Abstract
Soybean is a promising biomass resource for generation of second-generation biofuels. Despite the utility of soybean cellulosic biomass and post-processing residues in biofuel generation, there is no comprehensive information available on cell wall loosening and degradation related gene families. In order to achieve enhanced lignocellulosic biomass with softened cell walls and reduced recalcitrance, it is important to identify genes involved in cell wall polymer loosening and degrading. Comprehensive genome-wide analysis of gene families involved in cell wall modifications is an efficient stratagem to find new candidate genes for soybean breeding for expanding biofuel industry. We report the identification of 505 genes distributed among 12 gene families related to cell wall loosening and degradation. 1262 tandem duplication events contributed towards expansion and diversification of studied gene families. We identified 687 Simple Sequence Repeat markers and 5 miRNA families distributed on 316 and 10 genes, respectively. Publically available microarray datasets were used to explore expression potential of identified genes in soybean plant developmental stages, 68 anatomical parts, abiotic and biotic stresses. Co-expression networks revealed transcriptional coordination of different gene families involved in cell wall loosening and degradation process.
Introduction
Soybean (Glycine max (L.) Merrill), a model legume species, is one of the world’s main sources of protein and edible oil1, 2. It occupies first rank as edible oil source in the world with an estimated global production of 336.62 million metric tons by 20173. Being an energy-intensive crop, energy output from soybean must be increased. The utility of soybean molasses along with post-processing residues in ethanol production has already been considered in many countries, especially in Brazil4, 5 and is it has been reported as a promising and viable alternative in terms of usable energy and reduced greenhouse gases as compared to many other crops6, 7. Such a superiority makes soybean a potential alternative resource to provide agro-industrial by-products such as molasses and residual field biomass after oil production5. One ton of a de-oiled soybean yields 190.8 Kg molasses, which upon fermentation can produce 18.4 Kg ethanol8. Such a yield can be improved by increasing amount of polymeric compounds used in biofuel generation by employing functional genomics tools and molecular biology techniques.
Recently, the focus of biofuel industry has been shifted towards second generation bioethanol to overcome the future fuel demands. Second generation biofuel production targets plant biomass which is the most abundant organic raw material9, 10. Most of this raw material come from grasses i.e. sugarcane, maize, and fodder sorghum. None of the current crops fulfills the demands of an ideal biofuel crop, shifting the attention of scientists towards other crops such as legumes i.e. soybean4, 6. However, an optimized and enough bioethanol production from soybean is yet to be achieved. The first step in the production of bioethanol is to convert the biomass into fermentable sugars. Fermentable sugars can be generated by accessing polysaccharides which form plant cell walls11. The lignocellulosic material of plant cell walls is mainly composed of cellulose, hemicellulose, lignin, and pectin12. To aid the conversion of these polysaccharides, it is important to understand the mechanisms involved in cell wall loosening, reassembly and degradation13–15. The assembly and range of potential cross-links between polymers, (cellulose, hemicellulose, and pectins) outlines the cell-wall architecture which enforces several restrictions on its degradability10.
Degradation of cell wall polymers can be achieved by various strategies such as biomass pre-treatment with chemicals and the production of less-recalcitrant cell walls in planta by engineering the genes involved in cell wall reassembly and degradation (CWRD). However, each method comes at a cost. Pre-treatment can interfere with macro- and microfibril associations, resulting in the removal of certain soluble hemicelluloses. On the other hand, engineering plants with less recalcitrant cell walls is an effective strategy however reduced biomass yield, decreased germination frequency with less viable seed production and the unwanted aftermaths. Whatever the case is, it should not pose any serious consequences on plant health by conflicting with principle biological activities of the cell wall. Although some negative after effects have been witnessed in some experiments16 but thanks to recent developments in understanding cell wall synthesis, assembly, loosening, and degradation processes, modification of cell walls has helped genetic engineers to recover the biomass yields17–19. Clearly, a better understanding of the processes underlying the interactions between pathogens and the cell wall will support the development of plants with optimized lignocellulosic characteristics, without negatively affecting disease resistance. However, the recent studies on over-expression of pectin modifying enzymes resulted in increased amount of released sugars20, 21 suggesting that it is essential to understand the mechanistic aspects underlying some natural cell-wall degrading processes. This kind of investigations could be achieved by understanding co-expression networks of gene modules related to cell wall reassembly and degradation22, 23. These networks will in-turn lead towards a more detailed explanation of step-wise cell wall modification and degradation events from cell separation to programmed cell death and lysis of the cell wall polymers10, 24. Ultimately, plants could be engineered with modified cell walls by manipulating the key players which upon activation will give swollen cell walls before harvesting. Such a biomass would be more promising raw material for bioethanol industry.
Plants share several common features in relation to cell wall disassembly and degradation. This involves co-action of several enzymes and a step-by-step procedure which starts with expansion of the cell followed by separation and then hydrolysis of cell wall polymers (particularly the hydrolysis of pectins). Some of the natural cell wall degradation processes depend on cell targeting to start carbohydrate breakdown and then continue degradation to the surrounding cells where first the hemicelluloses are degraded followed by cellulose11. Cell wall loosening involves expansins (EXP), yieldins and xyloglucan endotransglucosylases/hydrolases (XTH)25–27. Expansin proteins provoke cell-wall relaxation by destabilizing hydrogen bonds which in turn ease the action of glycoside hydrolases (GHs) on cellulose13, 25, 28, which further promote slippage of xyloglucans on the cellulose microfibrils surface. The endogenous production and secretion of plant GHs is sequential and it follows spatial and temporal characteristics and is always compatible with architecture of particular cell wall. This in turn recruits enzymes (expansins and pectinases) for separation and expansion and xyloglucanases and cellulases to loosen cell wall. While degradation requires a range of pectin modifying and glycoside hydrolases such as endo-1, 4-b-glucanases, endo-xylanases, glucan1, 3-b-glucosidases, polygalacturonases, b-galactosidases, pectate and pectin lyases (PLs), rhamnogalacturonan I lyases, pectin methyl esterases and pectin acetyl esterases10, 11. Physiological roles of plant GHs has been well reviewed elsewhere29, 30. Theses enzymes act accordingly in a relatively conserved sequence of events in planta during fruit ripening, leaf abscission, pathogen attack, under stress, aerenchyma formation and transport of cell wall storage proteins10. Details of mechanism of cell wall degradation for biofuel has been discussed elsewhere10, 14.
The competitive and simultaneous demand of food and biofuel from soybean is increasing which demands a clear and thorough understanding of mechanisms underlying cell wall degradation and loosening by the application of genomics and computation biology tools. The availability of whole genome sequence, microarrays, co-expression network platforms, proteome and transcriptome data has enabled large-scale investigations in order to understand and examine gene families involved in soybean CWRD. It will greatly help to increase the efficiency of polymer digestibility and scarification and production of less-recalcitrant in planta cell walls. The present study focused on mining publically available soybean genome for identification and comprehensive analysis of gene families involved in soybean cell wall loosening and degradation. Phylogenetic analysis, physical mapping of genes, duplication analysis, synteny analysis and gene co-expression analysis was done to get insight into evolution, functional relationship, and transcriptional association and coordination. To further aid future molecular breeding and biotechnological applications, all the identified genes were subjected to predict the presence of SSR markers and miRNA target sites. Furthermore, publically available microarray datasets for various soybean plant developmental stages, anatomical parts as well as under different biotic and abiotic stresses were analyzed to study the expression potential of all gene families involved in CWRD. Finally, gene co-expression networks were studied to visualize transcriptional coordination within studied gene families.
Results
Identification of CWRD related genes from soybean
Understanding about the gene families related to CWRD in soybean can broaden the utility of soybean as a bioethanol resource. Several gene families are involved in the dynamic process of CWRD. A total of 505 genes were identified from 12 soybean CWRD related gene families (Table 1; Supplementary Table S1; Fig. 1).
Table 1.
Summary of identified cell wall reassembly and degradation related gene families in soybean.
| Gene families involved in reassembly and degradation of cell wall | ||||
|---|---|---|---|---|
| Substrate Category | Gene family | Pfam Domain(s) | CAZy ID | No. of Genes |
| Cell wall loosening | Expansins (EXP) | PF03330, PF01357 | 77 | |
| Yeildins (YLD) | PF00704 | GH18 | 25 | |
| Xyloglucan endotransglucosylases/hydrolases (XTH) | PF00722, PF06955 | GH16 | 58 | |
| Glycoside hydrolases | Endo-1, 4-β-glucanases (EGL) | PF00759 | GH9 | 37 |
| Endo-xylanases (EXL) | PF00331 | GH10 | 11 | |
| Glucan 1, 3- β-glucosidases (GGL) | PF00332, PF07983 | GH17 | 78 | |
| Polygalacturonases (PGL) | PF00295, PF12708 | GH28 | 84 | |
| β -Galactosidases (GL) | PF01301 | GH35 | 41 | |
| Pectin modifying | Pectate and pectin lyases | PF00544, PF04431 | PL1 | 37 |
| Rhamnogalacturonana l lyases | PF06045, PF14686, PF14683 | PL4 | 11 | |
| Pectin methyl esterases (PME) | PF01095 | CE8 | 19 | |
| Pectin acetyl esterases (PAE) | PF03283 | CE13 | 27 | |
| 505 | ||||
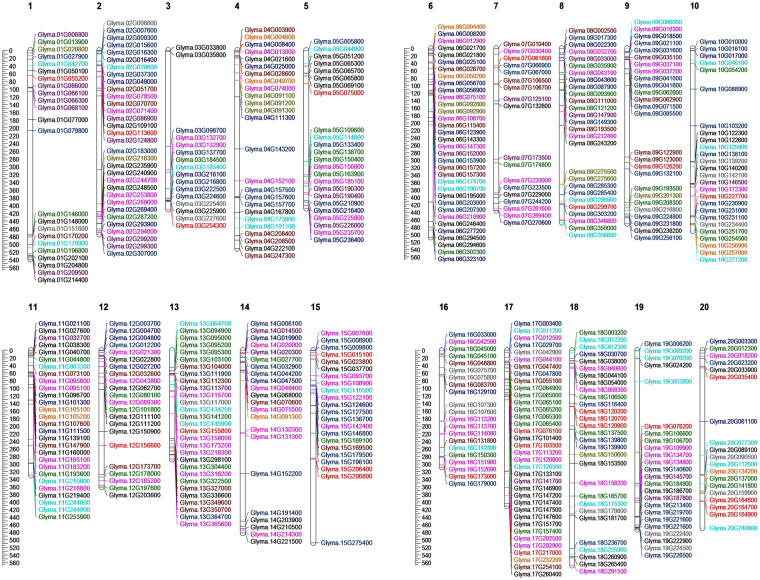
Figure 1.
Map showing the chromosomal locations of cell wall reassembly and degradation related genes in soybean. A scale in left represents length of chromosomes in 100- kilobases. Expansins (black), Yieldins (red), Xyloglucan endotransglucosylases/hydrolases (green), Endo-1, 4-β-glucanases (dark blue), Endo-xylanases (orange), Glucan 1, 3- β-glucosidases (hot pink), Polygalacturonases (central blue), β –Galactosidases (brown), Pectate and pectin lyases (cyan), Rhamnogalacturonana l lyases (olive), Pectin methyl esterases (blue violet), and Pectin acetyl esterases (grey).
Cell wall loosening gene families
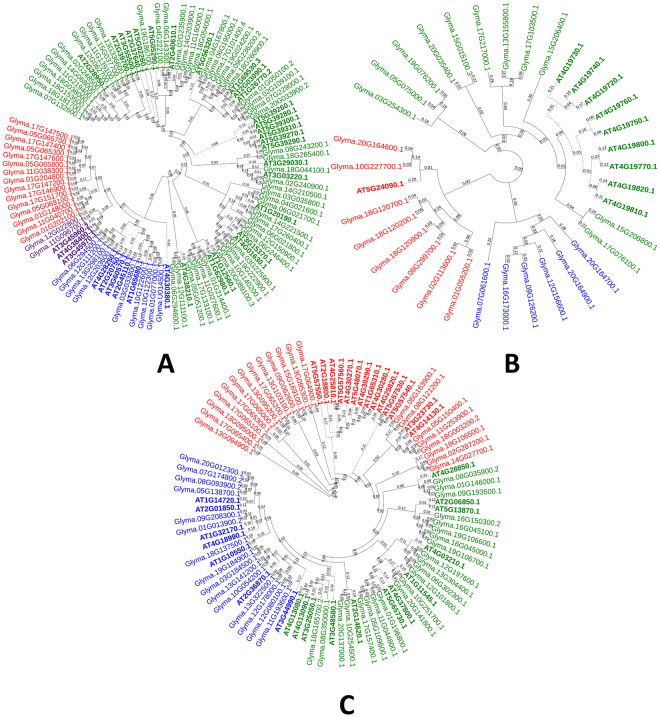
A total of 77, 25 and 58 genes were identified as the members of EXP, yieldins and XTH gene families, respectively. These gene families are principally involved in cell wall loosening (Table 1). The membership of the identified genes was primarily confirmed on the basis of presence of conserved domains i.e. PF03330 and PF01357 for EXPs, PF00704 for yieldins and PF00722 and PF06955 for XTHs. The phylogenetic tree with Arabidopsis EXPs is shown in Fig. 2A. The phylogeny showed four subfamilies i.e. EXPA, EXPB, EXLA and EXLB as previously studied31. Yieldins formed three major clades and the largest clade contained all Arabidopsis yieldins (except At5G24090.1) and grouped with 11 soybean yieldins. 8 soybean yieldins formed a clade with At5G24090.1, while remaining genes clustered into a third separate clade. Physical map revealed their presence on all chromosomes except Chr. 16. XTH family members were clustered in three main phylogenetic groups. All Arabidopsis XTHs and 54 soybean XTHs were grouped in one large phylogenetic group having 12 clades. XTHs showed distinct clustering with those of Arabidopsis (Fig. 2C). This classification was different from that of studied in monocots (rice and sorghum) where the family has been reported to be subdivided into three subfamilies12, 32. Maximum number of XTHs i.e. 7 was found on Chr. 6 (Fig. 1). Additionally, one scaffold gene i.e. Glyma.U014500 was also found in this gene family. Phylogenetic tree of soybean yieldins with those of Arabidopsis showed no distinct clustering pattern (Fig. 2B). Yieldins were distributed on 16 soybean chromosomes except on Chr. 4, 6, 11 and 14. Maximum yieldins i.e. 4 were located on Chr. 20. XTHs were found on all chromosomes except Chr. 4 and the maximum number of 8 XTHs were located on Chr. 13 (Fig. 1).
Figure 2.
Phylogenetic tree of cell wall loosening related gene families in soybean. (A) Expansins, (B) yieldins and (C) Xyloglucan endotransglucosylases/hydrolases. Tree was constructed using Neighbour-Joining method with 500 times bootstrap values. Arabidopsis genes are represented with bold letters and dotted lines.
Glycoside hydrolases
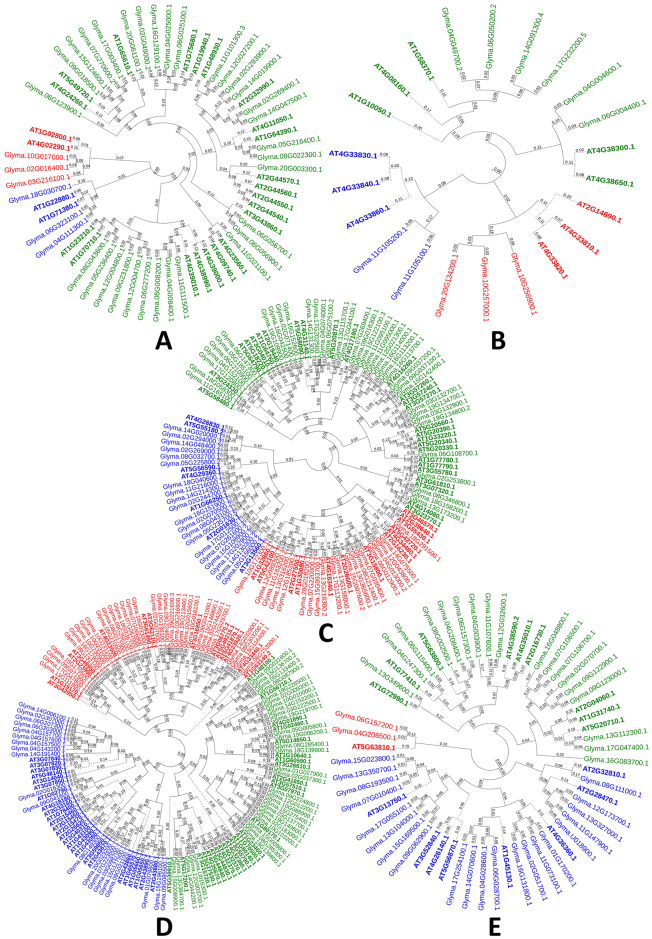
GHs are important class of enzymes involved in the hydrolysis of glycosidic bond between two or more carbohydrates. The cocktail of GHs is used in bioethanol industry in hydrolysis of cell wall polymers after chemical pre-treatment10. In our study we focused on 7 soybean gene families i.e. GH9, GH10, GH16, GH17, GH8 and GH35. Two of GH gene families i.e. GH16 and GH18 are also involved in cell wall loosening process, therefore these families have been described with cell wall loosening gene families. With 84 genes GH28 was the largest gene family followed by GH17 with 78 genes, GH35 with 41 genes, GH9 with 37 genes and GH10 with 11 genes (Table 1; Supplementary Table S1). Based on conserved Pfam domains given in Table 1, the family membership was confirmed. Phylogenetic classification of GH9, GH10, GH17, GH28 and GH35 gene families showed the formation of distinct clade formation with those of Arabidopsis homologues (Fig. 3A–E). Soybean GH9s were divided into three subfamilies with largest subfamily containing 31 genes. GH10 gene family members grouped into three clusters. GH17 family of soybean was divided into three clusters and in each cluster there was unique clustering of soybean GH17s with Arabidopsis homologues. Among GH28s, three phylogenetic groups were formed which further subdivided into 4, 3 and 5 clusters, respectively. Soybean GH35 gene family members grouped into three main phylogenetic groups which subdivided into 1, 3 and 3 clades, respectively. Physical map of GH9 family showed the distribution of GH9 members 17 chromosomes except Chr. 1, 13 and 19, GH10 genes on Chr. 4, 6, 10, 11, 14, 17 and 20, GH17 genes on all chromosomes except Chr. 1, GH28 genes on all chromosomes except Chr. 11, and GH35 genes on 14 chromosomes except on Chr. 3, 5, 10, 18, 19 and 20.
Figure 3.
Phylogenetic tree representing five glycoside hydrolase gene families. (A) Endo-1, 4-β-glucanases, (B) Endo-xylanases, (C) Glucan 1, 3- β-glucosidases, (D) Polygalacturonases, and (E) β –Galactosidases. Tree was constructed using Neighbour-Joining method with 500 times bootstrap values. Arabidopsis genes are represented with bold letters and dotted lines.
Pectin modifying gene families
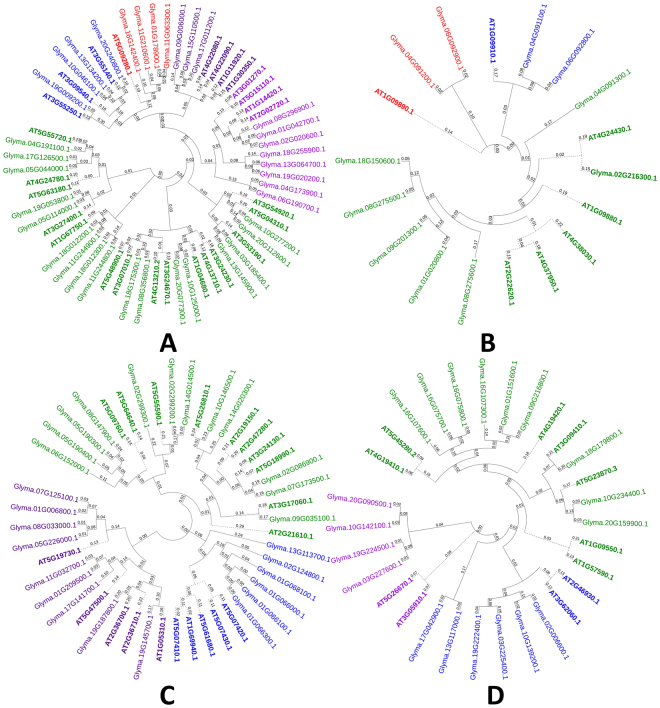
The first hurdle to hydrolysis is the selective cell-wall accessibility to GHs. This porosity feature is determined mainly by the pectin domain. To digest cell wall polymers for efficient ethanol generation, it is important to modify pectin through the action of pectin modifying enzymes33. A total of 94 gene members belonging to four pectin modifying gene families i.e. PL1, PL4, PME, and PAE were identified. Among pectin-related lyases (PLs) we found 37 and 11 genes for P1 and PL4 gene families, respectively. The membership of the identified genes to the respective families was confirmed by validating the presence of Pfam domain i.e. PF00544 and PF04431 for PL1 and PF06045, PF14686 and PF14683 for PL4 (Table 1). Phylogenetic analysis of PL1 grouped the genes into three groups with the largest containing 23 genes divided into five clades. PL4s showed their distribution into 3 main phylogenetic groups, third being the largest with 9 genes (Fig. 4A,B). Chromosomal distribution of members of PL1 gene family showed presence on all chromosomes except on Chr. 7, 12 and 14, while PL4 members were distributed on Chr. 1, 2, 4, 6, 8, 9 and 18.
Figure 4.
Phylogenetic tree representing pectin modifying gene families in soybean. (A) Pectate and pectin lyases, (B) Rhamnogalacturonana l lyases, (C) Pectin methyl esterases, and (D) Pectin acetyl esterases. Trees were constructed using Neighbour-Joining method with 500 times bootstrap values. Arabidopsis genes are represented with bold letters and dotted lines.
The other two gene families i.e. PME and PAE, belonged to pectin esterases. We identified 19 PMEs and 27 PAEs in soybean based on their similarity with Arabidopsis pectin esterases and presence of conserved domains i.e. PF01095 and PF03283, respectively. The NJ trees showed that PAE and PME family members form distinct clades with Arabidopsis PMEs and were divided into three phylogenetic groups each (Fig. 4C,D).
Gene structure analysis
Gene structure provides essential information related to gene function and conservation. Chromosomal locations, peptide sizes, protein molecular weights and isoelectric points for all gene families are given in Supplementary Table S1. To gain insight of evolution of CWRD gene families the intron/exon diversification was studied by knowing the exon/intron boundaries and intron phases. Among the cell wall loosening gene families maximum no. of introns was 5, 6 and 4 in EXP, yieldins and XTH, respectively. EXP subfamilies were characterized by similar intron/exon organization as reported elsewhere31, 34. However, among the whole EXP family, no conserved exons/intron was observed (Supplementary Table S2). Members of EXP gene family were found in phase 1; 2; 1, 2; and 0, 1, 2 implying that either during the course of evolution some member genes have experienced either gain or loss of introns, resulting into evolution of subfamilies. Interestingly, among analyzed gene families only yieldins showed intronless genes (8 genes). Because the ratio of the intronless gene to genes with introns was 8: 17 so it is possibly due to intron-loss. Furthermore, only three soybean yieldins had introns in phase 0, 1, 2 while all other were in different phases i.e. 0, 1 or 2. Average peptide length of EXPs, yieldins and XTHs was 256, 372 and 297 amino acids (Supplementary Table S2). Among GHs, exons length conservation was observed within subfamilies/phylogenetic groups of each gene family. GH35 family members showed a higher number of conserved exons within the family and subfamilies/phylogenetic groups. This might be because of longer gene sizes with a higher number of introns and exons. Maximum no. of introns in GH9, GH10, GH17, GH28 and GH35 (with 1 intronless gene) were 10, 7, 13, 11 and 20, respectively. Most of GH9 family member introns were either in phase 0 or 0, 1 except two genes having introns in 0, 1, 2 phases. Intron phase was not conserved in GH10 and GH17 gene families. On the other hand, GH28 gene family members tended to have an intron in phase 0, 1, 2. Strong conservation of intron phase was observed in GH35 gene family i.e. all introns were in phase 0, 1, 2. Average peptide sizes were 532, 650, 436, 451 and 802 residues for GH9, GH10, GH17, GH28 and GH35 gene families. Pectin modifying genes showed variation in maximum intron number, PL1, PL4, PAE and PME having 6, 16, 16 and 5, respectively. Intron phase was highly conserved (all genes having introns with phase 0, 1, 2) in PAE gene family members while the members of PME gene family also showed intron phase conservation with some variations. Average peptide size of PL1, PL4, PAE and PME members was 412, 642, 422 and 370 amino acids.
Duplication and Synteny analysis
We analyzed all CWRD related gene families for the presence of tandem duplication (TD) in order to reveal any expansion during evolution. Interestingly, out of 505 identified genes, we witnessed 1262 TDs in 401 genes (~80%). Maximum number of TDs (253) were observed in GH28 gene family and minimum number of TDs (17) were found in GH10 gene family (Fig. 5). Among cell wall loosening gene families, EXPs (254 events/55 genes), yieldins (33 events/16 genes) and XTHs (131 evens/45 genes) were observed. In GHs a total of 611 events were observed in 208 genes while in pectin modifying gene families 70 genes experienced 233 TD events (Supplementary Table S3). Chromosome 2 experienced maximum number of duplications i.e. 86 and Chr. 20 experienced minimum duplication events i.e. 32.
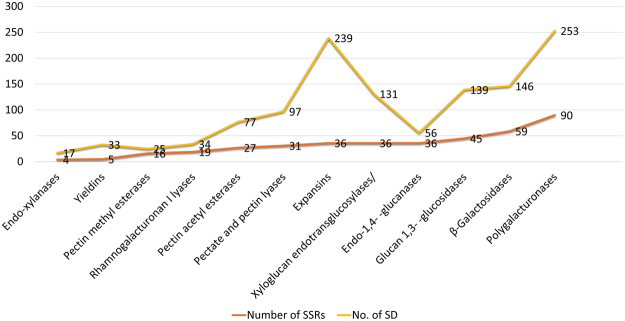
Figure 5.
Details of identified SSR markers and Tandem duplications in 505 cell wall reassembly and degradation related genes of soybean. Gene family wise distribution of identified SSR markers and Tandem duplications.
The estimated time of duplication for all CWRD related gene families ranged from 0 to ~300 mya. The most old duplication events took place in members of GH28 i.e. 300.82 mya followed by PL4 i.e. 296.72 mya. Among the cell wall loosening gene family members, EXPs duplicated around 61.47 to 177.05 mya, yieldins duplicated around 7.37 to 222.95 mya and XTHs duplicated around 4.92 to 282.78 mya. While those of GHs duplicated around 0 to ~300 mya. The pectin modifying gene family PL1 duplicated around 0–280.32 mya, PL4 duplicated around 0–296.72 mya, PAE duplicated around 0–165.57 mya and PME duplicated around 3.27 to 177.86 mya. Synteny within each soybean CWRD gene family members is given in Supplementary Fig. S1. The comparative synteny relationship map revealed a high degree of similarity within soybean CWRD related gene family members.
Simple Sequence Repeat Markers in CWRD related genes in soybean
Microsatellites are efficient molecular markers which are distributed across the eukaryotic genomes2, 15, 35. These are markers of choice mainly because of their genome-wide distribution and ability to detect polymorphism. They are considered an efficient marker system due to taxon-specific variation in motif structure, the frequency of occurrence, and genomic location and are an important tool for marker assisted selection. Such a tool can aid selection of parents for developing soybean lines and mutants for biofuel applications12, 15. Keeping in view their utility in soybean improvement stratagems, simple to score length variations and ease of mining, we analyzed soybean CWRD related gene family members for the presence of SSRs. Out of 505 genes, 316 had 687 SSR markers (Table 2; Supplementary Table S4). Mining for SSR markers resulted in the identification of 189 dinucleotides, 165 trinucleotides, 230 tetranucleotide, 74 pentanucleotides, and 29 hexanucleotide repeats (Table 2). Dinucleotide repeats were the most abundant SSRs in soybean CWRD. We found SSR markers in all 12 studies gene families. Maximum number of SSRs (166) was found in GH28 gene family.
Table 2.
Summary of putative SSRs identified in soybean CWDE.
| Parameter | Details |
|---|---|
| Total number of sequences examined | 505 |
| Total size of examined sequences (bp) | 1981887 |
| Total number of identified SSRs | 687 |
| Number of SSR containing sequences | 316 |
| Number of sequences containing more than 1 SSR | 316 |
| Distribution of different repeat type classes | |
| Dinucleotide repeats | 189 |
| Trinucleotide repeats | 165 |
| Tetranucleotide repeats | 230 |
| Pentanucleotide repeats | 74 |
| Hexanucleotide repeats | 29 |
| Number of Sequences with successful primer pairs | 319 |
| Number of sequences without primer pair picked | 31 |
| Total primer pairs picked | 695 |
Soybean CWRD genes with putative miRNA target sites
MicroRNAs (~21-nt long) regulate gene expression at transcriptional and post-transcriptional levels. Post-transcriptional regulation can contribute considerably to phenotypic modifications during plant development36. Here we explored the putative miRNA target sites in soybean CWRD gene family members. A total of 10 genes belonging to EXP (2), GH9 (4), GH10 (1), and GH17 (3) contained putative miRNA target sites. The BLAST alignment algorithm identified 13 query sequences with high statistical confidence and sequence homology to the short sequences present in the degradome database. On the other hand, 18 predicted miRNA targets didn’t retrieve any significant hits. Overall, our in silico analysis followed by a further validation in an independent dataset identified potential novel miRNA targets in the studied CWRD gene families. (Supplementary Table S5).
Expression potential of soybean CWRD genes
Digital expression in soybean plant development and anatomical parts
Recent developments in bioinformatics has facilitated availability of whole genome, proteome, metabolome and microarray data for major crops. To determine expression potential of CWRD genes in soybean development and 68 anatomical parts we used GENEVESTIGATOR and accessed the spatial expression profiles. The expression potential in five soybean plant developmental stages and different anatomical parts is presented as heat maps for individual gene families to better visualize and understand (Fig. 6; Supplementary Fig. S2). Among cell wall loosening gene families, EXP gene members showed differential expression potential with notable expression of Glyma.07G229000 and Glyma.17G260400 in germination stage. Interestingly one gene i.e. Glyma.20G033900 showed higher expression during bean development stage. All EXPs had notable expression during cell culture and primary cell development. Glyma.20G033900 was highly expressed throughout the anatomical parts of soybean plant except flowering stage. Among yieldins, all genes showed varied and relatively lower expression throughout five developmental stages. Similar expression profile was observed in all 68 anatomical parts. All XTHs showed higher expression potential in germination stage and lower or varied expression in remaining four developmental stages (Fig. 6A). Interestingly, same expression pattern was observed in anatomical parts of seedlings i.e. shoot apix (shoot apical meristem), hypocotyl and radicle (maturation zone and root hair). Glyma.12G197600 and Glyma.13G141200 had 100% expression potential in cotyledon, epidermis (abaxial and adaxial), hilum and endothelium (Supplementary Fig. S2a). The GH gene families GH9, GH17, GH28 and GH35 showed varied expression potential throughout the plant developmental stages. GH9 family members showed differential expression in all 68 anatomical parts with maximum expression potential in seedling (radicle; maturation zone and root hair). GH10 genes showed very low expression throughout the plant anatomical parts except the different compartments of seed where the expression was maximum. GH17s showed varied but relatively higher expression throughout plant anatomical parts except in inflorescence and flower compartments and leaf. GH28s had varied expression potential in all anatomical parts of soybean plant. Among GH35s, Glyma.04G003900 showed 100% expression potential in inflorescence (raceme, flower, androecium, stamen, anther and pollen). Glyma.13G10400, Glyma.13G349600, Glyma.14G070600, Glyma.17G254100 and Glyma.U018900 had higher expression in seed (testa, outer integument, palisade layer, hourglass-cell layer, inner-integument and parenchyma) (Supplementary Fig. S2b).
Figure 6.
Expression potential of soybean CWRD genes in soybean plant development. (A) Digital expression potential of cell wall loosening genes, (B) digital expression potential of glycoside hydrolases, and (C) digital expression potential of pectin modifying genes in five soybean plant developmental stages i.e. germination, main shoot growth, flowering, fruit formation and bean development.
Pectin modifying enzymes showed consistent but lower expression throughout five soybean plant development stages. Two PL1 genes i.e. Glyma.13G64700 and Glyma.20G240800 showed 100% expression potential in shoot development stage (Fig. 6C). Among PL1s, two genes i.e. Glyma.05G04400 and Glyma.19G09200 had 100% expression potential in cell culture and primary cell development (leaf cell, mesophyll cell; paraveinal mesophyll cell and palisade parenchyma cell). Interestingly, two PL1s (Glyma.13G064700 and Glyma.20G240800) showed maximum expression in inflorescence related anatomical parts. All pectin modifying enzyme related genes showed maximum expression during hourglass-cell layer of seed testa (Supplementary Fig. S2c).
Digital expression under abiotic stress
Plants exposed to abiotic stresses display morphological changes that in turn modify cell wall such as decrease in thickness and increased level of cell wall bound phenolics37–39. We determined expression potential of CWRD related gene family members under heat stress and salt stress. It was notable that expression of EXPs was same in plants with 20% and 30% photosynthesis inhibition (at 42 °C) and 22 °C as compared to control. However, four EXPs (Glyma.01G05100, Glyma.02G109100, Glyma.19G024200 and Glyma.20G033900) was relatively higher than other members of the same gene family (Supplementary Fig. S3a). Yieldins also displayed no difference in expression potential under heat stress and control conditions. Two XTH members i.e. Glyma.09G062600 and Glyma.15G169100 showed increased expression potential under heath stress (42 °C) in comparison to controls. Among the GH families, GH9 (except Glyma.4G025000, and Glyma.9G018500), GH17 (except Glyma.11G095000, Glyma.11G095100, Glyma.12G021300, and Glyma.14G020000), GH28 (except Glyma.15G127500 and Glyma.18G116400) and GH35 (except Glyma.08G1000, Glyma.09G062900 and Glyma.U018900) had similar expression pattern under heat stress and control (Supplementary Fig. S3a). Among pectin modifying gene families, PL4 and PME gene members showed no variations in expression potential however, members of PAE and PL1 showed variations in expression under heat stress and control. Notable expression potential was observed in Glyma.19G009200 (PL1) and Glyma.10G142100 and Glyma.16G107300 (PAE) (Supplementary Fig. S3a).
In Alkaline stress of 50 Mm NaHCO3 for 3, 6 and 12 hours, EXP gene family members showed an increase in expression potential with respect to time of exposure. A similar pattern was observed in yieldins and XTHs (Supplementary Fig. S3b). Among GHs, GH9s especially Glyma.02G269400 and Glyma.04G025000 showed higher expression potential when exposed to 3 and 6 hours alkaline stress as compared to 12 hours exposure time. GH10s showed very low but increasing expression pattern in response to increasing exposure time. GH17 gene family members expressed at higher levels when the exposure time was increased to 12 hours. GH28 and GH35 family member genes showed expression potential similar to those of XTHs (Supplementary Fig. S3b). Among pectin modifying enzymes, PL1 and PL2 and PME showed increasing expression potential in response to increasing exposure time while the members of PAE expressed at very higher levels during 3 and 6 hours exposure time as compared to 12 hours (Supplementary Fig. S3b).
Digital expression under biotic stress
The dynamic structure of cell wall determines the outcome of the plant-pathogen interactions40. The microbial pathogens exploit cell wall reassembly and degradation machinery of the host to facilitate pathogenesis. In order to understand the response of CWRD related gene family members again pathogen attack, we used Affymetrix data41 and https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc = GSE7124) during Bradyrhizobium japonicum and Phytophthora sojae infestations, respectively. All CWRD related gene family members showed increased expression potential as compared to control (Supplementary Fig. S3c) in B. japonicum infected plants. Interestingly, EXPs, XTHs, and GH9s showed maximum expression potential (up to 100%). Among GH gene families, lowest expression potentials were observed in gene members of GH10 while among pectin modifying gene families, PL4 had the lowest expression potential. Interestingly, the expression pattern of genes of CWRD families was higher in early infection period i.e. 6 and 12 hours and either decreased or remained unchanged with increased infection period (18 to 24 hours) (Supplementary Fig. S3c). The expression pattern of CWRD genes under the infection of P. sojae is given in Supplementary Fig. S3d. Among the cell wall loosening gene families, the expression potential was similar. It was notable that expression potential increased when the infection period was increased from 72 to 120 hours. All gene family members of GHs showed higher expression under prolonged infection time as compared to control. Among pectin modifying enzymes, the expression of PL2 and PME gene family members was lowest but comparable among treatments. PL1 and PAE gene families members were highly expressed under prolonged infection period as compared to lower infection time and control (Supplementary Fig. S3d).
Co-expression networks of soybean CWRD gene families
Availability of postgenomic technologies supported with genome sequences and expression data has made it easier to understand how biological systems work by analyzing co-expression networks of functionally related genes which are transcriptionally coordinated22. To broaden our understanding of transcriptional coordination of soybean CWRD related gene families, we developed co-expression networks of individual gene families by using PlaNet platform which uses available Affymetrix GeneChip microarray datasets (Supplementary Fig. S4 and Supplementary Table S6).
Co-expression network of EXP gene family showed 54 nodes involved in 36 biological processes, two cellular compartments, and 11 molecular functions. Most importantly, it contained nodes related to biological processes in response to different biotic and abiotic stimuli, regulation of cellular process, plant cell wall organization, and cell wall polysaccharide metabolic process etc. (Supplementary Table S6b). Among CWRD related gene families, PME and GH28 gene members had co-expression with EXPs (Supplementary Fig. S4a). Thirty-four nodes co-expressed with yieldins (query gene glyma03g41510). The co-expressed genes were involved in 14 biological processes and 14 molecular functions. All the co-expressed nodes belonged to different gene families involved in multiple biological processes but none was the member of CWRD related gene family (Supplementary Fig. S4b). Fifty nodes co-expressed with XTH gene family member glyma09g34140 which were involved in 13 biological processes with notably in cellular glucan metabolic process. The co-expressed genes were found to be involved in 10 molecular functions in five cellular compartments (Supplementary Fig. S4c, Supplementary Table S6a,b).
Among GHs, GH9 (query gene glyma04g02740; glyma06g02760) was co-expressed with 163 nodes involved in 21 different biological processes having 44 different molecular functions within 27 cellular compartments (Supplementary Fig. S4d, Supplementary Table S6a). Among molecular functions, microtubule motor activity, structural constituent of the cytoskeleton, cellulose synthase activity and lyase activity were notable (Supplementary Table S6b). PL1, GH17, GH28, GH35, XTHs, PAE, PME, cellulose synthase and COBRA gene families were observed in co-expression network of GH9 gene family (Supplementary Table S6a). GH10 showed co-expression with members of yieldins gene family (Supplementary Fig. S4e). Co-expression network of GH17 family member showed that GH28 had some functional correlation (Supplementary Fig. S4f). Similarly, GH28 and GH35 gene family members showed co-expression with PL1, GH35, PAE, and PAE (Supplementary Fig. S4g and Fig. 3H).
Among pectin modifying enzymes, co-expression networks revealed that GH9, PL1, PAE, and GH28 (Supplementary Fig. S4i–k). The biological processes, cellular components and molecular functions of co-expressed genes are given in Supplementary Table S6b. The annotations and labels of co-expressed genes are given in Supplementary Table S6a. Notably, Tubulin Tubulin_C and cellulose synthases were found in co-expression network of most of CWRD related gene families (Supplementary Table S6a).
Discussion
Among the key processes involved in second generation biofuel production, cell wall hydrolysis is an essential step, which demands identification and characterization of enzymes dealing with the cell wall loosening, hydrolysis, and degradation. The possible two ways to find possible candidates related to CWRD enzymes are either identify genes and proteins in different microorganisms which they use to break plant cell walls during pathogen attack or by controlling in planta mechanisms involved in CWRD12. It has been suggested previously that controlling early and over production of GHs and pectin modifying enzymes within plants will greatly aid the step-wise cell wall degradation process10. Successful production of biofuels from plant cell wall polymers calls for a comprehensive understanding of the genes and their transcriptional coordination. Soybean has been proposed as a resourceful biofuel crop6, however, a comprehensive report on CWRD related genes was missing in soybean. Identification of genes and their distribution will greatly comprehend our understanding about the natural processes involved in CWRD in soybean. Such a knowledge will help to improve the efficiency of degradation of cell walls and reduction of recalcitrance by modifying biomass composition. On the long run, such an inventory of genes would augment the plant breeders and biotechnologists to design and engineer plants having the ability to change or modify their cell walls in order to produce endogenous biological pre-treatments. In this study, 505 genes belonging to 12 CWRD related gene families have been identified and characterized in silico.
The identified genes were distributed in to 3 groups i.e. cell wall loosening genes families, GHs and pectin modifying gene families (Table 1). Conserved domain analysis and phylogeny constructed with Arabidopsis CWRD proteins suggested that these proteins are conserved (Figs 1, 2 and 3) and greatly supported by other studies11, 34. Gene structure analysis supported with intron or exon boundaries and intron phases greatly help to identify conserved exons and clarify the conservation or divergence of gene structure and functions within gene families42. High conservation of intron phases in GH35 and PAE gene families suggests the possibility of shuffling of symmetrical exons between the same phase-introns. The correlation of intron-exon boundaries with three dimensional structure of proteins has also been proposed43. Relatively higher number of CWRD related genes in soybean is most probably due to its late linage-specific duplication. Two rounds of whole genome duplications, shuffling of symmetric exons among introns having same phases further clarifies the expansion of CWRD gene families of soybean34, 44. Incredibly larger number of TD i.e. 1262 duplication events (~80%), greatly contributed towards expansion and diversification and implies that majority of studied genes are duplicated. This further suggests the expansion of these gene families has been mainly achieved by the retention of gene copies after two rounds of whole genome duplications. This expansion feature has been previously reported in EXP gene family of soybean, Arachis duranensis and Arachis ipaensis 31, 34
Such gene retentions and diversifications of genes copies in soybean CWRD gene families further suggests sub-functionalization of these gene families, which may result in different cell wall compositions. This evolutionary process may also support neofunctionalization as already established in cell wall compositional differences in eudicots and grasses45. Higher numbers of CWRD genes in soybean as compared to Arabidopsis advocates a selective advantage to preserve these multiple copies after a series of duplication. Abundance of CWRD genes were distributed on all 20 soybean chromosomes forming small clusters of same gene family members. Such clusters could be considered as hotspot targets for breeders and molecular biologists to engineer soybean for in planta loosening of cell walls which is strongly needed during pre-treatment of molasses for bioethanol production6, 10, 11.
Co-dominance, genome-wide distribution, ease of handling and detection and multi-allelic nature makes SSR markers a good candidate to support marker assisted breeding, map-based cloning and detection of specific genes15, 46. Engineering and breeding plants for higher biomass production and or in planta cell wall loosening could be accelerated by identifying molecular markers linked with the CWRD related genes. Such a strategy has been successfully used in alfalfa where marker-assisted selected was carried out to identify QTLs for enhanced biomass yield and cell wall compositional variations (Li et al. 2011). Identification of soybean CWRD related SSRs with greatly comprehends the selection of suitable cultivars or breeding lines for enhanced biomass production for the biofuel industry. Furthermore, these makers would help to screen and identify mutants46. In silico SSR identification along with a genome-wide analysis of CWRD related genes has been successfully adopted elsewhere12, 47. The identified SSR markers in our study showed a higher proportion of tetranucleotide repeats which is in line with our previous report related to cellulose synthase gene superfamily in soybean15 as well as reported in coffee48 (Table 2). Identification of cell wall-related genes based on molecular markers has also been successfully done in Arabidopsis, rice49, 50. Additional to SSRs, our study identified the presence of putative miRNA target sites (Supplementary Table S5). These miRNAs are one of the major types of endogenous non-coding RNAs involved in post-transcriptional/translational modifications in gene expression. The role of miRNAs during cell wall biogenesis has been confirmed in plants51. Shen et al.52 characterized miRNAs in response to the pathogenic attack of Verticillium longisporum in alfalfa. The expression is obviously related to cell defense structure i.e. cell wall. The identified families of miRNAs in our studies has been previously reported to be involved in different plant defence mechanism related to cell wall response to abiotic and biotic stress responses in soybean and other plants53–56. These studies greatly support that the expression these miRNAs greatly effect plant cell wall composition under different stimuli. Such a knowledge of miRNA target sites in uncharacterized CWRD related genes will increase our understanding of post-transcriptional/translational modifications.
Cell wall compositional variations accompanied by varied gene expression levels has been reported in different plant tissues29, 57. Expression potential analysis of CWRD related genes provided essential knowledge to determine spatiotemporal role of these genes and will help to decide at which plant developmental stage it could yield higher biomass. A similar approach was reported in sorghum11. We determined expression potential of soybean CWRD related genes at five plant developmental stages and 68 anatomical parts (Fig. 6 and Supplementary Fig. S1). Which in turn highlighted many tissue/anatomical part specific genes involved in CWRD, these could be potential candidates to engineer these tissues/anatomical parts for achieving higher biomass yields. A recent study on expression patterns of pectin, cellulose, cell, wall, lignin and fatty acid related genes was done in different soybean tissues5. In our studies, varied expression potential of all members of 12 studies gene families was observed in five soybean plant developmental stages. Relatively higher expression potential was observed at seedling stage for all gene families suggesting extensive cell wall modification during soybean germination stage occur (Fig. 6). During later four plant growth stages, all CWRD related genes were expressed. A more detailed expression potential analysis of studied gene families of soybean revealed that among the studied gene family groups GHs had relatively higher expression potential (Supplementary Fig. S1b). This could shed light upon spatiotemporal expression of GHs relative to other plant CWRD related gene families, especially those involved in cell wall loosening. Plant cell wall growth is resulted simultaneously by synthesis of cell wall polysaccharides as well as the expansion of existing polysaccharides29 as observed by Ohmiya et al.58 while studying members of GH9. Among cell wall loosening gene families higher expression potential of EXPs and XTHs in cell culture, shoot apical meristem, radicle and maturation zone during germination stages and endosperm, testa and integuments at pod elongation stages suggests their roles in the shoot and pod elongation (Supplementary Fig. S1a). A similar correlation of expression of a soybean EXP gene i.e. GmEXP1 with root elongation was observed59. A detailed review on mechanism and action of different enzymes was recently published by Cosgrove60 who explained about the cell wall role of EXPs. Higher expression of PMEs and PAEs in growing anatomical parts such as seedling (radicle, root hairs, and meristems), cotyledon, testa (hourglass-cell layer and parenchyma cells) and primary roots were in line with previous studies61, 62 (Supplementary Fig. S1c).
It is evident from the genetic studies that analogous gene family members (e.g., XTH, EXPs, and PLs) are frequently involved in the response to different stresses63. However, the intrinsic complexity of the cell wall and a large number of genes involved in its biosynthesis, assembly, loosening, reassembly, and degradation means that many details remain unclear regarding the genetic and biochemical basis for cell wall responses to stress64 Meides et al. 2014). Plants respond to abiotic stresses by various morphological, physiological and biochemical mechanisms65–67. The improvement of genomics and proteomics technologies with the availability of massive publically available data. Many studies have compared cell wall modification under abiotic stress with controls5, 64, 68. Higher expression potential of EXPs and XTHs under heat stress in our studies is strongly in line with the previously established fact that XTHs act as tethering polymers between cellulose fibrils64, 68–70. EXPs and XTHs showing higher expression potential could be strong candidates for engineering soybean for abiotic stress tolerance as well as for in planta cell wall loosening (Supplementary Fig. S2a). A soybean EXP gene i.e. GmEXPB2 was characterized and against abiotic stresses and was involved in root system architecture39. Similarly, those GHs which showed higher expression potential can be selected for breeding soybean with higher abiotic stress tolerance because GHs cover a broader range of molecular functions and also interact with plants hormones for their activation and inactivation29, 68. PAEs showed higher expression potentials in heat stress conditions as compared to controls (Supplementary Fig. S2a). Statistically significant variations in expression of PAEs, PMEs and PLs in barley caryopses exposed to heat stress have been previously reported71, 72. The change in expression potential of CWRD genes in response to salt stress is in line with previous reports in Arabidopsis, tobacco, and Jatropha73–76. Under salt stress conditions, the upregulation of XTHs and GHs was witnessed in Arabidopsis 77. Thus, many abiotic stress conditions lead to an altered response of CWRD related genes64.
Remodeling of cell wall under continuous threat of pathogen attack is a key response of plants78. The dynamic structure of cell wall determines the outcome of the plant-pathogen interactions40. The increased expression potential of soybean CWRD genes was observed in our in silico digital expression analysis by using available Affymetrix microarray data (Supplementary Fig. S2c,d). Previous transcriptome profiling studies revealed that 182 cell wall-related genes were differentially regulated in response to B. japonicum infection in soybean79. Decreased susceptibility to different pathogenic infections was observed by overexpressing PMEs in Arabidopsis, wheat, tomato, Populus, strawberry, broccoli and pepper80. Indeed most of CWRD genes especially GHs play significant roles in plant defense mechanisms and cell wall modification in response to pathogen attack29. The notable higher expression potential of XTHs in response to B. japonicum and P. sojae infection presented many potential candidates for engineering cell wall for biotic stresses as well as for bioethanol production.
Recent developments in bioinformatics and availability of genomics and proteomics data have enabled to explore more about the networks of genes regulating plant cell wall modification, loosening, degradation and hydrolysis63. Previously, co-expression networks of cell wall synthesis related genes have been explored in Arabidopsis, rice, and soybean15, 22, 81. We explored co-expressed networks of soybean CWRD genes to facilitate identification of genes for larger biomass production and in planta cell wall loosening (Supplementary Fig. S3a–k). This, in turn, shed light on correlated molecular functions in different biological processes related to cell wall synthesis, loosening, degradation. Such studies will aid in vitro and in vivo studies target to identify co-expressed genes under biotic and abiotic stresses as recently done in Arabidopsis 82. The transcriptional coordination of EXPs with PMEs and member of GH28 gene family is of great interest and could be the foundation of future studies targeted to investigate their coordination. The co-expression of cellulose synthases with all gene families is previously explored in many studies as discussed by Houston et al.63 and Miedes et al.78. The combined role of PLs with EXPs and GHs has already been under discussion and many reports showed the interaction of XTHs, EXPs with pectin modifying gene family members63.
Methods
Data retrieval and identification of CWRD related gene families
For the identification and analysis of soybean CWRD related gene families, publically available genomic, cds and peptide sequences were downloaded from Phytozome 11 database83 (https://phytozome.jgi.doe.gov/pz/portal.html) by conducting a BLASTp 2.2.28+ search. BLASTp was performed by using Arabidopsis CWRD related gene family members as queries to retrieve soybean CWRD related gene family members with an E-value of 10−5. All identified putative CWRD related proteins were further confirmed the presence of family specific conserved domains using NCBI’s Conserved Domain Database (CDD); (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) and EMBL InterProScan84 (https://www.ebi.ac.uk/interpro/). After confirming the CDDs for all CWRD related putative proteins, we further obtain gene IDs, functional annotation, chromosome locations, chromosome number, genomic coordinates and peptide sizes from Phytozome database and used for further analysis. Protein molecular weight and pI-values for each identified proteins were calculated using ExPASy online tool Compute pI/MW85 (http://web.expasy.org/compute_pi/).
Phylogeny, gene structure analysis, and physical mapping
Protein sequences of individual Arabidopsis and soybean CWRD gene families were aligned in ClustalW program of MEGA 7.086 to construct respective phylogenetic trees using neighbour-joining (NJ) method with a bootstrap value of 500 iterations. All positions containing gaps and missing data were excluded in order to achieve phylogenetic trees. Finally, the trees were visualized and managed in iTOL87 (http://itol.embl.de/). To generate a physical map of CWRD related gene member on soybean chromosomes, we used chromosome numbers and genomic coordinates in Mapchart 2.3088. An intron-exon map of all individual gene families was generated in accordance with previous reports. Gene Structure Display Server89 (GSDS; (http://gsds.cbi.pku.edu.cn/) was used to determine intron phase and number of introns and exons. The exon-intron boundaries were determined by employing Analyze feature of PhytoMine (https://phytozome.jgi.doe.gov/phytomine/).
Duplication and Synteny analysis
All identified members of CWRD related gene families were subjected to duplication analysis within the genome using Plant Genome Duplication Database59 (PGDD); http://chibba.agtec.uga.edu/). To avoid saturation anchors of >1.0 identity were rejected within a 100-kb range. Approximate dates of duplication events were calculated according to the method described by Lynch and Conery90. Briefly Ka (ratio of nonsynonymous substitutions per nonsynonymous sites) and Ks (synonymous substitutions per synonymous site) were retrieved from PGDD and were then used to calculate estimated duplication date in million years ago (mya) based on a rate of 6.1 × 10 − 9 substitutions per site per year, we calculated the divergence time (T) as T = Ks/(2 × 6.1 × 10 − 9) × 10 − 6 Mya87. Next, to identify syntenic regions within each soybean CWRD related gene family, we used Circoletto91 (http://tools.bat.infspire.org/circoletto/).
In silico SSR Markers and miRNA target sites prediction
Genomic sequences of 505 soybean CWRD related gene members were subjected to identification of SSRs in MIcroSAtellite identification tool (MISA(, http://pgrc.ipk-gatersleben.de/misa/misa.html)). We included only those repeats which had stretches of a minimum of five repeats. We searched for repeat units for di, tri, tetra, penta and hexa nucleotides and the maximum distance between two markers was set to 100. To confirm the number and exact motifs for each SSR in all CWRD related genes and design primers, we further used BatchPrimer3 v1.0 (http://batchprimer3.bioinformatics.ucdavis.edu/). To detect the presence of putative miRNA target sites, the CDS sequences of CWRD related gene families of soybean were analyzed using psRNATarget server92 (http://plantgrn.noble.org/psRNATarget/) with default parameters. To confirm the presence of predicted miRNA target sites, we utilized the degradome data set of Song et al.93 to construct a custom reference database for the multiple sequence alignment using the BLAST algorithm version 2.6.0 on a local standalone command line-based environment94. First, we formatted the reference database using the BLAST embedded function of makeblastdb then the query sequences were aligned using the blastn function optimized for short sequences. The statistical significance was pre-set to E-value < 10.
Digital Expression Analysis in soybean plant development, anatomical parts, biotic and abiotic stresses
Publically available microarray data was used to determine the expression of CWRD gene families of soybean in five soybean developmental stages, i.e. germination, main shoot growth, flowering, fruit formation and bean development and 68 anatomical parts. Heat maps were generated in GENEVESTIGATOR (https://genevestigator.com/gv/). We also performed digital expression profiling of CWRD gene families in soybean root hair cell in response to B. japonicum inoculation41, and P. sojae infection for 72 h and 120 h (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE7124), in soybean roots under 50 mM NaHCO3 treatment for 3 h, 6 h, and 12 h95 and under heat shock treatment from 22 °C to 42 °C96.
Co-expression networks of soybean CWRD related gene families
Co-expression networks of soybean CWRD related gene families were developed by using PlaNet22, 97 (http://aranet.mpimp-golm.mpg.de/index.html). To visualize functional association between gene families related to CWRD in soybean, we used FamNet database built in PlaNet (http://aranet.mpimp-golm.mpg.de/famnet.html). Briefly, Pfam labels of each gene family were used as a query to access the label specific networks and then a single probeset was select to redirect to co-expression network and ontology analysis. The networks containing all nodes supported by ELA and all genes two steps away from the query gene were drawn and exported along with the tables containing annotations and labels of genes found in co-expression networks98. We considered gene networks of a single query gene having maximum co-occurrences in order to expand our understanding about co-expressed CWRD gene families of soybean.
Data availability statement
All data generated or analyzed during this study are included in this published article (and its Supplementary Information files).
Electronic supplementary material
Acknowledgements
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (NRF-2015R1D1A1A09060925). Authors acknowledge guidance provided by Professor Chan Ting Fung and Alan Lin from School of Life Sciences, Chinese University of Hong Kong regarding synteny analysis. We also acknowledge help provided by Ali Mohamad Moustafa from Department of Medical Biochemistry and Cell Biology, University of Gothenburg for miRNA prediction.
Author Contributions
M.A.N., J.D.L., and S.H.Y. laid out the study. H.M.R. and M.I. retrieved the data and F.S.B. prepared figures. M.A.N. wrote the manuscript. G.C. and S.I.L. supervised the study. All authors reviewed the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-11495-4
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Soo In Lee, Email: silee@korea.kr.
Gyuhwa Chung, Email: chung@chonnam.ac.kr.
References
- 1.Chung G, Singh RJ. Broadening the genetic base of soybean: A multidisciplinary approach. Crit. Rev. Plant Sci. 2008;27:295–341. doi: 10.1080/07352680802333904. [DOI] [Google Scholar]
- 2.Nawaz, M. A. Genetic diversity and population structure of Korean wild soybean (Glycine soja Sieb. and Zucc) inferred from microsatellite markers. Biochem. Syst. Ecol. 71, 87–96, doi:10.1016/j.bse.2017.02.002 (2017a).
- 3.United States Department of Agriculture. https://www.fas.usda.gov/data/south-korea-oilseeds-and-products-annual-1 (last time accessed on April 11, 2017).
- 4.Letti LAJ, Karp SG, Woiciechowski AL, Soccol CR. Ethanol production from soybean molasses by Zymomonas mobilis. Biomass and Bioenergy. 2012;44:80–86. doi: 10.1016/j.biombioe.2012.04.023. [DOI] [Google Scholar]
- 5.Pestana-Calsa MC, et al. Cell wall, lignin and fatty acid-related transcriptome in soybean: Achieving gene expression patterns for bioenergy legume. Genet. Mol. Biol. 2012;35:322–330. doi: 10.1590/S1415-47572012000200013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hill J, Nelson E, Tilman D, Polasky S, Tiffany D. Environmental, economic, and energetic costs and benefits of biodiesel and ethanol biofuels. Proc. Nat. Acad. Sci. 2006;103:11206–11210. doi: 10.1073/pnas.0604600103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Manuel J. Battle of the Biofuels. Envrion. Heath Perspect. 2007;115:A92–A95. doi: 10.1289/ehp.115-a92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Siqueira PF, et al. Production of bio-ethanol from soybean molasses by Saccharomyces cerevisiae at laboratory, pilot and industrial scales. Bioresour Technol. 2008;99:8156–63. doi: 10.1016/j.biortech.2008.03.037. [DOI] [PubMed] [Google Scholar]
- 9.Rubin EM. Genomics of cellulosic biofuels. Nature. 2008;454:841–845. doi: 10.1038/nature07190. [DOI] [PubMed] [Google Scholar]
- 10.Tavares EQP, De Souza AP, Buckeridge MS. How endogenous plant cell-wall degradation mechanisms can help achieve higher efficiency in saccharification of biomass. J. Exp. Bot. 2015;66:4133–4143. doi: 10.1093/jxb/erv171. [DOI] [PubMed] [Google Scholar]
- 11.Grandis, A., De Souza, A. P., Tavares, E. Q. P., & Buckeridge, M. S. Using natural plant cell wall degradation mechanisms to improve second generation bioethanol. In: McCann, M. C., Buckeridge, M. S., Carpita, N. C., eds. Plants and bioenergy. New York: Springer, 211–230 (2014).
- 12.Rai, K. M. et al. Identification, Characterization, and Expression Analysis of Cell Wall Related Genes in Sorghum bicolor (L.) Moench, a Food, Fodder, and Biofuel Crop. Front. Plant Sci. doi:10.3389/fpls.2016.01287 (2016). [DOI] [PMC free article] [PubMed]
- 13.Cosgrove DJ. Catalysts of plant cell wall loosening. F1000 Faculty Rev. 2016;5:119. doi: 10.12688/f1000research.7180.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kubicek CP, Starr TL, Glass NL. Plant cell wall degrading enzymes and their secretion in plant-pathogenic fungi. Ann. Rev. Phytopath. 2014;52:427–451. doi: 10.1146/annurev-phyto-102313-045831. [DOI] [PubMed] [Google Scholar]
- 15.Nawaz, M. A. et al. Genome and transcriptome-wide analysis of cellulose synthase gene superfamily in soybean. J. Plant Physiol. 215, 163-175, http://dx.doi.org/10.1016/j.jplph.2017.04.009 (2017b). [DOI] [PubMed]
- 16.Bonawitz ND, Chapple C. Can genetic engineering of lignin deposition be accomplished without an unacceptable yield penalty? Curr. Opin. Plant Biol. 2013;24:336–343. doi: 10.1016/j.copbio.2012.11.004. [DOI] [PubMed] [Google Scholar]
- 17.Petersen PD, et al. Engineering of plants with improved properties as biofuels feedstocks by vessel-specific complementation of xylan biosynthesis mutants. Biotechnol. Biofuels. 2012;26:84. doi: 10.1186/1754-6834-5-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang F, et al. Engineering secondary cell wall deposition in plants. Plant Biotechnol. J. 2013;11:325–335. doi: 10.1111/pbi.12016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bonawitz ND, et al. Disruption of mediator rescues the stunted growth of a lignin-deficient Arabidopsis mutant. Nature. 2014;15:376–380. doi: 10.1038/nature13084. [DOI] [PubMed] [Google Scholar]
- 20.Lionetti V, et al. Engineering the cell wall by reducing de-methyl-esterified homogalacturonan improves saccharification of plant tissues for bioconversion. Proc. Nat. Acad. Sci. USA. 2010;107:616–621. doi: 10.1073/pnas.0907549107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Biswal AK, et al. Aspen pectate lyase PtxtPL1-27 mobilizes matrix polysaccharides from woody tissues and improves saccharification yield. Biotechnology for Biofuels. 2014;7:11. doi: 10.1186/1754-6834-7-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mutwil M, et al. PlaNet: combined sequence and expression comparisons across plant networks derived from seven species. Plant Cell. 2011;23:895–910. doi: 10.1105/tpc.111.083667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mutwil M, et al. Transcriptional wiring of cell wall-related genes in Arabidopsis. Mol. Plant. 2009;2:1015–1024. doi: 10.1093/mp/ssp055. [DOI] [PubMed] [Google Scholar]
- 24.Bouranis DL, Chorianopoulou SN, Siyiannis VF, Protonotarios VE, Hawkesford MJ. Lysigenous aerenchyma development in roots-triggers and cross-talks for a cell elimination program. International Journal of Plant Developmental Biology. 2007;1:127–140. [Google Scholar]
- 25.Cosgrove. DJ. Loosening of plant cell walls by expansins. Nature. 2000;407:321–326. doi: 10.1038/35030000. [DOI] [PubMed] [Google Scholar]
- 26.Okamoto-Nakazato A, Takahashi K, Katoh-Semba R, Katou K. Distribution of yieldin, a regulatory protein of the cell wall yield threshold, in etiolated cowpea seedlings. Plant Cell Physiol. 2001;42:952–958. doi: 10.1093/pcp/pce121. [DOI] [PubMed] [Google Scholar]
- 27.Imoto K, Yokoyama R, Nishitani K. Comprehensive approach to genes involved in cell wall modifications in Arabidopsis thaliana. Plant Mol. Biol. 2005;58:177–192. doi: 10.1007/s11103-005-5344-7. [DOI] [PubMed] [Google Scholar]
- 28.Arantes V, Saddler JN. Access to cellulose limits the efficiency of enzymatic hydrolysis: the role of amorphogenesis. Biotechnology for Biofuels. 2010;3:4. doi: 10.1186/1754-6834-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Minic Z. Physiological role of plant glycoside hydrolases. Planta. 2008;227:723. doi: 10.1007/s00425-007-0668-y. [DOI] [PubMed] [Google Scholar]
- 30.Minic Z, Jouanin L. Plant glycoside hydrolases involved in cell wall polysaccharides degradation. Plant Plysiol. Biochem. 2006;44:435–449. doi: 10.1016/j.plaphy.2006.08.001. [DOI] [PubMed] [Google Scholar]
- 31.Zhu Y, et al. Soybean (Glycine max) expansin gene superfamily origins: segmental and tandem duplication events followed by divergent selection among subfamilies. BMC Plant Biol. 2014;14:93. doi: 10.1186/1471-2229-14-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yokoyama R, Nishitani K. Genomic Basis for Cell-Wall Diversity in Plants. A Comparative Approach to Gene Families in Rice and Arabidopsis. Plant Cell Physiol. 2004. 2004;45:1111–1121. doi: 10.1093/pcp/pch151. [DOI] [PubMed] [Google Scholar]
- 33.Baron-Epel O, Gharyal PK, Schindler M. Pectins as mediators of wall porosity in soybean cells. Planta. 1988;175:389–395. doi: 10.1007/BF00396345. [DOI] [PubMed] [Google Scholar]
- 34.Guimaraes, L. A. et al. Genome-wide analysis of expansin superfamily in wild Arachis discloses a stress-responsive expansin-like B gene. Plant Mol. Biol. 1–18 10.1007/s11103-017-0594-8 (2017). [DOI] [PMC free article] [PubMed]
- 35.Nawaz MA, Sadia B, Awan FS, Zia MA, Khan IA. Genetic diversity in Hyper Glucose Oxidase Producing Aspergillus niger UAF mutants by using molecular markers. Int. J. Agri. Biol. 2013;15:362–366. [Google Scholar]
- 36.Sunkar R, Chinnusamy V, Zhu J, Zhu JK. Small RNAs as big players in plant abiotic stress responses and nutrient deprivation. Trends Plant Sci. 2007;12:301–9. doi: 10.1016/j.tplants.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 37.Balducci L, et al. How do drought and warming influence survival and wood traits of Picea mariana saplings? J. Exp. Bot. 2014;4:1–13. doi: 10.1093/jxb/eru431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hura T, et al. An increase in the content of cell wall-bound phenolics correlates with the productivity of triticale under soil drought. J. Plant Physiol. 2012;169:1728–1736. doi: 10.1016/j.jplph.2012.07.012. [DOI] [PubMed] [Google Scholar]
- 39.Guo W, et al. A soybean β-expansin gene GmEXPB2 intrinsically involved in root system architecture responses to abiotic stresses. Plant J. 2011;66:541–552. doi: 10.1111/j.1365-313X.2011.04511.x. [DOI] [PubMed] [Google Scholar]
- 40.Bellincampi D, Cervone F, Lionetti V. Plant cell wall dynamics and wall-related susceptibility in plant pathogen interactions. Fron. Plant. Sci. 2014;5:228. doi: 10.3389/fpls.2014.00228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Libault M, et al. Complete transcriptome of the soybean root hair cell, a single-cell model, and its alteration in response to Bradyrhizobium japonicum infection. Plant Physiol. 2010;152:541–52. doi: 10.1104/pp.109.148379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Betts MJ, Guigo R, Agarwal P, Russell RB. Exon structure conservation despite low sequence similarity: a relic of dramatic events in evolution? EMBO J. 2001;20:5354–5360. doi: 10.1093/emboj/20.19.5354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.De Roos, A. D. G. Conserved intron positions in ancient protein modules. Biol. Direct. 2, doi: 10.1186/1745-6150-2-7 (2007). [DOI] [PMC free article] [PubMed]
- 44.Schmutz J, et al. Genome sequence of the palaeopolyploid soybean. Nature. 2010;463:178–83. doi: 10.1038/nature08670. [DOI] [PubMed] [Google Scholar]
- 45.Sampedro J, Guttman M, Li LC, Cosgrove DJ. Evolutionary divergence of β-expansin structure and function in grasses parallels emergence of distinctive primary cell wall traits. Plant J. 2015;81:108–120. doi: 10.1111/tpj.12715. [DOI] [PubMed] [Google Scholar]
- 46.Mun, J. et al. Distribution of Microsatellites in the Genome of Medicago truncatula: A Resource of Genetic Markers That Integrate Genetic and Physical Maps. 2541–2555 doi:10.1534/genetics.105.054791 (2006). [DOI] [PMC free article] [PubMed]
- 47.Muthamilarasan M, et al. Integrative analysis and expression profiling of secondary cell wall genes in C4 biofuel model Setaria italica reveals targets for lignocellulose bioengineering. Front. PlantSci. 2015;6:965. doi: 10.3389/fpls.2015.00965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hendre, P. S., Phanindranath, R., Annapurna, V., Lalremruata, A., & Aggarwal, R. K. Development of new genomic microsatellite markers from robusta coffee (Coffea canephora Pierre ex A. Froehner) showing broad cross-species transferability and utility in genetic studies 19, doi:10.1186/1471-2229-8-51 (2008). [DOI] [PMC free article] [PubMed]
- 49.Zuo J, et al. KORRIGAN, an Arabidopsis endo-1, 4-β-Glucanase, localizes to the cell plate by polarized targeting and is essential for cytokinesis. Plant Cell. 2000;12:1137–1153. doi: 10.1105/tpc.12.7.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Inukai, Y. et al. Root growth inhibiting, a rice endo-1, 4-β-D-glucanase, regulates cell wall loosening and is essential for root elongation. J. Plant Growth Regul. 31, 373–381. doi:10.1007/s00344-011-9247-3.
- 51.Ong SS, Wickneswari R. Characterization of microRNAs Expressed during Secondary Wall Biosynthesis in Acacia mangium. PLoS ONE. 7. 2012;11:e49662. doi: 10.1371/journal.pone.0049662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shen D, et al. Identification and characterization of microRNAs in oilseed rape (Brassica napus) responsive to infection with the pathogenic fungus Verticillium longisporum using Brassica AA (Brassica rapa) and CC (Brassica oleracea) as reference genomes. New Phytologist. 2014;204:577–594. doi: 10.1111/nph.12934. [DOI] [PubMed] [Google Scholar]
- 53.Xie F, Wang Q, Sun R, Zhang B. Deep sequencing reveals important roles of microRNAs in response to drought and salinity stress in cotton. J. Exp. Bot. 2015;66:789–804. doi: 10.1093/jxb/eru437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yin ZJ, et al. Difference in miRNA expression profiles between two cotton cultivars with distinct salt sensitivity. Mol. Biol. Rep. 2012;39:4961–4970. doi: 10.1007/s11033-011-1292-2. [DOI] [PubMed] [Google Scholar]
- 55.Goettel W, et al. Systems and Evolutionary Characterization of MicroRNAs and Their Underlying Regulatory Networks in Soybean Cotyledons. PLoS ONE. 2014;9(1):e86153. doi: 10.1371/journal.pone.0086153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yan Z, et al. Identification and functional characterization of soybean root hair microRNAs expressed in response to Bradyrhizobium japonicum infection. Plant Biotechnol. J. 2016;14:332–41. doi: 10.1111/pbi.12387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McKinley, B., Rooney, W., Wilkerson, C., & Mullet, J. Dynamics of biomass partitioning, stem gene expression, cell wall biosynthesis, and sucrose accumulation during development of Sorghum bicolor. Plant. J, doi:10.1111/tpj.13269 (2016). [DOI] [PubMed]
- 58.Ohmiya Y, et al. Evidence that endo-1, 4-beta-glucanases act on cellulose in suspension-cultured poplar cells. Plant J. 2000;24:147–158. doi: 10.1046/j.1365-313x.2000.00860.x. [DOI] [PubMed] [Google Scholar]
- 59.Lee DK, Ahn JH, Song SK, Choi YD, Lee JS. Expression of an Expansin Gene Is Correlated with Root Elongation in Soybean. Plant Physiol. 2003;131:985–997. doi: 10.1104/pp.009902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cosgrove DJ. Plant cell wall extensibility: connecting plant cell growth with cell wall structure, mechanics, and the action of wall modifying enzymes. J. Exp. Bot. 2016;67:463–476. doi: 10.1093/jxb/erv511. [DOI] [PubMed] [Google Scholar]
- 61.Micheli F. Pectin methyl esterases: cell wall enzymes with an important role in plant physiology. Trend. Plant Sci. 2001;6:414–419. doi: 10.1016/S1360-1385(01)02045-3. [DOI] [PubMed] [Google Scholar]
- 62.De Souza A, Hull PA, Gille S, Pauly M. Identification and functional characterization of the distinct plant pectin esterases PAE8 and PAE9 and their deletion mutants. Planta. 2014;240:1123–38. doi: 10.1007/s00425-014-2139-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Houston K, Tucker MR, Chowdhury J, Shirley N, Little L. The Plant Cell Wall: A Complex and Dynamic Structure As Revealed by the Responses of Genes under Stress Conditions. Front. Plant Sci. 2016;7:984. doi: 10.3389/fpls.2016.00984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tenhaken R. Cell wall remodeling under abiotic stress. Front. Plant Sci. 2014;5:771. doi: 10.3389/fpls.2014.00771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gall, H. L. et al. Cell Wall Metabolism in Response to Abiotic Stress. Plants. 4, 112–166, doi: 10.3390/plants4010112 (2015). [DOI] [PMC free article] [PubMed]
- 66.Santos AP, et al. Transcription regulation of abiotic stress responses in rice: a combined action of transcription factors and epigenetic mechanisms. OMICS. 2011;15:839–57. doi: 10.1089/omi.2011.0095. [DOI] [PubMed] [Google Scholar]
- 67.Gehan MA, Greenham K, Mockler TC, McClung CR. Transcriptional networks-crops, clocks, and abiotic stress. Curr. Opin. Plant. Biol. 2015;24:39–46. doi: 10.1016/j.pbi.2015.01.004. [DOI] [PubMed] [Google Scholar]
- 68.Komatsu S, Yanagawa Y. Cell wall proteomics of crops. Front. Plant Sci. 2013;4:17. doi: 10.3389/fpls.2013.00017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rose JK, Braam J, Fry SC, Nishitani K. The XTH family of enzymes involved in xyloglucan endotransglucosylation and endohydrolysis: current perspectives and a new unifying nomenclature. Plant Cell Physiol. 2002;43:1421–35. doi: 10.1093/pcp/pcf171. [DOI] [PubMed] [Google Scholar]
- 70.Hayashi T. Xyloglucans in the primary-cell wall. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1989;40:139–168. doi: 10.1146/annurev.pp.40.060189.001035. [DOI] [Google Scholar]
- 71.Gachon CM, Langlois-Meurinne M, Saindrenan P. Plant secondary metabolism glycosyltransferases: the emerging functional analysis. Trends Plant. Sci. 2005;10:542–549. doi: 10.1016/j.tplants.2005.09.007. [DOI] [PubMed] [Google Scholar]
- 72.Mangelsen E, et al. Transcriptome analysis of high-temperature stress in developing barley caryopses: early stress responses and effects on storage compound biosynthesis. Mol. Plant. 2011;4:97–115. doi: 10.1093/mp/ssq058. [DOI] [PubMed] [Google Scholar]
- 73.Lu P, et al. RhEXPA4, a rose expansin gene, modulates leaf growth and confers drought and salt tolerance to Arabidopsis. Planta. 2013;237:1547–1559. doi: 10.1007/s00425-013-1867-3. [DOI] [PubMed] [Google Scholar]
- 74.Han Y, Li A, Li F, Zhao M, Wang W. Characterization of a wheat (Triticum aestivum L.) expansin gene, TaEXPB23, involved in the abiotic stress response and phytohormone regulation. Plant Physiol. Biochem. 2012;54:49–58. doi: 10.1016/j.plaphy.2012.02.007. [DOI] [PubMed] [Google Scholar]
- 75.Kwon YR, et al. Ectopic expression of Expansin3 or Expansinbeta1 causes enhanced hormone and salt stress sensitivity in Arabidopsis. Biotechnol. Lett. 2008;30:1281–1288. doi: 10.1007/s10529-008-9678-5. [DOI] [PubMed] [Google Scholar]
- 76.Zhang L, et al. Global analysis of gene expression profiles in physic nut (Jatropha curcas L.) seedlings exposed to salt stress. PLoS One. 2014;9:e97878. doi: 10.1371/journal.pone.0097878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nishiyama R, et al. Transcriptome analyses of a salt-tolerant cytokinin-deficient mutant reveal differential regulation of salt stress response by cytokinin deficiency. PLoS One. 2012;7(2):e32124. doi: 10.1371/journal.pone.0032124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Miedes E, Vanholme R, Boerjan W, Molina A. The role of the secondary cell wall in plant resistance to pathogens. Front. Plant Sci. 2014;5:358. doi: 10.3389/fpls.2014.00358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Brechenmacher L, et al. Transcription Profiling of Soybean Nodulation by Bradyrhizobium japonicum. MPMI. 2008;21:631–645. doi: 10.1094/MPMI-21-5-0631. [DOI] [PubMed] [Google Scholar]
- 80.Pogorelko G, Lionetti V, Bellincampi D, Zabotina O. Cell wall integrity. Plant Signaling & Behavior. 2013;8:e25435. doi: 10.4161/psb.25435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ruprecht C, et al. Large-scale co-expression approach to dissect secondary cell wall formation across plant species. Front. Plant Sci. 2011;2:23. doi: 10.3389/fpls.2011.00023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Taylor-Teeples M, et al. An Arabidopsis gene regulatory network for secondary cell wall synthesis. Nature. 2015;517:571–5. doi: 10.1038/nature14099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Goodstein, D.M. et al. Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 40 (2012). [DOI] [PMC free article] [PubMed]
- 84.Marchler-Bauer A, et al. CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 2011;39:D225–D229. doi: 10.1093/nar/gkq1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gasteiger, E. et al. “Protein identification and analysis tools on the ExPASy Server,” in The Proteomics Protocols Handbook, ed J.M. Walker (Totowa, N J: Humana Press), 571–607 (2005).
- 86.Stecher G, Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016;33:1870–4. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Letunic I, Bork P. Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics. 2016;23:127–128. doi: 10.1093/bioinformatics/btl529. [DOI] [PubMed] [Google Scholar]
- 88.Voorrips RE. MapChart: software for the graphical presentation of linkage maps and QTLs. J. Hered. 2002;93:77–8. doi: 10.1093/jhered/93.1.77. [DOI] [PubMed] [Google Scholar]
- 89.Hu B, et al. GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics. 2015;31:1296–1297. doi: 10.1093/bioinformatics/btu817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lynch M, Conery JS. The evolutionary fate and consequences of duplicate genes. Science. 2000;290:1151–115. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- 91.Darzentas N. Circoletto: Visualizing sequence similarity with Circos. Bioinformatics. 2010;26:2620–2621. doi: 10.1093/bioinformatics/btq484. [DOI] [PubMed] [Google Scholar]
- 92.Dai X, Zhao PX. psRNATarget: a plant small RNA target analysis server. Nucleic Acids Res. 2011;39:W155–W159. doi: 10.1093/nar/gkr319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Song, Q. X. et al. Identification of miRNAs and their target genes in developing soybean seeds by deep sequencing. BMC Plant Biol. 11, doi:10.1186/1471-2229-11-5 (2011). [DOI] [PMC free article] [PubMed]
- 94.Altschul, S.F. et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nuc. Acid. Res. 25, 3389–3402 (1997). [DOI] [PMC free article] [PubMed]
- 95.Ge Y, et al. Global transcriptome profiling of wild soybean (Glycine soja) roots under NaHCO3 treatment. BMC Plant Biol. 2010;26:153. doi: 10.1186/1471-2229-10-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Weston DJ, et al. Comparative physiology and transcriptional networks underlying the heat shock response in Populus trichocarpa, Arabidopsis thaliana and Glycine max. Plant Cell Environ. 2011;34:1488–506. doi: 10.1111/j.1365-3040.2011.02347.x. [DOI] [PubMed] [Google Scholar]
- 97.Proost, S., & Mutwil, M. PlaNet: Comparative Co-Expression Network Analyses for Plants. Plant Genomics Databases. 1533 of the series Methods in Molecular Biology pp 213–227 (2017). [DOI] [PubMed]
- 98.Ruprecht, C. et al. Phylogenomic analysis of gene co-expression networks reveals the evolution of functional modules. Plant J., 10.1111/tpj.13502 (2017). [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article (and its Supplementary Information files).