Abstract
The shortening of human telomeres has two opposing effects during cancer development. On the one hand, telomere shortening can exert a tumour-suppressive effect through the proliferation arrest induced by activating the kinases ATM and ATR at unprotected chromosome ends. On the other hand, loss of telomere protection can lead to telomere crisis, which is a state of extensive genome instability that can promote cancer progression. Recent data, reviewed here, provide new evidence for the telomere tumour suppressor pathway and has revealed that telomere crisis can induce numerous cancer-relevant changes, including chromothripsis, kataegis and tetraploidization.
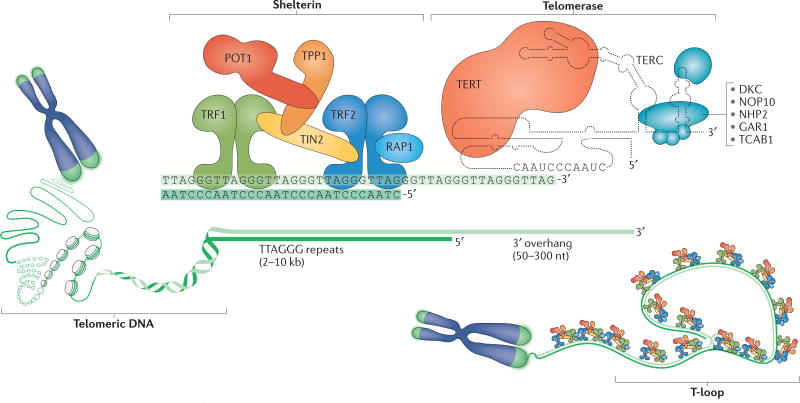
Telomeres have an essential role in ensuring that the natural ends of chromosomes are not mistaken for sites of DNA damage. Telomere function depends on three factors: telomeric DNA, the shelterin complex and the telomerase complex (FIG. 1). Human and mouse telomeres are composed of a long double-stranded array of TTAGGG repeats bound by the six-subunit shelterin complex (BOX 1; FIG. 1). Shelterin represses the DNA damage response (DDR) at telomeres, thereby preventing the activation of the kinases ATM and ATR that can induce cell cycle arrest in response to DNA double- strand breaks (DSBs) and other types of DNA damage. In addition, shelterin ensures that telomeres are not processed by several DSB repair pathways, including the non-homologous end joining (NHEJ) pathway that could lead to chromosome end fusions. Shelterin partially protects telomeres by forming the t-loop structure by which the telomere terminus is hidden (FIG. 1). T-loops are formed through strand invasion of the long 3′ overhang at the telomere end into the double-stranded telomeric DNA. This 3′ overhang is recreated after DNA replication through exonucleolytic degradation of the 5’ ends of the telomeres (FIG. 2a). As a result of this processing and the inability of DNA polymerases to duplicate the ends of linear DNA molecules, human telomeres shorten by ~50 bps per cell division. This telomere attrition can be counteracted by telomerase reverse transcriptase (TERT) (FIG. 1), which adds GGTTAG repeats to the chromosomal 3′ DNA terminus at the end of the chromosome.
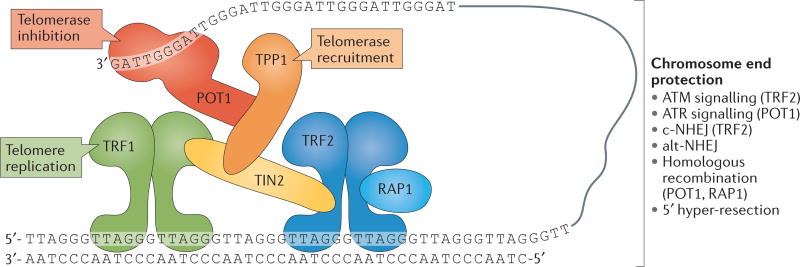
Figure 1. Composition and structure of the human telomere system.
Human telomeres comprise three components: telomeric DNA, the shelterin complex and the telomerase complex. Telomeric DNA consists of a long array of double-stranded TTAGGG repeats that culminates in a 50–300 nucleotide (nt) single-stranded 3′ overhang. This 3′ overhang invades double-stranded telomeric repeats to form a t-loop structure that is crucial for telomere function. Telomeric DNA protects chromosome ends through its association with the six-subunit shelterin complex. The length of telomeric repeats can be maintained by telomerase, which is composed of telomerase reverse transcriptase (TERT), telomerase RNA template component (TERC) and several accessory proteins (blue). TERT synthesizes telomeric DNA de novo using TERC as a template, whereas the accessory factors contribute to the biogenesis and nuclear trafficking of telomerase. DKC, dyskerin; NHP2, non-histone protein 2; NOP10, nucleolar protein 10; POT1, protection of telomeres 1; RAP1, repressor/activator protein 1; TCAB1, telomerase Cajal body protein 1; TIN2, TRF1-interacting nuclear factor 2; TRF, telomeric repeat-binding factor.
Box 1 | The shelterin complex and its functions.
Shelterin is a complex comprising the following six subunits: telomeric repeat-binding factor 1 (TRF1), TRF2, repressor/activator protein 1 (RAP1), TRF1-interacting nuclear factor 2 (TIN2), TPP1 (also known as adrenocortical dysplasia protein homologue) and protection of telomeres 1 (POT1) (see the figure). Shelterin associates with mammalian telomeres, where it regulates various aspects of telomere function175–177. Shelterin is recruited to telomeres through TRF1 and TRF2, which bind to double-stranded telomeric DNA and to TIN2. POT1 binds to single-stranded telomeric DNA and is linked to TRF1 and TRF2 through its binding partner TPP1, which associates with TIN2. RAP1 associates with TRF2.
Shelterin maintains telomere length and preserves genome integrity by regulating the access of telomerase to chromosome ends by controlling end-resection at newly replicated telomeres, and by masking telomeres from the DNA damage response (DDR). Specifically, TRF2 represses ATM-dependent DNA damage signalling and classical non-homologous end joining (c-NHEJ), whereas POT1 is responsible for repressing ATR signalling and cooperates with RAP1 in suppressing homologous recombination. Avoiding the DDR is partially mediated by TRF2-dependent t-loop formation. T-loops are formed through the invasion of the 3′ overhang at the telomere end into double-stranded telomeric DNA (FIG. 1), and is thought to prevent ATM activation by masking the chromosome end from the double-strand breaks sensor complex MRE11-RAD50-NBS1 (MRN) and by blocking c-NHEJ induction by preventing the loading of the Ku70-Ku80 heterodimer on the chromosome end. Repression of ATR signalling by POT1 involves occlusion of the single-stranded DNA sensor replication protein A (RPA). Importantly, this repression depends on the association of POT1 with the rest of shelterin via its interaction with TPP1. Telomere protection is compromised when telomeres become too short to support sufficient shelterin binding.
Shelterin also functions to facilitate telomere maintenance by the reverse transcriptase complex telomerase (FIG. 1), which is recruited to telomeres by the shelterin components TPP1 (REFS 178–181) and TIN2 (REFS 182,183). Shelterin also has a role in the regulation of telomerase-mediated telomere length maintenance (reviewed in REF. 184). Several shelterin subunits are negative regulators of telomere length, suggesting that shelterin subunits ‘count’ telomeric repeats to regulate telomerase activity and limit telomere length as part of a cis-acting negative-feedback loop (reviewed in REF. 185). This regulatory pathway may be important in the germ line, in which telomere length needs to be maintained within a narrow range to provide offspring with telomeres that are sufficiently long for normal development and tissue homeostasis, whereas at the same time are sufficiently short to suppress cell transformation by inducing replicative senescence.
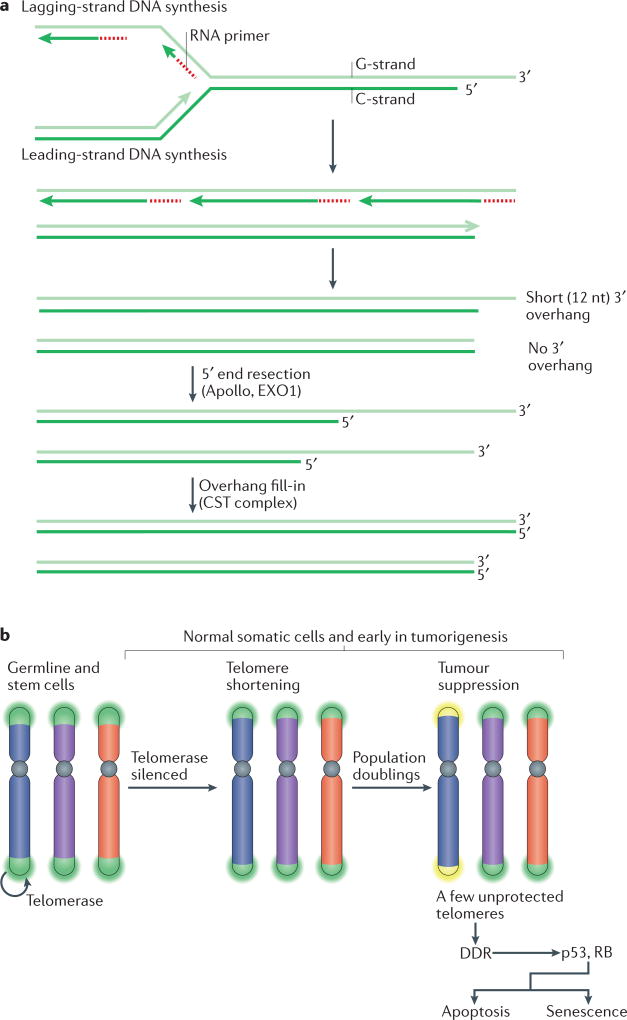
Figure 2. Telomere shortening as a barrier to tumorigenesis.
a | The molecular basis of telomere shortening. Incomplete DNA synthesis at the end of the lagging strand (at the site of the terminal RNA primer) leaves a short 3′ overhang. Additional loss of telomeric DNA occurs through the processing of the leading-strand ends of telomeres to regenerate the 3′ overhang, which is necessary for t-loop formation and the structural integrity of the telomere. This process is carried out by the nuclease Apollo, which is bound to telomeric repeat-binding factor 2 (TRF2). Both the leading end and the lagging end of telomeres are further resected by exonuclease 1 (EXO1) to generate transient long overhangs. The CST (CTC1–STN1–TEN1) complex then binds to shelterin and mediates fill-in synthesis of the cytosine-rich strand (C-strand) at both ends. b | During development, telomerase is switched off through telomerase reverse transcriptase (TERT) silencing. As a result, telomeres experience the gradual attrition described in part a. After numerous population doublings, a few telomeres become too short (yellow) and lose their protective function. As a result, the kinases ATM and ATR are activated at the unprotected chromosome ends and this DNA damage response (DDR) signalling induces replicative arrest and senescence or apoptosis. This process limits the proliferative capacity of incipient cancer cells, thus functioning as a tumour suppressor pathway. Cells lacking p53 and RB function can avoid this replicative arrest.
During human development, telomerase activity is downregulated through the silencing of TERT, which encodes the reverse transcriptase subunit of the complex. As a result, most human somatic cells (with the exception of certain stem cells) undergo programmed telomere shortening. Eventually, the loss of telomeric DNA leads to insufficient chromosome end protection and to the activation of the DDR, which will arrest cell proliferation and can induce senescence or apoptosis. The repression of telomerase in somatic cells and the resulting telomere proliferation barrier have the hallmarks of a tumour suppressor pathway that limits tumour cell outgrowth after a delay. New evidence, reviewed below, argues that telomere shortening indeed protects against tumour development. Eventually, however, in incipient cancer cells that lack the pathways necessary for cell cycle arrest, mounting telomere dysfunction becomes a source of genomic instability in a stage referred to as telomere crisis1–3. The escape from telomere crisis requires the activation of telomerase, which reconstitutes telomere function and restores proliferative capacity. The outcome of this scenario is a telomerase-positive, transformed cell with a heavily rearranged, but stabilized, genome that has attained new and potentially tumorigenic genetic mutations.
Cancer genomes are characterized by extensive chromosome rearrangements that facilitate oncogenic progression4. The unexpected extent and staggering complexity of these rearrangements, which include deletions and amplifications, translocations, chromothripsis, kataegis and tetraploidization, has only been appreciated in recent years5. Although there are many potential mechanisms underlying these rearrangements6, new data have linked telomere dysfunction to a near- comprehensive list of cancer-relevant genome alterations2,3,7–9, suggesting that telomere crisis contributes to the genetic disorder that is typical of cancer10. Here, we review these new data on the role of telomeres in genome instability in cancer and discuss new findings pertaining to the role of telomere shortening in tumour suppression.
Tumour suppression by short telomeres
Telomere shortening in human cells has long been thought to represent a tumour suppressor mechanism. Although mouse models have previously illustrated this potentially advantageous aspect of telomere attrition, recent data now provide evidence for this proliferative barrier in human cancer cells.
Silencing of telomerase and telomere shortening
Telomerase is a reverse transcriptase that synthesizes telomeric DNA de novo using integral RNA as the template and the 3′ end of the chromosome as the primer11–17 (FIG. 1). The core components of telomerase are the reverse transcriptase TERT and telomerase RNA component (TERC), which provides the template for the synthesis of telomeric DNA. Telomerase is associated with a set of accessory proteins, including dyskerin, nucleolar protein 10 (NOP10), non-histone protein 2 (NHP2), GAR1 and telomerase Cajal body protein 1 (TCAB1), that contribute to the biogenesis and trafficking of telomerase inside the nucleus18,19 (for reviews, see REFS 17,20,21).
TERT silencing downregulates telomerase activity in human somatic cells (reviewed in REF. 22). The other components of telomerase, including TERC, are expressed widely; thus, the expression of exogenous TERT is sufficient to activate telomerase in many primary human cells23. However, TERC expression can still be a limiting factor and the co-expression of TERT and TERC is needed for robust telomerase catalytic activity in some cell types21,24,25. Telomerase activation and the resulting telomere-length maintenance leads to the bypass of senescence and ultimately to cell immortalization23,26,27.
The programmed silencing of TERT, loss of telomerase activity and the resulting shortening of telomeres is not a universal phenomenon in mammals. Apparently, this tumour suppressor pathway is restricted to large animals with a reproductive strategy that requires a long lifespan28. For example, telomerase activity is repressed in somatic cells of elephants but not in mice.
In the absence of telomerase, each human telomere shortens at a rate of 50–100 bps per population doubling29. The rate of telomere attrition is partly due to the inability of DNA polymerases to copy the end of linear DNA (FIG. 2a). The 5’ end resection that generates the telomeric 3′ overhang contributes substantially to the rate of telomere shortening30 (FIG. 2a). Shelterin governs this processing and the formation of the correct structure of the telomere terminus in mouse cells and most likely in human cells. The process involves the initiation of resection by the shelterin-bound Apollo nuclease, further resection by exonuclease 1 (EXO1) and finally a fill-in synthesis step mediated by the shelterin-bound CST (CTC1–STN1–TEN1) complex31–38 (FIG. 2a). Owing to this regulated terminal sequence loss, the proliferative lifespan of primary human cells (known as the Hayflick limit) is partly determined by how shelterin controls resection and fill-in at telomeres. It will be interesting to determine whether the lifespan of human cells can be extended by diminishing the extent of 5’ end resection.
Telomere-induced senescence
Loss of telomere function at a few chromosome ends in a cell is sufficient to induce replicative arrest9,39,40 (FIG. 2b). The point at which telomere attrition results in the loss of telomere protection at one or a few chromosome ends is dependent on the rate of telomere shortening, the initial telomere length and, importantly, the length of the shortest telomeres in the cells41. Because human telomeres are heterogeneously sized, several very short telomeres can be present in cells with an apparently ample telomere reserve, which makes measurements of bulk telomere length an imprecise predictor of cellular proliferative potential.
Senescent human fibroblasts display the molecular hallmarks of an activated DDR39, including ATM and ATR signalling, and nuclear foci containing DNA damage markers, such as γ-H2AX, p53-binding protein 1 (53BP1) and mediator of DNA damage checkpoint protein 1 (MDC1). Upregulation of p53 and induction of the cyclin-dependent kinase (CDK) inhibitors p21 and p16 are also indicators of an activated DDR42,43. The inactivation of shelterin similarly activates DNA damage signalling pathways, results in the upregulation of p21 and p16, and leads to the accumulation of DDR factors at telomeres, thus linking telomere dysfunction and senescence39,42. Proof that replicative senescence is due to telomere shortening came from the bypass of senescence upon TERT expression23. Furthermore, over-expression of the shelterin subunit telomeric repeat-binding factor 2 (TRF2; also known as TERF2) can delay the onset of senescence44, arguing that the DDR in senescence is due to an insufficient loading of shelterin at the shortened telomeres.
Senescent cells are usually in G1 phase, consistent with p53 activation and induction of the CDK inhibitors p21 and p16. The upregulation of p16 and the accompanying hypophosphorylation of the tumour suppressor RB can contribute to telomere-induced senescence. Moreover, inactivation of both the RB and the p53 responses to dysfunctional telomeres is needed to completely circumvent this block to proliferation43. Because the complete bypass of telomere shortening-induced senescence in human cells requires the inactivation of multiple pathways, this mechanism of curbing the proliferation of transformed cells is likely to be robust43,45–48. By contrast, p53 inactivation alone is sufficient to avoid telomere-induced senescence and apoptosis in mice49,50.
Telomere shortening, telomerase downregulation and cancer prevention
Experiments in genetically altered mice support the view that telomere shortening can act as a strong barrier to tumorigenesis (FIG. 2b). Crosses of telomerase-deficient mice with various tumour model mice have demonstrated that critically short telomeres limit tumour formation when the p53 pathway is functional51–55. In addition, the original demonstration that telomerase is active in most human cancers, whereas the enzyme is undetectable in normal tissues, suggested that the telomere tumour suppressor pathway may operate in most cancer types56. However, the upregulation of telomerase expression could be an irrelevant consequence of transcriptional rewiring during tumorigenesis, perhaps reflecting a stem cell phenotype in cancer. The recent identification of activating mutations in the TERT promoter in several cancer types argues strongly that these tumours had undergone selection for the activation of telomerase57,58. Similarly, the amplification of TERT and other mutations that increase telomerase activity in some cancers argue in favour of a selected phenotype59–61.
A strong argument in support of the telomere tumour suppressor pathway emerged recently from a study of a large family with a predisposition to melanoma58. A linkage analysis and high-throughput sequencing identified an activating mutation in the TERT promoter that co-segregates with disease predisposition. This mutation (T>G; 57 bp upstream of the transcription start site) creates a binding motif for ETS (E26 transformation-specific) transcription factors. Thus, tissues that express ETS transcription factors are predicted to maintain telomerase activity, presumably resulting in the maintenance of telomere length and the consequent disruption of the telomere tumour suppressor pathway.
A similar example is provided by the recently identified melanoma-predisposing mutations in the gene encoding the protection of telomeres 1 (POT1) sub unit of shelterin62,63. These mutations alter POT1 mRNA splicing or compromise the oligonucleotide and/or oligosaccharide-binding folds in the single-stranded DNA-binding domains of POT1. As a consequence, the ability of POT1 to bind to single-stranded telomeric DNA is diminished. Carriers of these mutations have longer telomeres, presumably owing to the loss of POT1-mediated inhibition of telomerase (BOX 1). Because the increased telomere length is present at birth, these mutations are likely to push the onset of senescence to later population doublings, thus postponing the tumour-suppressive effects of telomere attrition to a point of irrelevance. However, whether the diminished POT1 function also promotes genome instability is currently unknown64.
Another observation in support of the telomere tumour suppressor pathway is that longer telomere length has been associated with an increased risk of B cell lymphoma and chronic lymphocytic leukaemia (CLL)65–67. In a recent study, single-nucleotide polymorphisms in telomere maintenance genes that are associated with telomere length68,69 were examined to determine the cancer risk of 95,568 individuals from the general population70. This analysis found that genetic determinants of long telomeres are associated with an increased overall cancer risk, especially lung cancer and melanoma. Collectively, these data are consistent with telomere shortening functioning as a tumour suppressor pathway.
Telomere crisis and genome instability
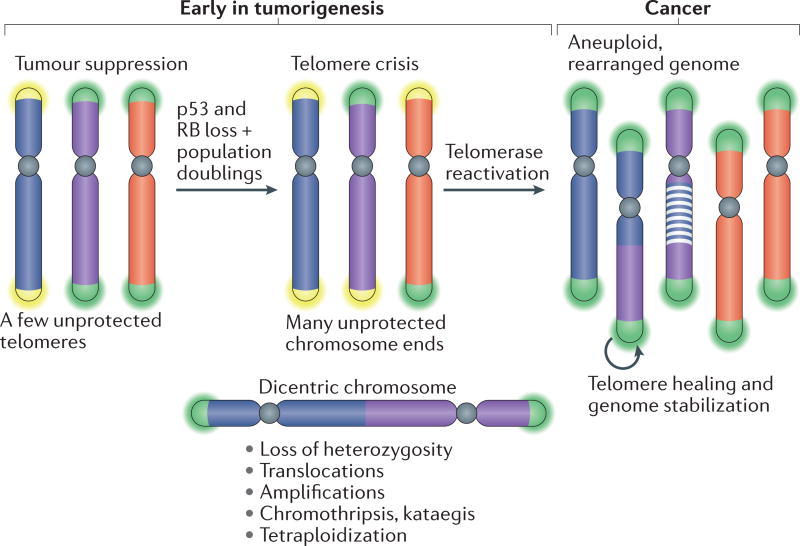
Although the telomere tumour suppressor pathway may be a powerful mechanism to limit cancer development, failure of transformed cells to undergo senescence can produce telomere crisis, during which the cell population does not expand. In telomere crisis, cells struggle with a high level of genome instability owing to the presence of many dysfunctional telomeres. Activation of telomerase provides a path out of telomere crisis, ultimately leading to the formation of a cancer clone with a heavily rearranged genome (FIG. 3).
Figure 3. Telomere crisis.
Loss of the RB and p53 tumour suppressor pathways disables the ability of cells to respond with cell cycle arrest to ATR and ATM signalling. As the cells continue to divide, their telomeres continue to shorten. Once many telomeres become too short to function, the unprotected chromosome ends generate end-to-end fusions and dicentric chromosomes, leading to many forms of genome instability. Ultimately, telomerase reactivation provides a route out of telomere crisis by healing critically shortened telomeres and improving genomic stability, thereby increasing cell viability. The resulting tumour will have active telomerase and a heavily rearranged genome.
Continued growth past the senescence barrier can occur in cells that lack the p53 and RB tumour suppressor pathways, rendering their cell cycle transitions impervious to inhibition through ATM and ATR signalling. Continued telomere shortening eventually leads to cells with numerous dysfunctional telomeres, thereby increasing the chance that one dysfunctional telomere becomes fused to another. Consequently, cells in telomere crisis have end-to-end fused dicentric chromosomes, which lead to mitotic mis-segregation and genomic instability. Cells in telomere crisis undergo frequent cell death. A common assumption is that this loss of viability is driven by chromosome breakage and mis-segregation, although it may also involve additional telomere deprotection during an extended mitotic arrest that occurs in some of the cells71,72.
Mammalian cells can use two types of end-joining pathways to repair DSBs: classical NHEJ (c-NHEJ) and alternative NHEJ (alt-NHEJ)73,74. c-NHEJ relies on the Ku70–Ku80 heterodimer and DNA ligase 4 and can either be accurate or result in small deletions. By contrast, alt-NHEJ, which is mediated by poly(ADP- ribose) polymerase 1 (PARP1) and DNA ligase 3, creates insertions and more extensive deletions. Telomere fusions formed during telomere crisis in cultured cells are mediated by alt-NHEJ and exhibit insertion of new sequences at the fusion point75,76. Similarly, alt-NHEJ has been implicated in telomere fusion in human cancer77,78 and in telomere fusions in mouse models79. By contrast, when telomeres are compromised through the loss of TRF2, their repair is carried out by c-NHEJ80–82. The reason for this difference is not yet clear.
Genome instability in cells undergoing telomere crisis was initially found to give rise to chromosome gains and losses (aneuploidy), translocations, gene loss (manifested as loss of heterozygosity (LOH)) and regional amplification through breakage–fusion–bridge (BFB) cycles1,80,83. However, it has recently become clear that the repertoire of genomic alterations that can be ascribed to telomere crisis is more extensive and includes whole-genome reduplication, chromothripsis and kataegis3,8,9.
BFB cycles and their associated chromosomal rearrangements
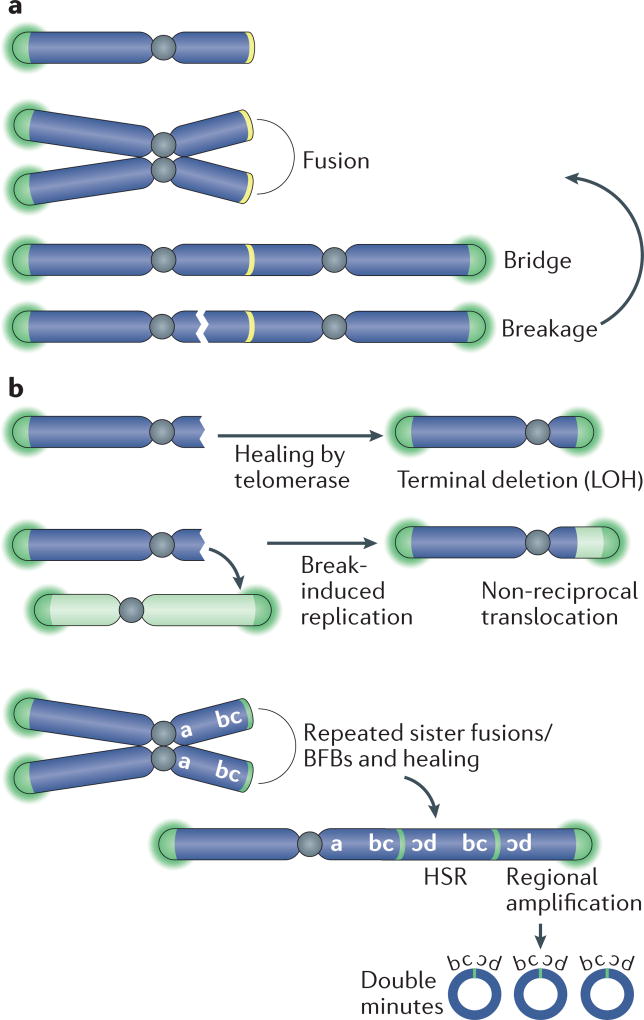
BFB cycles, first observed more than half a century ago by Barbara McClintock84, can occur when dicentric chromosomes (including those formed by telomere fusion) break, followed by a second fusion of the broken ends in the daughter cell85 (FIG. 4a). Telomere fusions can occur between different chromosomes or between sister chromatids after DNA replication, thus leading to different outcomes86. Collectively, BFB cycles can lead to three outcomes that are pertinent to cancer: LOH, non-reciprocal translocations and gene amplification.
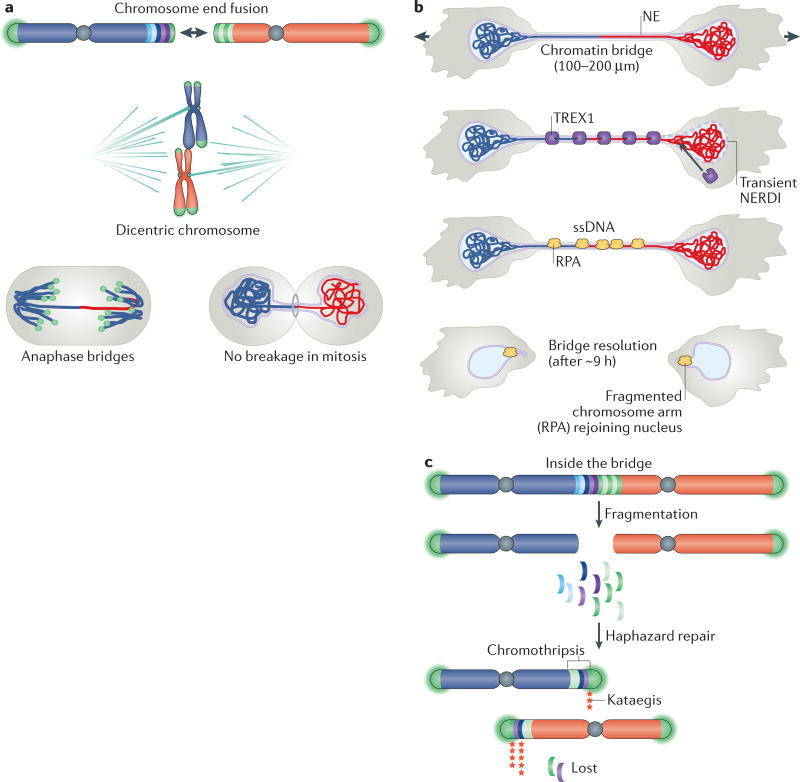
Figure 4. BFB cycles and chromosomal rearrangements during telomere crisis.
a | Breakage–fusion–bridge (BFB) cycles can occur when telomere fusion generates a dicentric chromosome. During anaphase, the mitotic spindle pulls this dicentric chromosome towards opposite spindle poles, thereby generating the widely observed anaphase bridges. During cell division, the dicentric chromosome undergoes breakage and the broken ends fuse again, giving rise to another dicentric chromosome. b | BFB cycles can be interrupted by telomerase-mediated telomere healing. If this process occurs following breakage, it can result in the formation of a terminal chromosome deletion and loss of heterozygosity (LOH). Alternatively, broken chromosomes can be repaired by break-induced replication, yielding a non-reciprocal translocation. Repeated cycles of BFB that occur between sister chromatids can result in regional amplification and the generation of a homogeneously staining region (HSR) following chromosome staining. This HSR consists of multiple amplicons of inverted repeats. Excision of the amplified sequences out of the chromosome will generate circular double-minute chromosomes.
LOH, which is frequent at cancer-relevant loci, can occur when a dicentric chromosome breaks and one of the daughter cells inherits a chromosome with a terminal deletion (FIG. 4b). Non-reciprocal trans-locations could arise when the DNA end of a broken chromosome invades another chromosome and copies part of this chromosome through a process called break-induced replication87,88. Non-reciprocal trans-locations occur during tumorigenesis in mice with shortening telomeres1 and are a frequent class of rearrangements in cancer89. Sequence analysis of more than 1,000 telomere fusion events has shown that a chromosome end lacking telomere protection can recombine with diverse chromosome-internal loci90.
Gene amplification can result when the telomere fusion event involves sister chromatids, thus creating a large palindrome (FIG. 4b). Subsequent asymmetric breakage of such an isochromosome and multiple BFB cycles can then generate amplicons that are organized in inverted repeats91. BFB cycles have been demonstrated to initiate gene amplification in human cancer cells and in hamster cells92–94. Moreover, the inverted amplicon arrangements that are typical of BFB cycles have been observed in many cancer types, including pancreatic cancer, oesophageal cancer, breast cancer and leukaemias91,95–98.
Chromothripsis
Recently, chromothripsis was shown to be one of the outcomes of experimentally induced telomere crisis. Chromothripsis is a mutagenic process whereby one or more chromosomal regions undergo catastrophic shattering in a single event, followed by an apparently haphazard repair of the DNA fragments. This process results in genomes in which one or few chromosome segments are affected by tens to hundreds of genomic rearrangements99. Chromothripsis has been observed in diverse tumour types, especially those with p53 loss100,101, and several studies have noted an association between BFB cycles and chromothripsis91,98,102. Consistent with these associations, chromothripsis was demonstrated to be the result of telomere crisis induced by the inactivation of the shelterin subunit TRF2 in p53-deficient and RB-deficient epithelial cells3. This study used live-cell imaging to determine the fate of dicentric chromosomes formed during telomere crisis and showed that dicentric chromosomes do not break during mitosis. This finding was in agreement with work in yeast cells and a subsequent analysis in human cells3,103,104 (FIG. 5a). These dicentric chromosomes invariably persist through mitosis and form long chromatin bridges that connect the daughter cells well into the next G1 phase3. These chromatin bridges contain a nuclear envelope that is contiguous with the nuclear envelop of the connected nuclei. However, when chromatin bridges are formed, there is frequent rupture of the nuclear envelope of the connected nuclei, resulting in mixing of the nuclear and cytoplasmic contents. Spontaneous nuclear envelope rupture has been observed in cancer cell lines and is frequent in micronuclei105,106. It is not clear how chromatin bridges induce nuclear envelope rupturing, but lamin depletion from the nuclear envelope may have a role as lamin B1 overexpression suppressed the ruptures, and lamin depletion can promote envelope rupture in cancer cell lines3,105. A second source of nuclear envelope rupture may be the deformation of the two nuclei connected by the stretching chromatin bridge. The dicentric chromosome in the bridge seems to exert pulling forces on the nuclear envelope, perhaps because it is attached to the nuclear lamins107. In support of this view, two recent studies have shown that cell migration through tight constrictions induces nuclear envelope rupture in the squeezed nuclei108,109.
Figure 5. Chromothripsis and kataegis in telomere crisis.
a | Dicentric chromosomes formed by telomere fusion rarely, if ever, break during mitosis and instead form chromatin bridges. b | Daughter nuclei connected by chromatin bridges undergo frequent nuclear envelope (NE) rupture in interphase (NERDI), resulting in the accumulation of 3′ repair exonuclease 1 (TREX1) on bridge DNA. TREX1-mediated resection of DNA leads to the formation of single-stranded DNA (ssDNA), which is bound by replication protein A (RPA), and bridge resolution. Bridge fragments are internalized into the nucleus where they remain associated with RPA for approximately 24 hours. c | Part of the dicentric chromosome that is present in the chromatin bridge undergoes extensive fragmentation followed by haphazard repair, which yields a chromothriptic chromosome in which many original chromosome fragments are lost and retained fragments are present in seemingly random order and orientation. Chromothriptic breakpoints are frequently associated with kataegis mutation clusters.
After persisting for many hours, the chromatin bridges are resolved by 3′ repair exonuclease 1 (TREX1) (FIG. 5b), a highly abundant and widely expressed 3′ exonuclease that degrades DNA species in the cytoplasm110–113. TREX1 seems to gain access to the chromatin bridge during nuclear envelope rupture3. The enzyme may preferentially attack the DNA in the chromatin bridges because they lack the nucleosomes that normally repress TREX1 activity3. The exonuclease creates extensive single-stranded DNA in chromatin bridges, leading to an accumulation of the single-stranded DNA-binding protein replication protein A (RPA) on the bridges (FIG. 5b). The nicks in the double-stranded DNA that allow TREX1 to initiate resection were shown to exist in chromatin bridges114, but their source is unknown. Eventually, TREX1 digestion is thought to resolve the chromatin bridge once the resection of the Watson and Crick strands of the DNA converges.
Following chromatin bridge resolution, the remnants of the dicentric chromosome are reincorporated back into the nuclear genome, but continue to be marked by the presence of RPA, indicating that single-stranded DNA persists3. As the RPA mark usually dissipates within one cell cycle, repair of the fragmented dicentric chromosome is presumed to occur during this period3. The exact repair pathways that are responsible for generating the chromothriptic product have not yet been determined.
Fragmentation of chromatin bridge DNA by TREX1 could explain the regional DNA breaks that are typical of chromothripsis because only the portion of the dicentric chromosome residing inside the chromatin bridge is attacked by TREX1. Moreover, the repair of these fragments in the primary nucleus is consistent with the catastrophic, but localized, rearrangements that are observed in chromothripsis (FIG. 5c).
The observation that dicentric chromosomes persist through mitosis intact, suffer extensive fragmentation and give rise to chromothripsis is not in conflict with a telomeric origin of BFB cycles. Chromatin bridges can be resolved even in the absence of TREX1, indicating that other mechanisms are at work3. Potentially a TREX1-independent pathway for bridge resolution could involve a nuclease that makes a single DSB and does not fragment the chromatin in the bridge. Such a broken dicentric chromosome could initiate BFB cycles by fusing the broken end with another broken end or, after DNA replication, by fusing with the sister chromatids. The genome rearrangements induced through this pathway may become clear from the analysis of genome instability in TREX1-deficient cells progressing through telomere crisis.
Another source of chromothripsis in cancer may be chromosomes that mis-segregate into micronuclei. The chromosome in a micronucleus experiences DNA damage, extensive fragmentation and subsequent repair, leading to shattering of the entire chromosome115,116. In addition, it was recently shown that simultaneous TRF2 depletion and inhibition of the spindle assembly checkpoint kinase MPS1 can also result in chromothripsis, but the precise molecular pathway is not clear117,118.
Kataegis
Chromothripsis induced by experimental telomere crisis is often accompanied by kataegis (FIG. 5c). Kataegis is a hypermutation pattern of clustered C>T and C>G changes at TpC dinucleotides119. Kataegis is thought to result from the activity of the apolipoprotein B mRNA-editing catalytic subunit (APOBEC) family of enzymes120–122, which can deaminate cytosine residues to generate uracil, and therefore act as mutators123. Many APOBECs are active in the cytoplasm, where they restrict RNA and DNA virus infection, most notably HIV, and other parasitic genomes, thereby contributing to an innate retroviral defence124. APOBECs preferentially target single-stranded DNA, and can produce a cluster of strand-coordinated mutations that affect cytosine bases in the same (Watson or Crick) strand. Consistent with APOBEC activity, kataegis is found at the breakpoints of chromothriptic rearrangements created by telomere crisis3. A possible explanation for this observation is that the extensive single-stranded DNA that accumulates following TREX1-mediated resection serves as a substrate for APOBEC deaminases.
Telomere-driven tetraploidy
Finally, telomere crisis can induce tetraploidization (doubling the set of chromosomes)8,9, which is inferred to be a frequent event during the development of human cancers125. Many human tumour cell lines have a near-tetraploid or hyper-triploid karyotype, which is indicative of past tetraploidization125. Tetraploidization can promote tumorigenesis9,125–133, and tetraploid cells have a high tolerance of chromosome mis-segregation and resilience to chromosomal instability134.
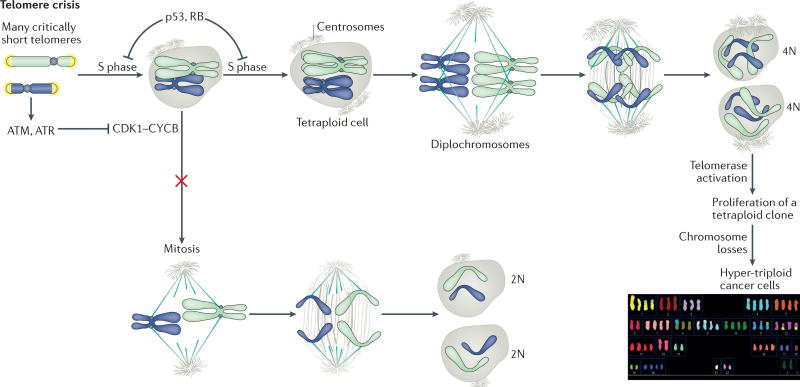
Tetraploidization can be induced in cells that lack the p53 and RB pathways, which have a high load of dysfunctional telomeres8,9 (FIG. 6). The mechanism of tetraploidization involves persistent ATM-dependent and/or ATR-dependent signalling induced by irreparably damaged telomeres. This signalling leads to a prolonged G2 phase and ultimately a bypass of mitosis and entry into a G1-like state. A second S phase then results in whole-genome reduplication and tetraploidy. Tetraploidization is observed following experimental inactivation of shelterin and in p53-deficient and RB-deficient human cells undergoing telomere crisis.
Figure 6. Tetraploidization during telomere crisis.
Telomere crisis can lead to persistent DNA damage signalling when repair fails to join all the unprotected ends and dysfunctional telomeres persist. The persistent ATM and ATR signalling and activation of their downstream effector kinases checkpoint kinase 2 (CHK2) and CHK1, respectively, results in prolonged inhibition of cyclin-dependent kinase 1 (CDK1)–cyclin B (CYCB), thus blocking entry into mitosis. Eventually, cells bypass mitosis, enter a G1-like state and then undergo a second S phase. The resulting tetraploid cells have diplochromosomes in the first mitosis following endoreduplication. Subsequently, the cells undergo frequent chromosome losses, leading to the hyper-triploid cells that are frequently observed in cancer. The example karyotype shown is from Capan-2, a hyper-triploid pancreatic cancer cell line (http://www.pawefish.path.cam.ac.uk/PancCellLineDescriptions/Capan-2.html), courtesy of Vorapan Sirivatanauksorn and Paul Edwards.
The prevalence of telomere crisis in cancer
Telomere crisis may be a frequent event during the development of human epithelial cancers, which initially lack telomerase. Shorter telomeres are frequently observed in cancer relative to their adjacent normal tissue135–142. Anaphase bridges, which can be formed by telomere– telomere fusion, have been observed in human cancer samples, including in early-stage colorectal tumours52. However, a consideration of anaphase bridges may overestimate the level of telomere dysfunction because they could also result from other defects, including errors in DNA decatenation143 and cohesin resolution144.
Telomere crisis in breast cancer has been particularly well documented. An analysis of genome instability and other features associated with telomere crisis, including anaphase and chromatin bridges, suggests that transition through telomere crisis in breast cancer occurs during progression from usual ductal hyperplasia (UDH) to ductal carcinoma in situ (DCIS)145. This phenomenon is consistent with the higher rate of chromosome aberrations in DCIS than in UDH146, the shortened telomeres found in DCIS147 and the activation of telomerase in DCIS148.
Methods to directly detect the scars of prior telomere crisis in cancer genomes have now been developed75,149,150. The telomere–telomere fusions that are typical of telomere crisis can be detected with a PCR-based assay that uses correctly oriented primers situated in the subtelomeric DNA of two (or more) chromosome ends75,150. Using this approach, evidence for past telomere crisis has been obtained in CLL as well as in breast cancer, colorectal adenomas and other solid tumours75,77,78. In colorectal cancer, telomere fusion occurs during the adenoma–carcinoma transition and may also be present before the occurrence of most somatic mutations78. These studies have also revealed a prognostic value to stratifying patients according to the length of the shortest telomeres — as determined by a PCR assay that measures individual telomere lengths — and the likelihood that telomere fusions will take place151,152. In CLL and invasive ductal carcinoma of the breast, overall survival is shorter when the telomeres are in a size range expected to result in telomere fusions151,152.
Telomerase activation
Telomerase activation is often accomplished through mutations in the TERT promoter57,58. These mutations are the most common mutations in non-coding sequences in cancer and are found in a long, and almost certainly growing, list of cancers153–158. Similar to the inherited TERT promoter mutations, the sporadic mutations (–57A>C, –124C>T and –146C>T) occur near the transcription start site where they create de novo binding sites for ETS transcription factors. An analysis of these mutations in urothelial cancers showed that they are correlated with higher levels of TERT mRNA and protein levels and enzymatic activity and with greater telomere length159. In glioblastomas, sporadic TERT mutations were shown to activate transcription by enabling the recruitment of the transcription factor GA-binding protein α-chain (GABPA)160. Introduction of these mutations into embryonic stem cells prevented TERT silencing upon differentiation and resulted in increased telomerase activity that counters telomere shortening25.
TERT promoter mutations are not the only mechanism by which telomerase activity can be restored or enhanced. In neuroblastoma, telomerase activity is increased by recurrent genomic rearrangements that pair the TERT coding sequence with strong enhancer elements, thereby defining a subgroup of patients with poor prognoses60. However, in many human cancers, the mechanism by which telomerase is upregulated is yet to be determined. Furthermore, although most human cancers (~90%) escape telomere crisis by activating telomerase56, a significant minority of cancers use an alternative telomere maintenance system, referred to as alternative lengthening of telomeres (ALT)161. ALT is associated with mutations in the chromatin remodeller α-thalassaemia/mental retardation syndrome X-linked (AT R X ) both in vitro and in several human cancers, including glioblastomas162–165. The observations that TERT promoter mutations are usually mutually exclusive with mutations in ATRX support the notion that ALT may provide a telomerase-independent escape from telomere crisis166,167.
Telomere dysfunction following telomerase activation
The types and severity of genome instability induced by dysfunctional telomeres can vary between transient and persistent telomere dysfunction. In mouse models, continued telomere dysfunction can constrain cancer progression, whereas telomerase reactivation alleviates intratumoural DNA damage and leads to more aggressive tumour progression and metastasis168.
However, some telomere dysfunction may persist even after telomerase activation and exit from telomere crisis. Changes in telomere length can occur owing to stochastic telomere loss, as has been demonstrated in squamous cell and bladder cell carcinoma cell lines169,170, and ongoing telomere dysfunction has been found in cancer cells with ALT162,171. Telomere loss at individual chromosome ends is sufficient to produce many of the rearrangements seen in telomere crisis and in cancer, including amplifications, LOH, translocations, chromosome non-disjunction during mitosis, and the formation of isochromosomes and ring chromosomes170,172. Even limited telomere dysfunction can wreak havoc on the genome because instability at individual chromosome ends can be transferred to other chromosomes through non-reciprocal translocations172,173. Telomeres in human cancer are often shorter than in normal tissues135. It is possible that this setting of short telomere length reflects selection for a telomere length distribution that affords a low level of genome instability without diminishing cell viability.
Perspectives
An attractive feature of telomere crisis as a source of genome instability in cancer is its transient nature. A mutator phenotype is favoured when extrinsic or intrinsic forces demand the generation of variants. This process can enable cancer cell populations to adapt rapidly to new challenges presented by shifting environments. However, persistence of a mutator phenotype comes at a cost because most mutations are deleterious to cellular fitness. The brief, or at least transient, episode of genomic instability offered by dysfunctional telomeres avoids a persistent mutator phenotype that might hamper cell proliferation10. Ultimately, telomerase reactivation provides a route out of telomere crisis, stabilizing the genome and rescuing cellular fitness.
Although much has been learned in recent years about the role of telomere crisis in cancer development, many basic questions remain. The current list of known genome rearrangements in cancer that follow telomere crisis is probably not comprehensive. For example, telomere crisis may contribute to chromoplexy, in which chains of translocations link several chromosomes in a temporally constrained event174. Bioinformatic methods to detect the remnants of telomere–telomere fusions in entire cancer genome sequencing data sets need to be developed to fully understand the relationship between chromosome fusion events and consequent chromosome rearrangements. Future studies will help to reveal the mechanisms underlying the complexity of the cancer genome, and with continued investment, these insights may be translated into valuable prognostic indicators and more effective treatments.
Acknowledgments
The authors thank S. Yu and the de Lange laboratory for discussions and help with this manuscript. The authors’ work is supported by grants from the US National Institutes of Health (CA181090, AG016642 and K99CA212290), the STARR Cancer Consortium and the Breast Cancer Research Foundation.
Glossary
- ATM
A PI3K–related protein kinase that initiates the response to double-strand breaks, with crucial roles in cell cycle regulation and DNA repair.
- ATR
A PI3K–related protein kinase that responds to the formation of single-stranded DNA, with a crucial role in the response to replication stress and double-strand breaks.
- Non-homologous end joining
A major double-strand break repair pathway that does not rely on sequence homology and can result in small insertions and deletions at the site of repair.
- Hayflick limit
The finite proliferation potential of primary human cells.
- Dicentric chromosomes
Abnormal chromosomes with two centromeres that can result from telomere–telomere fusion.
- Break-induced replication
An origin of replication-independent replication restart that is initiated by the invasion of resected DNA into homologous sequences.
- Micronuclei
Abnormal, small nuclei containing one or more chromosome (fragments);. often formed as a result of mitotic chromosome segregation defects.
- Lamin
An intermediate filament protein that imparts structural rigidity to the nucleus by assembling into a meshwork at the inner nuclear membrane.
- Hyper-triploid karyotype
A genome that contains more than three (3N) but less than four (4N) sets of chromosomes.
- Anaphase bridges
DNA bridges that connect chromatin masses undergoing separation during anaphase and can be observed with conventional DNA staining techniques.
- Usual ductal hyperplasia
A benign overgrowth of cells that line the ducts or milk glands and is associated with an elevated risk of breast cancer.
- Ductal carcinoma in situ
A noninvasive, early form of breast cancer characterized by proliferative, malignant cells that are confined to the milk duct.
- Alternative lengthening of telomeres
A telomere lengthening mechanism that relies on homologous recombination-mediated DNA copying to counteract telomere shortening.
- Chromoplexy
A class of complex DNA rearrangements frequently observed in prostate cancer, which is characterized by multiple chromatin rearrangements that arise in a highly interdependent manner.
Footnotes
Competing interests statement
The authors declare no competing interests.
References
- 1.Artandi SE, et al. Telomere dysfunction promotes non-reciprocal translocations and epithelial cancers in mice. Nature. 2000;406:641–645. doi: 10.1038/35020592. Demonstrates that telomere attrition in p53-mutant mice promotes epithelial cancers through the formation of chromosome rearrangements. [DOI] [PubMed] [Google Scholar]
- 2.Artandi SE, DePinho RA. Telomeres and telomerase in cancer. Carcinogenesis. 2010;31:9–18. doi: 10.1093/carcin/bgp268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maciejowski J, Li Y, Bosco N, Campbell PJ, de Lange T. Chromothripsis and kataegis induced by telomere crisis. Cell. 2015;163:1641–1654. doi: 10.1016/j.cell.2015.11.054. Shows that dicentric chromosomes formed during telomere crisis persist through mitosis, are fragmented by TREX1 in G1 phase and give rise to chromothripsis and kataegis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 5.Yates LR, Campbell PJ. Evolution of the cancer genome. Nat. Rev. Genet. 2012;13:795–806. doi: 10.1038/nrg3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Willis NA, Rass E, Scully R. Deciphering the code of the cancer genome: mechanisms of chromosome rearrangement. Trends Cancer. 2015;1:217–230. doi: 10.1016/j.trecan.2015.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.O’Hagan RC, et al. Telomere dysfunction provokes regional amplification and deletion in cancer genomes. Cancer Cell. 2002;2:149–155. doi: 10.1016/s1535-6108(02)00094-6. Demonstrates that tumours with telomere dysfunction have higher levels of genome instability, with frequent amplifications and deletions. [DOI] [PubMed] [Google Scholar]
- 8.Davoli T, Denchi EL, de Lange T. Persistent telomere damage induces bypass of mitosis and tetraploidy. Cell. 2010;141:81–93. doi: 10.1016/j.cell.2010.01.031. Finds that persistent telomere dysfunction and consequent DNA damage signalling lead to bypass of mitosis and tetraploidization. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davoli T, de Lange T. Telomere-driven tetraploidization occurs in human cells undergoing crisis and promotes transformation of mouse cells. Cancer Cell. 2012;21:765–776. doi: 10.1016/j.ccr.2012.03.044. Reports that telomere-driven tetraploidy occurs in human cells during telomere crisis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Lange T. Telomere-related genome instability in cancer. Cold Spring Harb. Symp. Quant. Biol. 2005;70:197–204. doi: 10.1101/sqb.2005.70.032. [DOI] [PubMed] [Google Scholar]
- 11.Blackburn EH, Collins K. Telomerase: an RNP enzyme synthesizes DNA. Cold Spring Harb Perspect. Biol. 2011;3:a003558. doi: 10.1101/cshperspect.a003558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lingner J, et al. Reverse transcriptase motifs in the catalytic subunit of telomerase. Science. 1997;276:561–567. doi: 10.1126/science.276.5312.561. [DOI] [PubMed] [Google Scholar]
- 13.Hamma T, Ferré-D’Amaré AR. The box H/ACA ribonucleoprotein complex: interplay of RNA and protein structures in post-transcriptional RNA modification. J. Biol. Chem. 2010;285:805–809. doi: 10.1074/jbc.R109.076893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nakamura TM, et al. Telomerase catalytic subunit homologs from fission yeast and human. Science. 1997;277:955–959. doi: 10.1126/science.277.5328.955. [DOI] [PubMed] [Google Scholar]
- 15.Meyerson M, et al. hEST2, the putative human telomerase catalytic subunit gene, is up-regulated in tumor cells and during immortalization. Cell. 1997;90:785–795. doi: 10.1016/s0092-8674(00)80538-3. [DOI] [PubMed] [Google Scholar]
- 16.Feng J, et al. The RNA component of human telomerase. Science. 1995;269:1236–1241. doi: 10.1126/science.7544491. [DOI] [PubMed] [Google Scholar]
- 17.Egan ED, Collins K. Biogenesis of telomerase ribonucleoproteins. RNA. 2012;18:1747–1759. doi: 10.1261/rna.034629.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Darzacq X, et al. Stepwise RNP assembly at the site of H/ACA RNA transcription in human cells. J. Cell Biol. 2006;173:207–218. doi: 10.1083/jcb.200601105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kiss T, Fayet-Lebaron E, Jády BE. Box H/ACA small ribonucleoproteins. Mol. Cell. 2010;37:597–606. doi: 10.1016/j.molcel.2010.01.032. [DOI] [PubMed] [Google Scholar]
- 20.Schmidt JC, Cech TR. Human telomerase: biogenesis, trafficking, recruitment, and activation. Genes Dev. 2015;29:1095–1105. doi: 10.1101/gad.263863.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hockemeyer D, Collins K. Control of telomerase action at human telomeres. Nat. Struct. Mol. Biol. 2015;22:848–852. doi: 10.1038/nsmb.3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shay JW. Role of telomeres and telomerase in aging and cancer. Cancer Discov. 2016;6:584–593. doi: 10.1158/2159-8290.CD-16-0062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bodnar AG, et al. Extension of life-span by introduction of telomerase into normal human cells. Science. 1998;279:349–352. doi: 10.1126/science.279.5349.349. Establishes a causal relationship between telomere shortening and cellular senescence. [DOI] [PubMed] [Google Scholar]
- 24.Cristofari G, Lingner J. Telomere length homeostasis requires that telomerase levels are limiting. EMBO J. 2006;25:565–574. doi: 10.1038/sj.emboj.7600952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chiba K, et al. Cancer-associated TERT promoter mutations abrogate telomerase silencing. eLife. 2015;4:e07918. doi: 10.7554/eLife.07918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ramirez RD, et al. Putative telomere-independent mechanisms of replicative aging reflect inadequate growth conditions. Genes Dev. 2001;15:398–403. doi: 10.1101/gad.859201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vaziri H, Benchimol S. Reconstitution of telomerase activity in normal human cells leads to elongation of telomeres and extended replicative life span. Curr. Biol. 1998;8:279–282. doi: 10.1016/s0960-9822(98)70109-5. [DOI] [PubMed] [Google Scholar]
- 28.Gomes NMV, et al. Comparative biology of mammalian telomeres: hypotheses on ancestral states and the roles of telomeres in longevity determination. Aging Cell. 2011;10:761–768. doi: 10.1111/j.1474-9726.2011.00718.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harley CB, Futcher AB, Greider CW. Telomeres shorten during ageing of human fibroblasts. Nature. 1990;345:458–460. doi: 10.1038/345458a0. Demonstrates that the telomeres of human fibroblasts shorten during growth in culture. [DOI] [PubMed] [Google Scholar]
- 30.Huffman KE, Levene SD, Tesmer VM, Shay JW, Wright WE. Telomere shortening is proportional to the size of the G-rich telomeric 3′-overhang. J. Biol. Chem. 2000;275:19719–19722. doi: 10.1074/jbc.M002843200. [DOI] [PubMed] [Google Scholar]
- 31.Miyake Y, et al. RPA-like mammalian Ctc1-Stn1-Ten1 complex binds to single-stranded DNA and protects telomeres independently of the Pot1 pathway. Mol. Cell. 2009;36:193–206. doi: 10.1016/j.molcel.2009.08.009. [DOI] [PubMed] [Google Scholar]
- 32.Surovtseva YV, et al. Conserved telomere maintenance component 1 interacts with STN1 and maintains chromosome ends in higher eukaryotes. Mol. Cell. 2009;36:207–218. doi: 10.1016/j.molcel.2009.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang C, Dai X, Chai W. Human Stn1 protects telomere integrity by promoting efficient lagging-strand synthesis at telomeres and mediating C-strand fill-in. Cell Res. 2012;22:1681–1695. doi: 10.1038/cr.2012.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dai X, et al. Molecular steps of G-overhang generation at human telomeres and its function in chromosome end protection. EMBO J. 2010;29:2788–2801. doi: 10.1038/emboj.2010.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu P, Takai H, de Lange T. Telomeric 3′ overhangs derive from resection by Exo1 and Apollo and fill-in by POT1b–associated CST. Cell. 2012;150:39–52. doi: 10.1016/j.cell.2012.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang F, et al. Human CST has independent functions during telomere duplex replication and C-strand fill-in. Cell Rep. 2012;2:1096–1103. doi: 10.1016/j.celrep.2012.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu P, van Overbeek M, Rooney S, de Lange T. Apollo contributes to G overhang maintenance and protects leading-end telomeres. Mol. Cell. 2010;39:606–617. doi: 10.1016/j.molcel.2010.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kasbek C, Wang F, Price CM. Human TEN1 maintains telomere integrity and functions in genome-wide replication restart. J. Biol. Chem. 2013;288:30139–30150. doi: 10.1074/jbc.M113.493478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.d’Adda di Fagagna F, et al. A DNA damage checkpoint response in telomere-initiated senescence. Nature. 2003;426:194–198. doi: 10.1038/nature02118. Finds that telomeres in senescent cells exhibit the hallmarks of DNA DSBs. [DOI] [PubMed] [Google Scholar]
- 40.Zou Y, Sfeir A, Gryaznov SM, Shay JW, Wright WE. Does a sentinel or a subset of short telomeres determine replicative senescence? Mol. Biol. Cell. 2004;15:3709–3718. doi: 10.1091/mbc.E04-03-0207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hemann MT, Strong MA, Hao LY, Greider CW. The shortest telomere, not average telomere length, is critical for cell viability and chromosome stability. Cell. 2001;107:67–77. doi: 10.1016/s0092-8674(01)00504-9. [DOI] [PubMed] [Google Scholar]
- 42.Takai H, Smogorzewska A, de Lange T. DNA damage foci at dysfunctional telomeres. Curr. Biol. 2003;13:1549–1556. doi: 10.1016/s0960-9822(03)00542-6. [DOI] [PubMed] [Google Scholar]
- 43.Jacobs JJL, de Lange T. Significant role for p16INK4a in p53-independent telomere-directed senescence. Curr. Biol. 2004;14:2302–2308. doi: 10.1016/j.cub.2004.12.025. [DOI] [PubMed] [Google Scholar]
- 44.Karlseder J, Smogorzewska A, de Lange T. Senescence induced by altered telomere state, not telomere loss. Science. 2002;295:2446–2449. doi: 10.1126/science.1069523. [DOI] [PubMed] [Google Scholar]
- 45.Shay JW, Pereira-Smith OM, Wright WE. A role for both RB and p53 in the regulation of human cellular senescence. Exp. Cell Res. 1991;196:33–39. doi: 10.1016/0014-4827(91)90453-2. [DOI] [PubMed] [Google Scholar]
- 46.Hara E, Tsurui H, Shinozaki A, Nakada S, Oda K. Cooperative effect of antisense-Rb and antisense-p53 oligomers on the extension of life span in human diploid fibroblasts, TIG-1. Biochem. Biophys. Res. Commun. 1991;179:528–534. doi: 10.1016/0006-291x(91)91403-y. [DOI] [PubMed] [Google Scholar]
- 47.Shay JW, Wright WE, Brasiskyte D, Van der Haegen BA. E6 of human papillomavirus type 16 can overcome the M1 stage of immortalization in human mammary epithelial cells but not in human fibroblasts. Oncogene. 1993;8:1407–1413. [PubMed] [Google Scholar]
- 48.Brown JP, Wei W, Sedivy JM. Bypass of senescence after disruption of p21CIP1/WAF1 gene in normal diploid human fibroblasts. Science. 1997;277:831–834. doi: 10.1126/science.277.5327.831. [DOI] [PubMed] [Google Scholar]
- 49.Chin L, et al. p53 deficiency rescues the adverse effects of telomere loss and cooperates with telomere dysfunction to accelerate carcinogenesis. Cell. 1999;97:527–538. doi: 10.1016/s0092-8674(00)80762-x. Shows that p53 is activated during telomere crisis to induce growth arrest and suppress transformation. [DOI] [PubMed] [Google Scholar]
- 50.Smogorzewska A, de Lange T. Different telomere damage signaling pathways in human and mouse cells. EMBO J. 2002;21:4338–4348. doi: 10.1093/emboj/cdf433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Greenberg RA, et al. Short dysfunctional telomeres impair tumorigenesis in the INK4aΔ2/3 cancer-prone mouse. Cell. 1999;97:515–525. doi: 10.1016/s0092-8674(00)80761-8. [DOI] [PubMed] [Google Scholar]
- 52.Rudolph KL, Millard M, Bosenberg MW, DePinho RA. Telomere dysfunction and evolution of intestinal carcinoma in mice and humans. Nat. Genet. 2001;28:155–159. doi: 10.1038/88871. [DOI] [PubMed] [Google Scholar]
- 53.Qi L, et al. Short telomeres and ataxia-telangiectasia mutated deficiency cooperatively increase telomere dysfunction and suppress tumorigenesis. Cancer Res. 2003;63:8188–8196. [PubMed] [Google Scholar]
- 54.Qi L, Strong MA, Karim BO, Huso DL, Greider CW. Telomere fusion to chromosome breaks reduces oncogenic translocations and tumour formation. Nat. Cell Biol. 2005;7:706–711. doi: 10.1038/ncb1276. [DOI] [PubMed] [Google Scholar]
- 55.Feldser DM, Greider CW. Short telomeres limit tumor progression in vivo by inducing senescence. Cancer Cell. 2007;11:461–469. doi: 10.1016/j.ccr.2007.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kim NW, et al. Specific association of human telomerase activity with immortal cells and cancer. Science. 1994;266:2011–2015. doi: 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- 57.Huang FW, et al. Highly recurrent TERT promoter mutations in human melanoma. Science. 2013;339:957–959. doi: 10.1126/science.1229259. Uses whole-genome sequencing in melanomas to identify activating mutations in the TERT promoter. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Horn S, et al. TERT promoter mutations in familial and sporadic melanoma. Science. 2013;339:959–961. doi: 10.1126/science.1230062. Identifies activating mutations in the TERT promoter through an analysis of melanoma-prone families. [DOI] [PubMed] [Google Scholar]
- 59.Beroukhim R, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Peifer M, et al. Telomerase activation by genomic rearrangements in high-risk neuroblastoma. Nature. 2015;526:700–704. doi: 10.1038/nature14980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Valentijn LJ, et al. TERT rearrangements are frequent in neuroblastoma and identify aggressive tumors. Nat. Genet. 2015;47:1411–1414. doi: 10.1038/ng.3438. [DOI] [PubMed] [Google Scholar]
- 62.Robles-Espinoza CD, et al. POT1 loss-of-function variants predispose to familial melanoma. Nat. Genet. 2014;46:478–481. doi: 10.1038/ng.2947. Links germline, loss-of-function variants of POT1 to melanoma susceptibility and increased telomere length. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shi J, et al. Rare missense variants in POT1 predispose to familial cutaneous malignant melanoma. Nat. Genet. 2014;46:482–486. doi: 10.1038/ng.2941. Reports the identification of unrelated, melanoma-prone families that carry variants of POT1 and show increased telomere lengths. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pinzaru AM, et al. Telomere replication stress induced by POT1 inactivation accelerates tumorigenesis. Cell Rep. 2016;15:2170–2184. doi: 10.1016/j.celrep.2016.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Machiela MJ, et al. Genetic variants associated with longer telomere length are associated with increased lung cancer risk among never-smoking women in Asia: a report from the female lung cancer consortium in Asia. Int. J. Cancer. 2015;137:311–319. doi: 10.1002/ijc.29393. Assesses telomere length using telomere length-associated single-nucleotide polymorphisms and finds that longer telomere length is associated with increased risk of non-Hodgkin lymphoma. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Machiela MJ, et al. Genetically predicted longer telomere length is associated with increased risk of B-cell lymphoma subtypes. Hum. Mol. Genet. 2016;25:1663–1676. doi: 10.1093/hmg/ddw027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ojha J, et al. Genetic variation associated with longer telomere length increases risk of chronic lymphocytic leukemia. Cancer Epidemiol. Biomarkers Prev. 2016;25:1043–1049. doi: 10.1158/1055-9965.EPI-15-1329. Shows that an inherited predisposition for longer telomeres is associated with an increased risk of CLL. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mangino M, et al. Genome-wide meta-analysis points to CTC1 and ZNF676 as genes regulating telomere homeostasis in humans. Hum. Mol. Genet. 2012;21:5385–5394. doi: 10.1093/hmg/dds382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Walsh KM, et al. Variants near TERT and TERC influencing telomere length are associated with high-grade glioma risk. Nat. Genet. 2014;46:731–735. doi: 10.1038/ng.3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rode L, Nordestgaard BG, Bojesen SE. Long telomeres and cancer risk among 95 568 individuals from the general population. Int. J. Epidemiol. 2016;45:1634–1643. doi: 10.1093/ije/dyw179. [DOI] [PubMed] [Google Scholar]
- 71.Hayashi MT, Cesare AJ, Fitzpatrick JAJ, Lazzerini Denchi E, Karlseder J. A telomere-dependent DNA damage checkpoint induced by prolonged mitotic arrest. Nat. Struct. Mol. Biol. 2012;19:387–394. doi: 10.1038/nsmb.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hayashi MT, Cesare AJ, Rivera T, Karlseder J. Cell death during crisis is mediated by mitotic telomere deprotection. Nature. 2015;522:492–496. doi: 10.1038/nature14513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Boboila C, Alt FW, Schwer B. Classical and alternative end-joining pathways for repair of lymphocyte-specific and general DNA double-strand breaks. Adv. Immunol. 2012;116:1–49. doi: 10.1016/B978-0-12-394300-2.00001-6. [DOI] [PubMed] [Google Scholar]
- 74.Smogorzewska A, Karlseder J, Holtgreve-Grez H, Jauch A, de Lange T. DNA ligase IV-dependent NHEJ of deprotected mammalian telomeres in G1 and G2. Curr. Biol. 2002;12:1635–1644. doi: 10.1016/s0960-9822(02)01179-x. [DOI] [PubMed] [Google Scholar]
- 75.Capper R, et al. The nature of telomere fusion and a definition of the critical telomere length in human cells. Genes Dev. 2007;21:2495–2508. doi: 10.1101/gad.439107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Oh S, et al. DNA ligase III and DNA ligase IV carry out genetically distinct forms of end joining in human somatic cells. DNA Repair. 2014;21:97–110. doi: 10.1016/j.dnarep.2014.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lin TT, et al. Telomere dysfunction and fusion during the progression of chronic lymphocytic leukemia: evidence for a telomere crisis. Blood. 2010;116:1899–1907. doi: 10.1182/blood-2010-02-272104. Shows that telomere shortening and fusions in CLL increase with advanced disease and correlate with large-scale genome rearrangements. [DOI] [PubMed] [Google Scholar]
- 78.Jones RE, et al. Escape from telomere-driven crisis is DNA ligase III dependent. Cell Rep. 2014;8:1063–1076. doi: 10.1016/j.celrep.2014.07.007. [DOI] [PubMed] [Google Scholar]
- 79.Maser RS, et al. DNA-dependent protein kinase catalytic subunit is not required for dysfunctional telomere fusion and checkpoint response in the telomerase-deficient mouse. Mol. Cell. Biol. 2007;27:2253–2265. doi: 10.1128/MCB.01354-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Roger L, et al. Extensive telomere erosion in the initiation of colorectal adenomas and its association with chromosomal instability. J. Natl Cancer Inst. 2013;105:1202–1211. doi: 10.1093/jnci/djt191. [DOI] [PubMed] [Google Scholar]
- 81.Celli GB, de Lange T. DNA processing is not required for ATM-mediated telomere damage response after TRF2 deletion. Nat. Cell Biol. 2005;7:712–718. doi: 10.1038/ncb1275. [DOI] [PubMed] [Google Scholar]
- 82.Celli GB, Denchi EL, de Lange T. Ku70 stimulates fusion of dysfunctional telomeres yet protects chromosome ends from homologous recombination. Nat. Cell Biol. 2006;8:885–890. doi: 10.1038/ncb1444. [DOI] [PubMed] [Google Scholar]
- 83.Riboni R, et al. Telomeric fusions in cultured human fibroblasts as a source of genomic instability. Cancer Genet. Cytogenet. 1997;95:130–136. doi: 10.1016/s0165-4608(96)00248-8. [DOI] [PubMed] [Google Scholar]
- 84.McClintock B. The behavior in successive nuclear divisions of a chromosome broken at meiosis. Proc. Natl Acad. Sci. USA. 1939;25:405–416. doi: 10.1073/pnas.25.8.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gisselsson D, et al. Chromosomal breakage-fusion-bridge events cause genetic intratumor heterogeneity. Proc. Natl Acad. Sci. USA. 2000;97:5357–5362. doi: 10.1073/pnas.090013497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Murnane JP. Telomeres and chromosome instability. DNA Repair. 2006;5:1082–1092. doi: 10.1016/j.dnarep.2006.05.030. [DOI] [PubMed] [Google Scholar]
- 87.Llorente B, Smith CE, Symington LS. Break-induced replication: what is it and what is it for? Cell Cycle. 2008;7:859–864. doi: 10.4161/cc.7.7.5613. [DOI] [PubMed] [Google Scholar]
- 88.Anand RP, Lovett ST, Haber JE. Break-induced DNA replication. Cold Spring Harb Perspect Biol. 2013;5:a010397. doi: 10.1101/cshperspect.a010397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Shih IM, et al. Evidence that genetic instability occurs at an early stage of colorectal tumorigenesis. Cancer Res. 2001;61:818–822. [PubMed] [Google Scholar]
- 90.Liddiard K, et al. Sister chromatid telomere fusions, but not NHEJ-mediated inter-chromosomal telomere fusions, occur independently of DNA ligases 3 and 4. Genome Res. 2016;26:588–600. doi: 10.1101/gr.200840.115. Uses single molecule analysis to demonstrate that a single dysfunctional telomere can fuse with diverse non-telomeric loci. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Li Y, et al. Constitutional and somatic rearrangement of chromosome 21 in acute lymphoblastic leukaemia. Nature. 2014;508:102. doi: 10.1038/nature13115. Uses whole-genome sequencing to show that, in leukaemia, dicentric chromosomes formed by telomere fusion or a Robertsonian translocation may precipitate chromothripsis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lo AWI, et al. DNA amplification by breakage/ fusion/bridge cycles initiated by spontaneous telomere loss in a human cancer cell line. Neoplasia. 2002;4:531–538. doi: 10.1038/sj.neo.7900267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ma C, Martin S, Trask B, Hamlin JL. Sister chromatid fusion initiates amplification of the dihydrofolate reductase gene in Chinese hamster cells. Genes Dev. 1993;7:605–620. doi: 10.1101/gad.7.4.605. [DOI] [PubMed] [Google Scholar]
- 94.Smith KA, Gorman PA, Stark MB, Groves RP, Stark GR. Distinctive chromosomal structures are formed very early in the amplification of CAD genes in Syrian hamster cells. Cell. 1990;63:1219–1227. doi: 10.1016/0092-8674(90)90417-d. [DOI] [PubMed] [Google Scholar]
- 95.Bignell GR, et al. Architectures of somatic genomic rearrangement in human cancer amplicons at sequence-level resolution. Genome Res. 2007;17:1296–1303. doi: 10.1101/gr.6522707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Campbell PJ, et al. The patterns and dynamics of genomic instability in metastatic pancreatic cancer. Nature. 2010;467:1113. doi: 10.1038/nature09460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Waddell N, et al. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature. 2015;518:495–501. doi: 10.1038/nature14169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Nones K, et al. Genomic catastrophes frequently arise in esophageal adenocarcinoma and drive tumorigenesis. Nat. Commun. 2014;5:5224. doi: 10.1038/ncomms6224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Stephens PJ, et al. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell. 2011;144:27–40. doi: 10.1016/j.cell.2010.11.055. Reports the discovery of chromothripsis using next-generation sequencing. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Rausch T, et al. Genome sequencing of pediatric medulloblastoma links catastrophic DNA rearrangements with TP53 mutations. Cell. 2012;148:59–71. doi: 10.1016/j.cell.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Jones MJK, Jallepalli PV. Chromothripsis: chromosomes in crisis. Dev. Cell. 2012;23:917. doi: 10.1016/j.devcel.2012.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Garsed DW, et al. The architecture and evolution of cancer neochromosomes. Cancer Cell. 2014;26:653–667. doi: 10.1016/j.ccell.2014.09.010. [DOI] [PubMed] [Google Scholar]
- 103.Lopez V, et al. Cytokinesis breaks dicentric chromosomes preferentially at pericentromeric regions and telomere fusions. Genes Dev. 2015;29:322–336. doi: 10.1101/gad.254664.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Pampalona J, et al. Chromosome bridges maintain kinetochore–microtubule attachment throughout mitosis and rarely break during anaphase. PLoS ONE. 2016;11:e0147420. doi: 10.1371/journal.pone.0147420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Vargas JD, Hatch EM, Anderson DJ, Hetzer MW. Transient nuclear envelope rupturing during interphase in human cancer cells. Nucleus. 2012;3:88–100. doi: 10.4161/nucl.18954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hatch EM, Fischer AH, Deerinck TJ, Hetzer MW. Catastrophic nuclear envelope collapse in cancer cell micronuclei. Cell. 2013;154:47–60. doi: 10.1016/j.cell.2013.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Guelen L, et al. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature. 2008;453:948–951. doi: 10.1038/nature06947. [DOI] [PubMed] [Google Scholar]
- 108.Raab M, et al. ESCRT III repairs nuclear envelope ruptures during cell migration to limit DNA damage and cell death. Science. 2016;352:7611–7362. doi: 10.1126/science.aad7611. [DOI] [PubMed] [Google Scholar]
- 109.Denais CM, et al. Nuclear envelope rupture and repair during cancer cell migration. Science. 2016;352:1–8. doi: 10.1126/science.aad7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lindahl T, Gally JA, Edelman GM. Properties of deoxyribonuclease 3 from mammalian tissues. J. Biol. Chem. 1969;244:5014–5019. [PubMed] [Google Scholar]
- 111.Höss M, et al. A human DNA editing enzyme homologous to the Escherichia coli DnaQ/MutD protein. EMBO J. 1999;18:3868–3875. doi: 10.1093/emboj/18.13.3868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Mazur DJ, Perrino FW. Structure and expression of the TREX1 and TREX2 3′-5′ exonuclease genes. J. Biol. Chem. 2001;276:14718–14727. doi: 10.1074/jbc.M010051200. [DOI] [PubMed] [Google Scholar]
- 113.Rice GI, Rodero MP, Crow YJ. Human disease phenotypes associated with mutations in TREX1. J. Clin. Immunol. 2015;35:235–243. doi: 10.1007/s10875-015-0147-3. [DOI] [PubMed] [Google Scholar]
- 114.Gisselsson D, et al. Telomere dysfunction triggers extensive DNA fragmentation and evolution of complex chromosome abnormalities in human malignant tumors. Proc. Natl Acad. Sci. USA. 2001;98:12683–12688. doi: 10.1073/pnas.211357798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Crasta K, et al. DNA breaks and chromosome pulverization from errors in mitosis. Nature. 2012;482:53–58. doi: 10.1038/nature10802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Zhang C-Z, et al. Chromothripsis from DNA damage in micronuclei. Nature. 2015;522:179–184. doi: 10.1038/nature14493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Santaguida S, Tighe A, D’Alise AM, Taylor SS, Musacchio A. Dissecting the role of MPS1 in chromosome biorientation and the spindle checkpoint through the small molecule inhibitor reversine. J. Cell Biol. 2010;190:73–87. doi: 10.1083/jcb.201001036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Mardin BR, et al. A cell-based model system links chromothripsis with hyperploidy. Mol. Syst. Biol. 2015;11:828–828. doi: 10.15252/msb.20156505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Nik-Zainal S, et al. Mutational processes molding the genomes of 21 breast cancers. Cell. 2012;149:979–993. doi: 10.1016/j.cell.2012.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Roberts SA, et al. Clustered mutations in yeast and in human cancers can arise from damaged long single-strand DNA regions. Mol. Cell. 2012;46:424–435. doi: 10.1016/j.molcel.2012.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Roberts SA, et al. An APOBEC cytidine deaminase mutagenesis pattern is widespread in human cancers. Nat. Genet. 2013;45:970–976. doi: 10.1038/ng.2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Chan K, et al. An APOBEC3A hypermutation signature is distinguishable from the signature of background mutagenesis by APOBEC3B in human cancers. Nat. Genet. 2015;47:1067–1072. doi: 10.1038/ng.3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Harris RS, Petersen-Mahrt SK, Neuberger MS. RNA editing enzyme APOBEC1 and some of its homologs can act as DNA mutators. Mol. Cell. 2002;10:1247–1253. doi: 10.1016/s1097-2765(02)00742-6. [DOI] [PubMed] [Google Scholar]
- 124.Harris RS, Dudley JP. APOBECs and virus restriction. Virology. 2015;480:131–145. doi: 10.1016/j.virol.2015.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Davoli T, de Lange T. The causes and consequences of polyploidy in normal development and cancer. Annu. Rev. Cell Dev. Biol. 2011;27:585–610. doi: 10.1146/annurev-cellbio-092910-154234. [DOI] [PubMed] [Google Scholar]
- 126.Shackney SE, et al. Model for the genetic evolution of human solid tumors. Cancer Res. 1989;49:3344–3354. [PubMed] [Google Scholar]
- 127.Galipeau PC, et al. 17p (p53) allelic losses, 4N (G2/tetraploid) populations, and progression to aneuploidy in Barrett’s esophagus. Proc. Natl Acad. Sci. USA. 1996;93:7081–7084. doi: 10.1073/pnas.93.14.7081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Olaharski AJ, et al. Tetraploidy and chromosomal instability are early events during cervical carcinogenesis. Carcinogenesis. 2006;27:337–343. doi: 10.1093/carcin/bgi218. [DOI] [PubMed] [Google Scholar]
- 129.Zack TI, et al. Pan-cancer patterns of somatic copy number alteration. Nat. Genet. 2013;45:1134–1140. doi: 10.1038/ng.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Nguyen HG, et al. Deregulated Aurora-B induced tetraploidy promotes tumorigenesis. FASEB J. 2009;23:2741–2748. doi: 10.1096/fj.09-130963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Duelli DM, et al. A virus causes cancer by inducing massive chromosomal instability through cell fusion. Curr. Biol. 2007;17:431–437. doi: 10.1016/j.cub.2007.01.049. [DOI] [PubMed] [Google Scholar]
- 132.Ganem NJ, Storchová Z, Pellman D. Tetraploidy, aneuploidy and cancer. Curr. Opin. Genet. Dev. 2007;17:157–162. doi: 10.1016/j.gde.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 133.Fujiwara T, et al. Cytokinesis failure generating tetraploids promotes tumorigenesis in p53-null cells. Nature. 2005;437:1043–1047. doi: 10.1038/nature04217. [DOI] [PubMed] [Google Scholar]
- 134.Dewhurst SM, et al. Tolerance of whole-genome doubling propagates chromosomal instability and accelerates cancer genome evolution. Cancer Discov. 2014;4:175–185. doi: 10.1158/2159-8290.CD-13-0285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.de Lange T, et al. Structure and variability of human chromosome ends. Mol. Cell. Biol. 1990;10:518–527. doi: 10.1128/mcb.10.2.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Hastie ND, et al. Telomere reduction in human colorectal carcinoma and with ageing. Nature. 1990;346:866–868. doi: 10.1038/346866a0. [DOI] [PubMed] [Google Scholar]
- 137.Furugori E, et al. Telomere shortening in gastric carcinoma with aging despite telomerase activation. J. Cancer Res. Clin. Oncol. 2000;126:481–485. doi: 10.1007/s004320000137. [DOI] [PubMed] [Google Scholar]
- 138.Mehle C, Ljungberg B, Roos G. Telomere shortening in renal cell carcinoma. Cancer Res. 1994;54:236–241. [PubMed] [Google Scholar]
- 139.Takagi S, et al. Telomere shortening and the clinicopathologic characteristics of human colorectal carcinomas. Cancer. 1999;86:1431–1436. [PubMed] [Google Scholar]
- 140.Sommerfeld HJ, et al. Telomerase activity: a prevalent marker of malignant human prostate tissue. Cancer Res. 1996;56:218–222. [PubMed] [Google Scholar]
- 141.Meeker AK, et al. Telomere shortening is an early somatic DNA alteration in human prostate tumorigenesis. Cancer Res. 2002;62:6405–6409. [PubMed] [Google Scholar]
- 142.Meeker AK, et al. Telomere length abnormalities occur early in the initiation of epithelial carcinogenesis. Clin. Cancer Res. 2004;10:3317–3326. doi: 10.1158/1078-0432.CCR-0984-03. [DOI] [PubMed] [Google Scholar]
- 143.Clarke DJ, Johnson RT, Downes CS. Topoisomerase II inhibition prevents anaphase chromatid segregation in mammalian cells independently of the generation of DNA strand breaks. J. Cell Sci. 1993;105:563–569. doi: 10.1242/jcs.105.2.563. [DOI] [PubMed] [Google Scholar]
- 144.Hauf S, Waizenegger IC, Peters JM. Cohesin cleavage by separase required for anaphase and cytokinesis in human cells. Science. 2001;293:1320–1323. doi: 10.1126/science.1061376. [DOI] [PubMed] [Google Scholar]
- 145.Chin K, et al. In situ analyses of genome instability in breast cancer. Nat. Genet. 2004;36:984–988. doi: 10.1038/ng1409. [DOI] [PubMed] [Google Scholar]
- 146.O’Connell P, et al. Analysis of loss of heterozygosity in 399 premalignant breast lesions at 15 genetic loci. J. Natl Cancer Inst. 1998;90:697–703. doi: 10.1093/jnci/90.9.697. [DOI] [PubMed] [Google Scholar]
- 147.Meeker AK, et al. Telomere shortening occurs in subsets of normal breast epithelium as well as in situ and invasive carcinoma. Am. J. Pathol. 2004;164:925–935. doi: 10.1016/S0002-9440(10)63180-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Herbert BS, Wright WE, Shay JW. Telomerase and breast cancer. Breast Cancer Res. 2001;3:146–149. doi: 10.1186/bcr288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Baird DM, Rowson J, Wynford-Thomas D, Kipling D. Extensive allelic variation and ultrashort telomeres in senescent human cells. Nat. Genet. 2003;33:203–207. doi: 10.1038/ng1084. [DOI] [PubMed] [Google Scholar]
- 150.Tanaka H, et al. Telomere fusions in early human breast carcinoma. Proc. Natl Acad. Sci. USA. 2012;109:14098–14103. doi: 10.1073/pnas.1120062109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Lin TT, et al. Telomere dysfunction accurately predicts clinical outcome in chronic lymphocytic leukaemia and even in patients with early stage disease. Br. J. Haematol. 2014;167:223. doi: 10.1111/bjh.13023. [DOI] [PubMed] [Google Scholar]
- 152.Simpson K, et al. Telomere fusion threshold identifies a poor prognostic subset of breast cancer patients. Mol. Oncol. 2015;9:1186–1193. doi: 10.1016/j.molonc.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Heidenreich B, Rachakonda PS, Hemminki K, Kumar R. TERT promoter mutations in cancer development. Curr. Opin. Genet. Dev. 2014;24:30–37. doi: 10.1016/j.gde.2013.11.005. [DOI] [PubMed] [Google Scholar]
- 154.Killela PJ, et al. TERT promoter mutations occur frequently in gliomas and a subset of tumors derived from cells with low rates of self-renewal. Proc. Natl Acad. Sci. USA. 2013;110:6026. doi: 10.1073/pnas.1303607110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Kinde I, et al. TERT promoter mutations occur early in urothelial neoplasia and are biomarkers of early disease and disease recurrence in urine. Cancer Res. 2013;73:7162–7167. doi: 10.1158/0008-5472.CAN-13-2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Remke M, et al. TERT promoter mutations are highly recurrent in SHH subgroup medulloblastoma. Acta Neuropathol. 2013;126:917–929. doi: 10.1007/s00401-013-1198-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Quaas A, et al. Frequency of TERT promoter mutations in primary tumors of the liver. Virchows Arch. 2014;465:673–677. doi: 10.1007/s00428-014-1658-7. [DOI] [PubMed] [Google Scholar]
- 158.Weinhold N, Jacobsen A, Schultz N, Sander C, Lee W. Genome-wide analysis of noncoding regulatory mutations in cancer. Nat. Genet. 2014;46:1160–1165. doi: 10.1038/ng.3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Borah S, et al. TERT promoter mutations and telomerase reactivation in urothelial cancer. Science. 2015;347:1006–1010. doi: 10.1126/science.1260200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Bell RJA, et al. The transcription factor GABP selectively binds and activates the mutant TERT promoter in cancer. Science. 2015;348:1036–1039. doi: 10.1126/science.aab0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Pickett HA, Reddel RR. Molecular mechanisms of activity and derepression of alternative lengthening of telomeres. Nat. Struct. Mol. Biol. 2015;22:875–880. doi: 10.1038/nsmb.3106. [DOI] [PubMed] [Google Scholar]
- 162.Lovejoy CA, et al. Loss of ATRX, genome instability, and an altered DNA damage response are hallmarks of the alternative lengthening of telomeres pathway. PLoS Genet. 2012;8:e1002772. doi: 10.1371/journal.pgen.1002772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Schwartzentruber J, et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature. 2012;482:226–231. doi: 10.1038/nature10833. [DOI] [PubMed] [Google Scholar]
- 164.Heaphy CM, et al. Altered telomeres in tumors with ATRX and DAXX mutations. Science. 2011;333:425. doi: 10.1126/science.1207313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Heaphy CM, et al. Prevalence of the alternative lengthening of telomeres telomere maintenance mechanism in human cancer subtypes. Am. J. Pathol. 2011;179:1608–1615. doi: 10.1016/j.ajpath.2011.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Ceccarelli M, et al. Molecular profiling reveals biologically discrete subsets and pathways of progression in diffuse glioma. Cell. 2016;164:550–563. doi: 10.1016/j.cell.2015.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Eckel-Passow JE, et al. Glioma groups based on 1p/19q, IDH, and TERT promoter mutations in tumors. N. Engl. J. Med. 2015;372:2499–2508. doi: 10.1056/NEJMoa1407279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Ding Z, et al. Telomerase reactivation following telomere dysfunction yields murine prostate tumors with bone metastases. Cell. 2012;148:896–907. doi: 10.1016/j.cell.2012.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Sprung CN, Sabatier L, Murnane JP. Telomere dynamics in a human cancer cell line. Exp. Cell Res. 1999;247:29–37. doi: 10.1006/excr.1998.4293. [DOI] [PubMed] [Google Scholar]
- 170.Fouladi B, Sabatier L, Miller D, Pottier G, Murnane JP. The relationship between spontaneous telomere loss and chromosome instability in a human tumor cell line. Neoplasia. 2000;2:540–554. doi: 10.1038/sj.neo.7900107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Murnane JP, Sabatier L, Marder BA, Morgan WF. Telomere dynamics in an immortal human cell line. EMBO J. 1994;13:4953–4962. doi: 10.1002/j.1460-2075.1994.tb06822.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Sabatier L, Ricoul M, Pottier G, Murnane JP. The loss of a single telomere can result in instability of multiple chromosomes in a human tumor cell line. Mol. Cancer Res. 2005;3:139–150. doi: 10.1158/1541-7786.MCR-04-0194. [DOI] [PubMed] [Google Scholar]
- 173.Gascoigne KE, Cheeseman IM. Induced dicentric chromosome formation promotes genomic rearrangements and tumorigenesis. Chromosome Res. 2013;21:407–418. doi: 10.1007/s10577-013-9368-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Baca SC, et al. Punctuated evolution of prostate cancer genomes. Cell. 2013;153:666–677. doi: 10.1016/j.cell.2013.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Palm W, de Lange T. How shelterin protects mammalian telomeres. Annu. Rev. Genet. 2008;42:301–334. doi: 10.1146/annurev.genet.41.110306.130350. [DOI] [PubMed] [Google Scholar]
- 176.Schmutz I, de Lange T. Shelterin. Curr. Biol. 2016;26:R397–R399. doi: 10.1016/j.cub.2016.01.056. [DOI] [PubMed] [Google Scholar]
- 177.Lazzerini Denchi E, Sfeir A. Stop pulling my strings — what telomeres taught us about the DNA damage response. Nat. Rev. Mol. Cell Biol. 2016;17:364–378. doi: 10.1038/nrm.2016.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Xin H, et al. TPP1 is a homologue of ciliate TEBP-β and interacts with POT1 to recruit telomerase. Nature. 2007;445:559–562. doi: 10.1038/nature05469. [DOI] [PubMed] [Google Scholar]
- 179.Nandakumar J, et al. The TEL patch of telomere protein TPP1 mediates telomerase recruitment and processivity. Nature. 2013;492:285–289. doi: 10.1038/nature11648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Nandakumar J, Cech TR. Finding the end: recruitment of telomerase to telomeres. Nat. Rev. Mol. Cell Biol. 2013;14:69–82. doi: 10.1038/nrm3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Sexton AN, et al. Genetic and molecular identification of three human TPP1 functions in telomerase action: recruitment, activation, and homeostasis set point regulation. Genes Dev. 2014;28:1885–1899. doi: 10.1101/gad.246819.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Abreu E, et al. TIN2-tethered TPP1 recruits human telomerase to telomeres in vivo. Mol. Cell. Biol. 2010;30:2971–2982. doi: 10.1128/MCB.00240-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Frank AK, et al. The shelterin TIN2 subunit mediates recruitment of telomerase to telomeres. PLoS Genet. 2015;11:e1005410. doi: 10.1371/journal.pgen.1005410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Greider CW. Regulating telomere length from the inside out: the replication fork model. Genes Dev. 2016;30:1483–1491. doi: 10.1101/gad.280578.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Smogorzewska A, de Lange T. Regulation of telomerase by telomeric proteins. Annu. Rev. Biochem. 2004;73:177–208. doi: 10.1146/annurev.biochem.73.071403.160049. [DOI] [PubMed] [Google Scholar]