Abstract
Background:
Despite the favourable survival rates of childhood acute lymphoblastic leukaemia (ALL), a significant number of patients present resistance to antileukaemic agents and dismal prognosis. In this study, we analysed miR-125b expression in childhood ALL and evaluated its clinical utility for patients treated with Berlin–Frankfurt–Münster (BFM) protocol.
Methods:
The study included 272 bone marrow specimens obtained on diagnosis and on BFM day 33 from 125 patients and 64 healthy children. Following extraction, RNA was polyadenylated and reverse transcribed. miR-125b levels were quantified by quantitative PCR. Cytogenetics, immunohistotype and MRD were analysed according to international guidelines.
Results:
Downregulated miR-125b levels were detected in childhood ALL patients and correlated with adverse prognosis. Following BFM induction, miR-125b levels were significantly increased, however, elevated day 33/diagnosis miR-125b ratio was associated with unfavourable disease features. Loss of miR-125b during diagnosis and higher day 33/diagnosis ratio were correlated with stronger risk for disease short-term relapse and patients’ worse survival. Moreover, multivariate regression models highlighted the independent prognostic value of miR-125b for childhood ALL. Finally, the combination of miR-125b with clinically used disease markers clearly enhanced the prediction of patients’ resistance to BFM chemotherapy.
Conclusions:
miR-125b significantly improves the prognosis of childhood ALL patients’ outcome under BFM treatment.
Keywords: ALL, childhood ALL, miRNAs, miR-125, ALL IC-BFM, Berlin–Frankfurt–Münster protocol, apoptosis, leukaemia
Acute lymphoblastic leukaemia (ALL) is the most commonly diagnosed malignancy in children, accounting for ∼25% of all paediatric cancers worldwide (Siegel et al, 2016). Over the past 50 years, the stratification of patients and the risk-adjusted treatment intensity have led to an outstanding improvement in event-free and overall survival (OS) rates, approaching 80% and 90% respectively, in developed countries (Pui, 2010).
Several conventional prognostic markers are currently in clinical use by the Berlin–Frankfurt–Münster (BFM) backbone protocol in order to categorise patients in risk groups with the appropriate therapeutic scheme, including white blood cells (WBC) count, immunophenotype, genetic abnormalities, peripheral blast cells (prednisone response), bone marrow (BM) blasts percentage (BM response) and minimal residual disease (MRD) (Moricke et al, 2008; Stary et al, 2014). Nonetheless, there are still challenges to overcome, as patients are sometimes overtreated, increasing toxicity phenomena or present resistance to antileukaemic agents (Pui et al, 2012; Bhojwani and Pui, 2013). The abovementioned facts highlight the need for the clinical evaluation of novel prognostic markers and therapeutic tools either to mitigate the risk for acute toxic and late side effects or to predict more effectively patients’ response to therapy and the risk for disease relapse.
MicroRNAs (miRNAs) are implicated in the vast majority of cellular processes, from proliferation and growth to differentiation and apoptosis (Bushati and Cohen, 2007). Over the past decade, researchers have been focusing on unravelling the role of miRNAs in the manifestation of different human diseases (Almeida et al, 2011), including cancer, as their deregulation is often responsible for tumour initiation and progression (Lu et al, 2005; Calin and Croce, 2006; Vasilatou et al, 2010). Numerous studies on ALL have revealed the implication of miRNAs in disease establishment and progression as well as their prognostic potential, making them significant tools for the prediction of patients’ response to treatment and survival outcome (Schotte et al, 2009; Wang et al, 2010; Schotte et al, 2011; Schotte et al, 2012).
The hsa-miR-125b-5p (miR-125b) has been the research topic of many studies, highlighting either an oncogenic or a tumour suppressive role (Sun et al, 2013). The miR-125b is encoded by two different genes, MIR125B1 (11q24.1) and MIR125B2 (21q21.1), which are transcribed into hsa-mir-125b-1 and hsa-miR-125b-2 stem-loop precursors, giving rise to the same mature sequence (Rodriguez et al, 2004). Abnormalities in miR-125b expression have been linked to various solid tumours and haematological malignancies (Sun et al, 2013). miR-125b has been reported to promote lymphoblastic leukaemia by targeting IRF4 tumour suppressor, necessary for B- and T-cell maturation (So et al, 2014), and ARID3a in progenitor B-cells enhancing cell proliferation and expression of pluripotency-associated factors OCT4, SOX2, KLF4, and NANOG (Puissegur et al, 2012). Moreover, miR-125b suppresses TNFAIP3, resulting to the NF-κB-mediated increase of B-cell proliferation and reduction of apoptosis in diffuse large B-cell lymphoma (DLBCL), as well as to inhibition of T-cell differentiation and metabolic reprograming in ALL (Kim et al, 2012; Liu et al, 2016).
In myeloid cell leukaemia, overexpression of miR-125b induces cells’ differentiation arrest, via targeting CBFβ and STAT3 transcription factors (Lin et al, 2011; Surdziel et al, 2011) and confers a proliferative advantage by suppressing ABTB anti-proliferative factor (Bousquet et al, 2012). In B-cell malignant multiple myeloma dexamethasone-induced miR-125b has been reported to stimulate chemotherapy resistance and to prevent cell death through suppression of p53 and p53/miR-34a/SIRT regulatory network (Murray et al, 2013). Moreover, miR-125b has been implicated in megakaryopoiesis and in the pathogenesis of trisomy 21/Down’s syndrome-associated megakaryoblastic leukaemia (DS-AMKL) and related transient leukaemia (DS-TL). The significant overexpression of miR-125b in DS-AMKL and DS-TL results to enhanced cells’ proliferation and self-renewal, an effect attributed to direct targeting of DICER1 and ST18 expression (Klusmann et al, 2010). Finally, the regulation of several pro-apoptotic genes supports miR-125b anti-apoptotic activity in haematological malignancies (Sun et al, 2013), while miR-125b loss has been involved in metabolic changes of chronic lymphoblastic leukaemia (CLL) cells leading to Warburg effect (Tili et al, 2012).
The other member of the miR-125 family, miR-125a, has been also studied in leukaemia and lymphomas. Methylation-mediated transcriptional suppression of MIR125A gene and decreased miR-125a levels have been reported in leukaemic myeloblasts of acute myeloid leukaemia (AML) patients, while ectopic miR-125a expression resulted to decreased cell proliferation and enhanced apoptosis of NB4 cells (Garzon et al, 2008; Ramsingh et al, 2010; Ufkin et al, 2014). Moreover, miR-125a is significantly overexpressed in DLBCL and in myelodysplastic syndrome patients, promoting NF-κB pathway activation and thus cells’ proproliferative and antiapoptotic phenotype, by targeting the negative NF-κB regulator TNFAIP3 (Kim et al, 2012; Ganan-Gomez et al, 2014).
Prompted by the involvement and crucial role of miR-125b in haematological malignancies, the present study aims to examine the expression profile of miR-125b in childhood ALL in order to evaluate for the first time its clinical significance for disease prognosis and prediction of patients’ response to BFM chemotherapy protocol.
Materials and methods
Study cohort
The present study enroled 125 children with newly diagnosed ALL, treated according to the BFM backbone protocol at ‘P&A Kyriakou’ Children’s Hospital, Athens, Greece between May 2006 and October 2016, as well as 64 children not suffering from any haematologic or other type of malignancy consisting our control cohort. Patients’ clinicopathological characteristics are presented in Table 1.
Table 1. Clinicopathological features of the childhood ALL patients.
| Variables | N (%) |
|---|---|
| Gender | |
| Male | 73 (58.4) |
| Female | 52 (41.6%) |
| Age | |
| 1–9 years | 84 (67.2) |
| <1 or ⩾10 years | 41 (32.8) |
| WBC | |
| <50 000 cells per μl | 102 (81.6) |
| ⩾50 000 cells per μl | 23 (18.4) |
| Immunophenotype | |
| Precursor B-ALL | 110 (88.0) |
| CD10+ | 104 (83.2) |
| CD10− | 6 (4.8) |
| T-ALL | 15 (12.0) |
| High hyperdiploidy (>50 chromosomes) | |
| Yes | 22 (17.6) |
| No | 102 (81.6) |
| Unknown | 1 (0.8) |
| Hypodiploidy (⩽45 chomosomes) | |
| Yes | 4 (3.2) |
| No | 120 (96.0) |
| Unknown | 1 (0.8) |
| TEL-AML1/t(12;21)(p13;q22) | |
| Negative | 93 (74.4) |
| Positive | 31 (24.8) |
| Unknown | 1 (0.8) |
| Philadelphia chromosome BCR-ABL1/t(9;22)(q34;q11) | |
| Negative | 122 (97.6) |
| Positive | 2 (1.6) |
| Unknown | 1 (0.8) |
| BM on day 15 | |
| M1 (blasts <5%) | 103 (82.4) |
| M2 (blasts 5–25%) | 16 (12.8) |
| M3 (blasts >25%) | 6 (4.8) |
| MRD on day 15 | |
| <0.01% | 22 (17.6) |
| >0.01% | 67 (53.6) |
| Unknown | 36 (28.8) |
| MRD on day 33 | |
| <0.01% | 50 (40.0) |
| >0.01% | 21 (16.8) |
| Unknown | 54 (43.2) |
| Prednisone response on day 8 | |
| Good (<1000 blasts) | 112 (89.6) |
| Poor (⩾1000 blasts) | 13 (10.4) |
| BFM risk groups | |
| Standard risk | 16 (12.8) |
| Intermediate risk | 87 (69.6) |
| High risk | 22 (17.6) |
| Treatment outcome | |
| Disease relapse | |
| CCR | 106 (84.8) |
| Relapse | 19 (15.2) |
| Patients’ survival | |
| Alive | 102 (81.6) |
| Dead | 23 (18.4) |
Abbreviations: ALL=acute lymphoblastic leukaemia; B-ALL=B-lineage acute lymphoblastic leukaemia; BFM=Berlin–Frankfurt–Münster; BM=bone marrow; CCR=complete continuous remission; CD10=cluster of differentiation 10 (common acute lymphoblastic leukaemia antigen, CALLA); MRD=minimal residual disease; T-ALL=T-lineage acute lymphoblastic leukaemia; TEL-AML1=TEL-AML1 rearrangement; WBC=white blood cell.
The biological samples included in the study consisted of 272 BM specimens. In particular, BM specimens were obtained from the 125 childhood ALL patients at disease diagnosis. In addition, matched BM samples on day 33 following BFM protocol induction were available from 83 patients of the abovementioned cohort and included in the analysis. Finally, the control group consisted of BM specimens from 64 children not suffering from any haematologic or other type of malignancy. All samples were collected by BM aspiration and undergone total RNA extraction without any cell fractionation or small RNA enrichment. All available matched day 33 samples following pathological evaluation were included in the analysis without any selection bias. Samples handling, preparation and experimental procedure was the same for leukaemic and normal specimens.
The BFM guidelines were followed for the risk group stratification of childhood ALL patients and their follow-up included the assessment of peripheral blast cells (prednisone response) on day 8, of BM blasts percentage (BM response) on day 15 and the measurement of MRD on days 15 and 33 of the induction protocol (Moricke et al, 2008; Stary et al, 2014). The study was approved by the Ethics Committee of ‘P&A Kyriakou’ Children’s Hospital, Athens, Greece, and performed with respect to the ethical standards of the Declaration of Helsinki, as revised in 2008. Informed consent was obtained from all parents and legal guardians of the participating patients.
Cytogenetic analysis
Cytogenetic analysis was performed by direct preparation of unstimulated BM cells, followed by 24 h of culture in RPMI 1540 culture medium with 25% foetal calf serum at 37 °C. The standard G-banding technique was carried out for the analysis of metaphase chromosomes, which were classified according to the International System for Cytogenetic Nomenclature (ISCN).
Immunohistotype analysis
Multiparametric flow cytometry by direct immunofluorescence was performed via CXP software on an FC-500 cytometer (Beckman Coulter Inc., Nyon, Switzerland). The monoclonal antibodies used were reactive with the following lymphoid differentiation antigens: CD19, CD20, CD22, CD24, CD10, CD34, CD38, CD58, CD11a, HLA-DR+, cTdt, cIgM and cCD79a for B-lineage ALL (B-ALL) and CD7, CD5, CD2, CD3, CD1a, CD4, CD8, CD99, CD38, CD44, CD34, CD45RA, CD45RO, CD56, TcR ab, TcR γδ, cTdt and cCD3 for T-lineage ALL (T-ALL). Lineage assignment was decided according to the EGIL recommendations. The presence of a surface antigen was confirmed when 20% of the cells expressed it, while for intracellular antigens the threshold was adopted at 10%. NEQAS programme was used for external quality assessment.
MRD analysis
MRD assessment was performed using a lineage-specific panel: CD45, CD10, CD19, CD20, CD22, CD34, CD38, CD58, CD9, CD11a for B-lineage ALL and CD4, CD8, cCD3 and sCD3, CD5, CD7, CD2, CD1a, CD99, CD38, cTtd and CD44 for T-lineage ALL according to BFM protocol guidelines. MRD was considered positive when leukaemic cells exceeded 0.01% of all marrow nucleated cells on days 15 and 33.
Isolation and polyadenylation of total RNA
Total RNA was isolated from BM specimens using TRI-Reagent BD (Molecular Research Center, Inc., Cincinnati, OH, USA) according to the manufacturer’s protocol. Total RNA concentration and purity were evaluated spectrophotometrically at 260 and 280 nm. RNA integrity was visually confirmed by agarose gel electrophoresis. Thereafter, 1.0 μg of total RNA was polyadenylated at the 3′-end in a 12.5 μl reaction containing 0.8 mM ATP and 1 U of E. coli poly(A) polymerase (New England Biolabs Inc., Ipswich, MA, USA), at 37 °C for 30 min. Enzyme heat inactivation was performed at 65 °C for 10 min.
First-strand cDNA synthesis
Polyadenylated RNA was reverse transcribed using the poly(T) adaptor 5′-GCGAGCACAGAATTAATACGACTCACTATAGGTTTTTTTTTTTTVN-3′ (V=G, A, C and N=G, A, T, C) in a 20 μl reaction containing 1 U MMLV reverse transcriptase (Invitrogen, Carlsbad, CA, USA), 0.2 U RNaseOUT recombinant ribonuclease inhibitor (Invitrogen) and 0.25 μM poly(T) adaptor, at 37 °C for 60 min. The poly(T) adaptor is hybridised to the 3′-poly(A) tail of the polyadenylated RNA template allowing the synthesis of a longer first-strand cDNA product able to be quantified by qPCR. The reverse transcriptase was inactivated by heating at 70 °C for 15 min.
Quantitative real-time PCR
The quantification of miR-125b levels was performed by a SYBR-Green fluorescent-based quantitative real-time PCR (qPCR) assay. Specific forward primers were designed according to published sequences and in silico specificity analysis for miR-125b-5p (NCBI RefSeq: NR_029671.1 and NR_029694.1) and the small nucleolar RNA, C/D box 48 (SNORD48), also known as RNU48 (NCBI RefSeq: NR_002745.1). The specific forward (hybridised to miR-125b- or RNU48-specific sequences of cDNA template) and universal reverse (hybridised to poly(T) adaptor RT primer sequence of cDNA template) primer sequences are presented in Supplementary Table 1.
The qPCR was performed in the 7500 Real-Time PCR System (Applied Biosystems, Carlsbad, CA, USA). The 10 μl reaction mixture consisted of 2X Kapa SYBR Fast Universal qPCR Master Mix (Kapa Biosystems Inc., Woburn, MA, USA), 200 nm of each qPCR primer and 4 ng of cDNA. Both targets were amplified in technical duplicates for each sample. The thermal protocol consisted of a 3 min polymerase activation step at 95 °C, followed by 40 cycles of denaturation at 95 °C for 15 s and the primer annealing and extension step at 60 °C for 1 min. Melting curve analysis and gel agarose electrophoresis were performed for the discrimination of specific amplicons from non-specific products or primer dimers. Following amplification, melting curve analysis of the products was performed from 60 to 95 °C by 0.3 °C temperature increment intervals for 30 s. Products melting temperature (Tm) was determined by plotting the negative first-derivative of the normalised fluorescence vs temperature. Only samples with a single peak at the appropriate Tm (temperature of the peak maximum) for both miR-125b and SNORD48 were included in the study.
The 2−ΔΔCT method was conducted for the analysis of miR-125b expression levels, using SNORD48 as an endogenous reference control for normalisation purposes. The amplification efficiencies of the target miR-125b-5p and the reference SNORD48 genes were assessed by a validation experiment, using serial dilutions of a control cDNA covering six orders of magnitude (1–10−5 ng cDNA) as template. The linear increases of miR-125b-5p (y=−3.181 × +37.142; r2=0.984) and SNORD48 (y=−3.225 × +34.368; r2=0.994) calibration curves highlight the 106.3% and 104.2% amplification efficiencies, respectively, as well as the absence of PCR inhibition by the template.
Statistical analysis
The non-parametric Mann–Whitney U test and Wilcoxon Signed Rank test were performed to evaluate the differences of miR-125b expression between leukaemic and healthy BM specimens, and between BM samples on diagnosis and on the 33rd day of the induction protocol, respectively. The discriminatory ability of miR-125b for childhood ALL was examined via ROC curve and logistic regression analysis. Moreover, the non-parametric Mann–Whitney U and Kruskal–Wallis tests were appropriately used to assess the correlation of miR-125b levels and day 33/diagnosis miR-125b expression levels ratio with patients’ clinicopathological features.
The X-tile algorithm was applied for the adoption of optimal cut-off values equal to the median (50th percentile) for miR-125b levels on disease diagnosis, on day 33 following protocol induction, and for the day 33/diagnosis miR-125b levels ratio. Logistic regression models were used to examine the ability of miR-125b expression levels to predict patients’ response to BFM treatment. Furthermore, Kaplan–Meier survival curves using log-rank test and Cox proportional regression analysis were conducted to estimate the prognostic utility of miR-125b for the disease-free survival (DFS) and OS of childhood ALL patients treated with the BFM protocol.
Results
Baseline clinical and experimental data
Median patients’ and healthy controls’ age was 5.0 and 4.0 years, respectively. The majority of the patients were males (58.4%), suffering from precursor B-ALL (83.2% CD10+ and 4.8% CD10−) and having WBC<50 000 cells per μl (81.6%). Focusing on cytogenetics, 17.6% of patients were characterised by high hyperdiploidy (>50 chromosomes) and 24.8% by t(12;21)(p13;q22) translocation (TEL-AML1). In order to evaluate the clinical significance of the blasts-related miR-125b expression in childhood ALL, the inclusion of patients with high % BM blast percentage at disease diagnosis was taken into account during study design and patients’ enrolment. The median and mean values of % BM blast percentage at disease diagnosis are 92.0% and 88.74%, respectively, for the total ALL patients cohort, and 95.0% and 88.79%, respectively, for the precursor B-ALL patients group. The histograms of % BM blast percentage are presented in Supplementary Figure 1. Regarding treatment response, poor prednisone response (>1000 blasts/μl) and M2-M3 BM response (⩾5% blasts) was observed in 13 (10.4%) and 22 patients (17.6%), respectively. MRD data were available for day 15 (89 patients) and day 33 (71 patients) of the BFM protocol, where 67 (75.3%) and 21 patients (29.6%) were found positive. During a median follow-up time (reverse Kaplan–Meier method) of 79 months, 19 patients relapsed (15.2%) and 23 died (18.4%), while the mean DFS and OS intervals were 105.7 months (95% CI: 98.23–113.2) and 103.1 months (95% CI: 95.42–110.8), respectively.
Patients excluded from miR-125b analysis on BFM day 33, due to absence of matched sample, did not differ from those included with respect to all clinicopathologic and disease prognostic parameters (gender, P=1.000; age, P=0.576; % blast percentage at diagnosis, P=0.216; WBC count, P=0.144; immunophenotype, P=0.831; high hyperdiploidy, P=0.088; hypodiploidy, P=0.299; TEL-AML1/t(12;21)(p13;q22) translocation, P=0.381; prednisone response, P=0.541; BM response, P=0.621; MRD on day 15, P=0.619; MRD on day 33, P=0.603; BFM risk groups, P=0.943; DFS, P=0.786 and OS, P=0.312).
The expression of miR-125b is significantly reduced in childhood ALL and increases following BFM protocol induction
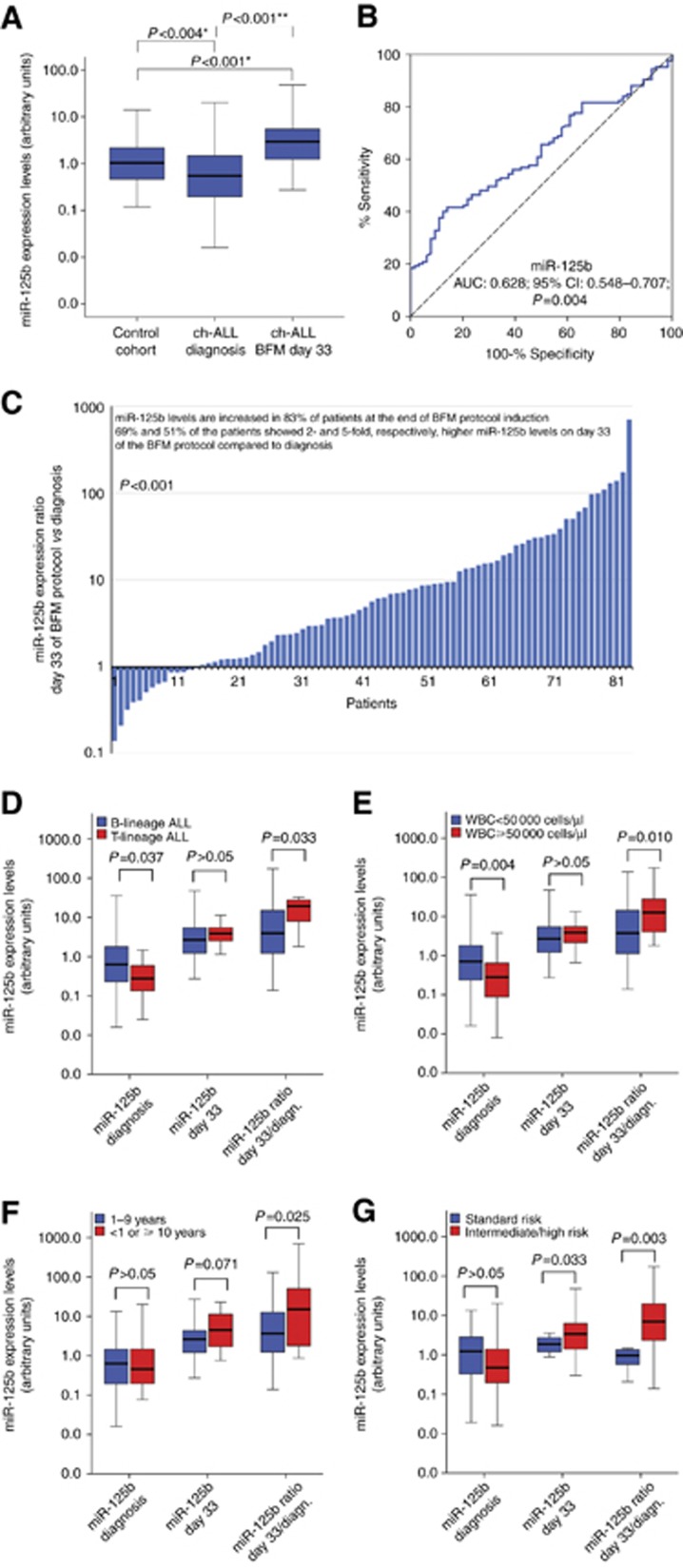
The descriptive statistics of miR-125b expression levels in childhood ALL patients and healthy control cohorts are presented in Supplementary Table 2. The expression analysis pointed out the significant downregulation of miR-125b levels in childhood ALL patients compared to the control group (P=0.004; Figure 1A). The discriminatory significance of miR-125b for childhood ALL from healthy control group was highlighted by the univariate logistic regression (OR: 0.477; 95% CI 0.288–0.790; P=0.004; Supplementary Table 3) and ROC analysis (AUC: 0.628; 95% CI 0.548–0.707; P=0.004; Figure 1B). Moreover, the discriminatory value of miR-125b revealed to be independent of patients’ age and gender by the adjusted multivariate logistic regression model (OR: 0.507; 95% CI 0.305–0.842; P=0.009; Supplementary Table 3).
Figure 1.
Evaluation of miR-125b expression levels in childhood ALL patients. (A) Box plots presenting miR-125b levels in BM specimens of childhood ALL patients on disease diagnosis and on BFM day 33, as well as of healthy controls. P-values calculated by Mann–Whitney U test (*) or Wilcoxon Singed Rank test (**). (B) ROC curve analysis of miR-125b levels for the discrimination of childhood ALL patients’ from healthy controls’ BM specimens. P-value calculated by Hanley and McNeil method. AUC=area under the curve; 95% CI=95% confidence interval. (C) Bar graph of the miR-125b levels ratio on BFM day 33 compared to disease diagnosis. P-value calculated by Wilcoxon Singed Rank test. (D–G) Box plots presenting the correlation of miR-125b expression levels on diagnosis and on BFM day 33, as well as of miR-125b levels ratio (BFM day 33/diagnosis) with patients’ (D) B-lineage ALL and T-lineage ALL, (E) WBC count, (F) age and (G) BFM risk group. P-values calculated by Mann–Whitney U test. A full colour version of this figure is available at the British Journal of Cancer journal online.
The evaluation of miR-125b expression levels from BM specimens obtained on day 33 after BFM induction demonstrated that miR-125b is significantly increased in 83.1% of patients at the end of the induction protocol (P<0.001; Figure 1A and C), which is also highlighted by the fact that 69% and 51% of the patients showed 2 and 5 times, respectively, higher miR-125b levels on day 33 compared to diagnosis. Finally, patients’ BM samples on BFM day 33 revealed to express significantly higher miR-125b related to healthy control cohort (P<0.001; Figure 1A), allowing us to hypothesise that miR-125b levels upregulation on BFM day 33 reflects patients’ response to treatment.
Downregulation of miR-125b levels on diagnosis and higher day 33/diagnosis expression ratio correlate with unfavourable clinicopathological prognostic features
Lower miR-125b expression levels on disease diagnosis were correlated with unfavourable prognostic features of childhood ALL (Figure 1). More precisely, reduced miR-125b levels were detected in T-lineage compared to B-lineage ALL (P=0.037) as well as in patients with WBC ⩾50 000 per μl related to WBC<50 000 per μl patients (P=0.004). The ratio of miR-125b levels at the end of BFM protocol induction (day 33) compared to disease diagnosis (day 33/diagnosis miR-125b ratio) revealed to be higher in T-ALL patients (P=0.033) and in WBC⩾50 000 per μl patients (P=0.010). Moreover, higher day 33/diagnosis miR-125b ratio was also detected in <1 or ⩾10 years old patients (P=0.025) and in high-/intermediate-risk group compared to standard-risk patients (P=0.003). Overall, our data reveal the association of the decreased miR-125b levels on disease diagnosis as well as of the higher day 33/diagnosis miR-125b ratio with unfavourable disease features and dismal prognosis. The correlation of patients’ clinicopathological features with miR-125b levels at disease diagnosis and with BFM day 33/diagnosis miR-125b levels ratio, as dichotomous variables, is presented in Supplementary Table 4.
Patients with reduced miR-125b on diagnosis and elevated day 33/diagnosis expression ratio are at a significantly higher risk for disease recurrence and poor survival outcome
The clinical value of miR-125b levels regarding the BFM chemotherapy treatment outcome was evaluated by Kaplan–Meier survival curves (Figure 2 and Supplementary Figure 2) and Cox regression analysis (Figure 3 and Supplementary Table 5). Kaplan–Meier analysis highlighted that patients with reduced miR-125b expression on disease diagnosis suffered from significantly shorter DFS (P=0.005) and OS (P=0.013) compared to patients with higher miR-125b levels. In addition, univariate Cox regression analysis confirmed the stronger risk for disease relapse (HR: 4.253; 95% CI 1.411–12.82; P=0.010) and death (HR: 3.072; 95% CI 1.209–7.806; P=0.018) of patients with loss of miR-125b on disease diagnosis related to those with miR-125b overexpression.
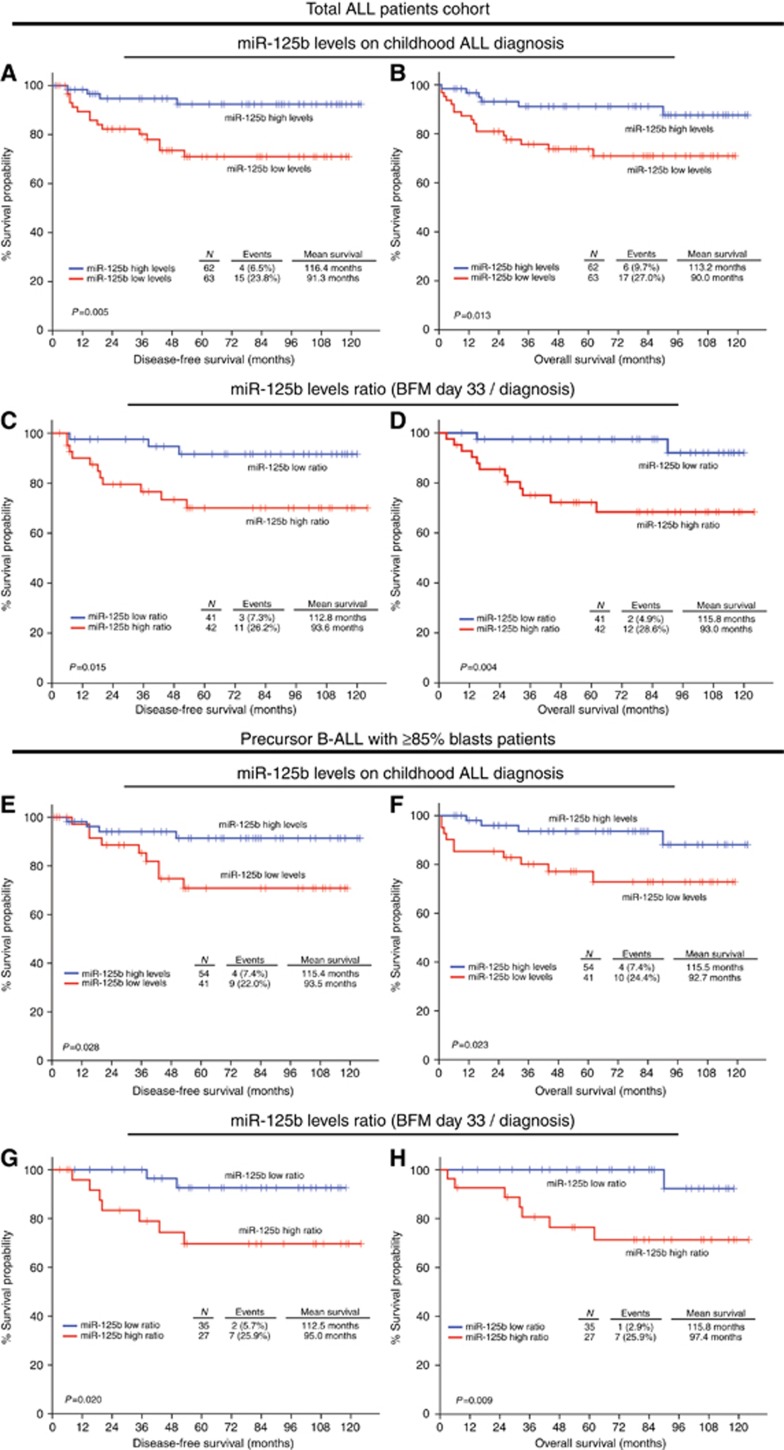
Figure 2.
Loss of miR-125b levels on disease diagnosis and overexpression of miR-125b on BFM day 33 are associated with poor response to BFM treatment. Kaplan–Meier survival curves for the DFS and OS according to miR-125 levels on disease diagnosis, and miR-125b levels ratio (BFM day 33/diagnosis) for (A–D) the total childhood ALL patients’ cohort, and for (E–H) the precursor B-ALL with ⩾85% blasts patients’ cohort. P-values calculated by log-rank test. A full colour version of this figure is available at the British Journal of Cancer journal online.
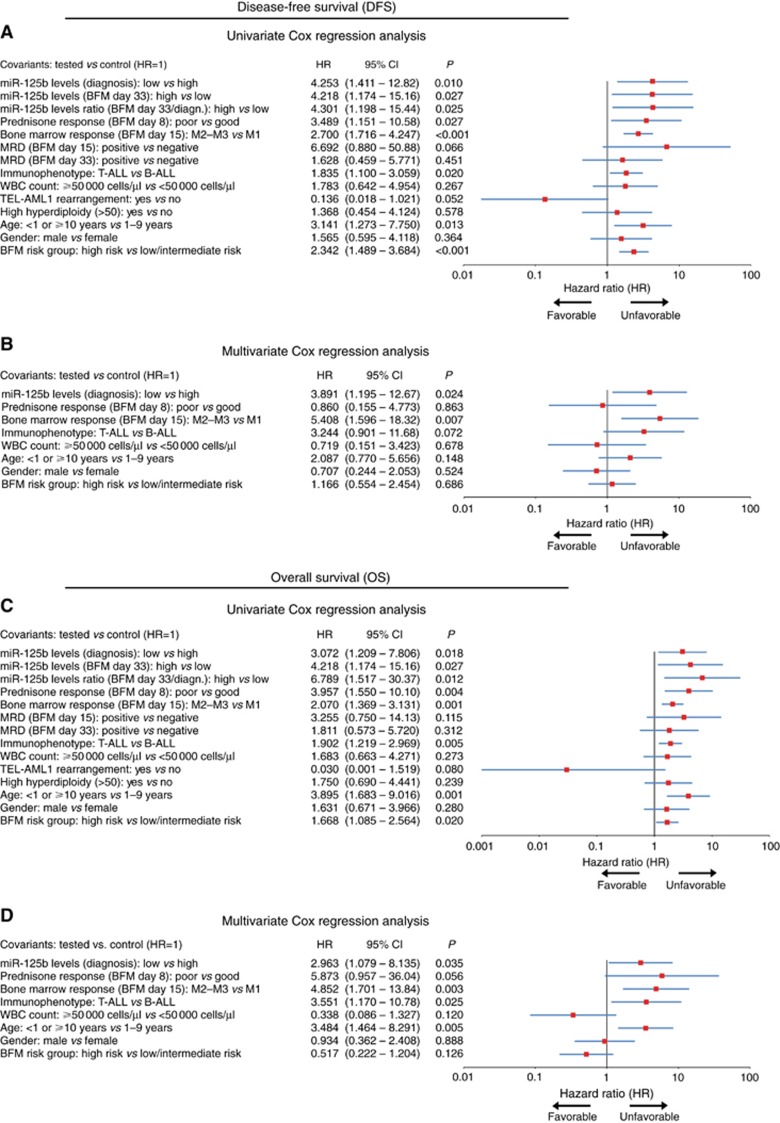
Figure 3.
Childhood ALL patients with lower miR-125b levels on disease diagnosis and overexpression of miR-125b following BFM protocol induction (BFM day 33) are at higher risk for disease short-term relapse and poor survival outcome. Forest plots of the (A and C) univariate and (B and D) multivariate Cox regression analysis for childhood ALL patients’ DFS and OS. Multivariate analysis adjusted for miR-125b expression on childhood ALL diagnosis, prednisone response on BFM day 8, BM response on BFM day 15, ALL immunophenotype, WBC count, BFM risk group, patients’ gender and age. P-values calculated by test for trend. 95% CI=95% confidence interval; HR=hazard ratio. A full colour version of this figure is available at the British Journal of Cancer journal online.
Following BFM induction, miR-125b levels were quantified on day 33 of treatment. Patients with elevated expression on day 33 presented significantly shorter DFS (P=0.016) and OS (P=0.016) (Supplementary Figure 2), as well as higher risk for disease recurrence (HR: 4.218; 95% CI 1.174–15.16; P=0.027) and death (HR: 4.218; 95% CI 1.174–15.16; P=0.027; Figure 3) compared to those with decreased miR-125b levels. To further enhance the clinical significance of miR-125b in childhood ALL, the same analysis was performed for the day 33/diagnosis miR-125b ratio. Patients with increased miR-125b ratio were found to suffer from significantly shorter DFS (P=0.015) and OS (P=0.004; Figure 2), while Cox regression analysis (Figure 3) revealed their stronger risk for short-term relapse (HR: 4.301; 95% CI 1.198–15.44; P=0.025) and death (HR: 6.789; 95% CI 1.517–30.37; P=0.012) despite BFM treatment.
To evaluate if the observed prognostic significance for BFM treatment outcome is caused by the BM blasts expression of miR-125b, we have repeated the analysis in the subset of precursor B-ALL patients with high % BM blast percentage at disease diagnosis. To classify patients to the high % BM blast percentage cohort, the ⩾85% blasts percentage was adopted as optimal cut-off point. Consequently, 95 precursor B-ALL patients were included in the high % BM blast percentage cohort. Kaplan–Meier survival curves highlighted that patients with lower miR-125b levels at disease diagnosis presented significantly worse DFS (P=0.028) and OS (P=0.023; Figure 2), which was confirmed also by univariate Cox regression analysis (Supplementary Table 6) either for disease recurrence (HR: 3.445; 95% CI 1.060–11.20; P=0.040) or death (HR: 3.532; 95% CI 1.107–11.27; P=0.033). Focusing on day 33/diagnosis miR-125b ratio, the increased miR-125b ratio was correlated with inferior DFS (P=0.020) and OS (P=0.009) by Kaplan–Meier analysis (Figure 2), and also with higher risk for BFM treatment resistance by univariate Cox analysis (Supplementary Table 6), in terms of disease relapse (HR: 5.279; 95% CI 1.097–25.42; P=0.038) or patients’ death (HR: 9.604; 95% CI 1.181–78.09; P=0.034). These findings highlight the prognostic significance of blasts’ miR-125b expression in BFM-treated childhood ALL and clearly support that the clinical value of BM miR-125b levels is induced by BM blasts expression.
Finally, multivariate Cox analysis was performed to evaluate the independent prognostic value of miR-125b for childhood ALL. The multivariate Cox models were adjusted for WBC count, prednisone response on day 8, BM response on day 15, immunophenotype, risk group according to BFM stratification, patients’ age and gender. Multivariate analysis (Figure 3 and Supplementary Table 5) highlighted the independent prognostic value of miR-125b levels on childhood ALL diagnosis both for disease relapse (HR: 3.891; 95% CI: 1.195–12.67; P=0.024) and death (HR: 2.963; 95% CI: 1.079–8135; P=0.035).
miR-125b enhances significantly the prognostic value of clinically established biomarkers for disease progression and patients’ survival outcome
Prompted by the significant prognostic value of miR-125b for patients’ survival, we further examined its ability in improving the prediction strength of the established and clinically used prognostic markers. In general, WBC ⩾50 000 cells per μl, poor prednisone response on day 8, M2-M3 BM response on day 15 and positive MRD represent independent unfavourable prognostic markers for patients’ relapse and survival expectancy.
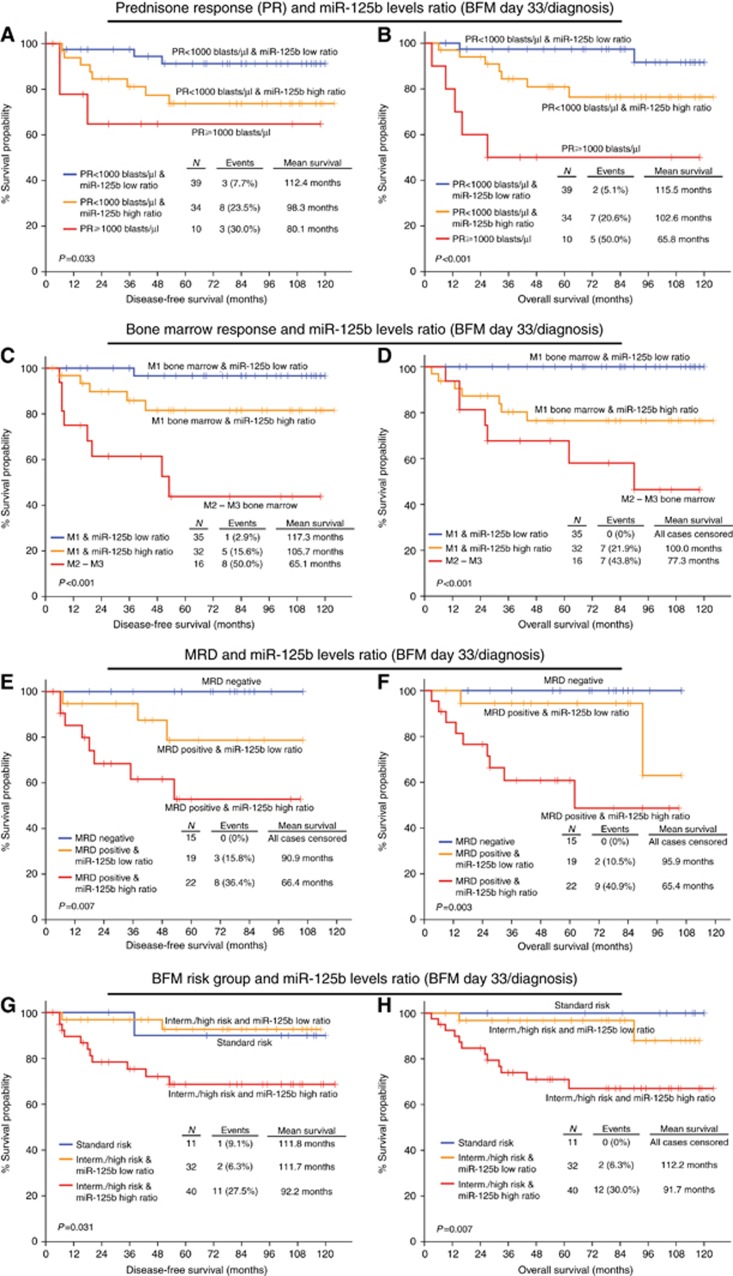
Combining the expression of miR-125b with the abovementioned markers, we revealed the improved stratification of patients regarding their BFM treatment outcome. More precisely, patients who underexpress miR-125b on disease diagnosis present significantly shorter DFS and OS (Figure 4), even if they have good prednisone response (P=0.003 and P=0.001, respectively) or M1 BM response (P<0.001 and P<0.001, respectively). The same conclusion arises for patients having an elevated day 33/diagnosis miR-125b expression ratio (Figure 5), when combined with prednisone response (P=0.033 and P<0.001, respectively) and BM response (P<0.001 and P<0.001, respectively). The study of miR-125b expression with patients’ WBC is presented in Supplementary Figure 3.
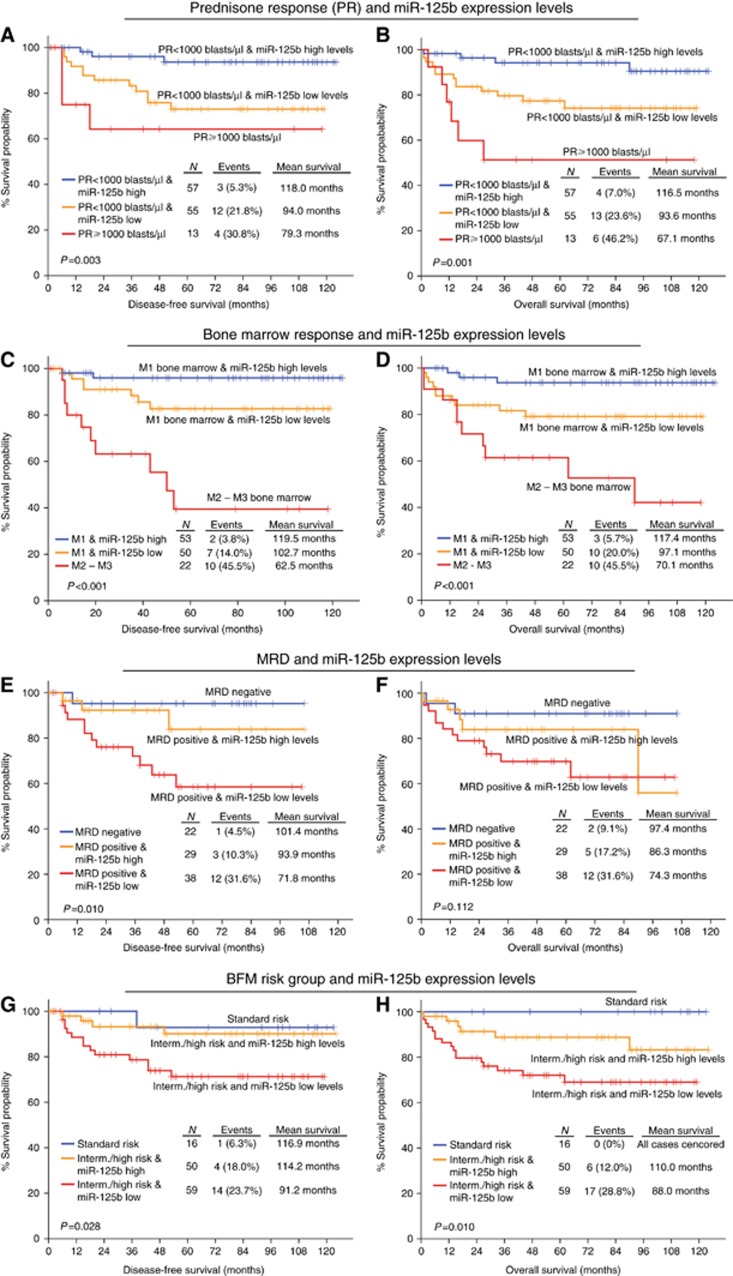
Figure 4.
Evaluation of miR-125b levels on disease diagnosis improves significantly the risk stratification of the childhood ALL patients according to the clinically established prognostic markers for the prediction of patients’ response to BFM treatment. Kaplan–Meier survival curves of the childhood ALL patients’ DFS and OS according to the combination of miR-125b levels on disease diagnosis with (A and B) peripheral blast cells (prednisone response/PR) on BFM day 8, (C and D) BM blasts percentage (BM response) on BFM day 15, (E and F) MRD measurement on BFM day 15, and (G and H) BFM risk groups. P-values calculated by log-rank test. A full colour version of this figure is available at the British Journal of Cancer journal online.
Figure 5.
Evaluation of miR-125b levels ratio (BFM day 33/diagnosis) following BFM protocol induction improves significantly the risk stratification of the childhood ALL patients according to the clinically established prognostic markers for the prediction of patients’ response to BFM treatment. Kaplan–Meier survival curves of the childhood ALL patients’ DFS and OS according to the combination of miR-125b levels ratio (BFM day 33/diagnosis) with (A and B) peripheral blast cells (prednisone response/PR) on BFM day 8, (C and D) BM blasts percentage (BM response) on BFM day 15, (E and F) MRD measurement on BFM day 15, and (G and H) BFM risk groups. P-values calculated by log-rank test. A full colour version of this figure is available at the British Journal of Cancer journal online.
MRD on day 15 is a well-established marker for adjusting treatment intensity. The implementation of the reduced miR-125b levels on diagnosis (Figure 4) ameliorated significantly the predicting ability of MRD for disease recurrence (P=0.010), distinguishing positive MRD patients with an even more adverse outcome. Overexpression of miR-125b on day 33 and thus, increased day 33/diagnosis miR-125b expression ratio (Figure 5), also allowed the distinction of MRD positive patients, characterised by significantly worse DFS (P=0.007) and OS (P=0.003). The study of miR-125b expression with patients’ MRD on day 33 (Supplementary Figure 4) highlighted a trend for unfavourable prognosis of MRD negative patients with reduced miR-125b levels on disease diagnosis and elevated day 33/diagnosis expression ratio. Finally, the combination of miR-125b with the BFM risk group stratification highlighted a subgroup of intermediate/high risk patients with considerably more aggressive leukaemia (Figures 4 and 5). The ability of miR-125b to improve childhood ALL prognosis according to the established markers was also verified in the subset of precursor B-ALL patients with high % BM blasts percentage (Supplementary Figures 5 and 6), confirming the blasts-mediated expression of BM miR-125b levels and strengthening its clinical value in BFM-treated childhood ALL.
Discussion
Over the past decades, there has been tremendous progress in treating paediatric ALL, increasing substantially the remission rates and the survival expectancy of the patients. However, there are still clinical concerns, as some patients are either being overtreated or experience adverse outcome, even if they present favourable clinical characteristics (Pui, 2010; Pui et al, 2012). It is, therefore, essential to discover new prognostic markers, capable of monitoring patients’ response to therapy and predicting their outcome. In recent years, miRNAs have emerged as important biomarkers in all types of cancer, including ALL, and several recent studies have highlighted their involvement in leukaemogenesis and disease morbidity and mortality (de Oliveira et al, 2012).
In this study, we performed the expression analysis of BM miR-125b in childhood ALL patients and we investigated its role as a prognostic tool of the disease. The expression of miR-125b was downregulated in childhood ALL patients compared to the control group, while ROC curve and regression analyses verified miR-125b ability for distinguishing normal from leukaemic BM samples. Moreover, patients’ follow-up revealed the significant overexpression of miR-125b on day 33 of BFM treatment protocol. In addition, reduced miR-125b expression on disease diagnosis, higher expression on day 33 of the induction protocol and elevated day 33/diagnosis expression ratio were correlated with unfavourable prognostic features, such as T-ALL, CD10(−) precursor B-ALL, WBC⩾50 000 cells per μl, increased age of diagnosis and higher BFM-risk stratification.
Numerous studies have supported the implication of miR-125b in various malignancies, acting either as oncogene or as tumour suppressor, depending on the cell context (Sun et al, 2013). Consistent with our findings, Schotte et al (2009) identified miR-125b downregulation in leukaemic mononuclear BM cells or peripheral blood cells in newly diagnosed childhood B-ALL patients being negative for TEL-AML1, BCR-ABL, E2A-PBX and hyperdiploidy compared to normal CD34+ cells, as well as in MLL-rearranged patients. Moreover, Tili et al (2012) reported the reduced expression of miR-125b in aggressive and indolent CLL patients and supported its role in the metabolic adaptation of the cells to a transformed state, allowing Warburg effect, possibly by targeting key enzymes, such as PCTP, LIPA, GSS, ACSS1, HK2, SCD1, AKT2 and PDK1. In addition, miR-125b has been reported to regulate p53 activity by targeting 5-lipoxygenase metabolic enzyme, a p53 inhibitor, in myeloid cells (Busch et al, 2015).
Overexpression of miR-125b was observed in progenitor B-ALL patients carrying the chromosomal translocation t(11;14)(q24;q32) (Sonoki et al, 2005; Chapiro et al, 2010; Tassano et al, 2010), and also in childhood ALL patients positive for TEL-AML1 (Gefen et al, 2010; Schotte et al, 2011). However, these studies focused on subsets of ALL patients with specific chromosomal abnormalities, and in the case of t(11;14)(q24.1;q32) patients, the higher expression of miR-125b can be explained by the fact that MIR125B1 gene is juxtaposed in cis to the strong IGH enhancer. Moreover, the ectopic expression of miR-125b was able to induce B-ALL, T-ALL and myeloproliferative neoplasm in nude mice as well as to promote the tumourigenicity of BCR-ABL induced leukaemia (Bousquet et al, 2010). However, the study was performed in mouse model using lineage-negative haematopoietic foetal liver cells, rather than lymphoid progenitors, while the majority of the transplanted mice developed T-cell rather than B-cell leukaemia, which characterised most of our patients. Nevertheless, the ectopic expression of miR-125b was 700-fold higher in peripheral blood than normal controls, and these immensely increased miR-125b levels may trigger different tumourigenic mechanisms than those in human ALL lymphoblasts, where miR-125b downregulation is observed.
Focusing on patients’ treatment outcome, decreased miR-125b levels on childhood ALL diagnosis and its overexpression on day 33 of chemotherapy were associated with significantly stronger risk for short-term relapse and poor OS of the treated patients. Moreover, multivariate Cox analysis highlighted the clinical significance of miR-125b levels for the childhood ALL prognosis independently of established prognostic markers and patients’ clinicopathological data. The clinical utility of BM miR-125b levels for the prognosis of BFM treatment outcome was confirmed also in the subset of precursor B-ALL patients with high % BM blast percentage at disease diagnosis, strengthening the miR-125b prognostic significance in BFM-treated childhood ALL and confirming the blasts-mediated expression of BM miR-125b levels.
Interestingly, the combination of miR-125b expression levels with established and clinically used disease prognostic markers improved significantly patients’ stratification for poor response to BFM chemotherapy either in the total patient cohort or the precursor B-ALL patients with high % BM blast percentage cohort. The downregulation of miR-125b on diagnosis and the higher values of day 33/diagnosis expression ratio were able to discriminate patients at higher risk for short-term relapse and worse survival outcome, despite the presence of favourable WBC <50 000 cells per μl, good prednisone response or good BM response. The same expression pattern of miR-125b can ameliorate the predicting value of MRD on day 15 and the current BFM-risk stratification system, as it can identify patients with even more aggressive disease among the MRD positive patients’ group or among intermediate/high risk patients’ cohorts.
The dismal prognosis of BFM-treated patients with increased miR-125b levels on day 33 compared to diagnosis is consistent with previous studies, where miR-125b levels correlated with chemotherapy resistance and inhibition of apoptosis. BFM treatment protocol includes several drugs that induce apoptosis, either directly via glucocorticoids (Schmidt et al, 2004), or as a secondary event via vincristine (Groninger et al, 2002) and daunorubicin (Richardson and Johnson, 1997). Schotte et al (2011) highlighted the significantly upregulated miR-125b levels in childhood precursor B-ALL patients resistant to vincristine and daunorubicin. Moreover, miR-125b was reported to promote survival against apoptotic stimuli and vincristine resistance in ETV6/RUNX1 leukaemic cells (Gefen et al, 2010; Akbari Moqadam et al, 2013), while a miR-125b-mediated resistance to vincristine was also observed in childhood acute megakaryoblastic leukaemia cells (Kandi et al, 2015). In addition, leukaemic cell lines resistance to daunorubicin has been induced by the miR-125b-mediated downregulation of GRK2 and PUMA (Zhou et al, 2014).
Focusing on apoptosis-related targets, miR-125b overexpression enhances drug resistance and inhibits apoptosis in acute promyelocytic leukaemia, by targeting BAK1 pro-apoptotic gene (Zhang et al, 2011), a network that is also observed in breast and ovarian cancers (Zhou et al, 2010; Kong et al, 2011). Apart from BAK1 (Shi et al, 2007), several other genes involved in the apoptotic pathway, such as BMF (Xia et al, 2009), TP53 (Le et al, 2009; Zeng et al, 2012; Murray et al, 2013), TP53INP1 (Enomoto et al, 2011; Bousquet et al, 2012), TNFAIP3 (Kim et al, 2012) and MAPK14 (p38a) (Tan et al, 2012) are also validated targets of miR-125b, while the ectopic overexpression of miR-125b has been found to block cell apoptosis through p53 pathway suppression, NF-κB pathway activation, or BAK1 downregulation. These studies clearly demonstrate a possible inhibitory role of miR-125b upon the activity of chemotherapeutic agents inducing apoptosis and support our findings concerning the poor treatment and survival outcome of the BFM-treated childhood ALL patients with increased post-treatment miR-125b levels.
In conclusion, our study revealed the downregulation of miR-125b in childhood ALL patients compared to the control group and its significant upregulation following induction of BFM chemotherapy protocol. Moreover, lower miR-125b levels on diagnosis, and higher expression on the 33rd day of treatment were associated with adverse disease features. Survival analysis clearly demonstrated the stronger risk for short-term relapse and worse OS of patients who underexpress miR-125b on diagnosis and overexpress it on the 33rd day, thus presenting elevated day 33/diagnosis expression ratio. Finally, the combination of miR-125b expression with the clinically used disease prognostic markers resulted in a superior positive prediction of childhood ALL patients’ poor response to BFM chemotherapy protocol compared to the study of the disease markers alone. To our knowledge this is the first study of miR-125b clinical value in the prognosis of childhood ALL and in prediction of patients’ response to BFM therapy.
Acknowledgments
We wish to sincerely thank Drs M. Varvoutsi, A. Pourtsidis and M. Servitzoglou for their valuable professional assistance in the characterisation and collection of our samples. We would also like to thank the nursing staff of the Department of Pediatric Oncology, ‘P & A Kyriakou’ Children’s Hospital for their expert help with the collection of samples.
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc)
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 4.0 Unported License.
The authors declare no conflict of interest.
Supplementary Material
References
- Akbari Moqadam F, Lange-Turenhout EA, Aries IM, Pieters R, den Boer ML (2013) MiR-125b, miR-100 and miR-99a co-regulate vincristine resistance in childhood acute lymphoblastic leukaemia. Leuk Res 37(10): 1315–1321. [DOI] [PubMed] [Google Scholar]
- Almeida MI, Reis RM, Calin GA (2011) MicroRNA history: discovery, recent applications, and next frontiers. Mutat Res 717(1-2): 1–8. [DOI] [PubMed] [Google Scholar]
- Bhojwani D, Pui CH (2013) Relapsed childhood acute lymphoblastic leukaemia. Lancet Oncol 14(6): e205–e217. [DOI] [PubMed] [Google Scholar]
- Bousquet M, Harris MH, Zhou B, Lodish HF (2010) MicroRNA miR-125b causes leukaemia. Proc Natl Acad Sci USA 107(50): 21558–21563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bousquet M, Nguyen D, Chen C, Shields L, Lodish HF (2012) MicroRNA-125b transforms myeloid cell lines by repressing multiple mRNA. Haematologica 97(11): 1713–1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch S, Auth E, Scholl F, Huenecke S, Koehl U, Suess B, Steinhilber D (2015) 5-lipoxygenase is a direct target of miR-19a-3p and miR-125b-5p. J Immunol 194(4): 1646–1653. [DOI] [PubMed] [Google Scholar]
- Bushati N, Cohen SM (2007) microRNA functions. Annu Rev Cell Dev Biol 23: 175–205. [DOI] [PubMed] [Google Scholar]
- Calin GA, Croce CM (2006) MicroRNA signatures in human cancers. Nat Rev Cancer 6(11): 857–866. [DOI] [PubMed] [Google Scholar]
- Chapiro E, Russell LJ, Struski S, Cave H, Radford-Weiss I, Valle VD, Lachenaud J, Brousset P, Bernard OA, Harrison CJ, Nguyen-Khac F (2010) A new recurrent translocation t(11;14)(q24;q32) involving IGH@ and miR-125b-1 in B-cell progenitor acute lymphoblastic leukaemia. leukaemia 24(7): 1362–1364. [DOI] [PubMed] [Google Scholar]
- de Oliveira JC, Brassesco MS, Scrideli CA, Tone LG, Narendran A (2012) MicroRNA expression and activity in pediatric acute lymphoblastic leukaemia (ALL). Pediatr Blood Cancer 59(4): 599–604. [DOI] [PubMed] [Google Scholar]
- Enomoto Y, Kitaura J, Hatakeyama K, Watanuki J, Akasaka T, Kato N, Shimanuki M, Nishimura K, Takahashi M, Taniwaki M, Haferlach C, Siebert R, Dyer MJ, Asou N, Aburatani H, Nakakuma H, Kitamura T, Sonoki T (2011) Emu/miR-125b transgenic mice develop lethal B-cell malignancies. leukaemia 25(12): 1849–1856. [DOI] [PubMed] [Google Scholar]
- Ganan-Gomez I, Wei Y, Yang H, Pierce S, Bueso-Ramos C, Calin G, Boyano-Adanez Mdel C, Garcia-Manero G (2014) Overexpression of miR-125a in myelodysplastic syndrome CD34+ cells modulates NF-kappaB activation and enhances erythroid differentiation arrest. PLoS One 9(4): e93404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garzon R, Volinia S, Liu CG, Fernandez-Cymering C, Palumbo T, Pichiorri F, Fabbri M, Coombes K, Alder H, Nakamura T, Flomenberg N, Marcucci G, Calin GA, Kornblau SM, Kantarjian H, Bloomfield CD, Andreeff M, Croce CM (2008) MicroRNA signatures associated with cytogenetics and prognosis in acute myeloid leukaemia. Blood 111(6): 3183–3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gefen N, Binder V, Zaliova M, Linka Y, Morrow M, Novosel A, Edry L, Hertzberg L, Shomron N, Williams O, Trka J, Borkhardt A, Izraeli S (2010) Hsa-mir-125b-2 is highly expressed in childhood ETV6/RUNX1 (TEL/AML1) leukaemias and confers survival advantage to growth inhibitory signals independent of p53. leukaemia 24(1): 89–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groninger E, Meeuwsen-De Boer GJ, De Graaf SS, Kamps WA, De Bont ES (2002) Vincristine induced apoptosis in acute lymphoblastic leukaemia cells: a mitochondrial controlled pathway regulated by reactive oxygen species? Int J Oncol 21(6): 1339–1345. [DOI] [PubMed] [Google Scholar]
- Kandi R, Gutti U, Saladi RG, Gutti RK (2015) MiR-125b and miR-99a encoded on chromosome 21 co-regulate vincristine resistance in childhood acute megakaryoblastic leukaemia. Hematol Oncol Stem Cell Ther 8(2): 95–97. [DOI] [PubMed] [Google Scholar]
- Kim SW, Ramasamy K, Bouamar H, Lin AP, Jiang D, Aguiar RC (2012) MicroRNAs miR-125a and miR-125b constitutively activate the NF-kappaB pathway by targeting the tumor necrosis factor alpha-induced protein 3 (TNFAIP3, A20). Proc Natl Acad Sci USA 109(20): 7865–7870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klusmann JH, Li Z, Bohmer K, Maroz A, Koch ML, Emmrich S, Godinho FJ, Orkin SH, Reinhardt D (2010) miR-125b-2 is a potential oncomiR on human chromosome 21 in megakaryoblastic leukaemia. Genes Dev 24(5): 478–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong F, Sun C, Wang Z, Han L, Weng D, Lu Y, Chen G (2011) miR-125b confers resistance of ovarian cancer cells to cisplatin by targeting pro-apoptotic Bcl-2 antagonist killer 1. J Huazhong Univ Sci Technolog Med Sci 31(4): 543–549. [DOI] [PubMed] [Google Scholar]
- Le MT, Teh C, Shyh-Chang N, Xie H, Zhou B, Korzh V, Lodish HF, Lim B (2009) MicroRNA-125b is a novel negative regulator of p53. Genes Dev 23(7): 862–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin KY, Zhang XJ, Feng DD, Zhang H, Zeng CW, Han BW, Zhou AD, Qu LH, Xu L, Chen YQ (2011) miR-125b, a target of CDX2, regulates cell differentiation through repression of the core binding factor in haematopoietic malignancies. J Biol Chem 286(44): 38253–38263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Smith KR, Khong HT, Huang J, Ahn EE, Zhou M, Tan M (2016) miR-125b regulates differentiation and metabolic reprogramming of T cell acute lymphoblastic leukaemia by directly targeting A20. Oncotarget 7(48): 78667–78679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, Downing JR, Jacks T, Horvitz HR, Golub TR (2005) MicroRNA expression profiles classify human cancers. Nature 435(7043): 834–838. [DOI] [PubMed] [Google Scholar]
- Moricke A, Reiter A, Zimmermann M, Gadner H, Stanulla M, Dordelmann M, Loning L, Beier R, Ludwig WD, Ratei R, Harbott J, Boos J, Mann G, Niggli F, Feldges A, Henze G, Welte K, Beck JD, Klingebiel T, Niemeyer C, Zintl F, Bode U, Urban C, Wehinger H, Niethammer D, Riehm H, Schrappe M (2008) Risk-adjusted therapy of acute lymphoblastic leukaemia can decrease treatment burden and improve survival: treatment results of 2169 unselected pediatric and adolescent patients enrolled in the trial ALL-BFM 95. Blood 111(9): 4477–4489. [DOI] [PubMed] [Google Scholar]
- Murray MY, Rushworth SA, Zaitseva L, Bowles KM, Macewan DJ (2013) Attenuation of dexamethasone-induced cell death in multiple myeloma is mediated by miR-125b expression. Cell Cycle 12(13): 2144–2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pui CH (2010) Recent research advances in childhood acute lymphoblastic leukaemia. J Formos Med Assoc 109(11): 777–787. [DOI] [PubMed] [Google Scholar]
- Pui CH, Mullighan CG, Evans WE, Relling MV (2012) Pediatric acute lymphoblastic leukaemia: where are we going and how do we get there? Blood 120(6): 1165–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puissegur MP, Eichner R, Quelen C, Coyaud E, Mari B, Lebrigand K, Broccardo C, Nguyen-Khac F, Bousquet M, Brousset P (2012) B-cell regulator of immunoglobulin heavy-chain transcription (Bright)/ARID3a is a direct target of the oncomir microRNA-125b in progenitor B-cells. leukaemia 26(10): 2224–2232. [DOI] [PubMed] [Google Scholar]
- Ramsingh G, Koboldt DC, Trissal M, Chiappinelli KB, Wylie T, Koul S, Chang LW, Nagarajan R, Fehniger TA, Goodfellow P, Magrini V, Wilson RK, Ding L, Ley TJ, Mardis ER, Link DC (2010) Complete characterization of the microRNAome in a patient with acute myeloid leukaemia. Blood 116(24): 5316–5326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson DS, Johnson SA (1997) Anthracyclines in haematology: preclinical studies, toxicity and delivery systems. Blood Rev 11(4): 201–223. [DOI] [PubMed] [Google Scholar]
- Rodriguez A, Griffiths-Jones S, Ashurst JL, Bradley A (2004) Identification of mammalian microRNA host genes and transcription units. Genome Res 14(10A): 1902–1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt S, Rainer J, Ploner C, Presul E, Riml S, Kofler R (2004) Glucocorticoid-induced apoptosis and glucocorticoid resistance: molecular mechanisms and clinical relevance. Cell Death Differ 11(Suppl 1): S45–S55. [DOI] [PubMed] [Google Scholar]
- Schotte D, Chau JC, Sylvester G, Liu G, Chen C, van der Velden VH, Broekhuis MJ, Peters TC, Pieters R, den Boer ML (2009) Identification of new microRNA genes and aberrant microRNA profiles in childhood acute lymphoblastic leukaemia. leukaemia 23(2): 313–322. [DOI] [PubMed] [Google Scholar]
- Schotte D, De Menezes RX, Akbari Moqadam F, Khankahdani LM, Lange-Turenhout E, Chen C, Pieters R, Den Boer ML (2011) MicroRNA characterize genetic diversity and drug resistance in pediatric acute lymphoblastic leukaemia. Haematologica 96(5): 703–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schotte D, Pieters R, Den Boer ML (2012) MicroRNAs in acute leukaemia: from biological players to clinical contributors. leukaemia 26(1): 1–12. [DOI] [PubMed] [Google Scholar]
- Shi XB, Xue L, Yang J, Ma AH, Zhao J, Xu M, Tepper CG, Evans CP, Kung HJ, deVere White RW (2007) An androgen-regulated miRNA suppresses Bak1 expression and induces androgen-independent growth of prostate cancer cells. Proc Natl Acad Sci USA 104(50): 19983–19988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel RL, Miller KD, Jemal A (2016) Cancer statistics, 2016. CA Cancer J Clin 66(1): 7–30. [DOI] [PubMed] [Google Scholar]
- So AY, Sookram R, Chaudhuri AA, Minisandram A, Cheng D, Xie C, Lim EL, Flores YG, Jiang S, Kim JT, Keown C, Ramakrishnan P, Baltimore D (2014) Dual mechanisms by which miR-125b represses IRF4 to induce myeloid and B-cell leukaemias. Blood 124(9): 1502–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoki T, Iwanaga E, Mitsuya H, Asou N (2005) Insertion of microRNA-125b-1, a human homologue of lin-4, into a rearranged immunoglobulin heavy chain gene locus in a patient with precursor B-cell acute lymphoblastic leukaemia. leukaemia 19(11): 2009–2010. [DOI] [PubMed] [Google Scholar]
- Stary J, Zimmermann M, Campbell M, Castillo L, Dibar E, Donska S, Gonzalez A, Izraeli S, Janic D, Jazbec J, Konja J, Kaiserova E, Kowalczyk J, Kovacs G, Li CK, Magyarosy E, Popa A, Stark B, Jabali Y, Trka J, Hrusak O, Riehm H, Masera G, Schrappe M (2014) Intensive chemotherapy for childhood acute lymphoblastic leukaemia: results of the randomized intercontinental trial ALL IC-BFM 2002. J Clin Oncol 32(3): 174–184. [DOI] [PubMed] [Google Scholar]
- Sun YM, Lin KY, Chen YQ (2013) Diverse functions of miR-125 family in different cell contexts. J Hematol Oncol 6: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surdziel E, Cabanski M, Dallmann I, Lyszkiewicz M, Krueger A, Ganser A, Scherr M, Eder M (2011) Enforced expression of miR-125b affects myelopoiesis by targeting multiple signaling pathways. Blood 117(16): 4338–4348. [DOI] [PubMed] [Google Scholar]
- Tan G, Niu J, Shi Y, Ouyang H, Wu ZH (2012) NF-kappaB-dependent microRNA-125b up-regulation promotes cell survival by targeting p38alpha upon ultraviolet radiation. J Biol Chem 287(39): 33036–33047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tassano E, Acquila M, Tavella E, Micalizzi C, Panarello C, Morerio C (2010) MicroRNA-125b-1 and BLID upregulation resulting from a novel IGH translocation in childhood B-Cell precursor acute lymphoblastic leukaemia. Genes Chromosomes Cancer 49(8): 682–687. [DOI] [PubMed] [Google Scholar]
- Tili E, Michaille JJ, Luo Z, Volinia S, Rassenti LZ, Kipps TJ, Croce CM (2012) The down-regulation of miR-125b in chronic lymphocytic leukaemias leads to metabolic adaptation of cells to a transformed state. Blood 120(13): 2631–2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ufkin ML, Peterson S, Yang X, Driscoll H, Duarte C, Sathyanarayana P (2014) miR-125a regulates cell cycle, proliferation, and apoptosis by targeting the ErbB pathway in acute myeloid leukaemia. Leuk Res 38(3): 402–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasilatou D, Papageorgiou S, Pappa V, Papageorgiou E, Dervenoulas J (2010) The role of microRNAs in normal and malignant hematopoiesis. Eur J Haematol 84(1): 1–16. [DOI] [PubMed] [Google Scholar]
- Wang Y, Li Z, He C, Wang D, Yuan X, Chen J, Jin J (2010) MicroRNAs expression signatures are associated with lineage and survival in acute leukaemias. Blood Cells Mol Dis 44(3): 191–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia HF, He TZ, Liu CM, Cui Y, Song PP, Jin XH, Ma X (2009) MiR-125b expression affects the proliferation and apoptosis of human glioma cells by targeting Bmf. Cell Physiol Biochem 23(4-6): 347–358. [DOI] [PubMed] [Google Scholar]
- Zeng CW, Zhang XJ, Lin KY, Ye H, Feng SY, Zhang H, Chen YQ (2012) Camptothecin induces apoptosis in cancer cells via microRNA-125b-mediated mitochondrial pathways. Mol Pharmacol 81(4): 578–586. [DOI] [PubMed] [Google Scholar]
- Zhang H, Luo XQ, Feng DD, Zhang XJ, Wu J, Zheng YS, Chen X, Xu L, Chen YQ (2011) Upregulation of microRNA-125b contributes to leukaemogenesis and increases drug resistance in pediatric acute promyelocytic leukaemia. Mol Cancer 10: 108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Bai H, Wang C, Wei D, Qin Y, Xu X (2014) microRNA125b promotes leukaemia cell resistance to daunorubicin by inhibiting apoptosis. Mol Med Rep 9(5): 1909–1916. [DOI] [PubMed] [Google Scholar]
- Zhou M, Liu Z, Zhao Y, Ding Y, Liu H, Xi Y, Xiong W, Li G, Lu J, Fodstad O, Riker AI, Tan M (2010) MicroRNA-125b confers the resistance of breast cancer cells to paclitaxel through suppression of pro-apoptotic Bcl-2 antagonist killer 1 (Bak1) expression. J Biol Chem 285(28): 21496–21507. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.