Abstract
Influenza A virus infection causes substantial morbidity and mortality in seasonal epidemic outbreaks, and more efficient treatments are urgently needed. Innate immune sensing of viral nucleic acids stimulates antiviral immunity, including cell-autonomous antiviral defense mechanisms that restrict viral replication. RNA oligonucleotide ligands that potently activate the cytoplasmic helicase retinoic-acid-inducible gene I (RIG-I) are promising candidates for the development of new antiviral therapies. Here, we demonstrate in an Mx1-expressing mouse model of influenza A virus infection that a single intravenous injection of low-dose RIG-I ligand 5′-triphosphate RNA (3pRNA) completely protected mice from a lethal challenge with influenza A virus for at least 7 days. Furthermore, systemic administration of 3pRNA rescued mice with pre-established fulminant influenza infection and prevented the fatal effects of a streptococcal superinfection. Type I interferon, but not interferon-λ, was required for the therapeutic effect. Our results suggest that the use of RIG-I activating oligonucleotide ligands has the clinical potential to confine influenza epidemics when a strain-specific vaccine is not yet available and to reduce lethality of influenza in severely infected patients.
Keywords: negative strand RNA virus, immunostimulatory oligonucleotides, RIG-I, innate immunity, influenza virus, antivirals, immunotherapy, 5’ triphosphate RNA, type I interferon, type III interferon
Coch, Hartmann, and colleagues show that systemic activation of the innate immune sensor RIG-I, a cytoplasmic helicase, with its specific ligand 5′-triphosphate RNA effectively protects mice expressing Mx1 from a lethal challenge with influenza A virus in both prophylactic and therapeutic settings, including additional challenge with bacterial superinfection.
Introduction
Influenza A virus causes severe respiratory infections, and worldwide pandemics occur as in 2009.1, 2 Current prophylactic vaccines and treatments bear significant limitations: vaccines induce antigen-specific immunity to hemagglutinin antigen, which is highly variable in different influenza A virus subtypes in humans and animals. Genomic variation through mutations or through reassortment of different genome segments (antigenic shift and drift) allows the virus to escape pre-established memory responses and requires a yearly adaptation of the commercial vaccine still with limited efficacy.3 Moreover, genetic instability leads to widespread resistance against conventional antiviral drugs targeting specific viral components (neuraminidase inhibitors and M2-proton channel inhibitors).4, 5 Therapeutic efficacy of neuraminidase and M2-protein inhibitors remains controversial.6 Therefore, new therapeutic principles that are less susceptible to mutational escape by the virus are highly desired.
The innate immune system provides an important barrier for viruses. A first barrier is the mucus of the respiratory epithelium containing defensins and cathelicidins. Once the virus enters cells, viral nucleic acids are detected by pattern recognition receptors, which trigger a signaling cascade that potently inhibits viral replication and in many cases eliminates the virus before a severe infection develops. Activation of nucleic-acid-sensing pattern recognition receptors (Toll-like receptor 3 [TLR3], TLR7, TLR8, TLR9) has been explored as a prophylactic strategy against influenza A virus replication in animal models.7, 8, 9 However, TLRs are expressed mainly on immune cell subsets, but not in target cells of viruses, such as epithelial cells. In contrast to TLRs, retinoic-acid-inducible gene I (RIG-I)-like receptors (RLRs; RIG-I, MDA5 [melanoma differentiation antigen 5], and LGP2 [Laboratory of Genetics and Physiology 2]) detect viral RNA in the cytosol of all somatic cells that are targeted by viruses, including epithelial cells.10, 11 Activation of RIG-I-like receptors in a cell before viral entry induces a broad spectrum of antiviral activities that provide potent protection of this cell from viral replication.
Because the RIG-I-like receptor MDA5 is activated by RNA ligand structures containing long, irregular double-stranded RNA (dsRNA), and because such dsRNA also activates a number of additional pathways, including TLR3 on endothelial cells, protein kinase R (PKR), oligoadenylate synthetase 1 (OAS1), and the inflammasome, the clinical use of MDA5 ligands such as polyinosinic:polycytidylic acid [poly(I:C)] is limited by toxicity.12, 13 In contrast, RIG-I can be selectively activated by short double-stranded blunt end 5′-triphosphate RNA (3pRNA).14, 15 Upon activation, RIG-I induces the expression of a set of antiviral cytokines such as type I interferon and type III interferon (interferon-λ), and upregulates the expression of antiviral genes with cell-autonomous antiviral effector functions (e.g., Mx proteins, interferon induced protein with tetratricopeptide repeats 1 [IFIT1], PKR, OAS1, and adenosine deaminase, RNA specific [ADAR]).16, 17
Influenza A virus is a member of the Orthomyxoviridae family of RNA viruses with a single-stranded negative sense RNA genome that forms blunt-end 5′-triphosphate panhandle structures that in principle are detected by RIG-I. It has been demonstrated that activation of RIG-I is critical to mount an effective antiviral immune response in the course of an influenza virus infection.16, 18 However, like all pathogenic negative strand RNA viruses, influenza A virus has evolved strategies to counteract detection by RIG-I. The non-structural protein 1 (NS1) of influenza A virus potently inhibits RIG-I activation and signaling.19, 20, 21 As a consequence, once a cell is infected by influenza A virus, RIG-I becomes non-functional with regard to virus detection and activation of antiviral effector mechanisms. However, if cells are preactivated by synthetic RIG-I ligands, they are protected. The rationale for RIG-I ligand treatment of influenza A virus infection is the protection of yet uninfected cells in vivo, thereby restricting viral spread from cell to cell. This is obviously the situation in a prophylactic setting where RIG-I activation occurs before viral infection, but RIG-I activation may also be effective in the course of an ongoing viral infection when the virus has not yet infected all potential target cells. Thus, therapeutic administration of a synthetic RIG-I ligand may substitute for insufficient innate immune activation by influenza A virus due to immune escape from innate immunorecognition.
RIG-I stimulation in the context of influenza A virus has been reported in the literature.22, 23, 24, 25, 26 However, in vivo data are limited because mostly surrogate parameters such as viral load rather than survival were used as endpoints and Mx1-negative mouse strains (C57BL/6 or BALB/c) were used. Mx proteins are interferon-induced antiviral proteins that interfere with virus replication in the cell at several levels and are highly conserved among vertebrates.27 Here, we make use of Mx1-positive B6.A2G-Mx1 mice, which in contrast to the often used Mx1-negative C57BL/6 strain more closely resemble the clinical situation in humans. We demonstrate that prophylactic treatment of Mx1-positive mice with a small dose of RIG-I agonist completely protects from an otherwise lethal challenge with influenza A virus for a minimum of 7 days prior to challenge. Furthermore, RIG-I ligand treatment up to 30 hr after infection still rescued mice from a lethal course of infection. We also found that systemic RIG-I ligand treatment improved the survival of influenza A virus-infected mice that were additionally challenged by bacterial superinfection, a major complication well-known to be responsible for influenza-associated morbidity and mortality in patients.
Results
Systemic Activation of RIG-I by Intravenous 3pRNA Application Induces CXCL10 in Lung Tissue and Ameliorates the Course of a Non-lethal Influenza A Virus Infection
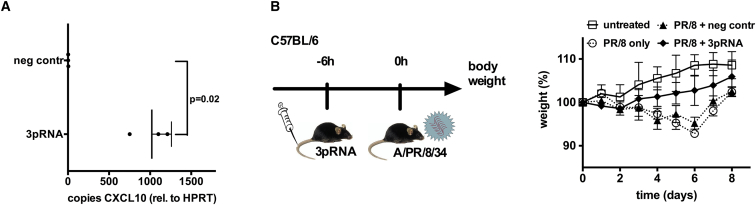
To investigate whether RIG-I activation protects from influenza A virus infection in vivo, 3pRNA was complexed to in vivo jetPEI and administered to C57BL/6 mice intravenously. The dosage was adjusted as a result of our previous in vitro data (data not shown). At 6 hr after injection, high levels of the type I interferon-stimulated gene CXCL10 mRNA were detected in lung tissue, the primary target tissue of influenza virus (Figure 1A). Next, C57BL/6 mice were intravenously injected with 3pRNA and after 6 hr they received a non-lethal dose of influenza virus A/PR/8/34. 3pRNA-treated mice demonstrated a milder clinical course of the infection as indicated by body weight (Figure 1B). Of note, protection in this setting occurred despite the absence of Mx1 protein, which is the mouse homolog of human MxA.28, 29 Mx1 in mice and MxA in humans are important antiviral effectors in the type I interferon pathway, and Mx1-positive mice represent a model that is much closer to the human situation.30, 31 Therefore, in all subsequent studies, we used Mx1-positive congenic B6.A2G-Mx1 mice as a well-established in vivo model of influenza infection.28, 32
Figure 1.
Systemic 3pRNA Induces CXCL10 in the Lungs and Ameliorates the Course of Non-lethal Influenza Virus Infection
(A) C57BL/6 mice were i.v. injected with 25 μg of 3pRNA or the control RNA polycytidylic, polyadenylic acid (polyCA). After 6 hr, expression of CXCL10 in lung tissue was analyzed by qPCR (n = 3 mice). Result shows mean with SD. (B) Left panel shows the experimental setup. Right panel shows that C57BL/6 mice were i.v. injected with 25 μg of 3pRNA or the control RNA polyCA 6 hr before intranasal infection with a non-lethal dose (105 PFU) A/PR/8/34. Body weight of mice was monitored daily. Results show the means and SD of n = 6 mice (ANOVA day 6: PR8 only versus PR8 + control [ctrl] RNA, not significant; PR8 only versus PR8 + 3pRNA, p < 0.01; PR8 + ctrl RNA versus PR8 + 3pRNA, p < 0.05).
Systemic 3pRNA Leads to Long-Term Protection of Mice from Lethal Influenza Challenge
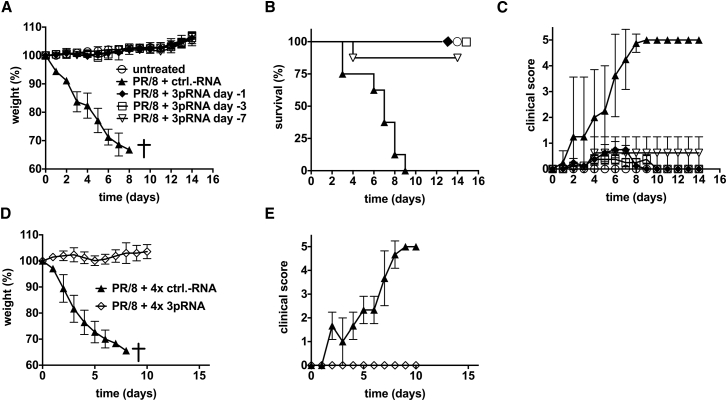
To study the time frame of protection, B6.A2G-Mx1 mice received a single injection of a low dose of 3pRNA (12.5 μg), 3 or 7 days before mice were challenged with a lethal dose of a highly virulent PR/8 variant (hvPR/8) that is replicating faster than the A/PR/8/34 strain.28 Although all mice without 3pRNA pre-treatment showed severe signs of infection and had to be sacrificed within 10 days (Figures 2A–2C), mice pre-treated with a single low dose of intravenous 3pRNA showed no weight loss or increased clinical disease score. 3pRNA pre-treatment resulted in an almost complete protection of mice for at least 7 days.
Figure 2.
Prophylactic Treatment with 3pRNA Leads to Long-Term Protection of Mice against a Lethal Challenge with Influenza Virus
B6.A2G-Mx1 mice received a single i.v. injection of 12.5 μg of 3pRNA at 1, 3, or 7 days before they were challenged with a lethal dose of 103 PFU of hvPR/8, a highly virulent variant of A/PR/8/34 (n = 8; untreated: n = 5). (A–C) Body weight (A), survival (B) (log-rank [Mantel-Cox] test: control [ctrl]-RNA versus PR/8 + 3pRNA day −3, p < 0.0001; ctrl-RNA versus PR/8 + 3pRNA day −7, p < 0.0009; PR/8 + 3pRNA day −7 versus PR/8 + 3pRNA day −3, p < 0.32), and clinical score (C) were monitored. (D and E) Mice received repeated i.v. injections of 3pRNA or control RNA on days 7, 5, 3, and 1 before they were challenged with a lethal dose of 103 PFU of hvPR/8. (D and E) Body weight (D) and clinical score (E) were measured (n = 8; ctrl-RNA: n = 3). Results are presented as mean values. Error bars represent the SD.
Repeated exposure to innate stimuli often leads to desensitization, which is a well-known phenomenon. Because repeated administration over a longer period of time would be required in case 3pRNA is used as prophylaxis against influenza, we investigated whether such repeated administration of 3pRNA would result in weaker protection. B6.A2G-Mx1 mice were intravenously injected four times with 3pRNA (days 7, 5, 3, and 1) before they were challenged with a lethal dose of influenza virus. Mice were still completely protected, indicating that repeated stimulation of RIG-I does not lead to desensitization (Figures 2D and 2E). Furthermore, repeated administration of 3pRNA in vivo was well tolerated with no clinical signs of side effects. There was no decrease in body weight and a normal clinical score at day of infection after repeated 3pRNA administration (Figures 2D and 2E; data not shown; body weight in grams; day −7 versus day 0; mean, SEM: 3pRNA-treated mice 23.4 ± 0.7 versus 23.6 ± 0.5 and non-3pRNA-treated mice 23.9 ± 1.3 versus 23.9 ± 1.4).
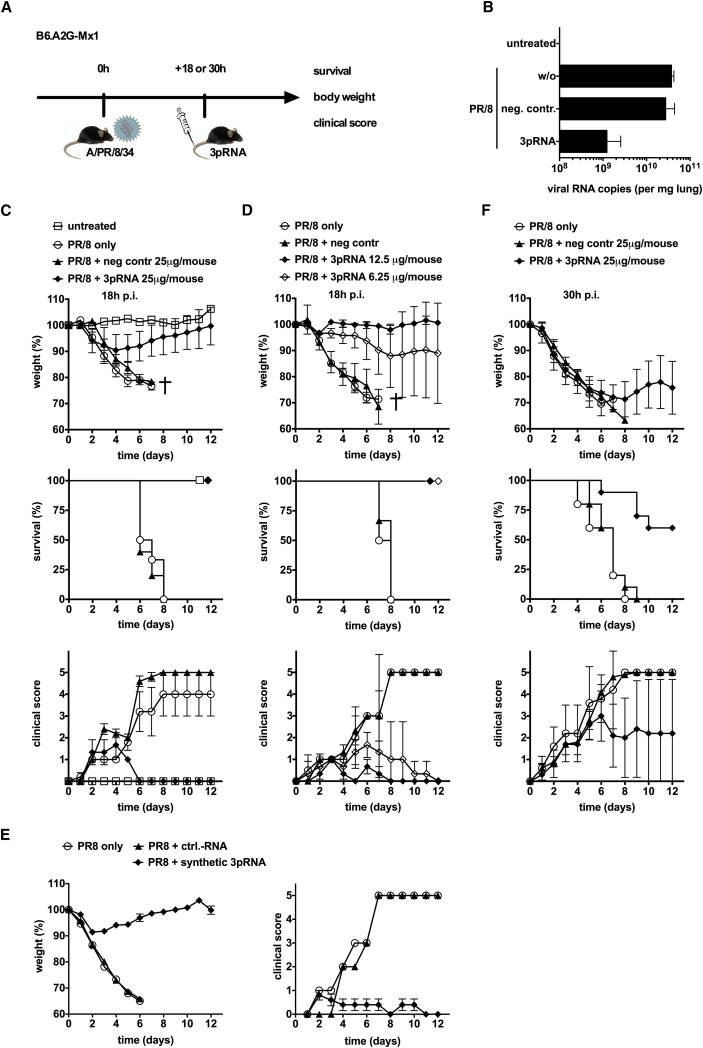
Systemic 3pRNA Rescues Mice from an Ongoing Lethal Influenza Virus Infection
Once the virus has successfully entered the epithelium, the protection of yet uninfected cells may still ameliorate the course of the infection. To study the therapeutic activity of RIG-I activation in pre-established infection, B6.A2G-Mx1 mice received a single intravenous injection of a low dose of 3pRNA (12.5 μg per injection) at 18 hr postinfection with a lethal dose of hvPR/8 (Figure 3A). 3pRNA, but not control RNA, strongly reduced the viral load in the lungs of mice on day 5 postinfection (Figure 3B). All mice receiving 3pRNA 18 hr after infection were rescued, whereas none of the mice in the control groups survived (Figure 3C). 3pRNA-treated infected mice showed a minor drop in body weight and a low clinical disease score compared with mice treated with control RNA that experienced a pronounced weight loss and a high clinical score (Figure 3C). This therapeutic effect was dose dependent, with the lowest dose used (6.25 μg per injection) still completely protecting mice from a fatal course of the infection (Figure 3D). Similar therapeutic activity was observed with a short synthetic 5′-triphosphate dsRNA oligonucleotide (Figure 3E). When the start of 3pRNA treatment was delayed to 30 hr after infection (instead of 18 hr), the therapeutic effect indicated by weight loss and clinical score was reduced, but survival rate was still at 60% compared with the untreated mice that all died (Figure 3F).
Figure 3.
Therapeutic Administration of 3pRNA Rescues Mice from a Pre-established Lethal Influenza Virus Infection
(A) Experimental setup: i.v. 3pRNA treatment at 18 or 30 hr after challenge with influenza virus. (B) B6.A2G-Mx1 mice were infected with 103 PFU of hvPR/8. 18 hr later, 3pRNA or control RNA was injected i.v. (both at 25 μg). Viral RNA was analyzed in lung tissue at day 5 postinfection (n = 4 mice). (C) B6.A2G-Mx1 mice were treated as in (B). Body weight (n = 6 mice; untreated: n = 2), survival (n = 6 mice; untreated: n = 2), and clinical score (n = 5 mice; PR8+3pRNA: n = 3; untreated: n = 2) are depicted (log-rank [Mantel-Cox test]: PR/8 only versus PR/8 + control [ctrl]-RNA, p = 0.65; PR/8 only versus PR/8 + 3pRNA, p = 0.0016; PR/8 + ctrl-RNA versus PR/8 + 3pRNA, p = 0.0015). (D) Mice were treated as in (B) except that 3pRNA was used at a dose of 12.5 and 6.25 μg per injection (means of n = 10 mice; PR/8 without treatment, n = 5 [log-rank (Mantel-Cox) test: PR/8 + ctrl-RNA versus PR/8 + 6.25 3pRNA, p = 0.03; PR/8 + ctrl-RNA versus PR/8 + 12.5 3pRNA, p = 0.03; PR/8 + 12.5 3pRNA versus PR/8 + 6.25 3pRNA, p = 1]). (E) Done as in (C), but chemically synthesized 3pRNA was used (n = 5). (F) Mice were treated as in (B) except injection of 3pRNA was at 30 hr postinfection (log-rank [Mantel-Cox] test: PR/8 only versus PR/8 + ctrl-RNA, p = 0.66; PR/8 only versus PR/8 + 3pRNA, p = 0.0005; PR/8 + ctrl-RNA versus PR/8 + 3pRNA, p = 0.0002). Results are presented as mean values. Error bars represent the SD.
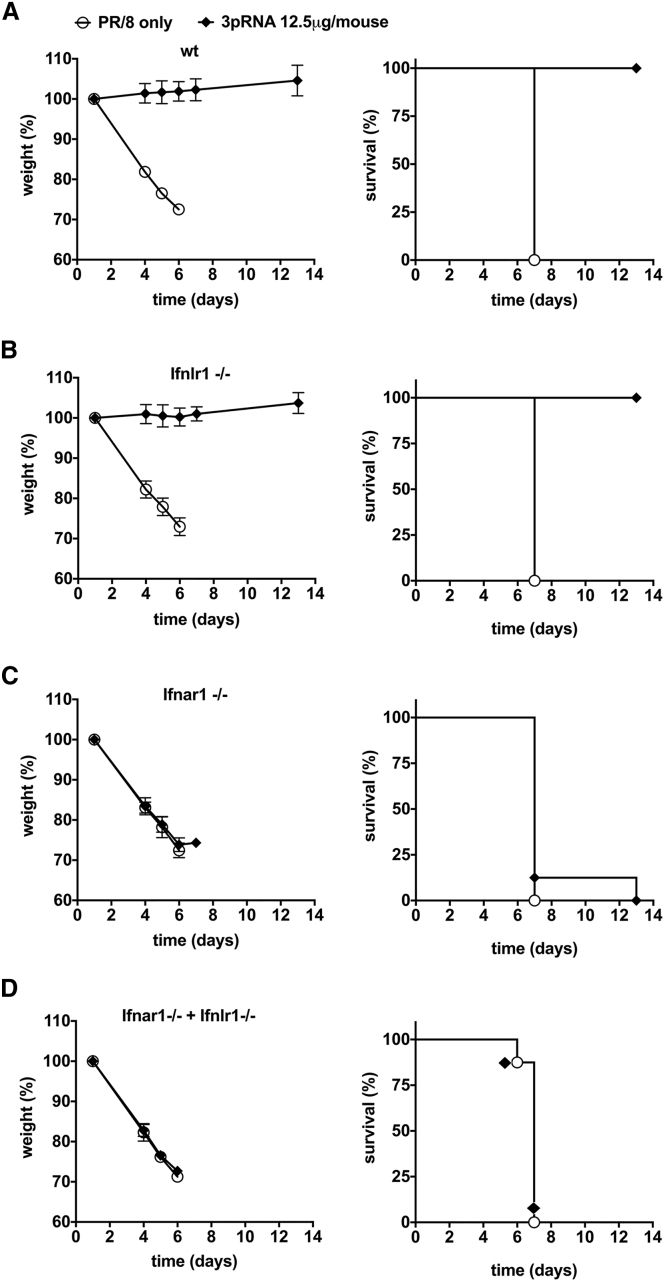
Therapeutic Activity of 3pRNA in Influenza Virus Infection Requires Type I Interferon, but Not Interferon-λ
Interferon-λ has been reported to be involved in the antiviral response against respiratory viruses, including influenza.33, 34 Whereas type I interferons all bind to the same type I interferon receptor, interferon-λ binds to a distinct receptor, IFNLR1. To examine the contribution of interferon-λ versus type I interferon, we studied B6.A2G-Mx1 mice that lack functional IFNLR (B6.A2G-Mx1-Ifnlr1−/− mice), interferon-α and -β receptor subunit 1 (IFNAR1) (B6.A2G-Mx1-Ifnar−/−), or both (B6.A2G-Mx1-Ifnar−/− Ifnlr−/−) (Figure 4). Mice were infected with a lethal dose of hv/PR/8 and 18 hr later intravenously injected with 12.5 μg of 3pRNA or control RNA. Body weight, survival, and clinical score were monitored. Only mice with functional IFNAR1 showed the 3pRNA-mediated survival benefit, whereas the presence of functional IFNLR was dispensable, indicating that in the absence of type I interferon function, RIG-I-induced interferon-λ is not sufficient to improve the clinical course of infection.
Figure 4.
Rescue from Lethal Influenza Virus Infection by Therapeutic Administration of 3pRNA Requires the Type I Interferon Receptor but Is Independent of Interferon-λ
(A–D) B6.A2G-Mx1 mice (A) (n = 8, p = 0.0003), B6.A2G-Mx1-Ifnlr1−/− mice lacking functional interferon-λ receptors (B) (n = 6, p = 0.0009), B6.A2G-Mx1-Ifnar1−/− mice lacking functional type I interferon receptors (C) (n = 8, p = 0.35), or B6.A2G-Mx1-Ifnar1−/−Ifnlr1−/− double-knockout mice lacking receptors for both interferon types (D) (n = 8, p = 1.00) were infected with 103 PFU hvPR/8. After 18 hr, mice were i.v. injected with 12.5 μg of 3pRNA or the control RNA polyCA. Body weight and survival were monitored. Results are presented as mean values. Error bars represent the SD. Log-rank (Mantel-Cox) test was used.
Comparison of 3pRNA with Established Oligonucleotide Ligands of TLR7/8 and TLR9
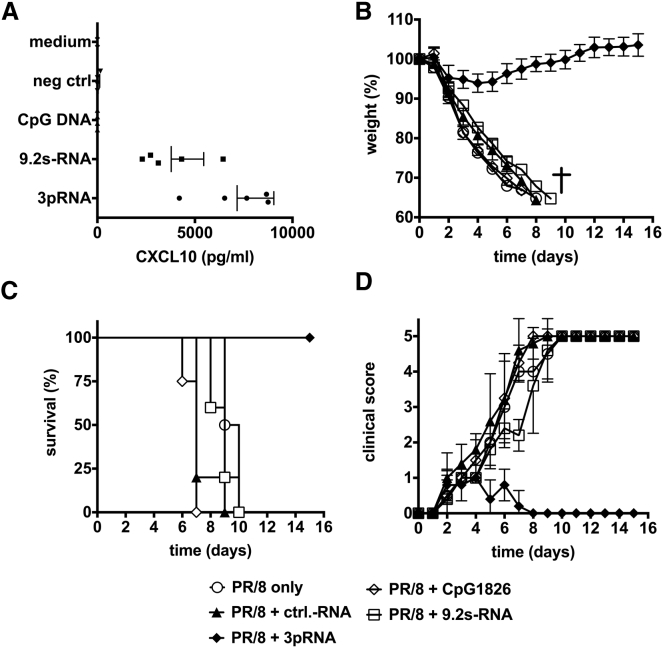
Activation of TLR7/8 and TLR9 has been described to provide protection against subsequent influenza virus infection.7, 8, 9 Because expression of these TLRs is mostly restricted to immune cells, the lack of direct protection of yet uninfected epithelial cells by TLR ligands may limit the therapeutic use of TLR ligands as compared with RIG-I ligands that directly induce antiviral activity in epithelial cells. Therefore, we compared the therapeutic activity of RIG-I activation with TLR7/8 and TLR9 activation in the therapeutic setting of influenza A virus infection. The TLR7/8 agonist 9.2 s-RNA35, 36 was complexed with N-[1-(2,3-dioleoyloxy)propyl]-N,N,N-trimethylammonium methyl-sulfate (DOTAP) and administered intravenously (i.v.) as described before.37 The TLR9 ligand CpG1826 was injected subcutaneously as previously established to treat influenza infection in mouse models and as approved for clinical trials.38, 39 First, we analyzed induction of CXCL10 in the serum as a systemic marker of a type I interferon response (Figure 5A). Although both the TLR7/8 ligand (9.2 s-RNA) and the RIG-I ligand (3pRNA) induced considerable amounts of CXCL10 in the serum of mice, the TLR9 ligand (CpG1826) did not induce systemic levels of CXCL10. Next, the three ligands were administered to mice at 18 hr after a lethal challenge with influenza A virus. Whereas the RIG-I-stimulating ligand 3pRNA showed the expected amelioration of the clinical course of infection, no effect was seen for the TLR9 ligand CpG1826. Furthermore, despite the induction of substantial levels of CXCL10 in the serum, the TLR7/8 ligand 9.2 s-RNA showed no improvement of clinical signs (Figures 5B–5D).
Figure 5.
Comparison of 3pRNA with Established Oligonucleotide Ligands of TLR7/8 and TLR9
B6.A2G-Mx1 mice were infected with 103 PFU of hvPR/8. 18 hr later, mice were i.v. injected with 3pRNA or the control RNA polyCA (both delivered with jetPEI), i.v. injected with the TLR7/8 ligand 9.2s RNA (delivered with DOTAP), or i.p. injected with CpG1826 (all 12.5 μg per injection). Infected mice without treatment served as control (PR/8 only). (A) At 6 hr after treatment, levels of CXCL10 were analyzed in serum of mice by ELISA (ANOVA: 3pRNA versus negative control [ctrl], p < 0.01; 3pRNA versus 9.2 s, p < 0.05; 9.2 s versus negative ctrl, p < 0.05). (B–D) Body weight (B), survival (log-rank [Mantel-Cox] test: PR/8 + ctrl-RNA versus PR/8 + 3pRNA, p = 0.0016; PR/8 + ctrl-RNA versus PR/8 + 9.2s RNA, p = 0.065; PR/8 + 3pRNA versus PR/8 + 9.2s RNA, p = 0.0019) (C), and clinical score (D) were monitored daily. Results show means of n = 4 or n = 5 animals and n = 2 for PR/8 only. Results are presented as survival curve or mean values with SD.
Systemic RIG-I Activation by 3pRNA Improves Outcome of Influenza Virus Infection with Bacterial Superinfection
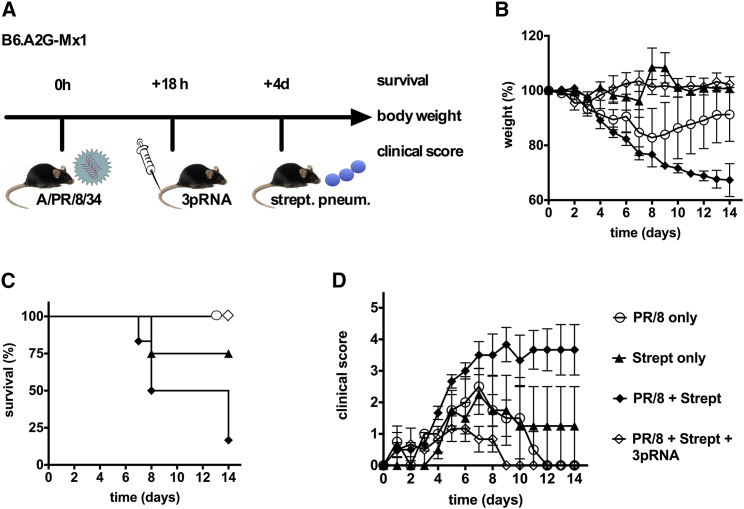
Bacterial superinfection causes severe complications during influenza virus infection.40, 41, 42 It has been proposed that induction of type I interferon may aggravate bacterial superinfection.43, 44 Although this point remains controversial,45 RIG-I-induced type I interferon may even promote bacterial superinfection. To evaluate the utility of 3pRNA in the case of bacterial superinfection, we infected B6.A2G-Mx mice with a non-lethal dose of influenza virus and 18 hr later treated them with 3pRNA. Four days after viral infection, mice were additionally challenged by superinfection with S. pneumoniae (Figure 6A). In this model, bacterial infection alone did not cause a substantial weight loss (Figure 6B, closed triangles), but it aggravated the weight loss induced by the non-lethal dose of influenza virus (Figure 6B, open circles versus closed diamonds). Intravenous treatment with 3pRNA at 18 hr after influenza infection not only prevented the moderate weight loss induced by the non-lethal dose of influenza virus, but also the aggravation of weight loss following bacterial superinfection (Figure 6B). All mice treated with 3pRNA survived, whereas 80% of the mice exposed to a non-lethal dose of influenza virus followed by bacterial superinfection died (Figures 6C and 6D). Together, these data demonstrate that RIG-I does not facilitate or aggravate bacterial (super)infection in the course of influenza virus infection.
Figure 6.
Therapeutic Administration of 3pRNA Improves the Outcome of Influenza-Infected Mice Superinfected with S. pneumoniae
B6.A2G-Mx1 mice were infected with 50 PFU of hvPR/8. 18 hr later, mice received an i.v. injection of 25 μg of 3pRNA. At 4 days after influenza virus infection, mice were challenged with 4.4 × 106 CFU of S. pneumoniae (strain TIGR4). Mice infected with either influenza or S. pneumoniae but without 3pRNA treatment served as controls (PR/8 only, Streptococcus pneumonia [strept] only, and PR/8 + strept). (A) Experimental setup. (B) Body weight. (C) Survival (log-rank [Mantel-Cox] test: strept versus PR/8 + strept, p = 0.11; strept versus PR/8 + strept + 3pRNA, p = 0.22; PR/8 + strept versus PR/8 + strept + 3pRNA, p = 0.005). (D) Clinical score. Results show mean values of n = 4–6 mice. Error bars represent the SD.
Discussion
The different pathways of innate immune sensing of viral nucleic acids heavily restrict the evolution of pathogenic viruses. As a consequence, successful pathogenic viruses need to develop molecular strategies to escape from the detection of their viral nucleic acids by the innate immune system. The RNA polymerase-based replicative principle of most negative strand RNA viruses, including influenza virus, implicates the formation of RNA molecules with triphosphate groups at the 5′ end. Because this nucleic acid structure represents a pathogen-associated molecular pattern (PAMPs) recognized by RIG-I, negative strand RNA viruses have to counteract RIG-I sensing in the cytoplasm. The negative strand RNA virus influenza A virus employs different strategies to evade detection by RIG-I, including encapsidation of the nascent RNA.46, 47 Furthermore, influenza A virus targets the type I interferon system and the immunorecognition of its RNA by innate immune sensors. influenza A virus encodes the non-structural protein NS120, 48, 49 and the PB1-F2 protein,19, 50 which inhibit the induction of type I interferon by RIG-I while maintaining NF-κB activation supporting viral replication.20, 51, 52 This highlights the fundamental role of RIG-I in antiviral defense against influenza A virus. Because inhibition of RIG-I by NS1 occurs only in infected cells, the induction of RIG-I-induced antiviral activities in yet uninfected cells provides a strong rationale for the use of RIG-I ligands not only for prophylaxis, but also to ameliorate the clinical signs of an ongoing influenza virus infection.
Our results support this rationale and the clinical development RIG-I ligands for the treatment of influenza virus infection. First, we demonstrate that a prophylactic small dose of the RIG-I ligand 3pRNA completely protects the host from a subsequent lethal challenge with influenza A virus for an extended period of time. Second, the same small dose of 3pRNA profoundly ameliorates the clinical signs and secures survival of mice if administered in the course of an ongoing influenza A virus infection that otherwise causes 100% lethality. The data demonstrate that therapeutic application of the RIG-I ligand 3pRNA is highly effective for a period of at least 30 hr after viral challenge. Notably, unlike other innate pathways such as TLR4, we did not observe desensitization or tachyphylaxis of RIG-I activation upon repeated dosing of 3pRNA. Third, we demonstrate that, unexpectedly, type I interferons, but not interferon-λ, were required for the therapeutic activity. And fourth, RIG-I activation had a positive effect on the course of influenza virus infection even in the situation of bacterial superinfection.
Our results are consistent with several studies in the literature reporting an effect of RIG-I activation on influenza virus replication, but these studies have a different focus and some technical limitations. Studies in the literature are in vitro studies or studies on the prophylactic treatment.23, 24, 25 Furthermore, these studies used C57BL/6 mice lacking functional Mx1, the mouse homolog to human MxA,28, 29 an important antiviral effector in the type I interferon pathway.30, 31 In our study, we used Mx1-positive congenic B6.A2G-Mx1 mice that are well established to resemble the human situation much more closely than C57BL/6 mice lacking Mx1.28, 29 Using this highly relevant model, we for the first time provide evidence for a long-lasting prophylactic, as well as therapeutic effect, in a severe and lethal influenza virus infection setting. The effective prophylactic period of 7 days as examined in this study may be further prolongable. Future studies will have to determine which dosing and which intervals of RIG-I activation are optimal for a complete and permanent protection against influenza virus infection. Because, unlike in our study, the viral challenge with influenza in natural habitats usually is much lower, and non-lethal, sufficient protection may be achieved on a low-dose and long-interval application scheme. Furthermore, it will be interesting to study whether the i.v. route can be replaced by subcutaneous (s.c.) or inhaled administration of 3pRNA, which would be a more practical approach for routine prophylactic use on a population basis.
An important technical issue that needs to be considered is the identity of RIG-I ligands in different studies.22, 26 Generation of 3pRNA by in vitro transcription leads to the formation of unexpected additional complementary short RNA sequences resulting in an unpredictable mixture of undefined RIG-I-stimulatory and non-stimulatory sequences, unless the in-vitro-transcribed RNA products are adequately purified. In addition to adequate purification of in-vitro-transcribed RNA, we used a chemically well-defined synthetic 5′-triphosphate dsRNA RIG-I ligand (20-mer), which is too short to activate MDA5, in order to confirm that the therapeutic activity is not dependent on the formation of in vitro transcription-dependent unintended longer RNA by-products.
Although in our study type I interferon is clearly required for the therapeutic activity, type I interferon may not be sufficient. It has been proposed that interferon-λ, as another RIG-I-inducible cytokine, has antiviral properties that are complementary to type I interferon, especially in case of respiratory infections such as influenza virus.33, 34, 53 Although the type I interferon receptor is expressed at the surface of all cells, expression of the type III interferon receptor is primarily restricted to epithelial cells in the respiratory and gastrointestinal tracts.34, 54 This differential expression of the type III interferon receptor might generate an antiviral response in airway epithelium while preventing exaggerated activation of immune cells and associated lung pathology.54 Moreover, it has been demonstrated that type III interferon, and not type I interferon, is the predominant interferon induced by respiratory viruses in nasal epithelial cells, and that type III interferon, rather than type I interferon, represents the main first-line defense via the RIG-I-dependent pathway.16 Our results do not contradict these findings, but they demonstrate that upon actual RIG-I ligand treatment in the situation of an established influenza A virus infection, the activity of type I interferon dominates over type III interferon, and RIG-I-induced type III interferon does not contribute to the therapeutic activity.
There are conflicting reports about increased susceptibility to bacterial infection in the context of a type I interferon-dominated response.43, 44, 45, 55 Therefore, one might speculate that the protection from influenza virus infection by RIG-I may come at the expense of a higher risk to develop bacterial superinfection. In our study, we find that RIG-I treatment improved the overall clinical outcome of a combined infection with influenza virus and bacterial superinfection by Streptococcus pneumoniae. Thus, a potential aggravation of bacterial superinfection in the presence of RIG-I-induced type I interferon seems to be outweighed by the protective antiviral activity. On the other hand, RIG-I may even exhibit antibacterial activities. The experimental dissection of antiviral and antibacterial activities of therapeutic RIG-I activation is challenging because viral infection of epithelial cells and bacterial outgrowth are tightly intertwined. RIG-I activation may positively or negatively interfere with multiple mechanisms reported to promote bacterial superinfection in the context of influenza infection: impaired NK cell response,56 depletion of alveolar macrophages,57 suppressed phagocytic bacterial clearance,58 Setdb2-mediated crosstalk between the type I interferons and NF-κB pathways,59 and downregulation of IL12 p70.60 Interestingly, there is recent evidence from the literature that death in influenza viral infection is due to endogenous bacterial burden, even in the absence of additional exogenous bacterial challenge. Pillai and colleagues61 conclusively demonstrate that mice infected with influenza virus died of bacterial bloom in the lungs on the basis of virus-induced tissue damage. Their work showed that mortality is due to bacterial burden, caspase-1/11, and neutrophil-dependent tissue damage, and that mortality was reversed by a functional blockade of the inflammasome despite even enhanced viral replication. In our experimental setting it is likely that the reduced viral load as a consequence of 3pRNA treatment is associated with reduced virus-induced epithelial damage leading to reduced bacteria-induced lethality. The future characterization of potential antibacterial activities of RIG-I in the absence of viral infection requires an elaborate set of experiments that are specifically designed to answer this question.
The fact that enhanced RIG-I activity obviously provides a robust protection against infection with respiratory viruses provokes the question why RIG-I is not permanently activated by nature. One could speculate that increased sensitivity of the RIG-I pathway for prolonged periods of time has negative effects for the host, because this may increase the detection of endogenous RIG-I ligands, and thus may lead to a chronic activity resulting in generalized inhibition of translation of proteins contributing to the normal homeostasis of the cell. Consequently, fine-tuning of RIG-I function most likely is a trade between protection from viral infection and proper biological homeostasis. Therefore, prophylactic treatment with RIG-I ligands should be limited to times of enhanced viral threat.
In conclusion, with this work we establish the activation of the innate immune sensor RIG-I as a promising strategy to treat influenza virus infection in both therapeutic and prophylactic settings. Specifically, for influenza, this novel therapeutic strategy has the advantage of acting independently of the specific virus strain and independently of viral resistance mechanisms to other established targeted antiviral treatments. Furthermore, our results suggest that activation of RIG-I may be useful to limit outbreaks of infections with other newly emerging RNA viruses such as Ebola or Zika before vaccination becomes available.
Materials and Methods
Pathogens
The influenza A virus variant hvPR/8 is closely related to the Cambridge strain of A/PR/8/34 (H1N1),62 which is moderately pathogenic for Mx1+/+ mice and was generated by serial lung passages in Mx1+/+ mice.63 hvPR/8 is closely related to the Mount Sinai strain of A/PR/8/34,64 which is non-pathogenic for Mx1+/+ mice even at high doses. Virus stocks were produced in embryonated chicken eggs. Streptococcus pneumoniae strain TIGR4 (provided by S. Hammerschmidt, Greifswald, Germany) was plated on Columbia sheep red blood agar and incubated at 37°C and 5% CO2 overnight. For infections, bacteria were resuspended in sterile NaCl 0.9% solution adjusted to an optical density of 1 McFarland unit, which corresponds to 3 × 108 CFU/mL in a Densimat (BD Biosciences), a standard method to measure the inoculum size.65
In Vivo Infection Models
C57BL/6 mice and Mx1-positive B6.A2G-Mx1 mice with or without defective receptors for type I interferon or interferon-λ28, 29 were treated according to animal welfare. Mice were anesthetized with isoflurane (Baxter) and intranasally inoculated with a non-lethal dose (105 plaque-forming units [PFU]) of A/PR/8/34 or a lethal dose (103 PFU) of hvPR/8 in 50 μL of PBS. Weight loss, survival, and a clinical score (0 = normal; 1 = slightly ruffled fur, cold sensation; 2 = ruffled fur, shivering; 3 = ruffled fur, inactivity, slowed movements; 4 = ruffled fur, inactivity, hunched; 5 = dead)66 was determined daily. 3pRNA was injected i.v. at concentrations and time points indicated using in vivo jetPEI in an N/P ratio of 8 as recommended by the manufacturer (Polyplus-transfection) CXCL10, chemokine (C-X-C motif) ligand 10. In case of bacterial superinfection, mice were intranasally inoculated with a non-lethal dose (50 PFU) of hvPR/8 and in addition with 4.4 × 106 CFU of S. pneumoniae (TIGR4).
Oligonucleotides
Synthetic 3pRNA was chemically synthesized by solid-phase synthesis using product-specific labeling as described.15, 67, 68 CA20-RNA (5′-CACACACACACACACACACA-3′), CpG 1826,69 and 9.2 s-RNA36 were purchased from Biomers. Base-paired in-vitro-transcribed 3pRNA was generated as described previously61 and purified by separation in a Quick Spin DNA/RNA column (Roche) to eliminate short oligonucleotides (<8 nt). Activity of in-vitro-transcribed 3pRNA was functionally monitored by type I interferon induction in human peripheral blood mononuclear cells (PBMCs), which were prepared as previously described70 and stimulated with 1.2 μg/mL 3pRNA complexed to Lipofectamine 2000 (Invitrogen).
Analysis of Viral Copy Number
RNA was purified by using Nucleo Spin RNA Virus kit (Macherey & Nagel) according to the manufacturer’s instructions. Purified RNA was quantified by RT-PCR using One Step RT-PCR Kit (QIAGEN). The following primers (TIB Molbiol) were used: 5′-AGA TGA GTC TTC TAA CCG AGG TCG-3′ and 5′-TGC AAA AAC ATC TTC AAG TCT CTG-3′ with the probe: 5′-FAM-TCA GGC CCC CTC AAA GCC GA-TAMRA-3′. A standard curve (7.95 × 109 copies/mL to 7.95 × 105 copies/mL) was used for quantification. Data were obtained using the LightCycler 480 Software (Roche).
Real-Time qPCR
cDNA synthesis was performed using VILO cDNA Synthesis Kit from Life Technologies (11754050) as described in the manual. For mouse CXCL10, cDNA was amplified in a total volume of 20 μL using LightCycler 480 System (Roche). Primer and probe designs were performed using Universal Probe Library (Roche). Used probes from Roche were 18 for mouse CXCL10 and 51 for mouse TATA box binding protein (TBP). The following primers were used: mCXCL10 fwd: 5′-gctgccgtcattttctgc-3′, mCXCL10 rev: 5′-tctcactggcccgtcatc-3′; and mTBP fwd: 5′-ccaatgactcctatgaccccta-3′, mTBP rev: 5′-cagccaagattcacggtagat-3′.
Cytokine Assays
CXCL10 was measured in the supernatant using ELISA (BD Biosciences) according to the manufacturer’s recommendations.
Statistics
Results of multiple donors are presented as means with error bars indicating SD. Statistical analysis was performed using two-sided paired Student’s t test for dependent samples. In case of multiple comparisons, ANOVA was applied with Tukey’s or Holm-Sidak’s test for multiple comparison. Survival curves were analyzed using log rank Mantel-Cox test. The p values less than 0.05 were considered statistically significant. GraphPad Prism 6 for Mac OS X was used for analysis.
Study Approval
Animal studies were performed after approval by the responsible animal welfare authority under approval number TVA 887-50103709110 and G13/54.
Author Contributions
C.C., G.H., M.S., and E.H. conceived and designed most of the experiments. C.C., E.H., V.L.-W., and J.P.S. performed most of the experiments. C.C., G.H., M.S., F.B., S.H., C.S.-W., V.L.-W., J.P.S., and E.H. analyzed the data. C.C., E.H., M.S., and N.G. supervised most of the experiments. C.C., E.H., and G.H. co-wrote the manuscript. All authors substantially revised the manuscript. B.M.K., D.W., W.B., and N.G. helped to conceive and design the in vivo experiments. J.L. chemically synthesized the RNA. P.S. and G.K. provided virus stocks and helped to conceive, design, perform, and analyze the experiments with knockout mice. I.B.-D. and A.H. helped to conceive, design, perform, and analyze the in vivo superinfection experiments. A.H. contributed the bacterial strain and helped to conceive, design, and analyze the co-infection experiments.
Conflicts of Interest
G.H. and C.S.-W. are founders and hold shares of the company Rigontec, GmbH.
Acknowledgments
We thank Bastian Putschli (Institute of Clinical Chemistry and Clinical Pharmacology, University Hospital Bonn, Germany) for skillful technical support. G.H., W.B., and M.S. received financial support from the DFG excellence cluster ImmunoSensation. This study was supported by grants from the German Center for Infection Research (DZIF) (to W.B. and G.H.) and the German Research Foundation (DFG) (SFB 670, SFB704, and KFO177 to G.H. and C.C.). This work is part of the thesis of V.L.-W. and J.P.S. at the University of Bonn and was supported by grants to E.H, V.L-W., and J.P.S. of the BONFOR program of the Medical Faculty Bonn.
References
- 1.Novel Swine-Origin Influenza A (H1N1) Virus Investigation Team. Dawood F.S., Jain S., Finelli L., Shaw M.W., Lindstrom S., Garten R.J., Gubareva L.V., Xu X., Bridges C.B., Uyeki T.M. Emergence of a novel swine-origin influenza A (H1N1) virus in humans. N. Engl. J. Med. 2009;360:2605–2615. doi: 10.1056/NEJMoa0903810. [DOI] [PubMed] [Google Scholar]
- 2.Writing Committee of the WHO Consultation on Clinical Aspects of Pandemic (H1N1) 2009 Influenza. Bautista E., Chotpitayasunondh T., Gao Z., Harper S.A., Shaw M., Uyeki T.M., Zaki S.R., Hayden F.G., Hui D.S. Clinical aspects of pandemic 2009 influenza A (H1N1) virus infection. N. Engl. J. Med. 2010;362:1708–1719. doi: 10.1056/NEJMra1000449. [DOI] [PubMed] [Google Scholar]
- 3.Glezen W.P. Clinical practice. Prevention and treatment of seasonal influenza. N. Engl. J. Med. 2008;359:2579–2585. doi: 10.1056/NEJMcp0807498. [DOI] [PubMed] [Google Scholar]
- 4.Hayden F.G., de Jong M.D. Emerging influenza antiviral resistance threats. J. Infect. Dis. 2011;203:6–10. doi: 10.1093/infdis/jiq012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ison M.G. Antivirals and resistance: influenza virus. Curr. Opin. Virol. 2011;1:563–573. doi: 10.1016/j.coviro.2011.09.002. [DOI] [PubMed] [Google Scholar]
- 6.Jefferson T., Demicheli V., Rivetti D., Jones M., Di Pietrantonj C., Rivetti A. Antivirals for influenza in healthy adults: systematic review. Lancet. 2006;367:303–313. doi: 10.1016/S0140-6736(06)67970-1. [DOI] [PubMed] [Google Scholar]
- 7.Nguyen D.N., Mahon K.P., Chikh G., Kim P., Chung H., Vicari A.P., Love K.T., Goldberg M., Chen S., Krieg A.M. Lipid-derived nanoparticles for immunostimulatory RNA adjuvant delivery. Proc. Natl. Acad. Sci. USA. 2012;109:E797–E803. doi: 10.1073/pnas.1121423109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.St Paul M., Mallick A.I., Read L.R., Villanueva A.I., Parvizi P., Abdul-Careem M.F., Nagy É., Sharif S. Prophylactic treatment with Toll-like receptor ligands enhances host immunity to avian influenza virus in chickens. Vaccine. 2012;30:4524–4531. doi: 10.1016/j.vaccine.2012.04.033. [DOI] [PubMed] [Google Scholar]
- 9.Wong J.P., Christopher M.E., Viswanathan S., Karpoff N., Dai X., Das D., Sun L.Q., Wang M., Salazar A.M. Activation of toll-like receptor signaling pathway for protection against influenza virus infection. Vaccine. 2009;27:3481–3483. doi: 10.1016/j.vaccine.2009.01.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barchet W., Wimmenauer V., Schlee M., Hartmann G. Accessing the therapeutic potential of immunostimulatory nucleic acids. Curr. Opin. Immunol. 2008;20:389–395. doi: 10.1016/j.coi.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 11.Schlee M., Hartmann G. Discriminating self from non-self in nucleic acid sensing. Nat. Rev. Immunol. 2016;16:566–580. doi: 10.1038/nri.2016.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Freeman A.I., Al-Bussam N., O’Malley J.A., Stutzman L., Bjornsson S., Carter W.A. Pharmacologic effects of polyinosinic-polycytidylic acid in man. J. Med. Virol. 1977;1:79–93. doi: 10.1002/jmv.1890010202. [DOI] [PubMed] [Google Scholar]
- 13.Cunningham C., Campion S., Teeling J., Felton L., Perry V.H. The sickness behaviour and CNS inflammatory mediator profile induced by systemic challenge of mice with synthetic double-stranded RNA (poly I:C) Brain Behav. Immun. 2007;21:490–502. doi: 10.1016/j.bbi.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 14.Hornung V., Ellegast J., Kim S., Brzózka K., Jung A., Kato H., Poeck H., Akira S., Conzelmann K.K., Schlee M. 5′-Triphosphate RNA is the ligand for RIG-I. Science. 2006;314:994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- 15.Schlee M., Roth A., Hornung V., Hagmann C.A., Wimmenauer V., Barchet W., Coch C., Janke M., Mihailovic A., Wardle G. Recognition of 5′ triphosphate by RIG-I helicase requires short blunt double-stranded RNA as contained in panhandle of negative-strand virus. Immunity. 2009;31:25–34. doi: 10.1016/j.immuni.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Okabayashi T., Kojima T., Masaki T., Yokota S., Imaizumi T., Tsutsumi H., Himi T., Fujii N., Sawada N. Type-III interferon, not type-I, is the predominant interferon induced by respiratory viruses in nasal epithelial cells. Virus Res. 2011;160:360–366. doi: 10.1016/j.virusres.2011.07.011. [DOI] [PubMed] [Google Scholar]
- 17.Ramos H.J., Gale M., Jr. RIG-I like receptors and their signaling crosstalk in the regulation of antiviral immunity. Curr. Opin. Virol. 2011;1:167–176. doi: 10.1016/j.coviro.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kato H., Takeuchi O., Sato S., Yoneyama M., Yamamoto M., Matsui K., Uematsu S., Jung A., Kawai T., Ishii K.J. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 19.Dudek S.E., Wixler L., Nordhoff C., Nordmann A., Anhlan D., Wixler V., Ludwig S. The influenza virus PB1-F2 protein has interferon antagonistic activity. Biol. Chem. 2011;392:1135–1144. doi: 10.1515/BC.2011.174. [DOI] [PubMed] [Google Scholar]
- 20.Guo Z., Chen L.-M., Zeng H., Gomez J.A., Plowden J., Fujita T., Katz J.M., Donis R.O., Sambhara S. NS1 protein of influenza A virus inhibits the function of intracytoplasmic pathogen sensor, RIG-I. Am. J. Respir. Cell Mol. Biol. 2007;36:263–269. doi: 10.1165/rcmb.2006-0283RC. [DOI] [PubMed] [Google Scholar]
- 21.Mibayashi M., Martínez-Sobrido L., Loo Y.-M., Cárdenas W.B., Gale M., Jr., García-Sastre A. Inhibition of retinoic acid-inducible gene I-mediated induction of beta interferon by the NS1 protein of influenza A virus. J. Virol. 2007;81:514–524. doi: 10.1128/JVI.01265-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chakravarthy K.V., Bonoiu A.C., Davis W.G., Ranjan P., Ding H., Hu R., Bowzard J.B., Bergey E.J., Katz J.M., Knight P.R. Gold nanorod delivery of an ssRNA immune activator inhibits pandemic H1N1 influenza viral replication. Proc. Natl. Acad. Sci. USA. 2010;107:10172–10177. doi: 10.1073/pnas.0914561107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goulet M.-L., Olagnier D., Xu Z., Paz S., Belgnaoui S.M., Lafferty E.I., Janelle V., Arguello M., Paquet M., Ghneim K. Systems analysis of a RIG-I agonist inducing broad spectrum inhibition of virus infectivity. PLoS Pathog. 2013;9:e1003298. doi: 10.1371/journal.ppat.1003298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hwang S.-Y., Sun H.-Y., Lee K.-H., Oh B.-H., Cha Y.J., Kim B.H., Yoo J.Y. 5′-Triphosphate-RNA-independent activation of RIG-I via RNA aptamer with enhanced antiviral activity. Nucleic Acids Res. 2012;40:2724–2733. doi: 10.1093/nar/gkr1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin L., Liu Q., Berube N., Detmer S., Zhou Y. 5′-Triphosphate-short interfering RNA: potent inhibition of influenza A virus infection by gene silencing and RIG-I activation. J. Virol. 2012;86:10359–10369. doi: 10.1128/JVI.00665-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ranjan P., Jayashankar L., Deyde V., Zeng H., Davis W.G., Pearce M.B., Bowzard J.B., Hoelscher M.A., Jeisy-Scott V., Wiens M.E. 5'PPP-RNA induced RIG-I activation inhibits drug-resistant avian H5N1 as well as 1918 and 2009 pandemic influenza virus replication. Virol. J. 2010;7:102. doi: 10.1186/1743-422X-7-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pavlovic J., Haller O., Staeheli P. Human and mouse Mx proteins inhibit different steps of the influenza virus multiplication cycle. J. Virol. 1992;66:2564–2569. doi: 10.1128/jvi.66.4.2564-2569.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grimm D., Staeheli P., Hufbauer M., Koerner I., Martínez-Sobrido L., Solórzano A., García-Sastre A., Haller O., Kochs G. Replication fitness determines high virulence of influenza A virus in mice carrying functional Mx1 resistance gene. Proc. Natl. Acad. Sci. USA. 2007;104:6806–6811. doi: 10.1073/pnas.0701849104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Staeheli P., Dreiding P., Haller O., Lindenmann J. Polyclonal and monoclonal antibodies to the interferon-inducible protein Mx of influenza virus-resistant mice. J. Biol. Chem. 1985;260:1821–1825. [PubMed] [Google Scholar]
- 30.Pavlovic J., Zürcher T., Haller O., Staeheli P. Resistance to influenza virus and vesicular stomatitis virus conferred by expression of human MxA protein. J. Virol. 1990;64:3370–3375. doi: 10.1128/jvi.64.7.3370-3375.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zürcher T., Pavlovic J., Staeheli P. Mechanism of human MxA protein action: variants with changed antiviral properties. EMBO J. 1992;11:1657–1661. doi: 10.1002/j.1460-2075.1992.tb05212.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Horisberger M.A., Staeheli P., Haller O. Interferon induces a unique protein in mouse cells bearing a gene for resistance to influenza virus. Proc. Natl. Acad. Sci. USA. 1983;80:1910–1914. doi: 10.1073/pnas.80.7.1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mordstein M., Kochs G., Dumoutier L., Renauld J.-C., Paludan S.R., Klucher K., Staeheli P. Interferon-lambda contributes to innate immunity of mice against influenza A virus but not against hepatotropic viruses. PLoS Pathog. 2008;4:e1000151. doi: 10.1371/journal.ppat.1000151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mordstein M., Neugebauer E., Ditt V., Jessen B., Rieger T., Falcone V., Sorgeloos F., Ehl S., Mayer D., Kochs G. Lambda interferon renders epithelial cells of the respiratory and gastrointestinal tracts resistant to viral infections. J. Virol. 2010;84:5670–5677. doi: 10.1128/JVI.00272-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Herberhold S., Coch C., Zillinger T., Hommertgen B., Busch N., Schuberth C., Hartmann E., Wimmenauer V., Hagmann C.A., Lüdenbach B. Delivery with polycations extends the immunostimulant Ribomunyl into a potent antiviral Toll-like receptor 7/8 agonist. Antivir. Ther. (Lond.) 2011;16:751–758. doi: 10.3851/IMP1822. [DOI] [PubMed] [Google Scholar]
- 36.Hornung V., Guenthner-Biller M., Bourquin C., Ablasser A., Schlee M., Uematsu S., Noronha A., Manoharan M., Akira S., de Fougerolles A. Sequence-specific potent induction of IFN-alpha by short interfering RNA in plasmacytoid dendritic cells through TLR7. Nat. Med. 2005;11:263–270. doi: 10.1038/nm1191. [DOI] [PubMed] [Google Scholar]
- 37.Bourquin C., Schmidt L., Lanz A.-L., Storch B., Wurzenberger C., Anz D., Sandholzer N., Mocikat R., Berger M., Poeck H. Immunostimulatory RNA oligonucleotides induce an effective antitumoral NK cell response through the TLR7. J. Immunol. 2009;183:6078–6086. doi: 10.4049/jimmunol.0901594. [DOI] [PubMed] [Google Scholar]
- 38.Manegold C., van Zandwijk N., Szczesna A., Zatloukal P., Au J.S.K., Blasinska-Morawiec M., Serwatowski P., Krzakowski M., Jassem J., Tan E.H. A phase III randomized study of gemcitabine and cisplatin with or without PF-3512676 (TLR9 agonist) as first-line treatment of advanced non-small-cell lung cancer. Ann. Oncol. 2012;23:72–77. doi: 10.1093/annonc/mdr030. [DOI] [PubMed] [Google Scholar]
- 39.Nichani A.K., Dar M.A., Mirakhur K.K., Krieg A.M., Booth J.S., Townsend H.G.G., Potter A.A., Babiuk L.A., Mutwiri G.K. Subcutaneous, but not intratracheal administration of the TLR9 agonist, CpG DNA transiently reduces parainfluenza-3 virus shedding in newborn lambs. Comp. Immunol. Microbiol. Infect. Dis. 2010;33:e111–e117. doi: 10.1016/j.cimid.2010.06.003. [DOI] [PubMed] [Google Scholar]
- 40.Ballinger M.N., Standiford T.J. Postinfluenza bacterial pneumonia: host defenses gone awry. J. Interferon Cytokine Res. 2010;30:643–652. doi: 10.1089/jir.2010.0049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peltola V.T., Murti K.G., McCullers J.A. Influenza virus neuraminidase contributes to secondary bacterial pneumonia. J. Infect. Dis. 2005;192:249–257. doi: 10.1086/430954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rothberg M.B., Haessler S.D., Brown R.B. Complications of viral influenza. Am. J. Med. 2008;121:258–264. doi: 10.1016/j.amjmed.2007.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li W., Moltedo B., Moran T.M. Type I interferon induction during influenza virus infection increases susceptibility to secondary Streptococcus pneumoniae infection by negative regulation of γδ T cells. J. Virol. 2012;86:12304–12312. doi: 10.1128/JVI.01269-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Navarini A.A., Recher M., Lang K.S., Georgiev P., Meury S., Bergthaler A., Flatz L., Bille J., Landmann R., Odermatt B. Increased susceptibility to bacterial superinfection as a consequence of innate antiviral responses. Proc. Natl. Acad. Sci. USA. 2006;103:15535–15539. doi: 10.1073/pnas.0607325103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stegemann S., Dahlberg S., Kröger A., Gereke M., Bruder D., Henriques-Normark B., Gunzer M. Increased susceptibility for superinfection with Streptococcus pneumoniae during influenza virus infection is not caused by TLR7-mediated lymphopenia. PLoS ONE. 2009;4:e4840. doi: 10.1371/journal.pone.0004840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Killip M.J., Fodor E., Randall R.E. Influenza virus activation of the interferon system. Virus Res. 2015;209:11–22. doi: 10.1016/j.virusres.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ye Q., Krug R.M., Tao Y.J. The mechanism by which influenza A virus nucleoprotein forms oligomers and binds RNA. Nature. 2006;444:1078–1082. doi: 10.1038/nature05379. [DOI] [PubMed] [Google Scholar]
- 48.Engel D.A. The influenza virus NS1 protein as a therapeutic target. Antiviral Res. 2013;99:409–416. doi: 10.1016/j.antiviral.2013.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hale B.G., Randall R.E., Ortín J., Jackson D. The multifunctional NS1 protein of influenza A viruses. J. Gen. Virol. 2008;89:2359–2376. doi: 10.1099/vir.0.2008/004606-0. [DOI] [PubMed] [Google Scholar]
- 50.Varga Z.T., Ramos I., Hai R., Schmolke M., García-Sastre A., Fernandez-Sesma A., Palese P. The influenza virus protein PB1-F2 inhibits the induction of type I interferon at the level of the MAVS adaptor protein. PLoS Pathog. 2011;7:e1002067. doi: 10.1371/journal.ppat.1002067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Flory E., Kunz M., Scheller C., Jassoy C., Stauber R., Rapp U.R., Ludwig S. Influenza virus-induced NF-kappaB-dependent gene expression is mediated by overexpression of viral proteins and involves oxidative radicals and activation of IkappaB kinase. J. Biol. Chem. 2000;275:8307–8314. doi: 10.1074/jbc.275.12.8307. [DOI] [PubMed] [Google Scholar]
- 52.Wurzer W.J., Ehrhardt C., Pleschka S., Berberich-Siebelt F., Wolff T., Walczak H., Planz O., Ludwig S. NF-kappaB-dependent induction of tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) and Fas/FasL is crucial for efficient influenza virus propagation. J. Biol. Chem. 2004;279:30931–30937. doi: 10.1074/jbc.M403258200. [DOI] [PubMed] [Google Scholar]
- 53.Jewell N.A., Cline T., Mertz S.E., Smirnov S.V., Flaño E., Schindler C., Grieves J.L., Durbin R.K., Kotenko S.V., Durbin J.E. Lambda interferon is the predominant interferon induced by influenza A virus infection in vivo. J. Virol. 2010;84:11515–11522. doi: 10.1128/JVI.01703-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sommereyns C., Paul S., Staeheli P., Michiels T. IFN-lambda (IFN-lambda) is expressed in a tissue-dependent fashion and primarily acts on epithelial cells in vivo. PLoS Pathog. 2008;4:e1000017. doi: 10.1371/journal.ppat.1000017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tian X., Xu F., Lung W.Y., Meyerson C., Ghaffari A.A., Cheng G., Deng J.C. Poly I:C enhances susceptibility to secondary pulmonary infections by gram-positive bacteria. PLoS ONE. 2012;7:e41879. doi: 10.1371/journal.pone.0041879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Small C.-L., Shaler C.R., McCormick S., Jeyanathan M., Damjanovic D., Brown E.G., Arck P., Jordana M., Kaushic C., Ashkar A.A., Xing Z. Influenza infection leads to increased susceptibility to subsequent bacterial superinfection by impairing NK cell responses in the lung. J. Immunol. 2010;184:2048–2056. doi: 10.4049/jimmunol.0902772. [DOI] [PubMed] [Google Scholar]
- 57.Ghoneim H.E., Thomas P.G., McCullers J.A. Depletion of alveolar macrophages during influenza infection facilitates bacterial superinfections. J. Immunol. 2013;191:1250–1259. doi: 10.4049/jimmunol.1300014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sun K., Metzger D.W. Influenza infection suppresses NADPH oxidase-dependent phagocytic bacterial clearance and enhances susceptibility to secondary methicillin-resistant Staphylococcus aureus infection. J. Immunol. 2014;192:3301–3307. doi: 10.4049/jimmunol.1303049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schliehe C., Flynn E.K., Vilagos B., Richson U., Swaminathan S., Bosnjak B., Bauer L., Kandasamy R.K., Griesshammer I.M., Kosack L. The methyltransferase Setdb2 mediates virus-induced susceptibility to bacterial superinfection. Nat. Immunol. 2015;16:67–74. doi: 10.1038/ni.3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Noone C.M., Lewis E.A., Frawely A.B., Newman R.W., Mahon B.P., Mills K.H., Johnson P.A. Novel mechanism of immunosuppression by influenza virus haemagglutinin: selective suppression of interleukin 12 p35 transcription in murine bone marrow-derived dendritic cells. J. Gen. Virol. 2005;86:1885–1890. doi: 10.1099/vir.0.80891-0. [DOI] [PubMed] [Google Scholar]
- 61.Pillai P.S., Molony R.D., Martinod K., Dong H., Pang I.K., Tal M.C., Solis A.G., Bielecki P., Mohanty S., Trentalange M. Mx1 reveals innate pathways to antiviral resistance and lethal influenza disease. Science. 2016;352:463–466. doi: 10.1126/science.aaf3926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Winter G., Fields S., Brownlee G.G. Nucleotide sequence of the haemagglutinin gene of a human influenza virus H1 subtype. Nature. 1981;292:72–75. doi: 10.1038/292072a0. [DOI] [PubMed] [Google Scholar]
- 63.Haller O. Inborn resistance of ice to orthomyxoviruses. Curr. Top. Microbiol. Immunol. 1981;92:25–52. doi: 10.1007/978-3-642-68069-4_3. [DOI] [PubMed] [Google Scholar]
- 64.Schickli J.H., Flandorfer A., Nakaya T., Martinez-Sobrido L., García-Sastre A., Palese P. Plasmid-only rescue of influenza A virus vaccine candidates. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2001;356:1965–1973. doi: 10.1098/rstb.2001.0979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Perry J.D., Morris K.A., James A.L., Oliver M., Gould F.K. Evaluation of novel chromogenic substrates for the detection of bacterial beta-glucosidase. J. Appl. Microbiol. 2007;102:410–415. doi: 10.1111/j.1365-2672.2006.03096.x. [DOI] [PubMed] [Google Scholar]
- 66.Humphreys I.R., Walzl G., Edwards L., Rae A., Hill S., Hussell T. A critical role for OX40 in T cell-mediated immunopathology during lung viral infection. J. Exp. Med. 2003;198:1237–1242. doi: 10.1084/jem.20030351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Goldeck M., Schlee M., Hartmann G., Hornung V. Enzymatic synthesis and purification of a defined RIG-I ligand. Methods Mol. Biol. 2014;1169:15–25. doi: 10.1007/978-1-4939-0882-0_2. [DOI] [PubMed] [Google Scholar]
- 68.Goldeck M., Tuschl T., Hartmann G., Ludwig J. Efficient solid-phase synthesis of pppRNA by using product-specific labeling. Angew. Chem. Int. Ed. Engl. 2014;53:4694–4698. doi: 10.1002/anie.201400672. [DOI] [PubMed] [Google Scholar]
- 69.Jiang T., Zhao H., Li X.-F., Deng Y.-Q., Liu J., Xu L.-J., Han J.F., Cao R.Y., Qin E.D., Qin C.F. CpG oligodeoxynucleotides protect against the 2009 H1N1 pandemic influenza virus infection in a murine model. Antiviral Res. 2011;89:124–126. doi: 10.1016/j.antiviral.2010.11.013. [DOI] [PubMed] [Google Scholar]
- 70.Coch C., Lück C., Schwickart A., Putschli B., Renn M., Höller T., Barchet W., Hartmann G., Schlee M. A human in vitro whole blood assay to predict the systemic cytokine response to therapeutic oligonucleotides including siRNA. PLoS ONE. 2013;8:e71057. doi: 10.1371/journal.pone.0071057. [DOI] [PMC free article] [PubMed] [Google Scholar]