Abstract
Oligonucleotide conjugates bearing two pyrene residues attached to 5′-phosphate through a phosphoramide bond were synthesised. Fluorescence spectra of the conjugates show a peak typical of monomer emission (λmax 382 nm) and a broad emission peak with λmax 476 nm, which indicates the excimer formation between the two pyrene residues. Conjugation of these two pyrene residues to the 5′-phosphate of oligonucleotides does not affect the stabilities of heteroduplexes formed by conjugates with the corresponding linear strands. A monomer fluorescence of the conjugates is considerably affected by the heteroduplex formation allowing the conjugates to be used as fluorescent hybridisation probes. The 5′-bis-pyrenylated oligonucleotides have been successfully used for investigation of affinity and kinetics of antisense oligonucleotides binding to the multidrug resistance gene 1 (PGY1/MDR1) mRNA. The changes of excimer fluorescence of the conjugates occurring during hybridisation depended on the structure of the binding sites: hybridisation to heavily structured parts of RNA resulted in quenching of the excimer fluorescence, while binding to RNA regions with a loose secondary structure was accompanied by an enhancement of the excimer fluorescence. Potentially, these conjugates may be considered as fluorescent probes for RNA structure investigation.
INTRODUCTION
Recognition of RNA sequences by complementary strands has been applied for specific down-regulation of gene expression by antisense RNA, antisense oligonucleotides and ribozymes (1). Results of the studies demonstrate that the structure of the target RNAs dramatically affects the performance of particular antisense agents (2). Gel-shift assay (3,4), RNase H mapping (5–7) and RNA hybridisation to oligonucleotide arrays (8,9) are used for identification of accessible hybridisation sites within RNA. Among these approaches, only gel-shift assay can provide quantitative evaluation of kinetics and thermodynamics of oligonucleotide binding to RNA, which is important for understanding the factors influencing antisense oligonucleotide activity. However, this technique is laborious especially for analysis of many oligonucleotides.
Fluorescence detection of hybridisation process using fluorescent oligonucleotide conjugates can be a powerful method for investigation of antisense oligonucleotide binding to target RNA (10,11). One of the most attractive dyes for the development of fluorescent oligonucleotide probes is pyrene (12). Pyrene molecules are able to form a bimolecular complex called an excimer, wherein one molecule is in an excited state and the other molecule is in a ground state (13). Because of spatial requirements, excimer formation is very sensitive to various interactions involving the label, which makes pyrene excimer fluorescence a powerful tool for investigation of nucleic acid interactions and structures. Fluorescent oligonucleotide conjugates with two (14–17) or multiple pyrene residues (18) were used for investigation of model DNA oligonucleotide hybridisation. However, the advantages of these probes have not yet been used for investigation of RNA oligonucleotide interactions or for investigation of RNA structure.
In the present study, we developed a simple and reliable procedure for synthesis of oligonucleotide conjugates containing two pyrenyl-1-methylamino residues attached to the 5′-phosphate of oligonucleotides via phosphoramide bonds. These conjugates display both pyrene monomer and excimer fluorescence and therefore can be used as sensitive non-perturbing oligonucleotide probes. Increase in pyrene monomer fluorescence intensities occurs upon binding of these conjugates to the complementary RNA sequences allowing us to study kinetics and thermodynamics of oligonucleotide binding to RNA. The changes in excimer fluorescence are affected by RNA structure within oligonucleotide binding sites that provide possibility to use such conjugates as probe for analysis of RNA secondary structure. The bis-pyrenylated oligonucleotide conjugates were applied for investigation of antisense oligonucleotides binding to the 5′-part of the human multidrug resistance gene 1 mRNA (PGY1/MDR1 mRNA).
MATERIALS AND METHODS
Nucleic acids
The RNA target was a 678-nt-long 5′-terminal fragment of PGY1/MDR1 mRNA (hereafter denoted as MDR 1 RNA). The SmaI-linearised pMDR-670 plasmid (19) was used as a template for RNA synthesis by T7 RNA polymerase (20). The RNA transcript was purified using Speen-column-30 (Sigma), ethanol precipitated, dissolved in MilliQ water and stored at –20°C.
Oligonucleotides
Oligodeoxyribonucleotides were synthesised using ASM-102U synthesiser (BIOSSET, Novosibirsk, Russia) by standard phosphoramidite chemistry. DNA oligomers were purified by consecutive ion-exchange and reverse-phase HPLC. Oligoribonucleotide r-tg-B was provided by TriLink Laboratories Inc., San-Diego. The homogeneity of synthesised oligonucleotides was assayed by 15% PAGE under denaturing conditions followed by staining with Stains-All (21).
Synthesis of bis-pyrenylated oligonucleotide conjugates
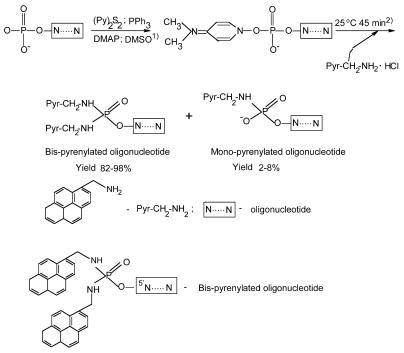
Two pyrene residues were attached to the 5′-terminal phosphate of the oligonucleotides (Scheme 1), which was activated by a mixture of triphenylphosphine (Ph3P) and 2,2′-dipyridyldisulfide (PyS)2 in the presence of 4-(N,N-dimethylamino)pyridine (DMAP) (22,23). Briefly, 1–10 A260 OD (5–100 nmol) of oligonucleotide in water (20–50 µl) was precipitated with N-cetyl-N,N,N-trimethylammonium bromide: after centrifugation, the oligonucleotide pellet was thoroughly dried in vacuum over P2O5. To activate the oligonucleotide phosphate, the reaction mixture [containing 5–100 nmol of the dried N-cetyl-N,N,N-trimethylammonium salt of the oligonucleotide, 0.05 ml of dried dimethylsulfoxide (DMSO), 10 mg (38 µmol) of Ph3P, 10 mg (50 µmol) of (PyS)2 and 10 mg (80 µmol) of DMAP] was thoroughly mixed, heated for 1 min at 50°C and intensively vortexed. The heating–mixing procedure was repeated four times followed by incubation at 25°C for 10 min. After adding 2 mg of pyrenyl-1-methylamine hydrochloride, the reaction mixture was incubated at 25°C for 40 min at constant mixing. The reaction was quenched by precipitation of the oligonucleotide conjugate with 1.5 ml of 2% lithium perchlorate solution in dry acetone. The bis-pyrenylated oligonucleotide was isolated by reverse-phase HPLC using a LiChrosorb RP-18, 10 µm (Merck), 4.6 × 250 mm column, Waters 600E chromatograph and Waters 484 tunable absorbance detector (USA). A linear (0–30%) gradient of acetonitrile in 0.05 M LiClO4 pH 7.5 (flow rate 2 ml/min) was used. Lithium salt of bis-pyrenylated oligonucleotide was precipitated with acetone, centrifuged, dried, and dissolved in water. Bis-pyrenylated oligonucleotides were stable under storage in water at –20°C for several months. The conjugates were characterised by polyacrylamide gel electrophoresis, reverse-phase HPLC, UV- and fluorescence spectroscopy, and liquid chromatography–mass spectrometry (LC–MS). LC–MS analysis was carried out by Dr Hans Gaus (ISIS Pharmaceutical). The Mr values of conjugate Bbis were as follows: expected, 5001.13, found, 4999.8 for the peak with retention time 21.13–22.71 min; and charge states –3 and –4.
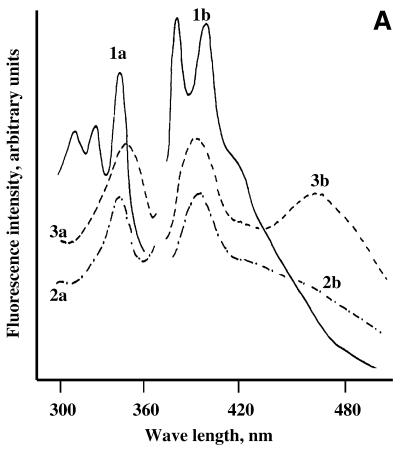
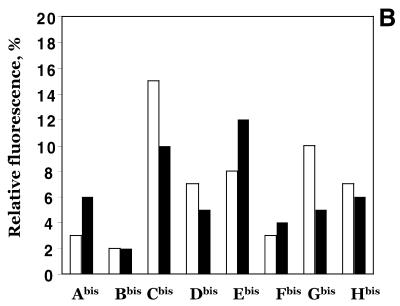
Figure 1.
Spectral properties of pyrene-labelled oligonucleotide conjugates. (A) Fluorescence excitation (a) and emission (b) spectra of pyrenyl-1-methylamine (1), 5′-mono-pyrenylated oligonucleotide B (2) and 5′-bis-pyrenylated oligonucleotide Bbis (3). (B) Fluorescence of bis-pyrenylated oligonucleotide conjugates Abis –Hbis as compared to pyrenyl-1-methylamine (Pyr). Open and closed bars show monomer (F382) and excimer (F476) fluorescence of the conjugates, respectively. Oligonucleotide conjugates (5 × 10–7 M) were dissolved in hybridisation buffer. 100% corresponds to the fluorescence of 5 × 10–7 M pyrenyl-1-methylamine in ethanol at λ = 382 nm. Pyrenyl-1-methylamine has equal fluorescence intensities in hybridisation buffer and ethanol (14,23).
Concentrations of targets and probes
Concentrations of RNAs, DNAs and unmodified oligonucleotides were determined by measuring UV absorbencies at 260 nm (A260) in 50 mM HEPES buffer, pH 7.5, containing 5 mM MgCl2, 200 mM KCl and 0.5 mM EDTA, using calculated molar extinction coefficients (24). The ɛ260 were assumed to be 149 × 103, 135 × 103, 134 × 103, 145 × 103, 141 × 103, 140 × 103, 153 × 103 and 132 × 103 µM–1 cm–1 for oligonucleotides A–H (see Table 1), respectively. Concentrations of the bis-pyrenylated probes were calculated from the absorption at λ = 260 nm of their oligonucleotide constituents as described (14,25). Contribution of the pyrene moieties to the total absorbance at 260 nm was calculated assuming that A260 of pyrene residues conjugated to oligonucleotide was the same as for pyrenyl-1-methylamine. The ɛ260 of pyrene used was 24 000 µM–1 cm–1 (23). A260 of the oligonucleotide portion was calculated using the following equation: A260oligonucleotide = A260 (Pyrene)2–oligonucleotide – 2A260Pyrene.
Hybridisation of bis-pyrenylated oligonucleotide to in vitro transcript of the MDR1 mRNA and to RNA and DNA oligonucleotides
Binding of bis-pyrenylated oligonucleotides to the RNA and DNA targets was studied at 37°C in a standard hybridisation buffer containing 50 mM HEPES pH 7.5, 5 mM MgCl2, 200 mM KCl and 0.5 mM EDTA. RNA or DNA targets at concentrations ranging from 1 × 10–7 to 1 × 10–5 M were added to the solution of bis-pyrenylated oligonucleotide conjugate (5 × 10–7 M). Before fluorescence measurement, reaction mixtures were allowed to set under hybridisation conditions until no fluorescent changes were detected.
Fluorescence measurements
Fluorescence spectra were recorded on a MPF4 Hitachi spectrofluorimeter using a bandwidth of 15 nm and 0.5 × 2 cm quartz cuvettes with a light pass of 1 cm. The cell holder was thermostated with circulating water controlled by an LKB-2219 Multitemp II Thermostatic Circulator. Pyrene was excited at 343 nm. Pyrene fluorescence emission was monitored at 382 nm for monomer and 476 nm for excimer fluorescence. Emission spectra were measured using reference dye (rodamine-B) to compensate for lamp fluctuations. In all the spectra, the background emission from the buffer alone was subtracted.
Thermal denaturation experiments
Thermal denaturation of oligonucleotide complexes was monitored in 50 mM cacodylate buffer pH 7.5, containing 5 mM MgCl2, 200 mM KCl and 0.5 mM EDTA. The concentration of each component was 1 × 10–5 M. The melting curves were recorded in a multiwave length mode; at least three different wavelengths were used. Absorption was measured for each oligonucleotide mixture as a function of temperature; the heating rate was 0.5–1°C/min; the thermoregulated sapphire cell (V = 2 µl) of the Millichrom liquid chromatograph (Econova, Co., Russia), which was specially designed for this purpose. Equilibrium optical melting curves were determined on the basis of more than 600 experimental points with the step 10 points/°C and were completely reversible in heating–cooling processes. The first derivatives of optical melting curves versus temperature were calculated using gradient of linear approximation by 10 experimental points. Melting temperature (Tm) of the duplex was determined as the average temperature corresponding to the maximum of first derivatives of optical melting curve of duplexes formed by stoichiometrical mixture of DNA or RNA target and the probe.
RESULTS
Synthesis and fluorescence properties of the 5′-bis-pyrenylated oligonucleotides
To develop fluorescent oligonucleotide probes displaying excimer fluorescence for real-time monitoring of the hybridisation process, we synthesised oligonucleotide conjugates with two pyrene residues, whose fluorescence emission spectra were considerably affected by microenvironment. We developed a simple and reliable synthetic procedure for preparation of 5′-bis-pyrenylated oligonucleotide conjugates displaying excimer fluorescence using well-known Mukaiyama reagents (26,27) and nucleophilic catalysis (22). Oligonucleotides A–H listed in Table 1 were used for synthesis of bis-pyrenylated conjugates Abis–Hbis. To synthesise 5′-bis-pyrenylated oligonucleotide conjugates, 5′-phosphate of the oligonucleotides was activated with dipyridyl-disulfide (Py2S2), triphenyl-phosphine (PPh3), in the presence of DMAP in dry DMSO and then treated with pyrenyl-1-methylamine hydrochloride (Scheme 1). Synthesis of pyrenyl-1-methylamine was described earlier (23). The presence of two pyrene residues in the conjugates was confirmed by polyacrylamide gel electrophoresis, reverse-phase HPLC, UV- and fluorescence spectroscopy, and LC–MS. Previously, a similar method was used for oligonucleotide derivatisation with N-methyl-N-(2-chloroethyl)-benzylamine (22) and pyrenyl-1-methylamine (23) to obtain alkylating oligonucleotide derivatives and mono-fluorophore labelled oligonucleotides. We used two modifications of the described protocols (22,23). To couple two pyrene residues to the 5′-phosphate of oligonucleotides, we used higher concentrations of activating agents (0.4 M PPh3, 0.5 M Py2S2 and 0.8 M DMAP) and fluorophore (0.08 M pyrenyl-1-methylamine). In addition, reaction mixtures were heated briefly (1 min) several times at 50°C during the activation step. The high concentration of DMAP permitted to use pyrenyl-1-methylamine hydrochloride instead of pyrenyl-1-methylamine base in the reaction without additional treatment with triethylamine. This synthetic procedure provides 82–98% yield of bis-pyrenylated oligonucleotides, while the mono-fluorophore-labelled oligonucleotide is formed as a byproduct with a yield of 2–8%. It was shown previously that di-anilide of pT(Ac) was formed in the reaction of pT(Ac) with Py2S2 and PPh3 in the presence of aniline (27). Recently, attachment of two alkylamine residues to the 5′-phosphate of oligonucleotides under the described conditions was confirmed by NMR studies (28).
Table 1. Effective association constants (Kx) and association rate constants (k+) for oligonucleotide probes binding to the in vitro transcript of mdr1 mRNA.
| Oligonucleotidesa 5′→3′ |
|
Complementary regionb |
Kx [M–1]c,d |
k+ [M–1 s–1]c,e |
| A |
pTGAGCTTGGAAGAGC |
17–31 |
(0.61 ± 0.18) × 106 |
(6.0 ± 0.3) × 102 |
| B |
pGACCTCGCGCTCCTTG |
122–137 |
(0.99 ± 0.28) × 106 |
(3.2 ± 0.25) × 103 |
| C |
pCATTGCGGTCCCCTTC |
150–165 |
(0.87 ± 0.14) × 107 |
(1.2 ± 0.13) × 103 |
| D |
pCATATACAACTTGTC3′ |
279–293 |
ndf |
ndf |
| E |
pGTCCAGCCCCATGGA3′ |
319–333 |
(0.77 ± 0.14) × 106 |
(3.2 ± 0.25) × 103 |
| F |
pGTCTTCCTCCAGATTC3′ |
455–470 |
(0.23 ± 0.2) × 106 |
(8.6 ± 0.12) × 102 |
| G |
pCCACTGTAATAATAG3′ |
485–499 |
nd |
nd |
| H |
pTATCTCCTGTCGCAT3′ |
606–620 |
(0.11 ± 0.15) × 106 |
(2.1 ± 0.15) × 102 |
| r-tg-B |
CCAAGGAGCGCGAGGTCG |
121–139g |
– |
– |
| tg-B |
CCAAGGAGCGCGAGGTCG |
121–139g |
– |
– |
| tg-F | TGAATCTGGAGGAAGACA | 454–470g | – | – |
aIn the text, oligonucleotide and oligonucleotide derivatives are named as follows. N, non-modified oligonucleotide; Nbis, 5′-bis-pyrenylated oligonucleotide conjugate; r, oligoribonucleotides; d in the names of oligodeoxyribonucleotides is omitted.
bNucleotide numbering is according to Kostenko et al. (19) where 1 corresponds to the first 5′-nucleotide of the MDR1 mRNA.
cData are presented for bis-pyrenylated oligonucleotide conjugates.
dEffective association constants (Kx) were determined from fluorescence monitoring titration experiments using modified Shtern–Folmer equation:
Where Fk represents fluorescence intensity of oligonucleotide conjugate bound to RNA; Fo represents fluorescence intensity of free oligonucleotide conjugate.
eAssociation rate constants k+ were estimated using two states model and the following equation:
fnd, binding not detected.
gshort oligonucleotide targets correspond to the indicated fragments of MDR1 mRNA.
UV spectra of the conjugates are consistent with the presence of two pyrene residues in their structure. Absorption spectra of bis-pyrenylated oligonucleotides (λmax = 344 nm) are similar to spectra of mono-pyrenylated oligonucleotide conjugates (λmax = 342 nm), suggesting additive contribution of two pyrene residues. This suggestion is in good agreement with the obtained A260/A342 ratio for bis-pyrenylated oligonucleotides. As expected, absorption intensity at 342 nm of the bis-pyrenylated oligonucleotide Bbis is enhanced twice as compared to mono-pyrenylated oligonucleotide Bmono (data not shown). Figure 1A shows the fluorescence excitation and emission spectra of bis-pyrenylated oligonucleotide Bbis that are typical for this type of oligonucleotide conjugate. The bis-pyrenylated oligonucleotides exhibit the typical pyrene monomer fluorescence emission with a maximum at 382 nm and a broad pyrene excimer emission band with a maximum at 476 nm (Fig. 1A, spectrum 3b), which is consistent with the proposed structure with two pyrene residues. The fluorescence spectrum of control mono-pyrenylated oligonucleotide (spectrum 2b) has no maximum at 476 nm.
The intensity of the monomer (F382) and the excimer (F476) fluorescence of bis-pyrenylated oligonucleotide conjugates Abis–Hbis varies depending on their sequences and structures. Figure 1B presents relative changes in the pyrene fluorescence upon conjugation to oligonucleotides as compared to pyrenyl-1-methylamine. The strong pyrene monomer fluorescence quenching (up to 50 times) was observed for conjugates Abis, Bbis and Fbis. Moderate quenching (7–14 times) was observed for all other oligonucleotide conjugates. Intensities of excimer fluorescence and ratios of monomer to excimer fluorescence emission intensities differed for different conjugates (Fig. 1B).
Hybridisation of bis-pyrenylated oligonucleotides to RNA
We studied the interaction of the bis-pyrenylated oligonucleotides with oligoribonucleotide and with the complementary sequences in the 678-nt-long 5′-fragment of the human multidrug resistance gene 1 mRNA (MDR1 mRNA) as a model (29). Binding sites for oligonucleotides were chosen in the RNA regions containing 4–12-nt-long single-stranded sequences, allowing nucleation complexes to be formed followed by invasion of the oligonucleotides into the RNA structure and extended heteroduplex formation (30–32). Figure 2 shows the secondary structures of oligonucleotide binding sites within MDR1 mRNA. Sequences of oligonucleotides A–H are shown in Table 1. Hybridisation of bis-pyrenylated oligonucleotides to the RNA was investigated by the fluorometric titration assay. Binding of pyrene labelled oligonucleotides to the target RNA resulted in characteristic changes in the fluorescence spectra of the conjugates (Fig. 3). Addition of the non-complementary RNA to bis-pyrenylated oligonucleotides did not affect hypochromicity and fluorescence emission spectra of the conjugates. Thus, the increase in monomer and excimer fluorescence of the conjugates in the presence of MDR1 mRNA can be attributed to duplex formation. Minor or no increase in the monomer and excimer fluorescence in the presence of MDR1 mRNA was observed for conjugates Gbis and Dbis that failed to form stable complexes with the RNA under the experimental conditions as shown by probing with RNase H and gel-shift assay. Although the target regions for oligonucleotide G and D include single-stranded tracts necessary for nucleation complex formation, these sites might be unfavourable for binding due to some tertiary interactions within the RNA molecule. All other oligonucleotide conjugates display an increase in the monomer fluorescence intensity (F382) upon the binding to the target RNA in a time- and concentration-dependent manner.
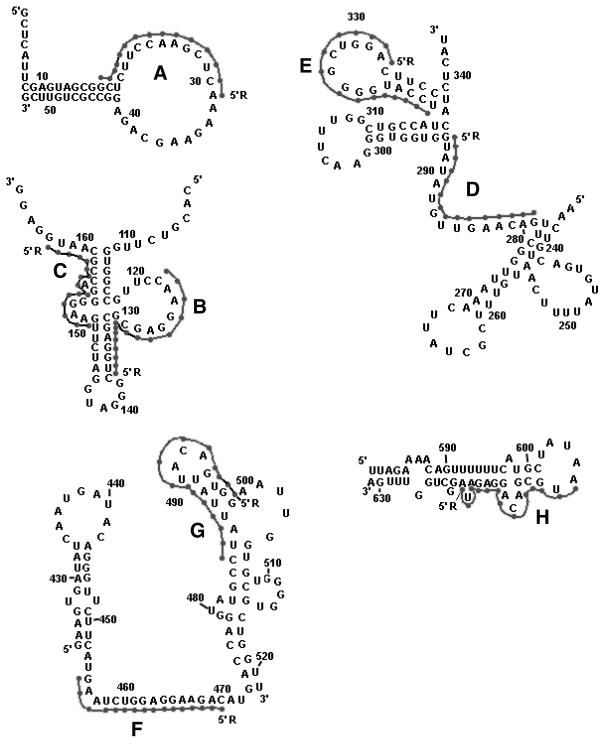
Figure 2.
Secondary structure of the target sites for oligonucleotides A–H within MDR1 mRNA (19,29). Two pyrene residues attached to the 5′-phosphate of oligonucleotides are indicated as R. Letters A–H indicate target sites of oligonucleotides A–H, respectively.
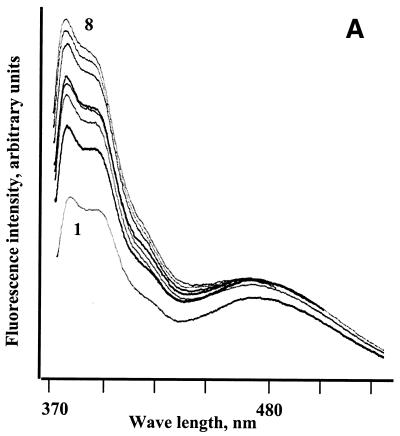
Figure 3.
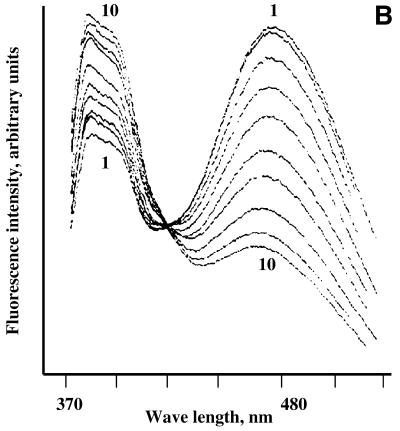
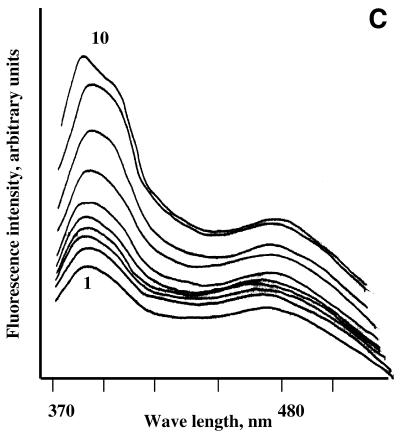
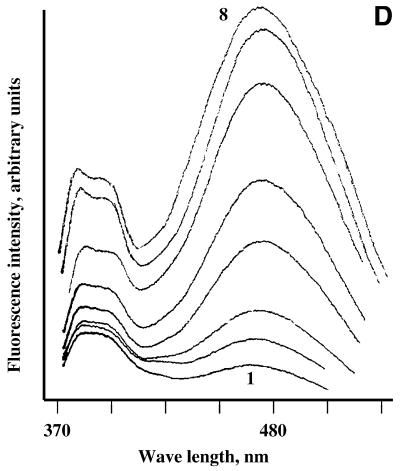
Fluorescence monitoring titration of bis-pyrenylated oligonucleotides Abis (A), Ebis (B) and Bbis (C and D) with their target RNAs in 50 mM HEPES, pH 7.0 containing 200 mM KCl, 5 mM MgCl2 and 0.1 mM EDTA at 37°C. In all figures, 1 corresponds to fluorescence emission spectrum of free conjugate (5 × 10–7 M) in hybridisation buffer; (A) 2–8 fluorescence emission spectra of 5 × 10–7 M conjugate Abis in the presence of 1 × 10–7, 3 × 10–7, 5 × 10–7, 7 × 10–7, 1 × 10–6, 2 × 10–6 and 2.5 × 10–6 M of the in vitro transcript of MDR1 mRNA, respectively. (B) 2–10, fluorescence emission spectra of 5 × 10–7 M conjugate Ebis in the presence of 1 × 10–7, 3 × 10–7, 5 × 10–7, 7 × 10–7, 1 × 10–6, 2 × 10–6, 2.5 × 10–6, 3 × 10–6 and 4 × 10–6 M of the in vitro transcript of MDR1 mRNA, respectively. (C) 2–10, fluorescence emission spectra of 5 × 10–7 M conjugate Bbis in the presence of 1 × 10–7, 3 × 10–7, 5 × 10–7, 7 × 10–7, 1 × 10–6, 2 × 10–6, 2.5 × 10–6, 3 × 10–6 and 4 × 10–6 M of the in vitro transcript of MDR1 mRNA, respectively. (D) 2–8, fluorescence emission spectra of 5 × 10–7 M conjugate Bbis in the presence of 1 × 10–7, 3 × 10–7, 5 × 10–7, 7 × 10–7, 1 × 10–6, 2 × 10–6 and 3 × 10–6 of r-tg-B, respectively.
The excimer fluorescence (F476) of oligonucleotide conjugates displayed a complex behaviour upon their binding to the RNA, which may be explained by the sensitivity of excimer fluorescence to the environment of the binding site. The fluorescent probes may be divided into four groups according to the characteristic changes in their excimer fluorescence emission upon hybridisation with their targets. Conjugates Cbis and Abis show little changes in the excimer fluorescence upon hybridisation to the MDR1 mRNA. Spectral changes occurring upon hybridisation of oligonucleotide conjugate Abis to the RNA are shown in Figure 3A. The conjugates Ebis and Hbis display quenching of excimer fluorescence upon binding to the RNA (Fig. 3B). Hybridisation of conjugates Bbis and Fbis to the RNA resulted in increase of their excimer fluorescence (Fig. 3C). The changes to the oligonucleotide conjugates excimer fluorescence in the presence of MDR1 mRNA can be determined by the features of the RNA secondary structure including the oligonucleotide binding sites. The effect of the target structure on the excimer fluorescence of the conjugates was tested by the experiments with conjugate Bbis. It was found that binding of the conjugate Bbis to the long structured MDR1 mRNA fragment and to the short oligoribonucleotide r-tg-B (Table 1) resulted in considerably different effects on the excimer fluorescence of the conjugate (Fig. 3D).
The binding of complementary bis-pyrenylated oligonucleotides to MDR1 mRNA was tested in parallel experiments by probing the RNA/conjugate hybrid duplexes with RNase H and by gel-shift assay. Figure 4 shows the results of RNase H footprint analysis of conjugates Bbis and Cbis binding to the RNA. It is evident that the conjugates bind to their target sequences 122–137 and 150–165: the main cleavages by RNase H occur in the regions 131–137 and 160–165 (Fig. 4, lanes 2 and 3 for Bbis and Cbis, respectively). The data for RNase H footprinting and gel-shift assay are in good agreement with fluorometric titration. According to RNase H assay, the conjugates Abis, Bbis, Cbis, Ebis, F bis and Hbis efficiently bind to the RNA at their complementary sequences, while the conjugates Dbis and Gbis do not form complexes with the RNA. The gel-mobility shift assay data confirmed the complexes formation (data not shown).
Figure 4.
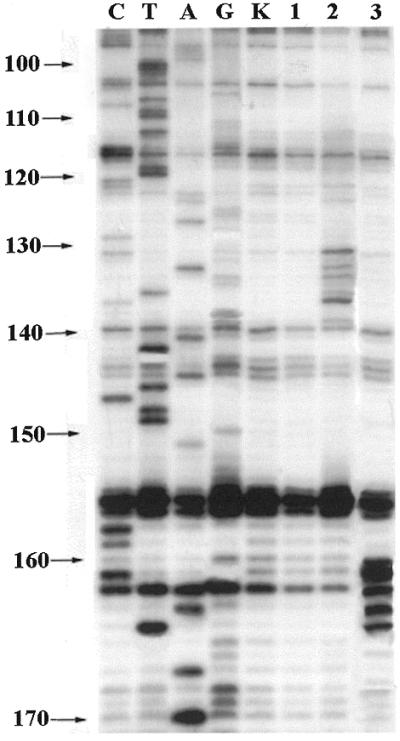
RNase H footprint analysis of hybridisation of oligonucleotide conjugates Bbis and Cbis with in vitro transcript of MDR1 mRNA in 50 mM HEPES, pH 7.0 containing 200 mM KCl, 5 mM MgCl2 and 0.1 mM EDTA at 37°C. Autoradiograph of 8% polyacrylamide/8 M urea gel with resolved products of the primer directed reverse transcription of MDR1 mRNA after treatment with RNase H. Lanes C, T, A and G, sequencing of MDR1 RNA. Lane K, control MDR1 RNA. Lane 1, MDR1 mRNA treated with ribonuclease H (1 U/ml). Lanes 2 and 3, 2 × 10–7 M MDR1 RNA treated with RNase H (1 U/ml) in the presence of 5 × 10–5 M of oligonucleotides Bbis and Cbis, respectively.
The time- and concentration-dependent changes of pyrene monomer fluorescence intensity upon binding of bis-pyrenylated conjugates to the MDR1 RNA allowed us to obtain kinetic and thermodynamic parameters of the hybridisation process. Kinetics of oligonucleotide hybridisation to the RNA was studied at 37°C in a standard hybridisation buffer. To determine the effective association constant, solution of 5 × 10–7 M oligonucleotide conjugate was titrated with increasing concentrations of MDR1 mRNA fragment. Since RNA unfolding is the limiting step of oligonucleotide hybridisation to structured RNA, the reaction mixture was incubated under hybridisation condition for 10–20 min after addition of each RNA portion, until no more changes of the fluorescence spectra occurred. Pyrene was excited at 343 nm and monomer and excimer fluorescence changes were measured as functions of time and RNA concentration. Second order rate constants and effective association constants of conjugate hybridisation to the MDR1 mRNA were calculated based on time- and concentration-dependent changes of pyrene monomer fluorescence. Kinetics and thermodynamic parameters of the binding process are summarised in Table 1. The constants obtained had a similar order of magnitude; the observed variations in association and rate constants can be explained by the differences in stability of RNA structure within the binding sites of the oligonucleotides.
Temperature melting studies
To exclude the possibility that presence of pyrene residues could affect the duplexes stabilities due to pyrene intercalation, we studied the hybridisation of the conjugates with their DNA and RNA targets in the independent UV-melting experiments. Stabilities of self-structures formed by bis-pyrenylated oligonucleotides themselves and stabilities of their complexes with the corresponding complementary targets were studied in the standard hybridisation solution (Materials and Methods). The effect of attachment of two pyrene residues to the 5′-end of oligonucleotides was determined by comparison of complexes formed by conjugates Bbis and Fbis and unmodified oligonucleotides B and F with their complementary DNA and RNA targets. Sequences of DNA targets tg-B and tg-F and RNA target r-tg-B used in the temperature melting experiments are listed in Table 1. The Tm values estimated from the UV-melting curves are summarised in Table 2. These data show that conjugation of pyrene does not significantly affect the hybridisation properties of the oligonucleotides. The ΔTm for duplexes of bis-pyrenylated oligonucleotides with target DNA compared to the unmodified duplexes were +2 and +3°C for oligonucleotides Bbis and Fbis, respectively. Thus, the pyrene residues attached to 5′-phosphate of oligonucleotide of this length had minor effects on the duplex stability. On the other hand, attachment of two pyrene residues to the 5′-phosphate of oligonucleotide caused rearrangement of the secondary structure of the oligonucleotide itself. For example, oligonucleotides B and F form hairpins with Tm 39 and 21°C, respectively, which are characterised by high co-operative transition upon melting. Conjugates Bbis and Fbis form structures with Tm 62 and 45°C, respectively, with low cooperativity and low activation energy of transition. Altogether, the UV-melting experiments provide evidence for the entropy nature of these self-structures formed by bis-pyrenylated oligonucleotides.
Table 2. Tm values of the duplexes formed by oligonucleotides and pyrene-labelled oligonucleotides with complementary DNA targets.
| Oligomersa |
Tm (°C) |
ΔTm (°C)b |
| B |
39 |
|
| Bbis |
62 |
+23 |
| F |
21 |
|
| Fbis |
45 |
+24 |
| B/tg-B |
73 |
|
| Bbis/tg-B |
75 |
+2 |
| B/r-tg-B |
76 |
|
| Bbis/r-tg-B |
76 |
0 |
| F/tg-F |
62 |
|
| Fbis/tg-F | 65 | +3 |
aDesignation of oligomers is in accordance with Table 1.
bΔTm is the difference in Tm values for duplexes formed by bis-pyrenylated oligonucleotides and by the parent unmodified oligonucleotides with corresponding DNA oligonucleotides. Tm values were determined with accuracy ±0.5°C.
DISCUSSION
Quantitative evaluation of the oligonucleotide hybridisation to the target RNA is essential to reveal the factors affecting activity of the antisense oligonucleotides. Few studies have been aimed at quantitative characterisation of affinity and kinetics of oligonucleotides binding to long natural RNA (3–5,11,33,34). The fluorescent assay, which will allow a quantitative, homogeneous and real-time monitoring of the hybridisation process, can be particularly useful for the investigation of the oligonucleotide binding to a high molecular weight RNA. Fluorescent correlation spectroscopy technique permits a rapid quantitative analysis of the antisense oligonucleotide binding to the RNA. However, this technique besides the fluorescent labelled oligonucleotides also requires an internal RNA labelling by enzymatic incorporation of the fluorophore-conjugated nucleotides (11).
Several types of single (35–41) and multiple pyrene-labelled oligonucleotide conjugates (14–18) have been described. The attached pyrene residue was used as a reporter group for investigation of the hybridisation of simple model oligonucleotides (14–18,35–41). Application of the pyrene monomer fluorescence has been generally limited since the pyrene residue displayed poor fluorescence when conjugated with DNA oligonucleotide (35–37). Pyrene excimer fluorescence could be potentially highly sensitive to environment. However, the results of the studies indicate that appropriate design of the oligonucleotide conjugates is needed to entirely use the advantages of the pyrene excimer fluorescence and to develop a non-perturbing oligonucleotide probe that displays the excimer fluorescence with pronounced fluorescence changes upon hybridisation to the complementary target. The bis-pyrene group attached to the 5′-end of an oligonucleotide through a five-atom linker was reported to perturb the duplex stability suggesting stacking interactions of the planar polyaromatic system with nucleobases (16). The interactions with nucleobases of the 5′-bis-pyrenyl group attached to the 5′-end of an oligonucleotide via the 15-atom-long linker arm prevents excimer formation and deteriorates the fluorescent properties of the conjugates (14). As expected, internal labelling of oligonucleotides with pyrene (including the 2′-position of ribonucleotides and various non-base pairing positions of the heterocyclic bases) tends to facilitate intercalation of the pyrene into the DNA duplex as confirmed by decreased fluorescence emission and higher melting temperatures (17,37–39).
In the present work we report the synthesis of the 5′-bis-pyrenylated oligonucleotide probes displaying excimer fluorescence and the results of the studies of the antisense oligonucleotide binding to MDR1 mRNA fragment using these oligonucleotide conjugates. We used an extremely short spacer between the fluorescent dye and 5′-phosphate of oligonucleotide to prevent the intercalation of the pyrene residues between base pairs within the duplex and to allow excimer formation. The UV-melting studies of duplexes of 5′-bis-pyrenylated oligonucleotides with target RNA and DNAs have revealed slight changes in the Tm values compared to unmodified duplexes and proved that the coupling of pyrene residues to oligonucleotides had a negligible effect on duplex stability. Thus, newly developed 5′-bis-pyrenylated oligonucleotides can serve as non-perturbing probes for the investigation of the hybridisation of nucleic acids in solution.
The synthetic procedure we developed is a simple and a reliable one-step and one-tube synthesis providing high yields of bis-pyrenylated oligonucleotide conjugates displaying the excimer fluorescence using commercially available pyrenyl-1-methyalmine as a precursor dye molecule. The oligonucleotide probes developed in this study showed strong quenching of the pyrene monomer fluorescence and appearance of the excimer fluorescence. Although it is known that pyrene fluorescence is strongly quenched upon conjugation to 5′-CMP and 5′-TMP (16,35) in our case the strongest quenching was observed for conjugates Bbis and Fbis with guanine in the first 5′ position. The UV-melting experiments and the analysis of these conjugate structures showed that strong quenching of pyrene fluorescence of conjugates Bbis and Fbis can be determined by their intrinsic structure with one or both pyrenes involved (Table 2). Thus, the fluorescence of pyrene conjugated to oligonucleotide is not determined solely by the nearest neighbour effect but rather represents a complex function of oligonucleotide sequences and structures.
The time- and concentration-dependent increase of pyrene monomer fluorescence intensity of 5′-bis-pyrenylated oligonucleotides upon binding to the target RNA allowed us to determine affinities and kinetics of antisense oligonucleotides binding to MDR1 mRNA (Table 1). Results of the experiments suggested that oligonucleotides B, C and E displaying the highest affinity to MDR1 mRNA in vitro could also efficiently target this mRNA in cells. Our preliminary studies have shown that these oligonucleotides can indeed efficiently decrease the content of MDR1 mRNA in KB-8-5 cells displaying the MDR phenotype. Correlation between the in vitro accessibility data and the intracellular antisense activity suggests that internal base pairing in MDR1 mRNA is most likely the reason for the lack of intracellular activity for some other previously tested oligonucleotides (42,43).
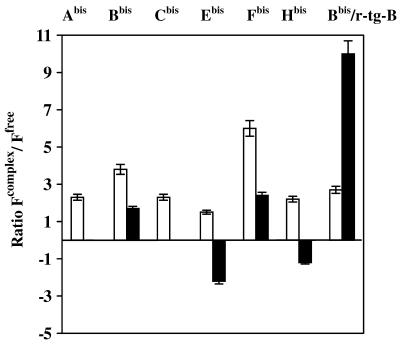
The variations in pyrene excimer fluorescence upon binding of different conjugates to their target sequences correlate with the secondary structure of RNA in the vicinity of the oligonucleotide binding sites (Figs 2 and 5). Binding of oligonucleotides Abis and Cbis to their targets delivers the pyrene residues to single-stranded regions of RNA, resulting in minor changes in the excimer fluorescence. Hybridisation of oligonucleotides Ebis and Hbis brings their 5′-bis-pyrenyl groups in the proximity of double-stranded regions of the RNA, resulting in quenching of excimer fluorescence apparently due to pyrene interaction with the double-stranded structure. Oligonucleotides Bbis and Fbis are complementary to stem–loop junctions. Binding of these conjugates to target RNA resulted in an increase in the excimer fluorescence. We found considerably less intensive changes in the pyrene excimer fluorescence of conjugate Bbis upon binding to the structured RNA. These changes were only 1.7-fold as compared to the 10-fold increase in excimer fluorescence of the same conjugate upon binding to a short RNA target (r-tg-B). These results prove that the pyrene excimer fluorescence of the conjugates is sensitive to the structure of target RNA.
Figure 5.
Changes in excimer (filled bars) and monomer (open bars) fluorescences of bis-pyrenylated oligonucleotide conjugates Abis –Hbis upon binding to RNA targets. Abis –Hbis corresponds to hybridisation of bis-pyrenylated oligonucleotides with fragment of MDR1 mRNA. Bbis/r-tg-B indicates binding of conjugate Bbis to oligoribonucleotide r-tg-B. Figures on the y-axis shows the ratios of fluorescence emission intensities of the target-bound (Fcomplex) and free (Ffree) bis-pyrenylated oligonucleotides.
Fluorescent secondary structure-specific oligonucleotide probes are particularly advantageous for fast screening of RNA structure to find open or double-stranded RNA regions. Sensitivity of the pyrene excimer fluorescence of the novel 5′-bis-pyrenylated oligonucleotides to the RNA secondary structure provides a possibility to use these oligonucleotide conjugates as specific probes for analysis of RNA structure in the vicinity of an oligonucleotide binding site. This structural information might be useful for the design of tight binding oligonucleotide derivatives for the targeting RNA.
The 5′-bis-pyrenylated oligonucleotides described in this study exhibit intensive characteristic changes in their fluorescence spectra upon hybridisation to the target RNAs. Pyrene monomer fluorescence increases in a time- and concentration-dependent manner upon binding of the conjugates to the target RNAs providing the possibility to determine thermodynamic parameters of the antisense oligonucleotides hybridisation to RNAs. Excimer fluorescence of the bis-pyrenylated conjugates is sensitive to the secondary structure of the oligonucleotide binding site.
Scheme 1. Synthesis of bis-pyrenylated oligonucleotide conjugates. Pyr-CH2-NH2, pyrenyl-1-methylamine; 1) reaction mixture was briefly heated (1 min) four times at 50°C followed by incubation at 25°C for 10 min; 2) pyrenyl-1-methylamine hydrochloride was added to the reaction mixture. No isolation steps take place between phosphate activation and condensation.
Acknowledgments
ACKNOWLEDGEMENTS
The authors are thankful to Dr Hans Gaus (ISIS Pharmaceutical) for LC–MS analysis of bis-pyrenylated oligonucleotide Bbis. We thank Drs M. Manoharan and A. Lebedev for helpful discussions. We acknowledge TriLink BioTechnologies for synthesis of r-tg-B. This work was supported by grants from INTAS (INTAS-RFBR 95-0653), the Russian Foundation for Basic Research (RFBR 99-04-49559, 00-15-97969) and the Siberian Branch of the Russian Academy of Sciences (Interdisciplinary Grant No. 26).
References
- 1.Crooke S.T. (2000) Progress in antisense technology: the end of the beginning. Methods Enzymol., 313, 3–45. [DOI] [PubMed] [Google Scholar]
- 2.Vickers T.A., Wyatt,J.R. and Freier,S.M. (2000) Effects of RNA secondary structure on cellular antisense activity. Nucleic Acids Res., 28, 1340–1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lima W.F., Monia,B.P., Ecker,D.J. and Freier,S.M. (1992) Implication of RNA structure on antisense oligonucleotide hybridisation kinetics. Biochemistry, 31, 12055–12061. [DOI] [PubMed] [Google Scholar]
- 4.Stull R.A., Zon,G. and Szoka,F.C. (1996) An in vitro messenger RNA binding assay as a tool for identifying hybridisation-competent antisense oligonucleotides. Antisense Nucleic Acid Drug. Dev., 6, 221–228. [DOI] [PubMed] [Google Scholar]
- 5.Lima W.F., Brown-Driver,V., Fox,M., Hanecak,R. and Bruice,T.W. (1997) Combinatorial screening and rational optimisation for hybridisation to folded hepatitis C virus RNA of oligonucleotides with biological antisense activity. J. Biol. Chem., 272, 626–638. [PubMed] [Google Scholar]
- 6.Matveeva O., Felden,B., Audlin,S., Gesteland,R.F. and Atkins,J.F. (1997) A rapid in vitro method for obtaining RNA accessibility patterns for complementary DNA probes: correlation with an intracellular pattern and known RNA structures. Nucleic Acids Res., 25, 5010–5016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ho S.P., Bao,Y., Lesher,T., Malhotra,R., Ma,L.Y., Fluharty,S.J. and Sakai,R.R. (1998) Mapping of RNA accessible sites for antisense experiments with oligonucleotide libraries. Nat. Biotechnol., 16, 59–63. [DOI] [PubMed] [Google Scholar]
- 8.Milner N., Mir,K.U. and Southern,E.M. (1997) Selecting effective antisense reagents on combinatorial oligonucleotide arrays. Nat. Biotechnol., 15, 537–541. [DOI] [PubMed] [Google Scholar]
- 9.Sohail M., Akhtar,S. and Southern,E.M. (1999) The folding of large RNAs studied by hybridisation to arrays of complementary oligonucleotides. RNA, 5, 646–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mansfield E.S., Worley,J.M., McKenzie,S.E., Surrey,S., Rappaport,E. and Fortina,P. (1995) Nucleic acid detection using non-radioactive labelling methods. Mol. Cell. Probes, 9, 145–156. [DOI] [PubMed] [Google Scholar]
- 11.Schwille P., Oehlenschlager,F. and Walter,N.G. (1996) Quantitative hybridisation kinetics of DNA probes to RNA in solution followed by diffusional fluorescence correlation analysis. Biochemistry, 35, 10182–10193. [DOI] [PubMed] [Google Scholar]
- 12.Birks J.B. (1970) Photophysics of Aromatic Molecules. John Wiley & Sons, London, UK, pp. 301–307.
- 13.Birks J.B. (1968) The pyrene excimer. Acta Phys. Pol., 34, 603–617. [Google Scholar]
- 14.Balakin K.V., Korshun,V.A., Prokhorenko,I.A., Maleev,G.V., Kudelina,I.A., Gontarev,S.V. and Berlin,Yu.A. (1997) Novel reagent for labelling of biomolecules-activated derivative of pyrene dichromophore with excimer fluorescence. Russian J. Bioorgan. Chem., 23, 33–41. [PubMed] [Google Scholar]
- 15.Balakin K.V., Korshun,V.A., Mickalev,I.I., Matveev,G.V., Malakhov,A.D., Prokopenko,I.A. and Berlin,Yu.A. (1998) Conjugates of oligonucleotides with polyaromatic fluorophores as promising DNA probes. Biosens. Bioelectron., 13, 771–778. [DOI] [PubMed] [Google Scholar]
- 16.Lewis F.D., Zhang,Y. and Letsinger,R. (1997) Bis-pyrenyl excimer fluorescence: a sensitive oligonucleotide probe. J. Am. Chem. Soc., 119, 5451–5452. [Google Scholar]
- 17.Kitamura M., Nimura,A., Yamana,K. and Shimidzu,T. (1991) Oligonucleotides with bis-pyrene adduct in the backbone: synthesis and properties of intramolecular excimer forming probe. Nucleic Acids Symp. Ser., 25, 67–68. [PubMed] [Google Scholar]
- 18.Tong G., Lawlor,J., Tregear,G. and Halambardis,J. (1995) Oligonucleotide-polyamide hybrid molecules containing multiple pyrene residues exhibit significant excimer fluorescence. J. Am. Chem. Soc., 117, 12151–12158. [Google Scholar]
- 19.Kostenko E.V., Beabealashvili,R.Sh., Vlassov,V.V. and Zenkova,M.A. (2000) Secondary structure of 5′-terminal region of mRNA of human multidrug resistance gene. Russian J. Mol. Biol., 34, 67–78. [PubMed] [Google Scholar]
- 20.Milligan J.F., Groebe,D.R., Witherell,G.W. and Uhlenbeck,O.C. (1987) Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res., 15, 8783–8798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.King L.E. and Morrison,M. (1976) The visualisation of human erythrocyte membrane proteins and glycoproteins in SDS polyacrylamide gels employing a single staining procedure. Anal. Biochem., 71, 223–230. [DOI] [PubMed] [Google Scholar]
- 22.Zarytova V., Ivanova,E. and Venyaminova,A. (1998) Design of functional diversity in oligonucleotides via zwitter-ionic derivatives of deprotected oligonucleotides. Nucl. Nucl., 17, 649–662. [Google Scholar]
- 23.Dobrikov M.I., Gaidamakov,S.A., Koshkin,A.A., Guinutdinov,T.I., Luk′ianchuk,N.P., Shishkin,G.V. and Vlasov,V.V. (1997) Sensitised photomodification by binary systems. I. Synthesis of oligonucleotide reagents, and the effect of their structure on the efficacy of target modification. Russian J. Bioorgan. Chem., 23, 191–199. [PubMed] [Google Scholar]
- 24.Fasman G.D. (1975) Handbook of Biochemistry and Molecular Biology. Nucleic Acids, 3rd Edn. CRC Press, Cleveland, OH, Vol. 1, p. 589.
- 25.Masuko M., Ohtani,H., Ebata,K. and Shimadzu,A. (1998) Optimisation of excimer-forming two-probe nucleic acid hybridisation method with pyrene as a fluorophore. Nucleic Acids Res., 26, 5409–5416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mukaiyama T. and Hoshimoto,M. (1971) Phosphorylation by oxidation-reduction condensation. Preparation of novel phosphorylating reagents. Bull. Chem. Soc. Jpn, 44, 2284. [Google Scholar]
- 27.Hashimoto M., Ueki,M. and Mukaiyama,T. (1976) S-S-Di-2-pyridyl di thiophosphate as a key intermediate in the phosphorylation by oxidation-reduction condensation. Chem. Lett., 2, 157–160. [Google Scholar]
- 28.Ryabinin V.A., Denisov,A.Yu., Pyshnyi,D.V., Abramova,T.V., Sinyakov,A.N. and Vlassov,V.V. (1999) Stabilisation of DNA-duplexes by minor groove ligands forming complexes of 1:2 type with parallel ligands orientation in minor groove. Dokl. Akad. Nauk., 368, 836–838. [PubMed] [Google Scholar]
- 29.Kostenko E.V., Beabealashvilly,R.S., Vlassov,V.V. and Zenkova,M.A. (2000) Secondary structure of the 5′-region of PGY1/MDR1 mRNA. FEBS Lett., 23, 181–186. [DOI] [PubMed] [Google Scholar]
- 30.Petyuk V.A., Zenkova,M.A., Giege,R. and Vlassov,V.V. (2000) Mechanism of oligonucleotides hybridisation with 3′ part of yeast tRNAPhe. Russian J. Mol. Biol., 34, 879–886. [Google Scholar]
- 31.Patzel V. and Sczakiel,G. (1999) Length dependence of RNA-RNA annealing. J. Mol. Biol., 294, 1127–1134. [DOI] [PubMed] [Google Scholar]
- 32.Reynaldo L.P., Vologodskii,A.V., Neri,B.P. and Lyamichev,V.I. (2000) The kinetics of oligonucleotide replacement. J. Mol. Biol., 297, 511–520. [DOI] [PubMed] [Google Scholar]
- 33.Schimmel P.R., Uhlenbeck,O.C., Lewis,J.B., Dickson,L.A., Eldred,E.W. and Schreier,A.A. (1972) Binding of complementary oligonucleotides to free and aminoacyl transfer ribonucleic acid synthetase bound transfer ribonucleic acid. Biochemistry, 11, 642–646. [DOI] [PubMed] [Google Scholar]
- 34.Zenkova M., Ehresmann,C., Caillet,J., Springer,M., Karpova,G., Ehresmann,B. and Romby,P. (1995) A novel approach to introduce site-directed specific cross-links within RNA-protein complexes. Application to the Escherichia coli threonyl-tRNA synthetase/translational operator complex. Eur. J. Biochem., 231, 726–735. [DOI] [PubMed] [Google Scholar]
- 35.Manoharan M., Kathleen,L.T., Zhao,M., Nafisi,K. and Netzel,L. (1995) Base-sequence dependence of emission lifetimes for DNA oligomers and duplexes covalently labeled with pyrene: relative electron-transfer quenching efficiencies of A, G and T nucleosides toward pyrene. J. Phys. Chem., 99, 17461–17472. [Google Scholar]
- 36.Mann J.S., Shibata,Y. and Meehan,T. (1992) Synthesis and properties of an oligodeoxynucleotide modified with a pyrene derivative at the 5′-phosphate. Bioconjugate Chem., 3, 554–558. [DOI] [PubMed] [Google Scholar]
- 37.Yamana K. and Letsinger,R.L. (1985) Synthesis and properties of oligonucleotides bearing a pendant pyrene group. Nucleic Acids Symp. Ser., 16, 169–172. [PubMed] [Google Scholar]
- 38.Yamana K., Ohashi,Y., Nunota,K., Aoki,M., Nakano,H. and Sangen,O. (1992) Fluorescent-labelled oligonucleotides that exhibit a measurable signal in the presence of complementary DNA. Nucleic Acids Symp. Ser., 27, 135–136. [PubMed] [Google Scholar]
- 39.Korshun V.A., Pestov,N.B., Birikh,K.R. and Berlin,Y.A. (1992) Reagent for introducing pyrene residues in oligonucleotides. Bioconjugate Chem., 3, 559–562. [DOI] [PubMed] [Google Scholar]
- 40.Yamana K., Iwase,R., Furutani,S., Tsuchida,H., Zako,H., Yamaoka,T. and Murakami,A. (1999) 2′-Pyrene modified oligonucleotide provides a highly sensitive fluorescent probe of RNA. Nucleic Acids Res., 27, 2387–2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Iwase R., Mahara,A., Yamana,K., Yamaoka,T. and Murakami,A. (1999) Study on RNA structure by pyrene-labelled 2′-O-methyloligoribonucleotides. Nucleic Acids Symp. Ser., 42, 115–116. [DOI] [PubMed] [Google Scholar]
- 42.Jaroszewski J., Kaplan,P., Syi,J.L., Sehested,M., Faustino,P. and Cohen,J. (1990) Concerning inhibition of multiple drug resistance genes. Cancer Comm., 2, 287–294. [DOI] [PubMed] [Google Scholar]
- 43.Thierry A., Rahman,A. and Dristchulo,A. (1993) Overcoming multidrug resistance in human tumour cells using free and liposomally encapsulated antisense oligodeoxynucleotides. Biophys. Biochem. Res. Commun., 190, 952–960. [DOI] [PubMed] [Google Scholar]