Abstract
Previous studies have shown that the expression of microRNA-4458 (miR-4458) is dysregulated in hepatocellular carcinoma and colon cancer. In this study, we investigated the direct target of miR-4458 and its biological functions in human lung cancer cells. By using the database TargetScan, we identified Lin28B, an oncogene, as a direct target gene of miR-4458. In dual-luciferase reporter assay, we found that miR-4458 mimics dose-dependently inhibited the luciferase activity of the wild-type 3′UTR of Lin28B in human lung cancer A549 and NCI-H1299 cell lines without affecting its mutant forms, whereas anti-miR-4458, an inhibitor of miR-4458, dose-dependently promoted the luciferase activity of the wild-type 3′UTR of Lin28B in A549 and NCI-H1299 cell lines without affecting its mutant forms. Overexpression of miR-4458 significantly decreased the protein levels of Lin28B in the cells, and inhibited the cell growth and colony formation. Conversely, knockdown of miR-4458 with anti-miR-4458 significantly increased the protein levels of Lin28B, and promoted the cell proliferation, which could be reverted by knockdown of Lin28B expression. In addition, we detected the expression of Lin28B using RT-PCR in 40 human lung cancer tissues and matched peritumoral tissues, and found that Lin28B was overexpressed in lung cancer tissues and negatively correlated with miR-4458 expression (r=-0.694, P<0.05). We conclude that miR-4458 is a tumor suppressor, and Lin28B is the direct target of miR-4458. These results suggest the modulation of miR-4458/Lin28B expression offers a potential therapeutic strategy for lung cancer.
Keywords: lung cancer, A549 cells, NCI-H1299 cells, miR-4458, Lin28B, cell proliferation
Introduction
Lung cancer is the most prevalent malignancy and the leading cause of cancer-related death in men and women worldwide. Most lung cancer cases (80%) are non-small cell lung cancer (NSCLC)1,2. Although great progress has been made in surgery, chemotherapy and radiotherapy treatment in recent years, the overall five-year survival rate for NSCLC has not been greatly improved and is still only 17.1%3. Late diagnosis accounts for the poor survival of advanced lung cancer patients and is the main obstacle in lung cancer management. Therefore, early diagnosis, safe and noninvasive detection methods, and understanding of the molecular mechanisms in lung cancer are in great demand.
MicroRNAs (miRNAs) are non-coding RNAs that are approximately 22 nucleotides in length. The main function of miRNAs is the regulation of gene expression through complementary binding to the 3′ untranslated regions (3′UTRs) of target mRNAs4,5,6. This post-transcriptional modification often leads to mRNA degradation or to translational repression. Over 60% of human protein-coding genes are predicted to be targeted and modulated by miRNAs. Numerous studies have shown that miRNAs are implicated in almost all cellular processes, including cell differentiation, proliferation, apoptosis, invasion and migration7,8,9,10,11. The dysregulation of miRNAs in cancer has been reported in different tumor types, including lung cancer. miRNAs can function as either oncogenes or tumor suppressors and play important roles in the initiation and progression of tumorigenesis10.
Previous studies have indicated that miR-4458 expression is dysregulated in hepatocellular carcinoma12 and colon cancer13. Those studies reported that miR-4458 is a tumor suppressor that is down-regulated in hepatocellular carcinoma and colon cancer, and that the high expression of miR-4458 suppresses the proliferation of tumor cells12,13. However, the role of miR-4458 in NSCLC remains unclear. Although certain target genes of miR-4458 have been predicted and detected in other types of cancer cells12,13, the direct target of miR-4458 and its function in lung cancer remains to be studied. Bioinformatics analysis predicted that miR-4458 could directly bind to the 3′UTR of Lin28B, which is frequently overexpressed in various cancers, including lung cancer, and is associated with the induction of tumorigenesis14,15,16. Therefore, we investigated the biological functions and direct target of miR-4458 in lung cancer in this study.
Materials and methods
Patients and tissue samples
Lung cancer tissues from a total of 40 cases and the adjacent normal tissues were obtained between the 1st of January 2012 and the 31st of December 2015 at the Second Hospital of Dalian Medical University. Detailed clinical information associated with the enrolled patients is listed in Supplementary Table S1. None of the 40 patients had received radiation therapy or chemotherapy before surgery. Tissue samples were stored in liquid nitrogen before use. The study was approved by the Ethics Committee of The Second Hospital of Dalian Medical University.
Cell lines and culture conditions
A549 and NCI-H1299 human lung cancer cell lines were acquired from the American Type Culture Collection (ATCC, Manassas, VA, USA). The lung cancer cells were maintained in RPMI-1640 (Invitrogen, Carlsbad, CA, USA) medium supplemented with 10% fetal bovine serum (FBS). Cells were cultured in a humidified incubator at 37 °C with 5% CO2.
Transfection of microRNA mimics, inhibitor oligonucleotides and siRNAs
The miR-4458 mimics, inhibitor (anti-miR-4458) oligonucleotides and siRNAs (si-Lin28B) were purchased from GenePharma (Shanghai, China). The miR-4458 mimics, anti-miR-4458 and negative controls were transfected into cells at concentrations of 50 nmol/L and 100 nmol/L using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions.
Cell proliferation assay
Cell proliferation was measured using a Click-iT EdU Microplate Assay (Invitrogen, Carlsbad, CA, USA). A549 and NCI-H1299 cells were transiently transfected with miR-4458 mimics, anti-miR-4458 or si-LinB28 siRNAs in six-well cell culture plates. In this assay, cells were treated with EdU, a thymidine analog that is incorporated into DNA during cell proliferation, and high rates of EdU incorporation correlate with high cell proliferation rates. For nucleic acid staining, Hoechst 33342 (Sigma, St Louis, MO, USA) was added to the culture medium; images were obtained with fluorescence microscopy using a filter at specific wavelengths.
Colony-formation assay
Twenty-four hours after transfection with miR-4458 mimics, anti-miR-4458, si-Lin28B or negative control oligonucleotides, the lung cancer cells were seeded in 6-well plates and grown for ten days for the colony-formation assay. The colonies were then washed with PBS, fixed with methyl alcohol, stained by crystal violet, photographed and counted.
Western blotting
Seventy-two hours after transfection, cells were harvested with RIPA lysis buffer, and protein lysates were separated using 12% SDS-PAGE, and transferred to PVDF membranes (Millipore, Billerica, MA, USA). The membranes were incubated with anti-Lin28B (Cell Signaling Technology, MA, USA) or anti-β-actin antibody (Cell Signaling Technology, MA, USA) followed by HRP-labeled goat anti-rabbit IgG or anti-mouse IgG antibody. The density of the bands was quantified using ImageQuant 5.2 software (GE Healthcare, Little Chalfont, UK).
RNA isolation and quantitative RT-PCR
Total RNA was extracted from cells using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's protocol. Total RNA was extracted from frozen tissues (up to 30 mg each) using a miRNeasy Minikit (Qiagen, Valencia, CA, USA). For Lin28B expression, 1 μg of total RNA samples was used for first strand cDNA synthesis in a 20 μL reaction system using PrimeScript RT Master Mix (TaKaRa Biotechnology, Dalian, China) following the manufacturer's instructions. Then, 1 μL of undiluted synthesized cDNA was used in the real-time quantitative PCR (RT-PCR) protocol. RT-PCR was performed using a QuantiNova SYBR Green PCR kit (Qiagen, Valencia, CA, USA). The relative Lin28B mRNA levels were analyzed by normalizing the threshold cycle (Ct) value to that of the internal loading control, GAPDH.
To quantify miR-4458, the expression level of miR-4458 was measured using a TaqMan MicroRNA Reverse Transcription Kit (Thermo Fisher, Carlsbad, CA, USA) and TaqMan gene expression master mix (Thermo Fisher, Carlsbad, CA, USA) according to the manufacturers' protocols. U6 snRNA was used as an internal loading control. The primers used in this study are listed in Supplementary Table S2. All PCR reactions were performed in triplicate for each sample.
Dual-luciferase reporter assay
The sequence of the Lin28B 3′UTR containing the predicted miR-4458 binding site and the sequence of the Lin28B 3′UTR with five mutations at the putative miR-4458 binding site were cloned into a pGL3 luciferase reporter vector (Promega, Madison, MI, USA). Then, cells seeded in 6-well plates were co-transfected with miR4458 mimics, anti-miR-4458 or negative control and reporter constructs (0.5 μg) using Lipofectamine 2000. Cell extracts were prepared 48 h after transfection, and luciferase activity was measured using a Dual-Luciferase Reporter Assay System (Promega).
Statistical analysis
All statistical analysis and plots were performed and generated using GraphPad Prism software (version 5.01; GraphPad Software, Inc, CA, USA). The data are presented as the mean values with standard error of the mean (SEM), and a P value of less than 0.05 was considered statistically significant. All experiments were performed independently at least three times. The statistical significance between two groups was measured using Student's t-test. One-way analysis of variance (ANOVA) was used to measure the significance of comparisons for more than two groups.
Results
Lin28B is a direct target gene of miR-4458
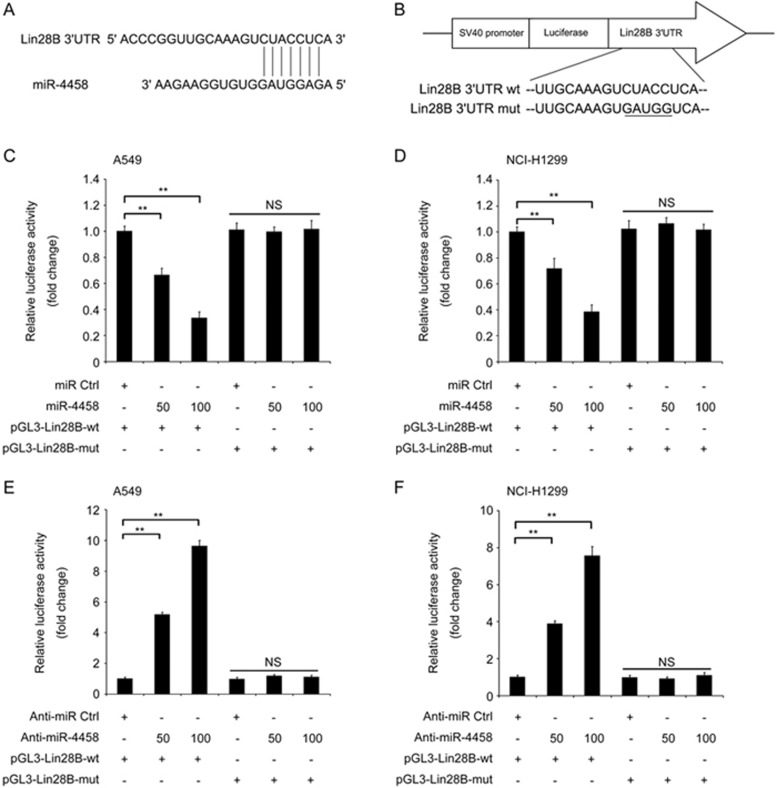
Potential miRNA targets were predicted using the database TargetScan (http://www.targetscan.org/), and Lin28B was identified as a direct target gene of miR-4458 in the in silico analysis. Lin28B is an oncogene that is overexpressed and associated with advanced disease and poor clinical outcome in diverse cancers16,17,18. Thus, we focused on Lin28B as a candidate for validation. The algorithm produces not only target genes but also potential conserved 8-mer or 7-mer sites matching the seed region of miRNAs. As shown in Figure 1A, the 3′UTR of Lin28B mRNA contains a putative binding site that is complementary to the seed region of miR-4458. To confirm that Lin28B is a direct target of miR-4458, a dual-luciferase reporter assay was performed. Both the putative miR-4458 binding site and the mutated binding site in the 3′UTR of Lin28B were synthesized and constructed into a pGL3 vector (Figure 1B). Mutation of 5 out of 7 nucleotides in the Lin28B 3′UTR region that is complementary to the seed region of miR-4458 was sufficient to abolish the regulation by miR-4458 in a previous report13,19. Our results showed that miR-4458 significantly inhibited the luciferase activity of the wild-type (wt) 3′UTR of Lin28B but had no inhibitory effect on the mutant form (mut) (Figure 1C and 1D). Similarly, knockdown of miR-4458 significantly increased the luciferase activity of the wild-type 3′UTR of Lin28B but had no inhibitory effect on the mutant form in both A549 and NCI-H1299 cell lines (Figure 1E and 1F). These results indicated that Lin28B is a direct target of miR-4458 in lung cancer cells.
Figure 1.
miR-4458 directly inhibits the expression of Lin28B by targeting its 3′UTR. (A) Target prediction for miR-4458 using Targetscan database (http://www.targetscan.org). (B) Sequences for plasmid construction of the wild-type (wt) and mutated-type (mut) 3′UTR of Lin28B mRNA. (C and D) miR-4458 significantly inhibited the luciferase activity of the wild-type (wt) 3′UTR of Lin28B in a dose-dependent manner in A549 and NCI-H1299 cell lines but had no inhibitory effect on the mutant form (mut). (E and F) An inhibitor of miR-4458, anti-miR-4458, significantly promoted the luciferase activity of the wild-type (wt) 3′UTR of Lin28B in a dose-dependent manner in A549 and NCI-H1299 cell lines but had no inhibitory effect on the mutant form (mut). *P<0.05; **P<0.01.
miR-4458 suppresses the expression of Lin28B
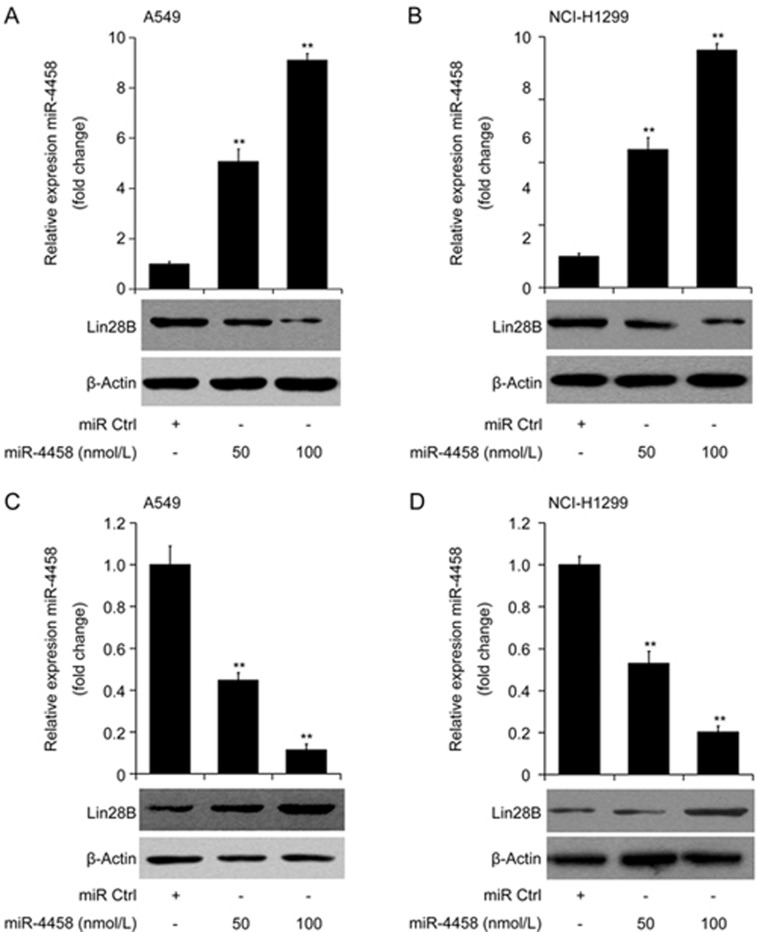
To validate and to further elucidate the regulation of Lin28B by miR-4458, the expression of Lin28B was detected using Western blotting in lung cancer cells with miR-4458 overexpressed or derepressed. Ectopic miR-4458 or anti-miR4458 was transfected into A549 and NCI-H1299 cells at concentrations of 50 nmol/L and 100 nmol/L. As shown in Figure 2A and 2B, the overexpression efficiency was sufficient to generate over four- and eight-fold upregulation in A549 and NCI-H1299 cells compared with the control group. Meanwhile, ectopic miR-4458 expression significantly suppressed the expression of Lin28B at the protein level in both A549 and NCI-H1299 cells in a dose-dependent manner. Consistently, knockdown of miR-4458 through the transfection of anti-miR-4458 significantly down-regulated the expression of miR-4458 in A549 and NCI-H1299 cell lines (Figure 2C and 2D), and the expression of Lin28B was significantly upregulated by anti-miR-4458 in a dose-dependent manner in lung cancer cells (Figure 2C and 2D).
Figure 2.
miR-4458 down-regulates Lin28B in lung cancer cells. (A and B) Transfection of A549 and NCI-H1299 cell lines with miR-4458 mimics significantly increased the expression of miR-4458 and inhibited the protein expression of Lin28B in a dose-dependent manner. (C and D) Transfection of A549 and NCI-H1299 cell lines with the miR-4458 inhibitor anti-miR-4458 significantly decreased the expression of miR-4458 and promoted the protein expression of Lin28B in a dose-dependent manner. *P<0.05; **P<0.01.
miR-4458 suppresses the proliferation of lung cancer cells through Lin28B
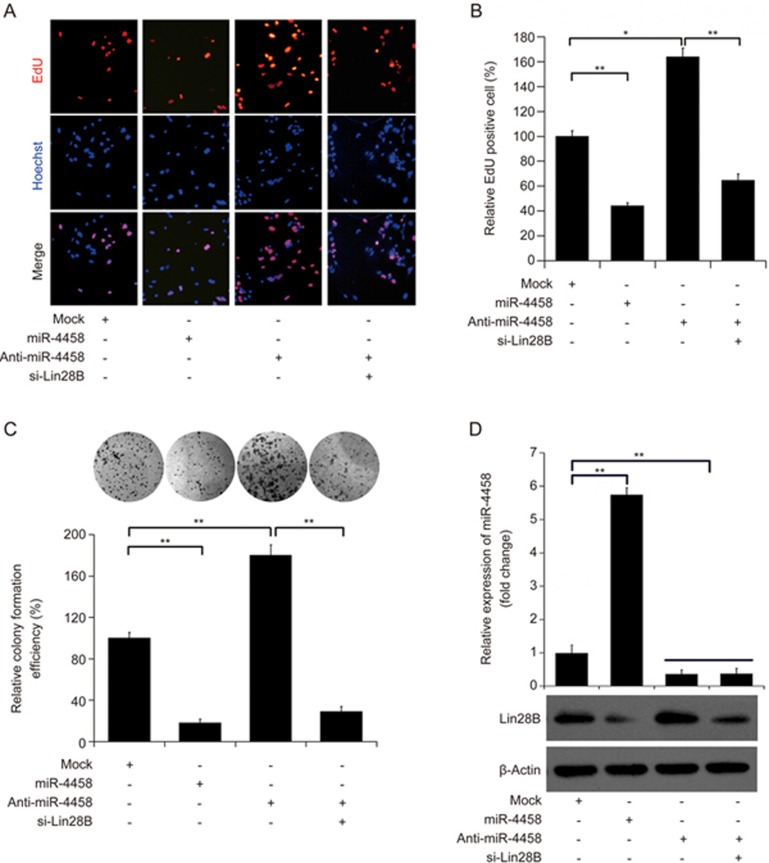
To characterize the function of miR-4458 in lung cancer, lung cancer cells were transiently transfected with miR-4458 mimics or anti-miR-4458. RT-PCR analysis confirmed that mature miR-4458 was successfully overexpressed in cells that were transfected with miR-4458 mimics (Figure 2). Ectopic miR-4458 expression markedly inhibited the growth of lung cancer cells, while knockdown of miR-4458 by anti-miR4458 significantly promoted the proliferation of lung cancer cells compared with control groups (Figure 3A and 3B). In the colony-formation assay, the colony-forming activity of both types of lung cancer cells transfected with miR-4458 was inhibited compared with that of the negative controls (Figure 3C). Consistently, knockdown of miR-4458 significantly promoted the colony-formation efficiency of lung cancer cells (Figure 3C).
Figure 3.
miR-4458 suppresses the proliferation of lung cancer cells through Lin28B. (A and B) EdU stain assay showed that miR-4458 significantly inhibited the proliferation of A549 cells, whereas anti-miR-4458 significantly promoted the proliferation of A549 cells. Furthermore, knockdown of Lin28B by siRNA transfection significantly reversed the effect of anti-miR-4458-induced promotion of proliferation in A549 cells. (C) Colony-formation assays showed that miR-4458 significantly inhibited the colony-formation efficiency of A549 cells, whereas anti-miR-4458 significantly promoted the efficiency of A549 cells, which could be reversed by si-Lin28B. (D) The transfection efficiency of miR-4458, anti-miR-4458 and si-Lin28B. *P<0.05; **P<0.01.
To further investigate whether the biological function of miR-4458 was mediated by Lin28B, we also knocked down the expression of Lin28B combined with miR-4458 through transient transfection with anti-miR-4458. The results showed that knockdown of Lin28B significantly reversed the effect of miR-4458 on the proliferation of lung cancer cells (Figure 3). As shown in Figure 3, compared with the anti-miR-4458 group, the growth ability and colony-formation efficiency of lung cancer cells were significantly decreased when the expression of Lin28B was inhibited. miR-4458 expression and Lin28B expression were also validated by RT-PCR and Western blotting after transfection (Figure 3D). Taken together, these data suggested that miR-4458 suppressed cell growth and colony formation by suppressing the expression of Lin28B.
miR-4458 is negatively correlated with Lin28B in human lung cancer tissues
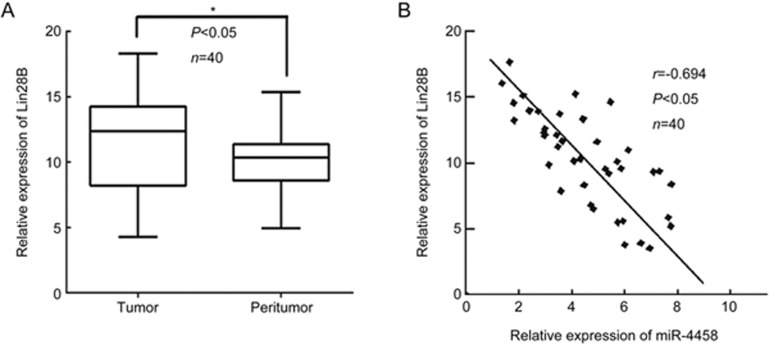
To investigate and further validate the pathophysiological relationship of miR-4458 and Lin28B in human lung cancer tissues, we detected the expression of miR-4458 and Lin28B in 40 frozen human lung cancer tissues and matched normal peritumoral tissues using RT-PCR. The expression level of Lin28B in tumor tissues was significantly higher than that in matched normal tissues (Figure 4A). Correlation analysis also showed that expression of miR-4458 was negatively correlated with Lin28B in 40 lung cancer tissues (r=-0.694, P<0.05) (Figure 4B). These findings suggested that upregulation of Lin28B was promoted by decreased miR-4458 expression and that this axis might be responsible for lung cancer cell growth.
Figure 4.
miR-4458 is negatively related to Lin28B expression in clinical lung cancer tissues. (A) The results of RT-PCR revealed that Lin28B was significantly overexpressed in 40 lung cancer tissues. (B) The RT-PCR results showed that the expression of miR-4458 was negatively correlated with the expression of Lin28B. *P<0.05.
Discussion
Dysregulation of microRNAs has commonly been observed in various cancer types. MicroRNAs play important roles in cancer development and progression by suppressing the expressing of genes involved in cell proliferation, apoptosis, differentiation, invasion and metastasis20,21,22.
MicroRNAs are emerging as potential targets for cancer treatment. Therefore, the identification of tumor-related miRNAs and their direct target genes is critical to understand the biological function of miRNAs in cancer and might offer novel biomarkers or therapeutic targets for the effective treatment of cancer.
Recently, two studies indicated that miR-4458 is a tumor suppressor that is down-regulated in human hepatocellular carcinoma and colon cancer and might influence cell viability in these two cancer types12,13. Moreover, miR-4458 was shown to be a potential prognosis predictor for hepatocellular carcinoma patients12 and inhibited glycolysis and lactate production in colon cancer cells13. Mechanistically, direct interaction between miR-4458 and the 3′UTR of IKBKE or HK2 mRNA was identified in cancer cells. However, the molecular mechanisms of miR-4458, along with its roles in tumors, have not been thoroughly studied, especially in lung cancer cells. In our study, miR-4458 is predicted to target the mRNA of Lin28B, which is an RNA binding protein that interacts with several microRNAs, especially inhibiting the expression of the Let-7 family. However, the regulation of Lin28B by microRNAs remains largely unstudied. Thus, we investigated the role of miR-4458 in regulating the expression and biological function of Lin28B in lung cancer cells.
Our results demonstrated that miR-4458 could directly target the 3′UTR of Lin28B mRNA and could suppress its protein expression. As a result, miR-4458 inhibited the proliferation ability of lung cancer cells. In miR-4458-inhibited cells, Lin28B expression, cell growth, and colony-formation efficiency were increased. Moreover, we validated the importance of the miR-4458/Lin28B axis via rescue experiments. Knockdown of Lin28B abolished anti-miR-4458-mediated cell growth promotion of lung cancer cells. Moreover, increased Lin28B expression was detected in lung cancer tissues compared with related normal tissues and was significantly negatively correlated with the expression of miR-4458 in lung cancer tissues.
Lin28B belongs to the Lin28 protein family, including its paralog Lin28A23. Lin28A and Lin28B are highly conserved RNA binding proteins with similar structure and functions23. It is worth noting that one major difference between Lin28A and Lin28B is that Lin28B predominantly resides in the nucleus, whereas Lin28A exclusively resides in the cytoplasm24. Dysregulation of Lin28B was first discovered in hepatocellular carcinoma, where it is overexpressed in tumor tissues17. Numerous studies have shown that both Lin28A and Lin28B are overexpressed in many cancer types. Notably, Lin28B overexpression is frequently observed in various cancers, such as hepatocellular carcinoma, colorectal cancer, pancreatic cancer, and NSCLC, and is associated with the induction of neuroblastoma14,16,17,25,26. Moreover, Lin28B can facilitate cellular transformation in vitro and is possibly linked to the repression of Let-7 family microRNAs and the derepression of Let-7 targets27. Therefore, Lin28B, acting as a post-transcriptional modulator, is usually considered to possess oncogenic properties. In return, our results demonstrated that the expression of Lin28B was regulated by miR-4458 through direct targeting of the 3′UTR of Lin28B mRNA. Down-regulation of miR-4458 promoted the expression of Lin28B, while overexpression of Lin28B inhibited the expression of other microRNAs. This interesting phenomenon highlights the hypothesis that a possible axis of miR-4458/Lin28B/microRNAs might be responsible for tumorigenesis and disease development in lung cancer cells. Future investigation of the role of the miR-4458/Lin28B axis in immortalized lung airway epithelial cells might uncover novel findings regarding tumor initiation. In conclusion, we demonstrated that miR-4458 inhibited cell growth and decreased cell viability and colony-formation efficiency in lung cancer cells. Furthermore, miR-4458 most likely exerted its biological effects on NSCLC cell growth and viability by directly targeting Lin28B. Our findings suggest that the modulation of miR-4458/Lin28B expression offers a potential therapeutic strategy for lung cancer.
Author contribution
Chang-hong LIU designed the research; Chang-hong LIU, De-sheng LV, Mo LI, and Ge SUN performed the research; Xue-fei ZHANG and Yu BAI analyzed the data; and all authors contributed to the manuscript preparation.
Footnotes
Supplementary information is available on the website of Acta Pharmacologica Sinica.
Supplementary Information
Clinical characteristics of lung cancer samples
List of primers used in this paper.
References
- Wood SL, Pernemalm M, Crosbie PA, Whetton AD. The role of the tumor-microenvironment in lung cancer-metastasis and its relationship to potential therapeutic targets. Cancer Treat Rev 2014; 40: 558–66. [DOI] [PubMed] [Google Scholar]
- How SH, Ng TH, Kuan YC, Jamalludin AR, Fauzi AR. Survival of lung cancer patients in a resource-limited country. Asia Pac J Clin Oncol 2015; 11: 221–7. [DOI] [PubMed] [Google Scholar]
- Siegel R, DeSantis C, Virgo K, Stein K, Mariotto A, Smith T, et al. Cancer treatment and survivorship statistics, 2012. CA Cancer J Clin 2012; 62: 220–41. [DOI] [PubMed] [Google Scholar]
- Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem 2010; 79: 351–79. [DOI] [PubMed] [Google Scholar]
- Shukla GC, Singh J, Barik S. MicroRNAs: processing, maturation, target recognition and regulatory functions. Mol Cell Pharmacol 2011; 3: 83–92. [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell 2009; 136: 215–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N, Olson EN. MicroRNA regulatory networks in cardiovascular development. Dev Cell 2010; 18: 510–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witwer KW, Halushka MK. Toward the promise of microRNAs–– Enhancing reproducibility and rigor in microRNA research. RNA Biol 2016; 13: 1103–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okajima W, Komatsu S, Ichikawa D, Miyamae M, Kawaguchi T, Hirajima S, et al. Circulating microRNA profiles in plasma: identification of miR-224 as a novel diagnostic biomarker in hepatocellular carcinoma independent of hepatic function. Oncotarget 2016; 7: 53820–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahay D, Leblanc R, Grunewald TG, Ambatipudi S, Ribeiro J, Clezardin P, et al. The LPA1/ZEB1/miR-21-activation pathway regulates metastasis in basal breast cancer. Oncotarget 2015; 6: 20604–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Jiang G, Zhang P, Fan J. Programmed cell death and its role in inflammation. Mil Med Res 2015; 2: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang D, Sun B, Yu H, Yang Z, Zhu L. Tumor-suppressing effect of miR-4458 on human hepatocellular carcinoma. Cell Physiol Biochem 2015; 35: 1797–807. [DOI] [PubMed] [Google Scholar]
- Qin Y, Cheng C, Lu H, Wang Y. miR-4458 suppresses glycolysis and lactate production by directly targeting hexokinase2 in colon cancer cells. Biochem Biophys Res Commun 2016; 469: 37–43. [DOI] [PubMed] [Google Scholar]
- Schnepp RW, Khurana P, Attiyeh EF, Raman P, Chodosh SE, Oldridge DA, et al. A LIN28B-RAN-AURKA signaling network promotes neuroblastoma tumorigenesis. Cancer Cell 2015; 28: 599–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manier S, Powers JT, Sacco A, Reagan MR, Moschetta M, Glavey S, et al. Lin28B/Let-7 axis regulates multiple myeloma proliferation by enhancing c-Myc and Ras survival pathways. Blood 2013; 122: 273. [Google Scholar]
- King CE, Cuatrecasas M, Castells A, Sepulveda AR, Lee JS, Rustgi AK. LIN28B promotes colon cancer progression and metastasis. Cancer Res 2011; 71: 4260–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Chen Y, Ito H, Watanabe A, Ge X, Kodama T, et al. Identification and characterization of lin-28 homolog B (LIN28B) in human hepatocellular carcinoma. Gene 2006; 384: 51–61. [DOI] [PubMed] [Google Scholar]
- Hsu KF, Shen MR, Huang YF, Cheng YM, Lin SH, Chow NH, et al. Overexpression of the RNA-binding proteins Lin28B and IGF2BP3 (IMP3) is associated with chemoresistance and poor disease outcome in ovarian cancer. Br J Cancer 2015; 113: 414–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao L, Wang L, Wei G, Wang Y, Wuyun G, Bo A. Role of microRNA-4458 in patients with non-small-cell lung cancer. Oncol Lett 2016; 12: 3958–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takashima Y, Terada M, Udono M, Miura S, Yamamoto J, Suzuki A. Suppression of lethal-7b and miR-125a/b maturation by Lin28b enables maintenance of stem cell properties in hepatoblasts. Hepatology 2016; 64: 245–60. [DOI] [PubMed] [Google Scholar]
- Powers JT, Tsanov KM, Pearson DS, Roels F, Spina CS, Ebright R, et al. Multiple mechanisms disrupt the let-7 microRNA family in neuroblastoma. Nature 2016; 535: 246–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv P, Zhang P, Li X, Chen Y. Micro ribonucleic acid (RNA)-101 inhibits cell proliferation and invasion of lung cancer by regulating cyclooxygenase-2. Thorac Cancer 2015; 6: 778–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, He Y, Zhu Y, Chen M, Weng M, Yang C, et al. Comparison of the expression and function of Lin28A and Lin28B in colon cancer. Oncotarget 2016; 7: 79605–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piskounova E, Polytarchou C, Thornton JE, LaPierre RJ, Pothoulakis C, Hagan JP, et al. Lin28A and Lin28B inhibit let-7 microRNA biogenesis by distinct mechanisms. Cell 2011; 147: 1066–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam M, Ahmad R, Rajabi H, Kufe D. MUC1-C induces the LIN28B-->LET-7-->HMGA2 axis to regulate self-renewal in NSCLC. Mol Cancer Res 2015; 13: 449–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helsmoortel HH, De Moerloose B, Pieters T, Ghazavi F, Bresolin S, Cave H, et al. LIN28B is over-expressed in specific subtypes of pediatric leukemia and regulates lncRNA H19. Haematologica 2016; 101: e240–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanathan SR, Powers JT, Einhorn W, Hoshida Y, Ng TL, Toffanin S, et al. Lin28 promotes transformation and is associated with advanced human malignancies. Nat Genet 2009; 41: 843–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Clinical characteristics of lung cancer samples
List of primers used in this paper.