Abstract
In this manuscript we use a two-dimensional coarse-grained model to study how amyloid fibrils grow towards an equilibrium state where they coexist with proteins dissolved in a solution. Free-energies to dissociate proteins from fibrils are estimated from the residual concentration of dissolved proteins. Consistent with experiments, the concentration of proteins in solution affects the growth rate of fibrils but not their equilibrium state. Also, studies of the temperature dependence of the equilibrium state can be used to estimate thermodynamic quantities, e.g., heat capacity and entropy.
Graphical abstract

INTRODUCTION
The self-assembly of proteins into amyloid-like fibrils is a ubiquitous process that arises for seemingly unrelated amino acid sequences when subjected to the right conditions [1, 2]. Segments of proteins incorporated into these fibrils adopt extended conformations, i.e., β-strands, which are hydrogen bonded to neighboring proteins forming a β-sheet. Stacking of β-sheets through side chain interactions accounts for the common pattern of amyloid fibrils, also known as cross-β structure [3, 4]–see Fig. 1a. Proteins that form fibrils at physiological conditions were found to be either functional, e.g., silk fibrils that are used by spiders to capture their prey, or related to plaque formation in amyloid diseases that include Alzheimer’s and Parkinson’s [5, 6]. Fibril formation proceeds through a nucleation phase followed by growth [7–9]. In the nucleation phase, some proteins form a disordered aggregate [10, 11] from which the fibril nucleates while in the growth phase proteins are added to the fibril increasing its length. Important questions regarding the pathway of fibril formation [12–14] as well as the structure of the disordered aggregates remain open [11]. Also, understanding the equilibrium of mature fibrils in solution is of great interest as it could enable the development of a thermodynamic framework to study these structures. In analogy with protein folding [15], thermodynamics may provide insights into the stability of fibrils and their underlying molecular mechanisms [16].
FIG. 1.
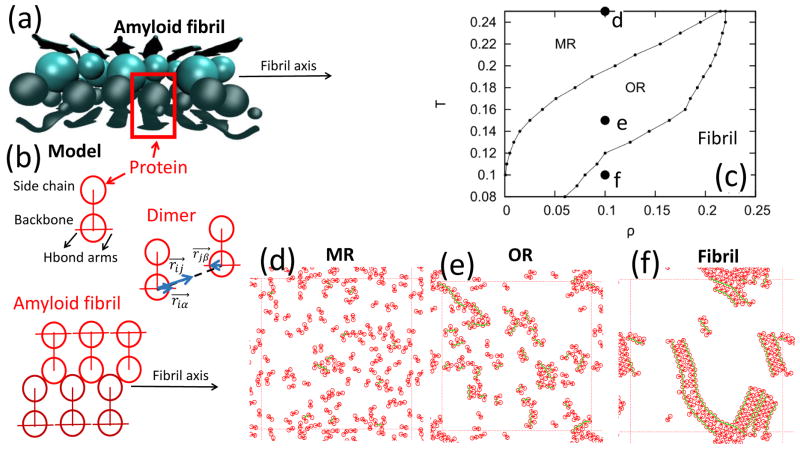
(a) Structure of a fibril made from residues 16–21 of the amyloid-β protein (PDB ID: 3OW9). Backbone and side chains of proteins are depicted using cartoon-like and bead representations, respectively. (b) Schematic representation of a protein in the model. Side chain and backbone beads are highlighted as well as Hbond arms. The representation of a dimer depicts the vectors (blue arrow) used in Eq. 3 to account for Hbonds. The representation of an amyloid fibril shows two β-sheets made of three peptides each stack on top of each other. The fibril axis is also shown. (c) Phase diagram showing monomeric (MR), oligomeric (OR) and fibrillar states as well as representative snapshots of the system at these states computed at the same density 0.1 and temperatures (d) 0.25, (e) 0.15, and (f) 0.1.
Equilibrium thermodynamic properties of amyloid fibrils are challenging to measure using conventional experimental methods. For example, heat capacities measured as a function of temperature in calorimetric experiments were found to be irreversible for most fibrils precluding the use of analytic frameworks to compute free-energies [17, 18]. Moreover, it is difficult to deconvolute contributions from molecular interactions between proteins and fibrils from these calorimetric heat capacities that depend on the overall morphology of the fibrillar network. Overcoming these limitations, recent “amyloid assembly equilibrium measurements” have shown that at least for some protein sequences mature fibrils can exist in equilibrium with dissolved proteins, i.e., monomers [19–23]. This equilibrium has been explored to measure free-energies required to dissociate a protein from a fibril, i.e., ΔG. Studies of the temperature dependence of this equilibrium can also provide insights of other thermodynamic quantities, i.e., change in enthalpy (ΔH), entropy (ΔS), and heat capacity (ΔCp). These quantities played a key role in determining the molecular driving forces of protein folding [24, 25] but they remain mostly unknown for amyloid fibrils. Only few experiments have estimated ΔCp for amyloid fibrils finding both positive and negative values [18, 26–28]. Currently, it is unknown how the magnitude and sign of ΔCp relate to the peptide sequence, fibril structure, and/or condition of the solvent. Notice that unfolding of proteins accounts for a large and positive (never negative) change in heat capacity [24, 29–36]. Timescales to equilibrate fibrils and proteins in a solution are of the order of hours and days which is beyond reach of all-atom computer simulations [20, 23, 37]. Thus, models to study these equilibrium reaction require a coarse-grained approach [38–40].
Here, we develop an off-lattice coarse-grained model to study how amyloid fibrils grow towards an equilibrium state wherein free-energies ΔF to add or dissociate a protein from a fibril can be measured. The initial density of proteins in a solution affects the kinetics of aggregation and the time required for mature fibrils to form. However, we show that mature fibrils that were generated from different densities of proteins can reach the same fibril-monomer equilibrium state defined by ΔF. Moreover, ΔH, ΔS, and ΔCp can be obtained by studying the temperature dependence of this equilibrium. We discuss how the greater stability of fibrils with respect to temperature when compared to native protein structures implies a small ΔCp (close to zero). Despite its small value, we show that ΔCp can be measured in our model. We also study ΔCp for the honeycomb lattice model of amyloid fibrils which has recently been developed to study fibril solubility [41]. We find that changes in the thickness of the fibril in the honeycomb lattice accounts for an increase in the magnitude of ΔCp. This suggests that measurements of ΔCp for amyloid fibrils might provide insights into the residues that are buried in the fibril core.
METHODOLOGY
Our coarse grained model accounts for the two length scales that are characteristic of amyloid fibrils and which emerge from backbone hydrogen bonding and side chain interactions. Proteins are described by two-dimensional dimers, i.e., two beads separated by a fixed distance l. Beads mimic side chain and backbone atoms (see Fig. 1b) and they interact with beads from neighboring proteins through a Lennard-Jones potential defined by εΦΨ (binding energy) and σ (distance parameter):
| (1) |
where riΦjΨ is the distance between bead Φ of protein i and bead Ψ of protein j.
In addition to Lennard-Jones interactions, backbone beads can also form two hydrogen bonds (Hbonds). The directionality of Hbonds in fibrils is accounted for by two arms separated by an angle of 180° and which are perpendicular to the line joining backbone and side chain beads–see Fig. 1b. If r⃗ij is the distance vector between centers of mass of backbone beads of proteins i and j, and r̂iα is the unit vector along arm α of backbone bead i (see Fig. 1b), then the Hbond energy in the Ben-Naim’s framework is [42–44]:
| (2) |
where εHB = −1 and RHB = 1 are the binding energy and equilibrium distance of the Hbond. The constant σHB = 0.085 RHB is the attenuation parameter of the interaction. Equation 2 favors configurations where the distance between backbone beads of molecules i and j is RHB, one arm of molecule i is aligned with the line joining the centers of mass of backbone beads, and the same for one arm of molecule j. Distances and energies in this work are given in units of RHB and εHB, respectively, and the Boltzmann constant kb is defined as one. In terms of these reduced units, the Lennard-Jones potential is defined by σ = 0.75 (same distance parameter for all beads) and εΦΨ = 0.1, 0.2, or 0.141 if beads Φ and Ψ correspond both to side chain, backbone, or a mixture of side chain and backbone beads, respectively. This set of binding energies for the Lennard-Jones potential was chosen by trial-and-error to account for fibril formation. In particular, a same εΦΨ value for pairs of side chain and backbone beads resulted in circular aggregates instead of linearly elongated fibrils. The binding energy for the interaction between side chain and backbone (i.e., εΦΨ =0.141) beads was obtained from the standard Lorentz-Berthelot rule. The fixed distance between beads is l = 1.
Here, we perform Monte Carlo (MC) simulations in the canonical (NVT) ensemble [45]. Periodic boundary conditions is used to mimic an infinite system. Initial conditions are chosen randomly. At each time-step, we tried to translate, rotate and switch (180° rotation) in random order a randomly chosen protein. A MC cycle which corresponds to N time-steps is used as our unit of time. Average densities are computed from 106 MC cycle simulations that are performed after an equilibration period of 105 cycles. Cluster analysis is used to classified the overall bonding state of the system. It uses an energy criteria wherein proteins are considered bonded when their potential energy is less than −0.05. Small variations of this energy cutoff did not account for significant differences in the bonding state of the system. The different states of the system are: the monomeric state when more than 50% of proteins are non-bonded, the fibril state when at least 50% of the proteins are bonded to one big cluster, and the oligomeric state which comprises all other situations, i.e., when less than 50% of proteins are non-bonded and less than 50% are not part of a big cluster.
RESULTS
In Fig. 1c we show the phase diagram of a system comprising N = 120 proteins. This diagram was obtained from simulations carried out at different temperatures (from 0.08 to 0.26 in steps of 0.01) and densities (from 0.01 to 0.25 in steps of 0.01). Characteristic configurations are shown in Fig. 1d–f [46]. The monomeric state occur for conditions of high temperature and low density which are unfavorable to bond formation. Accordingly, proteins in Fig. 1d are mostly dispersed in the simulation box. Conversely, the fibrillar state is favored by conditions of low temperature and high density. Under these conditions, proteins are mostly bonded to each other and fibrils in Fig. 1f are characterized by β-sheet structures, i.e., extended regions of Hbonded proteins stacked on top of each others via side chain interactions. In the oligomeric state, which occurs for intermediate values of temperature and density in Fig. 1c, Hbonded proteins do not form ordered β-sheets which is required for fibril formations.
In Fig. 2, we study how systems evolve towards equilibrium. Starting from a random initial conditions, the fraction of monomeric proteins fo in the system is shown as a function of time for systems with low and high protein concentrations. Notice that in this figure equilibrium is reached in about 4,000 MC cycles. For the low concentration system, the fraction of non-bonded proteins in equilibrium is greater than 50% and, therefore, the system is considered to be in the monomeric state–see Fig. 1c. Evolution towards equilibrium is a fast process which is fitted in Fig. 2 by a single exponential with a characteristic decay time of 30 MC cycles. The high concentration system reaches equilibrium in the oligomer state and a characteristic configuration is shown in Fig. 1e. The evolution of this system towards equilibrium proceeds in two steps: a very fast bonding process in which dimers and trimers are formed by local rearrangements of proteins, e.g., rotation, followed by a slower addition of proteins to β-sheets accounting for the extended but disordered Hbonded regions visible in Fig. 1e. The time dependence of this system is fitted by a double exponential with characteristic decay times of 1200 and 14 MC cycles.
FIG. 2.
Time dependence of the fraction of monomers fo in the system. Only a fraction of the whole trajectory is shown. Dashed and full lines correspond to systems in the monomeric (i.e., ρ = 0.02) and oligomeric (ρ = 0.1) states, respectively. The thermal energy of the system is 0.15 and each point in the figure is an average over 10 MC cycles. Lines correspond to exponential fits of the data.
Fig. 2 shows that systems evolve toward an equilibrium condition in which bonded and monomeric proteins coexist. Assuming that these two species interconvert, first-order rate equations predict a linear dependence of the equilibrium density of monomers on the overall density of proteins ρ [47]:
| (3) |
where kob and kbo are the rate constants for binding a monomer and dissolving a bonded protein, respectively. This equation is valid in the monomeric states of the phase diagram where all bonded proteins can be interconverted into monomers. However, in the fibril state interconvertion occurs only at the extremities of the fibril. This process is best described by a fibril dissociation reaction in which a protein bonded to the extremity of a fibril dissociates becoming a monomer. A first order rate equation for this reaction for detachment and second order for attachment of monomer predicts
| (4) |
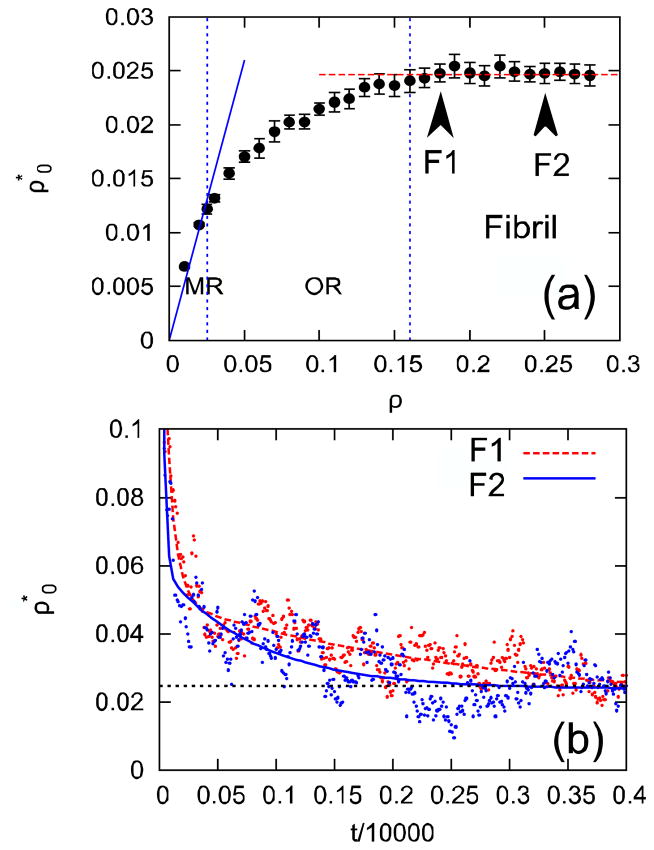
In Fig. 3a, we observe a constant density of monomers ( ) in the fibril region of the phase diagram [39]. Experiments aiming to study equilibrium properties of fibrils are performed with seeded fibrils for which ρf is a constant [19]. Thus, in our simulations and for seeded fibril experiments, ρo is independent of ρ in the fibril region of the phase diagram. The oligomeric phase provides cross-over for the dependence of ρo on ρ in monomeric (Eq. 3) and fibril (Eq. 4) regions. Fig. 3b shows that the kinetics of fibril formation for two fibril states (labeled F1 and F2 in Fig. 3a) is not the same despite both systems reaching the same density of monomers in equilibrium. Equilibrium is reached faster for the system (F2) with higher protein concentration [14].
FIG. 3.
(a) Dependence of the equilibrium density of monomers on the density of proteins ρ in the system at the reduced temperature 0.15. Vertical dashed lines separate monomeric (MR), oligomeric (OR), and Fibril regions. The full line corresponds to the fit of to Eq. 3 for data points in the monomeric region. The horizontal dashed line shows that the equilibrium density of monomers in the fibril state is independent of ρ. Error bars were estimated using block average in which simulations were divided in 20 blocks. (b) Time dependence of ρo compute at ρ = 0.17 (dashed line) and ρ = 0.25 (full line). Only a fraction of the whole trajectory is shown. The horizontal dashed line shows the equilibrium density of monomers.
The stability of mature fibrils can be quantified by measuring the free-energy ΔF required to dissociate a protein from a fibril. ΔF can be written in terms of the equilibrium dissociation constant Kd:
| (5) |
where [F] is the equilibrium concentrations of fibrils and where the standard concentration ρs = 1 (given in units of ) [48]. Accordingly, , where the equilibrium density of monomer is accessible both experimentally and in our simulations [49]. In experimental studies, ΔF was measured to unravel how mutations of the protein sequence and conditions of the solvent affect the stability of fibrils [19]. Here, we are interested in the temperature dependence of ΔF which, according to thermodynamics, can described by [15, 30, 50, 51]:
| (6) |
where ΔUo, ΔSo, and ΔCo are the change in energy, entropy, and heat capacity related to the dissociation of a protein from a fibril computed at temperature To. The temperature dependence of ΔF from our simulations is shown in Fig. 4 and the best fit of this dependence to Eq. 6 using To = 0.15 gives ΔUo = 0.740 ± 0.006, ΔSo = 1.22 ± 0.03, and ΔCo = −4.29 ± 0.14. While our model does not have enough detail to account for the magnitude and sign of these quantities in real proteins, it is interesting to observe that the free-energy is dominated by ΔUo close to To, i.e., ΔUo ≫ ToΔSo. This behavior is expected since fibrils are highly stable, i.e., more stable than native protein conformations [52], and internal energy (or enthalpy) favors the fibrillar state while entropy opposes it [51].
FIG. 4.
Dependence of the free-energy ΔF to dissociate a protein from a fibril on temperature. The best fit of the data point to Eq. 6 is shown.
In Eq. 6, ΔCo which is related to the curvature of ΔF is expected to be small (close to zero) for amyloid fibrils [51] on the basis that the stability of these structures is not strongly dependent on temperature. This justifies the use of van’t Hoff equation:
| (7) |
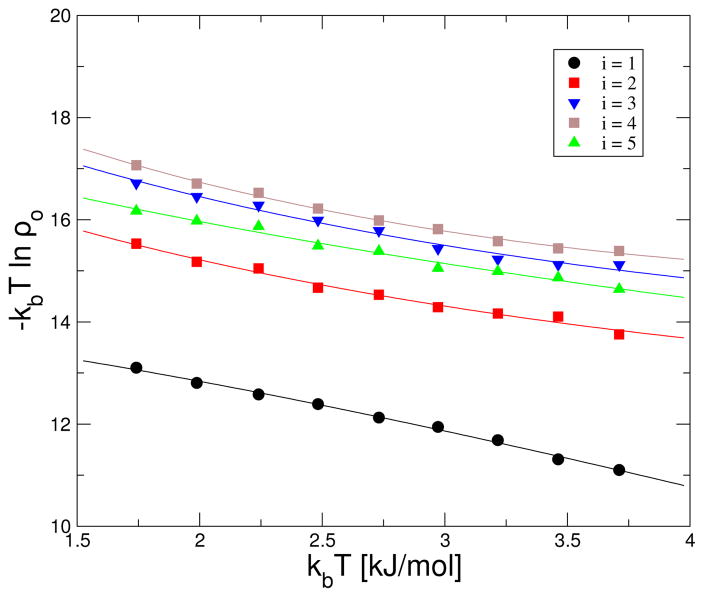
to study amyloid fibrils as it can be derived from Eq. 6 in the limit . According, Eq. 7 has recently been used to study the honeycomb-peptide lattice model of fibril formation [41]. In this model, peptides are represented by hexagons in a two-dimensional triangular lattice which can bind to each other along a straight line to form a β-sheet. Also, stacking of β-sheets account for fibrils with different thickness. Here, we show that despite the small value of ΔCo for the honeycomb lattice model, this quantity can be computed for amyloid fibrils. Symbols in Fig. 5 are results from Fig. 6b of reference [41] where i represents the thickness of the fibril, i.e., the number of β-sheet layers in the fibril. Results in reference [41] were obtained from Monte Carlo simulations. In particular, an initial fibril with fixed thickness and isolated peptides in solution (i.e., honeycombs) were placed in the simulation box. Allowed Monte Carlo moves were translation and rotation of peptides. Depending on the initial concentration of peptides, the fibril elongated or shrunk until a residual concentration of peptides (i.e., ρo in Fig. 6) in solution was reached. Lines in this figure are fits to Eq. 6 using To = 2 kJ/mol and ρs = 1. Quantities obtained from these fits, i.e., ΔUo, ΔSo, and ΔCo, are shown in Table 1. ΔUo and ΔSo increase with the thickness i of the fibril as more bonds need to break to dissociate peptides accounting for a greater dissociation entropy. ΔCo which is positive for i = 1 (i.e., ΔCo =0.36 kJ/mol/K) becomes increasingly more negative with increasing i implying that “thicker” fibrils are more stable to both high and low temperatures. In the honeycomb model, an increase in the thickess of the fibrils (i.e., an increase in i) accounts for more hydrophobic interactions between side chains. Accordingly, this interaction was shown to be responsible for the dependence of ΔUo on i [41] and it can also be expected to account for changes in ΔCo in the model.
FIG. 5.
Dependence of ΔF on temperature for the honeycomb peptide lattice model of fibril formation [41]. Lines correspond to best fit of the data point to Eq. 6.
TABLE I.
Changes in internal energy ΔUo, entropy ΔSo, and heat capacity ΔCo obtained from fits in Fig. 5 using To=2 kJ/mol. Errors were obtained from analysis of the fitting
| i | ΔUo kJ/mol | ΔSo kJ/mol/K | ΔCo kJ/mol/K |
|---|---|---|---|
| 1 | 14.62 ± 0.16 | 0.89 ± 0.07 | 0.36 ± 0.23 |
| 2 | 17.37 ± 0.26 | 1.08 ± 0.12 | −0.79 ± 0.32 |
| 3 | 17.79 ± 0.25 | 0.91 ± 0.11 | −0.42 ± 0.38 |
| 4 | 18.75 ± 0.30 | 1.15 ± 0.14 | −0.90 ± 0.45 |
| 5 | 19.15 ± 0.15 | 1.21 ± 0.06 | −1.16 ± 0.22 |
CONCLUSION
In conclusion, we provide computational proof of principle that fibrils can exist in equilibrium with proteins dissolved in solution and this equilibrium can be used to compute free-energies ΔF to dissociate a protein [40]. We show that conditions of density accounting for different fibril growth rates can lead to the same ΔF and studies of the temperature dependence of ΔF can be used to extract thermodynamic quantities of the system. These quantities remain mostly unknown but they have the potential to provide important insights into molecular mechanisms underlying fibril stability. For example, compensation between ΔHo and ΔSo in protein folding provides rationalization for the marginal stability of the native state [53]. This compensation is being explored in strategies to design drugs that target protein conformations [54, 55]. Thus, understanding enthalpy-entropy compensations in fibrils may be valuable to design more specific drugs for amyloid diseases. Also, ΔCo for protein unfolding depends linearly on the number of non-polar residues in the protein core [15]. It provides, therefore, insights into the position of non-polar amino acids in the native state. Similarly, if ΔCo for fibrils is found to be related to the position of residues, it may be used to complement other techniques to resolve the structure of fibrils. ΔCo may also be used to predict the stability of fibrils as a function of temperature [56]. Therefore, measurements of thermodynamic quantities will be a valuable addition to our understanding of amyloid fibrils.
Supplementary Material
Acknowledgments
T. U. are grateful for the support of the NIH (GM063592) and Slovenian Research Agency (P1 0103-0201, N1-0042) and the National Research, Development and Innovation Office of Hungary (SNN 116198).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Fändrich M, Dobson M. Christopher, EMBO J. 2002;21:5682. doi: 10.1093/emboj/cdf573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fändrich M, Fletcher MA, Dobson CM. Nature. 2001;410:165. doi: 10.1038/35065514. [DOI] [PubMed] [Google Scholar]
- 3.Eanes ED, Glenner GG. J Histochem Cytochem. 1968;16:673. doi: 10.1177/16.11.673. [DOI] [PubMed] [Google Scholar]
- 4.Geddes A, Parker K, Atkins E, Beighton E. J Mol Biol. 1968;32:343. doi: 10.1016/0022-2836(68)90014-4. [DOI] [PubMed] [Google Scholar]
- 5.Chiti F, Dobson CM. Annu Rev Biochem. 2006;75:333. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- 6.Dobson CM. Nature. 2003;426:884. doi: 10.1038/nature02261. [DOI] [PubMed] [Google Scholar]
- 7.Baftizadeh F, Pietrucci F, Biarnés X, Laio A. Phys Rev Lett. 2013;110:168103. doi: 10.1103/PhysRevLett.110.168103. [DOI] [PubMed] [Google Scholar]
- 8.Auer S, Ricchiuto P, Kashchiev D. Journal of molecular biology. 2012;422:723. doi: 10.1016/j.jmb.2012.06.022. [DOI] [PubMed] [Google Scholar]
- 9.Michaels TCT, Dear AJ, Kirkegaard JB, Saar KL, Weitz DA, Knowles TPJ. Phys Rev Lett. 2016;116:258103. doi: 10.1103/PhysRevLett.116.258103. [DOI] [PubMed] [Google Scholar]
- 10.Dahlgren KN, Manelli AM, Stine WB, Baker LK, Krafft GA, LaDu MJ. Journal of Biological Chemistry. 2002;277:32046. doi: 10.1074/jbc.M201750200. [DOI] [PubMed] [Google Scholar]
- 11.Cleary JP, Walsh DM, Hofmeister JJ, Shankar GM, Kuskowski MA, Selkoe DJ, Ashe KH. Nature neuroscience. 2005;8:79. doi: 10.1038/nn1372. [DOI] [PubMed] [Google Scholar]
- 12.Ni R, Abeln S, Schor M, Cohen Stuart MA, Bolhuis PG. Phys Rev Lett. 2013;111:058101. doi: 10.1103/PhysRevLett.111.058101. [DOI] [PubMed] [Google Scholar]
- 13.Mousseau N, Derreumaux P. Acc Chem Res. 2005;38:885. doi: 10.1021/ar050045a. [DOI] [PubMed] [Google Scholar]
- 14.Pellarin R, Caflisch A. Journal of Molecular Biology. 2006;360:882. doi: 10.1016/j.jmb.2006.05.033. [DOI] [PubMed] [Google Scholar]
- 15.Robertson AD, Murphy KP. Chemical reviews. 1997;97:1251. doi: 10.1021/cr960383c. [DOI] [PubMed] [Google Scholar]
- 16.Roland BP, Kodali R, Mishra R, Wetzel R. Peptide Science. 2013;100:780. doi: 10.1002/bip.22305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Doyle CM, Rumfeldt JA, Broom HR, Broom A, Stathopulos PB, Vassall KA, Almey JJ, Meiering EM. Archives of biochemistry and biophysics. 2013;531:44. doi: 10.1016/j.abb.2012.12.005. [DOI] [PubMed] [Google Scholar]
- 18.Morel B, Varela L, Conejero-Lara F. J Phys Chem B. 2010;114:4010. doi: 10.1021/jp9102993. [DOI] [PubMed] [Google Scholar]
- 19.Wetzel R. Accounts of Chemical Research. 2006;39:671. doi: 10.1021/ar050069h. [DOI] [PubMed] [Google Scholar]
- 20.O’Nuallain B, Shivaprasad S, Kheterpal I, Wetzel R. Biochemistry. 2005;44:12709. doi: 10.1021/bi050927h. [DOI] [PubMed] [Google Scholar]
- 21.Doran TM, Kamens AJ, Byrnes NK, Nilsson BL. Proteins: Structure, Function, and Bioinformatics. 2012;80:1053. doi: 10.1002/prot.24007. [DOI] [PubMed] [Google Scholar]
- 22.Williams AD, Shivaprasad S, Wetzel R. Journal of Molecular Biology. 2006;357:1283. doi: 10.1016/j.jmb.2006.01.041. [DOI] [PubMed] [Google Scholar]
- 23.Bhattacharyya AM, Thakur AK, Wetzel R. Proceedings of the National Academy of Sciences. 2005;102:15400. doi: 10.1073/pnas.0501651102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dill K. Biochemistry. 1990;29:7133. doi: 10.1021/bi00483a001. [DOI] [PubMed] [Google Scholar]
- 25.Kauzmann W. Adv Protein Chem. 1959;14:1. doi: 10.1016/s0065-3233(08)60608-7. [DOI] [PubMed] [Google Scholar]
- 26.Ikenoue T, Lee YH, Kardos J, Saiki M, Yagi H, Kawata Y, Goto Y. Angewandte Chemie International Edition. 2014;53:7799. doi: 10.1002/anie.201403815. [DOI] [PubMed] [Google Scholar]
- 27.Kardos J, Yamamoto K, Hasegawa K, Naiki H, Goto Y. Journal of Biological Chemistry. 2004;279:55308. doi: 10.1074/jbc.M409677200. [DOI] [PubMed] [Google Scholar]
- 28.Jeppesen MD, Hein K, Nissen P, Westh P, Otzen DE. Biophysical chemistry. 2010;149:40. doi: 10.1016/j.bpc.2010.03.016. [DOI] [PubMed] [Google Scholar]
- 29.Baldwin RL. Journal of Molecular Biology. 2007;371:283. doi: 10.1016/j.jmb.2007.05.078. [DOI] [PubMed] [Google Scholar]
- 30.Dias CL, Ala-Nissila T, Wong-ekkabut J, Vattulainen I, Grant M, Karttunen M. Cryobiology. 2010;60:91. doi: 10.1016/j.cryobiol.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 31.Edsall JT. Journal of the American Chemical Society. 1935;54:1506. [Google Scholar]
- 32.Bruscolini P, Casetti L. Phys Rev E. 2000;61:R2208. [Google Scholar]
- 33.Rios PDL, Caldarelli G. Phys Rev E. 2001;63:031802. doi: 10.1103/PhysRevE.63.031802. [DOI] [PubMed] [Google Scholar]
- 34.Dias CL, Ala-Nissila T, Karttunen M, Vattulainen I, Grant M. Physical Review Letters. 2008;100:118101. doi: 10.1103/PhysRevLett.100.118101. [DOI] [PubMed] [Google Scholar]
- 35.Dias CL. Physical Review Letters. 2012;109:048104. doi: 10.1103/PhysRevLett.109.048104. [DOI] [PubMed] [Google Scholar]
- 36.Bianco V, Franzese G. Physical review letters. 2015;115:108101. doi: 10.1103/PhysRevLett.115.108101. [DOI] [PubMed] [Google Scholar]
- 37.Nasica-Labouze J, Nguyen PH, Sterpone F, Berthoumieu O, Buchete NV, Coté S, De Simone A, Doig AJ, Faller P, Garcia A. Chemical reviews. 2015;115:3518. doi: 10.1021/cr500638n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Irbäck A, Linnemann N, Linse B, Wallin S, et al. Physical review letters. 2013;110:058101. doi: 10.1103/PhysRevLett.110.058101. [DOI] [PubMed] [Google Scholar]
- 39.Schmit J, Ghosh K, Dill KA. Biophys J. 2011;100:450. doi: 10.1016/j.bpj.2010.11.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ricchiuto P, Brukhno AV, Auer S. J Phys Chem B. 2012;116:5384. doi: 10.1021/jp302797c. [DOI] [PubMed] [Google Scholar]
- 41.Rizzi L, Auer S. The Journal of Physical Chemistry B. 2015;119:14631. doi: 10.1021/acs.jpcb.5b09210. [DOI] [PubMed] [Google Scholar]
- 42.Ben-Naim A. Statistical Thermodynamics for Chemists and Biochemists. Springer; 1992. pp. 459–559. [Google Scholar]
- 43.Urbič T, Vlachy V, Kalyuzhnyi YV, Southall NT, Dill KA. J Chem Phys. 2000;112:2843. [Google Scholar]
- 44.Urbic T, Vlachy V, Kalyuzhnyi YV, Southall NT, Dill KA. J Chem Phys. 2002;116:723. [Google Scholar]
- 45.Frenkel D, Smit B. Computational sciences series. 2002;1:1. [Google Scholar]
- 46.See Supplemental Material at [URL will be inserted by publisher] for an animation of the time evolution of the system.
- 47.Eq. 3 is obtained from the rate equation by assuming equilibrium (i.e., dNo/dt = 0) and the total number of proteins in the system is conserved. No and Nb are the number of monomers and bonded proteins.
- 48.Jamnik A. Fizikalna kemija. FKKT; Ljubljana: 2013. [Google Scholar]
- 49.Reif F. Fundamentals of statistical and thermal physics. Waveland Press; 2009. [Google Scholar]
- 50.Smeller L. Biochimica et Biophysica Acta (BBA) -Protein Structure and Molecular Enzymology. 2002;1595:11. doi: 10.1016/s0167-4838(01)00332-6. [DOI] [PubMed] [Google Scholar]
- 51.Jampani SR, Mahmoudinobar F, Su Z, Dias C. Proteins: Structure, Function, and Bioinformatics. 2015;83:19. [Google Scholar]
- 52.Baldwin AJ, Knowles TP, Tartaglia GG, Fitzpatrick AW, Devlin GL, Shammas SL, Waudby CA, Mossuto MF, Meehan S, Gras SL. Journal of the American Chemical Society. 2011;133:14160. doi: 10.1021/ja2017703. [DOI] [PubMed] [Google Scholar]
- 53.Sharp K. Protein Science. 2001;10:661. doi: 10.1110/ps.37801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chaires JB. Annu Rev Biophys. 2008;37:135. doi: 10.1146/annurev.biophys.36.040306.132812. [DOI] [PubMed] [Google Scholar]
- 55.Olsson TS, Williams MA, Pitt WR, Ladbury JE. Journal of molecular biology. 2008;384:1002. doi: 10.1016/j.jmb.2008.09.073. [DOI] [PubMed] [Google Scholar]
- 56.Mishra R, Winter R. Angw Chem Int Ed. 2008;47:6518. doi: 10.1002/anie.200802027. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.