Abstract
Tumor cells nearly invariably evolve sustained PI3K/Akt signaling as an effective means to circumvent apoptosis and maintain survival. However, for those tumor cells that do not acquire PI3K/Akt mutations to achieve this end, the underlying mechanisms have remained obscure. Here, we describe the discovery of a splice isoform–dependent positive feedback loop that is essential to sustain PI3K/Akt signaling in breast cancer. Splice isoform CD44s promoted expression of the hyaluronan synthase HAS2 by activating the Akt signaling cascade. The HAS2 product hyaluronan further stimulated CD44s-mediated Akt signaling, creating a feed-forward signaling circuit that promoted tumor cell survival. Mechanistically, we identified FOXO1 as a bona fide transcriptional repressor of HAS2. Akt-mediated phosphorylation of FOXO1 relieved its suppression of HAS2 transcription, with FOXO1 phosphorylation status maintained by operation of the positive feedback loop. In clinical specimens of breast cancer, we established that the expression of CD44s and HAS2 was positively correlated. Our results establish a positive feedback mechanism that sustains PI3K/Akt signaling in tumor cells, further illuminating the nearly universal role of this pathway in cancer cell survival.
Introduction
Resistance to cell death is a hallmark of cancer that tumor cells acquire to evade cancer therapies (1). This ability is commonly achieved through mutations of genes that result in sustained survival signals (2, 3). Independent of these mutations, however, cancer cells acquire the capacity to establish feedback circuits that allow for sustained survival signals (4–8). Therefore, disrupting these feedback-regulatory loops may provide an effective route to eradicate therapeutic resistance in cancer patients.
Aberrant activation of the PI3K/Akt signaling pathway drives tumor cell survival (9–11). Mutations that cause constitutive activation of PI3K/Akt are frequently found in advanced tumors and are associated with therapeutic resistance (9, 12, 13). However, many tumor cells displaying abnormal activity of PI3K/Akt do not harbor such mutations (9). The mechanisms by which these tumor cells sustain PI3K/Akt signaling were not fully understood.
Alternative splicing is an essential mechanism that generates protein diversity in mammals (14, 15). Emerging evidence suggested that dysregulation of alternative splicing plays a causal role in cancer progression (16, 17). One such example comes from our study of the cell surface protein and cancer stem cell marker CD44. Alternative splicing of CD44 gives rise to two groups of protein isoforms. The CD44v isoforms contain at least one of its nine variable exons, whereas the CD44s isoform is devoid of all variable exons. Work from our group and others showed that CD44s and CD44v are associated with distinct signaling activation. Although CD44v augments MAPK signaling and promotes cell proliferation (18–20), CD44s stimulates PI3K/Akt activation and renders tumor cells insensitive to drug-induced cell death (21, 22). We further showed that isoform switching from CD44v to CD44s is required for cells to undergo epithelial–mesenchymal transition (EMT; refs. 21, 23, 24), a developmental process that is abnormally activated in cancer therapeutic resistance (25, 26). These data suggest a functional role for CD44s in promoting therapeutic resistance by activating the PI3K/Akt signaling cascade.
In this study, we identify hyaluronan synthase 2 (HAS2) as a major downstream target of CD44s. We demonstrate a positive feedback loop that couples CD44s and HAS2, resulting in sustained Akt activation for cell death resistance in breast cancer cells. We found that FOXO1, a transcriptional repressor whose activity is inhibited by Akt-dependent phosphorylation, binds to the promoter of HAS2 and suppresses HAS2 transcription. CD44s-activated Akt signaling suppresses the transcriptional activity of FOXO1, causing de-repression of HAS2 transcription and thus an increase in HAS2 expression. The HAS2-synthesized product, hyaluronic acid (HA), is a ligand of CD44s. HA further augments CD44s-dependent Akt activation, resulting in formation of a positive feedback circuit. Disruption of this positive feedback loop inactivates Akt signaling and inhibits EMT and cancer cell survival. Hence, blocking this feed-forward circuit may serve as an effective strategy for the treatment of therapeutic-resistant breast cancers.
Materials and Methods
Cell lines and reagents
Human embryonic kidney 293FT cells, HT1080, MDA-MB-231, and LM2 cells were grown in DMEM supplemented with 10% FBS and 1% l-glutamine. Mes10A cell lines were derived from MCF10A cells as previously described (21) and used within eight passages. Stable cell lines were made to overexpress CD44s and CD44v3-10 cDNA or to silence CD44 by retro-viral or lentiviral infection. Stable cell lines were cultured in the media described above in the presence of the appropriate selectable drug markers. 293FT cells were obtained from Dr. Patrick Stern (MIT, Cambridge, MA, 2007). HT1080 cells were obtained from Dr. Bill Schnaper (Northwestern University, Boston, MA, 2012). MCF10A cells were obtained from Dr. Alexander Minella (Blood Center of Wisconsin, Milwaukee, WI, 2009). MDA-MB-231 and LM2 were obtained from Dr. Yibin Kang (Princeton University, Princeton, NJ, 2012). MDA-MB-231, LM2, and recurrent tumor cells were tested negative for the Mouse Essential CLEAR Panel conducted by Charles River Research Animal Diagnostic Services on November 13, 2014. All cells were expanded and stored in liquid nitrogen when received. Early passage vials (less than 10 passages) were thawed for the experiments described in this study. No further validation or authentication was performed. TrypLE (Gibco; 12605-010) was used for dissociating all aforementioned adherent cell lines before plating for assays. The PI3K inhibitor LY-294002 was obtained from Millipore (440202). The Akt inhibitor MK-2206 was obtained from Selleckchem (S1078). The FOXO1 inhibitor AS1842856 was obtained from Calbiochem (344355). Hyaluronidase Type I-S from bovine testes (Sigma-Aldrich H3506, 1 mg/mL stock concentration, which is equivalent to 400–1,000 U/mL) and HA sodium salt from rooster comb (Sigma-Aldrich H5388) were obtained from Sigma-Aldrich, and were freshly prepared before use. According to the manufacturer's report, the molecular weight of HA from rooster comb is expected to range from 1 to 4 million Da, which is considered high molecular weight HA. Cell culture plates were precoated with HA at either 0.1 mg/mL or 0.5 mg/mL in a tissue culture hood overnight. Plates were washed with cell culture medium to remove residual HA before plating cells. 4-methylumbelliferone (4-Mu) was obtained from Sigma-Aldrich (M1508) and used at a final concentration of 0.4 mmol/L in medium. Insulin was obtained from Sigma-Aldrich (I1882), and insulin stimulation was done using 10 μg/mL for 30 minutes unless otherwise specified. Maintenance of MCF10A and procedures for EMT induction were described previously (21).
Plasmids
The human CD44 shRNA that targets all CD44 isoforms and control luciferase shRNA were obtained from S. Godar, University of Cincinnati, Cincinnati, Ohio. Human CD44s and CD44v3-10 cDNA sequences were cloned into the EcoRI restriction site of pBabe-hygro vector. The HAS2 promoter region was PCR-amplified from genomic DNA isolated from human MDA-MB-231 cells (Qiagen Blood & Cell Culture Mini Kit). The HAS2 1.2-kb promoter region chosen for PCR amplification was based on a previously published study (27). PCR product was purified and digested with SacI and BgIII and cloned into the pGL3 luciferase vector. Ligation was carried out using the Roche Rapid DNA Ligation Kit according to the manufacturer's instructions. Sequences of primers used for HAS2 promoter cloning are as follows: Forward: 5′-CGCAATCTCCCAAGACCAAGTT-3′; Reverse: 5′-GAATTACCCAGTCCTGGCTTCG-3′. Expression plasmids pcDNA3-HA-myristoylated-Akt1, pcDNA3-Flag-FKHR (FOXO1), pECE-FLAG-FOXO3a, and pBR322-SV40-FLAG-FOXO4 were obtained from Addgene. pLX304 empty and pLX304-HAS2 (clone: HsCD00443505) plasmids were obtained from DNASU Plasmid Repository. The HAS2 shRNA–expressing plasmid (targeting sequence 5′-AATCCAGTGATAATCGCTTCG-3′) was kindly provided by Dr. Kounosuke Watabe at the Wake Forest School of Medicine, Winston-Salem, NC.
Luciferase assays
Luciferase reporter assays were performed using the Dual-Luciferase Reporter Assay System from Promega according to the manufacturer's instructions. Briefly, 293FT cells were plated in 24-well plates and transfected 24 hours after plating with Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Cells were cotransfected with the HAS2 promoter construct and transcription factors along with a Renilla luciferase construct serving as an internal control for transfection efficiency.
Quantitative RT-PCR
RNA was isolated from cells using E.Z.N.A. Total RNA Kit (Omega Bio-Tek). cDNA was generated by reverse transcription using GoScript RT (Promega). qRT-PCR was performed using GoTaq qPCR Master Mix (Promega), and mRNA levels were normalized to levels of TBP (TATA-binding protein). qRT-PCR primers are as follows: HAS2 (Forward: 5′-AGAGCACTGGGACGAAGTG-3′, Reverse: 5′-ATGCACTGAACACACCCAAA-3′); CD44 total (Forward: 5′-GATGGAGAAAGCTCTGAGCATC-3′, Reverse: 5′-TTGCTGCACAGATGGAGTTG-3′); CD44s (Forward: 5′-TACTGATGATGACGTGAGCA-3′, Reverse: 5′-GAATGTGTCTTGGTCTCTGGT-3′); CD44v5/6 (Forward: 5′-GTAGACAGAAATGGCACCAC-3′, Reverse: 5′-CAGCTGTCCCTGTTGTCGAA-3′); TBP (Forward: 5′GGAGAGTTCTGGGATTGTAC-3′, Reverse: 5′-CTTATCCTCATGATTACCGCAG-3′).
Antibodies
Antibodies used for Western blotting are as follows: phospho-Akt (4051S and 4056S), total Akt (9272S), and phospho-FOXO1 (9461S; Cell Signaling Technology), total FOXO1 (Santa Cruz Biotechnology; sc-11350), CD44 (R&D Systems; BBA10), HAS2 (Santa Cruz Biotechnology; sc-365263), GAPDH (Millipore; MAB374), N-cadherin (BD 610920), and occludin (Santa Cruz Biotechnology; sc-5562). Horseradish peroxidase–conjugated anti-mouse and anti-rabbit secondary antibodies were obtained from GE Healthcare. Blocking antibodies used for HA-CD44 binding experiments are as follows: Anti-human CD44 Clone 515 (BD550988), anti-mouse CD44 antibody KM81 (AB112178), anti-human CD44 (R&D Systems BBA10), and anti-human/mouse CD44 antibody IM7 (BioXcell BE0039).
Hyaluronic acid binding protein assay
Biotinylated hyaluronic acid binding protein (HABP) was obtained from EMD Millipore (385911). HABP was reconstituted in ddH2O upon arrival, aliquoted, and kept at −20°C until use. Cells were fixed with cold (−20°C) 100% methanol for 10 minutes followed by 1 × PBS wash 3 times. Endogenous biotin was blocked using endogenous Biotin-blocking kit (ThermoFisher E21390). Cells were then blocked with 5% BSA in 1 × PBS-0.1%Tween (PBS-T) for 20 minutes followed by addition of 1 μg/mL of HABP in 5% BSA in 1 × PBS-T overnight at 4°C The following day, cells were washed 3 times with 1 × PBS-T and then incubated with Alexa488-streptavdin and DAPI in 5% BSA in 1 × PBS-T for 1 hour before mounting on slides. Images were taken using Revolve microscope from Echo Laboratories (San Diego, CA).
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) assays were performed as previously described (23). Briefly, cells were crosslinked in 1% formaldehyde for 10 minutes at room temperature. Cross-linking was quenched with 0.125 mol/L glycine. Cells were collected by centrifugation at 1,000 rpm for 2 minutes and washed with 1×PBS. Nuclei were isolated by incubation in cell lysis buffer [10 mmol/L Tris, 10 mmol/L NaCl, 0.2% Nonidet P-40, 1 × protease inhibitor (Roche), pH 8.0] for 10 minutes on ice, followed by centrifugation at 2,500 rpm for 5 minutes. Nuclei were lysed in nuclei lysis buffer [50 mmol/L Tris, 10 mmol/L EDTA, 1% SDS, 1 × protease inhibitor (Roche), pH 8.1] for 10 minutes on ice. Lysates were sonicated using a Branson Sonifier 150 to obtain chromatin fragments with an average size of <500 bp. Sonicated chromatin was diluted with immunoprecipitation dilution buffer [20 mmol/L Tris, 2 mmol/L EDTA, 150 mmol/L NaCl, 1% Triton X-100, 0.01% SDS, 1 × protease inhibitor cocktail (Roche), pH 8.0] and precleared by the addition of Salmon sperm/DNA (Millipore). An aliquot of precleared chromatin was removed and served as “input” in the subsequent quantitative PCR analysis. The remaining chromatin was immunoprecipitated using 5 μg of FOXO1 (Santa Cruz Biotechnology; sc-11350) or rabbit IgG (The Jackson Laboratory) antibody prebound to protein-A beads overnight at 4°C. Beads were washed 7 times, and the bound proteins were eluted into 100 mmol/L NaHCO3 and 1% SDS. Samples were incubated overnight at 67°C to reverse cross-links and then incubated with Proteinase K (0.3 mg/mL) for 2 hours at 45°C to digest protein. DNA was purified using the QIAquick PCR Purification Kit (Qiagen). Quantitative PCR was performed using GoTaq qPCR Master Mix (Promega). Sequences of primers used are as follows: HAS2-promoter Forward: 5′-ACGAACTCTACCCAGCCTT-3′; HAS2-promoter Reverse: 5′-GCCATCTTGAAGGAAATGGCTT-3′. Intergenic control Forward: 5′-TGTGGATCGCTAGGAGTGGA-3′; Reverse: 5′TGACAAGGCTGCAGAGCAAT-3′.
Lactate dehydrogenase cell cytotoxicity assay
A total of 8,000 cells/well were plated in a 96-well cell culture plate. Twenty-four hours after plating, cells were treated with HAase for 6 hours followed by addition of 100 μmol/L cisplatin (Sigma-Aldrich) to induce apoptosis. Cisplatin-induced lactate dehydrogenase cytotoxicity in cells was analyzed 16 hours after cisplatin addition using the Cytotoxicity Detection Kit (LDH) (Roche) according to the manufacturer's instructions.
Mammosphere forming assay
Mes10A were grown in low attachment plates in serum-free DMEMF/12 supplemented with 2 ng/mL EGF, 2 ng/mL bFGF, 4 μg/mL heparin, 1% methylcellulose, and B27 supplement diluted 1:50. Cells were plated in each well of a low-attachment 96-well plate, and were fed every 3 or 4 days by addition of 50 μL of the aforementioned mammosphere media. Spheres were allowed to form for 10 to 14 days and then quantified under a microscope.
Statistical analysis
Statistical analyses, including two-tailed, paired Student t tests and Pearson correlation, were performed using GraphPad Prism software. P values < 0.05 were considered significant.
Results
HAS2 is a downstream target of CD44s
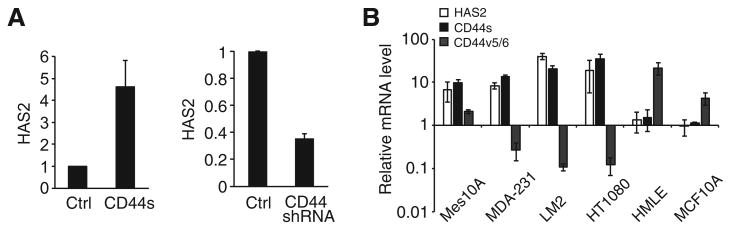
Because the CD44s splice isoform is functionally essential for EMT (21), a phenotype that promotes cancer cell survival and metastasis, we set out to investigate CD44s-mediated downstream pathways. We used Mes10A cells that were derived from MCF10A cells upon TGFβ treatment. CD44s expression in Mes10A cells promoted an EMT phenotype (21). Microarray analysis of control and CD44s-expressing Mes10A cells revealed that HAS2 is one of the most upregulated genes in response to expression of CD44s (Supplementary Fig. S1A). qRT-PCR validation showed that stable expression of CD44s resulted in a 3.5-fold induction in HAS2 expression (Fig. 1A). Conversely, shRNA knockdown of CD44 caused a 3-fold downregulation of HAS2 (Fig. 1A). The CD44-mediated increase in HAS2 expression is CD44s isoform specific, because stable expression of CD44v did not affect the levels of HAS2 (Supplementary Fig. S1B). Furthermore, examination of a number of cell lines showed that cells that expressed high levels of CD44s also expressed elevated levels of HAS2, whereas CD44v highly expressing cells exhibited lower levels of HAS2 expression (Fig. 1B).
Figure 1.
HAS2 expression is upregulated in response to CD44s overexpression. A, qRT-PCR results showing HAS2 mRNA levels in response to CD44s overexpression (left) and CD44 knockdown (right). B, qRT-PCR analysis showing levels of HAS2, CD44s, and CD44v that contain the v5- and v6- exons in breast cancer cell lines. Error bars, SD; n = 3.
CD44s and HAS2 positively correlate in clinical breast cancer samples
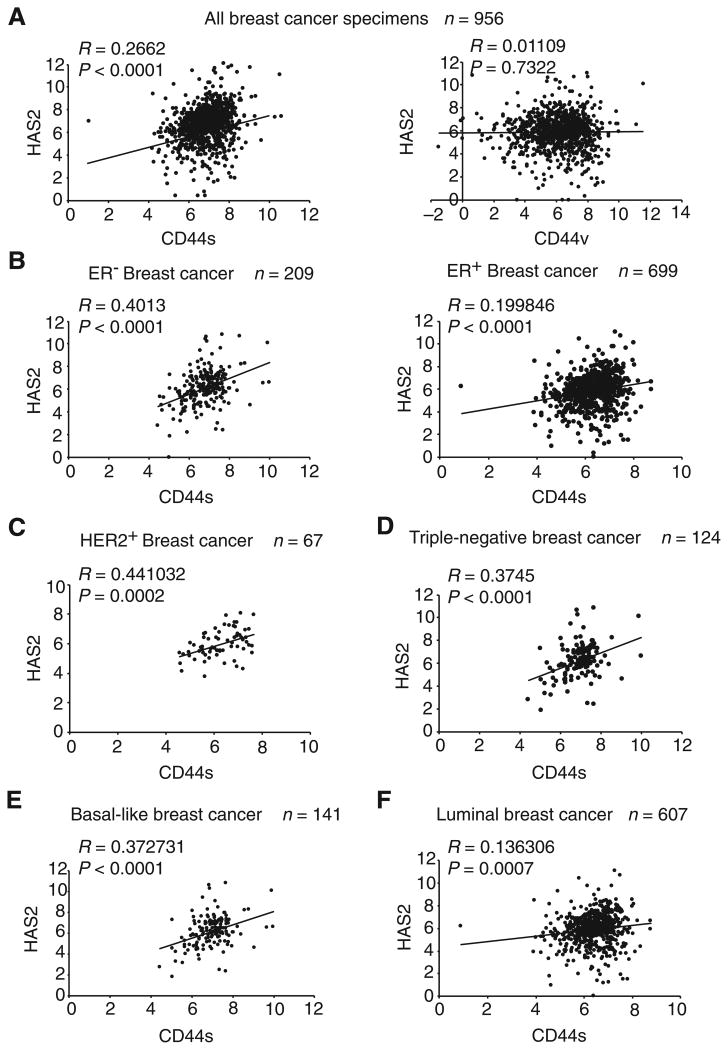
The above results indicate that CD44s, but not CD44v, promotes HAS2 expression in breast cancer cells. We next examined whether this relationship also exists in patient specimens of breast cancer. We analyzed the breast cancer the Cancer Genome Atlas (TCGA) database that includes RNA sequencing data of more than 1,000 patient breast tumor samples. We found a significant positive correlation between CD44s and HAS2 (Fig. 2A, left). By contrast, no significant correlation was found between CD44v and HAS2 (Fig. 2A, right). Close examination of different subtypes of breast cancers revealed that the expression levels of CD44s and HAS2 were strongly associated in estrogen receptor–negative (ER−) tumors (R = 0.40, P < 0.0001) than those in estrogen receptor–positive (ER+) tumors (Fig. 2B). This CD44s–HAS2-positive correlation was also seen in HER2+ tumors and in triple-negative tumors (Fig. 2C and D), suggesting that the positive correlation between CD44s and HAS2 exists in more aggressive breast cancer subtypes. In line with this observation, basal-like tumors exhibited stronger positive association between CD44s and HAS2 than that of luminal tumors (Fig. 2E and F). We also analyzed the relationships between CD44v and HAS2 in breast cancer subtypes and found no significant correlations (Supplementary Fig. S2). Together, these results demonstrate that the CD44s isoform, but not CD44v, positively correlates with HAS2 in clinical breast tumors.
Figure 2.
CD44s and HAS2 expression levels positively correlate in aggressive breast cancer subtypes from TCGA database. A, CD44s (left), but not CD44v (right), positively correlate with HAS2 in breast cancer specimens. B–F, Correlations of CD44s and HAS2 expression are shown in different subtypes of breast cancer.
To determine whether the CD44s–HAS2 relationship is a general phenomenon in cancers, we analyzed the TCGA database in two additional cancer types, lung cancer and pancreatic cancer. In agreement with results from the TCGA breast cancer analysis, CD44s levels significantly correlated with HAS2 expression (Supplementary Fig. S3), suggesting that the CD44s–HAS2 relationship exists as a general phenomenon in cancer cells.
HAS2 expression is elevated by CD44s-mediated Akt signaling
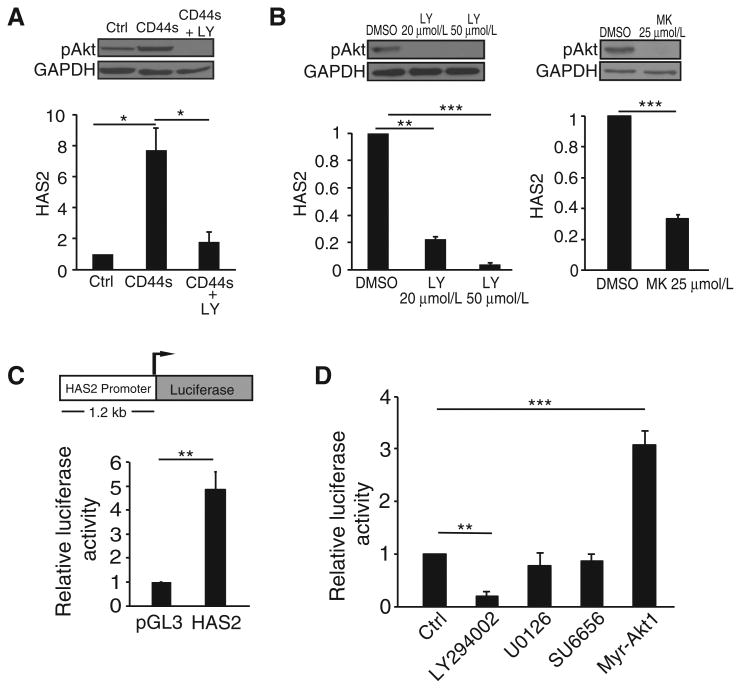
The above results prompted us to investigate the molecular mechanism by which CD44s stimulates HAS2 expression. Because CD44s is a cell surface protein that facilitates signaling activation, in particular the PI3K/Akt pathway (21), we examined whether CD44s-mediated Akt activation is required for HAS2 expression. As shown in Fig. 3A, overexpressing CD44s in Mes10A cells led to a significant increase in HAS2 expression. This increase was abolished when these cells were treated with a PI3K/Akt inhibitor, LY-294002. In addition, treatment of LM2 cells, which predominately express CD44s, with two different PI3K/Akt inhibitors, LY-294002 and MK-2206, also showed significant reductions on HAS2 expression (Fig. 3B), indicating that Akt signaling is required for HAS2 expression.
Figure 3.
CD44s-mediated HAS2 transcription is dependent on Akt signaling. A, qRT-PCR analysis showing that CD44s-dependent HAS2 upregulation is abolished by the PI3K/Akt inhibitor LY-294002 (LY; 20 μmol/L). Control and CD44s-overexpressing Mes10A cells were collected 20 hours after treatment. Immunoblot images on the top plot showing that LY-294002 effectively inhibited Akt activity. B, qRT-PCR analysis showing that inhibition of Akt signaling by LY-294002 (LY; 20 μmol/L and 50 μmol/L, left) or MK-2206 (MK; 25 μmol/L, right) suppressed expression of endogenous HAS2 in LM2 cells. Cells were treated for 20 hours. Immunoblot images on the top plot showing that LY-294002 and MK-2206 inhibited Akt activity in LM2 cells. C, Top, a schematic of the HAS2 promoter–driven luciferase reporter construct. Bottom, relative luciferase activity in 293FT cells transfected with the HAS2 luciferase reporter construct. D, Relative luciferase activity in 293FT cells transfected with the HAS2 luciferase reporter construct and treated with LY-294002 (20 μmol/L), U0126 (25 μmol/L), SU6656 (5 μmol/L), or cotransfected with Myr-Akt1. For each luciferase activity assay, ratios of firefly-to-Renilla luciferase activities were normalized to cells cotransfected with an empty vector control. Error bars, SD; n = 3. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
To determine whether CD44s-mediated Akt activation elevates HAS2 expression through transcriptional regulation, we cloned a 1.2-kilobase fragment of the HAS2 promoter that was previously shown to be active (27) in a luciferase reporter construct (Fig. 3C, top). Transfection of 293FT cells with the HAS2 promoter construct caused a 4-fold increase in luciferase activity compared with control (Fig. 3C, bottom), suggesting that this region represents a functional segment of the HAS2 promoter. Addition of the PI3K/Akt inhibitor LY-294002 abolished HAS2 promoter luciferase activity (Fig. 3D), and cotransfection of a constitutively activated Akt1 construct, myristoylated-Akt1, resulted in a 3-fold increase in luciferase activity (Fig. 3D). This HAS2 transcriptional regulation was specifically downstream of the PI3K/Akt pathway because MAPK and Src signaling inhibitors, U0126 and SU6656, respectively, showed no significant effects on luciferase activity (Fig. 3D). Together, these results demonstrate that HAS2 transcription is dependent on Akt activation.
HAS2/HA activates Akt signaling in a CD44s-dependent manner
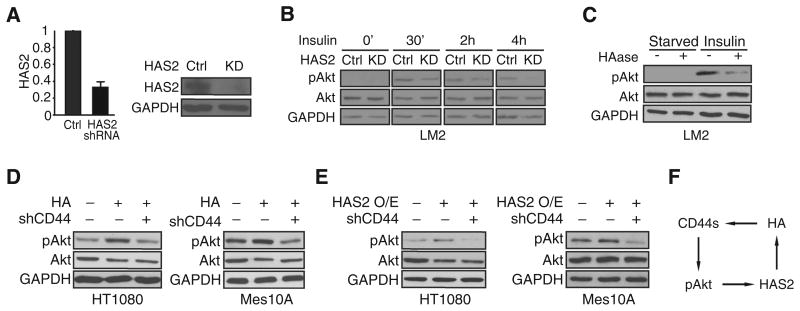
Previous studies have implicated HA in the PI3K/Akt signaling pathway (28, 29). Considering that the enzymatic activity of HAS2 is to produce HA, a ligand of CD44, we examined whether HAS2 promotes Akt activation and whether this involves CD44s. We treated cells with insulin, a growth factor that activates the PI3K/Akt pathway, to monitor Akt phosphorylation in response to HAS2 depletion. Knockdown of HAS2 (Fig. 4A) attenuated Akt activation at early time points, and this attenuation lasted for hours after insulin stimulation (Fig. 4B). Similarly, hyaluronidase (HAase) treatment to degrade HA (Supplementary Fig. S4A) also inhibited Akt phosphorylation, indicating that HAS2 and its HA product are required for Akt signaling (Fig. 4C). We then used control and CD44s-depleted cells (Supplementary Fig. S4B) and stimulated them with HA. We used high-molecular weight HA, as this is the native form produced by HAS2. HA addition augmented Akt phosphorylation in control cells, but this effect was abolished when CD44s was depleted, a phenotype that was consistently observed in three different cell lines (Fig. 4D and Supplementary Fig. S4C). In addition, HAS2 overexpression promoted Akt phosphorylation, and CD44s-depletion abolished this activity (Fig. 4E). These results reveal that HAS2, through its product HA, activates Akt signaling in a CD44s-dependent manner. These findings, together with the fact that CD44s-mediated Akt activation promotes HAS2 expression (Fig. 3), led us to postulate the existence of a positive feedback loop (Fig. 4F). In this model, CD44s promotes Akt signaling. Activated Akt stimulates HAS2 transcription, resulting in increased production of HA. HA further augments CD44s-dependent Akt activation, completing the loop and resulting in sustained Akt signaling.
Figure 4.
HAS2/HA promotes Akt activation in a CD44s-dependent manner. A, qRT-PCR and immunoblot analyses showing that HAS2 was silenced in HAS2 shRNA– expressing (KD) LM2 cells. Error bars, SD; n = 3. B, Immunoblot analysis of pAkt levels showing that HAS2 knockdown inhibits Akt activity. Control and HAS2 knockdown LM2 cells were stimulated with insulin (10 μg/mL) and collected at different time intervals. C, Immunoblot analysis of pAkt levels showing that HAase treatment impairs Akt phosphorylation. Control and HAase-treated (100 μg/mL, 6 hours) LM2 cells were stimulated with insulin (10 μg/mL) and collected 30 minutes after stimulation. D, Immunoblot analysis of pAkt levels showing that HA stimulates Akt activation in a CD44-dependent manner. HT1080 and Mes10A cells were grown on HA-coated plates (0.5 mg/mL HA for HT1080; 0.1 mg/mL HA for Mes10A). HT1080 and Mes10A cells were collected 24 hours after plating on HA-coated plates. E, Immunoblot analysis of pAkt levels showing that CD44 is required for HAS2-promoted Akt activation. Control and HAS2-overexpressing HT1080 cells were cultured in regular media for 24 hours after plating and collected for immunoblot analysis. Control and HAS2-overexpressing Mes10A cells were starved overnight and stimulated with insulin (10 μg/mL, 30 min) before lysate collection. F, A model of a positive feedback loop, involving CD44s and HAS2/HA, that sustains Akt activation.
HAS2/HA is required for CD44s-mediated Akt activation and autoregulates its mRNA expression
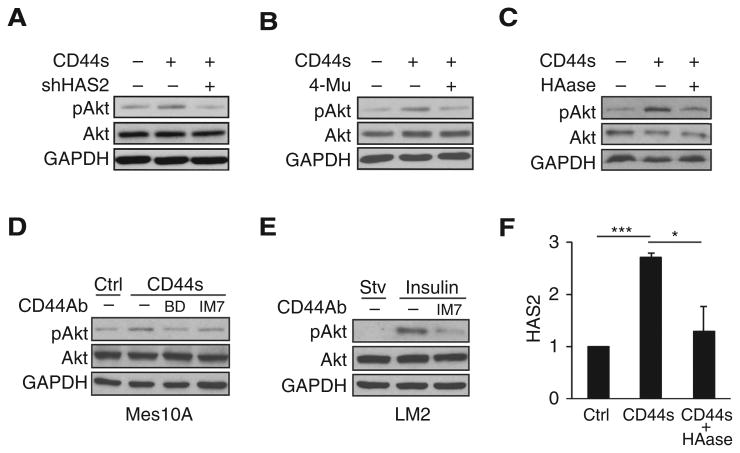
To directly examine the positive feedback model, we tested whether CD44s-stimulated Akt activation depends on HAS2/HA, downstream targets of Akt signaling. In agreement with previously published results (21), forced expression of CD44s in Mes10A cells (Supplementary Fig. S5A) resulted in elevated levels of Akt phosphorylation (Fig. 5A). Depletion of HAS2 in these cells markedly suppressed CD44s-stimulated Akt activation (Fig. 5A). Inhibiting HAS2 activity by a HAS2 inhibitor 4-methylumbelliferone (4-Mu) or depleting HA by HAase treatment (Supplementary Fig. S5B) also showed reduction of CD44s-dependent Akt signaling (Fig. 5B and C). These results indicate that CD44s-mediated Akt activation requires Akt downstream targets HAS2/HA. In support of this finding, addition of CD44 antibodies that block HA–CD44 interaction (30–33) resulted in inhibition of Akt activation, a phenotype that was observed in four different cell lines (Fig. 5D and E, Supplementary Fig. S5C–S5E).
Figure 5.
CD44-mediated Akt activation is dependent on HAS2/HA. A–C, Immunoblot analysis of pAkt levels showing that knockdown of HAS2 (A), treatment with HAS2 inhibitor 4-Mu (0.4 mmol/L; B), or treatment with HAase (35 μg/mL; C) abolished Akt activation in CD44s-overexpressing Mes10A cells. Cells were stimulated with insulin 10 μg/mL for 30 minutes. D, Immunoblot analysis showing that addition of CD44 antibodies abolishes CD44s-promoted Akt activation in CD44s-overexpressing Mes10A cells. BD clone 515 and IM7 antibodies were added at 1 μg/mL concentration in starvation media overnight. Cells were then washed with PBS and stimulated with insulin (10 μg/mL; 30 minutes). E, Immunoblot analysis showing that the CD44 antibody IM7 blocks Akt activation in LM2 cells. LM2 cells were starved overnight and then treated with IM7 antibody (10 μg/mL) for 3 hours, followed by insulin stimulation (10 μg/mL; 30 minutes). F, qRT-PCR analysis of HAS2 in control, CD44s-overexpressing, and HA-depleted CD44s-overexpressing Mes10A cells. Treatment of CD44s-overexpressing Mes10A cells with HAase (90 μg/mL) for 8 hours reduced CD44s-mediated HAS2 transcription. Error bars, SD; n = 3. *, P < 0.05; ***, P < 0.001.
If HAS2/HA are components of the positive feedback loop, the expression of HAS2 should depend on its product HA, because HA is required for CD44s to activate Akt signaling and thus to promote HAS2 expression. Accordingly, depletion of HA would be expected to disrupt the positive loop, causing repression of HAS2 expression. To test this hypothesis, we ectopically expressed CD44s in Mes10A cells to induce HAS2 expression (Fig. 5F). We then treated these cells with HAase to remove HA and measured endogenous HAS2 transcript levels by qRT-PCR. We observed a significant decrease in HAS2 expression to levels comparable with those in control cells (Fig. 5F). Collectively, these results support the existence of a positive feedback loop involving CD44s, HAS2, and Akt activation.
HAS2 transcription is suppressed by the Akt downstream target FOXO1
Our next goal was to determine the mechanism by which Akt signaling stimulates HAS2 transcription. In silico analysis of the HAS2 promoter revealed several putative transcription factor binding sites located within 2 kilobases upstream and 500 bases downstream of the HAS2 transcription start site (Fig. 6A). The consensus motifs recognized by the Forkhead family of proteins were particularly interesting because these sites show evolutionary conservation and because FOXOs are known targets of Akt signaling. In the Forkhead family, the FOXO subgroup contains FOXO1, FOXO3a, FOXO4, and FOXO6. FOXO1, FOXO3a, and FOXO4 are expressed to varying degrees in all tissues, and FOXO6 is predominantly expressed in the developing brain (34). Akt signaling was identified as a main regulator of FOXO's function (34). Phosphorylation of FOXOs by Akt causes their translocation to the cytoplasm, obviating FOXO's transcription activity (34).
Figure 6.
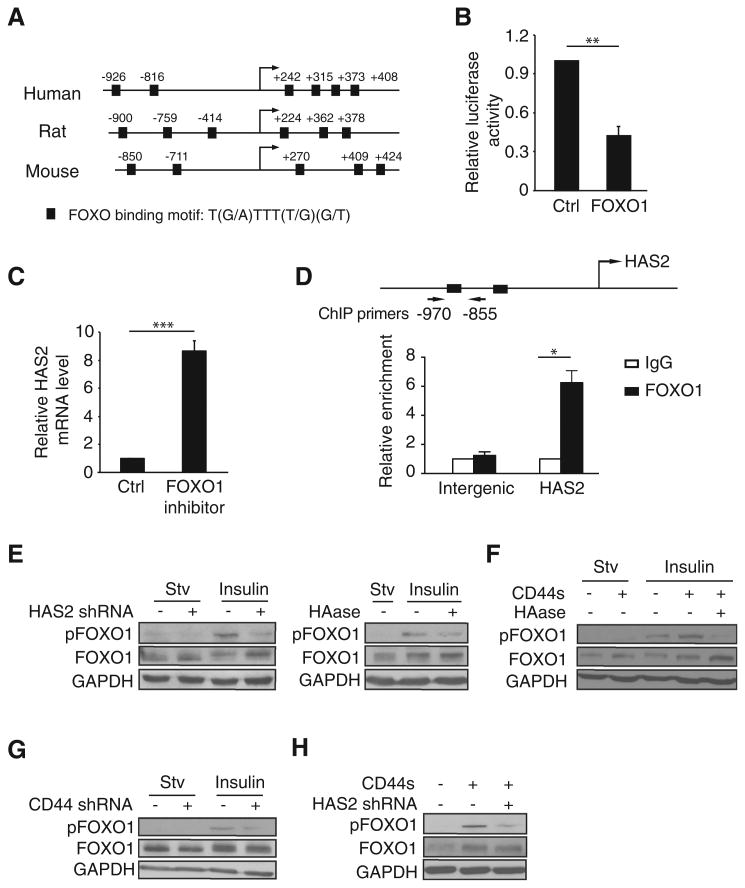
HAS2 transcription is repressed by FOXO1, whose activity is inhibited by CD44s and HAS2. A, Schematic of the HAS2 promoter showing conserved FOXO1 putative binding sites in human, rat, and mouse. Arrow, transcription start site. B, Relative luciferase activity in 293FT cells cotransfected with FOXO1 and the HAS2 promoter luciferase reporter constructs. C, qRT-PCR analysis of endogenous HAS2 levels in control and FOXO1 inhibitor-treated cells (AS1842856, 0.5 μmol/L). D, Quantitative ChIP analysis of the relative occupancy of FOXO1 at the HAS2 promoter in 293FT cells overexpressing FOXO1. As a negative control, enrichment at an amplicon located in an intergenic region was assayed. Bar graphs show averages of three independent ChIP experiments. E, Immunoblot analysis of pFOXO1 in HAS2-depleted (left) or HAase-treated (100 μg/mL) LM2 cells, followed by insulin stimulation (10 μg/mL; 30 minutes). F, Immunoblot analysis of pFOXO1 in Mes10A cells, CD44s-overexpressing Mes10A cells, and HAase-treated (35 μg/mL) CD44s-overexpressing Mes10A cells, followed by insulin stimulation (10 μg/mL; 30 minutes). G, Immunoblot analysis of pFOXO1 levels in control and CD44s-depleted Mes10A cells upon insulin stimulation (10 μg/mL; 30 minutes). H, Immunoblot analysis of pFOXO1 levels in control, CD44s-overexpressing, and CD44s-overexpressing and HAS2-depleted Mes10A cells, followed by insulin stimulation (10 μg/mL; 30 minutes). Error bars (B, C, and D), SD; n = 3. *, P < 0.05; ***, P < 0.001.
Cotransfection of individual FOXO1, FOXO3a, and FOXO4 with the HAS2 promoter luciferase reporter showed that forced expression of FOXO1 gave rise to the most drastic reduction of luciferase activity (Fig. 6B and Supplementary Fig. S6A). Furthermore, addition of a FOXO1 inhibitor, AS1842856, led to an increase in HAS2 level by approximately 8-fold (Fig. 6C). These results indicate that FOXO1 suppresses HAS2 transcription. To examine whether FOXO1 directly binds to the promoter of HAS2, we performed a ChIP assay and found that FOXO1 was enriched at the HAS2 promoter by 6.25-fold compared with a nonspecific control (Fig. 6D and Supplementary Fig. S6B). Thus, these results demonstrate that FOXO1 is a bona fide transcriptional repressor of HAS2.
We next investigated the relationship between FOXO1 and the positive feedback loop. We found that HAS2 and its product HA are required for FOXO1 phosphorylation. Depletion of HAS2 in LM2 cells resulted in a decrease in FOXO1 phosphorylation (Fig. 6E, left), a phenomenon that was also observed when treating these cells with HAase to remove the HAS2 product HA (Fig. 6E, right).
Similarly, ectopically expressing CD44s elevated the levels of FOXO1 phosphorylation (Fig. 6F), and knockdown of CD44 attenuated FOXO1 phosphorylation (Fig. 6G). Moreover, this CD44s-stimulated FOXO1 phosphorylation is dependent on HAS2 because treatment with HAase or knockdown of HAS2 both abolished the CD44s-mediated FOXO1 phosphorylation (Fig. 6F and H). Collectively, these results provide additional evidence that supports the existence of the positive feedback loop in which HAS2, the downstream target repressed by FOXO1, stimulates FOXO1 phosphorylation, thus abolishing FOXO1 transcription activity.
HAS2 knockdown reverses the mesenchymal phenotype and inhibits CD44s-mediated cell survival
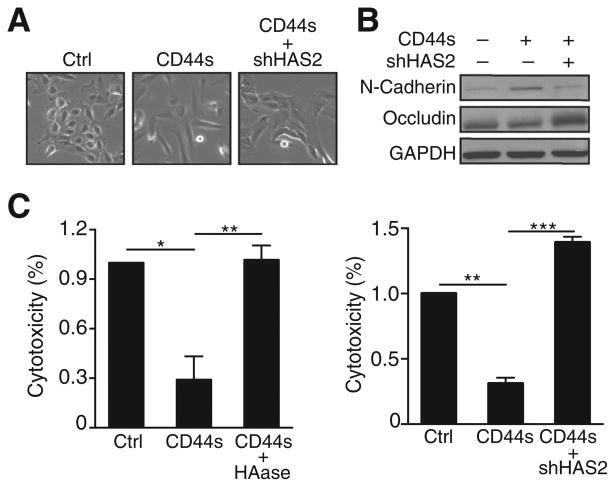
One of the key features of metastasis is that tumor cells exhibit survival advantages, which can be imparted by a mesenchymal phenotype that is dependent on Akt activation (21, 35, 36). Thus, the positive feedback loop presented here could serve as a mechanism to sustain Akt signaling, thus promoting EMT. To test this hypothesis, we examined whether disruption of the positive feedback loop causes reversal of an EMT phenotype. Treatment with TGFβ in MCF10A cells results in a partial EMT, and ectopically expressing CD44s further promotes EMT (21). We therefore examined whether disrupting the positive feedback loop by knocking down HAS2 would at least partially reverse EMT in CD44s-expressing Mes10A cells. As predicted, HAS2-depleted cells (Supplementary Fig. S7A) showed a greater tendency to form cell clusters, resembling the morphology in control cells (Fig. 7A). Western blot analysis revealed downregulation of the mesenchymal marker N-cadherin and upregulation of the epithelial marker occludin in HAS2-depleted cells (Fig. 7B), suggesting a role of this positive feedback loop in promoting an EMT phenotype.
Figure 7.
Depletion of HAS2 abolishes the effect of CD44s on promoting the mesenchymal phenotype and cell survival. A, Representative images of Mes10A, CD44s-overexpressing Mes10A, CD44s-overexpressing, and HAS2 knocked down Mes10A cells. B, Immunoblot analysis of EMT markers showing that HAS2 knockdown in CD44s-overexpressing Mes10A cells inhibited N-cadherin expression and enhanced occludin expression. C, Cytotoxicity analysis showing that HAase treatment (90 μg/mL) or depleting HAS2 by shRNA abolished CD44s-mediated cell death resistance in cisplatin (100 μmol/L)-treated CD44s-overexpressing Mes10A cells. Data are averages of three independent experiments. All error bars indicate SEM; n = 3. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Sustained PI3K/Akt activation is necessary to maintain cancer cell survival. To examine the contribution of the positive feedback loop to cell survival, we expressed CD44s cDNA in Mes10A cells and observed an inhibition of cisplatin-induced cytotoxicity (Fig. 7C). We then depleted HA by treating the CD44s-expressing Mes10A cells with HAase or silencing HAS2 by shRNA and found that both HA depletion and HAS2 knockdown greatly sensitized the CD44s-expressing cells to cisplatin-induced cell death (Fig. 7C), supporting the interplay between CD44s and HA in cell survival. Moreover, forced expression of CD44s enhanced mammosphere-forming potential, and this ability was significantly abolished when cells were treated with the HAS2 inhibitor 4-Mu (Supplementary Fig. S7B, left) or when HAS2 was knocked down (Supplementary Fig. S7B, right). These results reveal the connection between the positive feedback loop and a mesenchymal phenotype that confers a survival advantage.
Discussion
The present study demonstrates that the splice isoform CD44s promotes HAS2 transcription through activating Akt signaling. HAS2 further stimulates Akt activation in a CD44s-dependent manner, thus forming a positive regulatory circuit that sustains Akt signaling. We identify a mechanism by which Akt signaling stimulates HAS2 transcription by phosphorylating and inactivating FOXO1, a transcription repressor that binds to the HAS2 promoter and suppresses HAS2 transcription. Functionally, we describe that this positive feedback circuit is necessary for promoting EMT and cell death resistance. Our results also revealed that the levels of CD44s and HAS2 are most significantly correlated in basal-like and triple-negative breast tumor specimens, suggesting an important role of this feedback loop in advanced breast tumors. Thus, disruption of this positive feedback loop may offer an effective strategy for breast cancer therapeutic intervention.
Aberrant expression of HAS2 in tumor cells results in accumulation of HA in tumor-adjacent extracellular matrix, leading to breast cancer invasiveness and bone metastasis (37–40). In this regard, HAS2 enhances the interaction between tumor cells and tumor-associated macrophages. This communication is mediated by the HA–CD44 interaction, where HA is produced by HAS2 from tumor cells and CD44 is expressed on the surface of tumor-associated macrophages (41). Interestingly, our TCGA analysis indicates positive correlations of CD44s and HAS2 in aggressive breast cancers, a phenomenon that was also suggested by previous studies (41, 42). Furthermore, our work suggests that CD44s produced by tumor cells also interacts with HA secreted from tumor cells, resulting in autocrine signaling that plays a critical role in activating Akt signaling and promoting an aggressive phenotype. To further strengthen this conclusion, we performed HAS2 overexpression experiments and showed that HAS2 promotes CD44s-dependent Akt activation, emphasizing the importance of HAS2-synthesized HA on driving Akt signaling and mitigating any concerns on the possible impurity when using exogenous HA. Therefore, targeting HAS2 and/or CD44s may allow for suppression of tumor growth through both autonomous and nonautonomous means.
Despite the fact that HAS2 expression is elevated in breast cancers, little is known about how HAS2 is regulated at the transcriptional level, although a few transcription factors including CREB1, RARγ, and hepatocyte nuclear factor HNF4G have been shown to bind to the HAS2 promoter and promote HAS2 transcription (43–45). In this study, we identified a signaling-driven transcriptional regulation mechanism by which FOXO1, a tumor-suppressive transcription factor, represses HAS2 transcription. This suppression is removed by Akt-mediated FOXO1 phosphorylation, which in turn hinders transcriptional activity of FOXO1, allowing for increased HAS2 transcription.
Mounting evidence has shown that CD44 promotes tumor growth and metastasis (19, 21, 46–49). However, CD44 was also reported to suppress tumorigenesis (50, 51). We suspect that these contradictory results are, at least in part, explained by the existence of CD44 splice isoforms, which exhibit divergent functional consequences (18, 19, 21, 52, 53). In line with this observation, alternative splicing of other genes has also been connected to carcinogenesis. For example, alternative splicing of caspase-9 produces caspase-9a and caspase-9b that differ by a four-exon cassette. Caspase-9a is a member of the caspase family of proteases important for apoptosis, whereas caspase-9b lacks its catalytic activity and acts as an inhibitor of caspase-9a, resulting in increased tumorigenicity (54). These results stress the importance of investigating the function of different splice isoforms in addition to the function of a gene in the context of cancer phenotypes. The former may lead to the development of more effective therapeutic interventions that target cancer-specific isoforms.
The positive feedback circuit detailed in this study can sustain Akt signaling in the absence of genetic mutations. This mechanism helps explain a long-standing question of how Akt signaling is maintained in tumors lacking mutations or amplifications in the PI3K/Akt pathway. This finding is also significant given the fact that aberrant activation of Akt signaling is observed in the majority of tumors. Although attempts to develop Akt inhibitors have been intense, only a limited number of compounds have emerged. Thus far, no Akt inhibitor has been approved for cancer treatment due to low efficacy (55). Our findings suggest that a combinatory therapeutic approach using Akt inhibitors and CD44s and HAS2 antagonists could be a more effective strategy for cancer treatment.
Supplementary Material
Acknowledgments
We thank Dr. Kounosuke Watabe (Wake Forest School of Medicine) for providing the HAS2 shRNA–expressing plasmid.
Grant Support: This research was supported in part by grants from the US National Institutes of Health T32CA080621 to S. Liu and R01CA182467 to C. Cheng. C. Cheng is a CPRIT Scholar in Cancer Research.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Disclosure of Potential Conflicts of Interest: No potential conflicts of interest were disclosed.
Authors' Contributions: Conception and design: S. Liu, C. Cheng
Development of methodology: S. Liu
Acquisition of data (provided animals, acquired and managed patients, provided facilities, etc.): S. Liu
Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): S. Liu, C. Cheng
Writing, review, and/or revision of the manuscript: S. Liu, C. Cheng
Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): S. Liu, C. Cheng
Study supervision: C. Cheng
References
- 1.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Sordella R, Bell DW, Haber DA, Settleman J. Gefitinib-sensitizing EGFR mutations in lung cancer activate anti-apoptotic pathways. Science. 2004;305:1163–7. doi: 10.1126/science.1101637. [DOI] [PubMed] [Google Scholar]
- 3.Adams JM, Cory S. The Bcl-2 protein family: arbiters of cell survival. Science. 1998;281:1322–6. doi: 10.1126/science.281.5381.1322. [DOI] [PubMed] [Google Scholar]
- 4.Meredith JE, Jr, Fazeli B, Schwartz MA. The extracellular matrix as a cell survival factor. Mol Biol Cell. 1993;4:953–61. doi: 10.1091/mbc.4.9.953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carracedo A, Ma L, Teruya-Feldstein J, Rojo F, Salmena L, Alimonti A, et al. Inhibition of mTORC1 leads to MAPK pathway activation through a PI3K-dependent feedback loop in human cancer. J Clin Invest. 2008;118:3065–74. doi: 10.1172/JCI34739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li H, Yang B, Huang J, Lin Y, Xiang T, Wan J, et al. Cyclo-oxygenase-2 in tumor-associated macrophages promotes breast cancer cell survival by triggering a positive-feedback loop between macrophages and cancer cells. Oncotarget. 2015;6:29637–50. doi: 10.18632/oncotarget.4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Su S, Liu Q, Chen J, Chen J, Chen F, He C, et al. A positive feedback loop between mesenchymal-like cancer cells and macrophages is essential to breast cancer metastasis. Cancer Cell. 2014;25:605–20. doi: 10.1016/j.ccr.2014.03.021. [DOI] [PubMed] [Google Scholar]
- 8.O'Reilly KE, Rojo F, She QB, Solit D, Mills GB, Smith D, et al. mTOR inhibition induces upstream receptor tyrosine kinase signaling and activates Akt. Cancer Res. 2006;66:1500–8. doi: 10.1158/0008-5472.CAN-05-2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hennessy BT, Smith DL, Ram PT, Lu Y, Mills GB. Exploiting the PI3K/AKT pathway for cancer drug discovery. Nat Rev Drug Discov. 2005;4:988–1004. doi: 10.1038/nrd1902. [DOI] [PubMed] [Google Scholar]
- 10.Kang S, Bader AG, Vogt PK. Phosphatidylinositol 3-kinase mutations identified in human cancer are oncogenic. Proc Natl Acad Sci USA. 2005;102:802–7. doi: 10.1073/pnas.0408864102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Page C, Lin HJ, Jin Y, Castle VP, Nunez G, Huang M, et al. Overexpression of Akt/AKT can modulate chemotherapy-induced apoptosis. Anticancer Res. 2000;20:407–16. [PubMed] [Google Scholar]
- 12.Cheng JQ, Godwin AK, Bellacosa A, Taguchi T, Franke TF, Hamilton TC, et al. AKT2, a putative oncogene encoding a member of a subfamily of protein-serine/threonine kinases, is amplified in human ovarian carcinomas. Proc Natl Acad Sci USA. 1992;89:9267–71. doi: 10.1073/pnas.89.19.9267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun M, Wang G, Paciga JE, Feldman RI, Yuan ZQ, Ma XL, et al. AKT1/PKBalpha kinase is frequently elevated in human cancers and its constitutive activation is required for oncogenic transformation in NIH3T3 cells. Am J Pathol. 2001;159:431–7. doi: 10.1016/s0002-9440(10)61714-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Graveley BR. Alternative splicing: increasing diversity in the proteomic world. Trends Genet. 2001;17:100–7. doi: 10.1016/s0168-9525(00)02176-4. [DOI] [PubMed] [Google Scholar]
- 15.Nilsen TW, Graveley BR. Expansion of the eukaryotic proteome by alternative splicing. Nature. 2010;463:457–63. doi: 10.1038/nature08909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu S, Cheng C. Alternative RNA splicing and cancer. Wiley Interdiscip Rev RNA. 2013;4:547–66. doi: 10.1002/wrna.1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.David CJ, Manley JL. Alternative pre-mRNA splicing regulation in cancer: pathways and programs unhinged. Genes Dev. 2010;24:2343–64. doi: 10.1101/gad.1973010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cheng C, Yaffe MB, Sharp PA. A positive feedback loop couples Ras activation and CD44 alternative splicing. Genes Dev. 2006;20:1715–20. doi: 10.1101/gad.1430906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao P, Damerow MS, Stern P, Liu AH, Sweet-Cordero A, Siziopikou K, et al. CD44 promotes Kras-dependent lung adenocarcinoma. Oncogene. 2013;32:5186–90. doi: 10.1038/onc.2012.542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matter N, Herrlich P, Konig H. Signal-dependent regulation of splicing via phosphorylation of Sam68. Nature. 2002;420:691–5. doi: 10.1038/nature01153. [DOI] [PubMed] [Google Scholar]
- 21.Brown RL, Reinke LM, Damerow MS, Perez D, Chodosh LA, Yang J, et al. CD44 splice isoform switching in human and mouse epithelium is essential for epithelial-mesenchymal transition and breast cancer progression. J Clin Invest. 2011;121:1064–74. doi: 10.1172/JCI44540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Okabe H, Ishimoto T, Mima K, Nakagawa S, Hayashi H, Kuroki H, et al. CD44s signals the acquisition of the mesenchymal phenotype required for anchorage-independent cell survival in hepatocellular carcinoma. Br J Cancer. 2014;110:958–66. doi: 10.1038/bjc.2013.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reinke LM, Xu Y, Cheng C. Snail represses the splicing regulator epithelial splicing regulatory protein 1 to promote epithelial-mesenchymal transition. J Biol Chem. 2012;287:36435–42. doi: 10.1074/jbc.M112.397125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu Y, Gao XD, Lee JH, Huang H, Tan H, Ahn J, et al. Cell type-restricted activity of hnRNPM promotes breast cancer metastasis via regulating alternative splicing. Genes Dev. 2014;28:1191–203. doi: 10.1101/gad.241968.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zheng X, Carstens JL, Kim J, Scheible M, Kaye J, Sugimoto H, et al. Epithelial-to-mesenchymal transition is dispensable for metastasis but induces chemoresistance in pancreatic cancer. Nature. 2015;527:525–30. doi: 10.1038/nature16064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fischer KR, Durrans A, Lee S, Sheng J, Li F, Wong ST, et al. Epithelial-to-mesenchymal transition is not required for lung metastasis but contributes to chemoresistance. Nature. 2015;527:472–6. doi: 10.1038/nature15748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Monslow J, Williams JD, Guy CA, Price IK, Craig KJ, Williams HJ, et al. Identification and analysis of the promoter region of the human hyaluronan synthase 2 gene. J Biol Chem. 2004;279:20576–81. doi: 10.1074/jbc.M312666200. [DOI] [PubMed] [Google Scholar]
- 28.Torre C, Wang SJ, Xia W, Bourguignon LY. Reduction of hyaluronan-CD44-mediated growth, migration, and cisplatin resistance in head and neck cancer due to inhibition of Rho kinase and PI-3 kinase signaling. Arch Otolaryngol Head Neck Surg. 2010;136:493–501. doi: 10.1001/archoto.2010.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim MS, Park MJ, Moon EJ, Kim SJ, Lee CH, Yoo H, et al. Hyaluronic acid induces osteopontin via the phosphatidylinositol 3-kinase/Akt pathway to enhance the motility of human glioma cells. Cancer Res. 2005;65:686–91. [PubMed] [Google Scholar]
- 30.Tian X, Azpurua J, Hine C, Vaidya A, Myakishev-Rempel M, Ablaeva J, et al. High-molecular-mass hyaluronan mediates the cancer resistance of the naked mole rat. Nature. 2013;499:346–9. doi: 10.1038/nature12234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nandi A, Estess P, Siegelman MH. Hyaluronan anchoring and regulation on the surface of vascular endothelial cells is mediated through the functionally active form of CD44. J Biol Chem. 2000;275:14939–48. doi: 10.1074/jbc.275.20.14939. [DOI] [PubMed] [Google Scholar]
- 32.Girbl T, Hinterseer E, Grossinger EM, Asslaber D, Oberascher K, Weiss L, et al. CD40-mediated activation of chronic lymphocytic leukemia cells promotes their CD44-dependent adhesion to hyaluronan and restricts CCL21-induced motility. Cancer Res. 2013;73:561–70. doi: 10.1158/0008-5472.CAN-12-2749. [DOI] [PubMed] [Google Scholar]
- 33.Aguirre-Alvarado C, Segura-Cabrera A, Velazquez-Quesada I, Hernandez-Esquivel MA, Garcia-Perez CA, Guerrero-Rodriguez SL, et al. Virtual screening-driven repositioning of etoposide as CD44 antagonist in breast cancer cells. Oncotarget. 2016;7:23772–84. doi: 10.18632/oncotarget.8180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Calnan DR, Brunet A. The FoxO code. Oncogene. 2008;27:2276–88. doi: 10.1038/onc.2008.21. [DOI] [PubMed] [Google Scholar]
- 35.Paoli P, Giannoni E, Chiarugi P. Anoikis molecular pathways and its role in cancer progression. Biochim Biophys Acta. 2013;1833:3481–98. doi: 10.1016/j.bbamcr.2013.06.026. [DOI] [PubMed] [Google Scholar]
- 36.Larue L, Bellacosa A. Epithelial-mesenchymal transition in development and cancer: role of phosphatidylinositol 3′ kinase/AKT pathways. Oncogene. 2005;24:7443–54. doi: 10.1038/sj.onc.1209091. [DOI] [PubMed] [Google Scholar]
- 37.Bernert B, Porsch H, Heldin P. Hyaluronan synthase 2 (HAS2) promotes breast cancer cell invasion by suppression of tissue metalloproteinase inhibitor 1 (TIMP-1) J Biol Chem. 2011;286:42349–59. doi: 10.1074/jbc.M111.278598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Urakawa H, Nishida Y, Wasa J, Arai E, Zhuo L, Kimata K, et al. Inhibition of hyaluronan synthesis in breast cancer cells by 4-methylumbelliferone suppresses tumorigenicity in vitro and metastatic lesions of bone in vivo. Int J Cancer. 2012;130:454–66. doi: 10.1002/ijc.26014. [DOI] [PubMed] [Google Scholar]
- 39.Li Y, Li L, Brown TJ, Heldin P. Silencing of hyaluronan synthase 2 suppresses the malignant phenotype of invasive breast cancer cells. Int J Cancer. 2007;120:2557–67. doi: 10.1002/ijc.22550. [DOI] [PubMed] [Google Scholar]
- 40.Chanmee T, Ontong P, Mochizuki N, Kongtawelert P, Konno K, Itano N. Excessive hyaluronan production promotes acquisition of cancer stem cell signatures through the coordinated regulation of Twist and the transforming growth factor beta (TGF-beta)-Snail signaling axis. J Biol Chem. 2014;289:26038–56. doi: 10.1074/jbc.M114.564120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Okuda H, Kobayashi A, Xia B, Watabe M, Pai SK, Hirota S, et al. Hyaluronan synthase HAS2 promotes tumor progression in bone by stimulating the interaction of breast cancer stem-like cells with macrophages and stromal cells. Cancer Res. 2012;72:537–47. doi: 10.1158/0008-5472.CAN-11-1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Udabage L, Brownlee GR, Waltham M, Blick T, Walker EC, Heldin P, et al. Antisense-mediated suppression of hyaluronan synthase 2 inhibits the tumorigenesis and progression of breast cancer. Cancer Res. 2005;65:6139–50. doi: 10.1158/0008-5472.CAN-04-1622. [DOI] [PubMed] [Google Scholar]
- 43.Makkonen KM, Pasonen-Seppanen S, Torronen K, Tammi MI, Carlberg C. Regulation of the hyaluronan synthase 2 gene by convergence in cyclic AMP response element-binding protein and retinoid acid receptor signaling. J Biol Chem. 2009;284:18270–81. doi: 10.1074/jbc.M109.012492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schiller M, Dennler S, Anderegg U, Kokot A, Simon JC, Luger TA, et al. Increased cAMP levels modulate transforming growth factor-beta/Smad-induced expression of extracellular matrix components and other key fibroblast effector functions. J Biol Chem. 2010;285:409–21. doi: 10.1074/jbc.M109.038620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Okegawa T, Ushio K, Imai M, Morimoto M, Hara T. Orphan nuclear receptor HNF4G promotes bladder cancer growth and invasion through the regulation of the hyaluronan synthase 2 gene. Oncogenesis. 2013;2:e58. doi: 10.1038/oncsis.2013.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yu Q, Stamenkovic I. Localization of matrix metalloproteinase 9 to the cell surface provides a mechanism for CD44-mediated tumor invasion. Genes Dev. 1999;13:35–48. doi: 10.1101/gad.13.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ouhtit A, Abd Elmageed ZY, Abdraboh ME, Lioe TF, Raj MH. In vivo evidence for the role of CD44s in promoting breast cancer metastasis to the liver. Am J Pathol. 2007;171:2033–9. doi: 10.2353/ajpath.2007.070535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Godar S, Ince TA, Bell GW, Feldser D, Donaher JL, Bergh J, et al. Growth-inhibitory and tumor-suppressive functions of p53 depend on its repression of CD44 expression. Cell. 2008;134:62–73. doi: 10.1016/j.cell.2008.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhao P, Xu Y, Wei Y, Qiu Q, Chew TL, Kang Y, et al. The CD44s splice isoform is a central mediator for invadopodia activity. J Cell Sci. 2016;129:1355–65. doi: 10.1242/jcs.171959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lopez JI, Camenisch TD, Stevens MV, Sands BJ, McDonald J, Schroeder JA. CD44 attenuates metastatic invasion during breast cancer progression. Cancer Res. 2005;65:6755–63. doi: 10.1158/0008-5472.CAN-05-0863. [DOI] [PubMed] [Google Scholar]
- 51.Gao AC, Lou W, Dong JT, Isaacs JT. CD44 is a metastasis suppressor gene for prostatic cancer located on human chromosome 11p13. Cancer Res. 1997;57:846–9. [PubMed] [Google Scholar]
- 52.Cheng C, Sharp PA. Regulation of CD44 alternative splicing by SRm160 and its potential roleintumor cell invasion. Mol Cell Biol. 2006;26:362–70. doi: 10.1128/MCB.26.1.362-370.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ponta H, Sherman L, Herrlich PA. CD44: from adhesion molecules to signalling regulators. Nat Rev Mol Cell Biol. 2003;4:33–45. doi: 10.1038/nrm1004. [DOI] [PubMed] [Google Scholar]
- 54.Goehe RW, Shultz JC, Murudkar C, Usanovic S, Lamour NF, Massey DH, et al. hnRNP L regulates the tumorigenic capacity of lung cancer xenografts in mice via caspase-9 pre-mRNA processing. J Clin Invest. 2010;120:3923–39. doi: 10.1172/JCI43552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nitulescu GM, Margina D, Juzenas P, Peng Q, Olaru OT, Saloustros E, et al. Akt inhibitors in cancer treatment: the long journey from drug discovery to clinical use (Review) Int J Oncol. 2016;48:869–85. doi: 10.3892/ijo.2015.3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.