Abstract
The APOBEC3 family of cytosine deaminase enzymes are able to restrict replication of retroelements, such as LINE-1. However, each of the seven APOBEC3 enzymes have been reported to act differentially to prevent LINE-1 retrotransposition and the mechanisms of APOBEC3-mediated LINE-1 inhibition has not been well understood. The prevailing view for many years was that APOBEC3-mediated LINE-1 inhibition was deamination-independent and relied on APOBEC3s blocking the LINE-1 reverse transcriptase DNA polymerization or transport of the LINE-1 RNA into the nucleus. However, recently it was shown that APOBEC3A can deaminate cytosine, to form uracil, on transiently exposed single-stranded LINE-1 cDNA and this leads to LINE-1 cDNA degradation. In this study, we confirmed that APOBEC3A is a potent deamination-dependent inhibitor of LINE-1 retrotransposition, but show that in contrast, A3H haplotype II and haplotype V restrict LINE-1 activity using a deamination-independent mechanism. Our study supports the model that different APOBEC3 proteins have evolved to inhibit LINE-1 retrotransposition through distinct mechanisms.
Introduction
Transposable elements are DNA sequences that can move from one location of the genome to another. Approximately 45% of the human genome is recognized as being derived from transposable elements1, 2. Transposable elements can be grouped into two major classes based on whether they mobilize via a DNA intermediate (i.e., DNA transposons) or an RNA intermediate (i.e., retrotransposons)1, 2. DNA transposons, although not being able to mobilize in the current human genome, have played a major role during the evolution of the eukaryotic genome3–6. Retrotransposons transpose through mRNA intermediates followed by reverse transcription, synthesis of double-stranded (ds) DNA, and integration at new genomic locations. Retrotransposons can be divided into two classes: Long Terminal Repeat (LTR) retrotransposons (also known as endogenous retroviruses) and the non-LTR retrotransposons. The replication strategy of endogenous retroviruses resembles those of retroviruses transmitted person to person, except that they remain in one host and are transmitted vertically2. The present day endogenous retroviruses have acquired so many mutations that they cannot produce infectious virus2.
Non-LTR retrotransposons are found throughout the eukaryotes, and examples of non-LTR retrotransposons include LINE-1 (Long Interspersed Nuclear Element-1, L1) and SINE (Short Interspersed Nuclear Element). Human L1s account for ~17% of the genomic DNA with ~500,000 copies identified, and they represent the only autonomous transposable elements that are currently active in humans7, 8. Although the vast majority of L1s are inactive due to accumulated mutations or 5′ truncations, the human genome still contains ~80–100 copies of intact L1s that are retrotransposition competent1, 2, 9, 10. Full-length L1 contains a 5′ UTR, two Open Reading Frames (ORFs) that are separated by a 63 nt inter-ORF region, and a 3′ UTR that ends with a polyadenosine-rich sequence (poly(A) tail)11. Proteins encoded by ORF1 and ORF2 are both required for efficient L1 retrotransposition. ORF1 encodes a nucleic acid binding protein with demonstrated nucleic acid chaperone activity; ORF2 encodes proteins with endonuclease (EN) activities and reverse transcriptase (RT) activities12–14. L1-encoded EN preferentially cleaves ssDNA at an AT-rich consensus sequence (5′ TTTT/A, where “/” dictates the cleavage site). L1 RT is a DNA/RNA dependent DNA polymerase, but lacks RNaseH activity, a trait that distinguishes itself from the retroviral RTs15. As a result, during cDNA synthesis the RNA strand of the resulting L1 RNA/DNA hybrid could be either degraded by a yet unidentified cellular protein or displaced during (+) strand cDNA synthesis16. Cellular RNaseH2 cooperates with flap endonuclease 1 (FEN-1) to remove RNA primers during lagging strand DNA synthesis and serves as a good candidate17. Unlike retroviruses and LTR retrotransposons which undergo reverse transcription in cytoplasm, L1 RNA is reverse-transcribed in the nucleus18, 19. L1 relies on a mechanism termed the Target Primed Reverse Transcription (TPRT) for retrotransposition20.
Despite the positive effects of retroelement mobilizations, such as contributing to genome evolution and gene diversity, excessive transposition events need to be suppressed, as several genetic diseases have been associated with L1-mediated insertional mutagenesis in germline and somatic cells9, 10. The retroelement suppression mechanisms include DNA methylation21, 22, RNA interference23, 24, and cellular DNA repair factors25, 26. Also, studies suggest that several individual proteins are involved in L1 restriction. Examples of these proteins include: Three primer repair Exonuclease 1 (TREX1)27, Zinc finger Antiviral Protein (ZAP)28, RNA helicase Mov1029, RNaseL30, SAM domain and HD domain-containing protein 1(SAMHD1)31, 32, and the APOBEC family of cytosine deaminases33–41.
The APOBEC enzyme family emerged at the origin of vertebrates and have diversified throughout the vertebrate lineage42, 43. The most ancient APOBEC family members include Activation Induced cytidine Deaminase (AID) and APOBEC2 (A2) proteins that are identified in jawless and cartilaginous fish43, 44. AID initiates class-switch recombination and somatic hypermutation, two processes that occur during antibody maturation, by deaminating cytosine in single-stranded (ss)DNA to form promutagenic uracil. The ssDNA is available during transcription and initiates an error-prone DNA repair process that diversifies the antibody genes and enables a specialized recombination to occur for immunoglobulin class switching45. The A2 enzyme is involved in cardiac and skeletal muscle development46, 47. APOBEC4 (A4) and APOBEC5 (A5) then emerged in amphibians, although their functions are yet to be identified48, 49. During tetrapod evolution, duplication of AID led to the emergence of APOBEC1 (A1)3, 48, 50. A1 plays an important role in lipid transport by editing the apolipoprotein B mRNA in intestinal cells by introducing a uracil that results in a stop codon and truncated version of Apolipoprotein B that has a different function than the full length form51. A1 can also deaminate cytosine in ssDNA52. In placental mammals the APOBEC3 (A3) locus evolved and greatly expanded, which is thought to partly be due to the increasing number of retrotransposons in the genome43. While only one A3 gene is present in mice, up to seven A3 genes (A3A, A3B, A3C, A3D, A3F, A3G, A3H) are present in primates43. A3 genes have likely acquired the anti-L1 activities from the Activation Induced Cytidine Deaminase (AICDA)/A2-like gene from which they have evolved. Several studies have demonstrated that in a cell culture system that A3 family members could inhibit a number of mammalian L1s33–41. Since A3 genes are only limited to mammalian lineages, but L1s are far more ancient, dating back to the emergence of vertebrates, it was not surprising to find that APOBEC proteins, such as pre-mammalian AID and reptilian A1 from green anole lizard, are indeed capable of inhibiting L1 retrotransposition36, 53.
A3s have been reported to restrict L1 by both deamination -dependent or -independent mechanisms34, 35, 38, 40, 41, 54–57. For a deamination-dependent mechanism, during the reverse transcription of the L1 RNA, the (-)cDNA gets transiently exposed after RNA template degradation, which renders it susceptible to A3-mediated cytosine to uracil (C → U) deaminations57. The uracils present on L1 cDNA could trigger cDNA degradation through the actions of the host base excision repair pathway. Specifically, cellular uracil DNA N-glycosylase (UNG) excises the uracils from ssDNA, generating abasic sites and apurinic/apyrimidinic endonuclease (APE) produces nicks on the DNA which would induce L1 cDNA degradation. Alternatively, a uracil-containing cDNA may escape the degradation, become replicated to form a dsDNA, and integrate into the host chromosome, but be functionally inactivated through A3 induced C/G → T/A transition mutations that result from the polymerase using uracil as a template. For a deamination-independent mechanism of L1 restriction, the A3s can sequester L1 ribonucleoprotein (RNP) complexes to cytoplasmic compartments such as stress granules or physically interact with L1 RT and inhibit DNA polymerization during TPRT41, 58, 59. There is no clear agreement in the literature as to whether A3s utilize the deamination -dependent or -independent mode for L1 restriction. These ambiguities arose because no G → A mutations on the coding strand, which would indicate the occurrence of C → U deamination on the L1 cDNA, could be detected. However, none of these cell culture studies blocked the action of UNG34–36, 60. Richardson et al., reasoned that the unsuccessful attempts to uncover A3-induced mutations were likely due to the degradation of uracil-containing cDNA intermediates by the UNG and APE-mediated DNA repair proteins57. In their study, using the Uracil Glycosylase Inhibitor (UGI) protein of Bacillus subtilis bacteriophage PBS161, they demonstrated that A3A restricted L1 through a deamination-dependent mechanism, suggesting that the mechanism by which A3 enzymes restricted L1 needed to be readdressed. These data also explained how mammalian L1 sequences analyzed on larger genome-wide scales could show more evidence of cytosine deamination and is in agreement with the known restriction mechanism of several endogenous retroviruses that show a G → A mutation bias62–65.
To address whether the deamination-dependent mechanism of L1 inhibition is unique to A3A or common to other A3s, we examined how A3H restricted L1 retrotransposition in cell culture. There are at least seven A3H haplotypes that exist in humans. Three of the haplotypes are stable and catalytically active (II, V, VII), three are thermodynamically unstable and have no discernable activity (III, IV, VI), and one has an intermediate stability and is catalytically active (I)66–70. Since haplotype II (hap II) and haplotype V (hap V) comprise the majority of the stable haplotypes we examined how they restricted L1 retrotransposition. Previous research has demonstrated that stable A3H could inhibit L1, however, a mechanism was not identified67, 71. In this study, we demonstrate that both A3H hap II and hap V suppress L1 to a similar level but the inhibition of L1 does not involve DNA deamination, in contrast to A3A.
Results
To establish a context with which to interpret our results with A3H, we also included A3A and A3G in our study. These additional A3s were chosen based on previous publications indicating that A3A was a strong inhibitor of L1 retrotransposition and A3G was unable or weakly able to inhibit of L1 retrotransposition34, 39, 40, 54.
Effect of A3A, A3G, and A3H on L1 retrotransposition
To study the influence of A3 proteins on L1 retrotransposition, a pre-established cell line based retrotransposition assay was performed72, 73 (Fig. 1a). To detect retrotransposition events, the 3′ untranslated regions (3′UTRs) of a human L1 retrotransposon vector (pJM101/L1.3) is marked with a neomycin phosphotransferase (neo) reporter cassette, which, when expressed, can convey resistance to G418. The neo reporter cassette has its own promoter and polyadenyation signal, and it consists of an antisense neo gene disrupted by an intron of the γ-globin gene, which is in the same transcriptional orientation as the L1 element (Fig. 1a). Because the neo gene is rendered inactive by the presence of an intron, G418-resistant (G418R) cells will only arise when a transcript initiated from the L1 promoter is spliced, reverse transcribed, its cDNA is re-inserted into the host chromosome, and the neo gene is transcribed from an internal promoter (Fig. 1a). The L1 assay relies on neo expression as a readout and the rise of a G418R colony demonstrates a successful L1 retrotransposition event. Hela cells were utilized in this assay, as these cells contain a low endogenous L1 copy number (~25 copies per cell)38, can accommodate high levels of L1 retrotransposition events from ectopically expressed L1 reporter plasmids73, 74 and do not express endogenous A3A, A3G and A3H proteins33, 38.
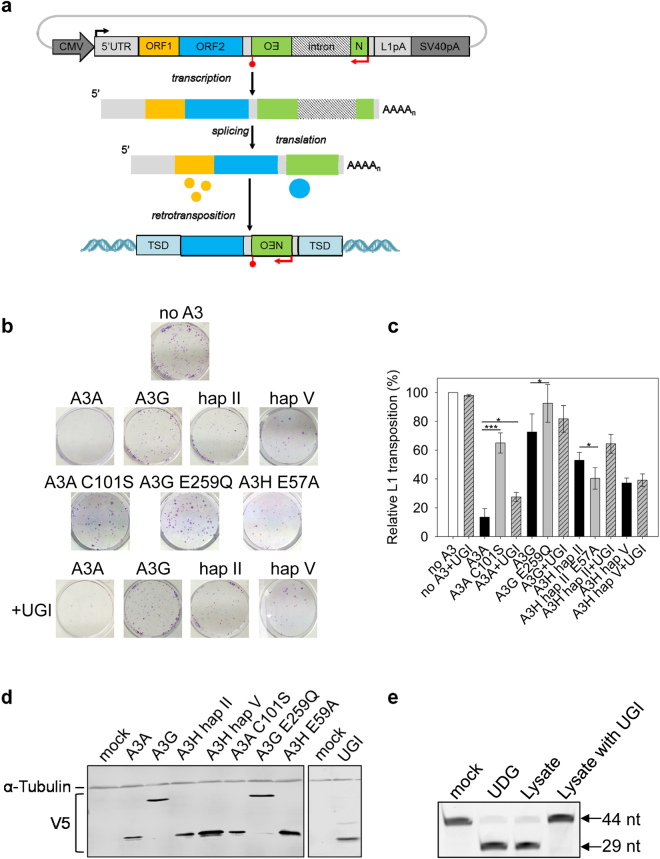
Figure 1.
Restriction of L1 retrotransposition by A3A, A3H, and A3G. (a) Schematic for L1 retrotransposition assay. A full-length retrotransposition-competent L1 (~6 Kb) that contains a neo-reporter cassette (green box) in the 3′UTR region was used. The reporter gene, neomycin phosphotransferase (backwards NEO), is in the opposite transcription orientation with respect to L1. Only upon reverse transcription and integration into a genomic locus can the neo gene be expressed to confer resistance to G418. The retrotransposed L1 is usually 5′ truncated and integration sites are usually flanked by target site duplications (TSD) sequences. (b) Representative experimental results of the neo-based L1 retrotransposition assay. (c) Quantification of the effects of various A3s on L1 transposition efficiencies in the presence or absence of UGI. Results from experiments in the absence of UGI are normalized to L1 with empty vector (no A3). Results from experiments in the presence of UGI are normalized to L1 + UGI. For comparison, on the graph the L1 + UGI is normalized to no A3 to demonstrate that UGI alone does not increase the restriction of L1. Results are the average of at least three independent experiments. Error bars represent the standard deviation. Designations for significant difference of values determined by a T-test were p ≤ 0.001 (***), p ≤ 0.01(**), or p ≤ 0.05 (*). (d) The cellular expression of V5-tagged A3 enzymes and UGI in HeLa cells was confirmed by immunoblotting using anti-V5 antibodies. The α-tubulin expression was used as a loading control and detected in parallel with the V5 tag. Detection of V5 for A3 enzymes and UGI was done on two separate blots. The UGI blot was cropped from a blot containing lanes with additional experimental replicates for conciseness. (e) A cell lysate based assay determined that UGI-V5 expression in HeLa cells inhibited endogenous UNG. From left to right, the lanes show the 44 nt deoxyuridine containing ssDNA oligonucleotide (mock), the ssDNA with commercially available UDG which generates a 29 nt band after uracil removal and heating of ssDNA under alkaline conditions, the ssDNA with HeLa cell lysate, and the ssDNA with HeLa cell lysate from cells that were transiently transfected with UGI.
In order to study the effect of A3 proteins on L1 retrotransposition, HeLa cells were co-transfected with the L1 reporter plasmid pJM101/L1.3 in the presence and absence of plasmids expressing V5-tagged A3s. Three days post transfection, the cells were subjected to G418 selection for 11 days and the G418R colonies were stained and counted. Transfection of L1 vector alone with no A3 expression plasmid generates a quantifiable amount of G418R colonies (Fig. 1b, no A3). In the presence of A3A and A3G, L1 retrotransposition efficiency decreased to 13.5% and 72.6%, respectively (Fig. 1b,c). These data are in agreement with previous studies that demonstrated A3A potently inhibits and A3G does not have a significant inhibitory effect on L1 retrotransposition in cultured cells34, 38, 40, 54. A3H hap II and hap V decreased the L1 retrotransposition efficiency to 53.0% and 37.2%, respectively, demonstrating that the two A3H haplotypes can restrict L1 retrotransposition, but their effect is 3- to 4 -fold less than A3A (Fig. 1b,c).
A3H restricts L1 by a deamination-independent mechanism
We investigated whether the inhibition of L1 by A3H involved deamination by creating a catalytic mutant, E57A that prevents coordination of water with the Zn2+ molecule in the active site75, 76. Similar mutants were made in A3A (C102S, disruption of Zn2+ coordination) and A3G (E259Q, disruption of water coordination) according to the previously published catalytic mutants34, 77, 78. In this experiment we tested only A3H hap II since both hap II and hap V restricted L1 retrotransposition similarly (Fig. 1c). Consistent with previous data, the efficient inhibition of L1 by A3A required deaminase activity, since the retrotransposition frequency in the presence of A3A C102S was 65.0%, a 4.8-fold increase in retrotransposition efficiency from the wild type A3A condition (Fig. 1b,c)57. The L1 retrotransposition efficiency in the presence of A3G E259Q was similar to wild type A3G, and although a statistically significant difference was found, it was only a 1.3-fold change, further confirming that A3G only imposes a minimal effect on L1 (Fig. 1b,c, retrotransposition efficiencies of 72.6% and 92.4%). Similarly, the A3H hap II E57A mutant had a significant, although only 1.3-fold, difference from wild type A3H hap II in restricting L1 retrotransposition, providing evidence that deaminase activity is not required for A3H-mediated L1 restriction (Fig. 1b,c, 53.0% for hap II and 40% for A3H hap II E57A). To exclude the possibility that the observed differences in retrotransposition efficiencies were due to different cellular A3 levels, we used immunoblotting to detect the A3 expression levels in the HeLa cells (Fig. 1d). The wild type and catalytic mutant A3 enzymes were expressed at equivalent levels (Fig. 1d). As a result, these data further confirm that A3A restriction of L1 is deamination-dependent and A3H restriction of L1 is deamination-dependent.
To extend these results we also cotransfected an expression plasmid for UGI into the HeLa cells during the experiment. The UGI is a small 9.5 kDa protein that inhibits UDG/UNG from bacterial and human cells by reversible protein binding at a 1:1 stoichiometry79, 80. The UGI-V5 expressed well in the HeLa cells (Fig. 1d). We also tested the UNG activity by lysing HeLa cells that were or were not transfected with UGI and incubating them with a 44 nt single-stranded DNA oligonucleotide containing a single deoxyuridine (Fig. 1e). The endogenous UNG in the cell lysates was able to completely excise the uracil from the ssDNA and the UGI was able to completely inhibit this process (Fig. 1e). Consistent with a previous study, the inhibition of L1 retrotransposition by A3A was reduced 2-fold when UGI was expressed (Fig. 1b,c, from 13.5% to 27.5%), indicating the L1 inhibition imposed by A3A indeed involves deamination57. Since the UGI was able to completely inhibit the UNG in cell lysates (Fig. 1e), the transient transfection of the UGI likely resulted in incomplete inhibition of UNG due to not every transfected cell receiving all three expression vectors (Fig. 1b,c), which is why we could not recover higher amounts of retrotransposition57. In the presence of A3G, the UGI expression did not cause a significant increase in L1 retrotransposition efficiency (Fig. 1b,c). For A3G, the retrotransposition efficiency in the presence and absence of UGI was 82.8% and 72.6%, respectively (Fig. 1c). For A3H hap II and hap V, the retrotransposition efficiencies in the presence and absence of UGI were also similar (Fig. 1b,c, A3H hap II, 64.4% and 53.0%; A3H hap V, 39.2% and 37.2%).
A3-induced mutagenesis of transposed neo gene
To determine if A3-mediated deaminations in transposed L1 were occurring we amplified a segment of the L1 construct for DNA sequencing. The detection of L1 retrotransposition events in the assay relies on the expression of the spliced neo gene. However, the neo-cassette is located within the 3′ UTR and would remain single-stranded for the longest time during reverse transcription since it is furthest from the priming site for the second DNA strand, which would make this region most vulnerable to mutagenesis. To avoid mutated clones being lost in the G418-resistance selection process, we extracted total cellular DNA three days post-transfection and prior to G418 selections. Because an intron was inserted into the neo gene and a spliced neo gene could be produced only after an L1 integration event (Fig. 1a), we designed primers that annealed to exon regions spanning the intron, enabling amplification of PCR products with different lengths that correspond to the spliced and unspliced products (Fig. 2a). The UGI expression plasmid was also transfected during some of these experiments to promote the recovery of mutations. Several bands were observed in the PCR amplification products even with the primers that were designed to eliminate the amplification of intron sequences (Fig. 2b), in agreement with the previous observation that retrotransposition is a rare event34. Sequencing confirmed the 588 bp species corresponded to the spliced, transposed neo products, whereas the 1496 bp species corresponded to the PCR amplification of the unspliced, full-length neo from the plasmid DNA. Interestingly, during sequence analysis of the spliced neo gene we noticed a high percentage of A → C transversion mutations (Figs 2 and 3). These A → C mutations were persistently observed across different transfection conditions, suggesting that this may be an insertion bias of the L1 reverse transcriptase (Fig. 2 and Fig. 3). Even in the absence of A3 enzymes, G → A transition mutations accounted for the second most frequent mutation type (Fig. 2c, no A3, 0.24 mutations/kb).
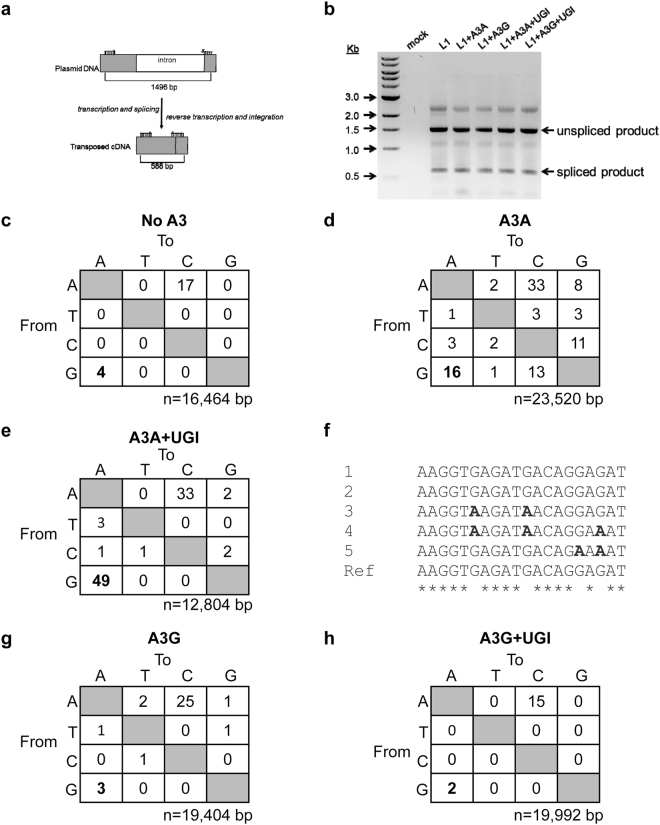
Figure 2.
A3A, but not A3G, promotes G→A mutations in the transposed neo gene. (a) Schematic of the neo-cassette before and after L1 retrotransposition with specific primers and the size of the expected PCR fragments indicated. (b) A representative agarose gel of the PCR products amplified using the primers shown in (a). (c) Sequencing of neo spliced product from cells where no A3 was transfected demonstrated the background G→A mutation frequency was 0.24 mutations/kb. (d) In the presence of A3A, the G→A mutation frequency was 0.68 mutations/kb. (e) The UGI expression increased the A3A G→A mutation frequency to 3.8 mutations/kb. (f) DNA sequencing data from a subset of clones shows the sequence context of G→A mutations induced by A3A. Asterisks denote homology. (g–f) A3G G→A mutation frequencies in the (g) absence (0.16 mutations/kb) and (h) presence (0.10 mutations/kb) of UGI were similar. The value of n is the total number of nucleotides analyzed.
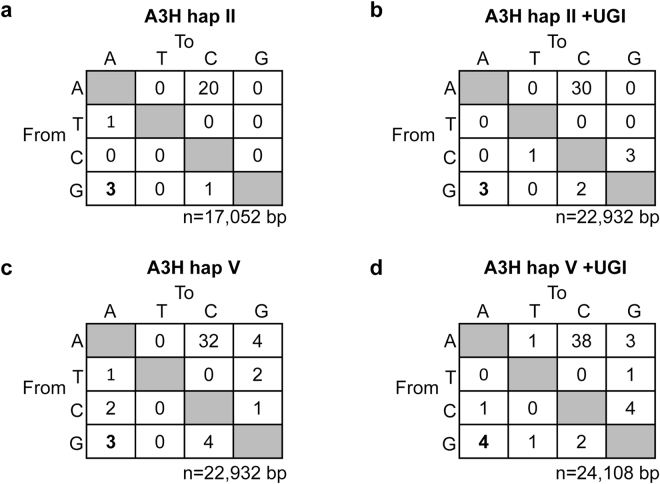
Figure 3.
A3H hap II and hap V do not induce G→A mutations in the transposed neo gene. (a,b) In the (a) absence (0.18 mutations/kb) and (b) presence (0.13 mutations/kb) of UGI, A3H hap II induced a similar G→A mutation frequency. (c,d) Similarly, for A3H hap V, there was no difference in the G→A mutation frequency in the (c) absence (0.13 mutations/kb) or (d) presence (0.16 mutations/kb) of UGI. The value of n is the total number of nucleotides analyzed.
In the presence of A3A, the G → A mutation rate increased ~3-fold to 0.68 mutations/kb (Fig. 2d). When UGI was co-transfected along with L1 and A3A, a ~6-fold increase in G → A mutation rate was observed from A3A alone (Fig. 2e, A3A + UGI, the G → A mutation rate is 3.8 mutations/kb). This suggests that introducing UGI blocked UNG and allowed more G → A mutations to be uncovered, supporting that restriction of L1 retrotransposition is at least in part due to degradation of uracil-containing L1 intermediates. However, since A3A-induced G → A mutations were recovered in the absence of UGI, some L1 intermediates can still escape the degradation resulting from DNA repair enzymes and integrate into the host chromosome. Consistent with A3A ssDNA cytosine deamination activity, very few C → T mutations were observed on the transposed L1 coding strands, excluding the possibility of A3A deaminating L1 mRNA (Fig. 2d,e). Further, sequencing of the unspliced neo gene did not recover any mutations confirming that A3A is not deaminating the transfected plasmid DNA before mRNA splicing (data not shown), although this has been observed under other experimental conditions81, and that indeed the mutations were acquired during integration of the L1 by TPRT, consistent with Richardson et al.57. Because A3A prefers to deaminate C → U within a 5′TC or 5′CC context (underlined C is deaminated)82, 83, we determined whether these G → A mutations on the genomic strand occurred within the preferred deamination motif as a confirmation of A3A deamination activity. Deamination of these motifs in the cDNA will result in 5′GA → 5′AA or 5′GG → AG mutations in the coding strand. Out of the 16 G → A mutations we recovered in the neo gene, two mutations were within the 5′GA → 5′AA context and 1 mutation was within the 5′GG → AG context (data not shown). In addition, other mutations at 5′GC → 5′AC and 5′GT → 5′AT motifs were observed that may or may not be due to A3A catalytic activity (data not shown). However, in the presence of UGI, the majority of the G → A mutations occurred within the 5′GA → 5′AA context, confirming that UGI blocked removal of uracils formed by A3A catalytic activity (Fig. 2f). Consistent with the previous observation that A3G has little inhibitory effect on L1 activity (Fig. 1b,c), a G → A mutation frequency similar to that of the background, was observed in neo gene in the presence and absence of UGI (Fig. 2g,h, G → A mutation frequencies of 0.16 and 0.10 mutations/kb). These data suggest that any minor inhibitory effect of L1 retrotransposition by A3G (Fig. 1c, 72.6%) was not caused by A3G-mediated deamination. Taken together, these data strengthen the findings that A3A inhibits L1 retrotransposition using a deamination-dependent mode, whereas A3G does not seem to affect L1 retrotransposition.
For A3H, we expected that similar levels of G → A mutations would be observed regardless of the presence of UGI since the A3H hap II wild type and E57A restricted L1 retrotransposition similarly (Fig. 1c). Consistent with transposon efficiency data, no enhanced rates of G → A mutations were detected in the neo gene in the presence of A3H hap II (Fig. 3a,b, mutation frequencies of 0.18 mutations/kb and 0.13 mutations/kb). Similar results were found for A3H hap V (Fig. 3c,d). Furthermore, none of the G → A mutations occurred within the preferred 5′GA → 5′AA context, which represents A3H deamination at 5′TC sites (data not shown)66. A3H hap II and hap V also did not appear to be able to deaminate L1 mRNA, since no C → T mutations with the correct sequence context were found (Fig. 3).
Discussion
In this study, we showed that different A3 family members have varying abilities and mechanisms to inhibit the replication of the retrotransposon L1. Specifically, A3A acts as a potent inhibitor for L1 retrotransposition using primarily a deamination-dependent mechanism, A3G does not appear to inhibit L1, and A3H does inhibit L1, consistent with previous studies34, 35, 38, 39, 54, 60. Unique to our study is that we determined A3H hap II and hap V inhibit L1 using a deamination-independent mechanism.
Numerous studies have reported the inhibitory effects of different A3 family members on L1, and in general a deamination-independent L1 restriction model was proposed due to an inability to detect G → A mutations34, 35, 38–40, 54–56. Richardson et al., studied the effects of A3A on L1 retrotransposition in a cell line that stably expressed UGI, and this allowed the recovery of A3A-mediated deamination events in the retrotransposed L1 sequences57. However, our study shows that deamination-dependent L1 restriction is not a common property for all A3 enzymes, since A3H hap II and hap V did not induce G → A mutations, even in the presence of UGI (Fig. 3).
A3A has been demonstrated to be the most potent inhibitor of L1 replication and can restrict L1 retrotransposition frequencies by 75% to 95%, depending on the transfection conditions34, 38, 40, 54, 57. In our system A3A decreased L1 retrotransposition frequency by 86.5% and the catalytic mutant, A3A C101S, could only decrease L1 retrotransposition frequency by 35.0% (Fig. 1b,c), indicating that the deamination-dependent mechanism of A3A contributed the majority of the L1 restriction activity. This is consistent with DNA sequencing where we found that UGI expression alleviated A3A-mediated L1 inhibition by 2-fold (Fig. 1b,c, from 13.5% to 27.5%). These data are also in agreement with Richardson et al.57.
We found A3H hap II and hap V are able to suppress L1 retrotransposition to a moderate level and there were no apparent differences in their L1 restriction abilities (Fig. 1b,c). Three lines of evidence suggest both A3H haplotypes inhibit L1 mobilization in a manner that does not involve deamination. First, the L1 retrotransposition efficiency was the same in the absence or presence of UGI when either A3H hap II or hap V were expressed (Fig. 1b,c). Second, we found that hap II and hap V produced a mutation profile with G → A mutations at the background level either in the absence or presence of UGI (Figs 2c and 3). Third, the A3H hap II catalytic mutant E57A decreased L1 retrotransposition to an equivalent degree as the wild type A3H hap II (Fig. 1b,c). Although there is no prerequisite for a specific cellular localization for an A3 to restrict L1, there does appear to be different mechanism38. For A3A that localizes to the nucleus when ectopically expressed, a deamination-dependent mechanism is involved84 (Fig. 1b,c). However, for A3H hap II and V that are primarily cytoplasmic, a deamination-independent mechanism is involved85 (Fig. 1b,c). Cytoplasmic A3s can inhibit L1 by binding ORF1 in an RNA-dependent manner and preventing the nuclear import of the L1 RNP complex41. In contrast, A3s that locate in nucleus have been reported to inhibit L1’s TPRT process either by deaminating (-)cDNA or physically impair RT’s activity56, 57, 86. Based on these observations and that both A3H Hap II and Hap V are primarily cytoplasmic proteins67, we hypothesize A3H could inhibit L1 mobility by binding to the L1 RNP in the cytoplasm, similar to what has been reported for A3C and A3D41, 59.
The analysis of the L1 restriction ability of A3A, A3G, and A3H hap II and hap V have demonstrated that each of the A3s has a unique effect on L1. In particular, although L1 restriction by A3s can be deamination-dependent, as shown for A3A, this does not appear to be applicable to all A3 enzymes. These data suggest that reanalysis of A3-mediated L1 restriction in the presence of UGI for all A3 family members is warranted to fully understand their mechanism of activity.
Materials and Methods
Plasmids, cloning and site-directed mutagenesis
L1 expression plasmid pJM101/L1.3-Neo was kindly provided by Dr. John V. Moran (University of Michigan). The cDNA of A3A, A3G and A3H have been previously reported82, 87, 88 and were cloned into pcDNA6-V5/His.A using XbaI and XhoI cloning sites giving the enzymes a C-terminal V5-His fusion tag. Mutants of A3 enzymes (A3A C101S, A3G E259Q, A3H E57A) were made using the wild type construct for site-directed mutagenesis (QuickChange site-directed mutagenesis protocol, Agilent). The Uracil DNA glycosylase inhibitor gene was synthesized by GenScript with codon optimization for human cells and subcloned into pcDNA6, but was expressed without the tags for the experiments or subcloned into pcDNA3.1 with a C-terminal V5 tag for checking expression by immunoblotting. All constructed plasmids were verified by sequencing.
Retrotransposition assay
HeLa cells were seeded in each well of a 6-well tissue culture plate at a density of 2 × 105 cells/well in Dulbecco’s Modified Eagle Medium (DMEM) with 10% FBS. Approximately 22 h after plating, Hela cells were co-transfected with 1 μg of L1 retrotransposon encoding plasmid (pJM101/L1.3) and 1 μg of an A3 encoding plasmid or empty vector. Transfections were performed using GeneJuice (EMD Millipore) according to the manufacturer’s protocol. Media was replaced the following day. Approximately 48 h post-transfection, cells were re-seeded into 6-well plates (at 1 in 5 dilution) and the next day attached cells were subjected to selection with 400 μg/mL G418 (Life Technologies). During the reseeding process, cells were checked for viability by trypan blue staining. We consistently observed a cell viability > 95% indicating that expression of A3A, A3G, or A3H did not cause cellular toxicity. G418 selection was carried out for 11 days and the selection media were replaced every other day. Afterwards, G418R colonies were washed twice with PBS, fixed with 2% paraformaldehyde/0.4% glutaraldehyde, and stained with 0.1% crystal violet solution.
Immunoblotting
To detect A3 expression levels, cells were washed in 1x PBS, harvested in Laemmli sample buffer (58 mM Tris (pH 6.8), 5% glycerol, 2% sodium dodecyl sulphate (SDS), 1.5% dithiothreitol (DTT)), and 40 μg of total protein was resolved by SDS-PAGE. Proteins were transferred to a nitrocellulose membrane and A3 or UGI in the cell lysates were probed with primary antibodies (mouse monoclonal V5, 1:1000 (Sigma) and rabbit monoclonal α-tubulin, 1:1000 (Sigma)). After incubation with the secondary antibodies (A3 proteins: IRDye 800-labeled goat anti-mouse; α-tubulin: IRDye 680-labeled goat anti-rabbit) the tagged proteins and loading control was detected simultaneously on the same membrane by using the Licor/Odyssey system.
Nucleic acid extraction and PCR
Hela cells were co-transfected with 1 μg of A3 and L1 plasmids in the presence or absence of 1 μg UGI vector as described above. Cellular DNA from the transfected cells was extracted 72 hr post-transfection using DNAzol reagent (Thermo Fisher). Extracted DNA was used as a template for PCR amplification of the spliced neo gene using the forward primer (G418Neoseq For) 5′-TCGGGAGCGGCGATACCGT-3′ and the reverse primer (G418Neoseq Rev) 5′-CGGTGCCCTGAATGAGCTTCA-3′ with the Q5 High-Fidelity polymerase (New England Biolabs). The PCR cycle used had an initial denaturation step at 98 °C (3 min) followed by 35 cycles of amplification (30 sec at 98 °C, 30 sec at 65.5 °C and 18 sec at 72 °C). PCR products were electrophoresed to separate unspliced (of plasmid origin) and spliced (of integrated L1 origin) PCR products. The spliced product was gel purified with the GenElute Gel Extraction kit (Sigma) and cloned into pJET1.2/blunt vector (CloneJet PCR cloning kit, Thermo Scientific). After transformation into Escherichia coli DH5α cells, clones were picked and sent for sequencing using a kit-specific primer (pJET1.2 Forward) at the National Research Council DNA sequencing Facility (Saskatoon, SK). Sequence alignments were performed using Clustal Omega.
Uracil DNA Glycosylase Inhibitor assay
HeLa cells were transfected with 1 μg of L1 retrotransposon encoding plasmid (pJM101/L1.3) and either 1 μg empty vector or UGI vector as described above. Cells were harvested and lysed 72 h after the transfection in buffer containing 50 mM Tris-Cl pH 7.4, 1% Nonidet-P40, 0.1% sodium deoxycholate, 10% glycerol, 150 mM NaCl, 1 mM DTT, 20 μg/mL RNaseA and EDTA-free protease inhibitor (Roche). To test for UGI activity, 8 μL of the lysate was incubated with 100 nM of a 44 nt ssDNA oligonucleotide with a single deoxyuridine and a 5′ Fluorescein (5′ (Fluorescein)-AAA GAG AAA GTG ATA AAC AAA GAG TAA AGU AGA TAG AGA GTG ATA 3′) for 15 min at 37 °C. To cleave the DNA at abasic sites generated by the action of UNG, the DNA was then heated at 95 °C for 10 min in the presence of 0.2 M NaOH. Purified UDG from E. coli (New England Biolabs) was used as a control. For DNA visualization the samples were mixed with an equal volume of formamide/EDTA loading dye, resolved on a urea denaturing 16% v/v polyacrylamide gel, and scanned using a Typhoon Trio multipurpose scanner (GE Healthcare).
Data Availability
No datasets were generated or analyzed during the current study.
Acknowledgements
We thank Robin P. Love for technical assistance during this study. This work was supported by a Natural Sciences and Engineering Research Council of Canada (NSERC) Discovery Grant (L.C.), NSERC Undergraduate Student Research Award (M.H.G.), and University of Saskatchewan College of Medicine Graduate Research Fellowship (Y.F.).
Author Contributions
Y.F., M.H.G., and T.B.F. conducted experiments. Y.F., M.H.G., T.B.F., and L.C. analyzed data. Y.F. and L.C. wrote the paper.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Richardson SR, et al. The Influence of LINE-1 and SINE Retrotransposons on Mammalian Genomes. Microbiol Spectr. 2015;3:MDNA3-0061-2014. doi: 10.1128/microbiolspec.MDNA3-0061-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Koito A, Ikeda T. Intrinsic immunity against retrotransposons by APOBEC cytidine deaminases. Front Microbiol. 2013;4:28. doi: 10.3389/fmicb.2013.00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Conticello SG, Thomas CJ, Petersen-Mahrt SK, Neuberger MS. Evolution of the AID/APOBEC family of polynucleotide (deoxy)cytidine deaminases. Mol Biol Evol. 2005;22:367–377. doi: 10.1093/molbev/msi026. [DOI] [PubMed] [Google Scholar]
- 4.Feschotte C, Pritham EJ. DNA transposons and the evolution of eukaryotic genomes. Annu Rev Genet. 2007;41:331–368. doi: 10.1146/annurev.genet.40.110405.090448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pace JK, 2nd, Feschotte C. The evolutionary history of human DNA transposons: evidence for intense activity in the primate lineage. Genome Res. 2007;17:422–432. doi: 10.1101/gr.5826307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jarmuz A, et al. An anthropoid-specific locus of orphan C to U RNA-editing enzymes on chromosome 22. Genomics. 2002;79:285–296. doi: 10.1006/geno.2002.6718. [DOI] [PubMed] [Google Scholar]
- 7.Lander ES, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 8.Brouha B, et al. Hot L1s account for the bulk of retrotransposition in the human population. Proc Natl Acad Sci USA. 2003;100:5280–5285. doi: 10.1073/pnas.0831042100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beck CR, Garcia-Perez JL, Badge RM, Moran JV. LINE-1 elements in structural variation and disease. Annu Rev Genomics Hum Genet. 2011;12:187–215. doi: 10.1146/annurev-genom-082509-141802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hancks DC, Kazazian HH., Jr. Active human retrotransposons: variation and disease. Curr Opin Genet Dev. 2012;22:191–203. doi: 10.1016/j.gde.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dombroski BA, Mathias SL, Nanthakumar E, Scott AF, Kazazian HH., Jr. Isolation of an active human transposable element. Science. 1991;254:1805–1808. doi: 10.1126/science.1662412. [DOI] [PubMed] [Google Scholar]
- 12.Feng Q, Moran JV, Kazazian HH, Jr., Boeke JD. Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell. 1996;87:905–916. doi: 10.1016/S0092-8674(00)81997-2. [DOI] [PubMed] [Google Scholar]
- 13.Mathias SL, Scott AF, Kazazian HH, Jr., Boeke JD, Gabriel A. Reverse transcriptase encoded by a human transposable element. Science. 1991;254:1808–1810. doi: 10.1126/science.1722352. [DOI] [PubMed] [Google Scholar]
- 14.Martin SL, Branciforte D, Keller D, Bain DL. Trimeric structure for an essential protein in L1 retrotransposition. Proc Natl Acad Sci USA. 2003;100:13815–13820. doi: 10.1073/pnas.2336221100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Piskareva O, Schmatchenko V. DNA polymerization by the reverse transcriptase of the human L1 retrotransposon on its own template in vitro. FEBS Lett. 2006;580:661–668. doi: 10.1016/j.febslet.2005.12.077. [DOI] [PubMed] [Google Scholar]
- 16.Kurzynska-Kokorniak A, Jamburuthugoda VK, Bibillo A, Eickbush TH. DNA-directed DNA polymerase and strand displacement activity of the reverse transcriptase encoded by the R2 retrotransposon. J Mol Biol. 2007;374:322–333. doi: 10.1016/j.jmb.2007.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zheng L, Shen B. Okazaki fragment maturation: nucleases take centre stage. J Mol Cell Biol. 2011;3:23–30. doi: 10.1093/jmcb/mjq048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luan DD, Korman MH, Jakubczak JL, Eickbush TH. Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: a mechanism for non-LTR retrotransposition. Cell. 1993;72:595–605. doi: 10.1016/0092-8674(93)90078-5. [DOI] [PubMed] [Google Scholar]
- 19.Cost GJ, Feng Q, Jacquier A, Boeke JD. Human L1 element target-primed reverse transcription in vitro. EMBO J. 2002;21:5899–5910. doi: 10.1093/emboj/cdf592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goodier JL, Kazazian HH., Jr. Retrotransposons revisited: the restraint and rehabilitation of parasites. Cell. 2008;135:23–35. doi: 10.1016/j.cell.2008.09.022. [DOI] [PubMed] [Google Scholar]
- 21.Yu F, Zingler N, Schumann G, Stratling WH. Methyl-CpG-binding protein 2 represses LINE-1 expression and retrotransposition but not Alu transcription. Nucleic Acids Res. 2001;29:4493–4501. doi: 10.1093/nar/29.21.4493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoder JA, Yen RW, Vertino PM, Bestor TH, Baylin SB. New 5′ regions of the murine and human genes for DNA (cytosine-5)-methyltransferase. J Biol Chem. 1996;271:31092–31097. doi: 10.1074/jbc.271.49.31092. [DOI] [PubMed] [Google Scholar]
- 23.Yang N, Kazazian HH., Jr. L1 retrotransposition is suppressed by endogenously encoded small interfering RNAs in human cultured cells. Nat Struct Mol Biol. 2006;13:763–771. doi: 10.1038/nsmb1141. [DOI] [PubMed] [Google Scholar]
- 24.Malone CD, Hannon GJ. Small RNAs as guardians of the genome. Cell. 2009;136:656–668. doi: 10.1016/j.cell.2009.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Suzuki J, et al. Genetic evidence that the non-homologous end-joining repair pathway is involved in LINE retrotransposition. PLoS Genet. 2009;5:e1000461. doi: 10.1371/journal.pgen.1000461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gasior SL, Roy-Engel AM, Deininger PL. ERCC1/XPF limits L1 retrotransposition. DNA Repair (Amst) 2008;7:983–989. doi: 10.1016/j.dnarep.2008.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stetson DB, Ko JS, Heidmann T, Medzhitov R. Trex1 prevents cell-intrinsic initiation of autoimmunity. Cell. 2008;134:587–598. doi: 10.1016/j.cell.2008.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moldovan JB, Moran JV. The Zinc-Finger Antiviral Protein ZAP Inhibits LINE and Alu Retrotransposition. PLoS Genet. 2015;11:e1005121. doi: 10.1371/journal.pgen.1005121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goodier JL, Cheung LE, Kazazian HH., Jr. MOV10 RNA helicase is a potent inhibitor of retrotransposition in cells. PLoS Genet. 2012;8:e1002941. doi: 10.1371/journal.pgen.1002941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang A, et al. RNase L restricts the mobility of engineered retrotransposons in cultured human cells. Nucleic Acids Res. 2014;42:3803–3820. doi: 10.1093/nar/gkt1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao K, et al. Modulation of LINE-1 and Alu/SVA retrotransposition by Aicardi-Goutieres syndrome-related SAMHD1. Cell Rep. 2013;4:1108–1115. doi: 10.1016/j.celrep.2013.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hu S, et al. SAMHD1 Inhibits LINE-1 Retrotransposition by Promoting Stress Granule Formation. PLoS Genet. 2015;11:e1005367. doi: 10.1371/journal.pgen.1005367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wissing S, Montano M, Garcia-Perez JL, Moran JV, Greene WC. Endogenous APOBEC3B restricts LINE-1 retrotransposition in transformed cells and human embryonic stem cells. J Biol Chem. 2011;286:36427–36437. doi: 10.1074/jbc.M111.251058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Muckenfuss H, et al. APOBEC3 proteins inhibit human LINE-1 retrotransposition. J Biol Chem. 2006;281:22161–22172. doi: 10.1074/jbc.M601716200. [DOI] [PubMed] [Google Scholar]
- 35.Esnault C, Millet J, Schwartz O, Heidmann T. Dual inhibitory effects of APOBEC family proteins on retrotransposition of mammalian endogenous retroviruses. Nucleic Acids Res. 2006;34:1522–1531. doi: 10.1093/nar/gkl054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.MacDuff DA, Demorest ZL, Harris RS. AID can restrict L1 retrotransposition suggesting a dual role in innate and adaptive immunity. Nucleic Acids Res. 2009;37:1854–1867. doi: 10.1093/nar/gkp030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schumacher AJ, Hache G, Macduff DA, Brown WL, Harris RS. The DNA deaminase activity of human APOBEC3G is required for Ty1, MusD, and human immunodeficiency virus type 1 restriction. J Virol. 2008;82:2652–2660. doi: 10.1128/JVI.02391-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kinomoto M, et al. All APOBEC3 family proteins differentially inhibit LINE-1 retrotransposition. Nucleic Acids Res. 2007;35:2955–2964. doi: 10.1093/nar/gkm181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Turelli P, Vianin S, Trono D. The innate antiretroviral factor APOBEC3G does not affect human LINE-1 retrotransposition in a cell culture assay. J Biol Chem. 2004;279:43371–43373. doi: 10.1074/jbc.C400334200. [DOI] [PubMed] [Google Scholar]
- 40.Chen H, et al. APOBEC3A is a potent inhibitor of adeno-associated virus and retrotransposons. Curr Biol. 2006;16:480–485. doi: 10.1016/j.cub.2006.01.031. [DOI] [PubMed] [Google Scholar]
- 41.Horn AV, et al. Human LINE-1 restriction by APOBEC3C is deaminase independent and mediated by an ORF1p interaction that affects LINE reverse transcriptase activity. Nucleic Acids Res. 2014;42:396–416. doi: 10.1093/nar/gkt898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sawyer SL, Emerman M, Malik HS. Ancient adaptive evolution of the primate antiviral DNA-editing enzyme APOBEC3G. PLoS Biol. 2004;2:E275. doi: 10.1371/journal.pbio.0020275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.LaRue RS, et al. The artiodactyl APOBEC3 innate immune repertoire shows evidence for a multi-functional domain organization that existed in the ancestor of placental mammals. BMC Mol Biol. 2008;9:104. doi: 10.1186/1471-2199-9-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liao W, et al. APOBEC-2, a cardiac- and skeletal muscle-specific member of the cytidine deaminase supergene family. Biochem Biophys Res Commun. 1999;260:398–404. doi: 10.1006/bbrc.1999.0925. [DOI] [PubMed] [Google Scholar]
- 45.Peled JU, et al. The biochemistry of somatic hypermutation. Annu Rev Immunol. 2008;26:481–511. doi: 10.1146/annurev.immunol.26.021607.090236. [DOI] [PubMed] [Google Scholar]
- 46.Sato Y, et al. Deficiency in APOBEC2 leads to a shift in muscle fiber type, diminished body mass, and myopathy. J Biol Chem. 2010;285:7111–7118. doi: 10.1074/jbc.M109.052977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ohtsubo H, et al. APOBEC2 negatively regulates myoblast differentiation in muscle regeneration. Int J Biochem Cell Biol. 2017;85:91–101. doi: 10.1016/j.biocel.2017.02.005. [DOI] [PubMed] [Google Scholar]
- 48.Severi F, Chicca A, Conticello SG. Analysis of reptilian APOBEC1 suggests that RNA editing may not be its ancestral function. Mol Biol Evol. 2011;28:1125–1129. doi: 10.1093/molbev/msq338. [DOI] [PubMed] [Google Scholar]
- 49.Rogozin IB, Basu MK, Jordan IK, Pavlov YI, Koonin EV. APOBEC4, a new member of the AID/APOBEC family of polynucleotide (deoxy)cytidine deaminases predicted by computational analysis. Cell Cycle. 2005;4:1281–1285. doi: 10.4161/cc.4.9.1994. [DOI] [PubMed] [Google Scholar]
- 50.Blanc V, Davidson NO. APOBEC-1-mediated RNA editing. Wiley Interdiscip Rev Syst Biol Med. 2010;2:594–602. doi: 10.1002/wsbm.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Young SG. Recent progress in understanding apolipoprotein B. Circulation. 1990;82:1574–1594. doi: 10.1161/01.CIR.82.5.1574. [DOI] [PubMed] [Google Scholar]
- 52.Harris RS, Petersen-Mahrt SK, Neuberger MS. RNA editing enzyme APOBEC1 and some of its homologs can act as DNA mutators. Mol Cell. 2002;10:1247–1253. doi: 10.1016/S1097-2765(02)00742-6. [DOI] [PubMed] [Google Scholar]
- 53.Lindic N, et al. Differential inhibition of LINE1 and LINE2 retrotransposition by vertebrate AID/APOBEC proteins. Retrovirology. 2013;10:156. doi: 10.1186/1742-4690-10-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bogerd HP, Wiegand HL, Doehle BP, Lueders KK, Cullen BR. APOBEC3A and APOBEC3B are potent inhibitors of LTR-retrotransposon function in human cells. Nucleic Acids Res. 2006;34:89–95. doi: 10.1093/nar/gkj416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hulme AE, Bogerd HP, Cullen BR, Moran JV. Selective inhibition of Alu retrotransposition by APOBEC3G. Gene. 2007;390:199–205. doi: 10.1016/j.gene.2006.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stenglein MD, Harris RS. APOBEC3B and APOBEC3F inhibit L1 retrotransposition by a DNA deamination-independent mechanism. J Biol Chem. 2006;281:16837–16841. doi: 10.1074/jbc.M602367200. [DOI] [PubMed] [Google Scholar]
- 57.Richardson SR, Narvaiza I, Planegger RA, Weitzman MD, Moran JV. APOBEC3A deaminates transiently exposed single-strand DNA during LINE-1 retrotransposition. Elife. 2014;3:e02008. doi: 10.7554/eLife.02008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chiu YL, et al. High-molecular-mass APOBEC3G complexes restrict Alu retrotransposition. Proc Natl Acad Sci USA. 2006;103:15588–15593. doi: 10.1073/pnas.0604524103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liang W, et al. APOBEC3DE Inhibits LINE-1 Retrotransposition by Interacting with ORF1p and Influencing LINE Reverse Transcriptase Activity. PLoS One. 2016;11:e0157220. doi: 10.1371/journal.pone.0157220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Esnault C, et al. APOBEC3G cytidine deaminase inhibits retrotransposition of endogenous retroviruses. Nature. 2005;433:430–433. doi: 10.1038/nature03238. [DOI] [PubMed] [Google Scholar]
- 61.Mol CD, et al. Crystal structure of human uracil-DNA glycosylase in complex with a protein inhibitor: protein mimicry of DNA. Cell. 1995;82:701–708. doi: 10.1016/0092-8674(95)90467-0. [DOI] [PubMed] [Google Scholar]
- 62.Esnault C, Priet S, Ribet D, Heidmann O, Heidmann T. Restriction by APOBEC3 proteins of endogenous retroviruses with an extracellular life cycle: ex vivo effects and in vivo “traces” on the murine IAPE and human HERV-K elements. Retrovirology. 2008;5:75. doi: 10.1186/1742-4690-5-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jern P, Stoye JP, Coffin JM. Role of APOBEC3 in genetic diversity among endogenous murine leukemia viruses. PLoS Genet. 2007;3:2014–2022. doi: 10.1371/journal.pgen.0030183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lee YN, Malim MH, Bieniasz PD. Hypermutation of an ancient human retrovirus by APOBEC3G. J Virol. 2008;82:8762–8770. doi: 10.1128/JVI.00751-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Carmi S, Church GM, Levanon EY. Large-scale DNA editing of retrotransposons accelerates mammalian genome evolution. Nat Commun. 2011;2:519. doi: 10.1038/ncomms1525. [DOI] [PubMed] [Google Scholar]
- 66.Starrett GJ, et al. The DNA cytosine deaminase APOBEC3H haplotype I likely contributes to breast and lung cancer mutagenesis. Nat Commun. 2016;7:12918. doi: 10.1038/ncomms12918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.OhAinle M, Kerns JA, Li MM, Malik HS, Emerman M. Antiretroelement activity of APOBEC3H was lost twice in recent human evolution. Cell Host Microbe. 2008;4:249–259. doi: 10.1016/j.chom.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang X, et al. Analysis of human APOBEC3H haplotypes and anti-human immunodeficiency virus type 1 activity. J Virol. 2011;85:3142–3152. doi: 10.1128/JVI.02049-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Harari A, Ooms M, Mulder LC, Simon V. Polymorphisms and splice variants influence the antiretroviral activity of human APOBEC3H. J Virol. 2009;83:295–303. doi: 10.1128/JVI.01665-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ooms M, Majdak S, Seibert CW, Harari A, Simon V. The localization of APOBEC3H variants in HIV-1 virions determines their antiviral activity. J Virol. 2010;84:7961–7969. doi: 10.1128/JVI.00754-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tan L, Sarkis PT, Wang T, Tian C, Yu XF. Sole copy of Z2-type human cytidine deaminase APOBEC3H has inhibitory activity against retrotransposons and HIV-1. FASEB J. 2009;23:279–287. doi: 10.1096/fj.07-088781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kopera HC, et al. LINE-1 Cultured Cell Retrotransposition Assay. Methods Mol Biol. 2016;1400:139–156. doi: 10.1007/978-1-4939-3372-3_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Moran JV, et al. High frequency retrotransposition in cultured mammalian cells. Cell. 1996;87:917–927. doi: 10.1016/S0092-8674(00)81998-4. [DOI] [PubMed] [Google Scholar]
- 74.Wei W, Morrish TA, Alisch RS, Moran JV. A transient assay reveals that cultured human cells can accommodate multiple LINE-1 retrotransposition events. Anal Biochem. 2000;284:435–438. doi: 10.1006/abio.2000.4675. [DOI] [PubMed] [Google Scholar]
- 75.Carlow DC, Short SA, Wolfenden R. Role of glutamate-104 in generating a transition state analogue inhibitor at the active site of cytidine deaminase. Biochemistry. 1996;35:948–954. doi: 10.1021/bi951498y. [DOI] [PubMed] [Google Scholar]
- 76.Betts L, Xiang S, Short SA, Wolfenden R, Carter CW., Jr. Cytidine deaminase. The 2.3 A crystal structure of an enzyme: transition-state analog complex. J Mol Biol. 1994;235:635–656. doi: 10.1006/jmbi.1994.1018. [DOI] [PubMed] [Google Scholar]
- 77.Navarro F, et al. Complementary function of the two catalytic domains of APOBEC3G. Virology. 2005;333:374–386. doi: 10.1016/j.virol.2005.01.011. [DOI] [PubMed] [Google Scholar]
- 78.Mitra M, et al. Sequence and structural determinants of human APOBEC3H deaminase and anti-HIV-1 activities. Retrovirology. 2015;12:3. doi: 10.1186/s12977-014-0130-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang Z, Mosbaugh DW. Uracil-DNA glycosylase inhibitor gene of bacteriophage PBS2 encodes a binding protein specific for uracil-DNA glycosylase. J Biol Chem. 1989;264:1163–1171. [PubMed] [Google Scholar]
- 80.Bennett SE, Schimerlik MI, Mosbaugh DW. Kinetics of the uracil-DNA glycosylase/inhibitor protein association. Ung interaction with Ugi, nucleic acids, and uracil compounds. J Biol Chem. 1993;268:26879–26885. [PubMed] [Google Scholar]
- 81.Stenglein MD, Burns MB, Li M, Lengyel J, Harris RS. APOBEC3 proteins mediate the clearance of foreign DNA from human cells. Nat Struct Mol Biol. 2010;17:222–229. doi: 10.1038/nsmb.1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Love RP, Xu H, Chelico L. Biochemical analysis of hypermutation by the deoxycytidine deaminase APOBEC3A. J Biol Chem. 2012;287:30812–30822. doi: 10.1074/jbc.M112.393181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Byeon IJ, et al. NMR structure of human restriction factor APOBEC3A reveals substrate binding and enzyme specificity. Nat Commun. 2013;4:1890. doi: 10.1038/ncomms2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Land AM, et al. Endogenous APOBEC3A DNA cytosine deaminase is cytoplasmic and nongenotoxic. J Biol Chem. 2013;288:17253–17260. doi: 10.1074/jbc.M113.458661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Li MM, Emerman M. Polymorphism in human APOBEC3H affects a phenotype dominant for subcellular localization and antiviral activity. J Virol. 2011;85:8197–8207. doi: 10.1128/JVI.00624-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lackey L, Law EK, Brown WL, Harris RS. Subcellular localization of the APOBEC3 proteins during mitosis and implications for genomic DNA deamination. Cell Cycle. 2013;12:762–772. doi: 10.4161/cc.23713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chelico L, Pham P, Calabrese P, Goodman MF. APOBEC3G DNA deaminase acts processively 3′–>5′ on single-stranded DNA. Nat Struct Mol Biol. 2006;13:392–399. doi: 10.1038/nsmb1086. [DOI] [PubMed] [Google Scholar]
- 88.Feng Y, et al. Natural Polymorphisms and Oligomerization of Human APOBEC3H Contribute to Single-stranded DNA Scanning Ability. J Biol Chem. 2015;290:27188–27203. doi: 10.1074/jbc.M115.666065. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No datasets were generated or analyzed during the current study.