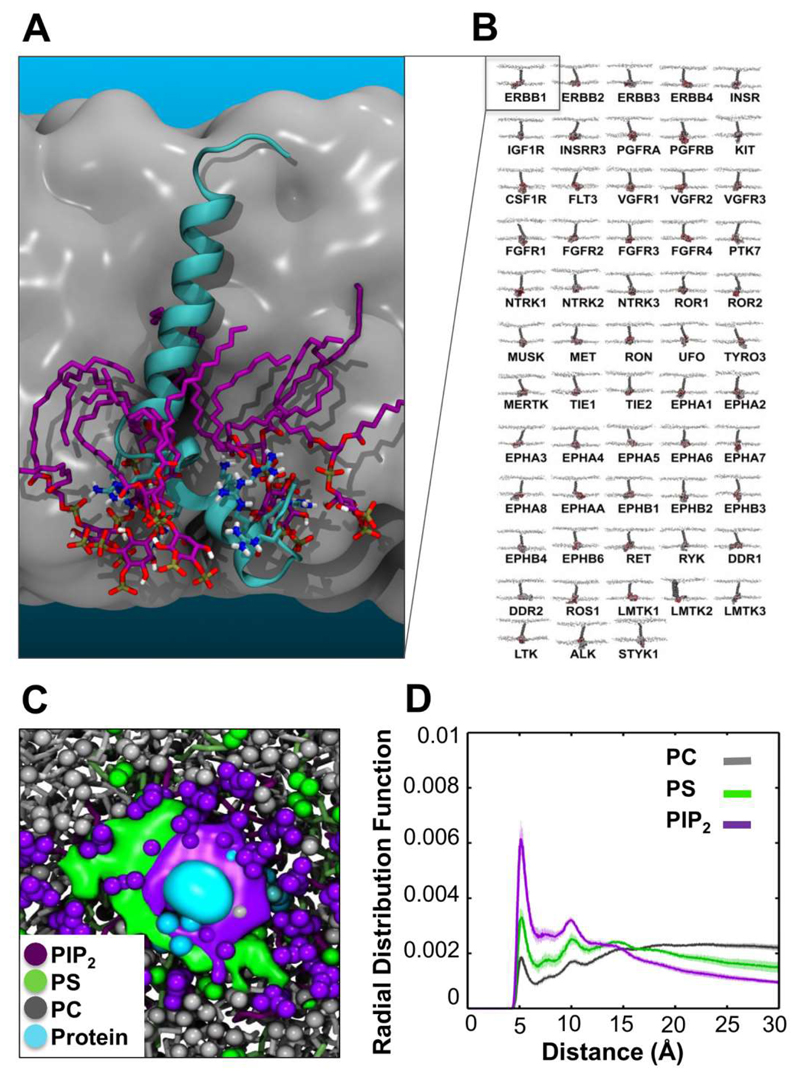
Recently we undertook a systematic comparative study of lipid interactions with models of the transmembrane helix plus juxtamembrane region (TM+JM) of all 58 human RTKs [35]. This revealed conservation of the interactions of the JM region with PIP2 (Figure 8). Furthermore, these conserved interactions were seen to induce local bilayer reorganisation and anionic lipid clustering (Figure 8C) seen previously for the TM+JM regions of the gp130 cytokine receptor [34]. This behaviour is likely to extend to other single transmembrane domain proteins besides RTKs and related receptors. However, it is of especial interest for RTKs as PIP2 lipids have been suggested to modulate the activity and cellular distribution of certain members of the family ([8, 121, 122] by interactions with the TM-JM domain.
Figure 8.
Conserved interaction of PIP2 lipids with TM-JM models of the human RTK superfamily [35]. (A) TM-JM model of the EGFR monomer (cyan) surrounded by a cluster of PIP2 lipids (purple). (B) The 58 known members of the human RTK superfamily. (C) Clustering of anionic lipids around the monomeric TM-JM domain of the Insulin receptor. The image shows the average spatial occupancy of PIP2 (purple), phosphatidylserine (green), and phosphatidylcholine (grey) lipids over the inner leafleft surface. The JM region density is indicated in cyan. (D) Radial distribution function for each lipid species relative to the protein. Figure modified from [35].
This comparative simulation study demonstrated that simulations can now be used to compare protein/lipid interactions across whole families of membrane proteins (as has also proved possible for e.g. aquaporins [123]). Conservation of such interactions across a family of proteins is an indicator of their likely biological importance.
2.4. Transporter proteins
Lipids have been shown to play a critical role in the function of a number of solute transporters, and structural studies have revealed lipid binding sites on several transporter proteins [5]. Simulation based approaches have made a recent valuable contribution to identification and characterisation of sites on transporters including the ADP/ATP carrier [124], the UraA H+-Uracil symporter [60], LacY [125], SERCA, the canonical P-type ATPase [126], and the dopamine transporter [127]
2.4.1. ADP/ATP carrier (AAC/ANT)
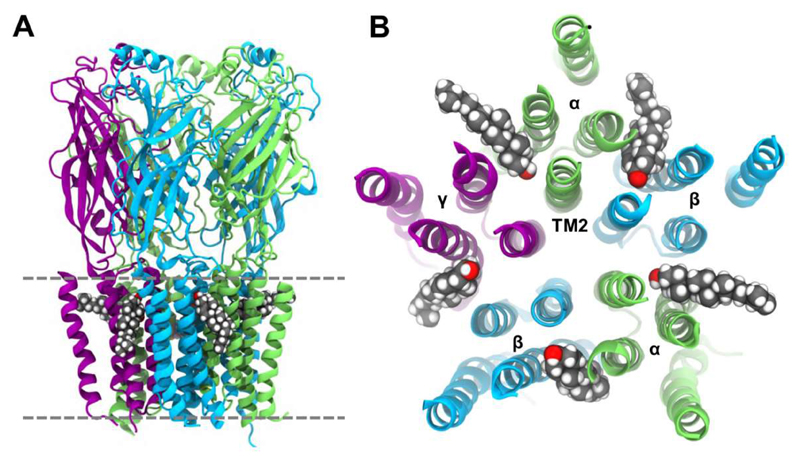
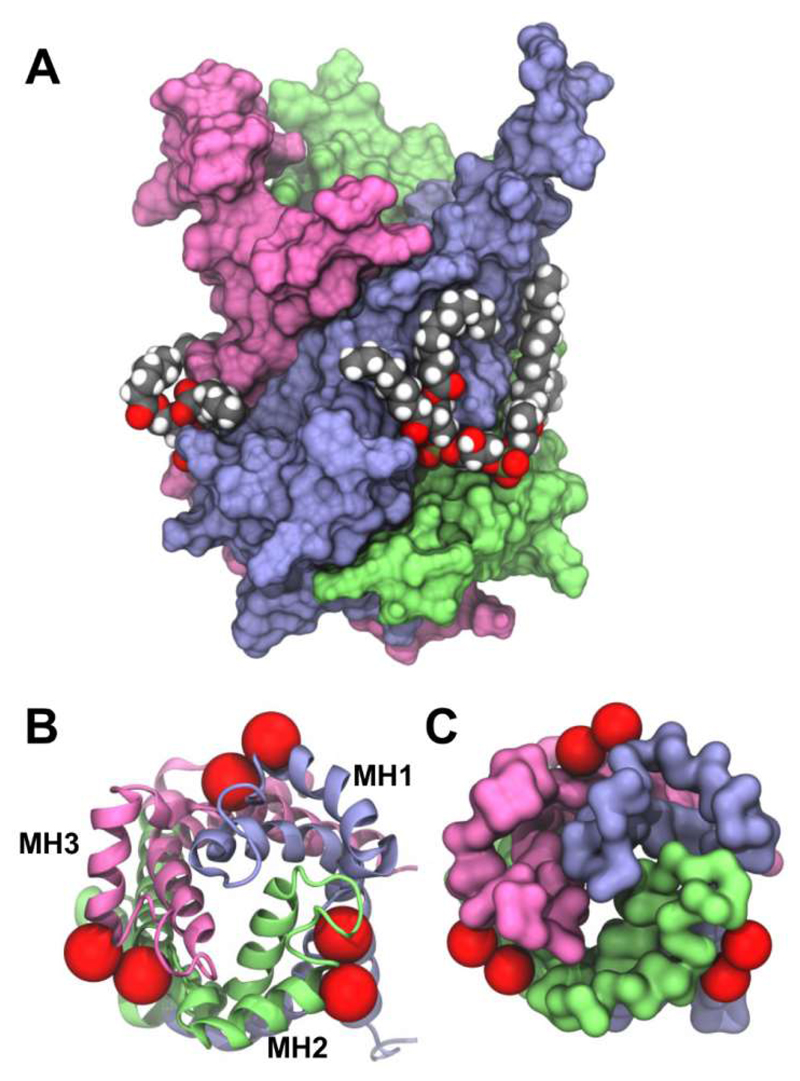
The ADP/ATP carrier (AAC/ANT) is the best studied member of the mitochondrial carrier family (MCF) of transporters, which enable the exchange of solutes across mitochondrial inner membranes [128]. In common with other MCF transporters, AAC/ANT consists of six transmembrane helices arranged around a central cavity, with three amphipathic matrix helices positioned perpendicular to the membrane normal. AAC/ANT requires the presence of cardiolipin for interactions with other proteins [28, 129], and the presence of cardiolipin is a requirement for reconstitution and crystallization of this protein. Simulations in a simple PC bilayer revealed three binding sites which coincide with the crystallographic locations of three quasi-equivalent cardiolipin (CL) binding sites [124]. Recently we have explored in more detail interactions with cardiolipin by combined CG and atomistic simulations (Rouse et al., unpublished data). Multiple CG simulations of microscecond duration of bovine AAC/ANT (PDB: 1OKC) embedded in a PC:PE:CL lipid bilayer (molar ratio 6:6:1) were conducted to assess cardiolipin interactions with the transporter. In all five simulations cardiolipin molecules were seen to bind into three ‘grooves’ on the protein between the amphipathic matrix helices. These three sites were in excellent agreement with previous X-ray structures containing co-crystallised cardiolipin molecules [36, 130, 131] (Figure 9). Simulation of a recently published structure of yeast AAC2 (PDB: 4C9G[131]) revealed the same three cardiolipin binding sites. This conservation of binding sites reinforces their likely biological significance. A parallel independent CG simulation study identified the same three sites on bovine and yeast AAC/ANT (Duncan et al., unpublished data), and showed the bound cardiolipin molecules remained stable during subsequent 20 ns atomistic simulation. The reproduction of these same three sites between multiple crystal structures, and two independent simulation studies is strongly suggestive of their biological significance. Indeed, cardiolipin is thought to serve a role in mitochondrial membranes as a ‘glue’ or ‘molecular filler’ in mediating certain protein-protein interactions. These studies of cardiolipin interactions with the AAC/ANT transporter protein in the inner mitochondrial membrane nicely complement landmark studies of on cardiolipin interactions with components of the electron transport chain (namely cytochrome bc1 [23, 132] and cytochrome c oxidase [133]), and suggest how simulations of lipid-protein interactions might aid development of models of larger scale organization [134–136] of the mitochondrial inner membrane [137].
Figure 9.
The ADP/ATP carrier binds cardiolipin with a 1:3 stoichiometry at specific sites identified by experiment and CG simulation. (A) Side view of the bovine transporter (PDB: 2C3E) open to the intermembrane space, with crystallographically-resolved [130] cardiolipin molecules shown as van der Waals spheres. The protein surface is coloured according to its three fold pseudo-symmetry. (B) Crystallographic (PDB: 2C3E) and (C) simulation (Rouse et al., unpublished data) views onto the base of the transporter from the matrix. The approximate position of cardiolipin phosphate groups are indicated as red spheres (each cardiolipin molecule contains two phosphate groups). The matrix-facing helices at the bilayer surface are labelled MH1 to MH3.
2.4.2. UraA H+-Uracil symporter
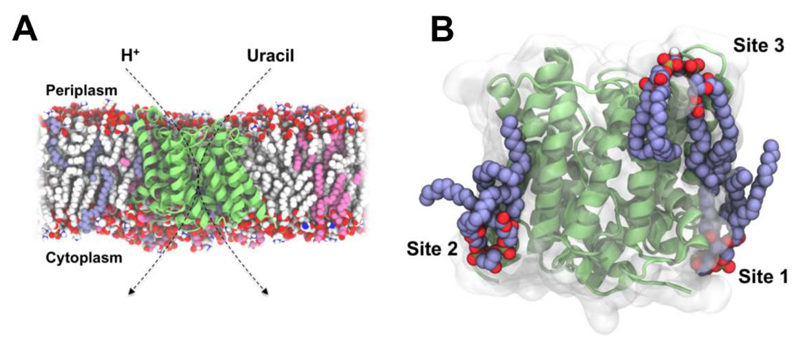
The Escherichia coli UraA H+-Uracil symporter is a member of the nucleobase-ascorbate transporter (NAT) family of transporters [138] which use proton and Na+ gradients to enable uptake of xanthine, uric acid, and uracil, as well as vitamin C in mammals [138, 139]. A recent MD study explored lipid interactions of the bacterial UraA transporter (PDB: 3QE7) in a bilayer, [60] the lipid composition of which mimicked that of the bacterial inner membrane, containing PE (75 mol %), PG (20 mol %) and CL (5 mol %). CG simulations of up to 10 μs duration revealed an enrichment of the anionic lipids PG and CL within the lipid annulus. In particular, CL exhibited preferential interactions with the transporter at three sites (Figure 10). A periplasmic site was seen to be positioned in the vicinity of the transport pathway. Given CL may act as a proton donor and acceptor at physiological pH, this suggests a putative proton-donating role for this cardiolipin site. Analysis of protein-lipid contact patterns during the simulation showed cardiolipin molecules at each site interacted predominantly via electrostatic interactions between negatively charged cardiolipin headgroups and basic sidechains, whilst in common with observations from other simulation studies [23, 25] the tail groups exhibited more dynamic interactions. The possible importance of interactions seen in identified lipid binding sites may be tested via in silico mutagenesis of binding site residues. Using this approach sidechains critical for cardiolipin binding were identified [60]. No lipid molecules have been seen in X-ray structures of UraA, which was crystallised in the presence of a detergent (n-nonyl-β-D-glucopyranoside), one molecule of which was observed bound in the crystal structure in the centre of the protein on the presumed transport pathway of the solute. This suggests possible weak association of cardiolipin with UraA, which may dissociate during detergent treatment.
Figure 10.
Cardiolipin binding sites on the UraA H+-Uracil symporter [60]. (A) Cross-section through a model of an E. coli inner membrane containing UraA (lime), PE (white), PG (pink), and cardiolipin (blue). (B) A representative simulation snapshot showing three predicted cardiolipin binding sites on the transporter. We thank Dr. Antreas Kalli for coordinates used to produce the figure.
2.4.3. Ca2+ ATPase: SERCA
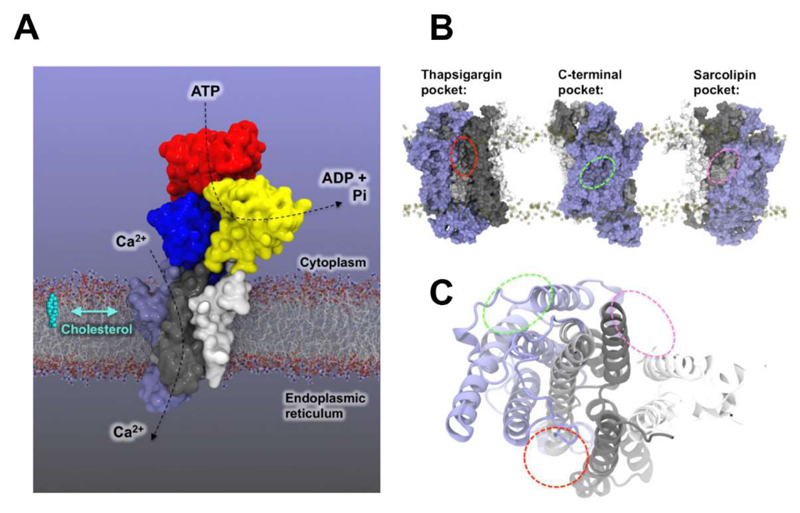
SERCA is a canonical P-type ATPase which enables ATP-driven reuptake of Ca2+ ions into the endo or sarcoplasmic reticulum during calcium signalling. Enrichment of cholesterol in ER membranes of macrophages has been shown to inhibit SERCA2b [30], and lipid-like drugs such as thapsigargin modulate SERCA. Further, phospholipids and other hydrophobic compounds have been observed in x-ray crystal structures [140, 141]. In a study by Autzen et al., thirty independent CG simulations of SERCA embedded in a PC:cholesterol bilayer (molar ratio 10:1) were conducted to address possible cholesterol binding and modulation of SERCA [126]. Mapping the frequency of cholesterol contacts for each residue of the protein onto the 3D structure revealed two major cholesterol binding pockets (Figure 11), one of which co-localized to a position known to bind sarcolipin, a small single TM helix protein which modulates SERCA activity. However, from the location of cholesterol within these sites and comparison of its exchange dynamics to bulk phospholipids it has proved difficult to rationalise how cholesterol binding may alter SERCA activity. Consideration of shared chemical features of thapsigargin and cholesterol lead to an expectation that cholesterol might bind in the thapsigargin binding pocket, but this was not observed in the simulations, in which the pocket instead was occupied by a phospholipid. A series of 100 ns all-atom simulations initiated from cholesterol manually positioned in the thapsigargin binding pocket (based on thapsigargin bound X-ray structure, PDB: 2C8K) did not result in a single stable configuration over the simulated time course, and instead exhibited a very dynamic cholesterol pose. This suggests that simulations may be used to rule out possible binding modes for lipids, although further experimental and computational studies will be needed to explore this in more detail.
Figure 11.
SERCA architecture and putative cholesterol binding pockets. (A) Architecture and domain organisation of SERCA in the E2 state (a simulated structure [126] derived from PDB 2C8K is shown). (B) Approximate locations of the thapsigargin (red line), C-terminal (green line), and sarcolipin (pink line) cholesterol binding pockets on the SERCA transmembrane domain (TM1-2: white, TM3-4: black, TM5-6: grey, TM7-10: blue). We are grateful to Dr. Henriette E. Autzen for providing coordinates for this diagram.
2.5. Other membrane proteins
A number of simulation studies not described in detail in this review have also addressed lipid interaction sites of membrane proteins. These include phospholipid binding to LacY [125], Kv channels [142], aquaporins [123, 143], the fungal lipid scrambalase TMEM [124], and the dopamine transporter [127]; cardiolipin binding to respiratory chain complexes [23, 132, 133]; and cholesterol binding to rhodopsin [98, 99], VDAC [144], β1AR [95] and β2AR [94].
3. Summary and Conclusions
From a number of studies it is evident that molecular dynamics simulations enable us to identify lipid binding and/or interaction sites on integral membrane proteins, and in particular on a number of channels, receptors, and transporters. Molecular simulations provide a valuable complement to experimental approaches, especially X-ray diffraction and mass spectrometry in allowing us to analyse membrane protein/lipid interactions in detail. MD simulations can not only identify interaction sites but also may provide details of selectivity and can be applied to explore competition effects by including multiple lipid species in simulations [55, 56]. Furthermore, the energetics of lipid binding may be estimated, as has been seen for e.g. Kir-cholesterol [22] and Kir-PIP2 interactions [145], A2A adenosine receptor-cholesterol interactions [24], and cytochrome bc1-cardiolipin interactions [23]. Simulations also enable comparison of lipid interactions with different conformational states of a protein, providing insights into function, e.g. for mechanosensitive ion channels [27].
Simulations of large ensembles of proteins in mixed lipid bilayers are starting to provide insights into protein/lipid interactions in membrane models which take increased account of the lipid compositional complexity of membranes in vivo [54]. Recent examples of complex lipid mixtures containing large ensembles of protein include simulations of the S1P1 receptor [47], the bacterial outer membrane proteins OmpF and BtuB [146], the membrane envelope of the influenza virion [147], mitochondrial respiratory chain complexes [137], and of the A2A adenosine and dopamine D2 receptors embedded in membranes with either a ‘healthy’ (i.e. high omega-3 fatty acid content) or a ‘disease state’ (i.e. a low omega-3 fatty acid content) lipid composition [108]. Such investigations benefit enormously from continuing developments in the MARTINI coarse-grained force field [148, 149], especially for complex mixtures of lipids [53, 55] which allows binding sites to be identified simply by random diffusion of lipid molecules in large systems with multiple copies of a given membrane protein. This approach is not routinely possible yet in atomistic detail due to the need for extended (multi-microsecond) simulations on large systems and the concomitant computational demands. Rather, atomistic simulations may be coupled with CG or docking methods, using these approaches to initially survey the protein surface for candidate lipid interaction sites, before applying atomistic simulations to evaluate and refine these sites.
In parallel with developments in CG simulations there have also been ongoing advances in atomistic parameters for lipids (e.g. [150–156]). Of particular importance is the development of accurate parameters for a greater diversity of lipids [157], including e.g. phosphatidyl inositols [158, 159], and e.g. bacterial lipopolysaccharide models [160].
As the number of membrane protein structures continues to increase (see http://blanco.biomol.uci.edu/mpstruc/) there are considerable opportunities for systematic and comparative analysis of lipid-protein interactions across all membrane proteins of known structure. In parallel there have also been ongoing developments in enabling high throughput simulations for membrane proteins [105, 161]. With the emergence of high-throughput technologies for studying lipid-protein interactions experimentally [162], such simulation approaches will have increasing scope in complementing structural and biophysical studies. In particular, it will be interesting to incorporate computational tools to identify lipid binding and interaction sites within existing simulation pipelines and databases such as MemProtMD [124]. Such automation would provide a larger scale survey of the existing database of membrane protein structures, which will enable general patterns and trends in lipid interactions to be identified.
Highlights.
Lipid molecules can selectively interact with specific sites on membrane proteins
Lipid binding can modulate protein structure and function
Molecular dynamics (MD) simulations provide a powerful tool for site identification
MD has been used to identify sites on a wide range of membrane proteins
Acknowledgements
Research in M. S. P. S.’s group is funded by the BBSRC (BB/L002558/1), and the Wellcome Trust (WT092970). G.H. is funded by the MRC and the University of Oxford. We thank Dr. Anna Duncan and other members of the M.S.P.S laboratory for fruitful discussions. Thanks to Dr. Chris Neale and Professor Angel E. García for simulation structures of β2AR used in the table of contents graphic.
Abbreviations
- MD
molecular dynamics
- CG
coarse-grained
- NMR
nuclear magnetic resonance
- ESR
electron spin resonance
- EM
electron microscopy
- POPC
1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine
- POPG
1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoglycerol
- POPE
1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine
- PIP2
phosphatidylinositol-4,5-bisphosphate
- Kir
inward rectifying potassium ion channel
- nAChR
nicotinic acetylcholine receptor
- GABAAR
y-aminobutyric acid receptor
- GPCR
G-protein coupled receptor
- β2AR
β2-adrenergic receptor
- ANT/AAC
ADP/ATP carrier
- SERCA
sarco/endoplasmic reticulum Ca2+ ATPase.
Footnotes
For BBA Biomembranes, BBAMEM-16-23 (Special Issue)
References
- [1].Shevchenko A, Simons K. Lipidomics: coming to grips with lipid diversity. Nature Rev Molec Cell Biol. 2010;11:593–598. doi: 10.1038/nrm2934. [DOI] [PubMed] [Google Scholar]
- [2].Pedersen BP, Nissen P. Membrane proteins - do we catch up with the breathless pace of soluble protein structural biology? Biochim Biophys Acta. 2015;1850:447–448. doi: 10.1016/j.bbagen.2015.01.001. [DOI] [PubMed] [Google Scholar]
- [3].Vinothkumar KR. Membrane protein structures without crystals, by single particle electron cryomicroscopy. Curr Opin Struct Biol. 2015;33:103–114. doi: 10.1016/j.sbi.2015.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Fahy E, Subramaniam S, Murphy RC, Nishijima M, Raetz CRH, Shimizu T, Spener F, van Meer G, Wakelam MJO, Dennis EA. Update of the LIPID MAPS comprehensive classification system for lipids. J Lipid Res. 2009;50:S9–S14. doi: 10.1194/jlr.R800095-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Koshy C, Ziegler C. Structural insights into functional lipid–protein interactions in secondary transporters. Biochim Biophys Acta. 2015;1850:476–487. doi: 10.1016/j.bbagen.2014.05.010. [DOI] [PubMed] [Google Scholar]
- [6].Lopes CMB, Zhang HL, Rohacs T, Jin TH, Yang J, Logothetis DE. Alterations in conserved Kir channel-PIP2 interactions underlie channelopathies. Neuron. 2002;34:933–944. doi: 10.1016/s0896-6273(02)00725-0. [DOI] [PubMed] [Google Scholar]
- [7].Romanenko VG, Rothblat GH, Levitan I. Modulation of endothelial inward-rectifier K+ current by optical isomers of cholesterol. Biophys J. 2002;83:3211–3222. doi: 10.1016/S0006-3495(02)75323-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Michailidis IE, Rusinova R, Georgakopoulos A, Chen Y, Iyengar R, Robakis NK, Logothetis DE, Baki L. Phosphatidylinositol-4,5-bisphosphate regulates epidermal growth factor receptor activation. Pflugers Arch. 2011;461:387–397. doi: 10.1007/s00424-010-0904-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Zocher M, Zhang C, Rasmussen SGF, Kobilka BK, Muller DJ. Cholesterol increases kinetic, energetic, and mechanical stability of the human beta(2)-adrenergic receptor. Proc Natl Acad Sci USA. 2012;109:E3463–E3472. doi: 10.1073/pnas.1210373109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Saxena R, Chattopadhyay A. Membrane cholesterol stabilizes the human serotonin1A receptor. Biochim Biophys Acta. 2012;1818:2936–2942. doi: 10.1016/j.bbamem.2012.07.032. [DOI] [PubMed] [Google Scholar]
- [11].Barrantes FJ. Cholesterol effects on nicotinic acetylcholine receptor. J Neurochem. 2007;103:72–80. doi: 10.1111/j.1471-4159.2007.04719.x. [DOI] [PubMed] [Google Scholar]
- [12].Sooksawate T, Simmonds MA. Effects of membrane cholesterol on the sensitivity of the GABAA receptor to GABA in acutely dissociated rat hippocampal neurones. Neuropharmacol. 2001;40:178–184. doi: 10.1016/s0028-3908(00)00159-3. [DOI] [PubMed] [Google Scholar]
- [13].Laganowsky A, Reading E, Allison TM, Ulmschneider MB, Degiacomi MT, Baldwin AJ, Robinson CV. Membrane proteins bind lipids selectively to modulate their structure and function. Nature. 2014;510:172–175. doi: 10.1038/nature13419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Yeagle PL. Non-covalent binding of membrane lipids to membrane proteins. Biochim Biophys Acta. 2014;1838:1548–1559. doi: 10.1016/j.bbamem.2013.11.009. [DOI] [PubMed] [Google Scholar]
- [15].Hansen SB, Tao X, Mackinnon R. Structural basis of PIP2 activation of the classical inward rectifier K+ channel Kir2.2. Nature. 2011;477:495–498. doi: 10.1038/nature10370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Durrant J, McCammon JA. Molecular dynamics simulations and drug discovery. BMC Biology. 2011;9:71. doi: 10.1186/1741-7007-9-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Caffrey M. A comprehensive review of the lipid cubic phase or in meso method for crystallizing membrane and soluble proteins and complexes. Acta Cryst F. 2015;71:3–18. doi: 10.1107/S2053230X14026843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Stansfeld PJ, Sansom MSP. Molecular simulation approaches to membrane proteins. Structure. 2011;19:1562–1572. doi: 10.1016/j.str.2011.10.002. [DOI] [PubMed] [Google Scholar]
- [19].Powl AM, East JM, Lee AG. Different effects of lipid chain length on the two sides of a membrane and the lipid annulus of MscL. Biophys J. 2007;93:113–122. doi: 10.1529/biophysj.107.105130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Marsh D. Electron spin resonance in membrane research: protein-lipid interactions from challenging beginnings to state of the art. Eur Biophys J. 2010;39:513–525. doi: 10.1007/s00249-009-0512-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Das N, Murray DT, Cross TA. Lipid bilayer preparations of membrane proteins for oriented and magic-angle spinning solid-state NMR samples. Nature Protocols. 2013;8:2256–2270. doi: 10.1038/nprot.2013.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Rosenhouse-Dantsker A, Noskov S, Durdagi S, Logothetis DE, Levitan I. Identification of novel cholesterol-binding regions in Kir2 channels. J Biol Chem. 2013 doi: 10.1074/jbc.M113.496117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Arnarez C, Mazat J-P, Elezgaray J, Marrink S-J, Periole X. Evidence for cardiolipin binding sites on the membrane-exposed surface of the cytochrome bc1. J Amer Chem Soc. 2013;135:3112–3120. doi: 10.1021/ja310577u. [DOI] [PubMed] [Google Scholar]
- [24].Lee JY, Lyman E. Predictions for cholesterol interaction sites on the A2A adenosine receptor. J Amer Chem Soc. 2012;134:16512–16515. doi: 10.1021/ja307532d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Stansfeld PJ, Hopkinson RJ, Ashcroft FM, Sansom MSP. The PIP2 binding site in Kir channels: definition by multi-scale biomolecular simulations. Biochem. 2009;48:10926–10933. doi: 10.1021/bi9013193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Schmidt MR, Stansfeld PJ, Tucker SJ, Sansom MSP. Simulation-based prediction of phosphatidylinositol 4,5-bisphosphate binding to an ion channel. Biochem. 2013;52:279–281. doi: 10.1021/bi301350s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Pliotas C, Dahl ACE, Rasmussen T, Mahendran KR, Smith TK, Marius P, Gault J, Banda T, Rasmussen A, Miller S, Robinson CV, et al. The role of lipids in mechanosensation. Nature Struct Mol Biol. 2015;22:991–998. doi: 10.1038/nsmb.3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Claypool SM, Oktay Y, Boontheung P, Loo JA, Koehler CM. Cardiolipin defines the interactome of the major ADP/ATP carrier protein of the mitochondrial inner membrane. J Cell Biol. 2008;182:937–950. doi: 10.1083/jcb.200801152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Robinson NC, Zborowski J, Talbert LH. Cardiolipin-depleted bovine heart cytochrome c oxidase: binding stoichiometry and affinity for cardiolipin derivatives. Biochem. 1990;29:8962–8969. doi: 10.1021/bi00490a012. [DOI] [PubMed] [Google Scholar]
- [30].Li Y, Ge M, Ciani L, Kuriakose G, Westover EJ, Dura M, Covey DF, Freed JH, Maxfield FR, Lytton J, Tabas I. Enrichment of endoplasmic reticulum with cholesterol inhibits sarcoplasmic-endoplasmic reticulum calcium ATPase-2b activity in parallel with increased order of membrane lipids: implications for depletion of endoplasmic reticulum calcium stores and apoptosis in cholesterol-loaded macrophages. J Biol Chem. 2004;279:37030–37039. doi: 10.1074/jbc.M405195200. [DOI] [PubMed] [Google Scholar]
- [31].Hanson M, Peach R. In: Sphingosine-1-Phosphate Signaling in Immunology and Infectious Diseases. Oldstone MBA, Rosen H, editors. Springer International Publishing; 2014. pp. 23–53. [Google Scholar]
- [32].Marsh D, Pali T. The protein-lipid interface: perspectives from magnetic resonance and crystal structures. Biochim Biophys Acta. 2004;1666:118–141. doi: 10.1016/j.bbamem.2004.08.006. [DOI] [PubMed] [Google Scholar]
- [33].Marsh D, Horváth LI. Structure, dynamics and composition of the lipid-protein interface. Perspectives from spin-labelling. Biochim Biophys Acta. 1998;1376:267–296. doi: 10.1016/s0304-4157(98)00009-4. [DOI] [PubMed] [Google Scholar]
- [34].Koldsø H, Sansom MSP. Local lipid reorganization by a transmembrane protein domain. J Phys Chem Lett. 2012;3:3498–3502. doi: 10.1021/jz301570w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Hedger G, Sansom MSP, Koldsø H. The juxtamembrane regions of human receptor tyrosine kinases exhibit conserved interaction sites with anionic lipids. Sci Reports. 2015;5:9198. doi: 10.1038/srep09198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Pebay-Peyroula E, Dahout-Gonzalez C, Kahn R, Trezeguet V, Lauquin GJ, Brandolin G. Structure of mitochondrial ADP/ATP carrier in complex with carboxyatractyloside. Nature. 2003;426:39–44. doi: 10.1038/nature02056. [DOI] [PubMed] [Google Scholar]
- [37].Shinzawa-Itoh K, Aoyama H, Muramoto K, Terada H, Kurauchi T, Tadehara Y, Yamasaki A, Sugimura T, Kurono S, Tsujimoto K, Mizushima T, et al. Structures and physiological roles of 13 integral lipids of bovine heart cytochrome c oxidase. EMBO J. 2007;26:1713–1725. doi: 10.1038/sj.emboj.7601618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Sengupta D, Chattopadhyay A. Molecular dynamics simulations of GPCR–cholesterol interaction: An emerging paradigm. Biochim Biophys Acta. 2015;1848:1775–1782. doi: 10.1016/j.bbamem.2015.03.018. [DOI] [PubMed] [Google Scholar]
- [39].Fantini J, Yahi N, Garmy N. Cholesterol accelerates the binding of Alzheimer's beta-amyloid peptide to ganglioside GM1 through a universal hydrogen-bond-dependent sterol tuning of glycolipid conformation. Front Physiol. 2013;4 doi: 10.3389/fphys.2013.00120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Planas-Iglesias J, Dwarakanath H, Mohammadyani D, Yanamala N, Kagan Valerian E, Klein-Seetharaman J. Cardiolipin interactions with proteins. Biophys J. 2015;109:1282–1294. doi: 10.1016/j.bpj.2015.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Dror RO, Dirks RM, Grossman JP, Xu HF, Shaw DE. Biomolecular simulation: a computational microscope for molecular biology. Ann Rev Biophys. 2012;41:429–452. doi: 10.1146/annurev-biophys-042910-155245. [DOI] [PubMed] [Google Scholar]
- [42].Marrink SJ, Tieleman DP. Perspective on the Martini model. Chem Soc Rev. 2013;42:6801–6822. doi: 10.1039/c3cs60093a. [DOI] [PubMed] [Google Scholar]
- [43].Voth GA. Coarse-Graining of Condensed Phase and Biomolecular Systems. CRC Press; 2008. [Google Scholar]
- [44].Stansfeld PJ, Sansom MSP. From coarse-grained to atomistic: a serial multi-scale approach to membrane protein simulations. J Chem Theor Comp. 2011;7:1157–1166. doi: 10.1021/ct100569y. [DOI] [PubMed] [Google Scholar]
- [45].Wassenaar TA, Pluhackova K, Böckmann RA, Marrink SJ, Tieleman DP. Going backward: a flexible geometric approach to reverse transformation from coarse grained to atomistic models. J Chem Theor Comput. 2014;10:676–690. doi: 10.1021/ct400617g. [DOI] [PubMed] [Google Scholar]
- [46].Ayton GA, Noid WG, Voth GA. Multiscale modeling of biomolecular systems: in serial and in parallel. Curr Opin Struct Biol. 2007;17:192–198. doi: 10.1016/j.sbi.2007.03.004. [DOI] [PubMed] [Google Scholar]
- [47].Koldsø H, Sansom MSP. Organization and dynamics of receptor proteins in a plasma membrane. J Amer Chem Soc. 2015;137:14694–14704. doi: 10.1021/jacs.5b08048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Dror RO, Mildorf TJ, Hilger D, Manglik A, Borhani DW, Arlow DH, Philippsen A, Villanueva N, Yang ZY, Lerch MT, Hubbell WL, et al. Structural basis for nucleotide exchange in heterotrimeric G proteins. Science. 2015;348:1361–1365. doi: 10.1126/science.aaa5264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Scott KA, Bond PJ, Ivetac A, Chetwynd AP, Khalid S, Sansom MSP. Coarse-grained MD simulations of membrane protein-bilayer self-assembly. Structure. 2008;16:621–630. doi: 10.1016/j.str.2008.01.014. [DOI] [PubMed] [Google Scholar]
- [50].Faraldo-Gómez JD, Smith GR, Sansom MSP. Setup and optimisation of membrane protein simulations. Eur Biophys J. 2002;31:217–227. doi: 10.1007/s00249-002-0207-5. [DOI] [PubMed] [Google Scholar]
- [51].Wolf MG, Hoefling M, Aponte-Santamaría C, Grubmüller H, Groenhof G. g_membed: Efficient insertion of a membrane protein into an equilibrated lipid bilayer with minimal perturbation. J Comput Chem. 2010;31:2169–2174. doi: 10.1002/jcc.21507. [DOI] [PubMed] [Google Scholar]
- [52].Jefferys E, Sands ZA, Shi J, Sansom MS, Fowler PW. Alchembed: a computational method for incorporating multiple proteins into complex lipid geometries. J Chem Theory Comput. 2015;11:2743–2754. doi: 10.1021/ct501111d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Wassenaar TA, Ingolfsson HI, Boeckmann RA, Tieleman DP, Marrink SJ. Computational lipidomics with Insane: a versatile tool for generating custom membranes for molecular simulations. J Chem Theor Comput. 2015;11:2144–2155. doi: 10.1021/acs.jctc.5b00209. [DOI] [PubMed] [Google Scholar]
- [54].van Meer G, de Kroon AIPM. Lipid map of the mammalian cell. J Cell Sci. 2011;124:5–8. doi: 10.1242/jcs.071233. [DOI] [PubMed] [Google Scholar]
- [55].Ingolfsson HI, Melo MN, van Eerden FJ, Arnarez C, Lopez CA, Wassenaar TA, Periole X, de Vries AH, Tieleman DP, Marrink SJ. Lipid organization of the plasma membrane. J Amer Chem Soc. 2014;136:14554–14559. doi: 10.1021/ja507832e. [DOI] [PubMed] [Google Scholar]
- [56].Koldsø H, Shorthouse D, Hélie J, Sansom MSP. Lipid clustering correlates with membrane curvature as revealed by molecular simulations of complex lipid bilayers. PLoS Comp Biol. 2014;10:e1003911. doi: 10.1371/journal.pcbi.1003911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Martinez L, Andrade R, Birgin EG, Martinez JM. PACKMOL: a package for building initial configurations for molecular dynamics simulations. J Comp Chem. 2009;30:2157–2164. doi: 10.1002/jcc.21224. [DOI] [PubMed] [Google Scholar]
- [58].Sommer B, Dingersen T, Gamroth C, Schneider SE, Robert S, Krueger J, Dietz K-J. CELLmicrocosmos 2.2 membrane editor: a modular interactive shape-based software approach to solve heterogeneous membrane packing problems. J Chem Inform Model. 2011;51:1165–1182. doi: 10.1021/ci1003619. [DOI] [PubMed] [Google Scholar]
- [59].Wu EL, Cheng X, Jo S, Rui H, Song KC, Davila-Contreras EM, Qi Y, Lee J, Monje-Galvan V, Venable RM, Klauda JB, et al. CHARMM-GUI membrane builder toward realistic biological membrane simulations. J Comp Chem. 2014;35:1997–2004. doi: 10.1002/jcc.23702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Kalli AC, Sansom MSP, Reithmeier RAF. Molecular dynamics simulations of the bacterial UraA H+-uracil symporter in lipid bilayers reveal a closed state and a selective interaction with cardiolipin. PLoS Comput Biol. 2015;11:e1004123. doi: 10.1371/journal.pcbi.1004123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comp Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Brannigan G, Hénin J, Law R, Eckenhoff R, Klein ML. Embedded cholesterol in the nicotinic acetylcholine receptor. Proc Natl Acad Sci USA. 2008;105:14418–14423. doi: 10.1073/pnas.0803029105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Hénin J, Salari R, Murlidaran S, Brannigan G. A predicted binding site for cholesterol on the GABAA receptor. Biophys J. 2014;106:1938–1949. doi: 10.1016/j.bpj.2014.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Valiyaveetil FI, Zhou Y, MacKinnon R. Lipids in the structure, folding and function of the KcsA channel. Biochem. 2002;41:10771–10777. doi: 10.1021/bi026215y. [DOI] [PubMed] [Google Scholar]
- [65].Deol SS, Domene C, Bond PJ, Sansom MSP. Anionic phospholipids interactions with the potassium channel KcsA: simulation studies. Biophys J. 2006;90:822–830. doi: 10.1529/biophysj.105.071407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Doupnik CA, Davidson N, Lester HA. The inward rectifier potassium channel family. Curr Opin Neurobiol. 1995;5:268–277. doi: 10.1016/0959-4388(95)80038-7. [DOI] [PubMed] [Google Scholar]
- [67].Suh BC, Hille B. PIP2 is a necessary cofactor for ion channel function: How and why? Ann Rev Biophys. 2008;37:175–195. doi: 10.1146/annurev.biophys.37.032807.125859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Hansen SB. Lipid agonism: The PIP2 paradigm of ligand-gated ion channels. Biochim Biophys Acta. 2015;1851:620–628. doi: 10.1016/j.bbalip.2015.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Zhang HL, He C, Yan XX, Mirshahi T, Logothetis DE. Activation of inwardly rectifying K+ channels by distinct PtdIns(4,5)P-2 interactions. Nature Cell Biology. 1999;1:183–188. doi: 10.1038/11103. [DOI] [PubMed] [Google Scholar]
- [70].Huang CL, Feng SY, Hilgemann DW. Direct activation of inward rectifier potassium channels by PIP2 and its stabilization by G beta gamma. Nature. 1998;391:803–806. doi: 10.1038/35882. [DOI] [PubMed] [Google Scholar]
- [71].Tao X, Avalos JL, Chen J, MacKinnon R. Crystal structure of the eukaryotic strong inward-rectifier K+ channel Kir2.2 at 3.1 Å resolution. Science. 2009;326:1668–1674. doi: 10.1126/science.1180310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Singh DK, Rosenhouse-Dantsker A, Nichols CG, Enkvetchakul D, Levitan I. Direct regulation of prokaryotic Kir channel by cholesterol. J Biol Chem. 2009;284:30727–30736. doi: 10.1074/jbc.M109.011221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Fürst O, Nichols CG, Lamoureux G, D'Avanzo N. Identification of a cholesterol-binding pocket in inward rectifier K+ (Kir) channels. Biophys J. 2014;107:2786–2796. doi: 10.1016/j.bpj.2014.10.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].D'Avanzo N, Hyrc K, Enkvetchakul D, Covey DF, Nichols CG. Enantioselective protein-sterol interactions mediate regulation of both prokaryotic and eukaryotic inward rectifier K+ channels by cholesterol. PLoS ONE. 2011;6:e19393. doi: 10.1371/journal.pone.0019393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Grouleff J, Irudayam SJ, Skeby KK, Schiøtt B. The influence of cholesterol on membrane protein structure, function, and dynamics studied by molecular dynamics simulations. Biochim Biophys Acta. 2015;1848:1783–1795. doi: 10.1016/j.bbamem.2015.03.029. [DOI] [PubMed] [Google Scholar]
- [76].Thompson AJ, Lester HA, Lummis SCR. The structural basis of function in Cys-loop receptors. Quart Rev Biophys. 2010;43:449–499. doi: 10.1017/S0033583510000168. [DOI] [PubMed] [Google Scholar]
- [77].Levitan I, Barrantes FJ. Cholesterol Regulation of Ion Channels and Receptors. John Wiley & Sons, Inc.; Hoboken, New Jersey: 2012. [Google Scholar]
- [78].Baenziger JE, Morris M-L, Darsaut TE, Ryan SE. Effect of membrane lipid composition on the conformational equilibria of the nicotinic acetylcholine receptor. J Biol Chem. 2000;275:777–784. doi: 10.1074/jbc.275.2.777. [DOI] [PubMed] [Google Scholar]
- [79].daCosta CJB, Dey L, Therien JPD, Baenziger JE. A distinct mechanism for activating uncoupled nicotinic acetylcholine receptors. Nature Chem Biol. 2013;9:701–707. doi: 10.1038/nchembio.1338. [DOI] [PubMed] [Google Scholar]
- [80].Cheng MH, Xu Y, Tang P. Anionic lipid and cholesterol interactions with α4β2 nAChR: insights from MD simulations. J Phys Chem B. 2009;113:6964–6970. doi: 10.1021/jp900714b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Baier CJ, Fantini J, Barrantes FJ. Disclosure of cholesterol recognition motifs in transmembrane domains of the human nicotinic acetylcholine receptor. Sci Reports. 2011;1 doi: 10.1038/srep00069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Hibbs RE, Gouaux E. Principles of activation and permeation in an anion-selective Cys-loop receptor. Nature. 2011;474:54–60. doi: 10.1038/nature10139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Rosenbaum DM, Rasmussen SGF, Kobilka BK. The structure and function of G-protein-coupled receptors. Nature. 2009;459:356–363. doi: 10.1038/nature08144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Oates J, Watts A. Uncovering the intimate relationship between lipids, cholesterol and GPCR activation. Curr Opin Struct Biol. 2011;21:802–807. doi: 10.1016/j.sbi.2011.09.007. [DOI] [PubMed] [Google Scholar]
- [85].Ganguly S, Clayton AHA, Chattopadhyay A. Organization of higher-order oligomers of the serotonin1A receptor explored utilizing homo-FRET in live cells. Biophys J. 100:361–368. doi: 10.1016/j.bpj.2010.12.3692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Harikumar KG, Puri V, Singh RD, Hanada K, Pagano RE, Miller LJ. Differential effects of modification of membrane cholesterol and sphingolipids on the conformation, function, and trafficking of the G protein-coupled cholecystokinin receptor. J Biol Chem. 2005;280:2176–2185. doi: 10.1074/jbc.M410385200. [DOI] [PubMed] [Google Scholar]
- [87].Gimpl G, Reitz J, Brauer S, Trossen C. In: Progress in Brain Research. Inga DN, Rainer L, editors. Elsevier; 2008. pp. 193–204. [DOI] [PubMed] [Google Scholar]
- [88].Cherezov V, Rosenbaum DM, Hanson MA, Rasmussen SG, Thian FS, Kobilka TS, Choi HJ, Kuhn P, Weis WI, Kobilka BK, Stevens RC. High-resolution crystal structure of an engineered human beta2-adrenergic G protein-coupled receptor. Science. 2007;318:1258–65. doi: 10.1126/science.1150577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Horsefield R, Norden K, Fellert M, Backmark A, Tornroth-Horsefield S, Terwisscha van Scheltinga AC, Kvassman J, Kjellbom P, Johanson U, Neutze R. High-resolution x-ray structure of human aquaporin 5. Proc Natl Acad Sci USA. 2008;105:13327–13332. doi: 10.1073/pnas.0801466105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Liu W, Chun E, Thompson AA, Chubukov P, Xu F, Katritch V, Han GW, Roth CB, Heitman LH, IJzerman AP, Cherezov V, et al. Structural basis for allosteric regulation of GPCRs by sodium ions. Science. 2012;337:232–236. doi: 10.1126/science.1219218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Wu H, Wang C, Gregory KJ, Han GW, Cho HP, Xia Y, Niswender CM, Katritch V, Meiler J, Cherezov V, Conn PJ, et al. Structure of a class C GPCR metabotropic glutamate receptor 1 bound to an allosteric modulator. Science. 2014;344:58–64. doi: 10.1126/science.1249489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Neale C, Herce Henry D, Pomès R, García Angel E. García, Can specific protein-lipid interactions stabilize an active state of the beta 2 adrenergic receptor? Biophys J. 2015;109:1652–1662. doi: 10.1016/j.bpj.2015.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Prasanna X, Chattopadhyay A, Sengupta D. Cholesterol modulates the dimer interface of the β2-adrenergic receptor via cholesterol occupancy sites. Biophys J. 2014;106:1290–1300. doi: 10.1016/j.bpj.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Cang X, Du Y, Mao Y, Wang Y, Yang H, Jiang H. Mapping the functional binding sites of cholesterol in β2-adrenergic receptor by long-time molecular dynamics simulations. J Phys Chem B. 2013;117:1085–1094. doi: 10.1021/jp3118192. [DOI] [PubMed] [Google Scholar]
- [95].Cang X, Yang L, Yang J, Luo C, Zheng M, Yu K, Yang H, Jiang H. Cholesterol-β1AR interaction versus cholesterol-β2AR interaction. Proteins: Struc Func Bioinf. 2014;82:760–770. doi: 10.1002/prot.24456. [DOI] [PubMed] [Google Scholar]
- [96].Sengupta D, Chattopadhyay A. Identification of cholesterol binding sites in the serotonin1A receptor. J Phys Chem B. 2012;116:12991–12996. doi: 10.1021/jp309888u. [DOI] [PubMed] [Google Scholar]
- [97].Shan JF, Khelashvili G, Mondal S, Mehler EL, Weinstein H. Ligand-dependent conformations and dynamics of the serotonin 5-HT2A receptor determine its activation and membrane-driven oligomerization properties. PLoS Comp Biol. 2012;8:e1002473. doi: 10.1371/journal.pcbi.1002473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Khelashvili G, Grossfield A, Feller SE, Pitman MC, Weinstein H. Structural and dynamic effects of cholesterol at preferred sites of interaction with rhodopsin identified from microsecond length molecular dynamics simulations. Proteins: Struc Func Bioinf. 2009;76:403–417. doi: 10.1002/prot.22355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Horn J, Kao T-C, Grossfield A. In: G Protein-Coupled Receptors - Modeling and Simulation. Filizola M, editor. Springer; Netherlands: 2014. pp. 75–94. [Google Scholar]
- [100].Hurst DP, Grossfield A, Lynch DL, Feller S, Romo TD, Gawrisch K, Pitman MC, Reggio PH. A lipid pathway for ligand binding is necessary for a cannabinoid G protein-coupled receptor. J Biol Chem. 2010;285:17954–17964. doi: 10.1074/jbc.M109.041590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Prasanna X, Chattopadhyay A, Sengupta D. Cholesterol modulates the dimer interface of the β2-adrenergic receptor via cholesterol occupancy sites. Biophys J. 2014;106:1290–1300. doi: 10.1016/j.bpj.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Hanson MA, Cherezov V, Griffith MT, Roth CB, Jaakola V-P, Chien EYT, Velasquez J, Kuhn P, Stevens RC. A specific cholesterol binding site is established by the 2.8 Å structure of the human β2-adrenergic receptor. Structure. 2008;16:897–905. doi: 10.1016/j.str.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Periole X, Huber T, Marrink SJ, Sakmar TP. G protein-coupled receptors self-assemble in dynamics simulations of model bilayers. J Amer Chem Soc. 2007;129:10126–10132. doi: 10.1021/ja0706246. [DOI] [PubMed] [Google Scholar]
- [104].Periole X, Knepp AM, Sakmar TP, Marrink SJ, Huber T. Structural determinants of the supramolecular organization of G protein-coupled receptors in bilayers. J Amer Chem Soc. 2012;134:10959–10965. doi: 10.1021/ja303286e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Wassenaar TA, Pluhackova K, Moussatova A, Sengupta D, Marrink SJ, Tieleman DP, Böckmann RA. High-throughput simulations of dimer and trimer assembly of membrane proteins. The DAFT approach. J Chem Theory Comput. 2015 doi: 10.1021/ct5010092. 150326134217003. [DOI] [PubMed] [Google Scholar]
- [106].Inagaki S, Ghirlando R, White JF, Gvozdenovic-Jeremic J, Northup JK, Grisshammer R. Modulation of the interaction between neurotensin receptor NTS1 and Gq protein by lipid. J Mol Biol. 2012;417:95–111. doi: 10.1016/j.jmb.2012.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Dawaliby R, Trubbia C, Delporte C, Masureel M, Van Antwerpen P, Kobilka BK, Govaerts C. Allosteric regulation of G protein-coupled receptor activity by phospholipids. Nature Chem Biol. 2015;12:35–39. doi: 10.1038/nchembio.1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Guixa-Gonzalez R, Javanainen M, Gomez-Soler M, Cordobilla B, Carles Domingo J, Sanz F, Pastor M, Ciruela F, Martinez-Seara H, Selent J. Membrane omega-3 fatty acids modulate the oligomerisation kinetics of adenosine A(2A) and dopamine D-2 receptors. Sci Reports. 2016;6 doi: 10.1038/srep19839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Lemmon MA, Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2010;141:1117–1134. doi: 10.1016/j.cell.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Luo B-H, Carman CV, Springer TA. Structural basis of integrin regulation and signaling. Ann Rev Immunol. 2007;25:619–647. doi: 10.1146/annurev.immunol.25.022106.141618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Endres NF, Das R, Smith AW, Arkhipov A, Kovacs E, Huang YJ, Pelton JG, Shan YB, Shaw DE, Wemmer DE, Groves JT, et al. Conformational coupling across the plasma membrane in activation of the EGF receptor. Cell. 2013;152:543–556. doi: 10.1016/j.cell.2012.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Bocharov EV, Lesovoy DM, Goncharuk SA, Goncharuk MV, Hristova K, Arseniev AS. Structure of FGFR3 transmembrane domain dimer: implications for signaling and human pathologies. Structure. 2013;21:2087–2093. doi: 10.1016/j.str.2013.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Manni S, Mineev KS, Usmanova D, Lyukmanova EN, Shulepko MA, Kirpichnikov MP, Winter J, Matkovic M, Deupi X, Arseniev AS, Ballmer-Hofer K. Structural and functional characterization of alternative transmembrane domain conformations in VEGF receptor 2 activation. Structure. 2014;22:1077–1089. doi: 10.1016/j.str.2014.05.010. [DOI] [PubMed] [Google Scholar]
- [114].Bragin P, Mineev K, Bocharova O, Volynsky P, Bocharov E, Arseniev A. HER2 transmembrane domain dimerization coupled with self-association of membrane-embedded cytoplasmic juxtamembrane regions. J Mol Biol. 2015;428:52–61. doi: 10.1016/j.jmb.2015.11.007. [DOI] [PubMed] [Google Scholar]
- [115].Coskun U, Grzybek M, Drechsel D, Simons K. Regulation of human EGF receptor by lipids. Proc Natl Acad Sci USA. 2011;108:9044–9048. doi: 10.1073/pnas.1105666108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Arkhipov A, Shan Y, Das R, Endres NF, Eastwood MP, Wemmer DE, Kuriyan J, Shaw DE. Architecture and membrane interactions of the EGF receptor. Cell. 2013;152:557–569. doi: 10.1016/j.cell.2012.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Kaszuba K, Grzybek M, Orlowski A, Danne R, Rog T, Simons K, Coskun U, Vattulainen I. N-Glycosylation as determinant of epidermal growth factor receptor conformation in membranes. Proc Natl Acad Sci USA. 2015;112:4334–4339. doi: 10.1073/pnas.1503262112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Prakash A, Janosi L, Doxastakis M. Self-association of models of transmembrane domains of ErbB receptors in a lipid bilayer. Biophys J. 2010;99:3657–3665. doi: 10.1016/j.bpj.2010.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Halim KBA, Koldsø H, Sansom MSP. Interactions of the EGFR juxtamembrane domain with PIP-containing lipid bilayers: Insights from multiscale molecular dynamics simulations. Biochim Biophys Acta. 2015;1850:1017–1025. doi: 10.1016/j.bbagen.2014.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Chavent M, Seiradake E, Jones EY, Sansom MSP. The structure of the EphA2 receptor in a membrane: role of lipid interactions. Structure. 2016;24:337–347. doi: 10.1016/j.str.2015.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Matsushita C, Tamagaki H, Miyazawa Y, Aimoto S, Smith SO, Sato T. Transmembrane helix orientation influences membrane binding of the intracellular juxtamembrane domain in Neu receptor peptides. Proc Natl Acad Sci USA. 2013;110:1646–1651. doi: 10.1073/pnas.1215207110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Wang Y, Gao J, Guo X, Tong T, Shi X, Li L, Qi M, Wang Y, Cai M, Jiang J, Xu C, et al. Regulation of EGFR nanocluster formation by ionic protein-lipid interaction. Cell Res. 2014;24:959–976. doi: 10.1038/cr.2014.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Stansfeld PJ, Jefferies EE, Sansom MSP. Multiscale simulations reveal conserved patterns of lipid interactions with aquaporins. Structure. 2013;21:810–819. doi: 10.1016/j.str.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Stansfeld PJ, Goose JE, Caffrey M, Carpenter EP, Parker JL, Newstead N, Sansom MSP. MemProtMD: automated insertion of membrane protein structures into explicit lipid membranes. Structure. 2015;23:1350–1361. doi: 10.1016/j.str.2015.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Lensink MF, Govaerts C, Ruysschaert J-M. Identification of specific lipid-binding sites in integral membrane proteins. J Biol Chem. 2010;285:10519–10526. doi: 10.1074/jbc.M109.068890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Autzen HE, Siuda I, Sonntag Y, Nissen P, Møller JV, Thøgersen L. Regulation of the Ca2+-ATPase by cholesterol: a specific or non-specific effect? Molec Memb Biol. 2015;32:75–87. doi: 10.3109/09687688.2015.1073382. [DOI] [PubMed] [Google Scholar]
- [127].Khelashvili G, Stanley N, Sahai MA, Medina J, LeVine MV, Shi L, De Fabritiis G, Weinstein H. Spontaneous inward opening of the dopamine transporter is triggered by PIP2-regulated dynamics of the N-terminus. ACS Chem Neurosci. 2015;6:1825–1837. doi: 10.1021/acschemneuro.5b00179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Kunji ERS. The role and structure of mitochondrial carriers. FEBS Lett. 2004;564:239–244. doi: 10.1016/S0014-5793(04)00242-X. [DOI] [PubMed] [Google Scholar]
- [129].Klingenberg M. Cardiolipin and mitochondrial carriers. Biochim Biophys Acta. 2009;1788:2048–2058. doi: 10.1016/j.bbamem.2009.06.007. [DOI] [PubMed] [Google Scholar]
- [130].Nury H, Dahout-Gonzalez C, Trezeguet V, Lauquin CJM, Brandolin G, Pebay-Peyroula E. Structural basis for lipid-mediated interactions between mitochondrial ADP/ATP carrier monomers. FEBS Lett. 2005;579:6031–6036. doi: 10.1016/j.febslet.2005.09.061. [DOI] [PubMed] [Google Scholar]
- [131].Ruprecht JJ, Hellawell AM, Harding M, Crichton PG, McCoy AJ, Kunji ERS. Structures of yeast mitochondrial ADP/ATP carriers support a domain-based alternating-access transport mechanism. Proc Natl Acad Sci USA. 2014;111:E426–E434. doi: 10.1073/pnas.1320692111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Poyry S, Cramariuc O, Postila PA, Kaszuba K, Sarewicz M, Osyczka A, Vattulainen I, Rog T. Atomistic simulations indicate cardiolipin to have an integral role in the structure of the cytochrome bc(1) complex. Biochim Biophys Acta. 2013;1827:769–778. doi: 10.1016/j.bbabio.2013.03.005. [DOI] [PubMed] [Google Scholar]
- [133].Arnarez C, Marrink SJ, Periole X. Identification of cardiolipin binding sites on cytochrome c oxidase at the entrance of proton channels. Sci Rep. 2013;3:1263. doi: 10.1038/srep01263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Baker LA, Watt IN, Runswick MJ, Walker JE, Rubinstein JL. Arrangement of subunits in intact mammalian mitochondrial ATP synthase determined by cryo-EM. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:11675–11680. doi: 10.1073/pnas.1204935109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Davies KM, Anselmi C, Wittig I, Faraldo-Gomez JD, Kuehlbrandt W. Structure of the yeast F1Fo-ATP synthase dimer and its role in shaping the mitochondrial cristae. Proc Natl Acad Sci USA. 2012;109:13602–13607. doi: 10.1073/pnas.1204593109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Jiko C, Davies KM, Shinzawa-Itoh K, Tani K, Maeda S, Mills DJ, Tsukihara T, Fujiyoshi Y, Kuhlbrandt W, Gerle C. Bovine F1Fo ATP synthase monomers bend the lipid bilayer in 2D membrane crystals. eLife. 2015;4:e06119–e06119. doi: 10.7554/eLife.06119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Ingolfsson HI, Arnarez C, Periole X, Marrink SJ. Computational ‘microscopy’ of cellular membranes. J Cell Sci. 2016 doi: 10.1242/jcs.176040. (online) [DOI] [PubMed] [Google Scholar]
- [138].Gournas C, Papageorgiou I, Diallinas G. The nucleobase-ascorbate transporter (NAT) family: genomics, evolution, structure-function relationships and physiological role. Molec BioSys. 2008;4:404–416. doi: 10.1039/b719777b. [DOI] [PubMed] [Google Scholar]
- [139].Kosti V, Lambrinidis G, Myrianthopoulos V, Diallinas G, Mikros E. Identification of the substrate recognition and transport pathway in a eukaryotic member of the nucleobase-ascorbate transporter (NAT) family. PLoS ONE. 2012;7:e41939. doi: 10.1371/journal.pone.0041939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Drachmann ND, Olesen C, Møller JV, Guo Z, Nissen P, Bublitz M. Comparing crystal structures of Ca2+-ATPase in the presence of different lipids. FEBS J. 2014;281:4249–4262. doi: 10.1111/febs.12957. [DOI] [PubMed] [Google Scholar]
- [141].Jensen AML, Sørensen TLM, Olesen C, Møller JV, Nissen P. Modulatory and catalytic modes of ATP binding by the calcium pump. EMBO J. 2006;25:2305–2314. doi: 10.1038/sj.emboj.7601135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Chen L, Zhang Q, Qiu Y, Li Z, Chen Z, Jiang H, Li Y, Yang H. Migration of PIP2 lipids on voltage-gated potassium channel surface influences channel deactivation. Scientific Reports. 2015;5:15079. doi: 10.1038/srep15079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Aponte-Santamaria C, Briones R, Schenk AD, Walz T, de Groot BL. Molecular driving forces defining lipid positions around aquaporin-0. Proc Natl Acad Sci USA. 2012;109:9887–9892. doi: 10.1073/pnas.1121054109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Weiser BP, Salari R, Eckenhoff RG, Brannigan G. Computational investigation of cholesterol binding sites on mitochondrial VDAC. J Phys Chem B. 2014;118:9852–9860. doi: 10.1021/jp504516a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145].D'Avanzo N, Lee S-J, Cheng WWL, Nichols CG. Energetics and location of phosphoinositide binding in human Kir2.1 channels. J Biol Chem. 2013;288:16726–16737. doi: 10.1074/jbc.M113.452540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Rassam P, Copeland NA, Birkholz O, Toth C, Chavent M, Duncan AL, Cross SJ, Housden NG, Kaminska R, Seger U, Quinn DM, et al. Supramolecular assemblies underpin turnover of outer membrane proteins in bacteria. Nature. 2015;523:333–336. doi: 10.1038/nature14461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].Reddy T, Shorthouse D, Parton DL, Jefferys E, Fowler PW, Chavent M, Baaden M, Sansom MS. Nothing to sneeze at: a dynamic and integrative computational model of an influenza A virion. Structure. 2015;23:584–97. doi: 10.1016/j.str.2014.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Marrink SJ, Risselada J, Yefimov S, Tieleman DP, de Vries AH. The MARTINI force field: coarse grained model for biomolecular simulations. J Phys Chem B. 2007;111:7812–7824. doi: 10.1021/jp071097f. [DOI] [PubMed] [Google Scholar]
- [149].de Jong DH, Singh G, Bennett WFD, Arnarez C, Wassenaar TA, Schafer LV, Periole X, Tieleman DP, Marrink SJ. Improved parameters for the Martini coarse-grained protein force field. J Chem Theor Comput. 2013;9:687–697. doi: 10.1021/ct300646g. [DOI] [PubMed] [Google Scholar]
- [150].Klauda JB, Venable RM, Freites JA, O’Connor JW, Tobias DJ, Mondragon-Ramirez C, Vorobyov I, MacKerell AD, Pastor RW. Update of the CHARMM all-atom additive force field for lipids: validation on six lipid types. J Phys Chem B. 2010;114:7830–7843. doi: 10.1021/jp101759q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [151].Dickson CJ, Madej BD, Skjevik AA, Betz RM, Teigen K, Gould IR, Walker RC. Lipid14: the Amber lipid force field. J Chem Theor Comput. 2014;10:865–879. doi: 10.1021/ct4010307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].Tjornhammar R, Edholm O. Reparameterized united atom model for molecular dynamics simulations of gel and fluid phosphatidylcholine bilayers. J Chem Theor Comput. 2014;10:5706–5715. doi: 10.1021/ct500589z. [DOI] [PubMed] [Google Scholar]
- [153].Kulig W, Pasenkiewicz-Gierula M, Rog T. Topologies, structures and parameter files for lipid simulations in GROMACS with the OPLS-aa force field: DPPC, POPC, DOPC, PEPC, and cholesterol. Data in brief. 2015;5:333–336. doi: 10.1016/j.dib.2015.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Madej BD, Gould IR, Walker RC. A parameterization of cholesterol for mixed lipid bilayer simulation within the Amber Lipid14 force field. J Phys Chem B. 2015;119:12424–12435. doi: 10.1021/acs.jpcb.5b04924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Ogata K, Nakamura S. Improvement of parameters of the AMBER potential force field for phospholipids for description of thermal phase transitions. J Phys Chem B. 2015;119:9726–9739. doi: 10.1021/acs.jpcb.5b01656. [DOI] [PubMed] [Google Scholar]
- [156].Papadimitriou NI, Kainourgiakis ME, Karozis SN, Charalambopoulou GC. Studying the structure of single-component ceramide bilayers with molecular dynamics simulations using different force fields. Molec Simul. 2015;41:1122–1136. [Google Scholar]
- [157].Khakbaz P, Klauda JB. Probing the importance of lipid diversity in cell membranes via molecular simulation. Chem Phys Lipids. 2015;192:12–22. doi: 10.1016/j.chemphyslip.2015.08.003. [DOI] [PubMed] [Google Scholar]
- [158].Li Z, Venable RM, Rogers LA, Murray D, Pastor RW. Molecular dynamics simulations of PIP2 and PIP3 in lipid bilayers: determination of ring orientation, and the effects of surface roughness on a Poisson-Boltzmann description. Biophys J. 2009;97:155–163. doi: 10.1016/j.bpj.2009.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [159].Rosen SAJ, Gaffney PRJ, Gould IR. A theoretical investigation of inositol 1,3,4,5-tetrakisphosphate. Phys Chem Chem Phys. 2011;13:1070–1081. doi: 10.1039/c0cp00956c. [DOI] [PubMed] [Google Scholar]
- [160].Wu EL, Engstrom O, Jo S, Stuhlsatz D, Yeom MS, Klauda JB, Widmalm G, Im W. Molecular dynamics and NMR spectroscopy studies of E. coli lipopolysaccharide structure and dynamics. Biophys J. 2013;105:1444–1455. doi: 10.1016/j.bpj.2013.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [161].Hall BA, Halim KA, Buyan A, Emmanouil B, Sansom MSP. Sidekick for membrane simulations: automated ensemble molecular dynamics simulations of transmembrane helices. J Chem Theor Comp. 2014;10:2165–2175. doi: 10.1021/ct500003g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [162].Saliba A-E, Vonkova I, Gavin A-C. The systematic analysis of protein-lipid interactions comes of age. Nat Rev Mol Cell Biol. 2015;16:753–761. doi: 10.1038/nrm4080. [DOI] [PubMed] [Google Scholar]