Abstract
Background:
Although distant metastasis (DM) in breast cancer (BC) is the most lethal form of recurrence and the most common underlying cause of cancer related deaths, the outcome following the development of DM is related to the site of metastasis. Triple negative BC (TNBC) is an aggressive form of BC characterised by early recurrences and high mortality. Athough multiple variables can be used to predict the risk of metastasis, few markers can predict the specific site of metastasis. This study aimed at identifying a biomarker signature to predict particular sites of DM in TNBC.
Methods:
A clinically annotated series of 322 TNBC were immunohistochemically stained with 133 biomarkers relevant to BC, to develop multibiomarker models for predicting metastasis to the bone, liver, lung and brain. Patients who experienced metastasis to each site were compared with those who did not, by gradually filtering the biomarker set via a two-tailed t-test and Cox univariate analyses. Biomarker combinations were finally ranked based on statistical significance, and evaluated in multivariable analyses.
Results:
Our final models were able to stratify TNBC patients into high risk groups that showed over 5, 6, 7 and 8 times higher risk of developing metastasis to the bone, liver, lung and brain, respectively, than low-risk subgroups. These models for predicting site-specific metastasis retained significance following adjustment for tumour size, patient age and chemotherapy status.
Conclusions:
Our novel IHC-based biomarkers signatures, when assessed in primary TNBC tumours, enable prediction of specific sites of metastasis, and potentially unravel biomarkers previously unknown in site tropism.
Keywords: distant metastasis, biomarkers, triple-negative breast cancer, site specificity
Breast cancer (BC) is the most common cancer and the second leading cause of cancer-related deaths among North American women (Group UCSW, 2016), and presents the largest overall cancer threat for women worldwide (Jemal et al, 2011). The vast majority of BC deaths result from dissemination of cancer to distant metastatic sites (Weigelt et al, 2005). BC, unlike prostate or sarcomas (Nguyen et al, 2009), shows significantly more organ variation in metastasis (Lee, 1983), making it very challenging to employ site-specific surveillance/preventative measures.
The molecular subtypes of BC have been shown to have very different underlying biology and distinct metastasis patterns (Smid et al, 2008). Triple negative BC (TNBC) is a subtype of BC characterised by an absence of the oestrogen receptor (ER), progesterone receptor (PR) and HER2 protein over-expression. TNBCs account for around 16% of invasive BCs (Rakha et al, 2007b) and are considered one of the most clinically aggressive subtypes, with over twice the risk of distant metastasis (DM) relative to other molecular subtypes (Anders and Carey, 2009). Compared with the other subtypes, TNBCs also show much higher frequencies of metastasis to the brain and lung—sites associated with higher mortality compared with bone and other sites (Luck et al, 2008; Kennecke et al, 2010). Predicting TNBC’s propensity for metastasis to those specific sites may allow preventive therapy and enable active surveillance to significantly improve outcomes.
The metastatic cascade is a multi-step process consisting of growth, vascularisation, detachment, invasion, evasion of host defenses and survival in circulation, extravasation and finally the ability to grow in the new organs’ microenvironment (Fidler, 2003; Gupta and Massagué, 2006). Few cells are successfully able to accomplish all of the steps, and require specific biological properties both for general metastasis (e.g., factors that trigger EMT) and site-specific metastasis (e.g., breaching the blood–brain barrier to colonise the brain; Luzzi et al, 1998; Gupta and Massagué, 2006; Nguyen et al, 2009). The similarity of genetic profiles between the primary and metastatic site tumours seems to suggest that many of the properties required for successful metastasis are developed early in the primary tumour cells well prior to the onset of metastasis (Weigelt et al, 2003). Therefore, the identification of the specific biomarker profile of a primary tumour that is primed to metastasise to a specific site would enable the development of preventative and surveillance strategies tailored specifically to that particular site.
Being able to predict site of metastasis has very tangible evidence of improving patient survival. For instance, Denosumab (Smith et al, 2012), a RANKL antibody, and Bonefos (Powles et al, 2006), an oral bisphosphonate clodronate, have shown significant effect in reducing bone-specific metastasis in clinical trials. However, both are currently recommended only for patients who already show evidence of bone metastasis (Hillner et al, 2003; Coleman et al, 2014). For brain metastases, where the blood–brain barrier makes targeting tumour areas very difficult (Steeg et al, 2011), there has been success pre-clinically, using Vorinostat (Palmieri et al, 2009), a histone deactylase inhibitor, and in clinical trials via sorafenib (Massard et al, 2010), a kinase inhibitor. For lung metastasis, there has been success of inhibiting metastasis by blocking-specific lung guiding molecules, S100A8 and S100A9 (Hiratsuka et al, 2006). Finally for liver, where COX-2 expression is increased, using etodolac markedly decreased invasive properties (Chen et al, 2001). Thus, if the site of metastasis could be identified in advance, active surveillance and the use of preemptive therapies could be implemented (Steeg et al, 2011).
Studies involving genomic data have attempted to identify signatures for metastatic tropism. Multiple studies using microarray data have characterised gene expression profiles of BC that preferentially metastasised to lung or bone in mice (Gupta et al, 2005; Minn et al, 2005). A retrospective study of transcriptomic data enabled the identification of a 6-gene prognostic classifier, which could significantly discriminate BC patients who developed DM to lung (Landemaine et al, 2008). Although the benefit of using genetic data is quite obvious, it also has pitfalls, the biggest of which is the lack of strong correlation between (a) gene expression and protein levels, and (b) protein levels and protein activity levels, the latter of which can be extensively modified post-translationally. In addition, although the price of sequencing a genome is decreasing exponentially, the price may still be prohibitive in a clinical setting (Caulfield et al, 2013).
To investigate protein signatures (in primary tumours) that are predictive of potential metastasis to specific anatomical sites, we evaluated a well-characterised cohort of clinically annotated TNBC with a long-term follow-up, utilising the immunohistochemical expression of 133 biomarkers with relevance to BC progression and metastasis. By taking into account the protein localisation (nuclear and/or cytoplasmic), the staining intensity, percentage of cells expressing the biomarker and standard clinicopathologic variables, we investigated over 400 variables to produce the most relevant statistical models. In this paper, we describe a method for step-wise filtering that yielded robust predictive models for four distinct sites of TNBC metastasis: bones, liver, lung and brain.
Materials and methods
Study population
This study was based on a well-characterised series of primary operable invasive breast carcinoma cases (TNM stage I–IIIA) diagnosed in Nottingham between 1989 and 1998 (N=1944), of which 322 were classified as TNBC (i.e., 0% IHC staining of PR, ER and HER2 0/1+ IHC staining or 2+ FISH non-amplified) (Supplementary Table S1). Patients’ clinical history and tumour characteristics, information on therapy, tumour recurrence and survival are described in previous publications (Abd El-Rehim et al, 2004, 2005; Rakha et al, 2006, 2007a, 2009; Luck et al, 2008). Data related to outcome including information on the development, site and time of DM and mortality were collected prospectively. Patients were treated according to a uniform protocol based on the Nottingham Prognostic Index (NPI) groups (Galea et al, 1992), ER and menopausal status. A systemic cyclophosphamide-methotrexate-5-fluorouracil (CMF) chemotherapy regimen was used if the patient was ER-negative, provided the patient was considered fit enough to withstand this regimen. None of the patients received neoadjuvant or anti-HER2-targeted therapy.
Antibody preparation and details for selected biomarkers is available in the online Supplementary Data. This study included 133 IHC-based biomarkers (Supplementary Table S2) of clinical and biological relevance to BC (Abd El-Rehim et al, 2004, 2005; Rakha et al, 2006, 2007a, 2009; Elsheikh et al, 2008; Luck et al, 2008; Rakha et al, 2009; Habashy et al, 2011; Mahmoud et al, 2011; Abduljabbar et al, 2015; Alshareeda et al, 2015; Jerjees et al, 2015). During the follow-up period (243 months), 197 patients (61.2%) remained disease-free, whereas 111 (34.5%) developed DM. Ethical approval was granted by Nottingham Research Ethics Committee 2 under the title ‘Development of a molecular genetic classification of breast cancer’ (C202313) and by The North West 7 Research Ethics Committee- Greater Manchester Central (10/H1008/72).
Statistical analysis
Statistical analysis was carried out with SAS 9.4 software (Cary, NC, USA) and Matlab version 9.2.0.556344 (R2017a, Natick, MA, USA). Patients were first grouped according to the site of DM or to a ‘no metastasis’ group. If a patient had multiple metastases, that patient would be included in all the relevant groups based on the sites of their multiple metastases. Differences between clinicopathological proportions were determined using the χ2 test. Differences between continuous clinicopatholgical variables were evaluated via a two-tailed t-test.
Biomarker feature selection
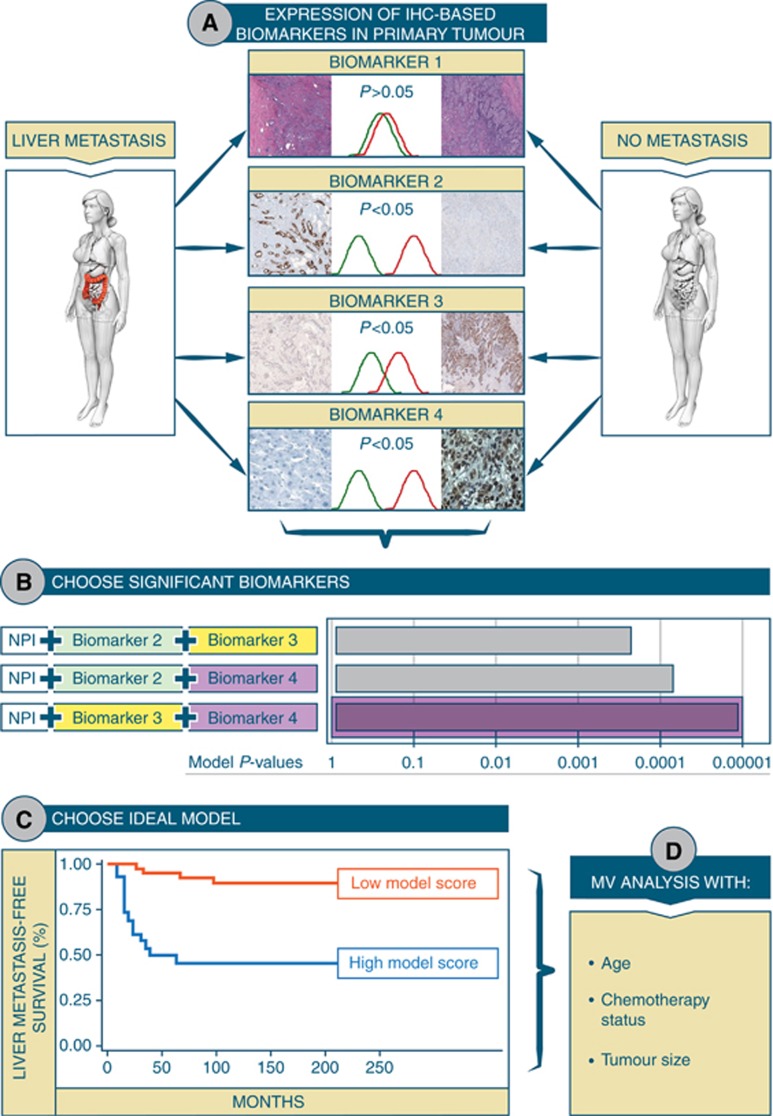
Owing to the variation of the number of biomarkers, which each patient in our data set was stained for (coefficient of variation=0.36, not shown), and the difference in number of cases with informative data for each stained biomarker, we chose to select two biomarkers for each DM model. This allowed us to preserve substantial n numbers and to keep the models clinically facile. Biomarker selection (Figure 1A) was done using three progressive significance tests for each site. First, two-tailed t-tests were performed between patients in whom DM occurred to that site vs patients who remained DM-free. This is to test for significant baseline differences between all biomarkers. Significantly different (P-value <0.05) biomarkers were displayed as waterfall plots, with the height of each bar representing the average difference between expression of that biomarker in the ‘site-specific metastasis’ group vs the ‘metastasis-free’ group. Further selection was done through logistic regression, with ‘yes vs no’ binary responses using all the biomarkers, one by one, as predictors. This selection appeared to be more stringent, as much fewer biomarkers were shown to be significant. Variables that were selected both by the t-test and the logistic regression are represented by asterisks on the waterfall plots. Finally, the selected biomarkers were run through a univariate Cox proportional hazard models for prognostic filtering, with Wald P-values <0.05 indicating significant variables (unless no biomarkers were found using this criterion, in which case it was relaxed to 0.1). Time to site-specific metastasis was considered as the time interval from date of surgery to date of DM to that particular site. Significant prognostic biomarkers were represented via arrows on the aforementioned waterfall plots.
Figure 1.
Schematic depicting sequence of steps leading to development of a model that predicts site-specific metastasis in TNBC. Briefly, a two-tailed t-test was used to compare the biomarker profile for each patient who developed a site-specific metastasis vs every patient who did not have any metastasis. The biomarkers that showed significant differences in expression were then compared prognostically, with a continuous univariate Cox model, for site-specific metastasis hazard. Those significant variables that had a P-value <0.1 were then all tested with each other to identify the best combination, alongside NPI.
Model building
Models were built by combining all previously selected prognostic biomarkers (in pairs), with the patient’s NPI. Each model used the Cox parameter of the respective biomarkers as weights, combined into a score, and was thresholded (Figure 1B) by using Contal’s and O’Quigley’s approach (Mandrekar et al, 2003). The model chosen, for each distant site studied, was the one that minimised the Cox and Wald’s P-values (Figure 1C). The NPI threshold for testing risk of metastasis to each site using our models, was determined by finding the highest NPI value, which would, regardless of the values for the IHC biomarkers in the relevant risk model, not allow the patient to have a score above the risk threshold (i.e., not allow the patient to fall into the high-risk group for that particular anatomical site). To evaluate whether their ability to predict risk of site-specific metastasis was robust regardless of the nature of model used, the selected biomarkers were also evaluated using two different machine learning algorithms: a support vector machine (SVM) and an Ensemble tree-based method. Hyperparameters for both types of models were found using the Bayesian optimisation, through maximisation of the ‘expected-improvement-plus’ (Bull, 2011; Gelbart et al, 2014) over 60 iterations (Supplementary Tables S3 and S4). The following parameters were optimised for the SVM algorithm: Box Constraint, Kernel Scale, Kernel type, Polynomial order (if polynomial kernel) and feature standardisation. For the Ensemble tree algorithm, the following hyperparameters were optimised: Ensemble method (Bagging, GentleBoost, LogitBoost, AdaBoost, RUSBoost’), maximum number of branch nodes, minimum number of leaf nodes and the split criteria. Both methods were also built with/without empirical prior data set probabilities for site-specific metastasis. The optimised hyperparameters for each model are detailed in the online Supplementary Data.
Model validation
All models (namely, our combined and then thresholded model, the optimised SVM, and the optimised Ensemble) to each site, were five-fold cross-validated for survival-risk evaluation. Kaplan–Meier survival curves were created by combining the five testing sets and then used to confirm significance and rank models. The comparison metric used to compare the cross validated models was the Akaike Information Criterion (AIC), a measure of fit. The model that granted the lowest AIC per site, was considered the optimal model for that site. Multivariate analysis was also performed to control for the effects of chemotherapy, tumour size and age.
Results
The ability of clinical variables to predict DM (Park et al, 2015a) and specifically in TNBCs (Pogoda et al, 2013) is well documented, with common features, as for instance tumour size and nodal stage providing significant prognostic ability. Our data corroborate these findings by showing tumour size (HR=0.002), age (P<0.048) and NPI (P<0.0001) as having significant univariate impact on DM-free survival. However, a comparison of the distribution of these clinical factors for specific metastasis sites (Supplementary Table S5) showed no difference in mean values of these variables or in the proportions of patients in each group. We also observed that chemotherapy did not affect recurrence patterns (Supplementary Table S6). This led us to investigate whether any of our biomarker models could provide the required specificity of being both prognostically relevant and unique to specific DM sites.
Bone metastasis
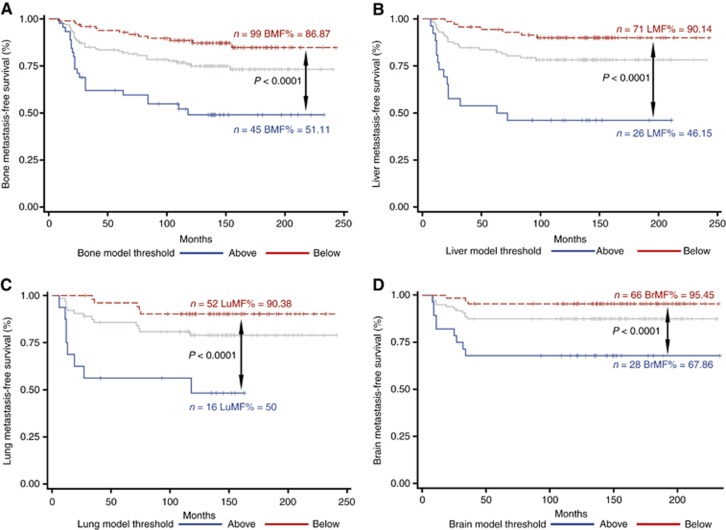
Among the protein biomarkers available in our TNBC data set, those whose expression was significantly different in patients who developed bone metastasis (Supplementary Figure S7A and B), included several that were overexpressed (blue lines) or highly underexpressed (red lines) in the primary tumour. The eight biomarkers that are eligible for inclusion into the final model, based on univariate prognostic significance, are indicated. S7B shows the results of the parameter selection, with the lowest P-value (P<0.0001) obtained combining the MTA1 nuclear H-score, KNPA2 nuclear percentage, in addition to NPI. NPI was included in all our models as a stand-in for a ‘generalised risk of metastasis’ as high-NPI patients have a higher risk of DM compared with low-NPI (metastasis HR=1.6, P<0.001). This model, detailed below, enables us to identify patients who have a five times higher risk of developing metastasis to bones (Figure 2A) and stayed significant after cross validation (Supplementary Figure S8A). Multivariate analysis (Table 1) confirmed the prognostic value of our model by having it independently associated with bone metastasis risk (P<0.0001) following adjustment for age, chemotherapy status, and tumour size.
Figure 2.
Model derived comparisons of high versus low risk patients for site specific metastasis. Kaplan–Meier survival curves showing patient stratification via our survival-based models for (A) bone (BMF=breast metastasis free), (B) liver (LMF=liver metastasis free), (C) lung (LuMF=lung metastasis free), and (D) brain sites (BrMF=brain metastasis free). All significances are measured via the log-rank test. Light grey lines represent baseline survival for the patients before stratification by the respective site-specific metastasis predictive models.
Table 1. Univariate and multivariate Cox regression analysis of common clinicopathological variables and IHC models affecting distant metastasis risk.
|
Univariate analysis |
Multivariate analysis |
||||||
|---|---|---|---|---|---|---|---|
| Variables | Hazard ratio | 95% Confidence interval | P-value | Hazard ratio | 95% Confidence interval | P-value | |
|
Bone | |||||||
| Age of diagnosis | 1.022 | 0.997–1.048 | 0.0813 | 0.998 | 0.959–1.038 | 0.9028 | |
| Chemotherapy | CMF vs none | 0.801 | 0.452–1.419 | 0.4471 | 0.741 | 0.301–1.824 | 0.5148 |
| Tumour size | Per CM | 1.397 | 1.001–1.951 | 0.0496* | 1.355 | 0.862–2.132 | 0.188 |
| Risk model | High vs low | 5.123 | 2.572–10.201 | <0.0001* | 4.939 | 2.281–10.692 | <0.0001* |
|
Liver | |||||||
| Age of diagnosis | 1.002 | 0.971–1.034 | 0.9042 | 0.969 | 0.925–1.016 | 0.1957 | |
| Chemotherapy | None vs CMF | 1.202 | 0.570–2.536 | 0.6294 | 0.57 | 0.185–1.757 | 0.3275 |
| Tumour size | Per CM | 1.688 | 1.171–2.432 | 0.005* | 1.134 | 0.592–2.172 | 0.7047 |
| Risk model | High vs low | 8.039 | 3.230–20.005 | <0.0001* | 9.156 | 3.376–24.837 | <0.0001* |
|
Lung | |||||||
| Age of diagnosis | 1.004 | 0.973–1.037 | 0.7878 | 0.972 | 0.902–1.047 | 0.4486 | |
| Chemotherapy | None vs CMF | 0.808 | 0.402–1.626 | 0.5507 | 0.686 | 0.156–3.016 | 0.618 |
| Tumour size | Per CM | 1.761 | 1.221–2.540 | 0.0025* | 1.735 | 0.712–4.226 | 0.225 |
| Risk model | High vs low | 7.661 | 2.491–23.564 | 0.0004* | 6.306 | 1.567–25.374 | 0.0095* |
|
Brain | |||||||
| Age of diagnosis | 1.005 | 0.971–1.040 | 0.7718 | 0.943 | 0.867–1.026 | 0.1721 | |
| Chemotherapy | None vs CMF | 1.261 | 0.572–2.778 | 0.565 | 0.035 | 0.003–0.390 | 0.0064* |
| Tumour size | Per CM | 1.717 | 1.129–2.610 | 0.0115* | 4.632 | 1.519–14.132 | 0.0071* |
| Risk model | High vs low | 8.506 | 2.299–31.471 | 0.0013* | 36.362 | 4.276–309.228 | 0.001* |
Abbreviations: CM = centimeter; CMF=cyclophosphamide-methotrexate-5-fluorouracil. *P<0.05.
 |
We also compared the performance of this model in the patient subgroup that received adjuvant CMF chemotherapy vs the subgroup that received no adjuvant chemotherapy, to determine whether the model’s prognostic value was affected by therapy. Results showed that the model for predicting bone-specific metastasis maintained significance regardless of whether chemotherapy was administered or not (Supplementary Table S9). Interestingly, the cross-validated AICs showed that this survival-based model slightly outperformed the SVM and Ensemble-based models (Supplementary Table S10). Notably, all models tested yielded statistically significant stratification.
Liver metastasis
For patients with liver metastases, we observed that the majority of differentially expressed biomarkers (P<0.05; 29 vs 9) were underexpressed in the patient subgroup with liver metastases compared with metastasis-free patients (Supplementary Figure S11A). Furthermore, we found that the underexpression of majority of these biomarkers (7 vs 1), was statistically significant in univariate analyses. The combination that yielded the lowest P-value (P <0.0001) involved N-cadherin H score, the cytoplasmic intensity of xeroderma pigmentosum complementation group D (XPD) and NPI (Supplementary Figure S11B). The model shown below can stratify patients into a high-risk group that shows ∼8 × higher risk of liver metastasis (Figure 2B), and retained significance after cross validation (Supplementary Figure S8B). Multivariate analysis indicated that this model is contributing predictive information for liver-specific metastasis independently of other factors (Table 1).
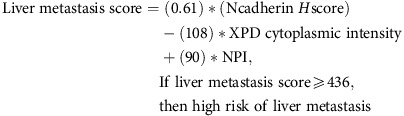 |
As with the bone model, the survival-based model for lung retained significance regardless of chemotherapy (Supplementary Table S9) and performed marginally better than machine learning approaches (Supplementary Table S10).
Lung metastasis
Unlike for liver, multiple IHC biomarkers, such as Fascin-1, Id1 and Id3 have been reported to mediate lung colonisation in invasive BC including TNBCs (Gupta et al, 2007; Ruiz de Garibay et al, 2015). Unlike the previously mentioned proteins (where overexpression was correlated with lung metastasis), our model filtering (Supplementary Figures S12A and B) led to the selection of two biomarkers that accorded a favourable prognosis when expressed at a high level. Combining TFF1 and RARa, as shown below, produced a high-risk group, which had over a seven times higher risk of developing lung metastasis (Figure 2C). In addition, this model retained its significance in cross validation (Supplementary Figure S8C) and multivariable analysis, independent of other factors (Table 1).
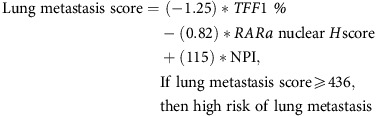 |
Unlike the previous two site-specific models, using an SVM to predict lung metastasis produced a marginally superior AIC, and thus fit (Supplementary Table S10), although all models retained significant stratification. Also, although the model showed powerful prognostic ability among CMF-treated patients, it lost significance in the patient subgroup that did not receive CMF (Supplementary Table S9, P=0.1); this was likely due to the low number of metastatic events and metastasis-free patients in that patient subgroup (4 and 27, respectively).
Brain metastasis
Although brain metastasis only accounts for around 10–16% of all breast metastasis sites (Barnholtz-Sloan et al, 2004), and is a relatively longer process due to the blood–brain barrier (Weil et al, 2005), it results in a very poor survival and a marked reduction in quality of life (Klos and O’Neill, 2004). The current paucity of biomarkers with the ability to predict metastasis to the brain (Arnold et al, 1999), coupled with lack of an effective targeted treatment (Deeken and Löscher, 2007) demonstrate that this is an area of urgent and unmet clinical need for BC patients. Recently, though, αB-crystallin, a chaperone protein predominantly expressed in brain metastasis, has shown promise as a TNBC site-specific IHC biomarker (Malin et al, 2014; Voduc et al, 2015). In our data set, we found only a few biomarkers that (a) showed significantly different expression between patients with metastasis to the brain and those with no metastases, and (b) had prognostic value in univariate analyses (Supplementary Figure S13A). Post-hoc survival analysis using a non-optimised (minimised Wald P-value) biomarker combination for brain metastasis patients yielded a very imbalanced high-risk group that included only two patients (not shown). We therefore combined the biomarkers whose combination had the second best P-value (Supplementary Figure S13B) to develop the model shown below. With this model, high-risk patients possessed more than a 7 × higher risk of brain metastasis (Figure 2D). This effect was maintained in multivariate analysis (Table 1) and cross validation (Supplementary Figure S8D).
 |
The prognostic value appears to result from significant stratification of the untreated patients (Supplementary Table S9), as only three treated patients, who were stained for both markers, had DM to the brain. Patient prognosis, while significant with our model, was predicted slightly better using an SVM (Supplementary Table S10).
We then addressed the question of whether every TNBC patient in the clinic should be prescribed the test for our panel of eight IHC-based biomarkers that are able to foretell risk of metastasis to specific sites. Interestingly, we found that the vast majority of TNBC patients in this data set had an NPI >4 regardless of whether they experienced metastasis or not (Supplementary Figure S14A), and thus would require testing for all eight biomarkers. We confirmed elevated NPI among TNBCs in a second independent data set (Supplementary Figure S14B). These data suggest that with the exception of a very small proportion of TNBC patients whose NPI is below 4, the majority of TNBCs may require testing for all eight biomarkers to determine risk of metastasis to these sites in the future.
Discussion
BC patients with DM have a median survival of only 2–3 years (Cardoso et al, 2012). Even more worrisome is the fact that both the time until DM and survival after metastasis is greatly reduced for TNBCs, especially among those with residual disease after neoadjuvant treatment (Liedtke et al, 2008; Cleere, 2010). However, metastasis to different sites is associated with distinct survival times after metastasis with some metastatic sites associated with poorer outcomes compared to others. Therefore, predicting DM before it occurs and identifying the potential sites of metastasis would have a significant impact in management of TNBC.
Previous studies investigating biomarkers predictive of the site of DM in BC have mainly utilised either global gene expression data using high-throughput techniques such as microarrays and next-generation sequencing or single proteins using IHC (Largillier et al, 2008; Hu et al, 2009; Lorusso and Ruegg, 2012). No studies have investigated DM using large groups of protein biomarkers in primary TNBC tumour samples. Using our novel models, we are able to introduce a clinically-facile IHC biomarker panel that can identify high-risk subgroups among TNBCs, with at least a 5 × increased risk of site-specific metastasis. The strength of the current study stems from (a) the large number of cases in our TNBC series, (b) their long-term follow-up and detailed clinical annotation, (c) the unique, and, to the best of our knowledge, largest IHC biomarker data set available for this cohort, and (d) the comprehensive analytical approaches.
Bone is the most studied BC DM site, with multiple steps of the metastasis cascade elucidated in substantial detail (Mundy, 2002; Roodman, 2004). Alongside this molecular knowledge, multiple bone metastasis-specific biomarkers have been proposed. For example, Winczura et al (2015) have found that a reduction of osteopontin was consistently observed in patients who developed bone metastasis, whereas Mihai et al (2006) have found that the calcium-sensing receptor (CaR) was commonly expressed in breast tumours, which metastasised to the bone. Although our models uncovered some proteins previously known to be associated with metastasis, it also uncovered several proteins that generally have not been studied in the context of BC tropism to specific metastatic sites/tissues, or have not been implicated directly in regulating metastasis. For example, upregulation of the high-risk biomarker MTA1 in our bone metastasis model, is seen in several aggressive cancers (Kumar et al, 2003) and has been linked to bone metastasis from prostate cancer (Kai et al, 2011). In BC, MTA1 upregulation has been shown to promote lung-specific metastasis in mice (Pakala et al, 2013). By contrast, we found that underexpression of the karyopherin, KPNA2, is associated with development of bone metastasis. KPNA2 has not been implicated in site specificity of metastasis; in fact, its overexpression was correlated with poorer recurrence-free overall survival in BC (Dahl et al, 2006; Dankof et al, 2007). More importantly, KPNA2 expression in patients with no metastases and patients with metastasis to sites other than bone, was higher than in patients with bone metastasis. These results suggest that TNBC patients (a) have high baseline expression of KPNA2 (Alshareeda et al, 2015), and (b) this high expression preferentially selects for all the other metastatic sites (seen in Supplementary Figure S15A, along with the other site-specific biomarker comparisons).
Although liver is one of the most common sites of metastasis for BC patients (Weigelt et al, 2005), there is little research into potential liver metastasis-specific IHC biomarkers. Interestingly, the biomarkers that were differentially expressed in patients with liver metastases showed a strong tendency to be overexpressed in patients with liver metastases (Supplementary Figure S11A). The best model included N-cadherin, whose upregulation has been associated with pro-migratory phenotypes (Cavallaro and Christofori, 2004), and is thus believed to contribute to the general risk of metastasis. There is also evidence suggesting a preference for tropism to liver in BCs that overexpress N-cadherin (Hazan et al, 2000; Aleskandarany et al, 2014), although the mechanistic underpinning of that preference is yet to be uncovered. The other biomarker selected, XPD (also known as ERCC2), has no previously published evidence of being involved in BC metastasis. In fact, most studies focus on the association between mutations in this gene and an increased risk of developing BC (Bernard-Gallon et al, 2008), or specifically TNBC (Smolarz et al, 2014).
We also found that the roles reported for some of the metastasis biomarkers in our models appear to differ between ER-positive and TNBC patients. For instance, in ER-positive BC cohorts, gene expression studies showed TFF1 to be very highly overexpressed in patients who had bone metastasis vs metastasis to another site (Smid et al, 2006). However, IHC data showed no significant difference between patients who developed bone metastasis and those with no metastasis (Bohn et al, 2009). There are also conflicting reports regarding the impact of TFF1 overexpression on BC prognosis with some studies suggesting that it may have an oncogenic role (Perry et al, 2008), whereas others indicate an association between its overexpression and a favourable prognosis (Buache et al, 2011). In our TNBC cohort, reduced TFF1 expression was associated with high risk of lung metastasis (Supplementary Figure S12A). However, studies in ER-positive BC suggest that high TFF1 levels could promote lung metastasis via TFF1’s role in enhancing chemotaxis (Prest et al, 2002). Another protein that has not previously studied with regard to promoting metastasis to any specific site is RARα. Our data suggest that TNBC patients who experience lung metastasis underexpress nuclear RARα (Supplementary Figure S6A). In ER-positive BC, the presence of ER both correlates with the number of RARα receptors and the ability of ER to inhibit cell growth in concert with RARα (Sheikh et al, 1993). In TNBCs that underexpress RARα, it is plausible that the brakes on proliferation are lifted; however, the molecular basis of the propensity of these low-RARα TNBCs to metastasise to the lung is currently unclear and merits further study.
An important feature that cancer cells require to metastasise to the lung, the ability to extravasation through non-fenestrated capillaries, is also vital for brain metastasis. In fact, multiple genetic similarities were shown between cells primed to metastasise to the brain and to the lungs, such as COX2 (Bos et al, 2009). In the brain metastasis model we derived, we observed an unexpected combination of overexpressed biomarkers (Supplementary Figure S13A) that were not significant for metastasis to any other site. A previous study of IHC biomarkers had shown that an increase in both PARP1 and nuclear BRCA2 expression is associated with a stark decrease in both OS and RFS, separately and when combined (Park et al, 2015b). By contrast though BRCA2 in our brain metastasis model was cytoplasmic; more studies are required to clarify the functions of cytoplasmic BRCA2 (Spain et al, 1999).
The aim of this retrospective study was exploratory, to identify IHC-based biomarkers, which held statistical significance in predicting TNBC metastasis to specific sites. Our study highlights the importance of evaluating protein subcellular localisation and identification of such ‘phenotypic’ biomarkers, as subcellular localisation can profoundly influence biological activities and prognostic significance of protein biomarkers. In fact, we found that in the majority of the cases, the nuclear-localised or cytoplasmic pools of the proteins in our signatures held prognostic significance, whereas the overall levels did not. This finding emphasises a key limitation of gene expression-based signatures where robust gene expression-based signatures would be limited to the subset of proteins whose cellular activities are directly proportional to mRNA expression levels.
It is also noteworthy that in our data set, among patients with metastases to multiple sites, the exact order of metastases is unknown and each metastasis was treated independently even though it is possible that some of these metastases may have arisen from other earlier metastases rather than from the primary tumour. In closing, our novel multi-parametric prognostic models allow for very significant identification of patients with TNBC who will experience DM to a specific site. Design of a cost-effective, clinically-facile IHC-based battery of tests to predict the most likely site of metastasis for TNBCs would (a) enable early detection of metastases through increased surveillance, (b) allow use of preventative therapy to prevent disease progression and (c) improve outcomes for TNBCs.
Acknowledgments
This work was supported by the National Institutes of Health (UO1CA179671 to RA).
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc)
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 4.0 Unported License.
The authors declare no conflict of interest.
Supplementary Material
References
- Abd El-Rehim DM, Ball G, Pinder SE, Rakha E, Paish C, Robertson JF, Macmillan D, Blamey RW, Ellis IO (2005) High-throughput protein expression analysis using tissue microarray technology of a large well-characterised series identifies biologically distinct classes of breast cancer confirming recent cDNA expression analyses. Int J Cancer 116(3): 340–350. [DOI] [PubMed] [Google Scholar]
- Abd El-Rehim DM, Pinder SE, Paish CE, Bell J, Blamey RW, Robertson JF, Nicholson RI, Ellis IO (2004) Expression of luminal and basal cytokeratins in human breast carcinoma. J Pathol 203(2): 661–671. [DOI] [PubMed] [Google Scholar]
- Abduljabbar R, Negm OH, Lai CF, Jerjees DA, Al-Kaabi M, Hamed MR, Tighe PJ, Buluwela L, Mukherjee A, Green AR, Ali S, Rakha EA, Ellis IO (2015) Clinical and biological significance of glucocorticoid receptor (GR) expression in breast cancer. Breast Cancer Res Treat 150(2): 335–346. [DOI] [PubMed] [Google Scholar]
- Aleskandarany MA, Negm OH, Green AR, Ahmed MA, Nolan CC, Tighe PJ, Ellis IO, Rakha EA (2014) Epithelial mesenchymal transition in early invasive breast cancer: an immunohistochemical and reverse phase protein array study. Breast Cancer Res Treat 145(2): 339–348. [DOI] [PubMed] [Google Scholar]
- Alshareeda AT, Negm OH, Green AR, Nolan CC, Tighe P, Albarakati N, Sultana R, Madhusudan S, Ellis IO, Rakha EA (2015) KPNA2 is a nuclear export protein that contributes to aberrant localisation of key proteins and poor prognosis of breast cancer. Br J Cancer 112(12): 1929–1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders CK, Carey LA (2009) Biology, metastatic patterns, and treatment of patients with triple-negative breast cancer. Clin Breast Cancer 9: S73–S81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold SM, Young AB, Munn RK, Patchell RA, Nanayakkara N, Markesbery WR (1999) Expression of p53, bcl-2, E-cadherin, matrix metalloproteinase-9, and tissue inhibitor of metalloproteinases-1 in paired primary tumors and brain metastasis. Clin Cancer Res 5(12): 4028–4033. [PubMed] [Google Scholar]
- Barnholtz-Sloan JS, Sloan AE, Davis FG, Vigneau FD, Lai P, Sawaya RE (2004) Incidence proportions of brain metastases in patients diagnosed (1973 to 2001) in the metropolitan detroit cancer surveillance system. J Clin Oncol 22(14): 2865–2872. [DOI] [PubMed] [Google Scholar]
- Bernard-Gallon D, Bosviel R, Delort L, Fontana L, Chamoux A, Rabiau N, Kwiatkowski F, Chalabi N, Satih S, Bignon YJ (2008) DNA repair gene ERCC2 polymorphisms and associations with breast and ovarian cancer risk. Mol Cancer 7: 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohn OL, Nasir I, Brufsky A, Tseng GC, Bhargava R, MacManus K, Chivukula M (2009) Biomarker profile in breast carcinomas presenting with bone metastasis. Int J Clin Exp Pathol 3(2): 139–146. [PMC free article] [PubMed] [Google Scholar]
- Bos PD, Zhang XH-F, Nadal C, Shu W, Gomis RR, Nguyen DX, Minn AJ, van de Vijver MJ, Gerald WL, Foekens JA (2009) Genes that mediate breast cancer metastasis to the brain. Nature 459(7249): 1005–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buache E, Etique N, Alpy F, Stoll I, Muckensturm M, Reina-San-Martin B, Chenard MP, Tomasetto C, Rio MC (2011) Deficiency in trefoil factor 1 (TFF1) increases tumorigenicity of human breast cancer cells and mammary tumor development in TFF1-knockout mice. Oncogene 30(29): 3261–3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bull AD (2011) Convergence rates of efficient global optimization algorithms. J Mach Learn Res 12: 2879–2904. [Google Scholar]
- Cardoso F, Costa A, Norton L, Cameron D, Cufer T, Fallowfield L, Francis P, Gligorov J, Kyriakides S, Lin N, Pagani O, Senkus E, Thomssen C, Aapro M, Bergh J, Di Leo A, El Saghir N, Ganz PA, Gelmon K, Goldhirsch A, Harbeck N, Houssami N, Hudis C, Kaufman B, Leadbeater M, Mayer M, Rodger A, Rugo H, Sacchini V, Sledge G, van’t Veer L, Viale G, Krop I, Winer E (2012) 1st International consensus guidelines for advanced breast cancer (ABC 1). Breast 21(3): 242–252. [DOI] [PubMed] [Google Scholar]
- Caulfield T, Evans J, McGuire A, McCabe C, Bubela T, Cook-Deegan R, Fishman J, Hogarth S, Miller FA, Ravitsky V, Biesecker B, Borry P, Cho MK, Carroll JC, Etchegary H, Joly Y, Kato K, Lee SSJ, Rothenberg K, Sankar P, Szego MJ, Ossorio P, Pullman D, Rousseau F, Ungar WJ, Wilson B (2013) Reflections on the cost of ‘low-cost’ whole genome sequencing: framing the health policy debate. PLoS Biol 11(11: e1001699). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavallaro U, Christofori G (2004) Cell adhesion and signalling by cadherins and Ig-CAMs in cancer. Nat Rev Cancer 4(2): 118–132. [DOI] [PubMed] [Google Scholar]
- Chen WS, Wei SJ, Liu JM, Hsiao M, Kou-Lin J, Yang WK (2001) Tumor invasiveness and liver metastasis of colon cancer cells correlated with cyclooxygenase-2 (COX-2) expression and inhibited by a COX-2-selective inhibitor, etodolac. Int J Cancer 91(6): 894–899. [DOI] [PubMed] [Google Scholar]
- Cleere DW (2010) Triple-negative breast cancer: a clinical update. Commun Oncol 7(5): 203–211. [Google Scholar]
- Coleman R, Body JJ, Aapro M, Hadji P, Herrstedt J (2014) Bone health in cancer patients: ESMO clinical practice guidelines. Ann Oncol 25(Suppl 3): iii124–iii137. [DOI] [PubMed] [Google Scholar]
- Dahl E, Kristiansen G, Gottlob K, Klaman I, Ebner E, Hinzmann B, Hermann K, Pilarsky C, Dürst M, Klinkhammer-Schalke M, Blaszyk H, Knuechel R, Hartmann A, Rosenthal A, Wild PJ (2006) Molecular profiling of laser-microdissected matched tumor and normal breast tissue identifies karyopherin α2 as a potential novel prognostic marker in breast cancer. Clin Cancer Res 12(13): 3950–3960. [DOI] [PubMed] [Google Scholar]
- Dankof A, Fritzsche FR, Dahl E, Pahl S, Wild P, Dietel M, Hartmann A, Kristiansen G (2007) KPNA2 protein expression in invasive breast carcinoma and matched peritumoral ductal carcinoma in situ. Virch Archiv 451(5): 877–881. [DOI] [PubMed] [Google Scholar]
- Deeken JF, Löscher W (2007) The blood–brain barrier and cancer: transporters, treatment, and trojan horses. Clin Cancer Res 13(6): 1663–1674. [DOI] [PubMed] [Google Scholar]
- Elsheikh SE, Green AR, Rakha EA, Samaka RM, Ammar AA, Powe D, Reis-Filho JS, Ellis IO (2008) Caveolin 1 and Caveolin 2 are associated with breast cancer basal-like and triple-negative immunophenotype. Br J Cancer 99(2): 327–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fidler IJ (2003) The pathogenesis of cancer metastasis: the ‘seed and soil’ hypothesis revisited. Nat Rev Cancer 3(6): 453–458. [DOI] [PubMed] [Google Scholar]
- Galea MH, Blamey RW, Elston CW, Ellis IO (1992) The Nottingham Prognostic Index in primary breast cancer. Br Cancer Res Treat 22: 207–219. [DOI] [PubMed] [Google Scholar]
- Gelbart MA, Snoek J, Adams RP (2014) Bayesian optimization with unknown constraints. Preprint at https://arxiv.org/abs/1403.5607.
- US Cancer Statistics Working Group (2016) United States Cancer Statistics: 1999-2013 Incidence and Mortality Web-Based Report. US Department of Health and Human Services, Centers for Disease Control and Prevention and National Cancer Institute: Atlanta, GA, USA.
- Gupta GP, Massagué J (2006) Cancer metastasis: building a framework. Cell 127(4): 679–695. [DOI] [PubMed] [Google Scholar]
- Gupta GP, Minn AJ, Kang Y, Siegel PM, Serganova I, Cordón-Cardo C, Olshen AB, Gerald WL, Massagué J (2005) Identifying site-specific metastasis genes and functions. Cold Spring Harb Symp Quant Biol 70: 149–158. [DOI] [PubMed] [Google Scholar]
- Gupta GP, Perk J, Acharyya S, de Candia P, Mittal V, Todorova-Manova K, Gerald WL, Brogi E, Benezra R, Massagué J (2007) ID genes mediate tumor reinitiation during breast cancer lung metastasis. Proc Natl Acad Sci 104(49): 19506–19511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habashy HO, Rakha EA, Aleskandarany M, Ahmed MA, Green AR, Ellis IO, Powe DG (2011) FOXO3a nuclear localisation is associated with good prognosis in luminal-like breast cancer. Breast Cancer Res Treat 129(1): 11–21. [DOI] [PubMed] [Google Scholar]
- Hazan RB, Phillips GR, Qiao RF, Norton L, Aaronson SA (2000) Exogenous expression of N-cadherin in breast cancer cells induces cell migration, invasion, and metastasis. J Cell Biol 148(4): 779–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillner BE, Ingle JN, Chlebowski RT, Gralow J, Yee GC, Janjan NA, Cauley JA, Blumenstein BA, Albain KS, Lipton A, Brown S (2003) American Society of Clinical Oncology 2003: update on the role of bisphosphonates and bone health issues in women with breast cancer. J Clin Oncol 21(21): 4042–4057. [DOI] [PubMed] [Google Scholar]
- Hiratsuka S, Watanabe A, Aburatani H, Maru Y (2006) Tumour-mediated upregulation of chemoattractants and recruitment of myeloid cells predetermines lung metastasis. Nat Cell Biol 8(12): 1369–1375. [DOI] [PubMed] [Google Scholar]
- Hu G, Kang Y, Wang XF (2009) From breast to the brain: unraveling the puzzle of metastasis organotropism. J Mol Cell Biol 1(1): 3–5. [DOI] [PubMed] [Google Scholar]
- Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D (2011) Global cancer statistics. CA Cancer J Clin 61(2): 69–90. [DOI] [PubMed] [Google Scholar]
- Jerjees DA, Negm OH, Alabdullah ML, Mirza S, Alkaabi M, Hameed MR, Abduljabbar R, Muftah A, Nolan CC, Green AR, Tighe PJ, Band V, Ellis IO, Rakha EA (2015) The mammalian target of rapamycin complex 1 (mTORC1) in breast cancer: the impact of oestrogen receptor and HER2 pathways. Breast Cancer Res Treat 150(1): 91–103. [DOI] [PubMed] [Google Scholar]
- Kai L, Wang J, Ivanovic M, Chung YT, Laskin WB, Schulze-Hoepfner F, Mirochnik Y, Satcher RL Jr, Levenson AS (2011) Targeting prostate cancer angiogenesis through metastasis-associated protein 1 (MTA1). Prostate 71(3): 268–280. [DOI] [PubMed] [Google Scholar]
- Kennecke H, Yerushalmi R, Woods R, Cheang MCU, Voduc D, Speers CH, Nielsen TO, Gelmon K (2010) Metastatic behavior of breast cancer subtypes. J Clin Oncol 28(20): 3271–3277. [DOI] [PubMed] [Google Scholar]
- Klos KJ, O’Neill BP (2004) Brain metastases. Neurologist 10(1): 31–46. [DOI] [PubMed] [Google Scholar]
- Kumar R, Wang R-A, Bagheri-Yarmand R (2003) Emerging roles of MTA family members in human cancers. Semin Oncol 30: 30–37. [DOI] [PubMed] [Google Scholar]
- Landemaine T, Jackson A, Bellahcène A, Rucci N, Sin S, Abad BM, Sierra A, Boudinet A, Guinebretière J-M, Ricevuto E, Noguès C, Briffod M, Bièche I, Cherel P, Garcia T, Castronovo V, Teti A, Lidereau R, Driouch K (2008) A six-gene signature predicting breast cancer lung metastasis. Cancer Res 68(15): 6092–6099. [DOI] [PubMed] [Google Scholar]
- Largillier R, Ferrero JM, Doyen J, Barriere J, Namer M, Mari V, Courdi A, Hannoun-Levi JM, Ettore F, Birtwisle-Peyrottes I, Balu-Maestro C, Marcy PY, Raoust I, Lallement M, Chamorey E (2008) Prognostic factors in 1,038 women with metastatic breast cancer. Ann Oncol 19(12): 2012–2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YTNM (1983) Breast carcinoma: pattern of metastasis at autopsy. J Surg Oncol 23(3): 175–180. [DOI] [PubMed] [Google Scholar]
- Liedtke C, Mazouni C, Hess KR, André F, Tordai A, Mejia JA, Symmans WF, Gonzalez-Angulo AM, Hennessy B, Green M, Cristofanilli M, Hortobagyi GN, Pusztai L (2008) Response to neoadjuvant therapy and long-term survival in patients with triple-negative breast cancer. J Clin Oncol 26(8): 1275–1281. [DOI] [PubMed] [Google Scholar]
- Lorusso G, Ruegg C (2012) New insights into the mechanisms of organ-specific breast cancer metastasis. Semin Cancer Biol 22(3): 226–233. [DOI] [PubMed] [Google Scholar]
- Luck AA, Evans AJ, Green AR, Rakha EA, Paish C, Ellis IO (2008) The influence of basal phenotype on the metastatic pattern of breast cancer. Clin Oncol 20(1): 40–45. [DOI] [PubMed] [Google Scholar]
- Luzzi KJ, MacDonald IC, Schmidt EE, Kerkvliet N, Morris VL, Chambers AF, Groom AC (1998) Multistep nature of metastatic inefficiency: dormancy of solitary cells after successful extravasation and limited survival of early micrometastases. Am J Pathol 153(3): 865–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmoud SM, Paish EC, Powe DG, Macmillan RD, Grainge MJ, Lee AH, Ellis IO, Green AR (2011) Tumor-infiltrating CD8+ lymphocytes predict clinical outcome in breast cancer. J Clin Oncol 29(15): 1949–1955. [DOI] [PubMed] [Google Scholar]
- Malin D, Strekalova E, Petrovic V, Deal AM, Ahmad AA, Adamo B, Miller CR, Ugolkov A, Livasy C, Fritchie K, Hamilton E, Blackwell K, Geradts J, Ewend M, Carey L, Shusta EV, Anders CK, Cryns VL (2014) αB-crystallin: a novel regulator of breast cancer metastasis to the brain. Clin Cancer Res 20(1): 56–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massard C, Zonierek J, Gross-Goupil M, Fizazi K, Szczylik C, Escudier B (2010) Incidence of brain metastases in renal cell carcinoma treated with sorafenib. Ann Oncol 21(5): 1027–1031. [DOI] [PubMed] [Google Scholar]
- Mandrekar JN, Mandrekar SJ, Cha SS (2003) Cutpoint Determination methods in Survival Analysis Using SAS. In Proceedings of the 28th SAS Users Group International (SUGI) Conference. pp 261–288.
- Mihai R, Stevens J, McKinney C, Ibrahim NBN (2006) Expression of the calcium receptor in human breast cancer—a potential new marker predicting the risk of bone metastases. Eur J Surg Oncol 32(5): 511–515. [DOI] [PubMed] [Google Scholar]
- Minn AJ, Kang Y, Serganova I, Gupta GP, Giri DD, Doubrovin M, Ponomarev V, Gerald WL, Blasberg R, Massagué J (2005) Distinct organ-specific metastatic potential of individual breast cancer cells and primary tumors. J Clin Investig 115(1): 44–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mundy GR (2002) Metastasis: Metastasis to bone: causes, consequences and therapeutic opportunities. Nat Rev Cancer 2(8): 584–593. [DOI] [PubMed] [Google Scholar]
- Nguyen DX, Bos PD, Massague J (2009) Metastasis: from dissemination to organ-specific colonization. Nat Rev Cancer 9(4): 274–284. [DOI] [PubMed] [Google Scholar]
- Pakala SB, Rayala SK, Wang RA, Ohshiro K, Mudvari P, Reddy SDN, Zheng Y, Pires R, Casimiro S, Pillai MR, Costa L, Kumar R (2013) MTA1 promotes STAT3 transcription and pulmonary metastasis in breast cancer. Cancer Res 73(12): 3761–3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmieri D, Lockman PR, Thomas FC, Hua E, Herring J, Hargrave E, Johnson M, Flores N, Qian Y, Vega-Valle E, Taskar KS, Rudraraju V, Mittapalli RK, Gaasch JA, Bohn KA, Thorsheim HR, Liewehr DJ, Davis S, Reilly JF, Walker R, Bronder JL, Feigenbaum L, Steinberg SM, Camphausen K, Meltzer PS, Richon VM, Smith QR, Steeg PS (2009) Vorinostat inhibits brain metastatic colonization in a model of triple-negative breast cancer and induces DNA double-strand breaks. Clin Cancer Res 15(19): 6148–6157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S, Han W, Kim J, Kim MK, Lee E, Yoo TK, Lee HB, Kang YJ, Kim YG, Moon HG, Noh DY (2015. a) Risk factors associated with distant metastasis and survival outcomes in breast cancer patients with locoregional recurrence. J Breast Cancer 18(2): 160–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SH, Noh SJ, Kim KM, Bae JS, Kwon KS, Jung SH, Kim JR, Lee H, Chung MJ, Moon WS, Kang MJ, Jang KY (2015. b) Expression of DNA damage response molecules PARP1, γH2AX, BRCA1, and BRCA2 predicts poor survival of breast carcinoma patients. Transl Oncol 8(4): 239–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry JK, Kannan N, Grandison PM, Mitchell MD, Lobie PE (2008) Are trefoil factors oncogenic? Trends Endocrinol Metabol 19(2): 74–81. [DOI] [PubMed] [Google Scholar]
- Pogoda K, Niwińska A, Murawska M, Pieńkowski T (2013) Analysis of pattern, time and risk factors influencing recurrence in triple-negative breast cancer patients. Med Oncol 30(1): 388–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powles T, Paterson A, McCloskey E, Schein P, Scheffler B, Tidy A, Ashley S, Smith I, Ottestad L, Kanis J (2006) Reduction in bone relapse and improved survival with oral clodronate for adjuvant treatment of operable breast cancer [ISRCTN83688026]. Breast Cancer Res 8(2): R13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prest SJ, May FEB, Westley BR (2002) The estrogen-regulated protein, TFF1, stimulates migration of human breast cancer cells. FASEB J 16(6): 592–594. [DOI] [PubMed] [Google Scholar]
- Rakha EA, El-Sayed ME, Green AR, Lee AH, Robertson JF, Ellis IO (2007. a) Prognostic markers in triple-negative breast cancer. Cancer 109(1): 25–32. [DOI] [PubMed] [Google Scholar]
- Rakha EA, El-Sayed ME, Green AR, Lee AHS, Robertson JF, Ellis IO (2007. b) Prognostic markers in triple-negative breast cancer. Cancer 109(1): 25–32. [DOI] [PubMed] [Google Scholar]
- Rakha EA, Elsheikh SE, Aleskandarany MA, Habashi HO, Green AR, Powe DG, El-Sayed ME, Benhasouna A, Brunet JS, Akslen LA, Evans AJ, Blamey R, Reis-Filho JS, Foulkes WD, Ellis IO (2009) Triple-negative breast cancer: distinguishing between basal and nonbasal subtypes. Clin Cancer Res 15(7): 2302–2310. [DOI] [PubMed] [Google Scholar]
- Rakha EA, Putti TC, Abd El-Rehim DM, Paish C, Green AR, Powe DG, Lee AH, Robertson JF, Ellis IO (2006) Morphological and immunophenotypic analysis of breast carcinomas with basal and myoepithelial differentiation. J Pathol 208(4): 495–506. [DOI] [PubMed] [Google Scholar]
- Roodman GD (2004) Mechanisms of bone metastasis. N Engl J Med 350(16): 1655–1664. [DOI] [PubMed] [Google Scholar]
- Ruiz de Garibay G, Herranz C, Llorente A, Boni J, Serra-Musach J, Mateo F, Aguilar H, Gómez-Baldó L, Petit A, Vidal A, Climent F, Hernández-Losa J, Cordero Á, González-Suárez E, Sánchez-Mut Jé V, Esteller M, Llatjós R, Varela M, López Jé I, García N, Extremera AI, Gumà A, Ortega R, Plà M, Fernández A, Pernas S, Falo C, Morilla I, Campos M, Gil M, Román A, Molina-Molina M, Ussetti P, Laporta R, Valenzuela C, Ancochea J, Xaubet A, Casanova Á, Pujana MA (2015) Lymphangioleiomyomatosis biomarkers linked to lung metastatic potential and cell stemness. PLoS ONE 10(7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheikh MS, Shao Z-M, Chen J-C, Hussain A, Jetten AM, Fontana JA (1993) Estrogen receptor-negative breast cancer cells transfected with the estrogen receptor exhibit increased RARα gene expression and sensitivity to growth inhibition by retinoic acid. J Cell Biochem 53(4): 394–404. [DOI] [PubMed] [Google Scholar]
- Smid M, Wang Y, Klijn JG, Sieuwerts AM, Zhang Y, Atkins D, Martens JW, Foekens JA (2006) Genes associated with breast cancer metastatic to bone. J Clin Oncol 24(15): 2261–2267. [DOI] [PubMed] [Google Scholar]
- Smid M, Wang Y, Zhang Y, Sieuwerts AM, Yu J, Klijn JGM, Foekens JA, Martens JWM (2008) Subtypes of breast cancer show preferential site of relapse. Cancer Res 68(9): 3108–3114. [DOI] [PubMed] [Google Scholar]
- Smith MR, Saad F, Coleman R, Shore N, Fizazi K, Tombal B, Miller K, Sieber P, Karsh L, Damião R, Tammela TL, Egerdie B, Van Poppel H, Chin J, Morote J, Gómez-Veiga F, Borkowski T, Ye Z, Kupic A, Dansey R, Goessl C (2012) Denosumab and bone metastasis-free survival in men with castration-resistant prostate cancer: results of a global phase 3, randomised, placebo-controlled trial. Lancet 379(9810): 39–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolarz B, Makowska M, Samulak D, Michalska MM, Mojs E, Wilczak M, Romanowicz H (2014) Single nucleotide polymorphisms (SNPs) of ERCC2, hOGG1, and XRCC1 DNA repair genes and the risk of triple-negative breast cancer in Polish women. Tumour Biol 35(4): 3495–3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spain BH, Larson CJ, Shihabuddin LS, Gage FH, Verma IM (1999) Truncated BRCA2 is cytoplasmic: implications for cancer-linked mutations. Proc Natl Acad Sci USA 96(24): 13920–13925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steeg PS, Camphausen KA, Smith QR (2011) Brain metastases as preventive and therapeutic targets. Nat Rev Cancer 11(5): 352–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voduc KD, Nielsen TO, Perou CM, Harrell JC, Fan C, Kennecke H, Minn AJ, Cryns VL, Cheang MC (2015) [alpha] B-crystallin expression in breast cancer is associated with brain metastasis. npj Breast Cancer 1: 15014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigelt B, Glas AM, Wessels LFA, Witteveen AT, Peterse JL, van't Veer LJ (2003) Gene expression profiles of primary breast tumors maintained in distant metastases. Proc Natl Acad Sci USA 100(26): 15901–15905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigelt B, Peterse JL, van't Veer LJ (2005) Breast cancer metastasis: markers and models. Nat Rev Cancer 5(8): 591–602. [DOI] [PubMed] [Google Scholar]
- Weil RJ, Palmieri DC, Bronder JL, Stark AM, Steeg PS (2005) Breast cancer metastasis to the central nervous system. Am J Pathol 167(4): 913–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winczura P, Sosińska-Mielcarek K, Duchnowska R, Badzio A, Lakomy J, Majewska H, Pęksa R, Pieczyńska B, Radecka B, Dębska-Szmich S, Adamowicz K, Biernat W, Jassem J (2015) Immunohistochemical predictors of bone metastases in breast cancer patients. Pathol Oncol Res 21(4): 1229–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.