Abstract
Traumatic life experiences are associated with alcohol use problems, an association that is likely to be moderated by genetic predisposition. To understand these interactions, we conducted a gene-by-environment genome-wide interaction study (GEWIS) of alcohol use problems in two independent samples, the Army STARRS (ASTARRS, N=16,361) and the Yale-Penn (N=8,084) cohorts. Because the two cohorts were assessed using different instruments, we derived separate dimensional alcohol misuse scales and applied a proxy-phenotype study design. In African-American subjects, we identified an interaction of PRKG1 rs1729578 with trauma exposure in the ASTARRS cohort and replicated its interaction with trauma exposure in the Yale-Penn cohort (discovery-replication meta-analysis: z=5.64, p=1.69*10−8). PRKG1 encodes cGMP-dependent protein kinase 1, which is involved in learning, memory, and circadian rhythm regulation. Considering the loci identified in stage-1 that showed same effect directions in stage-2, the gene ontology (GO) enrichment analysis showed several significant results, including calcium-activated potassium channels (GO:0016286; p=2.30*10−5), cognition (GO:0050890; p=1.90*10−6), locomotion (GO:0040011; p=6.70*10−5), and Stat3 protein regulation (GO:0042517; p=6.4*10−5). To our knowledge, this is the largest GEWIS performed in psychiatric genetics, and the first GEWIS examining risk for alcohol misuse. Our results add to a growing body of literature highlighting the dynamic impact of experience on individual genetic risk.
Introduction
Exposure to traumatic life events is associated with a variety of health risk behaviors, including alcohol use disorders (AUD).1, 2 The heritability of AUD is approximately 50%.3 Genome-wide association studies (GWAS) of AUD have identified several risk alleles.4 Individuals with AUD likely present some complex trait risk mechanisms that are different from those of the general population.5, 6 A recent phenome-wide analysis demonstrated that AUD risk alleles are associated with a wide range of physical and mental health consequences.7 The environment also contributes to the predisposition to AUD, moderating the effects of risk alleles8.
Traumatic events affect genome regulation via different mechanisms;9, 10 and it should be possible to identify specific genes that interact with traumatic experiences to moderate AUD risk. Previous studies focused on candidate stress-response genes, such as 5-HTTLPR, PER1, and FKBP5,11–13 and investigated how the exposure to life trauma interacts with risk alleles in relation to AUD. However, the candidate-gene approach has limited ability to identify the genetic basis of complex traits.14 Conversely, GWAS of complex traits conducted in large cohorts have identified numerous risk alleles and some of the pathogenic mechanisms underlying genetically complex diseases. Similarly, genome-wide gene-by-environment interaction studies (GEWIS) can be useful in understanding how environmental factors interact with an individual’s genetic background to regulate the predisposition to complex traits. However, to date, there have been few published GEWIS. One reason is that large cohorts that could be meta-analyzed are rarely ascertained using compatible criteria, and relevant differences can be present in the assessment of phenotypic outcome and environmental factors. Thus, it is difficult to investigate large cohorts evaluated with homogeneous assessments. Differences in phenotypic assessment can reduce the statistical power of meta-analysis and replication studies, especially for GEWIS, since they are performed using two kinds of phenotypic data with respect to both the outcome and an environmental factor.
Using data from a GWAS of AUD, we observed that risk alleles in ADH1B, one of the best-validated loci for alcohol drinking behaviors, show different associations with DSM-IV vs. DSM-5 AUD criteria.15 To reduce error related to different phenotypic assessments, we performed a genome-wide gene-by-trauma interaction study considering the proxy-phenotype approach proposed by Rietveld and colleagues in 2014.16 Specifically, we used the cohorts from the Army STARRS (ASTARRS) Initiative (N = 16,361) for stage-1 and the Yale-Penn sample (N = 8,084) for stage-2, which yielded a total sample of 24,445 individuals. To our knowledge this is the largest GEWIS performed in psychiatric genetics and the first examining risk for alcohol misuse.
Subjects and Methods
Army STARRS cohorts
Subjects investigated were selected from among the participants in the ASTARRS Initiative. All subjects gave written informed consent to participate. These procedures were approved by the Human Subjects Committees of all collaborating institutions.
Sample
Two study populations were included in the ASTARRS Initiative (Table 1). The New Soldier Study (NSS) includes soldiers at the start of their basic training at one of three Army installations. The Pre-Post Deployment Study (PPDS) is a multiple-wave panel survey that collected baseline data (time 0) from US Army soldiers in three brigade combat teams prior to their deployment to Afghanistan. Detailed information about the design and conduct of Army STARRS is available in a previous report.17
Table 1.
Characteristics of the ASTARRS and Yale-Penn cohorts investigated in the proxy-phenotype analysis.
| Characteristics | ASTARRS1 (n = 14,000) | ASTARRS2 (n = 2,361) | Yale-Penn1 (n = 5,546) | Yale-Penn2 (n = 2,538) |
|---|---|---|---|---|
| Age, mean (SD) | 24 (5) | 20 (3) | 40 (10) | 40 (12) |
| Sex (women), n (%) | 1,631 (12) | 510 (22) | 2,472 (45) | 989 (39) |
| African-Americans, n (%) | 2,056 (15) | 335 (14) | 3,215 (58) | 1,114 (44) |
| European-Americans, n (%) | 9,146 (65) | 1,586 (67) | 2,331 (42) | 1,424 (56) |
| Hispanic-Americans, n (%) | 2,798 (20) | 440 (19) | - | - |
| Lifetime Trauma (exposed), n (%) | 10,643 (76) | 1,874 (79) | 3,552 (64) | 1694 (67) |
Procedures
Phenotypes for ASTARRS were obtained using a self-administered questionnaire, which included a computerized version of the Composite International Diagnostic Interview Screening Scales (CIDI-SC).18 From the CIDI-SC assessment, we extracted information regarding lifetime trauma exposure and alcohol misuse.
Lifetime Trauma Assessment
Lifetime trauma exposure (i.e., exposed vs. unexposed) included reporting of any of the following experiences: serious physical assault; sexual assault or rape; witnessing someone being seriously injured or killed; discovering or handling a dead body; a life-threatening illness or injury; a disaster; any other experience that put the subject at risk of death or serious injury; murder of a close friend or relative; suicide of a close friend or relative; combat death of a close friend or relative; or accidental death of a close friend or relative. Further details on the trauma assessments were reported in our previous study.19
Alcohol Use Assessment
For the ASTARRS cohort, a dimensional measure of alcohol misuse was derived by summing responses to 13 items that assessed frequency and consequences of alcohol use including the array of alcohol misuse symptoms: 1) Frequency of Drinking; 2) Frequency of Binge Drinking; 3) Drinking Interfered with Responsibility; 4) Drinking Caused an Argument; 5) Drinking Resulted in Someone Getting Hurt; 6) Out of Control Drinking; 7) Arrested Due to Drinking; 8) Worried to Not Be Able to Drink; 9) Worried About Drinking; 10) Feel a Need to Cut Down; 11) Feel Annoyed by People Who Mention Drinking; 12) Feel Guilty About Drinking; 13) Drink First Thing in the Morning. Respondents rated each symptom on a 5-point frequency scale that ranged from “never” through “every or nearly every day”. Supplemental Table 1 summarizes the items related to lifetime trauma exposure and alcohol-related symptoms. We included only subjects who reported having ever consumed an alcoholic drink (i.e., alcohol exposed). For PPDS subjects, we considered trauma and alcohol information reported at time 0 (within approximately six weeks prior to deployment).
Genetics
The NSS and PPDS samples (ASTARRS1, N=14,000) underwent genotyping using the Illumina OmniExpress and Exome array with additional custom content. An additional 2,361 NSS samples (ASTARRS2) were genotyped on the Illumina PsychChip array. Methods for genotyping, imputation, ancestry assignment, and principal component (PC) analysis were described previously.19, 20
Yale-Penn Cohort
Sample
The subjects in the Yale-Penn cohort were recruited at five sites in the Eastern United States and were previously investigated in genetic studies of substance use disorders and other traits (Table 1).4, 21–24 The institutional review board at each participating site approved the study and we obtained written informed consent from each participant.
Trauma and Alcohol Use Assessment
The Yale-Penn participants were evaluated using the Semi-Structured Assessment for Drug Dependence and Alcoholism (SSADDA),25, 26 which yields DSM-IV and DSM-5 diagnoses of lifetime alcohol and drug dependence and other major psychiatric traits. Lifetime trauma exposure was defined as previous experience or witnessing of traumatic events including military combat; an assault, rape, or kidnapping; seeing someone seriously injured or killed; a flood, earthquake, large fire, or other disaster; an airplane crash or serious car accident; a shooting or bombing; or any situation where you feared there was a serious threat to your life or the life of another person. DSM-5 AUD criterion count was used as a dimensional measure based on the 11 DSM diagnostic criteria for AUD. Further details are reported in our previous study.15 In the analysis, we included only subjects who reported having ever consumed alcohol (i.e., were alcohol exposed).
Genetics
The Illumina HumanOmni1-Quad v1.0 microarray was used to genotype 5,546 subjects (Yale-Penn1) at the Center for Inherited Disease Research or the Yale Center for Genome Analysis; and the Illumina HumanCoreExome array was used to genotype 2,538 additional subjects at the Gelernter Lab at Yale (Yale-Penn2). Genotyping, Imputation, ancestry assignment, and PC analysis are described in our previous articles.4, 21–24
Data Analysis
A proxy-phenotype analysis was conducted considering the ASTARRS cohorts (ASTARRS1 and ASTARRS2) as the discovery sample (Stage-1) and the Yale-Penn cohorts (Yale-Penn1 and Yale-Penn2) as the replication sample (Stage-2). Proxy-phenotype analysis is a two-stage research strategy proposed by Rietveld and colleagues.15 In the first stage, a proxy phenotype (i.e., alcohol misuse) was used to identify a relatively small set of SNPs considering a suggestive statistical significance threshold (i.e., p < 5*10−5). In the second stage, this set of candidate SNPs was tested in an independent sample with respect to the phenotype of interest (i.e., AUD) at a significance threshold corrected for the number of proxy-associated SNPs. Consistent with the National Institute of Mental Health’s Research Domain Criteria (RDoC) initiative,27–29 we considered two-dimensional measures derived from symptom scales based on self-report information: an alcohol misuse score in the ASTARRS cohorts, and a DSM-5 AUD criterion count in the Yale-Penn cohorts. Dichotomous trauma exposure was considered as interactive factor, because it was available in both cohorts. A genome-wide gene-by-trauma interaction analysis was conducted in the ASTARRS cohorts (stage-1). Considering linkage disequilibrium (LD)-independent variants with p < 5*10−5 in the ASTARRS analysis, a replication analysis was performed in the Yale-Penn cohorts (stage-2). Independent variants were defined as more than 500 kb distant and with r2 < 0.2. SNPs that survived a Bonferroni correction that accounted for the number of independent loci were considered a replication. The statistical power of our GEWIS was calculated using QUANTO software (available at http://biostats.usc.edu/Quanto.html). In the ASTARRS sample, we have 92.7% statistical power to detect a moderate GxE effect (βGxE=0.7) at significance level p < 5*10−5 for alleles with frequency ≥ 5%. In the Yale-Penn sample, we have 88% statistical power to detect a moderate GxE effect (βGxE=0.7) at significance level p < 5*10−4 for alleles with frequency ≥ 5%.
Plink 1.930 was used to conduct the analysis in the ASTARRS cohort, and the interaction test was based on comparing the difference between regression coefficients in the trauma-exposed subjects vs. the trauma-unexposed subjects. Because the Yale-Penn cohorts include related individuals, we performed the interaction analysis using the R package GWAF31 to fit a generalized estimating equations (GEE) model to adjust for correlations among related individuals. To verify that no important differences between the two methods were present, we tested the GWAF package and Plink 1.9 in a cohort of unrelated subjects and observed negligible differences due to number approximations. In both analyses, we included SNPs with minor allele frequency ≥ 5% and high imputation quality (Info ≥ 0.8). Before being entered into the analysis, alcohol misuse dimensional scales (i.e., alcohol misuse count and DSM-5 AUD criterion count) were adjusted for age, sex, and the top-10 ancestry PCs, and then normalized using appropriate Box-Cox power transformations. In every analysis, the samples were stratified by genotyping array and ancestry, and the results were combined by meta-analysis using the program METAL.32 Functional annotation of the identified loci was conducted using HaploReg v4.1.33
Finally, we evaluated whether the number of loci that showed the same effect directions in the ASTARRS and the Yale-Penn cohorts was significantly different from chance, by conducting a permutation analysis; and then performed a gene ontology (GO)-enrichment analysis on the basis of the direction-replicated loci using DAVID 6.8 beta version (released May 2016; available at https://david-d.ncifcrf.gov/home.jsp). Fisher’s exact tests and Bonferroni multiple testing corrections were applied in the GO-enrichment analysis. We also used WebGestalt34 (available at http://www.webgestalt.org/) to conduct an enrichment analysis for KEGG pathways using hypergeometric statistical method and Benjamini-Hochberg multiple test adjustment with significance level set at 0.05.
Results
Table 1 reports the characteristics of the cohorts included in our GEWIS. Both ASTARRS and Yale-Penn samples include subjects genotyped with different platforms. Accordingly, we stratified the samples according to their genotyping platform and used a meta-analytic approach to integrate the results for Stage-1 (ASTARRS1 and ASTARRS2) and Stage-2 (Yale-Penn1 and Yale-Penn2) cohorts. Within each study population (ASTARRS and Yale-Penn), the cohorts have similar characteristics (age, sex, ancestry, and trauma exposure), except for sex between the ASTARRS cohorts (ASTARRS1 women = 12%; ASTARRS2 women = 22%). Considering the differences between the two study populations (ASTARRS vs. Yale-Penn), the ASTARRS cohorts are mainly constituted by young European-descent subjects, while the Yale-Penn cohorts mainly consist of older participants with both sexes and ancestries (European and African) almost equally represented. No Hispanic-American group is included in the Yale-Penn cohorts. Lifetime trauma exposure is slightly higher in the ASTARRS cohorts than that observed in the Yale-Penn cohorts (77% vs. 65%).
In the first stage, the genome-wide gene-by-trauma interaction analysis of alcohol misuse showed negligible inflation or deflation in the ancestry-stratified investigations of the ASTARRS cohorts (Supplemental Table 2). This confirmed that no systematic bias affected our GEWIS. GxE analysis can be biased by interactions among predictors and this can be hardly detected in a candidate gene study.35 In a GEWIS a systematic bias in the GxE model would have caused inflation in the distribution of the test statistics.
There was no genome-wide significant result in the ancestry-specific or the trans-population meta-analyses. Considering p < 5*10−5 as the significance threshold for follow-up in the second stage of the proxy-phenotype analysis, we identified 68, 49, and 45 independent loci in African-specific, European-specific, and trans-population meta-analyses, respectively (Supplemental Table 3). In African-American subjects, we identified an interaction of PRKG1 rs1729578 with trauma exposure in the ASTARRS cohort and replicated its interaction with trauma exposure in the Yale-Penn cohort (discovery-replication meta-analysis: z=5.64, p=1.69*10−8; ASTARRS AA: z=4.46, p = 8.09*10−6; Yale-Penn AA: z= 3.62, p = 2.98*10−4). Table 2 shows the details of the association of rs1729578 with alcohol misuse in trauma-exposed and unexposed subjects and its interaction with trauma exposure in relation to alcohol misuse. In the meta-analysis of the discovery (Stage-1) and replication (Stage-2) AA cohorts (N = 6,744), rs1729578 showed a genome-wide significant interaction with trauma-exposure in relation to alcohol misuse (z = 5.64, p = 1.69*10−8; Figure 1). Rs1729578*C allele was positively associated with alcohol misuse in trauma-exposed subjects (z=4.27, p=1.96*10−5); and was negatively associated in trauma-unexposed subjects (z=−5.29, p=1.21*10−7). No significant replication was observed in the EA or trans-population meta-analysis. The Hispanic-specific meta-analysis could not be included in the second stage because no Hispanic group was available for replication in the Yale-Penn cohorts.
Table 2.
Association of PRKG1 rs1729578*C allele with alcohol-related dimensional scales in subjects of African descent exposed and unexposed to lifetime trauma.
| Cohort | Allele Frequency | Lifetime Trauma | No Lifetime Trauma | Interaction | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||
| n | Beta | SE | P value | n | Beta | SE | P value | Beta | SE | P value | ||
| ASTARRS1 | 0.25 | 1,483 | 0.05 | 0.03 | 0.081 | 573 | −0.19 | 0.05 | 1.05*10−4 | 0.25 | 0.06 | 1.15*10−5 |
| ASTARRS2 | 0.21 | 265 | −0.02 | 0.09 | 0.775 | 70 | −0.25 | 0.13 | 0.068 | 0.22 | 0.16 | 0.161 |
| Yale-Penn1 | 0.23 | 1,986 | 0.20 | 0.07 | 3.49*10−3 | 1,229 | −0.21 | 0.08 | 7.79*10−3 | 0.10 | 0.03 | 8.92*10−5 |
| Yale-Penn2 | 0.22 | 752 | −0.02 | 0.13 | 0.846 | 362 | −0.12 | 0.16 | 0.436 | 0.02 | 0.05 | 0.625 |
|
| ||||||||||||
| Meta-analysis | 0.23 | 4,486 | Z=4.27 | 1.96*10−5 | 2,234 | Z=−5.29 | 1.21*10−7 | Z=5.64 | 1.69*10−8 | |||
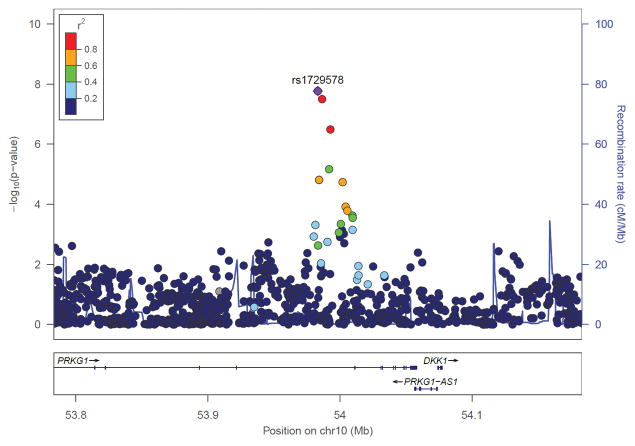
Figure 1.
Regional Manhattan Plots of PRKG1 rs1729578 in African American overall meta-analysis (ASTARRS cohorts + Yale-Penn cohorts).
Rs1729578 is located in an intron of PRKG1 (cGMP-dependent protein kinase 1) and in AAs it is in high LD (r2 > 0.8) with other two PRKG1 intronic variants (rs1194520 and rs871995). According to the data provided by the Roadmap Epigenomics Consortium and the ENCODE Project Consortium,36, 37 rs1729578 is associated with a MIZF motif change; rs1194520 is located in enhancer histone marks (6 tissues), DNAse sites (16 tissues), a CFOS protein bound site, and it is associated with 4 altered transcription factor (TF) motifs (Irf, SETDB1, STAT, YY1). Similarly, rs871995 is located in promoter histone marks (18 tissues), enhancer histone marks (8 tissues), DNAse sites (1 tissue), and is associated with 3 TF altered motifs (Foxp1, Hoxa10, Irf). Details on chromatin-state annotation of the PRKG1 variants are provided in Supplemental Table 4.
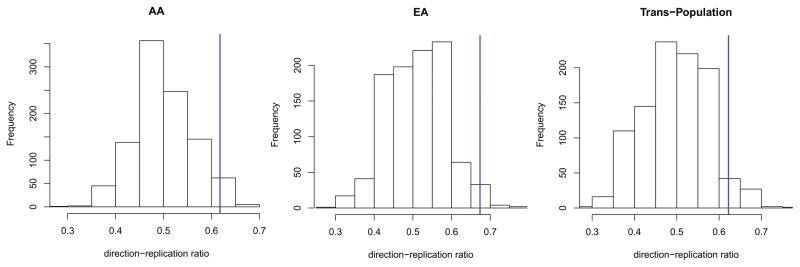
Finally, we verified whether the loci identified in Stage-1 had the same effect directions in the Stage-2 cohorts. In all of the analyses (African-specific, European-specific, and trans-population), we observed that the number of Stage1-identified loci with the same effect directions in the Stage-2 cohorts was higher than would be expected by chance (Figure 2): 42 loci in AAs (ppermutation = 0.022); 33 loci in EAs (ppermutation = 0.004); and 28 loci in the trans-population analysis (ppermutation = 0.041). Considering these loci with the same direction in both cohorts (Supplemental Table 5), we observed significant enrichment for: GO:0016286~small conductance calcium-activated potassium channel activity in AAs; GO:0040011~locomotion in EAs; GO:0050890~cognition and GO:0042517~positive regulation of tyrosine phosphorylation of Stat3 protein. Details regarding the results of enrichment analysis are reported in Table 3. The analysis based on KEGG pathways identified several significant enrichments (Supplemental Table 6). Calcium signaling pathway (KEGG ID: 04020) was also confirmed by this analysis. Other relevant molecular pathways were: Cytokine-cytokine receptor interaction (KEGG ID: 04060); Long-term potentiation (KEGG ID: 04720); Insulin signaling pathway (KEGG ID: 04910).
Figure 2.
Null distribution of direction-replication ratios generated through 1,000 random permutations. Blue lines indicate observed values.
Table 3.
Results from the enrichment analysis conducted considering the loci with same direction in discovery and replication cohorts in trans-population, AA, and EA analyses.
| Analysis | GO term | Genes | Fisher’s exact P value | Bonferroni-corrected P value |
|---|---|---|---|---|
| Trans-population | GO:0050890~cognition | CBR3, CTNS, DOPEY2, GRM5 | 1.9*10−6 | 7.1*10−4 |
| GO:0042517~positive regulation of tyrosine phosphorylation of Stat3 protein | CLCF1, IL23R, VEGFA | 6.4*10−5 | 1.6*10−2 | |
| AA | GO:0016286~small conductance calcium-activated potassium channel activity | KCNN2, KCNN3 | 2.3*10−5 | 5.4*10−3 |
| EA | GO:0040011~locomotion | ATP2B2, JPH3 | 6.7*10−5 | 5.4*10−3 |
Discussion
To our knowledge, this study, which included 24,445 individuals, is the largest GEWIS in psychiatric genetics, and the first examining risk for alcohol misuse. Because the cohorts investigated were assessed using different instruments and criteria intended to measure similar domains, we applied the proxy-phenotype approach.16
In AAs, our proxy-phenotype analysis uncovered a variant, rs1729578, identified in the ASTARRS cohorts and replicated in the Yale-Penn samples. The rs1729578*C allele showed an interaction with trauma exposure in relation to alcohol misuse symptoms. Although it has similar allele frequencies in European- and African-ancestry populations, no effect was observed in the European cohort, possibly due to a different haplotype structure in these groups.38 In African populations, the variant is in high LD (r2 > 0.8) with other two variants, rs1194520 and rs871995. These SNPs are located in an intron of PRKG1, the gene encoding cGMP-dependent protein kinase 1. Functional annotation from the Roadmap Epigenomics Consortium and the ENCODE Project Consortium indicated that these variants are located in genomic regulatory regions.36, 37 Specifically, they are located within multiple chromatin marks across different tissues. Previous studies have demonstrated that non-coding variants identified by GWAS are enriched for chromatin modifications.39 The PRKG1 protein product, cGMP-dependent protein kinase 1, corresponds to both the type I alpha and type I beta isoforms of cyclic guanosine monophosphate (cGMP)-dependent protein kinase, by alternative transcript splicing.40, 41 The gene is most strongly expressed in smooth muscle, platelets, cerebellar Purkinje cells, hippocampal neurons, and the lateral amygdala.40, 41 cGMP plays an important role in learning and memory,42 and a PRKG1 knockout mouse model showed alterations in the cerebellar phospho-proteome that suggests impaired cerebellar long-term depression at Purkinje cell synapses.43 PRKG1 mutant mice also show differences from wild-type mice in circadian rhythms, sleep and distinct aspects of learning.44 Mouse models also demonstrate that cGMP-dependent protein kinase 1 in the amygdala is critical for auditory-cued fear memory and long-term potentiation.45, 46 In vitro studies also show that cGMP-dependent protein kinase 1 isoforms are involved in serotonin transporter regulation.47 In Drosophila melanogaster, the homolog of the human PRKG1, the well-studied foraging (for) gene encodes a cGMP-dependent protein kinase (PKG). Two for variants have been observed in nature: rover allele (forR) with high PKG activity; and sitter allele (fors) with low PKG activity.48 In Drosophila, PKG activity interacts with early life stress in determining adult exploratory and fitness traits.49, 50 PKG activity seems to control synaptic transmission tolerance to acute stress at the Drosophila larval neuromuscular junction, where inhibition promotes functional protection, while activation increases susceptibility to neurotransmission breakdown.51 A GWAS of post-traumatic stress disorder identified PRKG1 as a risk locus in a military cohort independent from those investigated in the current studies,52 also supporting the role of PRKG1 in stress-response related traits in humans.
Finally, our proxy-phenotype analysis demonstrated that the enrichment for loci with same effect direction in stage-1 and stage-2 was unlikely to be due to chance. Investigating these loci, we observed indicative GO enrichments. The enrichment for GO:0016286~small conductance calcium-activated potassium channel activity confirmed previous findings of genome-wide genetic and epigenetic investigations on the role of potassium channels and calcium metabolism in substance use disorders.21, 22, 53 Both GO:0040011~locomotion and GO:0050890~cognition appear consistent with the role of genes involved in mobility and cognitive functions highlighted by the PRKG1 result. GO:0042517~positive regulation of tyrosine phosphorylation of Stat3 protein suggests that mechanisms related to STAT3 regulation are involved in the trauma response. STAT3 is involved in signal transduction and transcription activation of a wide range of genes in response to cell stimuli,54 but no studies have yet investigated its involvement in the behavioral stress response. Pathway-based enrichment analysis confirmed the GO result related to the calcium metabolism (Calcium signaling pathway, KEGG ID: 04020). Besides this confirmatory result, the most significant pathway enrichment was observed for Cytokine-cytokine receptor interaction (KEGG ID: 04060). This is a relevant pathway related to immune functions such as inflammatory host defenses, cell growth, differentiation, cell death, angiogenesis, and development and repair processes aimed at the restoration of homeostasis. Our previous GWAS of PTSD showed a strong overlapping with autoimmune diseases19 and our current finding confirm the role of immune functions in trauma response.
In conclusion, our study provides the first genome-wide evidence regarding a mechanism by which traumatic life events interact with human genetic variation in relation to alcohol misuse. However, although to our knowledge this is the largest GEWIS performed in psychiatric genetics, our current findings are still limited and further GEWIS with larger sample sizes together with translational studies will be needed to uncover more completely how traumatic experience interacts with the genetic predisposition to modify risk for alcohol use disorders. Our main result is the identification of PRKG1 rs1729578 as a risk locus interacting with trauma exposure in determining alcohol misuse. Our study design included different types of trauma within the “lifetime trauma exposure” category. Since certain traumatic experiences may be consequences of the alcohol abuse (e.g., individuals who abuse alcohol are more likely to have motor vehicle collisions than those who do not abuse alcohol), further studies are needed to dissect the complex interactive network of alcohol misuse, trauma experience, and genetics. Additional experiments are also necessary to understand the molecular mechanisms (e.g., epigenetic changes, transcriptional regulation) involved in this interactive process. However, the PRKG1 locus encodes a prospective target for the development of pharmacological treatments for stress-response related traits. Indeed, an in vivo study has shown that the blockade of a1-adrenergic receptors mitigates stress-disturbed cGMP signaling.55 Further studies should consider whether pharmacological treatments targeted at the cGMP signaling system are useful to reduce risk behaviors, such as alcohol misuse, in subjects exposed to traumatic events. Other potential targets for future translational investigations of trauma-related psychopathologies include potassium channels, calcium metabolism, and STAT3 regulation system.
Supplementary Material
Acknowledgments
This study was funded by National Institutes of Health grants R21 AA024404, U01 MH087981, RC2 DA028909, R01 DA12690, R01 DA12849, R01 DA18432, R01 AA11330, R01 AA017535, P50 AA012870, the VISN1 and VISN4 MIRECCs. We would like to thank an anonymous reviewer for the findings of the KEGG-pathway enrichment analysis.
Footnotes
Conflict of Interest
Dr. Stein has in the last three years been a consultant for Actelion Pharmaceuticals, Healthcare Management Technologies, Janssen, Pfizer, Resilience Therapeutics, Tonix Pharmaceuticals, and Oxeia Biopharmaceuticals. Dr. Kaufman has provided consultation to Pfizer and Merck Pharmaceutical Company to train investigators to assess bipolar disorder in youth. Dr. Kranzler has been an advisory board member, consultant, or CME speaker for Indivior, Lundbeck, and Otsuka. He is also a member of the American Society of Clinical Psychopharmacology’s Alcohol Clinical Trials Initiative, which is supported by AbbVie, Alkermes, Ethypharm, Indivior, Lilly, Lundbeck, Pfizer, and XenoPort. The other authors reported no biomedical financial interests or potential conflicts of interest.
References
- 1.Walsh K, Elliott JC, Shmulewitz D, Aharonovich E, Strous R, Frisch A, et al. Trauma exposure, posttraumatic stress disorder and risk for alcohol, nicotine, and marijuana dependence in Israel. Compr Psychiatry. 2014;55(3):621–630. doi: 10.1016/j.comppsych.2013.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kline A, Weiner MD, Ciccone DS, Interian A, St Hill L, Losonczy M. Increased risk of alcohol dependency in a cohort of National Guard troops with PTSD: a longitudinal study. J Psychiatr Res. 2014;50:18–25. doi: 10.1016/j.jpsychires.2013.11.007. [DOI] [PubMed] [Google Scholar]
- 3.Goldman D, Oroszi G, Ducci F. The genetics of addictions: uncovering the genes. Nat Rev Genet. 2005;6(7):521–532. doi: 10.1038/nrg1635. [DOI] [PubMed] [Google Scholar]
- 4.Gelernter J, Kranzler HR, Sherva R, Almasy L, Koesterer R, Smith AH, et al. Genome-wide association study of alcohol dependence:significant findings in African- and European-Americans including novel risk loci. Mol Psychiatry. 2014;19(1):41–49. doi: 10.1038/mp.2013.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Polimanti R, Zhang H, Smith AH, Zhao H, Farrer LA, Kranzler HR, et al. Genome-wide association study of body mass index in subjects with alcohol dependence. Addict Biol. 2015 doi: 10.1111/adb.12317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Polimanti R, Wang Q, Meda SA, Patel KT, Pearlson GD, Zhao H, et al. The Interplay Between Risky Sexual Behaviors and Alcohol Dependence: Genome-Wide Association and Neuroimaging Support for LHPP as a Risk Gene. Neuropsychopharmacology. 2016 doi: 10.1038/npp.2016.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Polimanti R, Kranzler HR, Gelernter J. Phenome-Wide Association Study for Alcohol and Nicotine Risk Alleles in 26394 Women. Neuropsychopharmacology. 2016;41(11):2688–2696. doi: 10.1038/npp.2016.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sartor CE, Wang Z, Xu K, Kranzler HR, Gelernter J. The joint effects of ADH1B variants and childhood adversity on alcohol related phenotypes in African-American and European-American women and men. Alcoholism, clinical and experimental research. 2014;38(12):2907–2914. doi: 10.1111/acer.12572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Turecki G, Ota VK, Belangero SI, Jackowski A, Kaufman J. Early life adversity, genomic plasticity, and psychopathology. Lancet Psychiatry. 2014;1(6):461–466. doi: 10.1016/S2215-0366(14)00022-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mehta D, Klengel T, Conneely KN, Smith AK, Altmann A, Pace TW, et al. Childhood maltreatment is associated with distinct genomic and epigenetic profiles in posttraumatic stress disorder. Proc Natl Acad Sci U S A. 2013;110(20):8302–8307. doi: 10.1073/pnas.1217750110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lieberman R, Armeli S, Scott DM, Kranzler HR, Tennen H, Covault J. FKBP5 genotype interacts with early life trauma to predict heavy drinking in college students. Am J Med Genet B Neuropsychiatr Genet. 2016;171(6):879–887. doi: 10.1002/ajmg.b.32460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baranger DA, Ifrah C, Prather AA, Carey CE, Corral-Frias NS, Drabant Conley E, et al. PER1 rs3027172 Genotype Interacts with Early Life Stress to Predict Problematic Alcohol Use, but Not Reward-Related Ventral Striatum Activity. Front Psychol. 2016;7:464. doi: 10.3389/fpsyg.2016.00464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaufman J, Yang BZ, Douglas-Palumberi H, Crouse-Artus M, Lipschitz D, Krystal JH, et al. Genetic and environmental predictors of early alcohol use. Biol Psychiatry. 2007;61(11):1228–1234. doi: 10.1016/j.biopsych.2006.06.039. [DOI] [PubMed] [Google Scholar]
- 14.Gelernter J. Genetics of complex traits in psychiatry. Biological psychiatry. 2015;77(1):36–42. doi: 10.1016/j.biopsych.2014.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hart AB, Lynch KG, Farrer L, Gelernter J, Kranzler HR. Which alcohol use disorder criteria contribute to the association of ADH1B with alcohol dependence? Addict Biol. 2016;21(4):924–938. doi: 10.1111/adb.12244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rietveld CA, Esko T, Davies G, Pers TH, Turley P, Benyamin B, et al. Common genetic variants associated with cognitive performance identified using the proxy-phenotype method. Proc Natl Acad Sci U S A. 2014;111(38):13790–13794. doi: 10.1073/pnas.1404623111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ursano RJ, Colpe LJ, Heeringa SG, Kessler RC, Schoenbaum M, Stein MB, et al. The Army study to assess risk and resilience in servicemembers (Army STARRS) Psychiatry. 2014;77(2):107–119. doi: 10.1521/psyc.2014.77.2.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kessler RC, Ustun TB. The World Mental Health (WMH) Survey Initiative Version of the World Health Organization (WHO) Composite International Diagnostic Interview (CIDI) Int J Methods Psychiatr Res. 2004;13(2):93–121. doi: 10.1002/mpr.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stein MB, Chen CY, Ursano RJ, Cai T, Gelernter J, Heeringa SG, et al. Genome-wide Association Studies of Posttraumatic Stress Disorder in 2 Cohorts of US Army Soldiers. JAMA Psychiatry. 2016 doi: 10.1001/jamapsychiatry.2016.0350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Polimanti R, Chen CY, Ursano RJ, Heeringa SG, Jain S, Kessler RC, et al. Cross-Phenotype Polygenic Risk Score Analysis of Persistent Post-Concussive Symptoms in U.S. Army Soldiers with Deployment-Acquired Traumatic Brain Injury. J Neurotrauma. 2016 doi: 10.1089/neu.2016.4550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gelernter J, Kranzler HR, Sherva R, Koesterer R, Almasy L, Zhao H, et al. Genome-wide association study of opioid dependence: multiple associations mapped to calcium and potassium pathways. Biol Psychiatry. 2014;76(1):66–74. doi: 10.1016/j.biopsych.2013.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sherva R, Wang Q, Kranzler H, Zhao H, Koesterer R, Herman A, et al. Genome-wide Association Study of Cannabis Dependence Severity, Novel Risk Variants, and Shared Genetic Risks. JAMA Psychiatry. 2016;73(5):472–480. doi: 10.1001/jamapsychiatry.2016.0036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gelernter J, Kranzler HR, Sherva R, Almasy L, Herman AI, Koesterer R, et al. Genome-wide association study of nicotine dependence in American populations: identification of novel risk loci in both African-Americans and European-Americans. Biol Psychiatry. 2015;77(5):493–503. doi: 10.1016/j.biopsych.2014.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gelernter J, Sherva R, Koesterer R, Almasy L, Zhao H, Kranzler HR, et al. Genome-wide association study of cocaine dependence and related traits: FAM53B identified as a risk gene. Mol Psychiatry. 2014;19(6):717–723. doi: 10.1038/mp.2013.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pierucci-Lagha A, Gelernter J, Chan G, Arias A, Cubells JF, Farrer L, et al. Reliability of DSM-IV diagnostic criteria using the semi-structured assessment for drug dependence and alcoholism (SSADDA) Drug Alcohol Depend. 2007;91(1):85–90. doi: 10.1016/j.drugalcdep.2007.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pierucci-Lagha A, Gelernter J, Feinn R, Cubells JF, Pearson D, Pollastri A, et al. Diagnostic reliability of the Semi-structured Assessment for Drug Dependence and Alcoholism (SSADDA) Drug Alcohol Depend. 2005;80(3):303–312. doi: 10.1016/j.drugalcdep.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 27.Cuthbert BN, Insel TR. Toward the future of psychiatric diagnosis: the seven pillars of RDoC. BMC Med. 2013;11:126. doi: 10.1186/1741-7015-11-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Insel T, Cuthbert B, Garvey M, Heinssen R, Pine DS, Quinn K, et al. Research domain criteria (RDoC): toward a new classification framework for research on mental disorders. Am J Psychiatry. 2010;167(7):748–751. doi: 10.1176/appi.ajp.2010.09091379. [DOI] [PubMed] [Google Scholar]
- 29.Montalvo-Ortiz JL, Gelernter J, Hudziak J, Kaufman J. RDoC and translational perspectives on the genetics of trauma-related psychiatric disorders. Am J Med Genet B Neuropsychiatr Genet. 2016;171B(1):81–91. doi: 10.1002/ajmg.b.32395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen MH, Yang Q. GWAF: an R package for genome-wide association analyses with family data. Bioinformatics. 2010;26(4):580–581. doi: 10.1093/bioinformatics/btp710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics (Oxford, England) 2010;26(17):2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ward LD, Kellis M. HaploReg v4: systematic mining of putative causal variants, cell types, regulators and target genes for human complex traits and disease. Nucleic Acids Res. 2016;44(D1):D877–881. doi: 10.1093/nar/gkv1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang J, Duncan D, Shi Z, Zhang B. WEB-based GEne SeT AnaLysis Toolkit (WebGestalt): update 2013. Nucleic Acids Res. 2013;41(Web Server issue):W77–83. doi: 10.1093/nar/gkt439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Keller MC. Gene x environment interaction studies have not properly controlled for potential confounders: the problem and the (simple) solution. Biol Psychiatry. 2014;75(1):18–24. doi: 10.1016/j.biopsych.2013.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roadmap Epigenomics Consortium. Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518(7539):317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Encode Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Polimanti R, Yang C, Zhao H, Gelernter J. Dissecting ancestry genomic background in substance dependence genome-wide association studies. Pharmacogenomics. 2015;16(13):1487–1498. doi: 10.2217/pgs.15.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Trynka G, Raychaudhuri S. Using chromatin marks to interpret and localize genetic associations to complex human traits and diseases. Curr Opin Genet Dev. 2013;23(6):635–641. doi: 10.1016/j.gde.2013.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hofmann F, Wegener JW. cGMP-dependent protein kinases (cGK) Methods Mol Biol. 2013;1020:17–50. doi: 10.1007/978-1-62703-459-3_2. [DOI] [PubMed] [Google Scholar]
- 41.Lohmann SM, Walter U. Tracking functions of cGMP-dependent protein kinases (cGK) Front Biosci. 2005;10:1313–1328. doi: 10.2741/1621. [DOI] [PubMed] [Google Scholar]
- 42.Kleppisch T, Feil R. cGMP signalling in the mammalian brain: role in synaptic plasticity and behaviour. Handb Exp Pharmacol. 2009;(191):549–579. doi: 10.1007/978-3-540-68964-5_24. [DOI] [PubMed] [Google Scholar]
- 43.Corradini E, Vallur R, Raaijmakers LM, Feil S, Feil R, Heck AJ, et al. Alterations in the cerebellar (Phospho)proteome of a cyclic guanosine monophosphate (cGMP)-dependent protein kinase knockout mouse. Mol Cell Proteomics. 2014;13(8):2004–2016. doi: 10.1074/mcp.M113.035154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Langmesser S, Franken P, Feil S, Emmenegger Y, Albrecht U, Feil R. cGMP-dependent protein kinase type I is implicated in the regulation of the timing and quality of sleep and wakefulness. PLoS One. 2009;4(1):e4238. doi: 10.1371/journal.pone.0004238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ota KT, Pierre VJ, Ploski JE, Queen K, Schafe GE. The NO-cGMP-PKG signaling pathway regulates synaptic plasticity and fear memory consolidation in the lateral amygdala via activation of ERK/MAP kinase. Learn Mem. 2008;15(10):792–805. doi: 10.1101/lm.1114808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Paul C, Schoberl F, Weinmeister P, Micale V, Wotjak CT, Hofmann F, et al. Signaling through cGMP-dependent protein kinase I in the amygdala is critical for auditory-cued fear memory and long-term potentiation. J Neurosci. 2008;28(52):14202–14212. doi: 10.1523/JNEUROSCI.2216-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang YW, Rudnick G. Myristoylation of cGMP-dependent protein kinase dictates isoform specificity for serotonin transporter regulation. J Biol Chem. 2011;286(4):2461–2468. doi: 10.1074/jbc.M110.203935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Osborne KA, Robichon A, Burgess E, Butland S, Shaw RA, Coulthard A, et al. Natural behavior polymorphism due to a cGMP-dependent protein kinase of Drosophila. Science. 1997;277(5327):834–836. doi: 10.1126/science.277.5327.834. [DOI] [PubMed] [Google Scholar]
- 49.Urquhart-Cronish M, Sokolowski MB. Gene-environment interplay in Drosophila melanogaster: chronic nutritional deprivation in larval life affects adult fecal output. J Insect Physiol. 2014;69:95–100. doi: 10.1016/j.jinsphys.2014.06.001. [DOI] [PubMed] [Google Scholar]
- 50.Burns JG, Svetec N, Rowe L, Mery F, Dolan MJ, Boyce WT, et al. Gene-environment interplay in Drosophila melanogaster: chronic food deprivation in early life affects adult exploratory and fitness traits. Proc Natl Acad Sci U S A. 2012;109(Suppl 2):17239–17244. doi: 10.1073/pnas.1121265109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Caplan SL, Milton SL, Dawson-Scully K. A cGMP-dependent protein kinase (PKG) controls synaptic transmission tolerance to acute oxidative stress at the Drosophila larval neuromuscular junction. J Neurophysiol. 2013;109(3):649–658. doi: 10.1152/jn.00784.2011. [DOI] [PubMed] [Google Scholar]
- 52.Ashley-Koch AE, Garrett ME, Gibson J, Liu Y, Dennis MF, Kimbrel NA, et al. Genome-wide association study of posttraumatic stress disorder in a cohort of Iraq-Afghanistan era veterans. J Affect Disord. 2015;184:225–234. doi: 10.1016/j.jad.2015.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cadet JL, Brannock C, Krasnova IN, Jayanthi S, Ladenheim B, McCoy MT, et al. Genome-wide DNA hydroxymethylation identifies potassium channels in the nucleus accumbens as discriminators of methamphetamine addiction and abstinence. Mol Psychiatry. 2016 doi: 10.1038/mp.2016.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.You L, Wang Z, Li H, Shou J, Jing Z, Xie J, et al. The role of STAT3 in autophagy. Autophagy. 2015;11(5):729–739. doi: 10.1080/15548627.2015.1017192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stojkov NJ, Baburski AZ, Bjelic MM, Sokanovic SJ, Mihajlovic AI, Drljaca DM, et al. In vivo blockade of alpha1-adrenergic receptors mitigates stress-disturbed cAMP and cGMP signaling in Leydig cells. Mol Hum Reprod. 2014;20(1):77–88. doi: 10.1093/molehr/gat052. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.