Abstract
Many antibiotics stop bacterial growth by inhibiting different steps of protein synthesis. However, no specific inhibitors of translation termination are known. Proline-rich antimicrobial peptides, a component of the antibacterial defense system of multicellular organisms, interfere with bacterial growth by inhibiting translation. Here we show that Api137, a derivative of the insect-produced antimicrobial peptide apidaecin, arrests terminating ribosomes using a unique mechanism of action. Api137 binds to the Escherichia coli ribosome and traps release factors 1 or 2 subsequent to release of the nascent polypeptide chain. A high-resolution cryo-EM structure of the ribosome complexed with release factor 1 and Api137 reveals the molecular interactions that lead to release factor trapping. Api137-mediated depletion of the cellular pool of free release factors causes the majority of ribosomes to stall at stop codons prior to polypeptide release, thereby resulting in a global shutdown of translation termination.
Keywords: ribosome, termination, release factors, antimicrobial peptides, antibiotic, stop codon bypass
Introduction
The release of the polypeptide from the ribosome is an essential step of protein synthesis. When the translating ribosome reaches the end of an open reading frame (ORF), it carries the completed protein chain attached to the P-site tRNA and has a stop codon in the A site. In bacteria, termination requires the action of three release factors (RFs), RF1, RF2 and RF3. RF1 or RF2 recognize the stop codon in the A site of the small (30S) subunit while their conserved GGQ motif is placed in the active site of the peptidyl transferase center (PTC) of the large (50S) subunit where it facilitates the hydrolysis of the peptidyl-tRNA ester bond, releasing the completed protein (reviewed in 1). Because the number of ribosomes in the cell greatly exceeds the number of RF1 and RF2 molecules 2,3, continuous translation relies upon the rapid turnover of these factors in order to serve the needs of all the cellular ribosomes. RF3 is a GTPase that facilitates recycling of RF1 and RF2 subsequent to polypeptide release 4,5. Finally, the ribosome recycling factor (RRF) together with the elongation factor G (EF-G) dislodge the ribosome from the mRNA and splits it into subunits 6. Inhibition of any of these reactions should reduce fitness and viability of the bacterial cell. Strikingly, in spite of the complexity and importance of the translation termination, no specific inhibitors of this key step in protein synthesis have so far been identified.
Antimicrobial peptides constitute an important component of the innate immune defense system of multicellular organisms against bacterial infection 7. While many antibacterial peptides lyse cells by disrupting their membrane, a specific class of non-lytic peptides, called proline-rich antimicrobial peptides (PrAMPs), act upon the intracellular target, the ribosome 8–13. Several investigated PrAMPs, such as oncocin 112 (Onc112) and others, whose size range from 15 to 20 amino acids, bind to the nascent peptide exit tunnel of the ribosome and, by encroaching upon the A site of the peptidyl transferase center (PTC), prevent binding of aminoacyl-tRNA10–13. This mode of action results in the arrest of the ribosome at the mRNA start codon before the first peptide bond can be formed10–13.
Among PrAMPs, the 18–20 amino acid long antimicrobial peptides called apidaecins, which are produced by bees, hornets and wasps, remain outliers. Compared to other PrAMPs, they compete with a different subset of ribosomal antibiotics for binding 14. Furthermore, while Onc112 and other PrAMPs readily inhibited protein synthesis in vivo and in vitro, apidaecins efficiently interfered with protein synthesis in living cells, but are poor inhibitors of in vitro translation 14–16. We sought to understand the mechanism of action of apidaecins using Api137 (Fig. 1a), an 18- amino acid derivative of the natural apidaecin 1b, which was optimized to have improved antibacterial properties and serum stability17.
Figure 1. Api137 stalls ribosomes at the termination step of translation.

a, Amino acid sequences of PrAMPs Api137 and Onc112. gu = N,N,N′,N′-tetramethylguanidino, O = L-ornithine, r = D-arginine. b, c, In vitro toeprinting analysis comparing the Onc112- or Api137- mediated translation arrest on model mRNA templates derived from the yrbA (b) or ermCL (c) genes. Positions of the toeprint bands (indicated on the gene sequence) are 16–17 nt downstream from the first nucleotide of the P-site codon. The P- and A- sites codons of the stalled ribosomes are in brackets. Toeprints in (c) were produced by wild-type ribosomes (wt) or by ribosomes with mutations in specific rRNA nucleotides (Supplementary Fig. 2a). Toeprint bands in (b) and (c) generated by Onc112-arrested ribosomes at the initiation codon are indicated with grey arrowheads; those from ribosomes arrested by Api137 at termination are marked with white arrowheads. The similar intensity of the PrAMP-independent toeprint bands marked with a white arrowhead with dotted outline in (c) shows that wt and mutant ribosomes translate with comparable efficiencies. Sequencing reactions are marked. The gels are representatives of (b) more than five and (c) two independent biological replicates.
Results
Api137 arrests translation at the stop codon of mRNAs
To identify the stage of translation inhibited by Api137, we used in vitro toeprinting analysis, which determines the location of stalled ribosomes on mRNA18. In contrast to Onc112, which arrests translation at the start codon12,13 (Fig. 1a), Api137 arrested translation when the stop codon entered the A site of the ribosome (Fig. 1b). Similar stalling at the stop codon was obtained with other tested mRNAs when translation was carried out in the presence of Api137 or the unmodified natural apidaecin 1a (Supplementary Fig. 1). These results show that Api137, unlike other ribosome-targeting PrAMPs or any other known antibiotic, has the unique ability to specifically arrest the terminating ribosome.
Mutations in RF1, RF2 and the ribosome confer resistance to Api137
In order to identify the components of the translation apparatus that are involved in the mechanism of Api137 action, we carried out an unbiased selection of spontaneous Api137-resistant mutants in two E. coli strains. We isolated three types of mutants. The resistance of the first type of mutants was caused by nonsense mutations in the sbmA gene (Supplementary Fig. 2a) encoding the transporter responsible for importing PrAMPs into the cell19.
Resistant mutants of the second type carried mutations in the prfA or prfB genes encoding release factors 1 and 2 (RF1 and RF2). RF1 and RF2 recognize the stop codon of the mRNA and facilitate hydrolysis of the peptidyl-tRNA ester bond, releasing the completed protein (reviewed in1). Mutants isolated using E. coli strain SQ110 carried the mutation in the prfA gene, which resulted in the replacement of Asp241 of the encoded RF1 with a glycine residue (Supplementary Fig. 2). The Api137-resistant mutant isolated with the E. coli strain BL21, had mutations in the prfB gene, resulting in substitutions of the RF2 residues Arg262 (with cysteine) or Gln280 (with leucine) (Supplementary Fig. 2). The difference in the results obtained with two strains likely reflects the fact that SQ110, being a derivative of the K12 strain, carries an alteration in the prfB gene which results in replacement of Ala246 of RF2 with a threonine 20 (Supplementary Fig. 2D). This mutation affects the properties of RF2 21 and conceivably, could alter the interactions of the K12-type RF2 with Api137. The RF1 and RF2 mutations found in Api137 resistant strains are located in close proximity to the catalytically important GGQ motif (Supplementary Fig. 2b, c), suggesting that Api137 interferes with the function of RF1 and RF2.
The third type of Api137 resistant mutants had a mutation in the gene rplP encoding ribosomal protein uL16 (Supplementary Fig. 2). Subsequent testing of other ribosomal protein mutants showed that the mutations in the proteins uL22 and uL4 that are located in the nascent peptide exit tunnel also increased resistance to Api137 (Supplementary Fig. 2). In agreement with this observation, mutations of nearby 23S rRNA nucleotides 2059 and 2503 rendered cells Api137-resistant (Supplementary Fig. 2). Consistently, Api137 failed to induced pronounced arrest of the A2059C or A2503G mutant ribosomes at the stop codons in vitro (Fig. 1c). Taken together, these results indicated that Api137 interferes with translation termination by influencing functional interactions between RF1 or RF2 and the ribosome.
Api137 inhibits turnover of RF1 and RF2
To understand the mode of inhibition of translation termination by Api137, we used a fully reconstituted in vitro translation system. We prepared a model termination complex corresponding to the state of the ribosome prior to hydrolysis of peptidyl-tRNA (pre-hydrolysis complex, PreHC)4,22 (Fig. 2a). Mixing the PreHC with RF1 or RF2 results in the hydrolysis of the ester bond linking fMet to the P-site tRNA, emulating the polypeptide release reaction. At a high concentration of RF1 or RF2, when recycling of the factors was not required for the reaction to progress to completion, rapid and complete hydrolysis of peptidyl-tRNA was observed even in the presence of high Api137 concentrations (Fig. 2b), suggesting that Api137 does not inhibit peptidyl-tRNA hydrolysis. In contrast, at limiting concentrations of RF1 or RF2, when multiple rounds of binding and dissociation of the factors from PreHC were needed to achieve termination on all PreHCs, the reaction was dramatically inhibited in the presence of as little as 1 μM Api137 (Fig. 2c). This result suggested that Api137 either competes with the RFs for binding to the PreHC or traps the RFs in the post-hydrolysis (PostHC) complex abolishing recycling of the factor.
Figure. 2. Api137 allows peptide hydrolysis but inhibits turnover of RF1 and RF2.
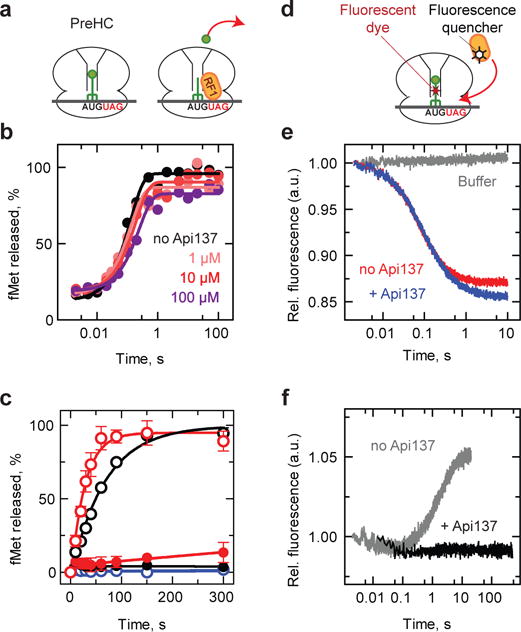
a, Schematics of the peptidyl-tRNA hydrolysis experiments. PreHC carrying f[3H]Met-tRNAfMet is reacted with RF1 (shown) or RF2 and the release of f[35S]Met is measured. b, Time courses of peptide hydrolysis in PreHC in the presence of excess of RF1 without (black) or with the indicated concentrations of Api137 (colored traces). c, Time courses of peptide hydrolysis in PreHC by RF1 (black circles) and RF2 (red circles) under turnover conditions in the absence (open circles) or presence of 1 μM Api137. RF3-GTP was present in all reactions. Control experiments (blue circles) lacked RF1 and RF2 in the absence (open circles) or presence (closed circles) of Api137. 100% corresponds to 10 cycles of RFs binding, catalysis and dissociation. Error bars represent the range of two independent replicates. d, Schematics of the RF1 binding experiments. PreHC carries fluorescein-labeled fMet-tRNA (PreHCFlu) and RF1 carries fluorescence quencher dye (RF1Qsy). e, Time courses of binding of RF1Qsy to PreHCFlu in the absence (red) or presence (blue) or Api137. Grey trace: no RF1. The traces represent the average of five to seven technical replicates. f, Time course of RF1 dissociation. RF1Qsy was incubated with PreHCFlu to generate PostHCFlu and then mixed with a 10-fold excess of unlabeled RF1 and RF3·GTP in the absence (grey) or in the presence (black) of Api137. The traces represent the average of up to seven technical replicates. See Methods for details.
To distinguish between these scenarios we directly examined the effect of Api137 on RF1 binding or dissociation using a fluorescent derivative of fMet-tRNAfMet (PreHCFlu), a quencher dye-labeled RF1 (RF1Qsy) and following changes in fluorescence resonance energy transfer (Fig. 2d). While Api137 did not affect binding of RF1 (Fig. 2e), it entirely blocked RF1 dissociation (Fig. 2f), demonstrating that Api137 prevents turnover of RF1 and RF2 by trapping them on the ribosome. When similar experiments were carried out with the Api137-resistant mutant of RF1 (Supplementary Fig. 2a), Api137 was unable to abolish RF1 dissociation (Supplementary Fig. 3a), indicating that the mutation allowed RF1 to escape Api137-mediated trapping in the PostHC complex. Similarly, the RF2 Ala246Thr mutation endemic in the K12 E. coli strain and located in the vicinity of the selected Api137 resistance mutations (Supplementary Fig. 2d), showed significantly increased tolerance to Api137 inhibition compared to the unaltered RF2 (Supplementary Fig. 3b). Collectively, these results showed that Api137 traps RF1 and RF2 on the ribosome after the release of the nascent protein, abolishes RF turnover and prevents disassembly of the termination complex and recycling of the ribosome for new rounds of translation.
Interactions of Api137 with the ribosome and RF1 illuminate molecular mechanisms of RF trapping
To obtain insights into the molecular mechanism of RF trapping, we determined a cryo-EM structure of Api137 bound to a terminating ribosome (Fig. 3a). The ribosome-nascent chain complex bearing a UAG stop codon in the A site was prepared by translating in vitro the model ermCL ORF in the presence of Api137, purified and subjected to cryo-EM analysis. In silico sorting of the cryo-EM data revealed a major subpopulation of ribosomes bearing a tRNA in the P site and RF1 bound in the A site (Supplementary Fig. 4 and Table 1). A final cryo-EM reconstruction with an average resolution of 3.4 Å enabled generation of a molecular model for the entire complex (Fig. 3a). In the Api137-stalled complex, the conformation of RF1 is similar to that observed previously in the PostHC during canonical termination23,24 (Supplementary Fig. 5a–c). Consistent with our kinetic data the P-site tRNA is deacylated, showing that RF1 has catalyzed hydrolysis of the polypeptide chain in the presence of Api137. A distinct electron density observed within the ribosomal exit tunnel could be unambiguously assigned to residues 5–18 of Api137 bound in extended conformation (Fig. 3b and Supplementary Fig. 4g, h). The orientation of Api137 within the tunnel matches that of a nascent peptide (Supplementary Fig. 5d) but is opposite from that observed for other investigated PrAMPs10–13. The C-terminal Arg17 and Leu18, which are critical for the activity of Api13716, are positioned close to the A and P sites of the PTC, respectively (Figs. 4a). However, in contrast to other PrAMPs that encroach upon the PTC A site, Api137 is positioned entirely within the exit tunnel, allowing it to bind when the A site is occupied by RF1 or RF2 (Supplementary Fig. 5e).
Figure. 3. Binding of Api137 to the terminating ribosome and its interactions with the exit tunnel.

a, Transverse section of the 50S subunit (grey) showing the binding site of Api137 (salmon) on the 70S ribosome (30S subunit in yellow) within the polypeptide exit tunnel relative to RF1 (orange) and the P-site tRNA (green). b, Cryo-EM density (mesh) and molecular model (salmon) for residues 5–18 of Api137. c, Placement of Api137 in the exit tunnel relative to RF1, P-site tRNA and ribosomal proteins uL4 and uL22. Boxed regions are zoomed in the panels d-f, showing interactions of Api137 with components of the exit tunnel, including (d, e) nucleotides of the 23S rRNA (grey) and (f) ribosomal protein uL4. In (e) sphere representation is used to approximate van der Waals interactions and in (f) a hydrogen bond is indicated with a dashed yellow line.
Table 1.
Cryo-EM data collection, refinement and validation statistics
| RF1-API (EMD 3730, PDB 5O2R) |
|
|---|---|
| Data collection | |
| Microscope | FEI Titan Krios |
| Camera | Falcon II |
| Magnification | 129,151 |
| Voltage (kV) | 300 |
| Electron dose (e−/Ǻ2) | 28 |
| Defocus range (μm) | −0.7 to −2.5 |
| Pixel size (Ǻ) | 1.084 |
| Initial particles (no.) | 116,212 |
| Final particles (no.) | 36,826 |
| Model composition | |
| Nonhydrogen atoms | 147,985 |
| Protein residues | 6,205 |
| RNA residues | 4,643 |
| Refinement | |
| Resolution (Å) | 3.4 |
| Map CC (around atoms) | 0.78 |
| Map CC (whole unit cell) | 0.76 |
| Map sharpening B factor (Ǻ2) | −73.07 |
| Average B factor (Ǻ2) | 52.5 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.0110 |
| Bond angles (°) | 1.30 |
| Validation | |
| MolProbity score | 1.99 |
| Clashscore | 7.00 |
| Poor rotamers (%) | 1.01 |
| Ramachandran plot | |
| Favored (%) | 87.90 |
| Allowed (%) | 11.58 |
| Disallowed (%) | 0.52 |
Figure 4. Inhibitory action of Api137 is mediated by its interactions with RF1 and P-site tRNA.

a, Position of Api137 (salmon) relative to RF1 (orange) and P-site tRNA (green). The boxed regions are enlarged in panels (b,c). b, Interactions of Api137 with RF1. Arg17 of Api137 is coordinated by bonding with 23S rRNA nucleotides C2452, G2505 and U2506 (grey) to form direct contacts with Gln235 of the GGQ motif of RF1 (orange). c, The C-terminal hydroxyl of Leu18 of Api137 interacts with the ribose of A76 of deacylated P-site tRNA (green). d, Dimethylsulfate (DMS) probing of Api137 interaction with PostHC 23S rRNA in the absence or presence of RF1. This gel is a representative of two independent experiments.
Api137 makes multiple interactions with the exit tunnel including stacking and van-der-Waals interactions with the 23S rRNA nucleotides (Fig. 3d, e) and a potential hydrogen bond with the ribosomal protein uL4 (Fig. 3f), clarifying how rRNA and ribosomal protein mutations could confer resistance (Supplementary Figs. 2 and 6).
The interactions of the central and N-terminal segments of Api137 with the tunnel elements help to place the functionally critical C-terminal amino acids of Api137 in the vicinity of the GGQ motif of RF1 in the PTC (Fig. 4a–c). The side chain of the penultimate Arg17 of Api137 is fixed in place by hydrogen bonding with the 2′ hydroxyl of the G2505 ribose and O2 of the C2452 nitrogen base (Fig. 4b). This network of hydrogen bonds with the nucleotides of 23S rRNA positions Arg17 for interaction with RF1. The Gln235 side chain carbonyl of RF1 is within hydrogen bonding distance from the terminal nitrogen of the Arg17 guanidinium group (Fig. 4b). The contact between the Arg17 side chain and RF1 is likely to be critical because mutations of the penultimate residue of Api137 to other amino acids decrease the affinity of the PrAMP for the ribosome and reduce its inhibitory activity 16. Also the backbone carboxyl of Arg17 of Api137 is also within hydrogen bond distance of the Gln235 side chain amine of RF1 (Fig. 4b). Interaction between Api137 and RF1 not only helps to trap the RF on the ribosome, but also significantly stabilize binding of Api137 itself. RNA probing experiments showed that in the absence of RF1, Api137 only minimally shielded A2058, A2059, and A2062 from modification, whereas the PrAMP readily protected these nucleotides when RF1 was present (Fig 4d).
The C-terminal hydroxyl of Api137 is within hydrogen bonding distance from the ribose hydroxyls of A76 of the deacylated P-site tRNA (Fig. 4c). These interactions could further contribute to RF1 (or RF2) trapping by preventing the ribosome from RF3-stimulated transitioning into the rotated state required for RF1 (or RF2) dissociation5,25.
The results of the structural analysis not only corroborate the findings of biochemical and genetic experiments, but also illuminate the possible molecular mechanism of trapping of RF1 (and RF2) on the terminating ribosome after the release of the nascent peptide.
Api137-mediated RF depletion inhibits peptide release and stimulates stop codon read-through
The number of ribosomes in the bacterial (E. coli) cell exceeds the number of RF2 and RF1 molecules by ~25-fold and ~200-fold, respectively2,3. Api137-mediated trapping of RF1 (or RF2) on a relatively small number of ribosomes should lead to a rapid depletion of the RFs. As a consequence, there would be no RF1 (or RF2) available to facilitate the peptide release when the remaining translating ribosomes reach a stop codon. Therefore, even though Api137 arrests the ribosome in a post-hydrolysis state, in the cells treated with Api137, most of the ribosomes should stall at stop codons in a pre-hydrolysis state carrying an intact peptidyl-tRNA.
We first tested this hypothesis in a cell-free translation system using the TnaC stalling peptide as a model. At high tryptophan concentrations (5 mM) the RF2-mediated release of TnaC peptide is impeded, leading to a well-documented accumulation of TnaC-tRNA26 (Fig. 5a). By contrast, at low concentrations of tryptophan (0.3 mM), the TnaC peptide is rapidly released at the RF2-specific UGA stop codon. Strikingly, when Api137 was present, TnaC-tRNA also accumulated at low tryptophan concentrations. A similar result was obtained with the tnaC template carrying a RF1-specific UAG stop codon (Fig. 5a). These results demonstrated that as a consequence of RF1 or RF2 depletion due to Api137-mediated trapping on a fraction of ribosomes, the majority of ribosomes are unable to release the TnaC peptide. Consistent with this conclusion, the Api137-induced accumulation of TnaC-tRNA was largely rescued by supplementing the reaction with a 5-fold molar excess of RF1 over the ribosomes (Fig. 5b).
Figure. 5. Api137 induces accumulation of peptidyl-tRNA and stop codon read-through.

a, Gel electrophoresis analysis of the [35S]-labeled products of the in vitro translation of the tnaC gene with its original UGA stop codon (lanes 1–6) or with the UAG stop codon (lanes 7–9), in the absence or presence of Api137. Where indicated, reaction products were treated with RNase. Lane 1, control reaction without mRNA. Lane 2 (labeled as M, marker) shows the band of RNase-sensitive TnaC-tRNA accumulated at high concentration of tryptophan. Lanes 5 and 8 show Api137-induced accumulation of TnaC-tRNA at low concentration of tryptophan. The bands of TnaC-tRNA and of the released TnaC peptide are shown with filled and open arrowheads, respectively. This gel is representative of five independent biological replicates. b, Excess of RF1 rescues Api137-induced accumulation of peptidyl-tRNA. Cell-free translation with low tryptophan was carried out in the standard conditions (lanes 1 and 3) or in the presence of a 5-fold molar excess of RF1 over the ribosomes (lane 2). This gel is representative of two independent biological replicates. c, In vivo expression of the chromosomal mutant lacZ with a premature stop codon mediated by the stop codon read-through stimulated by the miscoding antibiotic streptomycin (Str) or by Api137. The droplet of read-through inducing agents was placed at the indicated points on the lawn of the E. coli cells grown on an LB/agar plate supplemented with ampicillin, IPTG and X-Gal. This plate represents one of three independent experiments. d, e, The dual mode of Api137 action. d, Api137 binds to the ribosome after RF1 (or RF2) catalyzes the release of the complete protein and traps RF1 (or RF2) thereby preventing its turnover. e, Trapping of RF1 (or RF2) depletes their available pool causing stalling of most of the ribosomes at the stop codons unable to release the nascent proteins.
When the translating ribosome reaches a stop codon, the occasional binding of a near-cognate aminoacyl-tRNA instead of the RFs may promote a stop codon read-through event. The Api137-induced depletion of the pools of free RF1 and RF2 is expected to bias this competition in favor of aminoacyl-tRNA binding. Indeed, Api137 dramatically increased the read-through frequency in a reporter E. coli strain carrying a mutant lacZ allele with a premature UAG stop codon (Fig. 5c). Noteworthy, the efficiency of Api137-induced read-through was significantly higher than that induced by the miscoding antibiotic streptomycin (Fig. 5c). These results confirm that while Api137 traps RF1 and RF2 on the ribosome after the nascent protein release, the main downstream effect of Api137 action is the arrest of the ribosomes in the pre-hydrolysis state (Fig. 5d, e).
Discussion
Our biochemical, genetic and structural data reveal Api137 as the first known inhibitor that is specific for translation termination. While several inhibitors can potentially interfere with polypeptide release27,28, these antibiotics also target other steps of protein synthesis; in these cases, inhibition of termination is just a collateral effect of the antibiotic binding to the ribosomal centers critical for various ribosomal activities. In contrast, Api137 does not inhibit initiation or elongation of translation, but specifically arrests the ribosome at the stop codons. Api137 achieves its inhibitory action in two related, but functionally distinct ways. The primary effect of Api137 is to trap RF1 and RF2 on the ribosomes after the release of the nascent peptide (Fig 5d). This leads to depletion of the free RF pool and as a result, the majority of cellular ribosomes are arrested at the stop codons in the pre-hydrolysis state (Fig 5e). The arrested ribosome may additionally block other ribosomes on the same ORF from completing translation. Thus, treatment of cells with Api137 results in formation of two populations of ribosomes stalled at the stop codons: a small fraction is arrested in a post-hydrolysis state, whereas the majority carries unhydrolyzed peptidyl-tRNA.
Although Api137 belongs to the broad group of ribosome-targeting PrAMPs, its mode of binding is fundamentally different from those of the previously studied derivatives of oncocin, bactenecin, pyrrhocoricin and metalnikowin 10–14. While the binding sites of all PrAMPs overlap, the orientation of Api137 is opposite to that observed for other PrAMPs. Furthermore, the N-terminus of other PrAMPs encroaches upon the A site of the PTC, completely blocking it and hindering binding of any A site substrates 10–13, whereas Api137 binds entirely within the exit tunnel. Therefore, the binding of RF1 or RF2 to the A site is incompatible with the placement of oncocin and similar PrAMPs, whereas Api137 actually requires RF1 or RF2 for efficient binding.
Due to the spatial constraints of the tunnel, direct binding of Api137 promoted by its interactions with RF1 or RF2 is likely to occur only after the peptidyl-tRNA ester bond has been hydrolyzed and the newly synthesized protein has vacated the ribosome. Therefore, apidaecins have a rather narrow time window to exert their inhibitory action, namely, after departure of the newly made protein, but prior to RF1 or RF2 dissociation. Within this window, Api137 has to traverse the entire length of the exit tunnel to reach its binding site close to the PTC where it can establish interactions with the RF. Thus, Api137-dependent trapping of RF1 and RF2 is probably a fairly rare event in the context of the global cellular translation. However, the resulting complex is long-lived (Figure 2D), and the majority of RF1 and RF2 molecules will be eventually sequestered.
Recycling of RF1 and RF2 in the cell is facilitated by RF3, but RF3 does not prevent trapping of RF1 or RF2 by Api137 in vitro (Fig. 2f). Nevertheless, MIC testing shows that cells lacking RF3 are 8 times more sensitive to Api137 than those expressing RF3 (Supplementary Table 1). This suggests that RF3 can partly mitigate the Api137 effect, probably by speeding up RF1 or RF2 dissociation prior to Api137 binding or stimulating the dissociation of the already trapped factors.
Because of the unique mechanism of its action, Api137 and its analogs could serve as important tools for research and medicine. Api137 could find an application in synthetic biology where interference with peptide release at engineered stop codons could stimulate the incorporation of non-canonical amino acids via stop codon suppression29. The use of Api137 for medicine could go far beyond its known antibacterial action. Many human genetic disorders are caused by nonsense mutations. Although enabling premature stop codon read-through by using translation error-inducing compounds is one of the promising strategies, the decrease in translational accuracy makes such drugs highly toxic30. The ability of Api137 to dramatically stimulate read-through by interfering with the function of RFs provides new avenues for exploring this approach31 and our high resolution structure of Api137 complexed with the bacterial ribosome can serve as a starting point for the rational design of specific inhibitors of eukaryotic translation termination.
On-line Methods
Peptides and oligonucleotides
Api137 was synthesized by NovoPro Biosciences Inc. Onc112 was synthesized by GenScript. The ‘start-stop’ mRNA (Supplementary Table 2) was purchased from IBA GmbH. The 2XermCL_S10_UAG construct was synthesized by Eurofins. DNA oligonucleotides were synthesized by Integrated DNA Technologies.
Generation of templates for in vitro translation and toeprinting
The DNA templates for toeprinting (Supplementary Table 2) were generated by PCR using AccuPrime DNA Polymerase (Thermo Fisher Scientific) and primers listed in Supplementary Table 3. The synthetic template yrbA-fs15 was prepared using 3 overlapping primers (T7-IR-AUG, IR-yrbA-fs15-RF1 and posT-NV1) in a single PCR reaction. The ermCL template was created by PCR amplification of the gene from the plasmid pERMCT7-M32 using primers T7 and ermCL-UAG. The complete sequences of the templates are shown in Supplementary Table 2.
Toeprinting reactions were carried out in 5 μl of PURExpress transcription-translation system (New England Biolabs) as previously described 32,33. The reverse transcription on the ermCL template was carried out using primer ermCL-TP-term. The final concentrations of Api137 and Onc112 in the reactions were 50 μM; the PrAMPs were added as stock solutions in water.
Selection of Api137-resistant mutants
The first round of selection of Api137 resistant mutants was performed with the E. coli strain SQ110, derived from the K12 strain (Supplementary Table 4). An overnight culture grown in Luria-Bertani (LB) medium was diluted 100-fold into fresh medium containing subinhibitory concentration of Api137 (10 μM). After 24 h growth at 37°C, the culture was diluted 100-fold into 1 ml fresh LB medium containing 50 μM Api137. The culture was passaged one more time at 100 μM Api137 (8-fold MIC). The dilutions of cell culture were plated on LB agar. After overnight incubation, the sbmA gene was PCR amplified from 20 individual colonies using primers SbmA-seq-fwd and SbmA-seq-rev and sequenced. All but one clone had mutations in the sbmA gene. The Api137-resistant clone with the wt sbmA sequence (clone SQ110 ApiR21 in Supplementary Table 4) was grown in liquid culture; genomic DNA was isolated and prepared for sequencing using a Nextera XT kit (Illumina). Sequencing was performed on an Illumina NextSeq500 instrument (paired-end, 2×150 base reads) at the DNA Services facility at UIC. After mapping the reads to the genome of the strain SQ11034, the single mutation A722G in the prfA gene was identified. The presence of the mutation was verified by PCR-amplification of the prfA gene using primers PrfA-seq-fwd and PrfA-seq-rev from the parent and mutant strains and sequencing.
E. coli strain BL21(DE3) (Supplementary Table 4) was used in the second selection experiment. In order to avoid selection of sbmA mutants, prior to selection cells were transformed with the multicopy plasmid pZα-SbmA encoding the functional SbmA transporter. The pZα-SbmA plasmid was prepared by amplifying the E. coli sbmA gene using primers SbmA-seq-fwd and SbmA-EcoRI-rev, cutting the PCR product with restriction enzymes NdeI and EcoRI, and ligating the resulting DNA fragment into the pZα plasmid35 cut with the same enzymes. For selection of Api137-resistant mutants, the overnight culture of BL21(DE3)/pZα-SbmA cells was diluted 1:100 in LB medium containing ampicillin (100 μg/ml) and 0.1 μM isopropyl-b-D-1-thiogalactopyranoside (IPTG) and grown at 37°C until reaching A600 of 0.5. Two ml (approximately 109 cells) were plated on LB agar supplemented with 100 μg/ml ampicillin, 0.1 μM IPTG and 12 μM (4-fold MIC) Api137. After overnight incubation at 37°C, 10 colonies appeared. The prfA, prfB and prfC genes were PCR amplified using pairs of primers PrfA-seq-fwd with PrfA-seq-rev, PrfB-seq-fwd and PrfB-seq-rev, or PrfC-seq-fwd with PrfC-seq-rev, respectively, and sequenced. Five clones had mutations in the prfB gene: three of these had the C784T and two clones had the A839T mutation. The genome of one of the remaining five clones was sequenced and revealed the presence of the G241A mutation in the rplP gene encoding ribosomal protein uL16. The presence of this mutation in this and four remaining clones was verified by PCR-amplification of the rplP gene using primers RplP-seq-fwd and RplP-seq-rev and sequencing.
The minimal inhibitory concentrations (MICs) of Api137 for the parental strains and selected resistant mutants were determined by microbroth dilution technique in 96-well plates. Specifically, exponentially growing cells were diluted to the final density A600 = 0.002, 100 μl of the culture were placed in the wells and after addition of Api137 plates were incubated overnight at 37°C. The minimal Api137 concentration preventing appearance of the visible cell density was recoded as MIC.
Preparation of PreHC for fast kinetics experiments
All experiments were performed in buffer A (50 mM Tris-HCl, pH 7.5, 70 mM MgCl2, 30 mM KCl, 7 mM MgCl2) at 37°C if not stated otherwise. Ribosomes from the E. coli strain MRE600, E. coli initiation factors IF1, IF2 and IF3, f[3H]Met-tRNAfMet and its fluorescein-labeled version f[3H]Met-tRNAfMet(Flu) were prepared as described36,37. PreHC was assembled on the synthetic ‘start-stop’ mRNA (Supplementary Table 2) and purified through sucrose cushion as described38. The extent of f[3H]Met-tRNAfMet binding was better than 95% as determined by nitrocellulose filter binding. The pellets of PreHC were resuspended in buffer A, flash-frozen in liquid nitrogen, and stored at −80°C.
Single-cysteine mutants RF1(S167C), RF1(S167C/D241G) and the K12-type RF2(A246T) variant were generated by site-directed mutagenesis of the corresponding plasmids. C-terminally 6xHis-tagged RF1 and RF2 were purified and in vitro methylated by PrmC according to the published protocol22. RF3 was purified as described38.
Peptide hydrolysis assay
f[3H]Met-tRNAfMet hydrolysis was monitored at single round conditions, by mixing [3H]-PreHC (0.1 μM), preincubated with 0–100 μM Api137, with RF1 (1 μM) in a quench-flow apparatus at 37°C. Reactions were quenched with a 10% trichloroacetic acid (TCA) solution in 50% ethanol. The extent of hydrolysis was assessed by liquid scintillation counting of the supernatants after centrifugation for 30 min at 16000 x g at 4°C. For measuring peptide release under multi-turnover conditions, [3H]-PreHC (0.1 μM) was preincubated with RF3 (0.1 μM), GTP (1 mM), pyruvate kinase (0.1 mg/ml), and phosphoenol pyruvate (3 mM) for 15 min at 37°C. The concentration of Api137, when present, was 1 μM. Time courses were started by addition of RF1 or RF2 (10 nM) and after quenching the reactions with a 10% TCA solution in 50% ethanol, the samples were processed as described above.
Preparation of quencher-labeled RF1Qsy
Prior to labeling, RF1s containing a single cysteine was incubated for 30 min at room temperature with a 10-fold molar excess of Tris(2-carboxyethyl)phosphine (TCEP, Sigma). The quencher dye QSY9 (Thermo Fisher) was dissolved in dimethyl sulfoxide (DMSO) and added to the RF1 solution at a 10-fold molar excess. Labeling reaction was incubated for 1 h at room temperature with vigorous shaking and stopped by addition of 2 mM dithiothreitol (DTT). The excess dye was removed by gel filtration on a PD10 column (GE Healthcare) and protein purity was checked by SDS-PAGE. The extent of RF1 labeling (as analyzed by absorbance) was greater than 80%.
Measuring kinetics of RF1 binding and dissociation
Rapid kinetics measurements were performed on an SX-20MV stopped-flow apparatus (Applied Photophysics, Leatherhead, UK). Experiments were performed by rapidly mixing equal volumes (60 μl) of f[3H]Met-tRNAfMet(Flu)-carrying PreHC (0.05 μM), preincubated with Api137 for 2 min at room temperature and RF1Qsy (0.15 μM) at 37°C. Fluorescein was excited at 470 nm and fluorescence emission was monitored after passing a KV500 filter (Schott). Time courses were evaluated by fitting using exponential functions by GraphPad Prism software. Dissociation rates (koff) were determined by chase experiments: PreHCflu (0.05 μM) was preincubated with 0.15 μM RF1Qsy to generate PostHCflu in the absence or presence of 1 μM Api137. PreHC was then rapidly mixed with a 10-fold excess of unlabeled RF1 and RF3·GTP (1 mM); pyruvate kinase (0.1 mg/ml), and phosphoenol pyruvate (3 mM) were present in both syringes. The increase of fluorescence upon dissociation of RF1Qsy was monitored as described above.
Chemical probing of Api137 interaction with the ribosome
PostHC was prepared by incubating 70S ribosomes (9 μM) with tRNAfMet (18 μM) and start-stop mRNA (18 μM) at 37°C for 30 min in buffer A containing 20 mM MgCl2. PostHC (0.2 μM) was incubated in 50 μl of reaction buffer B (250 mM K-Borate, 50 mM MgCl2, 500 mM NH4Cl) with RF1 (1 μM) and/or Api137 (50 μM) at 37°C for 10 min. Modification with dimethylsulfate (Sigma-Aldrich) and quenching were carried out at 37°C for 10 min as described39. rRNA was isolated by phenol extraction and the distribution of modifications was analyzed by primer extension using primers L2667 and L2180.
Cell-free translation and analysis of peptidyl-tRNA accumulation
To prepare the templates for translation in the E. coli S30 Extract System for Linear Templates (Promega), the tnaC gene was first amplified by PCR from genomic DNA of E. coli MG1655 using primer Ptrc-tnaC-2 in combination with either tnaC-UGA-rev or tnaC-UAG-rev. These PCR fragments were cloned into the SmaI site of pUC18 and the tnaC template was re-amplified with primers Ptrc-eCLi and rev-44.
The transcription–translation reactions were carried out in a total volume of 5 μl. The reactions contained 0.5 pmol of the tnaC DNA template, 2 μCi [35S]-L-methionine (specific activity 1,175 Ci/mmol, MP Biomedicals). When needed, the reactions were supplemented with 50 μM of Api137 or 5 mM tryptophan, or 3.7 μM of purified RF1. The reactions were incubated at 37°C for 30 min and then, when needed, split in two aliquots, one of which was treated for 5 min at 37°C with 0.5 μg RNase A (Sigma-Aldrich). The translation products were precipitated with four volumes of cold acetone and resolved in a 16.5% Tris-Tricine gels that preserve the integrity of peptidyl-tRNA40. Gels were dried, exposed to the phosphoimager screen and scanned on a Typhoon scanner (GE).
In vivo suppression of premature stop codon
The E. coli strain with a premature stop codon in the lacZ gene was generated by subjecting the SQ171-ΔtolC strain (Supplementary Table 4) to chemical mutagenesis and selecting lacZ deficient mutants. For that, an overnight culture of SQ171-ΔtolC was diluted 1:200 into fresh LB medium supplemented with kanamycin (30 μg/ml), grown at 37°C until reaching A600 of 0.1 and then exposed to 0.1% of ethyl methanesulfonate (EMS) for 1 hr. Cells were washed twice with LB medium and plated at high density on LB-agar supplemented with kanamycin (50 μg/ml), X-gal (40 μg/ml), and IPTG (0.3 mM). White colonies were selected and re-streaked on fresh kanamycin (50 μg/ml), X-gal (40 μg/ml), and IPTG (0.3 mM) LB-agar plates. The presence of mutations was detected by PCR amplification of the lacZ gene and sequencing. The clone designated SQ171-ΔtolC/W3 (Supplementary Table 4) contained the C2035T mutation, which changed Gln679 of the encoded β-galactosidase to a UAG stop codon.
For testing the stop codon suppressing activity of Api137, SQ171-ΔtolC/W3 cells were grown in LB medium supplemented with 50 μg/ml of kanamycin. Upon reaching A600 of 1.0, 0.5 ml were mixed with 3.5 mL of LB agar (0.6%) kept at 50°C and poured on an LB-agar plate containing kanamycin (50 μg/ml), IPTG (0.2 mM) and X-gal (80 μg/ml). After solidification of the soft agar, 1 μl of 50 mg/ml solution of streptomycin (100 μg) or 1 μl of 2 mM solution of Api137 (4.6 μg) were spotted on top of the cell lawn. The plate was incubated overnight at 37°C. Stop-codon read-through activity was revealed by a blue halo around the spotted antibiotic.
Purification of RF1 for cryo-EM
N-terminally 6xHis-tagged E. coli RF1 was overexpressed in BL21 E. coli cells grown at 37°C from overnight culture in LB medium and in presence of 100 μg/mL Ampicillin. Protein expression was induced at A600 of 0.4 by adding IPTG to a final concentration of 1 mM. RF1 was expressed from pET28-plasmid kindly provided by Rachel Green (John Hopkins University, Baltimore). After 1 h of expression, cells were lysed using a microfluidizer. The cell lysate was cleared by centrifugation in a SS34 rotor (Sorval) at 4°C and 44,100 x g for 30 min. Purification of His-tagged RF1 was done with Protino Ni-NTA agarose beads (Macherey-Nagel). The final eluate was applied onto a Superdex HiLoad S75 16/600 column (GE Healthcare) to yield the final concentrated protein in gel filtration buffer (50 mM HEPES pH 7.4, 50 mM KCl, 100 mM NaCl, 2% glycerol and 5 mM 2-mercaptoethanol).
Sample preparation for cryo-electron microscopy
ErmCL_S10_UAG-SRCs (stalled ribosome complexes) were generated following the same disome purification procedure as previously described 41,42. The 2XermCL_S10_UAG template was based on the 2XermCL_disome construct described by 41 except that Ser10 codon was replaced by a UAG stop codon (Supplementary Table 2).
In vitro translation of the 2XermCL_S10_UAG template was performed using the Rapid Translation System RTS100 E. coli HY Kit (5PRIME) in the presence of 50 μM Api137. Disomes were isolated using sucrose density gradients (10–55% sucrose in buffer A, containing 50 mM HEPES-KOH, pH 7.4, 100 mM KOAc, 25 mM Mg(OAc)2, 6 mM 2-mercaptoethanol, 20 μM Api137 and one Complete EDTA-free Protease Inhibitor cocktail (Roche)) as previously described41,42. The final purified complex was re-incubated with a 2.5-fold excess of RF1 and 50 μM Api137 for 15 min at 37°C.
Cryo-electron microscopy and single particle reconstruction
A total of 5 A260/ml Api137-RF1 complex was applied to 2 nm pre-coated Quantifoil R3/3 holey carbon supported grids and vitrified using a Vitrobot Mark IV (FEI, Eindhoven). Data collection was performed using an FEI Titan Krios transmission electron microscope equipped with a Falcon II direct electron detector with a Falcon III chip (FEI, Eindhoven) at 300 kV using a pixel size of 1.084 Å and a defocus range of 0.7–2.5 μm. The data collection yielded a total number of 5132 micrographs. Each micrograph was recorded as a series of ten frames (2.5 e−/Å2 dose per frame). All frames (accumulated dose of 28 e-/Å2) were aligned using the Motion correction software43 and power-spectra, defocus values, astigmatism and estimation of micrograph resolution were determined by CTFFIND4 44. Micrographs showing Thon rings beyond 3.2 Å resolution were further manually inspected for good areas and power-spectra quality. Automatic particle picking was performed using SIGNATURE45 and single particles were processed using the FREALIGN Software package46. Initial alignment was performed with 116,212 particles using E. coli 70S ribosome as a reference structure. Subsequently, particles were subjected to 3D-classification resulting in six classes with a maximum resolution extending to <3.4 Å (0.143 FSC) for class 1 (Supplementary Fig. 4a–c). 3D classification and initial alignment was performed using three times decimated data. The local resolution of the final maps was computed using ResMap47 (Supplementary Fig. 4e–g). The final maps were sharpened by dividing the maps by the modulation transfer function of the detector and by applying an automatically determined negative B-factor to the maps using RELION48.
Molecular modeling and map-docking procedures
The molecular model of the 70S ribosome was based on E. coli-70S-EF-Tu structure49. Release factor 1 was modelled based on the previously reported RF1 structure from (PDB ID 5J3C)24. The Ile-tRNA model was generated based on the P-site tRNA from50. The models were initially adjusted and refined using Coot51. Api137 was modeled de novo into the map using Coot. The complete atomic model of the E. coli ribosome was refined using phenix.real_space_refine52 with secondary structure restraints calculated by PHENIX52. Cross-validation against overfitting (Supplementary Fig. 4d) was performed as described elsewhere53. The statistics of the refined model were obtained using MolProbity54 and are presented in Table 1.
Figure preparation
Figures showing electron densities and atomic models were generated using either UCSF Chimera55 or PyMol Molecular Graphic Systems (version 1.8 Schrödinger).
Data availability
The cryo-EM density map of the Api137-RF1-ribosome complex has been deposited in the Electron Microscopy Data Bank under accession code EMD-3739. The corresponding molecular model has been deposited in the Protein Data Bank under accession code 5O2R. All other data are available from the corresponding author upon reasonable request.
Supplementary Material
Acknowledgments
We thank I. Lomakin and M. Gagnon for helping us initiate this project. We also thank them and Y. Polikanov for critical discussions, T. Perez-Morales for experimental advice, A. Kefi for help with analysis of genome sequencing data, A. Bursy, O. Geintzer, S. Kappler, C. Kothe, T. Niese, S. Rieder, H. Sieber, T. Wiles, and M. Zimmermann for expert technical assistance. This work was supported by grant R01 GM 106386 from the National Institutes of Health (to A.S.M and N.V.-L), iNEXT project 2259 (to D.N.W.), grants of the Forschergruppe FOR 1805 (to D.N.W., R.B. and M.V.R.) and a project grant in the framework of the Sonderforschungsbereich SFB860 (to M.V.R.) from the Deutsche Forschungsgemeinschaft (DFG).
Footnotes
Author contributions:
A.S.M./N.V-L. lab (T.F., D.K., N.V.-L, A.S.M.) carried out biochemical, microbiological and genetic experiments, M.V.R. lab (C.M, P.K., M.V.R.) performed kinetic analysis, D.N.W. lab (M.G., O.B., R.B., D.N.W.) carried out structural studies. N.V-L., A.S.M., M.V.R. and D.W. designed the study and oversaw the experiments. T.F., C.M. and M.G. designed and performed the experiments, and analyzed the data. P.K. and D.K. performed the experiments. N.V-L., A.S.M., M.V.R., D.N.W., T.F., C.M. and M.G. wrote the paper.
Competing financial interests:
The authors declare no competing financial interests.
References
- 1.Korostelev AA. Structural aspects of translation termination on the ribosome. RNA. 2011;17:1409–1421. doi: 10.1261/rna.2733411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bremer H, Dennis P. In: Escherichia coli and Salmonella: Cellular and Molecular Biology. Neidhardt FC, et al., editors. Vol. 2. ASM Press; 1996. pp. 1553–1569. Ch. 97. [Google Scholar]
- 3.Schmidt A, et al. The quantitative and condition-dependent Escherichia coli proteome. Nat Biotech. 2016;34:104–110. doi: 10.1038/nbt.3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Koutmou KS, McDonald ME, Brunelle JL, Green R. RF3:GTP promotes rapid dissociation of the class 1 termination factor. RNA. 2014;20:609–620. doi: 10.1261/rna.042523.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shi X, Joseph S. Mechanism of translation termination: RF1 dissociation follows dissociation of RF3 from the ribosome. Biochemistry. 2016;55:6344–6354. doi: 10.1021/acs.biochem.6b00921. [DOI] [PubMed] [Google Scholar]
- 6.Kaji A, et al. The fourth step of protein synthesis: disassembly of the posttermination complex is catalyzed by elongation factor G and ribosome recycling factor, a near-perfect mimic of tRNA. Cold Spring Harb Symp Quant Biol. 2001;66:515–529. doi: 10.1101/sqb.2001.66.515. [DOI] [PubMed] [Google Scholar]
- 7.Zasloff M. Antimicrobial peptides of multicellular organisms. Nature. 2002;415:389–395. doi: 10.1038/415389a. [DOI] [PubMed] [Google Scholar]
- 8.Scocchi M, Mardirossian M, Runti G, Benincasa M. Non-membrane permeabilizing modes of action of antimicrobial peptides on bacteria. Curr Top Med Chem. 2016;16:76–88. doi: 10.2174/1568026615666150703121009. [DOI] [PubMed] [Google Scholar]
- 9.Li W, et al. Proline-rich antimicrobial peptides: potential therapeutics against antibiotic-resistant bacteria. Amino acids. 2014;46:2287–2294. doi: 10.1007/s00726-014-1820-1. [DOI] [PubMed] [Google Scholar]
- 10.Seefeldt AC, et al. Structure of the mammalian antimicrobial peptide Bac7(1-16) bound within the exit tunnel of a bacterial ribosome. Nucleic Acids Res. 2016;44:2429–2438. doi: 10.1093/nar/gkv1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roy RN, Lomakin IB, Gagnon MG, Steitz TA. The mechanism of inhibition of protein synthesis by the proline-rich peptide oncocin. Nat Struct Mol Biol. 2015;22:466–469. doi: 10.1038/nsmb.3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seefeldt AC, et al. The proline-rich antimicrobial peptide Onc112 inhibits translation by blocking and destabilizing the initiation complex. Nat Struct Mol Biol. 2015;22:470–475. doi: 10.1038/nsmb.3034. [DOI] [PubMed] [Google Scholar]
- 13.Gagnon MG, et al. Structures of proline-rich peptides bound to the ribosome reveal a common mechanism of protein synthesis inhibition. Nucleic Acids Res. 2016;44:2439–2450. doi: 10.1093/nar/gkw018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krizsan A, Prahl C, Goldbach T, Knappe D, Hoffmann R. Short proline-rich antimicrobial peptides inhibit either the bacterial 70S ribosome or the assembly of its large 50S subunit. Chembiochem. 2015;16:2304–2308. doi: 10.1002/cbic.201500375. [DOI] [PubMed] [Google Scholar]
- 15.Castle M, Nazarian A, Yi SS, Tempst P. Lethal effects of apidaecin on Escherichia coli involve sequential molecular interactions with diverse targets. J Biol Chem. 1999;274:32555–32564. doi: 10.1074/jbc.274.46.32555. [DOI] [PubMed] [Google Scholar]
- 16.Krizsan A, et al. Insect-derived proline-rich antimicrobial peptides kill bacteria by inhibiting bacterial protein translation at the 70S ribosome. Angew Chem Int Ed Engl. 2014;53:12236–12239. doi: 10.1002/anie.201407145. [DOI] [PubMed] [Google Scholar]
- 17.Berthold N, et al. Novel apidaecin 1b analogs with superior serum stabilities for treatment of infections by gram-negative pathogens. Antimicrob Agents Chemother. 2013;57:402–409. doi: 10.1128/AAC.01923-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hartz D, McPheeters DS, Traut R, Gold L. Extension inhibition analysis of translation initiation complexes. Methods Enzymol. 1988;164:419–425. doi: 10.1016/s0076-6879(88)64058-4. [DOI] [PubMed] [Google Scholar]
- 19.Mattiuzzo M, et al. Role of the Escherichia coli SbmA in the antimicrobial activity of proline-rich peptides. Mol Microbiol. 2007;66:151–163. doi: 10.1111/j.1365-2958.2007.05903.x. [DOI] [PubMed] [Google Scholar]
- 20.Uno M, Ito K, Nakamura Y. Functional specificity of amino acid at position 246 in the tRNA mimicry domain of bacterial release factor 2. Biochimie. 1996;78:935–943. doi: 10.1016/s0300-9084(97)86715-6. [DOI] [PubMed] [Google Scholar]
- 21.Dreyfus M, Heurgue-Hamard V. Termination troubles in Escherichia coli K12. Mol Microbiol. 2011;79:288–291. doi: 10.1111/j.1365-2958.2010.07468.x. [DOI] [PubMed] [Google Scholar]
- 22.Kuhlenkoetter S, Wintermeyer W, Rodnina MV. Different substrate-dependent transition states in the active site of the ribosome. Nature. 2011;476:351–354. doi: 10.1038/nature10247. [DOI] [PubMed] [Google Scholar]
- 23.Korostelev A, Zhu J, Asahara H, Noller HF. Recognition of the amber UAG stop codon by release factor RF1. EMBO J. 2010;29:2577–2585. doi: 10.1038/emboj.2010.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pierson WE, et al. Uniformity of peptide release Is maintained by methylation of release factors. Cell Rep. 2016;17:11–18. doi: 10.1016/j.celrep.2016.08.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gao H, et al. RF3 induces ribosomal conformational changes responsible for dissociation of class I release factors. Cell. 2007;129:929–941. doi: 10.1016/j.cell.2007.03.050. [DOI] [PubMed] [Google Scholar]
- 26.Gong F, Yanofsky C. Instruction of translating ribosome by nascent peptide. Science. 2002;297:1864–1867. doi: 10.1126/science.1073997. [DOI] [PubMed] [Google Scholar]
- 27.Uehara Y, Hori M, Umezawa H. Specific inhibition of the termination process of protein synthesis by negamycin. Biochim Biophys Acta. 1976;442:251–262. doi: 10.1016/0005-2787(76)90495-0. [DOI] [PubMed] [Google Scholar]
- 28.Svidritskiy E, Ling C, Ermolenko DN, Korostelev AA. Blasticidin S inhibits translation by trapping deformed tRNA on the ribosome. Proc Natl Acad Sci USA. 2013;110:12283–12288. doi: 10.1073/pnas.1304922110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Des Soye BJ, Patel JR, Isaacs FJ, Jewett MC. Repurposing the translation apparatus for synthetic biology. Curr Opin Chem Biol. 2015;28:83–90. doi: 10.1016/j.cbpa.2015.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Keeling KM, Xue X, Gunn G, Bedwell DM. Therapeutics based on stop codon readthrough. Annu Rev Genomics Hum Genet. 2014;15:371–394. doi: 10.1146/annurev-genom-091212-153527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roy B, et al. Ataluren stimulates ribosomal selection of near-cognate tRNAs to promote nonsense suppression. Proc Natl Acad Sci USA. 2016;113:12508–12513. doi: 10.1073/pnas.1605336113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vazquez-Laslop N, Thum C, Mankin AS. Molecular mechanism of drug-dependent ribosome stalling. Mol Cell. 2008;30:190–202. doi: 10.1016/j.molcel.2008.02.026. [DOI] [PubMed] [Google Scholar]
- 33.Orelle C, et al. Identifying the targets of aminoacyl-tRNA synthetase inhibitors by primer extension inhibition. Nucleic Acids Res. 2013;41:e144. doi: 10.1093/nar/gkt526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Quan S, Skovgaard O, McLaughlin RE, Buurman ET, Squires CL. Markerless Escherichia coli rrn deletion strains for genetic determination of ribosomal binding sites. G3. 2015;5:2555–2557. doi: 10.1534/g3.115.022301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bailey M, Chettiath T, Mankin AS. Induction of ermC expression by ‘non-inducing’ antibiotics. Antimicrob Agents Chemother. 2008;52:866–874. doi: 10.1128/AAC.01266-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rodnina MV, Wintermeyer W. GTP consumption of elongation factor Tu during translation of heteropolymeric mRNAs. Proc Natl Acad Sci USA. 1995;92:1945–1949. doi: 10.1073/pnas.92.6.1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Milon P, et al. Transient kinetics, fluorescence, and FRET in studies of initiation of translation in bacteria. Methods Enzymol. 2007;430:1–30. doi: 10.1016/S0076-6879(07)30001-3. [DOI] [PubMed] [Google Scholar]
- 38.Peske F, Kuhlenkoetter S, Rodnina MV, Wintermeyer W. Timing of GTP binding and hydrolysis by translation termination factor RF3. Nucleic Acids Res. 2014;42:1812–1820. doi: 10.1093/nar/gkt1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Merryman C, Noller HF. In: RNA:Protein Interactions, A Practical Approach. Smith CWJ, editor. Oxford University Press; 1998. pp. 237–253. [Google Scholar]
- 40.Schägger H, von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 41.Arenz S, Nguyen F, Beckmann R, Wilson DN. Cryo-EM structure of the tetracycline resistance protein TetM in complex with a translating ribosome at 3.9-A resolution. Proc Natl Acad Sci USA. 2015;112:5401–5406. doi: 10.1073/pnas.1501775112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Arenz S, et al. Molecular basis for erythromycin-dependent ribosome stalling during translation of the ErmBL leader peptide. Nat Commun. 2014;5:3501. doi: 10.1038/ncomms4501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li X, et al. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat Methods. 2013;10:584–590. doi: 10.1038/nmeth.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rohou A, Grigorieff N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J Struct Biol. 2015;192:216–221. doi: 10.1016/j.jsb.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen JZ, Grigorieff N. SIGNATURE: a single-particle selection system for molecular electron microscopy. J Struct Biol. 2007;157:168–173. doi: 10.1016/j.jsb.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 46.Grigorieff N. FREALIGN: high-resolution refinement of single particle structures. J Struct Biol. 2007;157:117–125. doi: 10.1016/j.jsb.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 47.Kucukelbir A, Sigworth FJ, Tagare HD. Quantifying the local resolution of cryo-EM density maps. Nat Methods. 2014;11:63–65. doi: 10.1038/nmeth.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Scheres SH. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fischer N, et al. Structure of the E. coli ribosome-EF-Tu complex at <3 A resolution by Cs-corrected cryo-EM. Nature. 2015;520:567–570. doi: 10.1038/nature14275. [DOI] [PubMed] [Google Scholar]
- 50.Huter P, et al. Structural basis for ArfA-RF2-mediated translation termination on mRNAs lacking stop codons. Nature. 2016;541:546–549. doi: 10.1038/nature20821. [DOI] [PubMed] [Google Scholar]
- 51.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 52.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brown A, et al. Tools for macromolecular model building and refinement into electron cryo-microscopy reconstructions. Acta Crystallogr D Biol Crystallogr. 2015;71:136–153. doi: 10.1107/S1399004714021683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen VB, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pettersen EF, et al. UCSF Chimera–a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 56.Laurberg M, et al. Structural basis for translation termination on the 70S ribosome. Nature. 2008;454:852–857. doi: 10.1038/nature07115. [DOI] [PubMed] [Google Scholar]
- 57.Svidritskiy E, Korostelev AA. Ribosome Structure Reveals Preservation of Active Sites in the Presence of a P-Site Wobble Mismatch. Structure. 2015;23:2155–2161. doi: 10.1016/j.str.2015.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Polikanov YS, Steitz TA, Innis CA. A proton wire to couple aminoacyl-tRNA accommodation and peptide-bond formation on the ribosome. Nat Struct Mol Biol. 2014;21:787–793. doi: 10.1038/nsmb.2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sohmen D, et al. Structure of the Bacillus subtilis 70S ribosome reveals the basis for species-specific stalling. Nat Commun. 2015;6:6941. doi: 10.1038/ncomms7941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wood WB. Host specificity of DNA produced by Escherichia coli: bacterial mutations affecting the restriction and modification of DNA. J Mol Biol. 1966;16:118–133. doi: 10.1016/s0022-2836(66)80267-x. [DOI] [PubMed] [Google Scholar]
- 61.Studier FW, Moffatt BA. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986;189:113–30. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 62.Bouck N, Adelberg EA. Mechanism of action of nalidixic acid on conjugating bacteria. J Bacteriol. 1970;102:688–701. doi: 10.1128/jb.102.3.688-701.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wittmann HG, et al. Biochemical and genetic studies on two different types of erythromycin resistant mutants of Escherichia coli with altered ribosomal proteins. Mol Gen Genet. 1973;127:175–89. doi: 10.1007/BF00333665. [DOI] [PubMed] [Google Scholar]
- 64.Chittum HS, Champney WS. Ribosomal protein gene sequence changes in erythromycin-resistant mutants of Escherichia coli. J Bacteriol. 1994;176:6192–8. doi: 10.1128/jb.176.20.6192-6198.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kannan K, Vázquez-Laslop N, Mankin AS. Selective protein synthesis by ribosomes with a drug-obstructed exit tunnel. Cell. 2012;151:508–520. doi: 10.1016/j.cell.2012.09.018. [DOI] [PubMed] [Google Scholar]
- 66.Baba T, et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol. 2006;2(2006):0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The cryo-EM density map of the Api137-RF1-ribosome complex has been deposited in the Electron Microscopy Data Bank under accession code EMD-3739. The corresponding molecular model has been deposited in the Protein Data Bank under accession code 5O2R. All other data are available from the corresponding author upon reasonable request.


