Abstract
RNase H1-dependent antisense oligonucleotides (ASOs) are active in reducing levels of both cytoplasmic mRNAs and nuclear retained RNAs. Although ASO activity in the nucleus has been well demonstrated, the cytoplasmic activity of ASOs is less clear. Using kinetic and subcellular fractionation studies, we evaluated ASO activity in the cytoplasm. Upon transfection, ASOs targeting exonic regions rapidly reduced cytoplasmically enriched mRNAs, whereas an intron-targeting ASO that only degrades the nuclear pre-mRNA reduced mRNA levels at a slower rate, similar to normal mRNA decay. Importantly, some exon-targeting ASOs can rapidly and vigorously reduce mRNA levels without decreasing pre-mRNA levels, suggesting that pre-existing cytoplasmic mRNAs can be cleaved by RNase H1-ASO treatment. In addition, we expressed a cytoplasm-localized mutant 7SL RNA that contains a partial U16 small nucleolar RNA (snoRNA) sequence. Treatment with an ASO simultaneously reduced both the nuclear U16 snoRNA and the cytoplasmic 7SL mutant RNA as early as 30 min after transfection in an RNase H1-dependent manner. Both the 5′ and 3′ cleavage products of the 7SL mutant RNA were accumulated in the cytoplasm. Together, these results demonstrate that RNase H1-dependent ASOs are robustly active in both the cytoplasm and nucleus.
Keywords: antisense, oligonucleotide, cytoplasm, nucleus, RNase H1, RNA
Liang et al. show that antisense oligonucleotide (ASO)-directed RNase H1 cleavage is robustly active in both the nucleus and cytoplasm. They found that ASOs can trigger rapid cleavage of both cytoplasmic and nuclear RNAs mediated by RNase H1, which is also present in the cytosol.
Introduction
RNase H1-dependent antisense oligonucleotides (ASOs) are commonly used as research tools and as therapeutics to downregulate gene expression.1, 2, 3 Second-generation ASOs are designed in a 5-10-5 gapmer configuration that contains 10 deoxynucleotides in the central portion flanked at both ends with five ribonucleotides modified with 2′-O-methoxyethol (MOE). The nucleotides are linked with phosphorothioate (PS) backbones. Without formulation, PS-ASOs can enter cells through endocytic pathways and enrich in the endosomes and lysosomes in the cytoplasm.4, 5, 6, 7 Once they are released from the endocytic organelles, ASOs distribute to both the nucleus and cytoplasm. Upon transfection, PS-ASOs are quickly enriched in the nucleus, following cytoplasmic release from the liposomes.8, 9, 10
ASOs utilize cellular RNase H1 to cleave target RNAs, although another RNase H enzyme (RNase H2) is expressed at higher levels than RNase H1 in mammalian cells.11, 12 In contrast with the nuclear localization of RNase H2, RNase H1 localizes in both the nucleus and cytoplasm.10, 13, 14 In the cytoplasm, RNase H1 is enriched in the mitochondria,13, 15 where it is involved in mitochondrial DNA replication and RNA processing, by removing the RNA/DNA hybrids during replication and transcription.16, 17, 18, 19
Consistent with the nuclear localization of RNase H1, it has been well demonstrated that ASOs can direct RNase H1 cleavage of target RNAs in this compartment.3, 20 Indeed, ASOs are effective in reducing the levels of targeted small nuclear RNAs (snRNAs), small cajal body RNAs (scaRNAs), small nucleolar RNAs (snoRNAs), and nucleoplasmic long non-coding RNAs (lncRNAs) and pre-mRNAs.10, 21, 22, 23, 24, 25 RNA reduction through the RNA-induced silencing complex (RISC) pathway is known to be highly active in the cytoplasm26 and several recent studies also demonstrated nuclear small interfering RNA (siRNA) activity,27, 28, 29, 30 although this nuclear activity was not observed in other studies.21, 31
Although both RNase H1 and ASOs are present in the cytoplasm and ASOs can reduce cytoplasmic mRNA levels, ASO-directed RNase H activity in this subcellular compartment is less well understood. Early studies performed in Xenopus oocytes suggested that RNase H-dependent ASOs are active in the nucleus and that cytoplasmic activity may only represent 5% of total ASO activity.32 Another study showed that co-transfection of synthesized mRNA and ASOs yielded no mRNA reduction; thus, the authors proposed the lack of cytoplasmic ASO activity, assuming that the transfected mRNA localized in the cytoplasm.33 A recent report showed that a higher confirmation rate of RNA reduction was observed for off-targeted RNAs by an ASO when targeting intronic regions rather than exonic regions.34 However, due to the small sample size (13 exon-targeted and 12 intron-targeted RNAs) and the quite different base-pairing potential of the ASO to the off-target sites (more mismatches for exon-targeted sites than intron-targeted sites), it is difficult to conclude that intron-targeting ASOs are more active than exon-targeting ASOs, particularly as ASO activity targeting pre-mRNAs can be largely affected by splicing.1, 35, 36
On the other hand, other reports show that ASOs might be active in the cytoplasm.11 For example, the degradation of cleaved mRNA fragments by RNase H1-dependent ASOs involved XRN1, a cytoplasm-localized exonuclease, whereas the degradation of cleavage fragments of nuclear RNAs largely depended on XRN2, a nuclear localized exonuclease.37, 38 In addition, cytoplasmic activity was also proposed based on observations that ASOs, upon gymnotic delivery (without transfection), caused an mRNA reduction without obvious nuclear localization of ASOs, as examined using confocal microscopy.39 Furthermore, a previous study showed ASOs can reduce the level of a spliced reporter mRNA by 50% within 3–4 hr after transfection, even though the mRNA half-life is longer than 6 hr.24 This observation suggests that RNase H1 cleavage is not solely a nuclear event, and the reduction of cytoplasmic mature mRNA is also possible. Moreover, a recent study compared the effects of RNA localization on siRNA and ASO activity and showed that the ASO-mediated reduction of cytoplasmic RNAs may be more effective than the RNAi-mediated reduction of nuclear lncRNAs, suggesting higher cytoplasmic activity of ASOs than previously appreciated.28 However, in that study, the levels of target RNAs were determined 24 hr after ASO or siRNA transfection. Although the ASO-mediated reduction of cytoplasmic lncRNAs supports the possibility of cytoplasmic RNase H1 cleavage, the results from a study that does not include a time course could not exclude the possibility that ASO-mediated cleavage occurred in the nucleus, leading to a gradual reduction of cytoplasmic RNAs through RNA decay pathways, since the mean half-life of lncRNAs in mammalian cells was reported at 4.8 hr.40 Despite these observations, it is unclear regarding the robustness of cytoplasmic ASO activity and therefore the relative contributions of nuclear and cytoplasmic cleavage to target RNA reduction.
In this report, we provide evidence that RNase H1-dependent ASOs are robustly active in both the nucleus and cytoplasm. Some ASOs can effectively reduce mature mRNA levels enriched in the cytoplasm without decreasing nuclear pre-mRNA levels. In addition, the ASO-mediated reduction of mature cytoplasmic mRNAs occurred within 0.5–2 hr after ASO treatment, which is much shorter than needed for normal mRNA decay; this is expected to happen if ASOs are only active in the nucleus. In addition, by using an engineered cytoplasmic RNA, we showed that cleavage of cytoplasmic and nuclear RNAs can have a similar, very early onset, and the reduction of the cytoplasmic RNA was accompanied by the accumulation of cleaved fragments in the cytoplasm.
Results
RNase H1-Dependent ASOs Reduced Cytoplasmic mRNAs Much Faster than Normal mRNA Decay
In mammalian cells, most nuclear-encoded mRNAs are transcribed in the form of pre-mRNAs, which contain multiple introns that are removed through splicing, as well as relatively short exons that are joined together and retained in the mature mRNAs. After nuclear maturation, mRNAs are quickly (within a few minutes) exported to the cytoplasm,41 where they are enriched and translated. If transcription or normal mRNA biosynthesis is inhibited, the pre-existing cytoplasmic mRNAs are gradually reduced through normal decay pathways.42 The balance of biogenesis and degradation of mRNAs determines steady-state mRNA levels. Thus, we reasoned that if ASOs are only active in the nucleus (where they reduce pre-mRNA and nuclear mRNAs, which represent a small portion of total mature mRNAs), the reduction of total mRNA levels should be at the normal rate of cytoplasmic decay for that mRNA, since the majority of mRNAs localize in the cytoplasm.43 For example, in liver cells and pancreatic beta cells, the mean cytoplasm/nucleus ratios of mRNAs were 6.5 ± 1.3 and 3.8 ± 0.05, respectively, although some mRNAs may have a higher level in the nucleus.43 On the other hand, if RNase H1-dependent ASOs could directly induce cytoplasmic cleavage of the pre-existing mature mRNAs, the rate of mRNA reduction should be faster than normal mRNA decay, since degradation by RNase H1 could rapidly decrease the levels of targeted RNAs, as shown previously.24
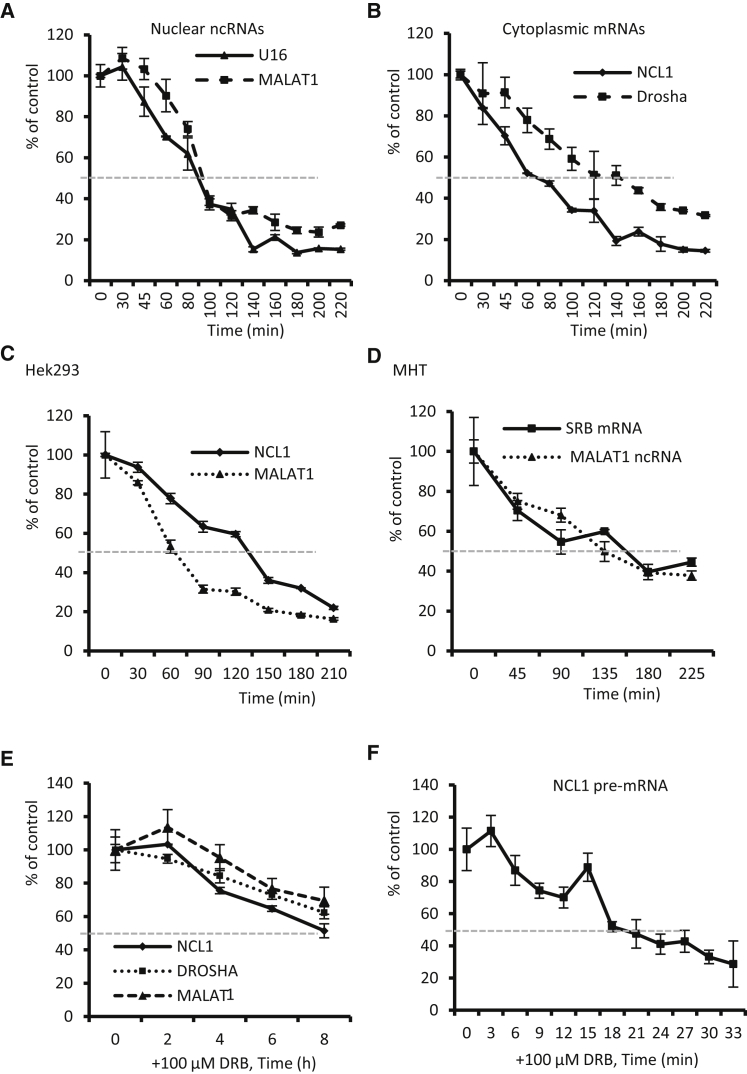
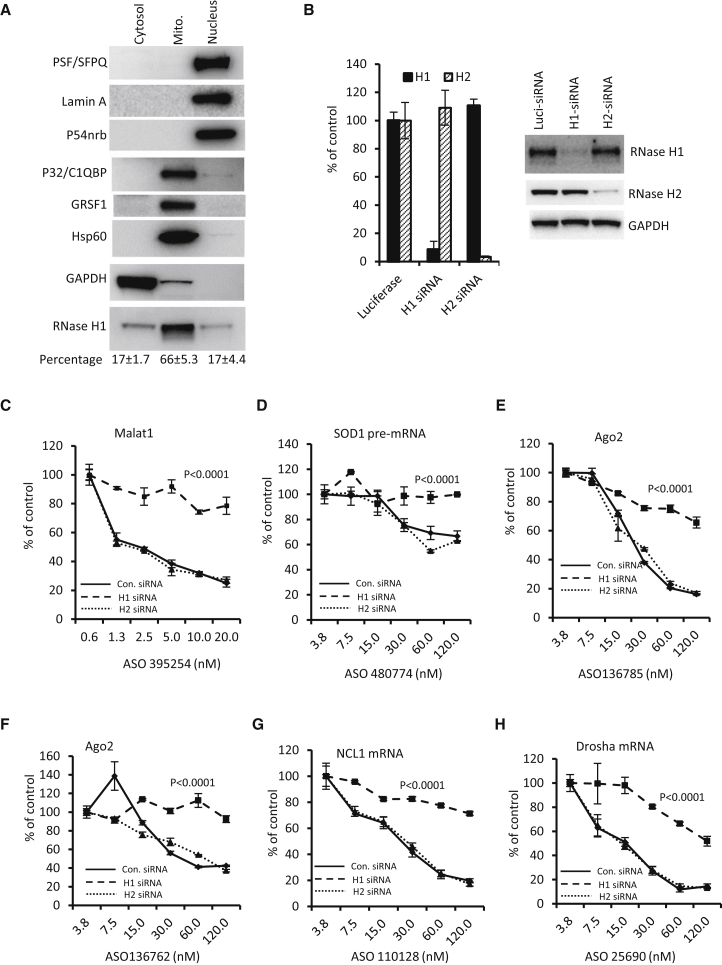
To assess the rates of mRNA loss in the presence or absence of ASOs, we transfected HeLa cells with ASOs targeting nuclear RNAs (U16 snoRNA and MALAT1 lncRNA) or the exonic regions of cytoplasmic mRNAs (NCL1 and Drosha). ASO treatment was performed for different times, and total RNA was prepared and subjected to qRT-PCR analyses to determine the levels of the targeted RNAs. As expected, the nuclear U16 and MALAT1 RNAs were quickly reduced by 50% within 110 and 130 min, respectively (Figure 1A), indicating robust nuclear ASO activity, which is consistent with previous results.21 Importantly, cytoplasmic NCL1 and Drosha mRNAs were also rapidly reduced by 50% within 75 and 180 min, respectively (Figure 1B). Similarly, a rapid reduction of both NCL1 mRNA and nuclear MALAT1 lncRNA by ASO treatment was found in HEK293 cells (Figure 1C). In mouse hepatocellular SV40 large T-antigen carcinoma cells (MHTs), transfection of ASOs targeting either SRB mRNA or MALAT1 lncRNA caused a similarly rapid degradation of these RNAs, with a 50% reduction within 180 min (Figure 1D). Together, these results showed that ASOs can rapidly reduce the targeted mRNAs, with an onset of action and rate of reduction similar to nuclear RNAs, in different cell types.
Figure 1.
ASOs Can Degrade mRNAs at a Rate Faster Than Normal mRNA Decay
(A) qRT-PCR for U16 and MALAT1 RNA levels in HeLa cells transfected with either 50 nM ASO462026 or 5 nM ASO395254, respectively, for different times. (B) qRT-PCR for NCL1 and Drosha mRNA levels in HeLa cells transfected with 50 nM ASO110074 (for NCL1) or 25690 (for Drosha), respectively, for different times. (C) qRT-PCR for NCL1 and MALAT1 RNA levels in HEK293 cells transfected with 50 nM ASO110074 or 5 nM ASO395254, respectively, for different times. (D) qRT-PCR for SRB and MALAT1 RNA levels in mouse MHT cells transfected with 30 nM ASO 205382 or 5 nM ASO 399479, respectively, for different times. (E) qRT-PCR for different RNA levels in HeLa cells treated with 100 μg/mL DRB for different times. (F) qRT-PCR for NCL1 pre-mRNA levels in HeLa cells treated with 100 μg/mL DRB for different times. The gray dashed lines indicate a 50% reduction of the RNAs. Error bars are SDs from three independent experiments.
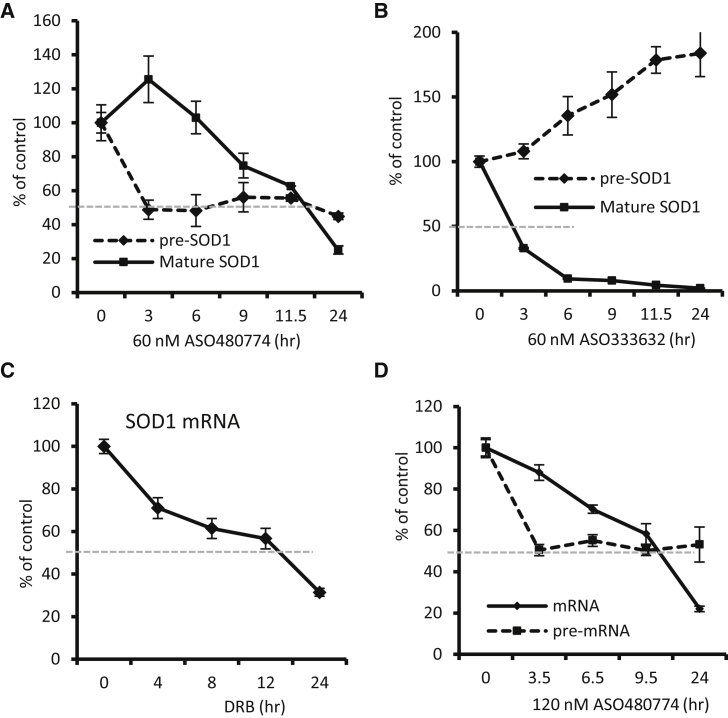
Next, we determined the half-lives of the NCL1 mRNA, Drosha mRNA, and MALAT1 lncRNA in HeLa cells. Cells were treated with a transcription inhibitor (5,6-dichloro-1-beta-D-ribofuranosylbenzimidazole [DRB]) for different times. qRT-PCR results showed that the half-lives of these three RNAs are all longer than 8 hr (Figure 1E), which is consistent with previous reports.44, 45 The effectiveness of DRB treatment was confirmed by the rapid reduction of NCL1 pre-mRNA (Figure 1F), which exhibited a half-life of less than 20 min. Together, these results indicate that RNase H1-dependent ASOs can cause mRNA reduction at a rate much faster than normal mRNA decay, consistent with previous observations,24 suggesting that ASOs may degrade pre-existing mRNAs that are localized mainly in the cytoplasm.
ASO Treatment Can Reduce mRNA Levels without Decreasing Pre-mRNA Levels
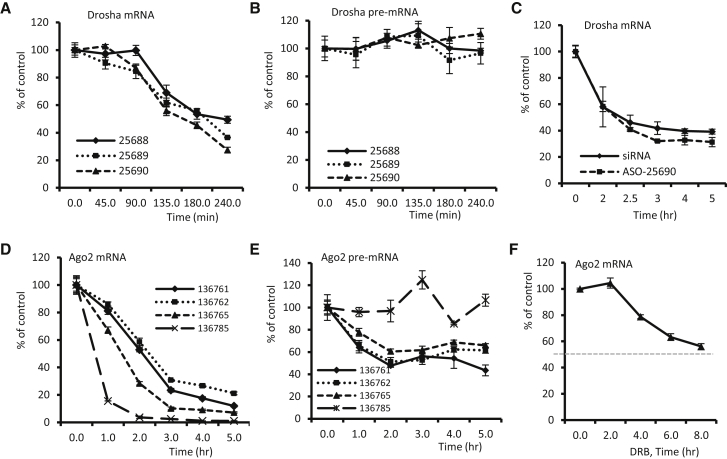
Although the tested mRNAs can be reduced by ASOs more rapidly than the normal decay rate, it is possible (although very unlikely) that ASOs could degrade the target pre-mRNA only in the nucleus, whereas the cytoplasmic mRNA reduction may be caused by other unknown effects. To exclude this possibility, we reasoned that if ASOs are only active in the nucleus, we would expect the reduction of pre-mRNA to occur more rapidly than the reduction of mature mRNAs. To this end, three different ASOs targeting exonic regions of Drosha mRNA were transfected into HeLa cells for different times. qRT-PCR results showed that all three ASOs reduced Drosha mRNA by 50% within 3 hr (Figure 2A), while the pre-mRNA level was not substantially affected by these ASOs even after 4-hr treatment (Figure 2B). These results strongly suggest that mRNAs can be reduced in the cytoplasm, without the need for reduction of pre-mRNAs in the nucleus. We observed a similar onset of target mRNA reduction after the transfection of a Drosha-specific ASO (25690) or an siRNA (Figure 2C), which is also consistent with the view that mRNA degradation occurs in the cytoplasm. siRNA is known to be active in the cytoplasm.26
Figure 2.
Some ASOs Can Rapidly Reduce Mature mRNAs without Reducing Pre-mRNA Levels
(A) qRT-PCR for Drosha mRNA levels in HeLa cells transfected with different ASOs at 40 nM for different times. (B) qRT-PCR for Drosha pre-mRNA levels in the same samples as in (A). (C) qRT-PCR for Drosha mRNA levels in HeLa cells transfected with either 40 nM ASO25690 or 3 nM Drosha siRNA for different times. (D) qRT-PCR for Ago2 mRNA levels in HeLa cells transfected with different ASOs at 50 nM for different times. (E) qRT-PCR for pre-mRNA levels of Ago2 in the same samples as in (D). (F) qRT-PCR for Ago2 mRNA levels in cells treated with 100 μg/mL DRB for different times. Error bars are SDs from three independent experiments.
However, we note that some ASOs can indeed reduce nuclear pre-mRNAs (as expected), which is likely dependent on the accessibility of the target sequence in pre-mRNA. Four ASOs targeting exonic regions of Ago2 mRNA could reduce the mRNA level by more than 50% within 1–3 hr (Figure 2D), and the pre-mRNA level of Ago2 were also substantially reduced by three of the four ASOs within 2 hr (Figure 2E). In this case, reduction of the pre-mRNA may contribute to, but is not required for, the reduction of Ago2 mRNA, since ASO136785 caused the greatest reduction of Ago2 mRNA but did not decrease the pre-mRNA level. It is possible that the faster reduction of Ago2 mRNA by ASO136785 than by other ASOs may result from a higher potency of this ASO, as can be seen from the greatest mRNA reduction by this ASO across all time points.
For the three ASOs that also reduced pre-mRNA, Ago2 mRNA was already decreased by more than 50% at 2–3 hr after transfection. This suggests that these ASOs may act on both pre-mRNA and mature mRNAs, since the half-life of Ago2 mRNA appeared to be more than 8 hr (Figure 2F). Assuming that the mRNA half-life is 8 hr and that all pre-mRNA molecules were reduced by ASO treatment at 2 hr after transfection, pre-mRNA degradation should lead to a less than 12.5% reduction of the mature mRNA due to normal mRNA decay. Compared with the ∼40% mRNA reduction by ASO136761 and ASO136762, as well as the ∼70% mRNA reduction by ASO136765 at this time point, pre-mRNA degradation should contribute to less than 18%–30% of the total ASO activity in reducing the mature mRNA. Thus, the major contribution of mRNA reduction (>70%) should occur at the mature mRNA level. This is consistent with a previous report.24 Considering that the cytoplasmic/nuclear ratio of Ago2 mRNA was ∼2.7 (see below), the cytoplasmic reduction of mRNA should account for at least 50% of total activity (70% × 2.7/3.7). Together, such a nuclear degradation is not obligatory for some exonic targeting ASOs that can degrade mature mRNAs, although they have the potential to base pair with and induce cleavage of pre-mRNA in the nucleus. It is likely that both cytoplasmic and nuclear RNA degradation occurs in some cases (e.g., for Ago2) but that degradation may occur primarily in the cytoplasm for some other mRNA species (Drosha) or ASOs (136785).
ASO Accessibility to the Same Target Sequence Present in Pre-mRNA and Processed RNA Can Be Different and Affects ASO Activity
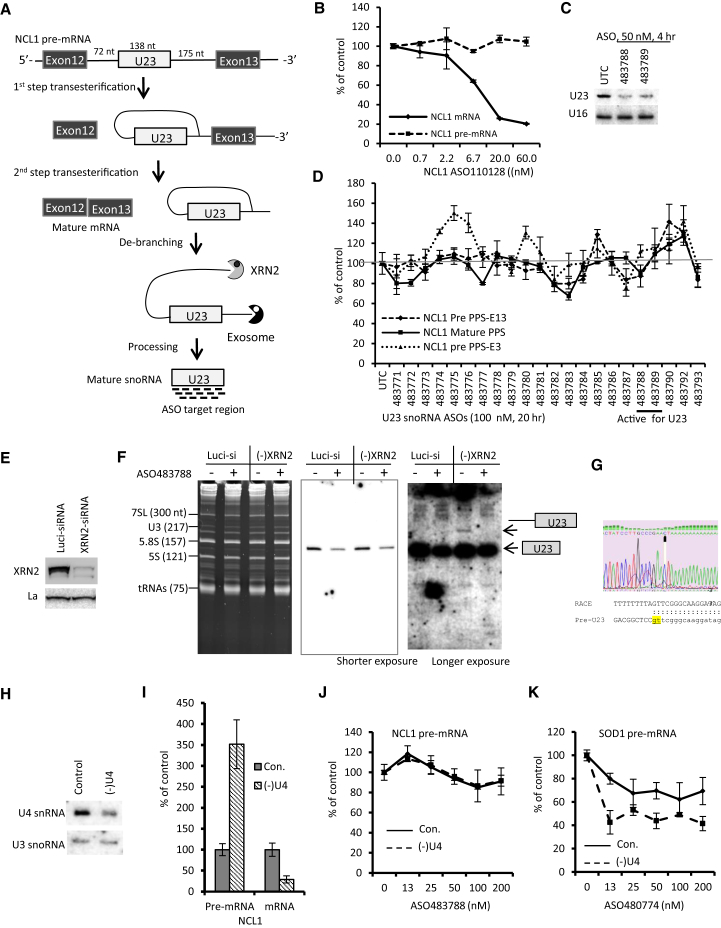
ASOs are shown to reduce pre-mRNA levels, even when targeting intron-exon junctions,35 demonstrating nuclear ASO activity. However, as described above, some ASOs reduce levels of mature mRNAs and not the corresponding pre-mRNAs. Similar observations were made in our previous studies,21 which showed that several tested ASOs that reduced intron-encoded snoRNAs did not decrease their host pre-mRNA levels, indicating the complexity of ASO action. These observations suggest that either some pre-mRNAs are difficult to reduce (e.g., due to a short half-life compared with mature mRNAs or processed snoRNAs) or the pre-mRNA may have different structure/protein binding than that of the processed mature RNAs, leading to reduced accessibility of pre-mRNA to the ASOs. To further evaluate these possibilities, we took advantage of our previous findings that an ASO targeting the intronic U23 snoRNA could reduce the snoRNA, but not the pre-mRNA, of the host gene.21 U23 snoRNA is encoded in an intron of NCL1 pre-mRNA, and the intronic snoRNAs are usually processed after splicing by the 5′→3′ and 3′→5′ exonucleases XRN2 and exosomes, respectively, to generate mature snoRNAs46, 47, 48 (Figure 3A).
Figure 3.
Pre-mRNA and Processed Mature RNA Can Have Different Accessibility to ASOs
(A) Schematic representation of partial NCL1 pre-mRNA and processing of the intronic snoRNA U23. U23 snoRNA and its flanking introns are marked with white boxes or lines, respectively. (B) qRT-PCR for mature and pre-mRNA levels of NCL1 in HeLa cells transfected for 4 hr with different concentrations of ASO110128. (C) Northern hybridization for U23 snoRNA in HeLa cells transfected for 4 hr with two different ASOs. U16 snoRNA was detected and served as a loading control. (D) qRT-PCR for mature or pre-mRNA levels of NCL1 in HeLa cells transfected with 100 nM ASOs for 20 hr. Two different primer probe sets were used to detect the pre-mRNA of NCL1. The two ASOs used, as in (C), are underlined. (E) Western blot analysis of XRN2 in HeLa cells transfected with a control luciferase siRNA or an siRNA specific to XRN2 mRNA for 72 hr. La protein was detected and served as the loading control. (F) Northern hybridization for U23 snoRNA in luciferase siRNA- or XRN2 siRNA-treated HeLa cells subsequently transfected with or without 50 nM ASO483788 for 4 hr. Ethidium bromide staining of the PAGE gel is shown in the left panel, and the sizes of known RNA bands are given. The northern blot was hybridized with probe XL099, and images with shorter or longer (middle and right panels, respectively) exposure time are shown. The identities of the hybridized bands are indicated. (G) Sequence of the 5′-RACE product for the U23 intermediate. The 5′ end of the product is highlighted. (H) Northern hybridization of U4 snRNA in cells treated for 24 hr with control ASO (129700) and U4-specific ASO (479333) at 30 nM. U3 snoRNA was detected and served as a loading control. (I) qRT-PCR for mature and pre-mRNA levels of NCL1 in control cells (con) or U4 reduced cells. (J) qRT-PCR for NCL1 pre-mRNA levels in different cells transfected with the U23 snoRNA ASO at different doses for 4 hr. (K) qRT-PCR for SOD1 pre-mRNA levels in different cells transfected with the SOD1 intron-targeting ASO at different doses for 4 hr. Error bars in different panels are SDs from three experiments.
NCL1 mRNA was able to be reduced using an ASO (110128) targeting an exonic region (Figure 3B), without a reduction of the NCL1 pre-mRNA level. In addition, two ASOs targeting the U23 snoRNA region reduced the mature snoRNA level when transfected at 50 nM for 4 hr (Figure 3C), suggesting that the ASO binding sites in the processed mature mRNA and U23 snoRNA are accessible. However, these two and an additional 20 ASOs targeting different regions of U23 snoRNA (and thus the pre-mRNA of NCL1) did not reduce the pre-mRNA level, even at 20 hr after transfection of ASOs at a 100-nM concentration (Figure 3D). Pre-mRNA levels were determined by qRT-PCR using two different primer probe sets that recognize 5′ (PPS-E3, Intron2/Exon3 junction) or 3′ (PPS-E13, Intron12/Exon13 junction) regions of the pre-mRNA. In addition, the NCL1 mature mRNA level was also not substantially reduced, further confirming that these U23-targeting ASOs did not degrade the pre-mRNA, consistent with our previous observation.21 Together, these results suggest that the processed mature RNAs and the pre-mRNA have distinct characteristics with regard to the effects of ASOs.
Compared with mature RNAs, pre-mRNAs are usually short lived, contain more sequence space, and bind more proteins that are required for RNA processing (e.g., splicing or miRNA and snoRNA processing).49, 50, 51 Thus, pre-mRNAs are likely to have structures that are different from mature RNAs. These differences may render the target sites in pre-mRNA less accessible than the processed mature RNAs. To further evaluate these possibilities and to determine whether short-lived precursors are accessible to ASOs, XRN2 protein was reduced by siRNA treatment (Figure 3E), which should lead to the accumulation of processing intermediate(s) of U23 snoRNA, as XRN2 is required for the processing of the 5′ flanking sequence of intronic snoRNAs.46, 47, 48 Control or XRN2 reduced cells were transfected with a U23 ASO (483788) for 4 hr, and an ∼60% reduction of the U23 snoRNA was detected by northern hybridization in this experiment (Figure 3F, middle panel). We note that the greater reduction by this ASO was achieved at a later time.21 As expected, a processing intermediate of U23 snoRNA was detected from cells depleted of XRN2 (Figure 3F, right panel), which was confirmed by 5′ rapid amplification of cDNA ends (RACE) and sequencing to contain the 5′ intron sequence stopped at a cryptic 5′ splice site (Figure 3G). Interestingly, this intermediate disappeared upon ASO treatment, indicating that this intermediate, which is normally short lived, is degradable by the ASO.
To further determine whether increasing pre-mRNA stability can lead to the reduction of NCL1 pre-mRNA by snoRNA ASOs, we reduced U4 snRNA by ASO treatment (Figure 3H) to inhibit pre-mRNA splicing, resulting in stabilization of NCL1 pre-mRNA (Figure 3I). Next, the U23 ASO (483788) was transfected for 4 hr. qRT-PCR results showed that no significant reduction of NCL1 pre-mRNA was detected in both control cells and in U4 reduced cells (Figure 3J). As a positive control, an ASO (480774) targeting an intronic region of SOD1 pre-mRNA, which is able to reduce SOD1 pre-mRNA (Tim Vickers, personal communication), exhibited increased activity upon splicing inhibition (Figure 3K), consistent with previous observations.24 These results suggest that the failure of U23 snoRNA ASOs to reduce NCL1 pre-mRNA is likely due to low accessibility of the target sequence in the pre-mRNA and is less likely a result of the short half-life of the transcript.
ASOs Can Reduce Cytoplasmic mRNA Levels
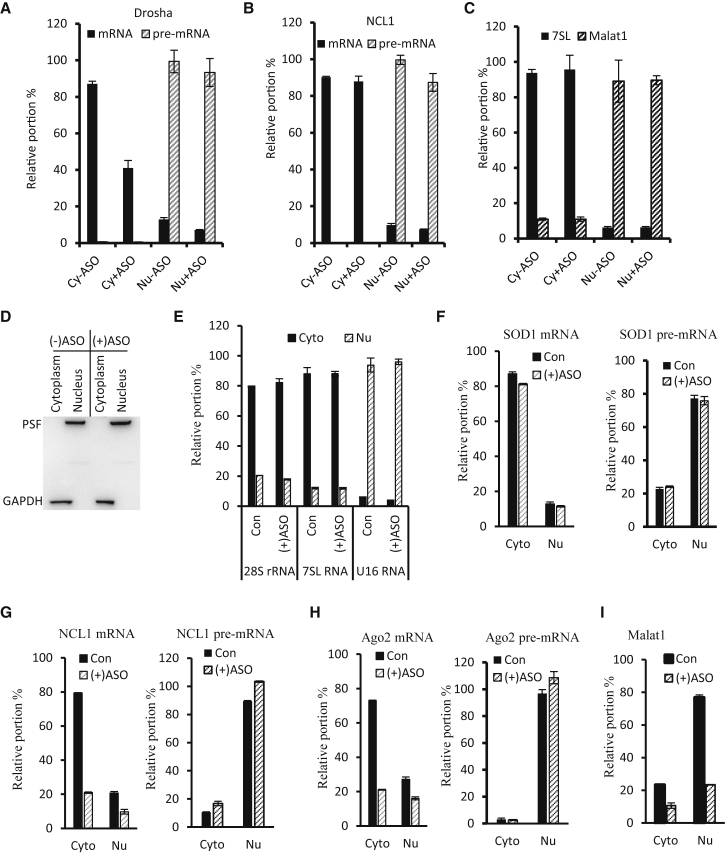
Typically, mRNAs are enriched in the cytoplasm where they are translated. If ASOs can directly degrade mRNA in the cytoplasm, a significant reduction of the mRNAs in the cytoplasmic fraction should be observed shortly after ASO treatment. To evaluate this possibility, HeLa cells were transfected with an ASO targeting Drosha mRNA for 2 hr at an approximate 50% of maximal inhibitory concentration (IC50) to better quantify changes in mRNA levels, followed by fractionation of nuclear and cytoplasmic RNAs. qRT-PCR analyses of Drosha mRNA showed that the majority of mRNA in control cells was enriched in the cytoplasmic fraction, with only a small portion detected in the nucleus. Pre-mRNA was mostly found in the nucleus, as expected (Figure 4A). ASO treatment significantly reduced the cytoplasmic mature mRNA, whereas the nuclear pre-mRNA level was not affected, consistent with the observation in Figure 2. As a control, NCL1 mRNA and pre-mRNA were enriched in the cytoplasm and nucleus, respectively, and the levels of these RNAs were not reduced by the Drosha ASO (Figure 4B). The fractionation quality was determined using the well-documented cytoplasmic 7SL RNA and nuclear MALAT1 RNA,52, 53 respectively (Figure 4C), which showed expected enrichment of these RNAs in the corresponding fractions.
Figure 4.
mRNA Reduction Can Occur in the Cytoplasm
(A) qRT-PCR for Drosha mature and pre-mRNA levels in different subcellular fractions prepared from HeLa cells treated with 5 nM ASO25690 for 2 hr. Cy and Nu indicate cytoplasmic and nuclear fractions, respectively. +ASO and −ASO indicate cells treated with or without ASO25690, respectively. (B) qRT-PCR for NCL1 mature and pre-mRNA levels in the same samples as used in (A). (C) qRT-PCR for cytoplasmic 7SL RNA and nuclear Malat1 RNA levels in the same samples as in (A). (D) Western blot analyses of nuclear protein PSF and cytoplasmic protein GAPDH in cytoplasmic and nuclear fractions prepared from HeLa cells treated with [(+)ASO] or without [(−)ASO] ASOs. (E) qRT-PCR for 28S rRNA, 7SL RNA, and U16 snoRNA levels in cytoplasmic or nuclear RNAs prepared from aliquots of cytoplasmic and nuclear samples as in (D). The same set of RNAs was also subjected to qRT-PCR analyses for mature and pre-mRNA levels of SOD1 (F), NCL1 (G), Ago2 (H), and MALAT1 (I) RNA. Error bars are SDs from three experiments.
A previous study reported that some mRNAs can be retained in the nucleus.43 To determine the subcellular distribution of other tested mRNAs and whether ASOs can directly reduce cytoplasmic mRNA levels, subcellular fractionation was performed 2 hr after simultaneous transfection of three ASOs targeting NCL1 mRNA (ASO110128, 20 nM), Ago2 mRNA (ASO136785, 20 nM), and nuclear MALAT1 RNA (ASO 395254, 5 nM). The fractionation quality was confirmed by western blot analyses of the nuclear protein PSF and the cytoplasmic protein GAPDH, which were distributed in the corresponding fractions without cross-contamination (Figure 4D). In addition, the fractionation quality was also confirmed by the distribution patterns of cytoplasmic RNAs (28S rRNA and 7SL RNA) and nuclear U16 snoRNA, which all showed enrichment in the expected fractions (Figure 4E).
Next, qRT-PCR analyses were performed to detect the levels and subcellular distribution of mature and pre-mRNAs for the untargeted SOD1 (Figure 4F) and targeted NCL1 (Figure 4G), Ago2 (Figure 4H), and MALAT1 lncRNA (Figure 4I). The results showed that in mock-transfected control cells, the mature mRNAs were enriched in the cytoplasmic fractions, with cytoplasmic/nuclear ratios of ∼6.7, 4, and 2.7 for SOD1, NCL1, and Ago2 mRNAs, respectively. As expected, pre-mRNAs of all of these genes and the lncRNA MALAT1 were enriched in the nucleus. Levels of targeted cytoplasmic NCL1 and Ago2 mRNAs, but not the untargeted SOD1 mRNA, were dramatically reduced in ASO-treated cells, indicating ASO specificity. More importantly, these findings also indicate that the cytoplasmic mRNAs can be rapidly reduced within 2 hr of ASO treatment. In addition, nuclearly distributed mature mRNA levels, although low, were also reduced by the corresponding ASOs. As expected, the targeted nuclear MALAT1 RNA was dramatically reduced in ASO-treated cells, in alignment with the nuclear activity of ASOs. Consistent with the above observations, NCL1 and Ago2 pre-mRNA levels were not decreased, suggesting that the reduction of these mRNAs did not stem from pre-mRNA degradation. Together, these results suggest that the targeted mRNAs are enriched in the cytoplasmic fraction (as expected) and that ASO treatment can quickly reduce cytoplasmic mRNA levels.
RNase H1 Protein Exists in the Cytosol
It has been well documented that RNase H1 protein is present in the nucleus.11 In the cytoplasm, RNase H1 protein is mainly localized in the mitochondria.13 Since nuclear-encoded mRNAs are translated in the cytosol outside of the mitochondria, ASO-mediated cleavage of cytoplasmic mRNAs requires the presence of RNase H1 protein in the cytosolic fraction of cells. To determine the subcellular distribution of RNase H1 protein, nuclear, mitochondrial, and cytosolic fractions were separated from HeLa cells. Proteins of each fraction from an equal number of cells were subjected to western blot analyses (Figure 5A).
Figure 5.
RNase H1 Protein Is Present in the Cytosol and the Reduction of RNase H1 Dramatically Decreases ASO Activity
(A) Cytosolic, mitochondrial, and nuclear fractions were prepared, and proteins of each fraction from an equal number of cells were analyzed by SDS-PAGE and western blot assay. The membrane was probed for the following, using specific antibodies: RNase H1; nuclear proteins PSF, P54nrb, and Lamin A; mitochondrial proteins P32, GRSF1, and Hsp60; and cytoplasmic protein GAPDH. The RNase H1 protein level was quantified using ImageJ software (NIH). The mean levels and SDs from three independent experiments are shown below the lanes. (B) qRT-PCR (left) and western blot (right) analyses for the levels of RNase H1 and RNase H2 mRNAs and proteins in HeLa cells treated with specific siRNAs or a control luciferase siRNA for 60 hr. GAPDH was probed in western blot analysis and served as a loading control. (C–H) Control, RNase H1-, and RNase H2-siRNA-treated cells, as used in (A), were treated for 4 hr with different ASOs, and the levels of the targeted MALAT1 (C), SOD1 pre-mRNA (D), Ago2 (E and F), NCL1 (G), and Drosha (H) RNAs were determined by qRT-PCR. Error bars are SDs from three experiments. p values were calculated using the F test (curve comparison between control and H1-reduced cells).
Although RNase H1 protein is not abundant in cells, it was clearly detected in both the cytosolic and nuclear fractions and was mainly enriched in the mitochondrial fraction. The detection of cytosolic RNase H1 was not caused by contamination from the mitochondria or nuclear fractions. Three abundant mitochondrial proteins (P32 [C1QBP], GRSF1, and Hsp60) were enriched in the mitochondrial fraction but were not detected in the cytosolic fraction. We note that a trace amount of these proteins was detected in the nuclear fraction, due to either a low level of mitochondrial contamination in the nuclear fraction or to partial localization of these proteins to the nucleus. In fact, the mitochondrial protein P32 that interacts with RNase H1 was reported to also be present in the nucleus.15, 54, 55 Importantly, three abundant nuclear proteins (PSF [SFPQ], P54nrb, and Lamin A) were only detected in the nuclear fraction and not in the mitochondrial or cytosolic fractions. Furthermore, the cytoplasmic protein GAPDH was detected mostly in the cytosolic fraction, with a low level in the mitochondrial fraction, consistent with previous reports that GAPDH enters the mitochondria.56 Fractionation experiments were performed three times and a similar distribution of RNase H1 was observed, with 17% ± 1.7%, 66% ± 5.3%, and 17% ± 4.4% in the cytosolic, mitochondrial, and nuclear fractions, respectively (data not shown). Together, our results indicate that RNase H1 protein is present in the cytosol, in addition to the mitochondria and nucleus.
ASO Activities Are Dependent on RNase H1
It was shown previously that RNase H1, and not RNase H2, is responsible for the activities of DNA-like gapmer ASOs in cells.11 To confirm that the ASO-induced reduction of the tested RNAs observed here is mediated by RNase H1, HeLa cells were treated with siRNAs targeting either RNASEH1 or RNASEH2 mRNAs. siRNA transfection specifically reduced the targeted mRNAs by more than 90% (Figure 5B, left panel), and protein levels were also dramatically reduced (>90%), compared with those in control siRNA-treated cells (Figure 5B, right panel).
ASOs targeting different RNAs were transfected into these test cells, and the levels of targeted RNAs were determined by qRT-PCR. As expected, the reduction of RNase H1, and not RNase H2, dramatically decreased ASO activity targeting the nuclear MALAT1 RNA (Figure 5C) and the SOD1 pre-mRNA (Figure 5D), supporting the nuclear activity of RNase H1. Similarly, reduced activity was also observed for ASOs targeting cytoplasmically enriched mRNAs, including Ago2 (Figures 5E and 5F), NCL1 (Figure 5G), and Drosha (Figure 5H) mRNAs. We note that residual amounts of RNase H1 protein could still be detected in RNase H1-reduced cells (Figure 5B), which may explain the residual activity of ASOs in these cells, as less efficient reduction of RNase H1 results in weaker effects on decreasing ASO activity.57 Moreover, we previously showed that no ASO activity was present in mice with RNase H1 knocked out specifically in hepatocytes.12 Together, these results indicate that the observed ASO activity in reducing cytoplasmic mRNAs and nuclear RNAs is mediated by RNase H1 and not by RNase H2.
ASOs Targeting Pre-mRNA Intron Regions Can Slowly Reduce mRNA Levels at a Rate Similar to mRNA Decay
As described above, if ASOs are active only in the nucleus, the loss of mRNA should mimic the intrinsic rate of cytoplasmic mRNA decay. To evaluate this hypothesis, we used the ASO (480774) targeting an intron region of SOD1 pre-mRNA, which should only act on the nuclear pre-mRNA but not mature mRNA. As a control, an exon-targeting ASO (333632), in theory, should be able to induce cleavage of both SOD1 mature and pre-mRNA. These ASOs were transfected into HeLa cells for different times, and SOD1 mRNA and pre-mRNA levels were determined by qRT-PCR. While the intron-targeting ASO reduced the pre-mRNA by more than 50% by 3 hr (Figure 6A), more than 11.5 hr was required to reduce the mature mRNA by 50%. A further reduction of the SOD1 mRNA level was observed at 24 hr after transfection, resulting in an elimination half-life of ∼13.7 hr. In contrast, the exon-targeting ASO reduced the mature mRNA by more than 70% within 3 hr (Figure 6B). This ASO did not reduce the SOD1 pre-mRNA; rather, the pre-mRNA level was increased, especially at later time points, likely due to compensatory upregulation. Nonetheless, these results again indicate that the rapid reduction of mature mRNA does not require a reduction of the pre-mRNA, and nuclear-only cleavage of a target pre-mRNA caused a much slower decrease of the mature mRNA levels. This is consistent with the intrinsic cytoplasmic decay rate for the mRNA, as determined by qRT-PCR for SOD1 mRNA upon DRB inhibition of transcription (Figure 6C).
Figure 6.
An Intron-Targeting ASO that Degrades Nuclear Pre-mRNAs Causes a Slow Reduction of mRNA Levels
(A) qRT-PCR for mature and pre-mRNA levels of SOD1 in HeLa cells transfected for different times with 60 nM intron-targeting ASO480774. (B) qRT-PCR for mature and pre-mRNA levels of SOD1 in HeLa cells transfected for different times with 60 nM exon-targeting ASO333632. (C) qRT-PCR for mature SOD1 mRNA levels in HeLa cells treated with 100 μg/mL DRB for the indicated times. (D) qRT-PCR for mature and pre-mRNA levels of SOD1 in HeLa cells transfected for different times with 120 nM ASO480774. The gray dashed lines indicate a 50% reduction of the RNAs. Error bars indicate SDs from three experiments.
We increased the dose of the intronic ASO to 120 nM, with the aim to achieve a better reduction of the pre-mRNA and thus an earlier onset of mRNA reduction. The results showed that the pre-mRNA level was again reduced by ∼50% within 3 hr (Figure 6D), and no further reduction was observed at later time points, consistent with the results of 60 nM ASO treatment (Figure 6A) and the results for Ago2 ASO (Figure 2D). A higher dose of the intronic ASO indeed caused an mRNA reduction at an earlier time point, more obviously at 6.5 hr after transfection. However, even with this earlier onset, it still took ∼11.5 hr to reduce 50% of the mRNA, which was much slower than the reduction rate by the exonic ASO.
An ASO Targeting a Nucleolar RNA Can Also Reduce an Engineered Cytoplasmic RNA Containing the ASO Binding Sequence
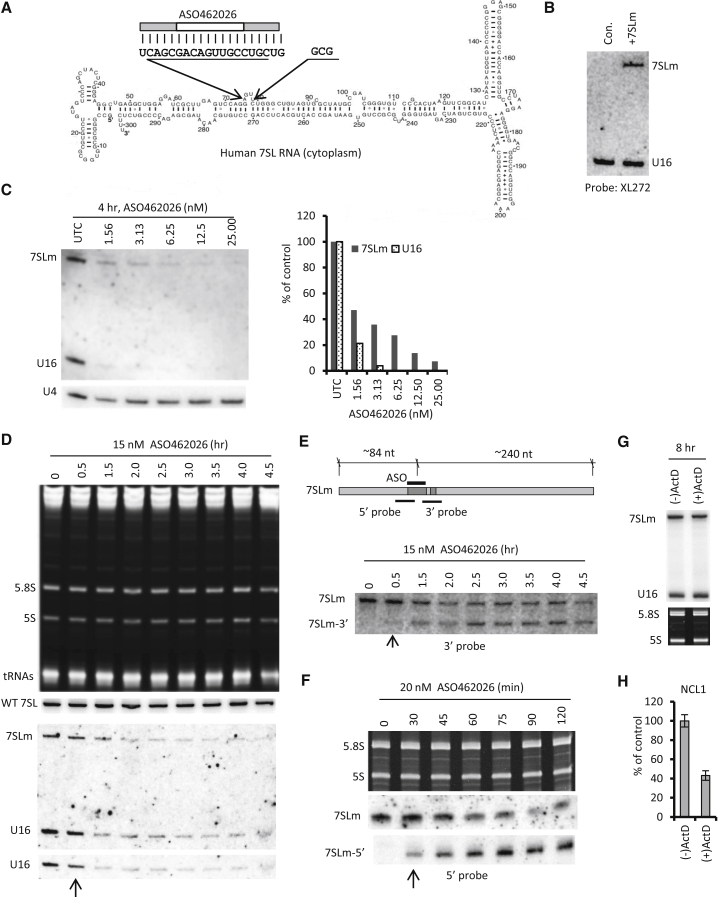
Pre-mRNA and mRNA differ not only in subcellular localization but also in length, stability, copy number, and whether or not they are translated. The difference in ASO potency/onset in reducing the levels of mRNA and pre-mRNA may be affected by many factors. To simplify the situation and further characterize the cytoplasmic activity of ASO-directed RNase H1 cleavage, we expressed a mutant non-coding RNA (ncRNA), 7SLm RNA, which was derived from 7SL RNA. 7SL RNA localizes in the cytoplasm in the form of a ribonucleoprotein (RNP) complex, the signal recognition particle (SRP).58 The mutant 7SLm RNA has an inserted sequence including the ASO462026 target site within the snoRNA U1621 (Figure 7A). Both U16 and 7SLm RNAs are ncRNAs present in RNP complexes, with similar sizes (101 and 324 nt, respectively). A single transfection of ASO462026 could simultaneously target both the nucleolar U16 RNA and the cytoplasmic 7SLm RNA, allowing better characterization of cytoplasmic ASO activity.
Figure 7.
A Mutant 7SLm RNA Could Be Rapidly Degraded by ASOs in HeLa Cells
(A) Schematic depiction of human 7SL RNA. The position of the insertion site, the insert sequences, and the ASO target site are shown. (B) Northern hybridization analysis of 7SLm RNA expression in HeLa cells transfected with the expression plasmid (+7SLm) or in mock-transfected cells (con). The northern membrane was hybridized with a 5′-end labeled probe XL272 that recognizes both U16 and the mutant 7SLm RNAs. (C) Northern hybridization analysis of 7SLm RNA and U16 snoRNA levels in HeLa cells transiently expressing 7SLm RNA and transfected with different concentrations of ASO462026 for 4 hr. The upper panel indicates the hybridization result using probe XL272. The membrane was re-hybridized with a probe specific to U4 snRNA, which served as a loading control. The hybridization signals for 7SLm and U16 RNAs were quantified using ImageJ, and the relative levels are plotted in the right panel. (D) Northern hybridization of different RNAs in HeLa cells transiently expressing 7SLm RNA and transfected with 15 nM ASO462026 for different times. The PAGE gel was stained with ethidium bromide and the identities of the abundant RNAs are indicated in the upper panel. The membrane was either probed for the wild-type 7SL RNA or hybridized with probe XL272, which recognizes both U16 snoRNA and 7SLm RNA (middle panel). An image with a shorter exposure time for U16 is shown in the lower panel. The 30-min time point is marked with an arrow. (E) 7SLm RNA, the ASO target site, and the expected sizes of the cleaved fragments are depicted in the upper panel. The positions of hybridization probes used in subsequent studies are indicated. The lower panel shows northern hybridization analysis of the same blot as in (D) using a 3′ probe (XL297) that detects both the full-length and 3′ cleaved fragment of the 7SLm RNA. The identities of detected bands are indicated. (F) Northern hybridization analysis of 7SLm RNA using a 5′ probe (XL299). HeLa cells expressing 7SLm RNA were transfected with 20 nM ASO462026 for different times and total RNA was prepared and subjected to northern hybridization analyses. The 5.8S and 5S rRNA visualized by ethidium bromide staining served as loading controls. (G) Northern hybridization analysis of U16 and 7SLm RNA in HeLa cells treated [(+)ActD] or not treated [(−)ActD] with 5 μg/mL Actinomycin D for 8 hr. The ethidium bromide staining of 5.8S and 5S rRNAs was used as a loading control. (H) qRT-PCR for NCL1 mRNA levels in the RNA samples as used in (G). Error bars indicate SDs from three experiments.
The plasmid was transfected into HeLa cells, and 7SLm RNA expression was confirmed by northern hybridization (Figure 7B). Although 7SL RNA is known to be highly structured and is bound by six proteins in the SRP complex,59 the inserted sequence was accessible to the ASO, since transfection of ASO462026 reduced the 7SLm RNA level, in addition to U16 snoRNA (Figure 7C). It appears that the potency of this ASO is higher when targeting the nuclear U16 snoRNA rather than 7SLm RNA, due to either the different RNase H1 concentrations in different compartments (see the Discussion) or to differences in the local structures of the host RNAs. However, the ASO is still very active in reducing 7SLm RNA, with an IC50 of less than 2 nM.
ASO Transfection Can Rapidly Degrade Both U16 snoRNA and 7SLm RNA
To determine the cleavage onset of 7SLm RNA, ASO462026 was transfected for different times at a high dose into HeLa cells transiently expressing 7SLm RNA. Total RNA was then prepared and U16 and 7SLm RNA levels were detected by northern hybridization using a probe that recognizes both RNAs (Figure 7D). A significant reduction of both U16 and 7SLm RNAs was observed 1.5 hr after ASO transfection. As expected, wild-type 7SL RNA, which lacks the target sequence, was not reduced by the ASO treatment. A reduction of U16 snoRNA was also confirmed using a different probe (data not shown). A modest reduction of U16 was observed as early as 30 min after ASO treatment. This is more obvious in a shorter exposure image (Figure 7D, lower panel, indicated by an arrow). This result suggests an early cleavage onset of U16 RNA in the nucleus. The reduction of 7SLm RNA was not obvious at 30 min after transfection when we examined the full-length RNA level. However, a weak but detectable cleaved 3′ fragment of 7SLm RNA was observed when the blot was re-hybridized using a probe specific to the 3′ portion of the mutant RNA (Figure 7E, arrow), suggesting that cleavage of 7SLm RNA had already commenced at this early time point. Accumulation of the 5′ cleavage product of 7SLm RNA was also detected in a different experiment at 30 min after ASO transfection (Figure 7F). We note that the accumulation of the cleavage fragments of 7SLm RNA may stem from protection by the binding of SRP proteins or from the highly structured nature of this ncRNA. In most cases, ASO-mediated cleavage does not cause accumulation of cleaved fragments37 (our unpublished data).
Actinomycin D treatment of HeLa cells for 8 hr to inhibit transcription did not reduce the 7SLm RNA level (Figure 7G). However, under the same condition, NCL1 mRNA was already reduced by more than 50% (Figure 7H), indicating that Actinomycin D treatment was effective and 7SLm RNA is stable. Together, these results show that ASOs can rapidly trigger the cleavage of both U16 snoRNA and 7SLm RNA, and the latter should localize in the cytoplasm.
Both the Full-Length 7SLm RNA and the Cleaved Fragments Are Accumulated in the Cytoplasm
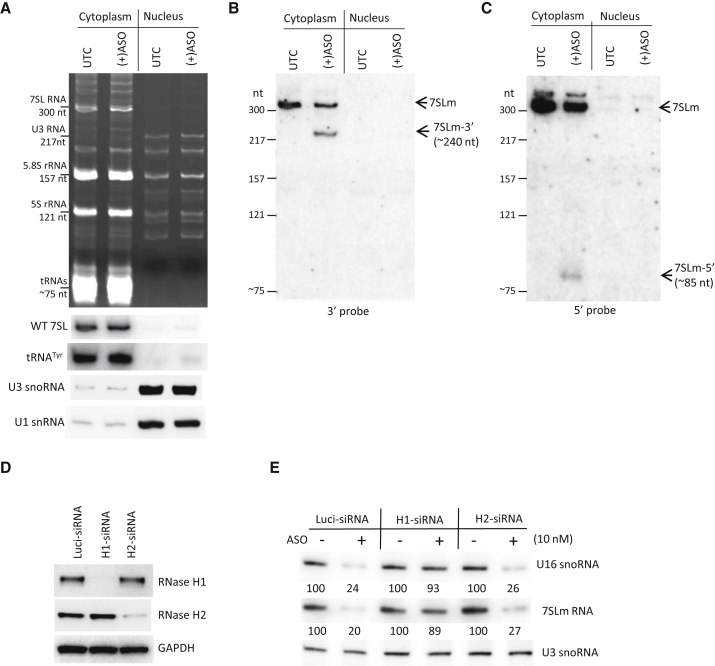
To determine whether 7SLm RNA is indeed properly localized in the cytoplasm and whether cleavage of 7SLm RNA occurs in this compartment, HeLa cells transiently expressing 7SLm RNA were either mock transfected or transfected with ASO462026 for 2 hr and subcellular fractionation was performed. Cytoplasmic and nuclear RNAs were analyzed by northern hybridization (Figure 8A). As expected, cytoplasmically localized endogenous 7SL RNA and tRNAtyr were enriched in the cytoplasmic fractions, whereas nuclear localized U3 snoRNA and U1 snRNA were highly enriched in the nuclear fraction, indicating little cross-contamination between the fractions. The fractionation quality was further confirmed by ethidium bromide staining (Figure 8A, upper panel), which showed that tRNAs accumulate in the cytoplasmic fractions and not in the nuclear fractions.
Figure 8.
The Full-Length and Cleaved Fragments of 7SLm RNA Are Accumulated in the Cytoplasm
(A) Northern hybridization analysis of RNAs prepared from cytoplasmic or nuclear fractions of cells treated with [(+)ASO] or without (UTC) 20 nM ASO462026 for 2 hr. The upper panel shows ethidium bromide staining of the PAGE gel. The identities and the sizes of known abundant RNAs are indicated, which served as size markers. The lower panels are northern hybridization for wild-type 7SL RNA and tRNAtyr, which served as markers for cytoplasmic RNAs, and U1 snRNA and U3 snoRNA, which served as markers for nuclear RNAs, respectively. (B) Northern hybridization of the membrane transferred from the gel in (A) using probe XL297, which detects the full-length and the 3′ cleaved fragments of 7SLm RNA. The size ladder was generated from the known RNAs as shown in (A). The cleaved 3′ fragment and its expected size are indicated. (C) Northern hybridization of the same membrane as in (B) using the 5′ probe XL299. The cleaved fragment and the expected size are indicated. (D) Western analyses of RNase H1 and RNase H2 in HeLa cells treated with either a control luciferase siRNA (Luci-siRNA) or specific siRNAs for 60 hr. GAPDH was detected and served as a loading control. (E) Northern hybridization of 7SLm RNA and U16 snoRNA in 7SLm RNA-expressing HeLa cells transfected with siRNAs for 60h, followed by transfection of 15 nM ASO462026 for 5 hr. U3 snoRNA was detected and served as a loading control. The relative levels of 7SLm and U16 were quantified and normalized to U3 RNA and are shown below the lanes.
Next, the membrane was hybridized using probes specific to either the 3′ (Figure 8B) or 5′ (Figure 8C) portion of 7SLm RNA relative to the predicted cleavage site. The results showed that full-length 7SLm RNA localized in the cytoplasm, as expected, in both control cells and in ASO-transfected cells that showed a reduced RNA level. The 3′ cleaved fragment was also detected only in the cytoplasmic fraction, at the expected size. Similarly, when the 5′ probe was used, both the full-length 7SLm RNA and the 5′ cleaved fragment were detected in the cytoplasm but not in the nucleus. These results strongly suggest that ASO-directed RNase H1 cleavage occurs rapidly in the cytoplasm.
Next, we evaluated whether ASO-induced cytoplasmic cleavage of 7SLm RNA was indeed mediated by RNase H1, similar to the other ASOs tested above (Figure 5). RNase H1 and RNase H2 proteins were reduced by siRNA treatment in HeLa cells expressing 7SLm RNA (Figure 8D), followed by transfection of ASO462026. Northern hybridization results showed that the ASO activity in degrading 7SLm or U16 RNA was dramatically decreased in RNase H1-depleted, but not RNase H2-depleted, cells (Figure 8E). This result confirms that ASO462026 indeed induces RNase H1 cleavage of the target 7SLm RNA and U16 snoRNA, like other tested ASOs (Figure 5). Altogether, our results indicate that RNase H1-dependent ASOs are robustly active in degrading target RNAs in both the cytoplasm and nucleus.
Discussion
Although ASOs have been well studied and developed (both as a drug and as a research tool) for more than 30 years,1 their mechanisms of action are still not fully understood. ASOs are shown to be active in the nucleus to reduce both nuclear ncRNAs and pre-mRNAs. The cytoplasmic activity of RNase H1-dependent ASOs has been debated, and it has been assumed that ASOs are only active in the nucleus.33, 60 Here we provide evidence showing that RNase H1-dependent ASOs are robustly active in both the cytoplasm and nucleus and, in many cases, cytoplasmic cleavage may be a major contributor to the target RNA reduction.
RNase H1 localizes in both the nucleus and cytoplasm.11 Therefore, it is not surprising that ASOs are active in the nucleus. However, in the cytoplasm, RNase H1 appears to accumulate in the mitochondria13, 15 (a membrane-enclosed organelle). In this study, we showed that RNase H1 protein is clearly present in the cytosolic fraction outside of the mitochondria, at an amount similar to that in the nucleus (Figure 5A). Since cytoplasmic volume is higher than nuclear volume in most mammalian cells,61 the concentration of RNase H1 protein should be higher in the nucleus than in the cytoplasm, which may explain why in many cases ASOs are very active in the nucleus.22, 62 As RNase H1 is also present in the cytosol, it is expected that gapmer ASOs are active in both the cytoplasm and nucleus. The potential biological function of cytosolic RNase H1 is currently unclear; however, based on its enzymatic properties, it is possible that this protein may play a role in cellular defense against invasion of foreign DNA virus.63
Although the possibility of non-RNase H-mediated mRNA reduction by an ASO was proposed based on sequencing data of 5′-RACE products,39 the many 5′ ends detected by RACE around the predicted cleavage site could be most simply explained by incomplete trimming of the cleaved fragments by exonucleases. In cells depleted of XRN1 or XRN2, ASO cleavage sites were detected by 5′-RACE for different mRNAs and nuclear RNAs to locate within the regions complementary to the DNA portion of the ASOs,37 further supporting the RNase H-mediated mechanism. The RNase H1 dependence of ASOs designed to recruit RNase H1 was further demonstrated recently using the liver-specific RNase H1 knockout mouse system, which showed a loss of ASO activity in the mouse liver.12 In the current study, we showed that the reduction of RNase H1, and not RNase H2, dramatically reduced ASO activity when targeting mRNAs or nuclear RNAs, further confirming other studies showing that RNase H1 is responsible for ASO-directed RNA cleavage.11 Since RNase H1 is an active ribonuclease, a more complete reduction of this protein might be required for better inhibition of its activity.
A previous study performed in Xenopus oocytes showed that cytoplasmic activity accounts for ∼5% of total oligonucleotide-induced RNase H activity,32 suggesting the presence of RNase H1 in this compartment where it is available to perform ASO-directed cleavage of target RNAs. However, Xenopus oocytes are unique cells that may not represent mammalian cells. In another study, the reduction of a co-transfected mRNA was not observed using an ASO that otherwise could reduce the level of the expressed mRNA.33 However, it is possible that the transfected mRNA might form a local structure(s) that makes the target site inaccessible to the ASO, whereas an mRNA expressed in cells could be regulated by cellular processes to exhibit a different, open structure(s).
In the current study, we showed that after ASO transfection, mature mRNAs could be rapidly reduced at a rate much faster than expected if ASOs only degraded the target RNAs in the nucleus, which is in alignment with our previous observations.24 A rapid reduction by ASO treatment was observed for five different mRNAs in both human and mouse cells as well as for a cytoplasmic ncRNA. If ASOs are active only in the nucleus, degradation of nuclear transcripts (pre-mRNA and processed mRNA) should mimic the situation of blocking mRNA biogenesis. In this case, reduction of the pre-existing cytoplasmic mRNAs should occur mainly through normal mRNA decay pathways. This was confirmed by the observations with the intron-targeting ASO for SOD1 pre-mRNA (Figure 6). However, we observed that the levels of different mRNAs could be rapidly and robustly reduced by exon-targeting ASOs in an RNase H1-dependent manner. Although previous studies show that some mRNAs can be retained in the nucleus,43 like most mRNAs, the mRNAs tested in this study were all enriched in the cytoplasm. In addition, nuclear re-import was reported for tRNAs and snRNAs as part of the maturation process;64, 65 however, to our knowledge, robust mRNA re-import has not yet been reported. Although low mRNA levels were detected in the nuclear fractions and ASO treatment reduced nuclear mRNA levels (as shown in Figure 4), mRNAs are enriched in the cytoplasm, and a rapid and robust reduction of cytoplasmic mRNA was observed shortly after ASO treatment. Together, these results from fractionation and kinetic studies indicate that the pre-existing cytoplasmic mRNAs could also be directly degraded by ASOs and that RNase H1-dependent ASOs are active in the cytoplasm.
In theory, a simultaneous reduction of both cytoplasmic mRNA and nuclear pre-mRNA levels (and nuclear processed mRNA) could occur for exon-targeting ASOs (Figures 2D and 2E). However, it appears that the nuclear cleavage event is not necessary and may not be a major contributor to the rapid, vast reduction of mature mRNAs by some ASOs. This is more evident for the three Drosha ASOs that rapidly reduced the level of the mature mRNA, but not the pre-mRNA (Figures 2A and 2B). In addition, an Ago2 ASO that did not reduce pre-mRNA levels caused an even greater and faster reduction of mRNA levels, compared with several other Ago2 ASOs that reduced pre-mRNA levels, likely due to its high potency (Figures 2D and 2E).
While an ASO may be active in directing cleavage of mature mRNA, it may not be active in degrading pre-mRNAs, even though pre-mRNAs contain the ASO binding sites, as observed for the Drosha and the SOD1 ASOs. A similar observation was also made for the U23 snoRNA ASOs. This difference in ASO activity could be attributed to different ASO accessibility and/or different half-lives between mRNAs and pre-mRNAs. However, although the short half-life of pre-mRNA can affect the activity of some ASOs (i.e., inhibition of splicing increased ASO activity,24 as shown previously and in this study), it seems that the short half-life is not the only determinant for ASO activity, as Ago2 pre-mRNA and the processing intermediate of U23 snoRNA, which are short lived, could be reduced by ASOs. In addition, stabilization of pre-mRNA by splicing inhibition did not increase U23 snoRNA ASO activity in reducing the level of its host, NCL1 pre-mRNA.
Pre-mRNAs contain much longer sequences (introns) that could form different local structures compared with mature mRNAs. In addition, pre-mRNAs could also interact with many proteins required for processing (e.g., spliceosomes and hnRNPs, as well as proteins for miRNA and snoRNA processing),49, 50, 51, 66, 67 which are not present on cytoplasmic mature mRNAs. Therefore, the target sites in pre-mRNAs may not be accessible to ASOs, which in most cases are screened for activity based on mRNA levels. The observed difference in target site accessibility in different compartments may in part explain the relatively weak nuclear activity of siRNAs;28 therefore, screening of different sequences when targeting nuclear RNAs may be helpful for siRNA design. Nonetheless, our observations suggest that robust cytoplasmic activity of RNase H1-dependent ASOs could be a major contributor to the rapid mRNA reduction for some ASOs and that cleavage of pre-mRNA is not absolutely required for targeted mRNA degradation. In addition, some ASOs may primarily act on pre-mRNAs in the nucleus, thus ultimately leading to the reduction of mRNAs, albeit at a slower rate.
Although we have conclusively shown that RNase H1 ASOs are active in the cytoplasm, a number of lines of evidence suggest that RNase H1 activity may be weaker in the cytoplasm than in the nucleus, as reported for Xenopus oocytes. We caution against over-interpreting such data, as the cytoplasmic volume of most cells is greater than the nuclear volume68 and we have shown that RNase H1 is limiting.24 Therefore, the apparent difference in efficiency in oocytes may simply reflect a lower concentration of the enzyme in the cytoplasm.
RNase H1-dependent cytoplasmic cleavage activity was more evident for 7SLm RNA. ASO treatment caused cleavage for both nuclear U16 snoRNA and cytoplasmic 7SLm RNA, with an early onset. The stabilized cleavage products of 7SLm were already detected 30 min after ASO transfection. Importantly, both the 5′ and 3′ cleavage fragments were accumulated in the cytoplasm but were not detected in the nucleus. Since full-length 7SLm RNA was predominantly accumulated in the cytoplasm and was barely detectable in the nucleus, cytoplasmic accumulation of the cleaved fragments should stem from the cleavage of cytoplasmic 7SLm RNAs. Thus, cytoplasmic RNA cleavage could have an early onset after ASO transfection, similar to the reduction of the nuclear U16 snoRNA. Together, our results indicate that RNase H1-dependent ASOs are robustly active in both the cytoplasm and nucleus. These findings may thus help with ASO design and screening when targeting RNAs in different subcellular compartments and may improve understanding of the potential mechanisms related to target reduction and the onset of phenotypes.
Our observations of the different activities of an ASO targeting cytoplasmic and nuclear RNAs are interesting. The difference in activity of a given ASO in different subcellular compartments may be a summation of many factors that contribute to target accessibility, such as different protein binding, RNA local structure, subcellular localization, RNA stability, and concentrations of the ASO, target RNA, and RNase H1 protein. Previous studies showed that local structure and protein binding affect ASO binding and RNase H1 recruitment,25, 69 and increasing the stability of pre-mRNA also enhanced ASO potency.24 This complexity in determining the activity of ASOs (and likely siRNA as well) makes it difficult to identify the best target sites through computational prediction. However, experimental screening could be more efficient with a better understanding of the factors that govern RNA accessibility and ASO availability, which requires further investigation.
Materials and Methods
ASOs, siRNAs, TaqMan primer probe sets for qRT-PCR, and probes for northern hybridization are listed in the Supplemental Materials and Methods.
Cell Culture and Transfection
HeLa, HEK293, and MHT cells were grown on plates in DMEM supplemented with 10% fetal calf serum (FCS) and 1% penicillin/streptomycin at 37°C in a 5% or 8% CO2 incubator. For siRNA transfection, cells were seeded at ∼50% confluence, incubated overnight (ON), and transfected with siRNAs at a final concentration of 3–5 nM for 36 hr using RNAiMax (Life Technologies) based on the manufacturer’s instructions. Cells were then re-seeded in 96-well plates, incubated ON, and transfected with ASOs for 4 hr in an ASO activity assay, as described previously.70 For ASO transfection, cells were seeded at 50%–70% confluence, grown ON, and transfected using Lipofectamine 2000 (Life Technologies) with ASOs for different time points and at different concentrations as indicated in the figure legends. For plasmid transfection, 1–2 μg plasmid DNA was transfected into a 10-cm dish of HeLa cells for 48 hr using Effectene (QIAGEN) based on the manufacturer’s instructions.
RNA Preparation and Detection
For qRT-PCR analyses, total RNA or RNA from different subcellular fractions was prepared using the RNeasy (QIAGEN) kit based on the manufacturer’s instructors. For northern hybridization analysis, RNA was prepared using TRIzol (Thermo Fisher Scientific). qRT-PCR was performed using TaqMan primer probe sets with the StepOne real-time PCR system, as described previously.71 Northern hybridization was carried out essentially as described.72 5′-RACE was performed for U23 intermediates using a second-generation 5′-/3′-RACE kit (Roche) following the manufacturer’s protocol using RNA from control and XRN1 reduced cells, with a gene-specific primer (5′-TGAGAAGACACATTAGACTC-3′) to synthesize cDNA and a gene-specific nest primer (5′-CTCAAGGTCTGCACAGAACA-3′) and the adaptor primer from the kit to amplify the cDNA. The PCR product was cloned into the pCR™4-Topo vector and sequenced.
Subcellular Fractionation
Cytoplasmic and nuclear fractions were prepared from ∼5 × 107 HeLa cells using a nuclear protein kit (QIAGEN) following the manufacturer’s instruction, supplemented with RNase inhibitor in all buffers, or using the PARIS kit (Thermo Fisher Scientific) as shown in Figures 4D–4I. An aliquot of cytoplasmic and nuclear fractions was analyzed by SDS-PAGE and western blot for the distribution of marker proteins. An aliquot of cytoplasmic and nuclear fractions was subjected to RNA preparation using either TRIzol for northern hybridization (Figure 8) or an RNeasy column for qRT-PCR (Figure 4). To compare RNA levels in different fractions, mRNA levels in cytoplasmic or nuclear fractions were normalized to the levels of cytoplasmic 7SL RNA or nuclear MALAT1 RNAs, respectively (Figures 4A–4C), or to the levels of cytoplasmic 7SL RNA or the nuclear U16 RNA, respectively (Figures 4F–4I). The cytoplasmic and nuclear marker RNAs (7SL, Malat1, and U16) were normalized within cytoplasmic or nuclear fractions, respectively, using the Quant-iT RiboGreen RNA Assay Kit (Thermo Fisher Scientific). The relative RNA levels in different fractions in ASO-treated cells were calculated as a percentage of the total RNA levels (sum of cytoplasmic and nuclear RNA) in control cells.
Cytosolic, mitochondrial, and nuclear fractions of HeLa cells were separated using the Cell Fractionation Kit–Standard (Abcam) based on the manufacturer’s protocol. Cytosolic, mitochondrial, and nuclear proteins were dissolved in an equal volume of lysis buffer, and an equal volume of proteins from each fraction was analyzed by SDS-PAGE and western blot analyses.
Construction of the 7SLm RNA Expression Plasmid
To express the mutant 7SL RNA (7SLm RNA) containing a sequence derived from U16 snoRNA, 7SL RNA cDNA was synthesized. Using the cDNA as template, PCR was performed with primers XL265 (5′-GAGGATCGCTTGAGTCCAGG TCAGCGACAGTTGCCTGCTGAGTT GCGCTGGGCTGTAGTGCGCTATG-3′; the inserted sequences are underlined) and XL266 (5′-CCGAATTC GAGAATATTCATCTCTTGAGAGTCC-3′, antisense; the EcoRI site is underlined) to synthesize the 3′ portion of 7SL RNA containing the inserted sequence. The PCR product was gel purified and used as a mega primer to perform PCR together with primer XL264 (5′-GG GGATCC ATACCACAGCTTCT AGTGC -3′, sense; the BamHI site is underlined). The PCR products were ligated into BamH1 and EcoRI sites of plasmid pSIREN-RetroQ (Promega), and the mutant 7SLm RNA was expressed from a U6 promoter.
Western Blot Analyses
Whole cell lysate was prepared using radioimmunoprecipitation assay (RIPA) buffer (Thermo Fisher Scientific) and ∼40 μg total protein was separated on 4%–12% gradient SDS-PAGE and transferred to membranes. Proteins were detected using primary antibodies specific to each protein. Antibodies for RNase H1 and RNase H2A were raised in rabbits, as described.15 Antibodies for Xrn2 (sc-365258), GAPDH (sc-32233), PSF (sc-374502), P54nrb (sc-376865), GRSF1 (sc-133638), and P32 (sc-271201) were purchased from Santa Cruz Biotechnology. Antibodies for La (ab75927), Lamin A (ab26300), and Hsp60 (ab110312) were purchased from Abcam. Anti-rabbit (1706515) or anti-mouse (1721011) secondary antibodies were purchased from Bio-Rad.
Author Contributions
X.-H.L. and S.T.C. designed the study and wrote the paper. X.-H.L., H.S., and J.G.N. performed the experiments. All authors analyzed the data.
Conflicts of Interest
All authors are employees of Ionis Pharmaceuticals, Inc.
Acknowledgments
This work was supported by internal funding from Ionis Pharmaceuticals, Inc. The authors thank Tim Vickers, Wen Shen, Shiyu Wang, and Jeff Bailey for discussions.
Footnotes
Supplemental Information includes Supplemental Materials and Methods and can be found with this article online at http://dx.doi.org/10.1016/j.ymthe.2017.06.002.
Supplemental Information
References
- 1.Crooke S.T., Vickers T., Lima W., Wu H. Mechanisms of antisense drug action, an introduction. In: Crooke S.T., editor. Antisense Drug Technology: Principles, Strategies, and Applications. Second Edition. CRC Press; 2008. pp. 3–46. [Google Scholar]
- 2.Dias N., Stein C.A. Antisense oligonucleotides: basic concepts and mechanisms. Mol. Cancer Ther. 2002;1:347–355. [PubMed] [Google Scholar]
- 3.Lima W., Wu H., Crooke S.T. The RNase H mechanism. In: Crooke S.T., editor. Antisense Drug Technology: Principles, Strategies, and Applications. Second Edition. CRC Press; 2008. pp. 47–74. [Google Scholar]
- 4.Juliano R.L., Ming X., Nakagawa O. Cellular uptake and intracellular trafficking of antisense and siRNA oligonucleotides. Bioconjug. Chem. 2012;23:147–157. doi: 10.1021/bc200377d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Juliano R.L., Carver K. Cellular uptake and intracellular trafficking of oligonucleotides. Adv. Drug Deliv. Rev. 2015;87:35–45. doi: 10.1016/j.addr.2015.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Geary R.S., Norris D., Yu R., Bennett C.F. Pharmacokinetics, biodistribution and cell uptake of antisense oligonucleotides. Adv. Drug Deliv. Rev. 2015;87:46–51. doi: 10.1016/j.addr.2015.01.008. [DOI] [PubMed] [Google Scholar]
- 7.Wang S., Sun H., Tanowitz M., Liang X.H., Crooke S.T. Annexin A2 facilitates endocytic trafficking of antisense oligonucleotides. Nucleic Acids Res. 2016;44:7314–7330. doi: 10.1093/nar/gkw595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bennett C.F., Chiang M.Y., Chan H., Shoemaker J.E., Mirabelli C.K. Cationic lipids enhance cellular uptake and activity of phosphorothioate antisense oligonucleotides. Mol. Pharmacol. 1992;41:1023–1033. [PubMed] [Google Scholar]
- 9.Liang X.H., Shen W., Sun H., Prakash T.P., Crooke S.T. TCP1 complex proteins interact with phosphorothioate oligonucleotides and can co-localize in oligonucleotide-induced nuclear bodies in mammalian cells. Nucleic Acids Res. 2014;42:7819–7832. doi: 10.1093/nar/gku484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shen W., Liang X.H., Crooke S.T. Phosphorothioate oligonucleotides can displace NEAT1 RNA and form nuclear paraspeckle-like structures. Nucleic Acids Res. 2014;42:8648–8662. doi: 10.1093/nar/gku579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu H., Lima W.F., Zhang H., Fan A., Sun H., Crooke S.T. Determination of the role of the human RNase H1 in the pharmacology of DNA-like antisense drugs. J. Biol. Chem. 2004;279:17181–17189. doi: 10.1074/jbc.M311683200. [DOI] [PubMed] [Google Scholar]
- 12.Lima W.F., Murray H.M., Damle S.S., Hart C.E., Hung G., De Hoyos C.L., Liang X.H., Crooke S.T. Viable RNaseH1 knockout mice show RNaseH1 is essential for R loop processing, mitochondrial and liver function. Nucleic Acids Res. 2016;44:5299–5312. doi: 10.1093/nar/gkw350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Suzuki Y., Holmes J.B., Cerritelli S.M., Sakhuja K., Minczuk M., Holt I.J., Crouch R.J. An upstream open reading frame and the context of the two AUG codons affect the abundance of mitochondrial and nuclear RNase H1. Mol. Cell. Biol. 2010;30:5123–5134. doi: 10.1128/MCB.00619-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liang X.H., Shen W., Sun H., Migawa M.T., Vickers T.A., Crooke S.T. Translation efficiency of mRNAs is increased by antisense oligonucleotides targeting upstream open reading frames. Nat. Biotechnol. 2016;34:875–880. doi: 10.1038/nbt.3589. [DOI] [PubMed] [Google Scholar]
- 15.Wu H., Sun H., Liang X., Lima W.F., Crooke S.T. Human RNase H1 is associated with protein P32 and is involved in mitochondrial pre-rRNA processing. PLoS ONE. 2013;8:e71006. doi: 10.1371/journal.pone.0071006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Akman G., Desai R., Bailey L.J., Yasukawa T., Dalla Rosa I., Durigon R., Holmes J.B., Moss C.F., Mennuni M., Houlden H. Pathological ribonuclease H1 causes R-loop depletion and aberrant DNA segregation in mitochondria. Proc. Natl. Acad. Sci. USA. 2016;113:E4276–E4285. doi: 10.1073/pnas.1600537113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holmes J.B., Akman G., Wood S.R., Sakhuja K., Cerritelli S.M., Moss C., Bowmaker M.R., Jacobs H.T., Crouch R.J., Holt I.J. Primer retention owing to the absence of RNase H1 is catastrophic for mitochondrial DNA replication. Proc. Natl. Acad. Sci. USA. 2015;112:9334–9339. doi: 10.1073/pnas.1503653112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reyes A., Melchionda L., Nasca A., Carrara F., Lamantea E., Zanolini A., Lamperti C., Fang M., Zhang J., Ronchi D. RNASEH1 mutations impair mtDNA Replication and cause adult-onset mitochondrial encephalomyopathy. Am. J. Hum. Genet. 2015;97:186–193. doi: 10.1016/j.ajhg.2015.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Uhler J.P., Falkenberg M. Primer removal during mammalian mitochondrial DNA replication. DNA Repair (Amst.) 2015;34:28–38. doi: 10.1016/j.dnarep.2015.07.003. [DOI] [PubMed] [Google Scholar]
- 20.Bennett C.F., Baker B.F., Pham N., Swayze E., Geary R.S. Pharmacology of antisense drugs. Annu. Rev. Pharmacol. Toxicol. 2017;57:81–105. doi: 10.1146/annurev-pharmtox-010716-104846. [DOI] [PubMed] [Google Scholar]
- 21.Liang X.H., Vickers T.A., Guo S., Crooke S.T. Efficient and specific knockdown of small non-coding RNAs in mammalian cells and in mice. Nucleic Acids Res. 2011;39:e13. doi: 10.1093/nar/gkq1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liang X.H., Vickers T.A., Crooke S.T. Antisense-mediated reduction of eukaryotic noncoding RNAs. In: Erdmann V.A., Barciszewski J., editors. From Nucleic Acid Sequences to Molecular Medicine. Springer; 2012. pp. 191–214. [Google Scholar]
- 23.Zong X., Huang L., Tripathi V., Peralta R., Freier S.M., Guo S., Prasanth K.V. Knockdown of nuclear-retained long noncoding RNAs using modified DNA antisense oligonucleotides. Methods Mol. Biol. 2015;1262:321–331. doi: 10.1007/978-1-4939-2253-6_20. [DOI] [PubMed] [Google Scholar]
- 24.Vickers T.A., Crooke S.T. The rates of the major steps in the molecular mechanism of RNase H1-dependent antisense oligonucleotide induced degradation of RNA. Nucleic Acids Res. 2015;43:8955–8963. doi: 10.1093/nar/gkv920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vickers T.A., Crooke S.T. Antisense oligonucleotides capable of promoting specific target mRNA reduction via competing RNase H1-dependent and independent mechanisms. PLoS ONE. 2014;9:e108625. doi: 10.1371/journal.pone.0108625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang J., Lu Z., Wientjes M.G., Au J.L. Delivery of siRNA therapeutics: barriers and carriers. AAPS J. 2010;12:492–503. doi: 10.1208/s12248-010-9210-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gagnon K.T., Li L., Chu Y., Janowski B.A., Corey D.R. RNAi factors are present and active in human cell nuclei. Cell Rep. 2014;6:211–221. doi: 10.1016/j.celrep.2013.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lennox K.A., Behlke M.A. Cellular localization of long non-coding RNAs affects silencing by RNAi more than by antisense oligonucleotides. Nucleic Acids Res. 2016;44:863–877. doi: 10.1093/nar/gkv1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Castel S.E., Martienssen R.A. RNA interference in the nucleus: roles for small RNAs in transcription, epigenetics and beyond. Nat. Rev. Genet. 2013;14:100–112. doi: 10.1038/nrg3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kalantari R., Chiang C.M., Corey D.R. Regulation of mammalian transcription and splicing by Nuclear RNAi. Nucleic Acids Res. 2016;44:524–537. doi: 10.1093/nar/gkv1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ploner A., Ploner C., Lukasser M., Niederegger H., Hüttenhofer A. Methodological obstacles in knocking down small noncoding RNAs. RNA. 2009;15:1797–1804. doi: 10.1261/rna.1740009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cazenave C., Frank P., Toulme J.J., Büsen W. Characterization and subcellular localization of ribonuclease H activities from Xenopus laevis oocytes. J. Biol. Chem. 1994;269:25185–25192. [PubMed] [Google Scholar]
- 33.Hasselblatt P., Hockenjos B., Thoma C., Blum H.E., Offensperger W.B. Translation of stable hepadnaviral mRNA cleavage fragments induced by the action of phosphorothioate-modified antisense oligodeoxynucleotides. Nucleic Acids Res. 2005;33:114–125. doi: 10.1093/nar/gki155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kamola P.J., Kitson J.D., Turner G., Maratou K., Eriksson S., Panjwani A., Warnock L.C., Douillard Guilloux G.A., Moores K., Koppe E.L. In silico and in vitro evaluation of exonic and intronic off-target effects form a critical element of therapeutic ASO gapmer optimization. Nucleic Acids Res. 2015;43:8638–8650. doi: 10.1093/nar/gkv857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Freier S.M., Watt A.T. Basic principles of antisense drug discovery. In: Crooke S.T., editor. Antisense Drug Technology: Principles, Strategies, and Applications. Second Edition. CRC Press; 2008. pp. 117–141. [Google Scholar]
- 36.Hodges D., Crooke S.T. Inhibition of splicing of wild-type and mutated luciferase-adenovirus pre-mRNAs by antisense oligonucleotides. Mol. Pharmacol. 1995;48:905–918. [PubMed] [Google Scholar]
- 37.Lima W.F., De Hoyos C.L., Liang X.H., Crooke S.T. RNA cleavage products generated by antisense oligonucleotides and siRNAs are processed by the RNA surveillance machinery. Nucleic Acids Res. 2016;44:3351–3363. doi: 10.1093/nar/gkw065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nagarajan V.K., Jones C.I., Newbury S.F., Green P.J. XRN 5′→3′ exoribonucleases: structure, mechanisms and functions. Biochim. Biophys. Acta. 2013;1829:590–603. doi: 10.1016/j.bbagrm.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Castanotto D., Lin M., Kowolik C., Wang L., Ren X.Q., Soifer H.S., Koch T., Hansen B.R., Oerum H., Armstrong B. A cytoplasmic pathway for gapmer antisense oligonucleotide-mediated gene silencing in mammalian cells. Nucleic Acids Res. 2015;43:9350–9361. doi: 10.1093/nar/gkv964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Clark M.B., Johnston R.L., Inostroza-Ponta M., Fox A.H., Fortini E., Moscato P., Dinger M.E., Mattick J.S. Genome-wide analysis of long noncoding RNA stability. Genome Res. 2012;22:885–898. doi: 10.1101/gr.131037.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oeffinger M., Zenklusen D. To the pore and through the pore: a story of mRNA export kinetics. Biochim. Biophys. Acta. 2012;1819:494–506. doi: 10.1016/j.bbagrm.2012.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ghosh S., Jacobson A. RNA decay modulates gene expression and controls its fidelity. Wiley Interdiscip. Rev. RNA. 2010;1:351–361. doi: 10.1002/wrna.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bahar Halpern K., Caspi I., Lemze D., Levy M., Landen S., Elinav E., Ulitsky I., Itzkovitz S. Nuclear retention of mRNA in mammalian tissues. Cell Rep. 2015;13:2653–2662. doi: 10.1016/j.celrep.2015.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schwanhäusser B., Busse D., Li N., Dittmar G., Schuchhardt J., Wolf J., Chen W., Selbach M. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 45.Tani H., Nakamura Y., Ijiri K., Akimitsu N. Stability of MALAT-1, a nuclear long non-coding RNA in mammalian cells, varies in various cancer cells. Drug Discov. Ther. 2010;4:235–239. [PubMed] [Google Scholar]
- 46.Petfalski E., Dandekar T., Henry Y., Tollervey D. Processing of the precursors to small nucleolar RNAs and rRNAs requires common components. Mol. Cell. Biol. 1998;18:1181–1189. doi: 10.1128/mcb.18.3.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Allmang C., Kufel J., Chanfreau G., Mitchell P., Petfalski E., Tollervey D. Functions of the exosome in rRNA, snoRNA and snRNA synthesis. EMBO J. 1999;18:5399–5410. doi: 10.1093/emboj/18.19.5399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu H., Yin Q.F., Luo Z., Yao R.W., Zheng C.C., Zhang J., Xiang J.F., Yang L., Chen L.L. Unusual processing generates SPA LncRNAs that sequester multiple RNA binding proteins. Mol. Cell. 2016;64:534–548. doi: 10.1016/j.molcel.2016.10.007. [DOI] [PubMed] [Google Scholar]
- 49.Yang P.K., Hoareau C., Froment C., Monsarrat B., Henry Y., Chanfreau G. Cotranscriptional recruitment of the pseudouridylsynthetase Cbf5p and of the RNA binding protein Naf1p during H/ACA snoRNP assembly. Mol. Cell. Biol. 2005;25:3295–3304. doi: 10.1128/MCB.25.8.3295-3304.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Richard P., Kiss T. Integrating snoRNP assembly with mRNA biogenesis. EMBO Rep. 2006;7:590–592. doi: 10.1038/sj.embor.7400715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Westholm J.O., Lai E.C. Mirtrons: microRNA biogenesis via splicing. Biochimie. 2011;93:1897–1904. doi: 10.1016/j.biochi.2011.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Marnef A., Jády B.E., Kiss T. Human polypyrimidine tract-binding protein interacts with mitochondrial tRNA(Thr) in the cytosol. Nucleic Acids Res. 2016;44:1342–1353. doi: 10.1093/nar/gkv1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wilusz J.E., Freier S.M., Spector D.L. 3′ end processing of a long nuclear-retained noncoding RNA yields a tRNA-like cytoplasmic RNA. Cell. 2008;135:919–932. doi: 10.1016/j.cell.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Muta T., Kang D., Kitajima S., Fujiwara T., Hamasaki N. p32 protein, a splicing factor 2-associated protein, is localized in mitochondrial matrix and is functionally important in maintaining oxidative phosphorylation. J. Biol. Chem. 1997;272:24363–24370. doi: 10.1074/jbc.272.39.24363. [DOI] [PubMed] [Google Scholar]
- 55.Brokstad K.A., Kalland K.H., Russell W.C., Matthews D.A. Mitochondrial protein p32 can accumulate in the nucleus. Biochem. Biophys. Res. Commun. 2001;281:1161–1169. doi: 10.1006/bbrc.2001.4473. [DOI] [PubMed] [Google Scholar]
- 56.Tarze A., Deniaud A., Le Bras M., Maillier E., Molle D., Larochette N., Zamzami N., Jan G., Kroemer G., Brenner C. GAPDH, a novel regulator of the pro-apoptotic mitochondrial membrane permeabilization. Oncogene. 2007;26:2606–2620. doi: 10.1038/sj.onc.1210074. [DOI] [PubMed] [Google Scholar]
- 57.Hori S., Yamamoto T., Obika S. XRN2 is required for the degradation of target RNAs by RNase H1-dependent antisense oligonucleotides. Biochem. Biophys. Res. Commun. 2015;464:506–511. doi: 10.1016/j.bbrc.2015.06.171. [DOI] [PubMed] [Google Scholar]
- 58.Leung E., Brown J.D. Biogenesis of the signal recognition particle. Biochem. Soc. Trans. 2010;38:1093–1098. doi: 10.1042/BST0381093. [DOI] [PubMed] [Google Scholar]
- 59.Nagai K., Oubridge C., Kuglstatter A., Menichelli E., Isel C., Jovine L. Structure, function and evolution of the signal recognition particle. EMBO J. 2003;22:3479–3485. doi: 10.1093/emboj/cdg337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kasuya T., Hori S., Watanabe A., Nakajima M., Gahara Y., Rokushima M., Yanagimoto T., Kugimiya A. Ribonuclease H1-dependent hepatotoxicity caused by locked nucleic acid-modified gapmer antisense oligonucleotides. Sci. Rep. 2016;6:30377. doi: 10.1038/srep30377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Su Lim C., Sun Kim E., Yeon Kim J., Taek Hong S., Jai Chun H., Eun Kang D., Rae Cho B. Measurement of the nucleus area and nucleus/cytoplasm and mitochondria/nucleus ratios in human colon tissues by dual-colour two-photon microscopy imaging. Sci. Rep. 2015;5:18521. doi: 10.1038/srep18521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tripathi V., Ellis J.D., Shen Z., Song D.Y., Pan Q., Watt A.T., Freier S.M., Bennett C.F., Sharma A., Bubulya P.A. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol. Cell. 2010;39:925–938. doi: 10.1016/j.molcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Moelling K., Broecker F. The reverse transcriptase-RNase H: from viruses to antiviral defense. Ann. N Y Acad. Sci. 2015;1341:126–135. doi: 10.1111/nyas.12668. [DOI] [PubMed] [Google Scholar]
- 64.Kramer E.B., Hopper A.K. Retrograde transfer RNA nuclear import provides a new level of tRNA quality control in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 2013;110:21042–21047. doi: 10.1073/pnas.1316579110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Will C.L., Lührmann R. Spliceosomal UsnRNP biogenesis, structure and function. Curr. Opin. Cell Biol. 2001;13:290–301. doi: 10.1016/s0955-0674(00)00211-8. [DOI] [PubMed] [Google Scholar]
- 66.Krecic A.M., Swanson M.S. hnRNP complexes: composition, structure, and function. Curr. Opin. Cell Biol. 1999;11:363–371. doi: 10.1016/S0955-0674(99)80051-9. [DOI] [PubMed] [Google Scholar]
- 67.Kiss T., Fayet E., Jády B.E., Richard P., Weber M. Biogenesis and intranuclear trafficking of human box C/D and H/ACA RNPs. Cold Spring Harb. Symp. Quant. Biol. 2006;71:407–417. doi: 10.1101/sqb.2006.71.025. [DOI] [PubMed] [Google Scholar]
- 68.Huber M.D., Gerace L. The size-wise nucleus: nuclear volume control in eukaryotes. J. Cell Biol. 2007;179:583–584. doi: 10.1083/jcb.200710156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lima W.F., Vickers T.A., Nichols J., Li C., Crooke S.T. Defining the factors that contribute to on-target specificity of antisense oligonucleotides. PLoS ONE. 2014;9:e101752. doi: 10.1371/journal.pone.0101752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Liang X.H., Sun H., Shen W., Crooke S.T. Identification and characterization of intracellular proteins that bind oligonucleotides with phosphorothioate linkages. Nucleic Acids Res. 2015;43:2927–2945. doi: 10.1093/nar/gkv143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liang X.H., Crooke S.T. RNA helicase A is not required for RISC activity. Biochim. Biophys. Acta. 2013;1829:1092–1101. doi: 10.1016/j.bbagrm.2013.07.008. [DOI] [PubMed] [Google Scholar]
- 72.Liang X.H., Liu Q., Fournier M.J. rRNA modifications in an intersubunit bridge of the ribosome strongly affect both ribosome biogenesis and activity. Mol. Cell. 2007;28:965–977. doi: 10.1016/j.molcel.2007.10.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.