Abstract
TGF-β1, a multifunctional regulator of cell growth and differentiation, is the most abundant bone matrix growth factor. During differentiation of human bone stromal cells (hBMSCs), which constitute bone marrow osteoblast (OS) and adipocyte (AD) progenitor cells, continuous TGF-β1 (10 ng/ml) treatment enhanced OS differentiation as evidenced by increased mineralised matrix production. Conversely, pulsed TGF-β1 administration during the commitment phase increased mature lipid-filled adipocyte numbers. Global gene expression analysis using DNA microarrays in hBMSCs treated with TGF-β1 identified 1587 up- and 1716 down-regulated genes in OS-induced, TGF-β1-treated compared to OS-induced hBMSCs (2.0 fold change (FC), p < 0.05). Gene ontology (GO) analysis revealed enrichment in ‘osteoblast differentiation’ and ‘skeletal system development-associated’ genes and up-regulation of several genes involved in ‘osteoblastic-differentiation related signalling pathways’. In AD-induced, TGF-β1-treated compared to AD-induced hBMSCs, we identified 323 up- and 369 down-regulated genes (2.0 FC, p < 0.05) associated with ‘fat cell differentiation’, ‘fatty acid derivative biosynthesis process’, ‘fatty acid derivative metabolic process’, and ‘inositol lipid-mediated’. Serpin peptidase inhibitor, clade B (ovalbumin), member 2 (SERPINB2) was down-regulated 3-fold in TGF-β1-treated hBMSCs. siRNA-mediated SERPINB2 inhibition enhanced OS and AD differentiation. Thus, TGF-β signalling is important for hBMSC OS and AD differentiation and SERPINB2 is a TGF-β-responsive gene that plays a negative regulatory role in hBMSC differentiation.
Introduction
Skeletal stem cells (also known as bone marrow-derived multipotent stromal cells or bone marrow mesenchymal stem cells (BMSC)) comprise multipotent stem cells that can differentiate into adipocytes (ADs or osteoblasts (OS) in response to micro-environmental signals including growth factors, cytokines, and epigenetic regulators1. An imbalance between OS and AD lineage commitment and differentiation has been implicated as a cause for age-related impaired bone formation; thus, a number of therapeutic interventions have been proposed for enhancing bone mass through the targeting of BMSC2, 3.
TGF-β1 constitutes one of the most abundant growth factor in the bone matrix (200 μg/kg)4 and is secreted by several cells associated with the skeleton; e.g. OS, endothelial cells, smooth muscle cells, and stromal cells, as well as by cells of the immune system5. TGF-β1 is produced in large precursor complexes that are composed of mature TGF-β1 and latency-associated protein (LAP). TGF-β1 is secreted and deposited in bone matrix as an inactive, latent complex owing to its non-covalent linkage to LAP, which masks the receptor-domains of the active TGF-β1. Osteoclast-mediated bone resorption activates TGF-β1 by cleavage of LAP and releases it from the bone matrix, creating a gradient of active TGF-β1 that signals to recruit osteoprogenitor cells to the bone remodelling sites and thus support bone formation6. TGF-β1 has been shown to regulate the proliferation and differentiation of osteoblastic cells7 and inhibition of TGF-β receptor signalling in OS has been reported to decrease bone remodelling and increase trabecular bone mass6.
In the current study, we examined the role of TGFβ-1 in OS and AD lineage commitment and the differentiation of human BMSC (hBMSC) and the dependency of these effects on the timing of induction as determined using a single pulse dose during the commitment phase of hBMSC versus continuous treatment during the whole differentiation period. In addition, we examined the molecular mechanisms of TGFβ-1-mediated differentiation of hBMSCs employing DNA microarrays. We identified one of the significantly (3-fold) down-regulated genes during TGFβ1 stimulation, serpin peptidase inhibitor, clade B (ovalbumin), member 2 (SERPINB2), as a novel TGF-β-responsive gene that plays a role in hBMSC differentiation. We demonstrated that inhibition of SERPINB2 in hBMSC led to enhanced OS and AD differentiation suggesting a negative regulatory role in OS and AD differentiation, downstream of TGF-β signalling.
Results
Continuous treatment with TGF-β1 enhances OS differentiation
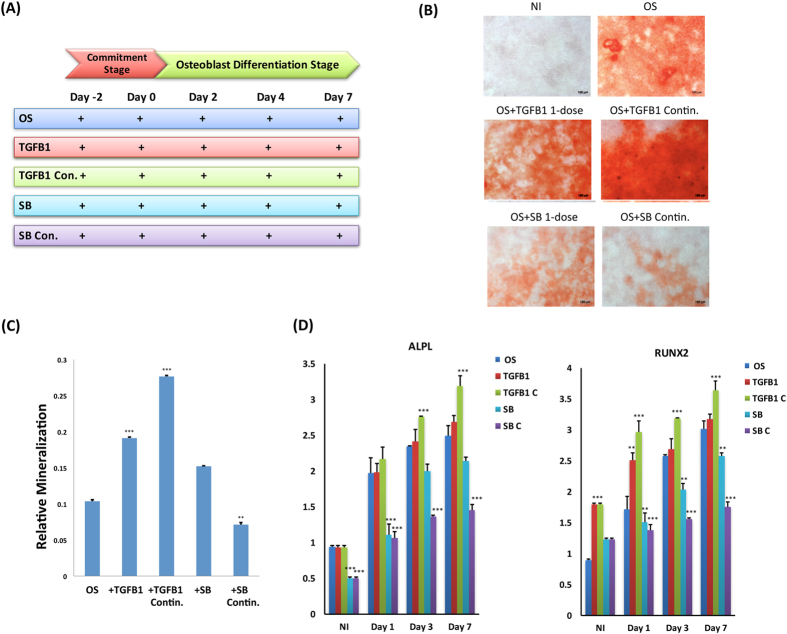
We compared the effect on hBMSC differentiation to OS when TGFβ1 (10 ng/ml) treatment was conducted as a single pulse dose during the commitment phase of differentiation (day −2 to day 0) versus continuous treatment during the whole course of differentiation (day −2 to day 7) (Fig. 1A). As judged by qualitative and quantitative alizarin red staining for mineralised matrix formation, continuous treatment with TGF-β1 induced a higher degree of OS differentiation (Fig. 1B,C, p < 0.01). These effects represented direct effects of TGF-β1, as they were reduced following exposure to the TGFβ receptor kinase inhibitor SB-431542 (10 M). Quantitative reverse transcription-polymerase chain reaction (RT-PCR) was further performed to assess gene expression of osteoblastic markers upon continuous application of TGFβ-1. Gene expression of alkaline phosphatase, liver/bone/kidney (ALPL) exhibited significant up-regulation at day 3, whereas runt-related transcription factor 2 (RUNX2) showed gradual up-regulation starting from day 1 up to day 3 (Fig. 1D).
Figure 1.
TGF-β1 promotes osteogenic differentiation. Human bone marrow stromal (skeletal) stem cells (hBMSC) were differentiated into osteoblasts (OS) using osteogenic induction mixture for 7 days. (A) Time line scheme of experimental setup illustrating TGF-β1 or SB-431542 (SB) treatment that was performed as either single pulse dose (TGFB1 1-dose or SB 1-dose) or continuous treatment (TGFB1 Contin. Or SB Contin.) at commitment and differentiation stages of in vitro OS differentiation (B) Mineralised calcium deposition was determined by Alizarin Red S staining, which is shown as microscopic images (20× magnification). (C) Alizarin Red Quantification under different experimental conditions: osteo-induced (OS), single dose of 10 ng/ml TGF-β1 (+TGFB1); continuous exposure to TGF-β1, TGFB1 con, SB 1-dose, and SB Con. Data are presented as the means ± SD of three independent experiments; n = 6; (D) qRT-PCR of ALPL (left panel) or RUNX2 (right panel) mRNA expression preformed on cells exposed to the indicated treatment on days 1, 3, and 7. Expression of each target gene was normalised to GAPDH. Data are presented as the means ± SD from three independent experiments, n = 6; *p < 0.05; **p < 0.01, ***p < 0.005.
Single pulse treatment with TGFβ-1 enhances AD differentiation
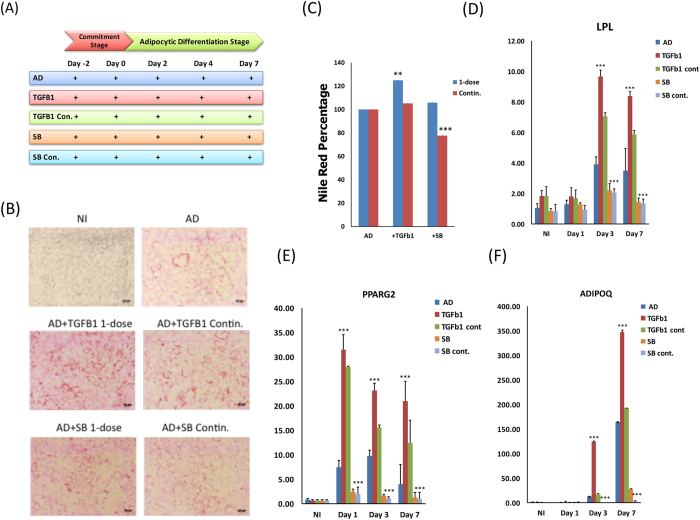
We next compared the effect on hBMSC differentiation to AD when TGFβ1 (10 ng/ml) treatment was conducted as a single pulse dose during the commitment phase of differentiation (day −2 to day 0) versus continuous treatment during the whole course of differentiation (day −2 to day 7) (Fig. 2A). As shown in Fig. 2A and B, qualitative and quantitative Nile red staining of mature adipocytes following single pulse treatment with TGFβ-1 led to a greater degree of AD differentiation enhancement compared with continuous treatment (Fig. 2B and C). SB-431542 treatment significantly reduced AD induction during continuous treatment and to a lesser degree in the single pulse dose. Gene expression analysis of the adipogenic markers lipoprotein lipase (LPL), peroxisome proliferator-activated receptor gamma 2 (PPARG2), and adiponectin or (ADIPOQ) exhibited similar effects (Fig. 2D–F).
Figure 2.
TGF-β1 promotes adipogenic differentiation. (A) Time line schematic model illustrating the dose of TGF-β1 or SB-431542 (SB) treatment for either single pulse or continuous treatment at commitment and differentiation stages during adipogenic induction of human bone marrow stromal (skeletal) stem cells (hBMSC) (B) hBMSCs were induced into adipocyte using adipocyte induction medium in the presence of single dose (AD + TGFB1 1-dose) or continuous (AD + TGFB1 Contin.) exposure to 10 ng/ml TGF-β1 or a single (AD + SB 1-dose) or continuous (AD + SB Contin.) treatment with 10 µM SB-431542. Cells were stained on day 7 using oil red O staining for adipocytes containing lipid droplets, and shown as microscopic images (20×, magnification), non-induced (NI), adipocytic induced (AD). (C) Nile red quantification under the indicated treatment conditions was performed on day 7 post adipocyte induction. qRT-PCR quantification for LPL (D), PPARG2 (E), and ADIPOQ (F) mRNA under the indicated experimental conditions: non-induced (CNT), adipo-induced (AD), single dose of TGF-β1 (+TGFB1), continuous exposure to TGF-β1 (+TGFB1 Contin.), single dose of SB-431542 (+SB), and continuous dose of SB-431542 (+SB Contin.). Expression of each target gene was normalised to GAPDH. Data are presented as the means ± SD from three independent experiments, n = 6; *p < 0.05; **p < 0.01, ***p < 0.005.
Molecular phenotype of TGF-β1-induces osteoblastic cells
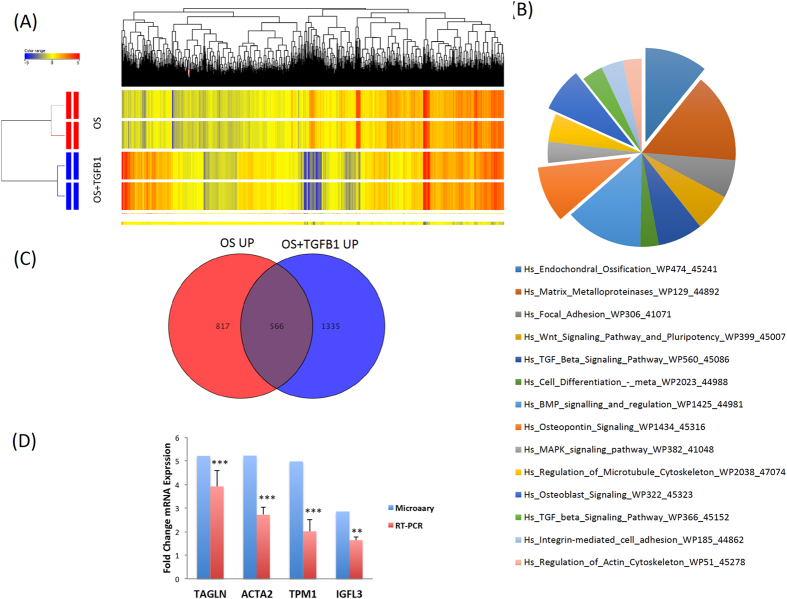
Global gene expression profiling was conducted on hBMSC treated with or without TGF-β1 (10 ng/ml) in the presence of OS induction medium. Hierarchical clustering based on differentially expressed genes revealed clear separation of control cells treated with OS induction medium alone (OS-differentiated) from those treated with TGF-βl (OS-differentiated-TGF-β1) (Fig. 3A). We identified 1587 up-regulated and 1716 down-regulated genes in OS-differentiated-TGF-β1 compared to OS-differentiated control (2.0 fold change (FC), p < 0.05; Supplementary Tables S1, S2). Pathway analysis of the up-regulated genes revealed significant enrichment in several pathways related to ‘endochondral ossification’, ‘matrix metalloproteinases’, ‘TGF-β signalling’, ‘WNT signalling’, ‘MAPK signalling’, ‘focal adhesion’, and’ regulation of actin cytoskeleton pathways’ (Fig. 3B, Supplementary Figs S1–S7). Table 1 lists the matched entities from the microarray data and the corresponding signalling pathway. Comparing the up-regulated genes in OS-differentiated cells with those in OS-differentiated-TGF-β1 cells revealed 566 common genes that were predicted to be regulated by TGF-β signalling and concurrently to be involved in OS-differentiation (Fig. 3C, Supplementary Table S3). Notably, we found that the selectively up-regulated genes in OS-differentiated-TGF-β1 cells were enriched in categories of bone formation, such as ‘skeletal system development’ (58 genes), ‘ossification’ (42 genes), and ‘osteoblast differentiation’ (23 genes) (Table 2). qRT-PCR results for selective up-regulated osteoblastic-related genes showed good concordance with microarray results (Fig. 3D).
Figure 3.
Gene expression profiling on hBMSC induced into osteoblasts in the presence of TGF-β1. (A) Hierarchical clustering of human bone marrow stromal (skeletal) stem cells (hBMSC) induced into osteoblasts (OS) (day 3) in the presence or absence of TGF-βl. Each row represents one replica sample and each column represents a transcript. Expression level of each gene in a single sample is depicted according to the colour scale. (B) Pie chart illustrating the distribution of the top 13 pathway designations for the up-regulated genes in TGF-β1 treated cells during OS differentiation (C) Venn diagram depicting the overlap between the up-regulated genes during OS differentiation of hBMSCs and the upregulated genes in hBMSC induced to OS in presence of TGF-β1. (D) qRT-PCR validation of selected genes (TAGLN1, ACTA2, TPM1, and IGFL3).
Table 1.
Up-regulated genes involved in osteogenic-related pathways in TGF-β treated cells after osteogenic induction.
| Endochondral Ossification | Matrix Metallo-proteinases | TGFB Signalling | WNT Signalling | MAPK Signalling | Focal Adhesion | Regulation of Actin Cytoskeleton |
|---|---|---|---|---|---|---|
| IGF1 | MMP1 | INHBA | WNT7B | IL1A | COL11A1 | ITGA1 |
| IGF1R | MMP2 | NOG | WNT11 | TGFB2 | COL3A1 | GSN |
| IGF2 | MMP11 | THBS1 | FZD2 | ACVR1C | COL4A1 | NRAS |
| BMP6 | MMP14 | LEFTY1 | FZD7 | CASP9 | COL4A2 | IGF1 |
| CDKN1C | MMP24 | SMAD7 | DVL1 | AKT3 | COL4A4 | PDGFA |
| TGFB2 | MMP23B | LIF | CTNNB1 | HSPB2 | COL5A1 | C11orf13 |
| COL10A1 | TIMP1 | CTNNB1 | TCF7 | MAPK8 | COL5A2 | BDKRB1 |
| C4ST1 | TIMP2 | RUNX3 | TCF7L1 | HSPA5 | COL1A1 | LIMK1 |
| RUNX3 | TIMP3 | NKD1 | MAPK8IP3 | THBS1 | PAK3 | |
| TIMP3 | PPP2R1B | MAP3K14 | THBS2 | |||
| PLAT | PLAU | NRAS | TNC | |||
| PLAU | PPARD | ARHGAP5 | ||||
| ADAMTS4 | AKT3 | |||||
| PAK3 | ||||||
| MAPK8 | ||||||
| IGF1R | ||||||
| ITGB1 | ||||||
| ITGB3 | ||||||
| ITGA11 | ||||||
| IGF1 | ||||||
| PDGFA | ||||||
| PDGFC | ||||||
| PGF |
Table 2.
Up-regulated biological processes and related genes in TGF-β1 treated cells during osteogenesis using GO analysis.
| Skeletal System Development | Ossification | Osteoblast Differentiation | |||
|---|---|---|---|---|---|
| Gene Symbol | Gene Name | Gene Symbol | Gene Name | Gene Symbol | Gene Name |
| CYTL1 | cytokine-like 1 | GLI1 | GLI family zinc finger 1 | GLI1 | GLI family zinc finger 1 |
| HOXB2 | homeobox B2 | SEMA7A | semaphorin 7 A, GPI membrane anchor (John Milton Hagen blood group) | SEMA7A | semaphorin 7 A, GPI membrane anchor (John Milton Hagen blood group) |
| PLEKHA1 | pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1 | FBXL15 | F-box and leucine-rich repeat protein 15 | SNAI1 | snail family zinc finger 1 |
| CSRNP1 | cysteine-serine-rich nuclear protein 1 | SNAI1 | snail family zinc finger 1 | VCAN | versican |
| SNAI1 | snail family zinc finger 1 | IGF1 | insulin-like growth factor 1 (somatomedin C) | IGF2 | insulin-like growth factor 2 (somatomedin A) |
| IGF1 | insulin-like growth factor 1 (somatomedin C) | VCAN | versican | TNC | tenascin C |
| CHST11 | carbohydrate (chondroitin 4) sulphotransferase 11 | IGF2 | insulin-like growth factor 2 (somatomedin A) | BMP6 | bone morphogenetic protein 6 |
| TIPARP | TCDD-inducible poly(ADP-ribose) polymerase | CDH11 | cadherin 11, type 2, OS-cadherin (osteoblast) | ITGA11 | integrin, alpha 11 |
| IGF2 | insulin-like growth factor 2 (somatomedin A) | TNC | tenascin C | GLI2 | GLI family zinc finger 2 |
| CDH11 | cadherin 11, type 2, OS-cadherin (osteoblast) | BMP6 | bone morphogenetic protein 6 | LRRC17 | leucine rich repeat containing 17 |
| TGFBI | transforming growth factor, beta-induced, 68 kDa | ITGA11 | integrin, alpha 11 | CYP24A1 | cytochrome P450, family 24, subfamily A, polypeptide 1 |
| VDR | vitamin D (1,25- dihydroxyvitamin D3) receptor | GLI2 | GLI family zinc finger 2 | IGF2 | insulin-like growth factor 2 (somatomedin A) |
| HHIP | hedgehog interacting protein | COL10A1 | collagen, type X, alpha 1 | RSPO2 | R-spondin 2 |
| EBP | emopamil binding protein (sterol isomerase) | LRRC17 | leucine rich repeat containing 17 | SMO | smoothened, frizzled class receptor |
| BMP6 | bone morphogenetic protein 6 | CYP24A1 | cytochrome P450, family 24, subfamily A, polypeptide 1 | HDAC5 | histone deacetylase 5 |
| GLI2 | GLI family zinc finger 2 | MAPK8 | mitogen-activated protein kinase 8 | WNT11 | wingless-type MMTV integration site family, member 11 |
| COL10A1 | collagen, type X, alpha 1 | TUFT1 | tuftelin 1 | DLX5 | distal-less homeobox 5 |
| HOXC9 | homeobox C9 | TEK | TEK tyrosine kinase, endothelial | BMP2 | bone morphogenetic protein 2 |
| LRRC17 | leucine rich repeat containing 17 | IGF2 | insulin-like growth factor 2 (somatomedin A) | COL1A1 | collagen, type I, alpha 1 |
| DLX2 | distal-less homeobox 2 | RSPO2 | R-spondin 2 | LRRC17 | leucine rich repeat containing 17 |
| COL5A2 | collagen, type V, alpha 2 | SMO | smoothened, frizzled class receptor | CYP24A1 | cytochrome P450, family 24, subfamily A, polypeptide 1 |
| ADAMTS4 | ADAM metallopeptidase with thrombospondin type 1 motif, 4 | HDAC5 | histone deacetylase 5 | NOG | noggin |
| TEK | TEK tyrosine kinase, endothelial | PTGS2 | prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) | TMEM119 | transmembrane protein 119 |
| DLG1 | discs, large homolog 1 (Drosophila) | WNT11 | wingless-type MMTV integration site family, member 11 | ||
| IGF2 | insulin-like growth factor 2 (somatomedin A) | GNAS | GNAS complex locus | ||
| RSPO2 | R-spondin 2 | MMP14 | matrix metallopeptidase 14 (membrane-inserted) | ||
| EXTL1 | exostosin-like glycosyltransferase 1 | ASPN | asporin | ||
| LEPRE1 | leucine proline-enriched proteoglycan (leprecan) 1 | CDH11 | cadherin 11, type 2, OS-cadherin (osteoblast) | ||
| SMO | smoothened, frizzled class receptor | DLX5 | distal-less homeobox 5 | ||
| THBS1 | thrombospondin 1 | MMP2 | matrix metallopeptidase 2 (gelatinase A, 72 kDa gelatinase, 72 kDa type IV collagenase) | ||
| PBX1 | pre-B-cell leukaemia homeobox 1 | BMP2 | bone morphogenetic protein 2 | ||
| WNT11 | wingless-type MMTV integration site family, member 11 | MAPK8 | mitogen-activated protein kinase 8 | ||
| GNAS | GNAS complex locus | SLC26A2 | solute carrier family 26 (anion exchanger), member 2 | ||
| TGFB2 | transforming growth factor, beta 2 | FOXS1 | forkhead box S1 | ||
| SOX4 | SRY (sex determining region Y)-box 4 | COL1A1 | collagen, type I, alpha 1 | ||
| COL3A1 | collagen, type III, alpha 1 | FSTL3 | follistatin-like 3 (secreted glycoprotein) | ||
| RUNX3 | runt-related transcription factor 3 | LRRC17 | leucine rich repeat containing 17 | ||
| CSRNP1 | cysteine-serine-rich nuclear protein 1 | CYP24A1 | cytochrome P450, family 24, subfamily A, polypeptide 1 | ||
| CDH11 | cadherin 11, type 2, OS-cadherin (osteoblast) | SPARC | secreted protein, acidic, cysteine-rich (osteonectin) | ||
| DLX5 | distal-less homeobox 5 | PDLIM7 | PDZ and LIM domain 7 (enigma) | ||
| MMP2 | matrix metallopeptidase 2 (gelatinase A, 72 kDa gelatinase, 72 kDa type IV collagenase) | NOG | noggin | ||
| BMP2 | bone morphogenetic protein 2 | TMEM119 | transmembrane protein 119 | ||
| CYTL1 | cytokine-like 1 | ||||
| WNT7B | wingless-type MMTV integration site family, member 7B | ||||
| COL11 A1 | collagen, type XI, alpha 1 | ||||
| FOXS1 | forkhead box S1 | ||||
| COL1A1 | collagen, type I, alpha 1 | ||||
| SGPL1 | sphingosine-1-phosphate lyase 1 | ||||
| LRRC17 | leucine rich repeat containing 17 | ||||
| SUFU | suppressor of fused homolog (Drosophila) | ||||
| HES7 | hes family bHLH transcription factor 7 | ||||
| SPARC | secreted protein, acidic, cysteine-rich (osteonectin) | ||||
| NOG | noggin | ||||
| THBS1 | thrombospondin 1 | ||||
| MATN3 | matrilin 3 | ||||
| CADM1 | cell adhesion molecule 1 | ||||
| POSTN | periostin, osteoblast specific factor | ||||
| CADM1 | cell adhesion molecule 1 | ||||
Molecular phenotype of TGF-β1-induces adipocytic cells
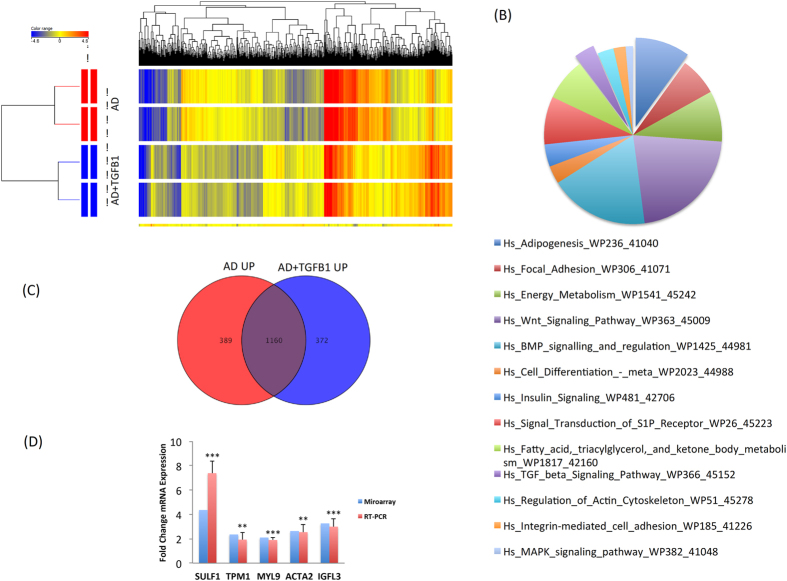
Global gene expression profiling was conducted on cells treated with or without TGF-β1 (10 ng/ml) (day 3) in the presence of adipogenic induction medium. As shown in Fig. 4A, hierarchical clustering based on differentially expressed genes revealed clear separation of control samples treated with adipocyte induction medium alone (AD-differentiated) and those induced in presence of TGF-βl (AD-differentiated-TGF-β1). We identified 323 up- and 369 down-regulated genes in AD-differentiated-TGF-β1 compared to AD-differentiated cells (2.0 FC, p < 0.05; Supplementary Table S4, S5). Pathway analysis on the up-regulated genes in AD-differentiated-TGF-β1 treated samples revealed significant enrichment in several pathways related to adipogenesis, e.g. ‘adipogenesis’, ‘energy metabolism’, and ‘insulin signalling’ (Fig. 4B and Table 3). Comparing the list of the up-regulated genes in AD-differentiated cells and those in AD-differentiated-TGF-β1 cells revealed 1160 common genes that are predicted to be regulated by TGF-β signalling and also involved in AD-differentiation (Fig. 4C, Supplementary Table S6). The selectively up-regulated genes in AD-differentiated-TGF-β1 were enriched in categories of ‘fat cell differentiation’, ‘fatty acid derivative biosynthesis process’, ‘fatty acid derivative metabolic process’, and ‘inositol lipid-mediated signalling’ (Table 4). qRT-PCR findings showed concordance with microarray data for a selected gene panel of the up regulated genes in TGF-β1 treated cells (Fig. 4D).
Figure 4.
Gene expression profiling of hBMSC induced into adipocytes in the presence of TGF-β1. (A) Hierarchical clustering of human bone marrow stromal (skeletal) stem cells (hBMSC) induced into adipocytes (AD) (day 3) in the presence or absence of TGF-βl. Each row represents one replica sample and each column represents a transcript. Expression level of each gene in a single sample is depicted according to the colour scale. (B) Pie chart illustrating the distribution of the top 13 pathway designations for the up-regulated genes in TGF-β1 treated cells during adipogenesis. (C) Venn diagram depicting the overlap between the up-regulated genes during AD induction of hBMSC and the upregulated genes in hBMSC induced to AD in presence of TGF-β1. (D) qRT-PCR validation of selected genes from the microarray data (SULF1, TPM1, MYL9, ACTA2, and IGFL3).
Table 3.
Up regulated genes involved in adipogenesis pathway in TGF-β1 treated cells after adipogenic induction.
| Probe Name | Gene Name | Gene Symbol | FC ([AD + TGFB1] vs. [AD]) |
|---|---|---|---|
| A_23_P53891 | Kruppel-like factor 5 (intestinal) | KLF5 | 6.4302254 |
| A_23_P13907 | insulin-like growth factor 1 (somatomedin C) | IGF1 | 5.5686526 |
| A_23_P46936 | early growth response 2 | EGR2 | 3.4646597 |
| A_23_P18447 | peroxisome proliferator-activated receptor gamma, coactivator 1 alpha | PPARGC1A | 3.062086 |
| A_24_P22079 | forkhead box O1 | FOXO1 | 2.6298373 |
| A_33_P3302295 | forkhead box C2 (MFH-1, mesenchyme forkhead 1) | FOXC2 | 2.3352392 |
| A_23_P137381 | inhibitor of DNA binding 3, dominant negative helix-loop-helix protein | ID3 | 2.3178163 |
| A_33_P3237150 | bone morphogenetic protein 2 | BMP2 | 2.2145567 |
| A_24_P38276 | frizzled class receptor 1 | FZD1 | 2.089059 |
| A_24_P154037 | insulin receptor substrate 2 | IRS2 | 2.0130024 |
| A_23_P211007 | nuclear receptor interacting protein 1 | NRIP1 | 2.0108652 |
Table 4.
Up-regulated biological processes and related genes in TGF-β1 treated cells during adipogenesis using GO analysis.
| Fat Cell Differentiation | Fatty Acid Derivative Biosynthesis Process | Fatty Acid Derivative Metabolic Process | Inositol Lipid-Mediated Signalling | ||||
|---|---|---|---|---|---|---|---|
| Gene Symbol | Gene Name | Gene Symbol | Gene Name | Gene Symbol | Gene Name | Gene Symbol | Gene Name |
| RGS2 | regulator of G-protein signalling 2, 24 kDa | EDN1 | endothelin 1 | EDN1 | endothelin 1 | IGF1 | insulin-like growth factor 1 (somatomedin C) |
| INHBB | inhibin, beta B | PTGS1 | prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) | PTGS1 | prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) | FGF1 | fibroblast growth factor 1 (acidic) |
| PPARGC1A | peroxisome proliferator-activated receptor gamma, coactivator 1 alpha | PTGS2 | prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) | ALOX15B | arachidonate 15-lipoxygenase, type B | EDN1 | endothelin 1 |
| RUNX1T1 | runt-related transcription factor 1; translocated to, 1 (cyclin D-related) | PTGS1 | prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) | PTGS2 | prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) | NPR3 | natriuretic peptide receptor 3 |
| EGR2 | early growth response 2 | GGT5 | gamma-glutamyltransferase 5 | PTGS1 | prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) | NRG1 | neuregulin 1 |
| FOXO1 | forkhead box O1 | C9orf3 | chromosome 9 open reading frame 3 | GGT5 | gamma-glutamyltransferase 5 | IRS2 | insulin receptor substrate 2 |
| PTGS2 | prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) | C9orf3 | chromosome 9 open reading frame 3 | FOXO1 | forkhead box O1 | ||
| FGF7 | fibroblast growth factor 7 | ||||||
Identification of SERPINB2 as a TGF-β-responsive gene
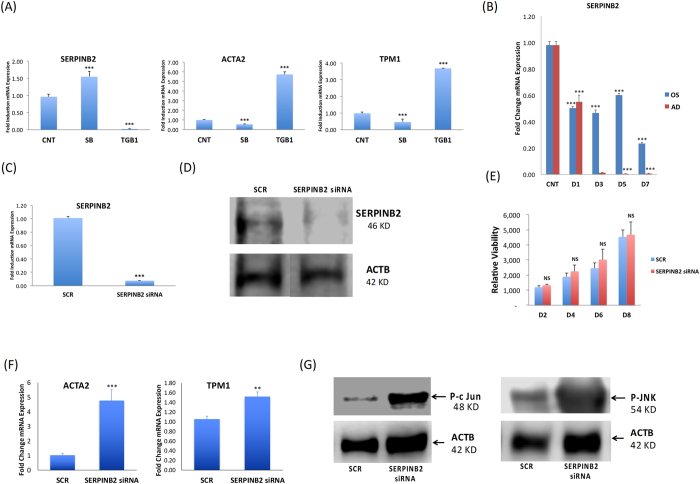
Among the genes highly regulated by TGF-β1 treatment, we identified SERPINB2 (significantly down-regulated), ACTA2 (6 fold up-regulated), and TPM1 (4 fold up-regulated) (Fig. 5A). The specificity of TGF-β1 induction of SERPINB2 was demonstrated by SB-431542 treatment, which exerted an opposite effect (Fig. 5A). As shown in Fig. 5B, the enhanced OS and AD differentiation was associated with reduced expression of SERPINB2 during the course of differentiation, which reached a minimum (close to zero) during AD differentiation. Further, we interestingly observed by gene expression analysis that SERPINB2 gene is negatively regulated by TGF-β1 and positively upon inhibition of TGF-β1 pathway by SB, and this finding was consistent both during osteoblast and adipocyte differentiation (Fig. s8)
Figure 5.
SERPINB2 is a TGF-β-target gene that is suppressed in human bone marrow stromal (skeletal) stem cells (hBMSC) during osteoblast and adipocyte differentiation. (A) qRT-PCR performed for TGF-β responsive genes: SERPINB2, ACTA2, and TPM2 for controls (CNT), as well as for cells treated with: SB-431542 (SB) and TGF-β1. Expression of each target gene was normalised to GAPDH. Data are shown as the SD of three independent experiments (B) qRT-PCR showing time course of SERPINB2 expression between day (D) 0, D1, D3, D5, and D7 for cells induced with either osteoblast (OS) or adipocyte (AD) induction medium. (C) qRT-PCR of SERPINB2 mRNA expression on day 3 post transfection with SERPINB2-specific or scrambled control siRNA. Data are presented as fold change mRNA expression ± SD from three independent experiments (D) Western blot analysis of SERPINB2 in SERPINB2-siRNA-depleted cells compared to scramble-siRNA transfected control cells (upper panel). B-Actin (ACTB, lower panel) was used as a loading control. (E) Cell viability measured using Alamar blue assay for hBMSCs on days 2, 4, 6, and 8 post transfection with scrambled (SCR) or SERPINB2-specific siRNA. (F) qRT-PCR for ACTA2, and TPM1 expression on day 3 post transfection with SERPINB2-specific or scramble (SCR) siRNA, in the presence or absence of SB-431542. Data are presented as mean fold change in mRNA expression ± SD, from three independent experiments. (G) Western blot analysis for P-c-JUN and P-JNK in SERPINB2-depleted cells compared to scramble transfected control cells (upper panel), whereas B-Actin (ACTB, lower panel) was used as a loading control. ***p < 0.005.
Silencing of SERPINB2 in hBMSC promotes OS and AD differentiation
As osteoblastic and adipocytic differentiation was associated with reduced SERPINB2 expression, we employed a loss-of-function approach to assess the specific role of SERPINB2 in the differentiation processes. siRNA targeting of SERPINB2 successfully down-regulated SERPINB2 gene expression (Fig. 5C) and protein expression as demonstrated by qRT-PCR and Western blot (Fig. 5D), respectively. No significant changes were observed in cell proliferation in SERPINB2-depleted cells as compared to siRNA-treated controls (Fig. 5E). The gene expression of ACTA2 and TPM1, which are TGF-β responsive genes, was up-regulated in SERPINB2-depleted cells (Fig. 5F). Phosphorylated c-Jun and phosphorylated JNK proteins, which are modulators of TGF-β signalling pathways, were found to be induced in SERPINB2-depleted cells (Fig. 5G).
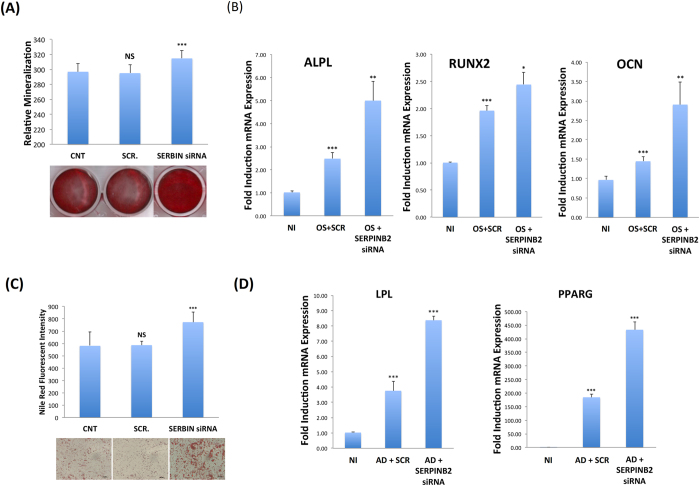
Notably, siRNA-mediated SERPINB2 inhibition led to enhanced OS and AD differentiation as evident by increased mineralised matrix formation (Fig. 6D), up-regulation of specific osteoblastic markers: ALPL, RUNX2, and OCN (Fig. 6E), increased numbers of mature adipocytes (Fig. 6F), and up-regulation of the adipogenic gene markers LPL and PPARG2 (Fig. 6G). These effects on hBMSCs differentiation were further confirmed by using primary human bone-marrow derived MSC, where similar results were obtained (Fig. s9).
Figure 6.
Down-regulation of SERPINB2 promotes osteoblastic and adipocytic differentiation of human bone marrow stromal (skeletal) stem cells (hBMSC) (A) mineralised matrix deposition was assessed using Alizarin Red S staining (lower panel). Quantification of mineralised matrix formation under control (CNT), cells transfected with either transfection with SERPINB2-specific (SERBIN siRNA) or scramble (SCR) siRNA (upper panel). Data are presented as relative mineralisation ± SD from three independent experiments. (B) qRT-PCR quantification of the osteoblastic markers (ALPL, RUNX2, and OCN) on day 7 under the indicated treatment conditions, OS (osteoblast differentiation). Expression of each target gene was normalised to GAPDH. Data are presented as the means ± SD from three independent experiments, hBMSCs under different experimental conditions were induced into adipocytes for 5 days and subsequently stained using Oil Red O (C, lower panel). Data are shown as microscopic images (40×). Nile red stain quantification is shown in the upper panel. Data are presented as mean relative Nile red staining intensity ± SD from three independent experiments. (D) qRT-PCR quantification for the adipogenic markers (PPARG2 and LPL) for hBMSCs under different treatment conditions. AD (adipocyte differentiation). Expression of each target gene was normalised to GAPDH. Data are presented as fold change in mRNA expression ± SD from three independent experiments. *p < 0.05; **p < 0.005, ***p < 0.0005.
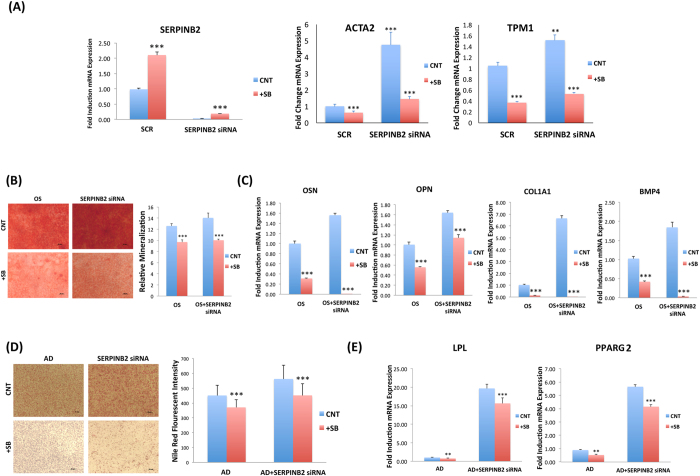
SB-431542 rescued SERPINB2-siRNA-induced enhanced differentiation hMSC
To investigate the effect of induced expression of SERPINB2 on hBMSC differentiation, we conducted rescue experiment using SB-431542 to induce the endogenous SERPINB2. Treating SERPINB2-defecient cells with SB-431542 significantly upregulated SERPINB2 gene expression (>6.0 FC; Fig. 7A). The gene expression of ACTA2 and TPM1, which are TGFB responsive genes, were up-regulated in SERPINB2-depleted cells. Enhanced OS and AD differentiation in SERPINB2-depleted cells were significantly reversed in the presence of SB-431542 as shown by reduced mineralized matrix formation (Fig. 7B) and down-regulation of osteoblastic gene markers: OCN, OPN, COL1a1, and BMP4 (Fig. 7C), and reduced number of mature adipocytes (Fig. 7D) as well as down-regulation of adipocytic gene markers: LPL and PPARG2 (Fig. 7E).
Figure 7.
Inhibition of TGFB signaling reversed the enhanced osteoblastic and adipocytic differentiation observed in SERPINB2-depleted human bone marrow stromal (skeletal) stem cells (hBMSC). (A) q-RT-PCR for SERPINB2, ACTA2, and TPM1 expression on day 3 post transfection with SERPINB2-specific-siRNA or Scramble siRNA (SCR), in the presence or absence of SB-431542 (SB). Data are presented as mean fold change in mRNA expression ± SD, from three independent experiments. (B) Mineralized matrix deposition was determined by Alizarin Red S staining presented as microscopic images (magnification 40X) of stained wells (left panel). Quantification of mineralized matrix formation under different treatment conditions is presented in the right panel, OS (osteoblast differentiation). Data are presented as mean relative mineralization ± SD from three independent experiments; (C) qRT-PCR for OSN, OSP, COL1A1 and BMP4 osteoblastic markers. Expression of each target gene was normalized to GAPDH. Data are presented as mean ± SD from three independent experiments. Cells were induced into adipocyte (AD) for 5 days in the presence or absence of SB-431542 (SB) and stained using Oil Red O (D) as shown in the representative microscopic images (left panel) as well as Nile Red stain quantification (right panel) under different experimental conditions: AD induced of scramble-siRNA (CNT), and adipo-induced SERPINB2-siRNA-depleted cells. (E) qRT-PCR quantification for PPARG2, and LPL adipocytic markers under the indicated treatments. Expression of each target gene was normalized to GAPDH. Data are presented as mean ± SD from three independent experiments. n = 6. **p < 0.005, ***P < 0.0005.
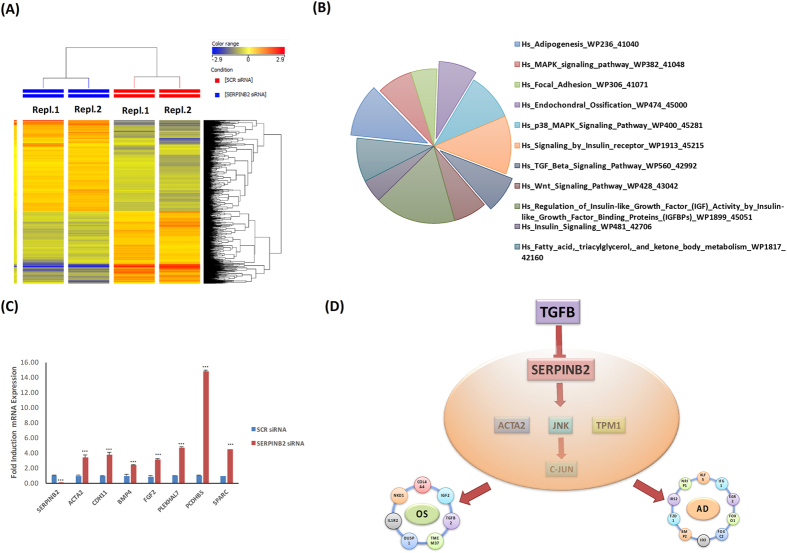
Molecular phenotype of SERPINB2-depleted hBMSCs
Global gene expression profiling was conducted on SERPINB2-depleted hBMSCs and compared with control scramble siRNA transfected cells. Hierarchical clustering based on differentially expressed genes revealed clear separation of control cells from SERPINB2-siRNA transfected cells (Fig. 8A). We identified 836 up-regulated and 670 down-regulated genes in SERPINB2-depleted cells compared to control cells (2.0 FC, p < 0.05; Supplementary Table S7). Pathway analysis performed on the up-regulated genes revealed significant enrichment in several genetic pathways related to ‘endochondral ossification’, ‘adipogenesis’, ‘TGF-β signalling’, ‘WNT signalling’, and ‘MAPK signalling’ pathways (Fig. 8B). Good concordance was observed between microarray and qRT-PCR results for a selected regulated gene panel (Fig. 8C).
Figure 8.
Gene expression profiling of SERPINB2-depleted human bone marrow stromal (skeletal) stem cells (hBMSC). (A) Hierarchical clustering of SERPINB2-depleted hBMSCs compared to scramble transfected control cells, based on differentially expressed mRNA transcripts. Each column represents one replica sample and each row represents a transcript. Expression level of each gene in a single sample is depicted according to the colour scale. (B) Pie chart illustrating the distribution of 11 pathways out of the top pathway designations for the de-regulated genes in SERPINB2-depleted hBMSCs. (C) The expression levels of selected genes from the microarray data were validated using qRT-PCR in SERPINB2-depleted hBMSC. Data are presented as the means ± SD from two independent experiments, n = 6 **p < 0.005; ***p < 0.0005. Scrambled cells were used as a control. (D) Proposed working model illustrating the biological role for TGF-β1 in promoting osteogenesis and adipogenesis through suppression of SERPINB2 and possible down-stream targets.
Discussion
The TGFβ signalling pathway plays a regulatory role in hBMSC biology; however, the molecular details of its action have not been elucidated. In the present study, we investigated the role of TGF-β1 during OS and AD differentiation of hBMSC and described a novel TGF-β1-responsive gene in hBMSC: SERPINB2.
We observed differences in the biological effects of TGF-β1 on hBMSC differentiation dependent on the temporal exposure of the cells. Continuous treatment with TGF-β1 was more effective in enhancing OS differentiation, whereas a single treatment with TGF-β1 during the commitment phase preferentially enhanced AD differentiation. However, these effects were quantitative rather than qualitative and suggest that TGF-β1 is important for both OS and AD differentiation. Moreover, the specificity of the effects was demonstrated by using the TGF-β receptor kinase inhibitor SB-431542.
We observed that TGF-β1 promoted OS differentiation and up-regulation of osteoblastic genes (ALPL, RUNX2, and OSC). Previous studies have also demonstrated that TGF-β1 regulates the differentiation of osteo-progenitor cells7–10, although the direction of such regulation has been disputed. For example, some studies reported the inhibition of OS differentiation in vitro by TGF-β18, 11. In contrast, it has also been reported that TGF-β1 stimulates bone matrix deposition and bone cell replication12, whereas inhibition of TGF-β1 signalling was shown to suppress OS differentiation13. Despite these contradictory data, most studies support a model in which TGF-β1 promotes bone formation by recruiting OS progenitors and inducing the formation of bone matrix at early stages of OS differentiation. However, at later stages of OS differentiation, TGF-β1 appears to inhibit OS differentiation and mineralisation11, 14, 15, probably secondary to regulation by other growth factors e.g. bone morphogenic proteins (BMPs)16, 17. The reported findings of TGF-β1 enhancing the expression of RUNX2 in combination with BMPs during the early stage of OS differentiation18 and its induction of SMAD2 at later stages of OS differentiation that, interacts with RUNX2 causing suppression of the expression of RUNX2 itself and of a consequently other osteoblastic genes including ALPL, collagen type 1, and oxidosqualene cyclase (OSC) by an auto-regulatory feedback mechanism15, 19–23, may explain this dual role of TGF-β1 in OS differentiation. These findings suggest that TGF-β1 effects are dependent on the differentiation stage of the target cells and/or presence of other interacting factors.
Although many studies have reported inhibitory effect of TGF-β1 on adipogenesis, some reports have demonstrated a pro-adipogenic effect of TGF-β124, 25, which is more consistent with the observed TGF-β1 enhancement of bone marrow adipocyte formation observed in the current study. For example, the treatment of mouse NIH/3T3-L1 fibroblasts with TGF-β1 significantly suppressed mature adipocyte formation, irrespective of whether the treatment constituted a single dose at the initiation of differentiation or a continuous exposure26. Continuous exposure to TGF-β1 during adipogenic induction, inhibited adipogenic differentiation of mesenchymal cells, through repression of the function of the key adipogenic transcription factors C/EBPβ and C/EBPδ27. Furthermore, it was demonstrated that continuous exposure to TGF-β inhibited adipogenesis due to significant reduction in TGF-β receptor levels, which was mediated through SMAD2 and SMAD328. In contrast, an early study of brown adipocytes in rat showed up-regulation of the expression of lipogenic enzymes upon treatment with 100 pM TGF-β125. TGF-β1-induced inhibition models used either a low single dose (approximately 2 ng/ml) at the time of differentiation induction or a continuous dose (ranging from 0.2–10 ng/ml). Therefore, the discrepancies observed in different studies might reflect different cell types employed and experimental setups.
Among the genes highly regulated by TGF-β1 treatment, we identified SERPINB2 as a novel gene supressed by TGF-β induction in hBMSC. The physiological role of SERPINB2 in bone biology has not previously been investigated. Plasminogen activator inhibitor-2 (SERPINB2), also called PAI-2, is a member of the Ov-serpin family of serine protease inhibitors29. It is expressed in particular in monocytes/macrophages, as well as in a wide range of other hematopoietic and non-hematopoietic cells30. SERPINB2 has been shown to bind several intracellular and extracellular proteins, indicating physiological function in both intracellular and extracellular compartments31. SERPINB2 functions as a coagulation factor that inactivates urokinase and as a tissue-type plasminogen activator in the extracellular space and on the cell surface29, 32. SERPINB2 is highly expressed in pregnancy, infection and inflammation32. Interestingly, SERPINB2 was downregulated in co-cultured hMSC with U87-MG (glioblastoma multiformae (GBM) cell line) suggesting a role in stroma-cancer cell interaction that needs to be clarified33. We have clearly shown that SERPINB2 is up-regulated when TGF-βpathway is blocked by SB which lead to inhibition of differention hBMSCs and the opposite was seen when we stimulated TGF-β pathway see Fig. 7.
In conclusion, our results provide additional details to the role of TGF-β intracellular signalling pathway in OS and AD differentiation of hBMSC and identify SERPINB2 as a negative regulator of these effects Fig. 8D illustrates our current working model for the role of TGF-β on hBMSC through the suppression of SERPINB2 gene and we provide a possible list of down-stream targets ACTA2, TPM1, c-JUN, and JNK. Future studies are required to clarify the mechanistic role of SERPINB2 in vivo skeletal biology. SERPINB2 may also serve as a target to develop strategies for regulating bone mass and bone marrow fat.
Methods
Cell Culture
As a model for primary human bone marrow MSCs, the hMSC-TERT cell line was established from normal human bone marrow MSCs though overexpression of the human telomerase reverse transcriptase gene (hTERT)34. hMSC-TERT cells have been extensively characterised as exhibiting a similar cellular and molecular phenotype to primary hBMSCs33. In the current study, we employed a sub-clone derived from hMSC-TERT termed hMSC-TERT-CL1, which has been characterised previously35. For simplicity, we refer to this line as hBMSC throughout the manuscript.
The cells were cultured in Dulbecco’s modified Eagle medium (DMEM) supplemented with 4500 mg/l D-glucose, 4 mM L-glutamine, 110 mg/l sodium pyruvate, 10% foetal bovine serum (FBS), 1× penicillin-streptomycin (Pen-strep), and non-essential amino acids (all purchased from Gibco-Invitrogen, Carlsbad, CA).
In vitro OS differentiation
Cells were grown in standard DMEM growth medium in 6-well plates at 0.3 × 106 cells/ml. When 70–80% confluence was reached, test cells were cultured in DMEM supplemented with OS induction mixture containing 10% FBS, 1% Pen-strep, 50 μg/ml L-ascorbic acid (Wako Chemicals, Neuss, Germany), 10 mM β-glycerophosphate (Sigma, St. Louis, MO), and 10 nM calcitriol (1α, 25-dihydroxyvitamin D3; Sigma), and 10 nM dexamethasone (Sigma). The media were replaced 3 times per week. Cells cultured in standard culture medium were considered as the negative control.
For dose response experiments, cells were seeded and treated at D -2 with 10 ng/ml TGF-β1 for 2 days commitment stage, then differentiation stage was initiated by adding osteoblastic induction medium at D0, while TGF-β1 treatment was continued till day 7 in TGF-β -continuous treated cells.
In vitro AD differentiation
Cells were grown in standard DMEM growth medium in 6-well plates at 0.3 × 106 cells/ml. At 90–100% confluence, cells were switched to DMEM supplemented with adipogenic induction mixture containing 10% FBS, 10% horse serum (Sigma), 1% Pen-strep, 100 nM dexamethasone, 0.45 mM isobutyl methyl xanthine36 (Sigma)], 3 μg/ml insulin (Sigma), and 1 μM rosiglitazone37 (Novo Nordisk, Bagsvaerd, Denmark). The media were replaced 3 times per week. Cells cultured in standard culture medium were considered as the negative control. For dose response experiments, cells were seeded and treated at D -2 with 10 ng/ml TGF-β1 for 2 days commitment stage, then differentiation stage was initiated by adding adipocytic induction medium at D0, while TGF-β1 treatment was continued till day 7 in TGF-β -continuous treated cells.
Cytochemical staining
Alizarin Red S staining for mineralised matrix
The cell layer was washed with phosphate buffered saline (PBS) and then fixed with 4% paraformaldehyde for 15 min at room temperature. After removing the fixative, the cell layer was rinsed in distilled water 3 times and stained using a 2% Alizarin Red S Staining Kit (Cat. No. 0223, ScienCell, Research Laboratories, Carlsbad, CA) for 20–30 min at room temperature. Excess dye was washed off with water 3–5 times; subsequently, the stained cells were kept in water to prevent drying out. Images were acquired using an inverted Zeiss microscope (Thornwood, NY). For quantification of Alizarin Red S staining, plates were air-dried and then the Alizarin Red S dye was eluted in 800 µl acetic acid incubated in each well for 30 min at room temperature as previously described38 and measured using an Epoch spectrophotometer (Bio-Tek, Winooski, VT) at 405 nm.
OsteoImage mineralisation assay
The in vitro-formed mineralised matrix was quantified using the OsteoImage Mineralization Assay Kit (Cat. No. PA-1503, LONZA, Allendale, NJ). Culture medium was removed and cells were washed once with PBS, then fixed with 70% cold ethanol for 20 min. An appropriate amount as recommended by the manufacturer of diluted staining reagent was added and plates were incubated in the dark for 30 min at room temperature. Cells were washed and staining intensity was assessed using a fluorescent plate reader at 492/520 excitation emission wavelengths.
Oil red-O staining for lipid droplets
Mature adipocytes filled with cytoplasmic lipid droplets were visualised by staining with Oil Red-O. After washing with PBS, the cells were fixed in 4% formaldehyde for 10 min at room temperature, then rinsed once with 3% isopropanol and stained for 1 h at room temperature using filtered Oil Red-O staining solution (prepared by dissolving 0.5 g Oil red-O powder in 60% isopropanol). To quantify the mature adipocytes formed, Oil Red O stain was eluted by adding 100% isopropanol to each well and absorbance was measured using an Epoch spectrophotometer at 510 nm.
Nile red fluorescence determination and quantification of mature adipocytes
Stock solution of Nile red (1 mg/ml) in DMSO was prepared and stored at −20 °C protected from light. Staining was performed after fixing the cells using 4% paraformaldehyde (Sigma) for 15 min at room temperature, then washed with PBS. The dye was added directly to the cells (5 μg/ml in PBS) and the cells were incubated for 10 min at room temperature. Fluorescent signal was measured with a SpectraMax/M5 fluorescence spectrophotometer plate reader (Molecular Devices Co., Sunnyvale, CA) using the bottom well-scan mode where nine readings were taken per well using excitation 485 nm and emission 572 nm spectra.
Real time qRT-PCR
Total RNA was extracted from differentiating cells either treated or not treated (control samples) with single dose of TGF-β1 at D5 of induction using a PureLink RNA mini isolation kit (Cat No: 12183018 A, Ambion by Life Technologies, Austin, TX) as recommended by the manufacturer. Total RNA was quantified using a Nanodrop spectrophotometer (Nanodrop 2000, Thermo Scientific, Waltham, MA). cDNA was synthesised from 1 μg of the RNA samples using a High Capacity cDNA Reverse Transcription kit (Applied Biosystems, Foster City, CA) using a Labnet, Multigene thermocycler (Edison, NJ) according to the manufacturer’s instructions. Relative levels of mRNA were determined from cDNA using real time PCR (Applied Biosystem-Real Time PCR Detection System) with a Power SYBR Green PCR kit (Applied Biosystems, UK), or with the TaqMan Universal master Mix II, no UNG (Applied Biosystems, Foster City, CA) according to the manufacturer’s instructions. Following normalisation to the reference gene GAPDH, quantification of gene expression was carried out using a comparative Ct method where ΔCT is the difference between the CT values of the target and reference gene. Primers (Supplementary Tables S8 and S9) were either obtained directly from Applied Biosystems (Foster City, CA) as TaqMan primers or were synthesised by Life Technologies based on previously published primer sequences.
DNA microarray global gene expression profiling
Total RNA was extracted from differentiating cells either treated or not treated (control samples) with single dose of TGF-β1 at D5 of induction using the PureLink RNA mini isolation kit as recommended by the manufacturer. Then, 150 ng total RNA was labelled and hybridised to the Agilent Human SurePrint G3 Human GE 8 × 60 k microarray chip (Agilent Technologies, Santa Clara, CA). All microarray experiments were conducted at the Microarray Core Facility (Stem Cell Unit, King Saud University College of Medicine). Normalisation and data analyses were conducted using GeneSpring GX software (Agilent Technologies). Pathway analysis was conducted using the Single Experiment Pathway analysis feature in GeneSpring 12.0 (Agilent Technologies). A two-fold cut off with p < 0.05 was used.
Western blotting
Whole cell lysates were prepared as previously described38. Soluble proteins were analysed by immunoblotting with antibodies anti-SERPINB2, anti-P-c Jun, and anti-P-UNK (ThermoFisher Scientific, diluted 1:5000), and anti-β-ACTIN (Sigma, A3854, diluted 1:10,000). Reactivity was detected with horseradish peroxidase-conjugated secondary antibodies (Santa-Cruz Biotechnology, Dallas, TX) and Clarity western ECL substrate (Bio-Rad Laboratories, Berkeley, CA) for chemiluminescence using a C-Digit Blot Scanner (LI-COR, Lincoln, NE).
siRNA transfection
For transfection, hBMSC cells in logarithmic growth phase were reverse-transfected with Silencer Select Pre-designed SERPINB2-siRNA (25 nM) (Ambion ID: s10016, s10017, and s10018, Cat. no. 4392420, Thermo Fisher Scientific Life Sciences) using Lipofectamine RNAiMAX Reagent (Invitrogen) plus serum-free Opti-MEM® I medium (Thermo Fisher Scientific Life Sciences) as per the manufacturer recommendation. On day 3 of transfection, the cells were induced into OSs or ADs for an additional 5 days.
Statistical analysis
All results presented are given as the means ± standard deviation (SD) of at least 3 independent experiments, unless indicated otherwise. A student’s t-test was used for testing differences between groups. p-values < 0.05 were considered statistically significant.
Electronic supplementary material
Acknowledgements
The authors would like to extend their sincere appreciation to the Deanship of Scientific Research at King Saud University for its funding of this research through the Research Group Project no RGP-1438-032.
Author Contributions
M.E. conducted part of the experimental part and drafted the manuscript. M.M. conducted some part of the experiments. M.A. did the data analysis and preparation of the figures. R.A.D. supervised part of the work and corrected part of the manuscript draft. S.A. conducted some part of the experiments. Z.A.K. helped in data analysis and corrected part of the manuscript. N.M.A. revised the manuscript. M.A. provided all the facilities and revised the manuscript. MK supervised the experimental work and revised the manuscript. A.M. supervised the work, participated in data analysis and participated in writing and revising the manuscript. All authors read the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-10983-x
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Mona Elsafadi, Email: monasafadi@gmail.com.
Amer Mahmood, Email: ammahmood@ksu.edu.sa.
References
- 1.Tencerova M, Kassem M. The bone marrow-derived stromal cells: commitment and regulation of adipogenesis. Front. Endocrinol. [Lausanne] 2016;7:127. doi: 10.3389/fendo.2016.00127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Watson L, Elliman SJ, Coleman CM. From isolation to implantation: a concise review of mesenchymal stem cell therapy in bone fracture repair. Stem Cell Res. Ther. 2014;5:51. doi: 10.1186/scrt439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ren G, et al. Concise review: mesenchymal stem cells and translational medicine: emerging issues. Stem Cells Transl. Med. 2012;1:51–58. doi: 10.5966/sctm.2011-0019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pfeilschifter J, Bonewald L, Mundy GR. Characterization of the latent transforming growth factor beta complex in bone. J. Bone Miner. Res. 1990;5:49–58. doi: 10.1002/jbmr.5650050109. [DOI] [PubMed] [Google Scholar]
- 5.Sheppard D. Transforming growth factor beta: a central modulator of pulmonary and airway inflammation and fibrosis. Proc. Am. Thorac. Soc. 2006;3:413–417. doi: 10.1513/pats.200601-008AW. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tang Y, et al. TGF-beta1-induced migration of bone mesenchymal stem cells couples bone resorption with formation. Nat. Med. 2009;15:757–765. doi: 10.1038/nm.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Janssens K, ten Dijke P, Janssens S, Van Hul W. Transforming growth factor-beta1 to the bone. Endocr. Rev. 2005;26:743–74. doi: 10.1210/er.2004-0001. [DOI] [PubMed] [Google Scholar]
- 8.Jian H, et al. Smad3-dependent nuclear translocation of beta-catenin is required for TGF-beta1-induced proliferation of bone marrow-derived adult human mesenchymal stem cells. Genes Dev. 2006;20:666–674. doi: 10.1101/gad.1388806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Erlebacher A, Filvaroff EH, Ye JQ, Derynck R. Osteoblastic responses to TGF-beta during bone remodeling. Mol. Biol. Cell. 1998;9:1903–1918. doi: 10.1091/mbc.9.7.1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Filvaroff E, et al. Inhibition of TGF-beta receptor signaling in osteoblasts leads to decreased bone remodeling and increased trabecular bone mass. Development. 1999;126:4267–4279. doi: 10.1242/dev.126.19.4267. [DOI] [PubMed] [Google Scholar]
- 11.Maeda S, Hayashi M, Komiya S, Imamura T, Miyazono K. Endogenous TGF-beta signaling suppresses maturation of osteoblastic mesenchymal cells. EMBO J. 2004;23:552–563. doi: 10.1038/sj.emboj.7600067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hock JM, Canalis E, Centrella M. Transforming growth factor-beta stimulates bone matrix apposition and bone cell replication in cultured fetal rat calvariae. Endocrinology. 1990;126:421–426. doi: 10.1210/endo-126-1-421. [DOI] [PubMed] [Google Scholar]
- 13.Ali D, et al. Epigenetic library screen identifies abexinostat as novel regulator of adipocytic and osteoblastic differentiation of human skeletal (mesenchymal) stem cells. Stem Cells Transl. Med. 2016;5:1036–1047. doi: 10.5966/sctm.2015-0331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Centrella M, McCarthy TL, Canalis E. Transforming growth factor beta is a bifunctional regulator of replication and collagen synthesis in osteoblast-enriched cell cultures from fetal rat bone. J. Biol. Chem. 1987;262:2869–2874. [PubMed] [Google Scholar]
- 15.Alliston, T., Choy, L., Ducy, P., Karsenty, G. & Derynck, R. TGF-beta-induced repression of CBFA1 by Smad3 decreases cbfa1 and osteocalcin expression and inhibits osteoblast differentiation. EMBO J. 20, 2254–2272 (2001). [DOI] [PMC free article] [PubMed]
- 16.Canalis E, Economides AN, Gazzerro E. Bone morphogenetic proteins, their antagonists, and the skeleton. Endocr. Rev. 2003;24:218–235. doi: 10.1210/er.2002-0023. [DOI] [PubMed] [Google Scholar]
- 17.Katagiri T, et al. Bone morphogenetic protein-2 converts the differentiation pathway of C2C12 myoblasts into the osteoblast lineage. J. Cell Biol. 1994;127:1755–1766. doi: 10.1083/jcb.127.6.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Spinella-Jaegle S, et al. Opposite effects of bone morphogenetic protein-2 and transforming growth factor-beta1 on osteoblast differentiation. Bone. 2001;29:323–330. doi: 10.1016/S8756-3282(01)00580-4. [DOI] [PubMed] [Google Scholar]
- 19.Ducy P, Zhang R, Geoffroy V, Ridall AL, Karsenty G. Osf2/Cbfa1: a transcriptional activator of osteoblast differentiation. Cell. 1997;89:747–754. doi: 10.1016/S0092-8674(00)80257-3. [DOI] [PubMed] [Google Scholar]
- 20.Birmingham E, et al. Osteogenic differentiation of mesenchymal stem cells is regulated by osteocyte and osteoblast cells in a simplified bone niche. Eur. Cell Mater. 2012;23:13–27. doi: 10.22203/eCM.v023a02. [DOI] [PubMed] [Google Scholar]
- 21.Runyan CE, Schnaper HW, Poncelet AC. Smad3 and PKCdelta mediate TGF-beta1-induced collagen I expression in human mesangial cells. Am. J. Physiol. Renal Physiol. 2003;285:F413–422. doi: 10.1152/ajprenal.00082.2003. [DOI] [PubMed] [Google Scholar]
- 22.Pan X, Chen Z, Huang R, Yao Y, Ma G. Transforming growth factor β1 induces the expression of collagen type I by DNA methylation in cardiac fibroblasts. PloS One. 2013;8:e60335. doi: 10.1371/journal.pone.0060335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bouvard B, et al. Hypoxia and vitamin D differently contribute to leptin and dickkopf-related protein 2 production in human osteoarthritic subchondral bone osteoblasts. Arthritis Res. Ther. 2014;16:459. doi: 10.1186/s13075-014-0459-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ignotz RA, Massagué J. Type beta transforming growth factor controls the adipogenic differentiation of 3T3 fibroblasts. Proc. Natl. Acad. Sci. USA. 1985;82:8530–8534. doi: 10.1073/pnas.82.24.8530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Teruel T, Valverde AM, Benito M, Lorenzo M. Transforming growth factor beta 1 induces differentiation-specific gene expression in fetal rat brown adipocytes. FEBS Lett. 1995;364:193–197. doi: 10.1016/0014-5793(95)00385-M. [DOI] [PubMed] [Google Scholar]
- 26.Tan JT, et al. Connective tissue growth factor inhibits adipocyte differentiation. Am. J. Physiol. Cell Physiol. 2008;295:C740–751. doi: 10.1152/ajpcell.00333.2007. [DOI] [PubMed] [Google Scholar]
- 27.Choy L, Derynck R. Transforming growth factor-beta inhibits adipocyte differentiation by Smad3 interacting with CCAAT/enhancer-binding protein (C/EBP) and repressing C/EBP transactivation function. J. Biol. Chem. 2003;278:9609–9619. doi: 10.1074/jbc.M212259200. [DOI] [PubMed] [Google Scholar]
- 28.Choy L, Skillington J, Derynck R. Roles of autocrine TGF-beta receptor and Smad signaling in adipocyte differentiation. J. Cell Biol. 2000;149:667–682. doi: 10.1083/jcb.149.3.667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Medcalf RL. Plasminogen activator inhibitor type 2: still an enigmatic serpin but a model for gene regulation. Methods Enzymol. 2011;499:105–134. doi: 10.1016/B978-0-12-386471-0.00006-7. [DOI] [PubMed] [Google Scholar]
- 30.Schroder WA, Major L, Suhrbier A. The role of SerpinB2 in immunity. Crit. Rev. Immunol. 2011;31:15–30. doi: 10.1615/CritRevImmunol.v31.i1.20. [DOI] [PubMed] [Google Scholar]
- 31.Rasmussen HH, et al. Microsequences of 145 proteins recorded in the two-dimensional gel protein database of normal human epidermal keratinocytes. Electrophoresis. 1992;13:960–969. doi: 10.1002/elps.11501301199. [DOI] [PubMed] [Google Scholar]
- 32.Lee JA, Cochran BJ, Lobov S, Ranson M. Forty years later and the role of plasminogen activator inhibitor type 2/SERPINB2 is still an enigma. Semin. Thromb. Hemost. 2011;37:395–407. doi: 10.1055/s-0031-1276589. [DOI] [PubMed] [Google Scholar]
- 33.Human mesenchymal stem cells exploit the immune response mediating chemokines to impact the phenotype of glioblastoma, Motaln H1, Gruden K, Hren M, Schichor C, Primon M, Rotter A, Lah TT [DOI] [PubMed]
- 34.Simonsen JL, et al. Telomerase expression extends the proliferative life-span and maintains the osteogenic potential of human bone marrow stromal cells. Nat. Biotechnol. 2002;20:592–596. doi: 10.1038/nbt0602-592. [DOI] [PubMed] [Google Scholar]
- 35.Elsafadi M, et al. Characterization of cellular and molecular heterogeneity of bone marrow stromal cells. Stem Cells Int. 2016;2016:9378081. doi: 10.1155/2016/9378081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hildebrand A, et al. Interaction of the small interstitial proteoglycans biglycan, decorin and fibromodulin with transforming growth factor beta. Biochem. J. 1994;302:527–534. doi: 10.1042/bj3020527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Serra R, Chang C. TGF-beta signaling in human skeletal and patterning disorders. Birth Defects Res. Part C Embryo Today. 2003;69:333–351. doi: 10.1002/bdrc.10023. [DOI] [PubMed] [Google Scholar]
- 38.Gregory CA, Gunn WG, Peister A, Prockop DJ. An Alizarin red-based assay of mineralization by adherent cells in culture: comparison with cetylpyridinium chloride extraction. Anal. Biochem. 2004;329:77–84. doi: 10.1016/j.ab.2004.02.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.