Abstract
Purpose:
Implementation of novel genetic diagnostic tests is generally driven by technological advances because they promise shorter turnaround times and/or higher diagnostic yields. Other aspects, including impact on clinical management or cost-effectiveness, are often not assessed in detail prior to implementation.
Methods:
We studied the clinical utility of whole-exome sequencing (WES) in complex pediatric neurology in terms of diagnostic yield and costs. We analyzed 150 patients (and their parents) presenting with complex neurological disorders of suspected genetic origin. In a parallel study, all patients received both the standard diagnostic workup (e.g., cerebral imaging, muscle biopsies or lumbar punctures, and sequential gene-by-gene–based testing) and WES simultaneously.
Results:
Our unique study design allowed direct comparison of diagnostic yield of both trajectories and provided insight into the economic implications of implementing WES in this diagnostic trajectory. We showed that WES identified significantly more conclusive diagnoses (29.3%) than the standard care pathway (7.3%) without incurring higher costs. Exploratory analysis of WES as a first-tier diagnostic test indicates that WES may even be cost-saving, depending on the extent of other tests being omitted.
Conclusion:
Our data support such a use of WES in pediatric neurology for disorders of presumed genetic origin.
Genet Med advance online publication 23 March 2017
Keywords: diagnostic yield, health-care resource use, Prospective Clinical Utility Study, pediatric neurology, whole-exome sequencing
Introduction
Pediatric neurologists aim to provide swift and precise diagnosis as well as appropriate therapeutic and supportive management for children with neurological disorders and their families. Many different, often rare and severe, acquired as well as genetic disorders of the brain, spinal cord, and neuromuscular system are encountered in everyday practice.1 A diagnosis may lead to rational therapeutic choices and, even if no medical treatment is available, provide insights into the prognosis and recurrence risk of the disorder within the family.
Establishing a diagnosis is, in many cases, a complex, expensive, and lengthy process involving burdensome procedures. This is especially the case for patients presenting with developmental delay, movement disorders, neuromuscular disorders, severe epilepsy, and/or combinations thereof that are expected to be of genetic origin.2 The armamentarium to reach a diagnosis includes tests such as magnetic resonance imaging, electroencephalography, biochemical and metabolic blood tests, urine tests, and cerebrospinal fluid tests, muscle biopsy, genomic microarrays, and Sanger sequencing of one or more genes and multiple consultations from various medical specialists.2 However, the diagnostic yield of each individual test is low.3,4 Despite all efforts of doctors and patients, a definitive diagnosis is reached only for a minority of all children with a neurological disorder of presumed genetic origin,3 leaving ample room to improve the diagnostic process.
Whole-exome sequencing (WES) has recently been proven to be of great potential value as a diagnostic tool in clinical practice. Successful application of WES in pediatric neurology has been confined mostly to single case reports and retrospective studies in which WES was used as a last-resource genetic test.5,6,7,8,9,10,11 However, prospective clinical utility studies of larger groups of patients have not yet been performed. Here, we present such data for a cohort of 150 patients who received both the standard diagnostic assessment and WES, allowing a direct comparison of the performance of both trajectories.
Materials and Methods
Patient recruitment, selection, and counseling
The Department of Pediatric Neurology at the Radboud University Medical Center is a tertiary referral center for pediatric neurology. The majority of patients are referred by pediatricians, neurologists, and psychiatrists for further diagnostics or second opinions.
A total of 150 consecutive patients with (nonacute) neurological symptoms of suspected genetic origin were selected. The group consisted of patients who were newly referred by either a general practitioner (n = 11) or a medical specialist (n = 55) and patients who had been referred to our center previously but still visited the outpatient clinic for diagnostic purposes because no conclusive diagnosis had been reached (n = 84). As such, the group was representative of pediatric patients presenting with complex neurological symptoms of suspected genetic origin seen in a tertiary referral center.
Family history was negative for 143 subjects. For the 7 others, diagnosis of a recessive disorder was expected based on affected siblings and/or parental consanguinity. Patients with well-known, clinically easily recognizable genetic disorders such as neurofibromatosis type 1 were not included in this study. A clinical geneticist counseled all 150 patients and their parents regarding the WES procedure, and all participants or their legal representatives gave written informed consent. This study was approved by the Medical Review Ethics Committee Arnhem-Nijmegen under the realm of clinical diagnostic genetic testing (2011/188).
Study design and classification of outcome of diagnostic workup
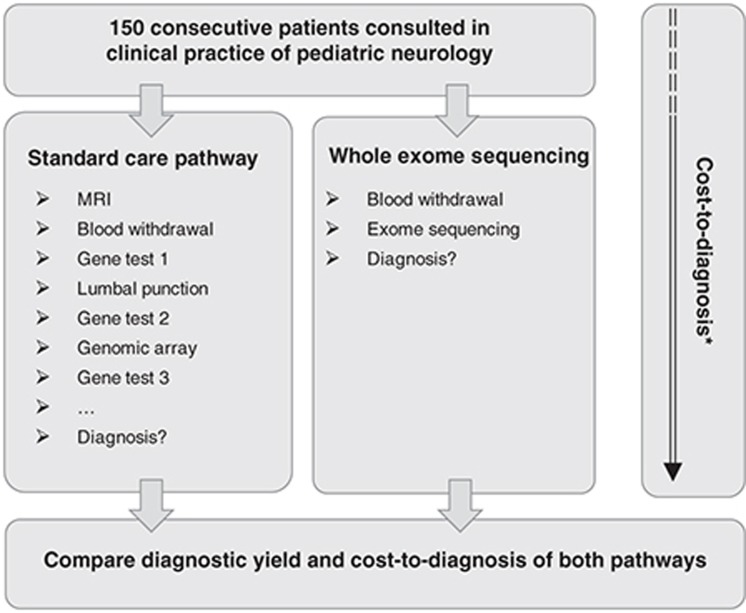
After inclusion, all patients received two diagnostic pathways in parallel, i.e., the standard diagnostic pathway and the WES pathway (Figure 1). Data regarding diagnostic tests and outcomes were not exchanged between the two pathways prior to reaching a conclusion for each of them separately. All patients were followed for a minimum of 6 months after starting WES (median, 17 months; range, 6–42 months). The primary end point for this study was the diagnostic yield of both diagnostic pathways.
Figure 1.
Schematic representation of the parallel study design. Schematic outline of the parallel design of our study including 150 patients with complex pediatric neurological problems of presumed genetic origin. *To determine the cost-to-diagnosis ratio, health-care resource use prior to inclusion was collected from the first visit to hospital clinic.
For the standard diagnostic pathway, the diagnostic workup was left to the discretion of the pediatric neurologists and was based on the clinical presentation of the patient. The outcome of this pathway was classified as follows:
1. There is no conclusive diagnosis. A clinical, descriptive, diagnosis can be reached, but genetic testing does not confirm this clinical descriptive diagnosis.
2. A conclusive diagnosis is reached. The clinical diagnosis is confirmed by the presence of a causative genetic variant (class 4 or 5)12 in a disease gene associated with the patient’s phenotype.
In the WES pathway, DNA of 150 patients and their parents (for 143 of 150 patients) was isolated from whole blood. Exome sequencing was performed according to our diagnostic procedures under ISO15189 certification (Supplementary Table S1 and Supplementary Figure S1 online). Sequencing was performed to an approximate read depth of 70- to 100-fold. Variant calling and interpretation were performed essentially as described previously (Supplementary Table S1 online).13,14 First, an in silico gene-panel analysis was performed using the WES data by referring to the situation when variants, both single-nucleotide variants (SNV) and copy-number variations (CNV), in known genes for the patient’s phenotype are evaluated for pathogenicity (Supplementary Table S1 and Supplementary Figure S1 online). In a second step, variants outside the gene panel were evaluated for pathogenicity and relevance for disease (both SNV and CNV). Patients who were sequenced as singletons received SNV analysis of the respective disease gene panel(s) as well as CNV analysis limited to genes within the relevant disease gene panels (Supplementary Table S1 and Supplementary Figure S1 online).
The results of the WES procedure were classified in accordance with ACMG guidelines:12,13
1. There is no conclusive diagnosis: no obvious pathogenic variant was detected that could potentially explain disease.
2. A possible diagnosis is obtained: variant(s) of unknown significance (class 3) in a gene(s) is identified in a disease gene that is associated with the patient’s phenotype or, alternatively, a pathogenic variant(s) (class 4 or 5) in a candidate disease gene(s) is identified with a potential relationship to the patient’s phenotype.
3. A conclusive diagnosis is obtained: a causative genetic variant (class 4 or 5) in a disease gene is associated with the patient’s phenotype.
Diagnostic end points for each of the care pathways were compared and discussed after completion of the diagnostic quest during monthly meetings and in the presence of molecular geneticists, clinical genetic laboratory specialists, clinical geneticists, and pediatric neurologists.
Diagnostic concordance between the standard care pathway and WES was measured by comparing diagnostic yield in the two pathways at individual patient levels.
Determining costs associated with the diagnostic pathways
To accurately estimate the costs associated with diagnostic tests in the standard diagnostic pathway, types and volumes of diagnostic tests used were determined for each patient individually. This included diagnostic tests that were performed after inclusion of the patient (until a diagnosis was established) and diagnostic tests that had been used prior to inclusion in our hospital or elsewhere. For the latter, data were extracted from medical records and from referral letters.
For the costs of genetic testing in the WES pathway, we used €3,500 for a trio analysis and €1,800 for a singleton; these costs represented prices for diagnostic exome sequencing in our laboratory (August 2016).15
To obtain an estimate of the costs associated with WES as a first-tier test for these patients, an exploratory analysis was conducted based on the following:
1. For children, when WES resulted in a conclusive diagnosis, conventional tests (and associated costs) performed in the standard pathway could have been precluded. To estimate the magnitude of the substitutive effect of WES, the pediatric neurologists indicated for all 44 patients receiving a conclusive diagnosis what conventional diagnostic tests would—in the context of clinical practice—still have been performed. Additionally, we assumed that these patients would still have two physician visits (pre- and post-WES test counseling) and one telephone consultation to inquire about patients’ general well-being.
2. For children, when WES did not result in a conclusive diagnosis (n = 106), the costs of the conventional diagnostic tests would have remained identical with the exception of costs associated with genetic testing, which were replaced by those of diagnostic WES.
Unit cost prices were obtained from the Dutch Healthcare Authority (https://www.nza.nl/ organisatie/sitewide/english/). Such costs cover the test, the interpretation of its results, and the physician fee. All unit costs were converted to 2016 Euros. Because cost and utilization data are generally skewed, bootstrap simulations with 1,000 replications were performed to estimate the mean costs and utilization and 95% confidence intervals.16
Results
Demographics
A total of 150 pediatric patients with complex neurological disorders of suspected genetic origin were included between November 2011 and January 2015. The median age of our study cohort at the time of inclusion was 5 years and 7 months (range, 5 months–18 year), and 53.3% of the patients were male (Supplementary Table S1 and Supplementary Figure S2 online). Patients were diagnosed according to the following major clinical subgroups: intellectual disability (ID; 78 patients), movement disorders (20 patients), neuromuscular disease (8 patients), epilepsy (5 patients), or a combination thereof, such as “ID+epilepsy” or “ID+movement disorder” (39 patients; Supplementary Table S1 online). This cohort thereby represented the broad phenotypic spectrum of disorders seen in daily routine pediatric neurology care at tertiary referral centers in the Netherlands.
Diagnostic yield in the standard diagnostic pathway
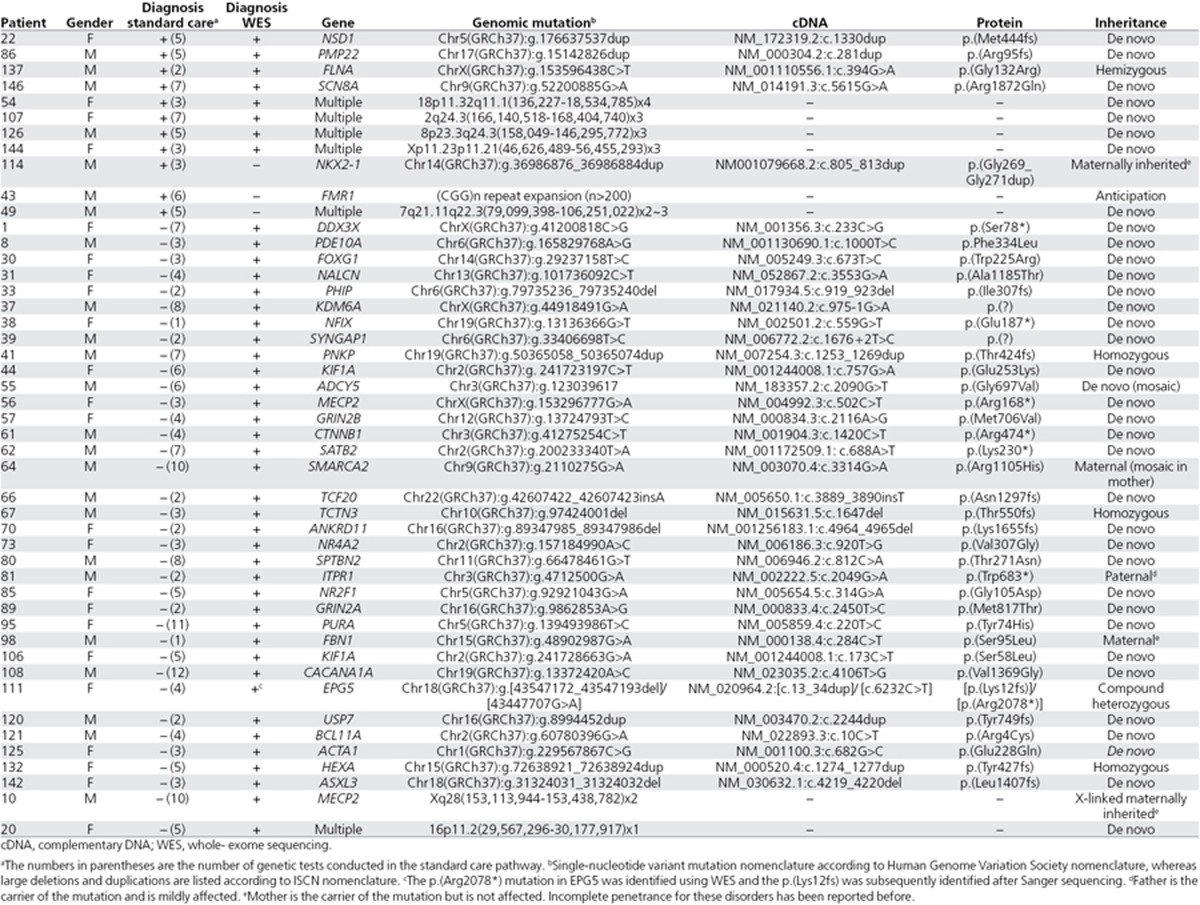
In the standard diagnostic pathway, 11 (7.3%) of 150 patients obtained a conclusive diagnosis confirmed by genetic testing (Table 1, Supplementary Table S2 online). These diagnoses included both well-known syndromes, such as Sotos syndrome (NSD1 point mutation) and fragile X syndrome ((CGG)n repeat expansion), and CNVs of various sizes ranging from a 2.2-Mb duplication to a mosaic whole chromosome 8 trisomy. On average, conclusive diagnoses in the standard pathway were reached after 4.6 genetic tests (range, 2–7 tests; Supplementary Table S3 online).
Table 1. Conclusive genetic diagnosis obtained during the standard of care pathway and/or WES pathway.
Because it may be expected that patients receiving a conclusive diagnosis underwent either less (“easy to diagnose”) or more testing (“perseverance of continued diagnostic testing”), we compared whether patients receiving a conclusive diagnosis underwent fewer or more genetic tests than those without a diagnosis. However, there was no difference in the number of genetic tests in the standard pathway performed between patients who received a diagnosis and patients without one (P = 0.58, two-tailed t-test).
Diagnostic yield using whole-exome sequencing
In the WES pathway, 44 patients (29.3%) received a conclusive diagnosis, including 26 clinically well-known syndromes (e.g., MECP2 syndrome, KBG syndrome; Table 1, Supplementary Table S2 online). For 13 patients, a de novo point mutation was identified in a recently reported disease gene (Table 1). In addition, five pathogenic CNVs were identified that ranged in size from 342 kb to a whole chromosome 8 trisomy. Of note, the 44 patients who obtained a diagnosis using WES had underwent an average of 4.7 genetic tests in the standard diagnostic pathway (range, 1–12), which was not different from those for patients who had tested negative according to WES (P = 0.23, two-tailed t-test).
In addition to 44 conclusive diagnoses, WES identified a possible diagnosis for 41 further patients (27.3%) by reporting on presumably disease-causing mutations in candidate disease genes (Supplementary Table S4 online).
Diagnostic concordance between the standard care pathway and WES
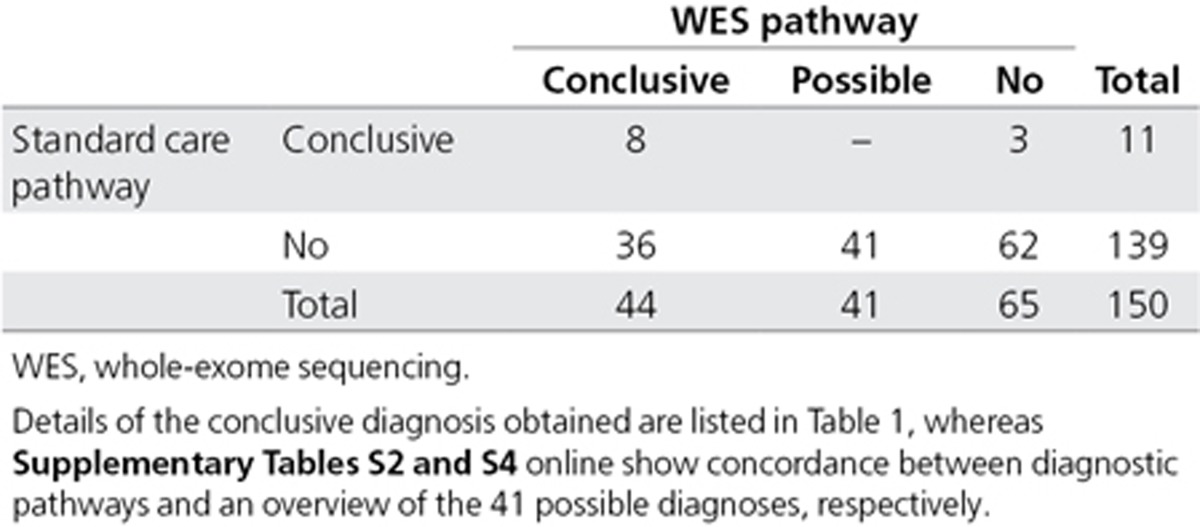
Next, we compared the diagnoses obtained at individual patient levels to determine the concordance between both pathways. In total, 47 patients (31.3%) obtained a diagnosis through either the standard or the WES pathway (Table 2; Supplementary Table S2 online). For eight patients (17.0%), the same conclusive diagnosis was obtained by both pathways. Three further patients (6.4%) received their genetic diagnosis only through the standard genetic care pathway, whereas 36 patients (76.6%) received their genetic diagnosis only through WES (Table 2; Supplementary Table S2 online). Statistical analysis comparing the overall diagnostic yields between both pathways indicated that the use of WES resulted in significantly more conclusive diagnoses than the standard care pathway (n = 11 vs. n = 44, P = 9.7 × 10–7, two-tailed Fisher’s exact test). The three genetic diagnoses that were not identified using WES included a 9-bp duplication event (patient 114), a repeat (CGG)n expansion in FMR1 (patient 43), and a mosaic 27 Mb duplication of chromosome 7 (patient 49) (Table 2; Supplementary Table S2 online).
Table 2. Diagnostic concordance between standard genetic care pathway and WES.
Health-care resource use
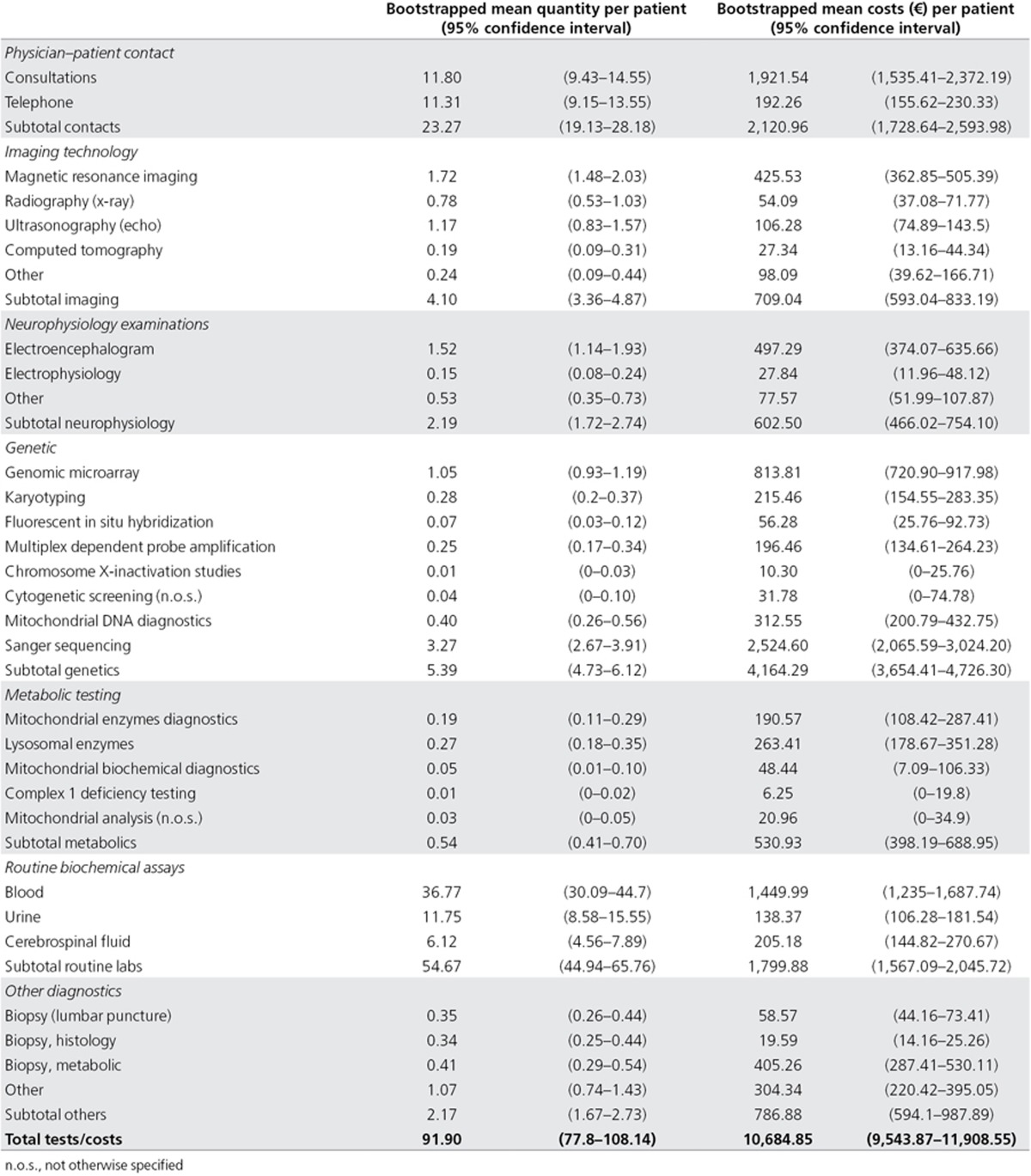
To gain insight into the cost-to-diagnosis ratio, we collected the health-care resource use during the diagnostic process both retrospectively (prior to inclusion to our study) and prospectively (after inclusion in our study). On average, patients had 23.3 physician–patient contacts and an extensive diagnostic workup including various forms of imaging technology (n = 4.1 tests), neurophysiology examinations (n = 2.2 tests), genetic diagnostic testing (n = 5.4 tests; Supplementary Table S3 online), (iv) metabolic assays (n = 0.5 assays), (v) basic clinical chemistry tests of blood, urine, and spinal fluid (n = 54.7 tests), and other diagnostic laboratory tests (n = 2.2; Table 3).
Table 3. Health-care resource use in the standard diagnostic trajectory.
Total costs for the standard diagnostic pathway, on average, amounted to €10,685 per patient (95% confidence interval: €9,544–€11,909), of which genetic testing was the major driver of diagnostic costs (€4,164); 150 patients received 806 genetic tests, with an average of 5.4 genetic tests per patient (range, 0–28 tests; Table 3; Supplementary Figures S3 and S4 and Supplementary Table S3 online). Of note, patients who were already in a diagnostic trajectory in our university hospital before inclusion, on average, underwent significantly more genetic tests (n = 6.6) than patients that were either referred by a general practitioner (n = 2.9; P < 0.01) or referred by a peripheral hospital (n = 4.1; P < 0.01).
In comparison, the average cost of WES-based genetic testing in this cohort was €3,420. Consequently, substitution of conventional genetic testing by WES in this cohort resulted in an average cost saving of €744 per patient. Notably, 17 patients (11.3%) received additional diagnostic testing as a consequence of the WES diagnostic report to support or decline the WES diagnosis (Supplementary Figure S5 and Supplementary Table S5 online).
Our exploratory analysis of the costs of diagnosis if WES were used as a first-tier test indicated that for 36 of 44 patients in whom WES led to a conclusive diagnosis, no additional tests would have been performed. The remaining eight patients would still have undergone additional (confirmatory) diagnostic tests ranging from a single cerebral imaging test or metabolic investigation to all diagnostic workup as performed before (Supplementary Figure S6 and Supplementary Table S6 online). Taking this into account, the mean costs of diagnostic tests in our patient cohort would have amounted to €8,356 (95% CI: €7,591–9,247) per patient. For patients with a conclusive WES diagnosis, those costs could be further reduced to €4,349 (95% CI: €3,994–4,777) because only limited other diagnostic testing would be required; however, for patients with a negative WES diagnosis, those costs would be €10,035 (95% CI: €9,312–10,797) because the remainder of the diagnostic pathway would remain the same (Supplementary Figure S6 online; Supplementary Table S6 online).
Discussion
Over the past few years, WES has been implemented in routine diagnostic genetic testing for clinically and genetically heterogeneous disorders.13,17,18,19,20 This implementation was technology-driven and based largely on the added diagnostic value obtained for patients who had reached the end stage in the standard diagnostic pathway. Here, we performed a clinical utility study using a design in which 150 consecutive patients with complex pediatric neurological disorders received both the standard diagnostic pathway as well as WES in parallel to determine the value of WES in pediatric neurology care. Using this study design, we were able to show that WES provides significantly more conclusive genetic diagnoses than the standard genetic care, without incurring higher costs.
WES was able to provide a conclusive diagnosis for 44 (29.3%) of 150 patients, whereas the standard diagnostic pathway was only able to provide a diagnosis for 11 patients (7.3%). The overall diagnostic yield for the different clinical entities as seen in pediatric neurology is in agreement with previous studies reporting on the diagnostic yield of WES for each of these entities (Supplementary Table S7 online). Despite the relatively high number of conclusive diagnoses by WES, three disease-causing mutations were identified only by standard genetic analysis. The three disease-causing mutations not identified by WES were directly related to technical pitfalls of WES. Two of three were not detected using the SNV detection tool, including a (CGG)n repeat expansion, which cannot be mapped because of the high repeat content of the expansion and a 9-bp duplication that was not mapped owing to relatively long duplication in an homopolymer (G) stretch in relation to the length of the sequence read. The third, a mosaic 27 Mb duplication, was not detected by the CNV detection tool. In retrospect, the latter mosaic duplication could be detected upon visual inspection of the data. Optimization of both the sequencing technology and the bioinformatic algorithms dedicated to the detection of such variants (both at SNV and CNV level) would eventually overcome these current technological pitfalls. In this respect, much progress is to be expected from genome (long-read) sequencing technologies because these will also capture (non-)coding variants and structural variation that currently remain undetected by WES.7,21,22
The standard care pathway did not identify 36 conclusive genetic diagnoses, including those of 21 known syndromes. This lack of conclusive diagnoses for known syndromes may be explained by three main factors. First, the median age of our patient cohort was low and young patients may not (yet) display all characteristic clinical features of the syndrome. Second, previous genetic testing may not have uncovered the causal variant. For two patients, we identified a CNV explaining the patients’ phenotype despite the fact that those patients underwent genomic microarray-based testing in their standard genetic pathway. Retrospective analysis of these original array data indicated that these two regions were poorly covered on the array, thereby explaining why these escaped detection (Supplementary Figure S7 online). Third, the clinical and genetic heterogeneity of the disorders encountered in pediatric neurology clinic is enormous.
Over the past few years, the use of next-generation sequencing technologies has led to the discovery of many novel disease-causing genes, especially for clinically and genetically heterogeneous disorders.23 This makes it virtually impossible to keep up to date on all genes involved in the respective diseases, thereby limiting the possibility to request the right gene for targeted mutation analysis. Patient 37, who received a clinical diagnosis of Kabuki syndrome, exemplified this situation. KMT2D, which causes Kabuki syndrome when mutated, was tested but no mutations were identified. Then, WES identified a disruptive mutation in KDM6A. Mutations in the KDM6A gene also cause Kabuki syndrome, but this was published24 only weeks after the patient was last seen by the medical specialist. This difficulty is further underscored by the fact that 13 of 44 (30%) of the conclusive WES diagnoses involved a gene that has been implicated in disease only in the past few years.
WES provided a possible genetic diagnosis for another 41 patients (27%). Although further clinical and genetic follow-up for larger cohorts of patients with similar phenotypic characteristics is required to establish the true nature of these diagnoses, our experience has shown that up to two-thirds of these possible diagnoses will be revised to conclusive diagnoses once additional patients with mutations in the same gene are identified and characterized.13,25,26 Moreover, with increasing knowledge over time and more genes being implicated in disease, WES data can be revisited and reinterpreted, and more conclusive diagnoses can be made. For instance, 3 of 106 WES-negative patients have already obtained a conclusive genetic diagnosis since our data freeze (July 2015), based on the availability of new clinical information and targeted reanalysis of existing WES data. Although speculative, it can be anticipated that the conclusive diagnostic yield of WES in this cohort may be as high as 50%, but this would require regular reanalysis of the available WES data.
In this study, we gathered data regarding health-care resource use to gain insight into the cost-to-diagnosis ratio in both the standard pathway and the WES genetic diagnostic tract. The standard care pathway results in a mean cost of €10,685 per patient, with €4,164 for genetic testing. Replacing all diagnostic efforts in this cohort by WES increased the diagnostic yield while reducing the expenses for genetic testing by €744. Our study thus determined the potential savings resulting from clinical implementation of WES, albeit for the Dutch health-care system. In this system, all patients (and parents) received complete insurance coverage for their diagnostic pathway, including the genetic tests. Patients who were already in a diagnostic trajectory in our hospital prior to study inclusion had undergone more genetic tests than patients who were newly referred. This suggests that the cost-to-diagnosis ratio could be reduced if clinicians perform WES instead of the conventional genetic tests during an earlier stage of the diagnostic trajectory. Moreover, it may be suggested that extensive (genetic) diagnostic workup after a negative WES result be refrained from in order to reduce the cost-to-diagnosis ratio even more. This is because our data show that only 2% of patients with a negative WES report obtained a conclusive diagnosis using the standard diagnostic workup and costs associated with these additional diagnoses are immense. Considering cost-effectiveness, it is also noteworthy that a reduction in the cost-to-diagnosis ratio in a WES-first approach may only be obtained for certain pediatric patients with complex phenotypes because patients with less complex phenotypes may be easier to diagnose with cheaper targeted testing such as gene-panel strategies. Additionally, given the worldwide differences in prices for diagnostic WES11 and other (genetic) diagnostic tests analyzed in our study, the extensibility and translation of our cost analyses to other health-care systems and countries require further studies.
A more precise (genetic) diagnosis may have a major impact on clinical management and facilitates personalized medicine. For instance, in patient 146, who had been diagnosed with an SCN8A mutation, the antiepileptic drug was changed to oxcarbazepine, which exerts its action by blocking voltage-sensitive sodium channels. In our study, we revisited the diagnostic care pathways of our patients and evaluated what changes could have been made if WES had been the first-tier diagnostic test. Importantly, for 36 of 150 patients (24%) the diagnostic process would have been limited to WES and patients would not have undergone additional (invasive) medical procedures. We have previously reported the time to diagnosis for 50 of the 150 patients in the conventional pathway,3 which showed that the mean duration is at least 40 months. Routine turnaround times of WES are 4–6 months worldwide, which can probably be reduced to weeks27,28 by further automation and streamlining of the laboratory and interpretation process.
One aspect of responsible health-care innovation and changes in diagnostic pathways is the perception, expectations, and needs of the end users of the novel product. We have previously explored the parents’ information and communication needs in the counseling strategies for diagnostic WES using in-depth semi-structured interviews with 15 families of our cohort.29 Additionally, we investigated the psychosocial effects of WES diagnoses.30 Whereas our interviews did not identify any major hurdles for widespread implementation of WES as a (first-tier) diagnostic test, which is in line with other studies,31 the interviews did emphasize that a diagnosis has a broader impact than just a medical one.29,30 The latter was highlighted by novel (psychosocial) uncertainties that parents face when their child is diagnosed with a rare genetic disease for which little is known and only a few patients have been diagnosed with worldwide. Also, parents reported the need for more postcounseling advice, which can be fulfilled by a genetic counselor who, after the diagnosis, maintains long-term contact with the family and helps with everyday problems associated with having a child with a (rare) genetic disease. Another important aspect of patient empowerment is the disclosure of incidental findings and related consent procedures.32,33,34,35 Several groups have reported incidental findings that vary between 1.2 and 4.6% for patients undergoing WES.36,37 However, our study did not reveal any. Nonetheless, the detailed interviews with parents did indicate that a tailor-made disclosure policy for incidental findings is warranted, which is in accordance with previous studies.38,39
In conclusion, we performed a study of the diagnostic yield and costs of WES in pediatric neurology as compared to the conventional diagnostic trajectory. Our data show that WES provides more conclusive genetic diagnoses than standard (genetic) care without incurring higher costs. Moreover, in this patient population, a reduction of costs associated with obtaining a molecular diagnosis may be achieved if WES were used as the first-tier test, thereby partially substituting imaging studies, biopsies, and lumbar punctures and replacing conventional genetic diagnostics. Our data support a change in clinical pediatric neurology practice toward WES as a first-tier test in the diagnostic trajectory of patients presenting with complex neurological disorders of presumed genetic origin.
Disclosure
The authors declare no conflict of interest.
Acknowledgments
The authors thank all families included in this study for their participation. In addition, they thank the Genomics Technology Platform of the Radboud University Medical Center for performing diagnostic WES for this study and Christian Gilissen and Jayne Hehir-Kwa for bioinformatic support of the diagnostic analyses pipelines for WES. The study was financially supported by the Netherlands organization of Health Research and Development (ZonMW 40-41200-98-9131).
Supplementary Material
References
- National Institute of Neurological Disorders and Stroke. https://www.ninds.nih.gov/Disorders/all-disorders.
- Gahl WA, Markello TC, Toro C, et al. The National Institutes of Health Undiagnosed Diseases Program: insights into rare diseases. Genet Med 2012;14:51–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Nimwegen KJ, Schieving JH, Willemsen MA, et al. The diagnostic pathway in complex paediatric neurology: a cost analysis. Eur J Paediatr Neurol 2015;19:233–239. [DOI] [PubMed] [Google Scholar]
- Olesen J, Gustavsson A, Svensson M, Wittchen HU, Jönsson B; CDBE2010 study group; European Brain Council. The economic cost of brain disorders in Europe. Eur J Neurol 2012;19:155–162. [DOI] [PubMed] [Google Scholar]
- Singleton AB. Exome sequencing: a transformative technology. Lancet Neurol 2011;10:942–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durr A. Autosomal dominant cerebellar ataxias: polyglutamine expansions and beyond. Lancet Neurol 2010;9:885–894. [DOI] [PubMed] [Google Scholar]
- Srivastava S, Cohen JS, Vernon H, et al. Clinical whole exome sequencing in child neurology practice. Ann Neurol 2014;76:473–483. [DOI] [PubMed] [Google Scholar]
- Nolan D, Carlson M. Whole exome sequencing in pediatric neurology patients: clinical implications and estimated cost analysis. J Child Neurol 2016;31:887–894. [DOI] [PubMed] [Google Scholar]
- Das AS, Agamanolis DP, Cohen BH. Use of next-generation sequencing as a diagnostic tool for congenital myasthenic syndrome. Pediatr Neurol 2014;51:717–720. [DOI] [PubMed] [Google Scholar]
- Iglesias A, Anyane-Yeboa K, Wynn J, et al. The usefulness of whole-exome sequencing in routine clinical practice. Genet Med 2014;16:922–931. [DOI] [PubMed] [Google Scholar]
- Lazaridis KN, Schahl KA, Cousin MA, et al. Outcome of whole exome sequencing for diagnostic Odyssey cases of an individualized medicine clinic: The Mayo Clinic Experience. Mayo Clin Proc 2016;91: 297–307. [DOI] [PubMed] [Google Scholar]
- Richards S, Aziz N, Bale S, et al.; ACMG Laboratory Quality Assurance Committee. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 2015;17:405–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Ligt J, Willemsen MH, van Bon BW, et al. Diagnostic exome sequencing in persons with severe intellectual disability. N Engl J Med 2012;367:1921–1929. [DOI] [PubMed] [Google Scholar]
- Pfundt R, Del Rosario M, Vissers LE, et al. Detection of clinically relevant copy-number variants by exome sequencing in a large cohort of genetic disorders. Genet Med 2016; e-pub ahead of print 27 October 2016. [DOI] [PMC free article] [PubMed]
- Genome Diagnostics Nijmegen. Radboudumc, Human Genetics. https://www.radboudumc.nl/Informatievoorverwijzers/Genoomdiagnostiek/en/Pages/Exomesequencing.aspx.
- Briggs AH, Wonderling DE, Mooney CZ. Pulling cost-effectiveness analysis up by its bootstraps: a non-parametric approach to confidence interval estimation. Health Econ 1997;6:327–340. [DOI] [PubMed] [Google Scholar]
- Neveling K, Feenstra I, Gilissen C, et al. A post-hoc comparison of the utility of sanger sequencing and exome sequencing for the diagnosis of heterogeneous diseases. Hum Mutat 2013;34:1721–1726. [DOI] [PubMed] [Google Scholar]
- Beaulieu CL, Majewski J, Schwartzentruber J, et al.; FORGE Canada Consortium. FORGE Canada Consortium: outcomes of a 2-year national rare-disease gene-discovery project. Am J Hum Genet 2014;94:809–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Muzny DM, Reid JG, et al. Clinical whole-exome sequencing for the diagnosis of mendelian disorders. N Engl J Med 2013;369:1502–1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauch A, Wieczorek D, Graf E, et al. Range of genetic mutations associated with severe non-syndromic sporadic intellectual disability: an exome sequencing study. Lancet 2012;380:1674–1682. [DOI] [PubMed] [Google Scholar]
- Huddleston J, Eichler EE. An incomplete understanding of human genetic variation. Genetics 2016;202:1251–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilissen C, Hoischen A, Brunner HG, Veltman JA. Unlocking Mendelian disease using exome sequencing. Genome Biol 2011;12:228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vissers LE, Gilissen C, Veltman JA. Genetic studies in intellectual disability and related disorders. Nat Rev Genet 2016;17:9–18. [DOI] [PubMed] [Google Scholar]
- Miyake N, Mizuno S, Okamoto N, et al. KDM6A point mutations cause Kabuki syndrome. Hum Mutat 2013;34:108–110. [DOI] [PubMed] [Google Scholar]
- Coe BP, Witherspoon K, Rosenfeld JA, et al. Refining analyses of copy number variation identifies specific genes associated with developmental delay. Nat Genet 2014;46:1063–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mencacci NE, Kamsteeg EJ, Nakashima K, et al. De novo mutations in PDE10A cause childhood-onset chorea with bilateral striatal lesions. Am J Hum Genet 2016;98:763–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soden SE, Saunders CJ, Willig LK, et al. Effectiveness of exome and genome sequencing guided by acuity of illness for diagnosis of neurodevelopmental disorders. Sci Transl Med 2014;6:265ra168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders CJ, Miller NA, Soden SE, et al. Rapid whole-genome sequencing for genetic disease diagnosis in neonatal intensive care units. Sci Transl Med 2012;4:154ra135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krabbenborg L, Schieving J, Kleefstra T, et al. Evaluating a counselling strategy for diagnostic WES in paediatric neurology: an exploration of parents’ information and communication needs. Clin Genet 2016;89:244–250. [DOI] [PubMed] [Google Scholar]
- Krabbenborg L, Vissers LE, Schieving J, et al. Understanding the psychosocial effects of WES test results on parents of children with rare diseases. J Genet Couns 2016;25:1207–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sie AS, Prins JB, van Zelst-Stams WA, Veltman JA, Feenstra I, Hoogerbrugge N. Patient experiences with gene panels based on exome sequencing in clinical diagnostics: high acceptance and low distress. Clin Genet 2015;87:319–326. [DOI] [PubMed] [Google Scholar]
- Green RC, Berg JS, Grody WW, et al.; American College of Medical Genetics and Genomics. ACMG recommendations for reporting of incidental findings in clinical exome and genome sequencing. Genet Med 2013;15:565–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ACMG Board of Directors. ACMG policy statement: updated recommendations regarding analysis and reporting of secondary findings in clinical genome-scale sequencing. Genet Med 2015;17: 68–9. [DOI] [PubMed] [Google Scholar]
- Regier DA, Peacock SJ, Pataky R, et al. Societal preferences for the return of incidental findings from clinical genomic sequencing: a discrete-choice experiment. CMAJ 2015;187:E190–E197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hehir-Kwa JY, Claustres M, Hastings RJ, et al. Towards a European consensus for reporting incidental findings during clinical NGS testing. Eur J Hum Genet 2015;23:1601–1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Muzny DM, Xia F, et al. Molecular findings among patients referred for clinical whole-exome sequencing. JAMA 2014;312:1870–1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorschner MO, Amendola LM, Turner EH, et al.; National Heart, Lung, and Blood Institute Grand Opportunity Exome Sequencing Project. Actionable, pathogenic incidental findings in 1,000 participants’ exomes. Am J Hum Genet 2013;93:631–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bredenoord AL, Onland-Moret NC, Van Delden JJ. Feedback of individual genetic results to research participants: in favor of a qualified disclosure policy. Hum Mutat 2011;32:861–867. [DOI] [PubMed] [Google Scholar]
- Christenhusz GM, Devriendt K, Van Esch H, Dierickx K. Focus group discussions on secondary variants and next-generation sequencing technologies. Eur J Med Genet 2015;58:249–257. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.