Abstract
Background:
Hypoxia is negatively associated with glioblastoma (GBM) patient survival and contributes to tumour resistance. Anti-angiogenic therapy in GBM further increases hypoxia and activates survival pathways. The aim of this study was to determine the role of hypoxia-induced autophagy in GBM.
Methods:
Pharmacological inhibition of autophagy was applied in combination with bevacizumab in GBM patient-derived xenografts (PDXs). Sensitivity towards inhibitors was further tested in vitro under normoxia and hypoxia, followed by transcriptomic analysis. Genetic interference was done using ATG9A-depleted cells.
Results:
We find that GBM cells activate autophagy as a survival mechanism to hypoxia, although basic autophagy appears active under normoxic conditions. Although single agent chloroquine treatment in vivo significantly increased survival of PDXs, the combination with bevacizumab resulted in a synergistic effect at low non-effective chloroquine dose. ATG9A was consistently induced by hypoxia, and silencing of ATG9A led to decreased proliferation in vitro and delayed tumour growth in vivo. Hypoxia-induced activation of autophagy was compromised upon ATG9A depletion.
Conclusions:
This work shows that inhibition of autophagy is a promising strategy against GBM and identifies ATG9 as a novel target in hypoxia-induced autophagy. Combination with hypoxia-inducing agents may provide benefit by allowing to decrease the effective dose of autophagy inhibitors.
Keywords: glioblastoma, autophagy, hypoxia, ATG9A, chloroquine, patient-derived xenografts
Despite considerable advancement in the molecular characterisation of glioblastoma (GBM), survival of patients under current treatment regimen remains disappointing. Treatment failure is partially due to the capacity of tumour cells to activate pro-survival pathways in an unfavourable microenvironment. The GBM vasculature is poorly functional, leading to insufficient oxygen supply and necrotic areas (Evans et al, 2004). Hypoxia and angiogenic factors are correlated with tumour grade and poor patient prognosis in brain tumours (Yang et al, 2012) and are linked to radiation- and chemotherapy resistance (Vaupel and Mayer, 2007). Although targeting angiogenesis has long been regarded as an attractive therapeutic approach, anti-angiogenic agents are incapable to halt tumour progression and improve patient survival (Gilbert, 2016). We have previously shown that administration of bevacizumab, an antibody against vascular endothelial growth factor (VEGF), resulted in an adaptive metabolic switch leading to an increased hypoxia and induction of glycolysis (Keunen et al, 2011; Fack et al, 2015). However, the exact mechanism of GBM cell survival and adaptation under hypoxia are still incompletely understood.
Solid tumours use autophagy as one of the survival mechanisms upon various stressors including metabolic stress and starvation (Yang et al, 2011), hypoxia (Rabinowitz and White, 2010; Rouschop et al, 2010), chemotherapy (Kanzawa et al, 2004; Ciechomska et al, 2013) and radiotherapy (Firat et al, 2012). In physiological situations, autophagy has an important role in organelle turnover, degradation of proteins, cellular differentiation and aging (Glick et al, 2010). During stress, autophagy protects cells by eliminating damaged organelles and proteins via autophagosomes. Autophagosomes fuse with lysosomes to form the autolysosome responsible for enzymatic self-digestion of cellular waste. Recycled cellular components may serve as an energy source during periods of starvation, hypoxia or high-energy demand. Under physiological hypoxia (0.1–3%O2), the autophagic response is HIF1α-dependent (Mazure and Pouyssegur, 2010) and relies on the induction of the pro-autophagic genes BNIP3 (BCL2/adenovirus E1B 19kDA interacting protein 3) and BNIP3L (BNIP3-like) (Pouyssegur et al, 2006; Bellot et al, 2009). Furthermore, autophagy is strongly dependent on the synchronised action of autophagy-related (ATG) genes. Although many ATG genes are modulated upon induction of autophagy (Gasch et al, 2000), their specific roles are not always fully elucidated. ATG9A is the only transmembrane autophagy-related protein and has been associated with the regulation of autophagosome formation (Jin and Klionsky, 2014). ATG9A cycles between the Golgi network, endosomes and the so called ‘ATG9A reservoir’, and ATG9A-containing vesicles in cytoplasm, creating a ready source to support autophagosome formation (Reggiori and Tooze, 2012). Although the detailed mechanism is poorly understood, it is thought to support the growth and maturation of autophagic membranes by recruiting membrane structures to the LC3-positive autophagosomes (Orsi et al, 2012; Yamamoto et al, 2012; Corcelle-Termeau et al, 2016; Lamb et al, 2016).
Following up on our earlier studies (Fack et al, 2015; Sanzey et al, 2015), we addressed the role of autophagy in enabling cell survival in severe hypoxia and during anti-angiogenic treatment. We show that GBM cells activate autophagy in hypoxia and that ATG9A has an essential role in the autophagic response of GBM.
Materials and methods
GBM patient material
Human GBMs were obtained from the Neurosurgery Department of the Centre Hospitalier in Luxembourg (CHL) (T16) or the Department of Neurosurgery, Haukeland University Hospital in Bergen (P3, P8), Norway. All patients had provided informed consent, tumour collection was approved by the National Research Ethics Committee for Luxembourg (CNER) or by the Regional Ethical Board at the Haukeland University Hospital in Bergen. All biopsies were primary GBM based on neuropathological diagnosis and genomic analysis (Supplementary Table S1). The original organotypic GBM spheroids from patient samples were prepared as previously described (Keunen et al, 2011; Golebiewska et al, 2013; Bougnaud et al, 2016) and maintained in spheroid medium (DMEM medium, 10% FBS, 2 mM L-Glutamine, 0.4 mM NEAA and 100 U ml−1 Pen-Strep; Lonza, Basel, Switzerland) in agar pre-coated flasks for 7–10 days.
Orthotopic patient-derived GBM xenografts
Serial transplantation of PDXs in eGFP-expressing NOD/SCID mice were used to expand the tumour material and prepare spheroids for in vitro assays, as previously described (Niclou et al, 2008; Bougnaud et al, 2016). For treatment experiments, P3 and T16 GBM spheroids expressing RFP were orthotopically implanted into the right frontal lobe of Swiss nude mice (6 per mice). Tumour growth was monitored by in vivo fluorescence imaging (IVIS Lumina Fluorescence system; PerkinElmer, Waltham, MA, USA). Three weeks post implantation mice were randomly allocated into treatment groups (6–7 mice per group). Bevacizumab, chloroquine and normal saline were delivered by intraperitoneal injections. The treatment schedule is summarised in Supplementary Table S2. NCH421k and NCH644 harbouring Scramble or ATG9A shRNA were stereotactically implanted in NOD/SCID mice (13 7500 NCH421k cells or 50 000 NCH644 cells per animal; 6–7 per group). Animals were monitored daily and the following criteria were evaluated: (1) loss of >10% of body weight, (2) exhibition of strong neurological signs (3) increased lordosis or (4) swollen belly. The criteria were scored as: 0=none, 1=early, 2=established, 3=severe signs and animals were killed when three criteria with grade 2 or 1 criteria with grade 3 were reached. All procedures were approved by the national authorities responsible for animal experiments in Luxembourg.
Immunohistochemistry
For mouse-specific CD31 staining cryostat sections (10 μm) of flash-frozen brains were fixed in ice-cold acetone and acetone : chloroform (1 : 1) for 5 min each. Sections were blocked for 1 h in TBS/2% FCS, followed by a 1 h incubation in rat anti-mouse CD31 antibody (Merck Millipore, Nottingham, UK, 1 : 200). Alexa Fluor 488-conjugated secondary antibodies were applied for 1 h. Sections were analysed by fluorescence microscopy. Quantification of vessel staining was done using ImageJ (NHS, Bethesda, MA, USA) from 3–4 mice per group (9–34 images per mice).
Western blotting
GBM cells were cultured in normoxia or 0.1% O2 hypoxia for 48 h. When indicated, 20 μM chloroquine was added 16 h before cell collection. Cultured cells or spheroids were lysed in RIPA buffer (Merck Millipore) with 0.1% SDS. Overall, 30 μg of proteins were loaded and separated in a NuPAGE Novex 4–12% Bis-Tris Gels (Life Technologies, Merelbeke, Belgium) followed by electroblot transfer to a PVDF membrane (Novex, Invitrolon PVDF, Life Technologies). Membranes were blocked with 2% non-fat milk in Tris-buffered saline containing 0.1% Triton-X before incubation with primary antibodies (LC3B: Cell Signaling Technology, Danvers, MA, USA, 1 : 2000; p62: BD Bioscience, Erembodegem, Belgium, 1 : 1000; Actin: Millipore, 1 : 10 000; Tubulin: Millipore, 1 : 5000). Secondary coupled to horseradish peroxidase were detected by enhanced chemiluminescence (ECL) (Lumigen TMA6, GE Healthcare) with luminescent image analyser (Image Quant LAS4000, GE Healthcare, Diegem, Belgium). Quantification was performed with the ImageQuant TL. Owing to the substantial normalisation problems linked to disturbed actin and tubulin signal in hypoxic cells upon induction of autophagy (Klionsky et al, 2016), WB signals were normalised to total protein content.
Cell viability in GBM spheroids
Cell viability after 72 h of treatment with inhibitors was assessed by double labelling with 2 μM Calcein AM and 4 μM Ethidium homodimer-1 (LIVE/DEAD Viability/Cytotoxicity assay kit, Molecular Probes, Eugene, OR, USA) for 6 h. Measurements of viable (‘green’) and dead (‘red’) cells were performed using fluorescence confocal microscopy (Zeiss LSM STO META, Zeiss, Zaventem, Belgium) by obtaining 20–25 stacks of two-dimensional images from successive focal planes (5 μm). Quantification was performed using IMARIS software (Bitplane, Belfast, UK). The volume of viable and dead cells within a spheroid was calculated by multiplying the surface area of each component per stack by the total height of the image stacks. The percentage of dead cell volume was calculated as: % dead cell in spheroids (volume)=Dead cell volume (‘red’)) × 100/Total spheroid volume (‘green’+‘red’). Experiments were carried out three times with at least five spheroids each.
Cell culture
The primary adherent P3 cells (P3A) was derived from patient xenograft-derived P3 3D spheroids grown in uncoated flasks until a confluent adherent culture was obtained. P3A, U87, U251 and T98G cells were cultured as monolayers in DMEM containing 10% FBS, 2 mM L-glutamine and 100 U ml−1 Pen-Strep (Lonza). The normal human astrocytes (NHA) (kindly provided by Dr Uros Rajcevic, Ljubljana, Slovenia) grew in DMEM, 10% FBS, 2 mM L-glutamine and 100 100 U ml−1 Pen-Strep (Lonza). GBM stem-like cultures (NCH421k, NCH660h, NCH465, NCH601 and NCH644) were kindly provided by Christel Herold-Mende (University of Heidelberg, Germany) and were cultured as previously described (Sanzey et al, 2015). Normoxic cultures were performed at 37 °C under 5% CO2 atmospheric oxygen. Hypoxic conditions at 0.1–0.5% O2 were maintained in the hypoxic incubator chamber (Galaxy 48R incubator, New Brunswick, Eppendorf, Rotsellar, Belgium).
Cytotoxicity assay
Cells were plated at semi-confluency in 96 well plates. NCH644 were attached on ECM Cell-Tak (VWR, Leuven, Belgium) precoated plates. Increasing concentrations of tested compounds (chloroquine diphosphate (Sigma, Overijse, Belgium; C6628) and mefloquine hydrochloride (Sigma, M2319)) were applied for 72 h. Induction of cell death was measured after 72 h with the Sulforhodamine (SRB) assay (In Vitro Toxicology Assay Kit, Sigma). The optical density was measured at 540 nm. The percentage inhibition of cell mass was determined as: % cell mass reduction=(Mean ODcontrol−mean ODsample) × 100/Mean ODcontrol. IC50 was determined with the GraphPad Prism 5 (GraphPad Software, La Jolla, CA, USA).
Gene expression analysis
The gene expression profiles were analysed as described previously (Sanzey et al, 2015). Lists of differentially expressed genes (DEGs) were obtained with ANOVA (false discovery rate (FDR)<0.01, any FC). The Ingenuity Pathway Analysis (IPA) (Ingenuity Systems) was used for data mining. Right-tailed Fisher’s exact test was used to calculate a P value for functional enrichment analysis (threshold: −log(P value) >1.3). Upstream regulator analysis was used to detect potential transcriptional regulators (an overlap of P value <0.05 and activation z-score >2). Venn diagram analysis was performed with the SUMO software (http://angiogenesis.dkfz.de/oncoexpress/software/). Microarray data are available in the ArrayExpress database (www.ebi.ac.uk/arrayexpress) under accession number E-MTAB-3085.
Real-time quantitative PCR
Overall, 1 μg of total RNA was reverse transcribed using iScript cDNA Synthesis Kit (BioRad, Temse, Belgium). Quantitative PCR (qPCR) was carried out using Fast SYBR Green Master Mix and the Viia 7 Real Time PCR System (Life Technologies) with ATG9A (F: GCCAGACGCCTTTTTGCCTGC; R: TAGGGATGCGCAGAGCGTGC) and EZRIN (F: TGCCCCACGTCTGAGAATC; R: CGGCGCATATACAACTCATGG) primers. Fold-change (FC) was calculated using the ΔΔCt method (QBase).
shRNA-mediated knockdown of ATG9A
A control shRNA (shScramble, Open Biosystems, RHS4346) or a shRNA targeting ATG9A (Open Biosystems, RHS4430-99150604) were introduced using lentiviral particles. Individual pGIPZ shRNAmir constructs were obtained as E. coli cultures in LB-lenox medium with 8% glycerol, 100 μg ml−1 carbenicillin and 25 μg ml−1 zeocin. Lentiviral particles were produced in HEK cells by co-transfection of the pGIPZ vector with the viral core packaging construct pCMVdeltaR8.74 and the VSV-G envelope protein vector pMD.G.2. Supernatant containing viral particles was used to transduce 100 000 cells and puromycine selection permitted to obtain 100% of stably transduced GFP-positive cells (0.5 μg ml−1 for NCH421k and U87, 1 μg ml−1 for NCH644 for at least 2 weeks). Cells were regularly verified for GFP expression via flow cytometry and puromycine selection was repeated, if required.
Transient transfection with LC3B
U87 and U251 were seeded in ibidi iTreat μ-Dish transfected using lipofectamine (Thermo Fisher, Illkirch, France) with 2 μg of LC3B-GFP or LC3-Tomato plasmid for 3 h. Transfected cells were incubated for 16 h in either normoxia or 0.1–0.5% O2 hypoxia in the presence of 20 μM chloroquine. Nuclei were visualised with Hoechst33342. Images were taken using fluorescence confocal microscopy (Zeiss LSM STO META) by obtaining 20–25 stacks of two-dimensional images from successive focal planes (10–15 μm total). Quantification of autophagosomes was performed with ImageJ. Experiments were performed twice, 35 individual cells were acquired in total for analysis.
Cell proliferation assay
shScramble and shATG9A transfected NCH421k, NCH644 (10 000 cells) and U87 (5000 cells) were plated in 6 well plates. Cells were cultured for 4, 7 and 11 days. At each time point, total number of viable cells was measured with a Countess cell counter (Thermo Fisher). Experiments were performed three times with three replicates each.
Statistical analysis
The data was analysed with unpaired independent-samples t-test (Excel software, Microsoft, Redmond, Seattle, WA, USA). Kaplan–Meier survival curves, log-rank test for survival analysis and IC50 were calculated with the GraphPad Prism5. Data were considered statistically significant with a P value <0.05.
Results
Bevacizumab sensitises GBM cells to anti-autophagy treatment in vivo in orthotopic patient-derived xenografts
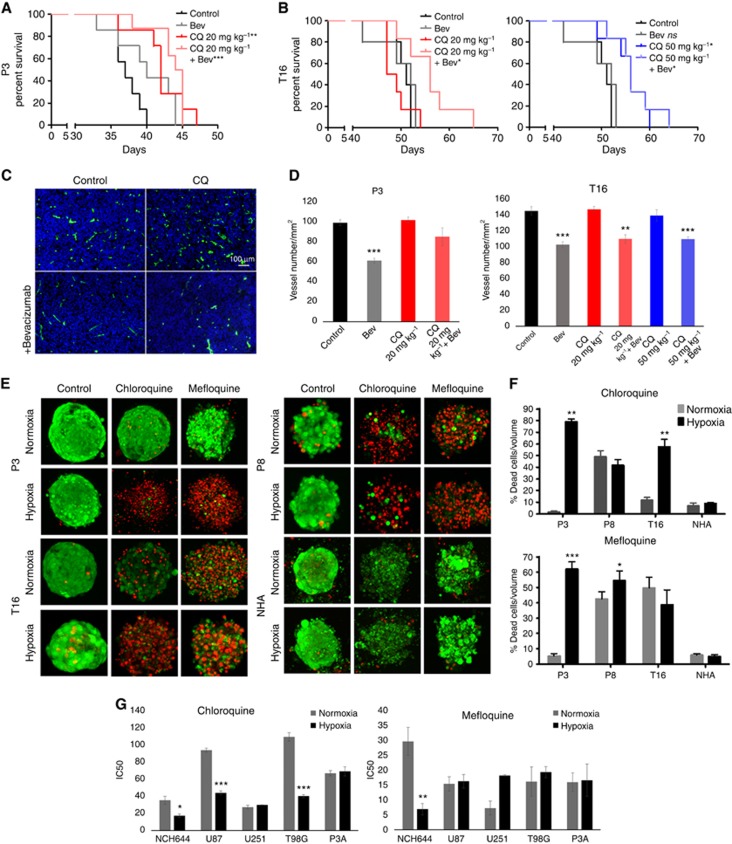
We showed previously that administration of bevacizumab (Bev), an anti-angiogenic agent, leads to a hypoxic signature in GBM patient-derived xenografts (PDXs) (Keunen et al, 2011; Demeure et al, 2015; Fack et al, 2015). As autophagy appears as an essential survival mechanism under hypoxia, we hypothesised that the combination of bevacizumab with an autophagy inhibitor would have an additional anti-tumour effect. We applied the well-known autophagy inhibitor chloroquine in vivo on two different PDXs. Organotypic P3 and T16 spheroids were orthotopically implanted into nude mice and treatment was started 3 weeks post implantation (Supplementary Table S2).
Chloroquine treatment (20 mg kg−1) significantly prolonged survival of P3 mice (+18.4% Figure 1A; Supplementary Table S2), whereas it had no effect in T16 xenografts; however, increasing the dose to 50 mg kg−1 (3 × -weekly) increased the survival (+9.6% Figure 1B; Supplementary Table S2). As previously shown (Keunen et al, 2011; Golebiewska et al, 2013) treatment with bevacizumab did not significantly prolong survival of mice with these PDXs (Figure 1A and B), despite the fact that vessel morphology was normalised. At an effective chloroquine concentration, bevacizumab did not lead to a statistically significant additive benefit in both PDXs. However, the addition of bevacizumab led to a synergistic effect in the low chloroquine dose in T16 (11.5% P=0.0095), at a concentration where chloroquine was not effective as a single agent. The effect was equivalent to the high-dose chloroquine treatment (CQ 50 mg kg−1 vs CQ 20 mg kg−1+Bev; P=0.85) indicating that the addition of bevacizumab allows to lower the effective chloroquine treatment dose.
Figure 1.
Hypoxia sensitises GBM cells to autophagy inhibitors. Chloroquine and bevacizumab were administered as single agents or simultaneously in P3 (A: 20 mg kg−1) and T16 (B: 20 and 50 mg kg−1) PDXs. Kaplan–Meier graphs show the survival of mice upon treatment. See Supplementary Table S1 for summary. Abbreviations: Bev=Bevacizumab; CQ=chloroquine; log-rank test, *P<0.05, **P<0.01, ***P<0.001. (C) Blood vessels from control and treated P3 PDXs were visualised by mouse-specific anti-CD31 (scale bars 100 μm). (D) Quantification of vessel number per mm2 upon treatment (mean±s.e.m., *P<0.05, **P<0.01, ***P<0.001). (E) The cytotoxic effect of inhibitors (chloroquine 20 μM, mefloquine 10 μM) was analysed for PDX-derived spheroids and NHA after 72 h treatment in normoxia and hypoxia. Representative images of treated spheroids are presented (‘green’=viable, ‘red’=dead). (F) Quantification of cell death upon treatment displayed as % of dead cells/volume (n⩾5, *P<0.05, **P<0.01, ***P<0.001). (G) Sensitivity of GBM cultures to chloroquine and mefloquine 72 h after treatment. Concentration gradients were used to determine the median inhibitory concentration (IC50). IC50 are expressed as mean±s.e.m. (n⩾3, *P<0.05, **P<0.01, ***P<0.001).
It has been shown that in melanoma chloroquine acts on the normalisation of tumour vessels independently of autophagy (Maes et al, 2014). We did not detect any direct effect of chloroquine on vessel normalisation: bevacizumab but not chloroquine significantly decreased vessel density, total vessel density reduced upon bevacizumab was not further affected by adding chloroquine (Figure 1C and D).
In conclusion, our data show that chloroquine has a therapeutic effect as a single agent in GBM PDXs, albeit the effective dose differing between GBM. Addition of bevacizumab allowed to lower the dose of chloroquine to reach the same survival benefit.
GBM cells exhibit increased sensitivity to chloroquine in hypoxia
To further confirm a role of hypoxia in the outcome of anti-autophagy treatment, we assessed the efficacy of two autophagy inhibitors, chloroquine and mefloquine, at different oxygen levels. We have first assessed the cytotoxic effects in primary PDX-derived 3D spheroids standardised for drug testing (Supplementary Figure S1), known to recapitulate well the genetic makeup of patient tumours (De Witt Hamer et al, 2008; Bougnaud et al, 2016) (Supplementary Table S1) and drug responses (Hirschhaeuser et al, 2010). Non-transformed human astrocytes (NHA) cultured under identical conditions were used as a control. Spheroids treated for 72 h with chloroquine (20 μM) or mefloquine (10 μM) in normoxia or severe hypoxia (0.1% O2; Figure 1E and F) displayed a heterogeneous response to autophagy inhibitors. Little cell death was observed within P3 and T16 spheroids treated with chloroquine in normoxia, whereas cell death was markedly increased in hypoxia (Figure 1E and F). P8 spheroids were already sensitive to chloroquine in normoxia and exhibited no further increase in sensitivity under hypoxia. Mefloquine, a more potent lysosomotropic agent, was generally more toxic already in normoxia. In P3 spheroids, sensitivity, however strongly increased in hypoxia, which appeared relatively resistant to mefloquine in normoxia. At the indicated concentration, chloroquine and mefloquine did not induce cell death in astrocytes (Figure 1E and F), suggesting that astrocytes are less dependent on autophagy compared to GBM cells.
We further determined the half maximal inhibitory concentration (IC50) for chloroquine and mefloquine in a panel of GBM cultures. Out of six cultures tested NCH644, U87 and T98G exhibited increased sensitivity to chloroquine in hypoxia (Figure 1G). U251 and P3A were already very sensitive under normoxia and no additive effect was observed in hypoxia (Figure 1G). Again, mefloquine was generally more potent in normoxia, and increased sensitivity in hypoxia was observed only for NCH644, which displayed highest IC50 at normal oxygen levels (Figure 1G).
Taken together, we show that hypoxia potentiates the cytotoxic effect of autophagy inhibitors in GBM spheroids and in GBM cultures. Similar to the in vivo situation, the GBM response is heterogeneous and the additive effect is observed in hypoxic cells only when the treatment reaches mild/moderate effect in normoxia.
Induction of autophagy at the transcript and protein level
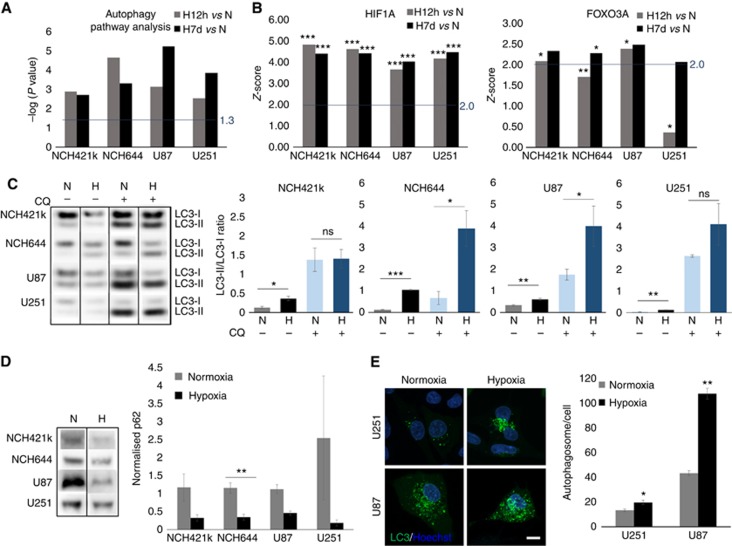
We have recently shown that GBM cells can survive under long-term severe hypoxia, undergoing transcriptional changes and increasing dependency on glycolysis (Sanzey et al, 2015). Although autophagy is known to be regulated mainly at the post-transcriptional level, transcriptional regulation has an important role in the induction of the process (Moussay et al, 2011). We therefore investigated transcriptional regulation of autophagy-associated genes. Gene expression patterns were obtained from a panel of GBM cultures including glioma stem-like cells (NCH421k, NCH644) and classical GBM lines (U87, U251), cultured under short (12 h) and long-term (7 days) hypoxia. Differentially expressed genes (FDR<0.01; any FC) were further subjected for functional enrichment analysis by IPA. As genes associated with the autophagy pathway genes regulating autophagy are poorly annotated in ontology databases we applied an in-house gene list (244 genes referred as ‘Autophagy pathway’) (Moussay et al, 2011), revealing strong modulation of the autophagy pathway upon both short- and long-term hypoxia (Figure 2A). As expected, the upstream regulator analysis by IPA predicted the hypoxia inducible factor 1-alpha (HIF1A) transcription network to be strongly activated upon hypoxia (P value<0.05; z-score>2; Figure 2B), as was FOXO3A – one of the transcription factors responsible for induction of autophagy (Figure 2B).
Figure 2.
Hypoxia activates autophagy in GBM cells. (A) DEG lists between hypoxia 12 h vs normoxia and hypoxia 7 days vs normoxia (FDR<0.01; any FC, n=3–6) were subjected to IPA. Autophagy-associated genes (Moussay et al, 2011) were significantly altered (threshold: −log(P value) >1.3). (B) Upstream Regulator analysis (IPA) predicted activation of HIF1α and FOXO3 network upon hypoxia (threshold: z-score >2 and P value of overlap <0.05; *P<0.05 **P<0.01, ***P<0.001). (C) Western blot analysis showing LC3-I/II. Increase of autophagy in hypoxia is visualised by increased LC3-II (lower band)/LC3-I (upper band) ratio in the presence of chloroquine (mean±s.e.m., n=3; *P<0.05, **P<0.01, ***P<0.001). Representative images were cropped from the same blot (CQ=chloroquine, N=normoxia, H: 0.1–0.5% O2 48 h hypoxia). (D) Western blot analysis showing p62 degradation upon hypoxia (mean normalised to total protein content±s.e.m., n=3; **P<0.01). Representative images were cropped from the same blots. Control cells (normoxia, no chloroquine) were used as an internal calibration (value=‘1’). (E) Representative images show an increase in autophagosome formation upon hypoxia. Cells were exposed to hypoxia for 16 h in the presence of chloroquine. Autophagosomes were counted as LC3B–GFP-positive vacuoles 24 h after transfection; (mean±s.e.m.; n=34; *P<0.05, **P<0.01).
Activation of autophagy was further visualised via increased conversion of LC3-I to LC3-II isoform under hypoxia (Figure 2C). To appropriately detect changes in the autophagic flux, experiments were performed in the absence and in the presence of the lysosomotropic agent chloroquine, which inhibits both the fusion of autophagosome with lysosome and lysosomal protein degradation. Contrary to the previous experiments where chloroquine was used as a treatment agent (Figure 1), the inhibition of the autophagic flux was detectable upon short chloroquine treatment (3–16 h) according to well-established protocols (Shintani and Klionsky, 2004; Klionsky et al, 2016). High levels of the LC3-II isoform were detected in all GBM cells treated with chloroquine upon hypoxia as reflected in the LC3-II/LC3-I ratios. Interestingly, NCH421k and U251 cells displayed high levels of LC3-II already in normoxia, suggesting their strong dependence on autophagy in normal conditions (Figure 2C). This is in accordance with the high sensitivity of U251 to chloroquine in both conditions (Figure1G). Induction of autophagy by hypoxia was further confirmed by a decrease in p62 (Figure 2D) and an increase in the number of autophagosomes visualised via transient LC3-GFP transfection (Figure 2E). In conclusion, these data indicate that autophagy is induced under severe hypoxia in GBM cells. The heterogeneous sensitivity to autophagy inhibition corroborates with the differential basal level of autophagy in normoxia and further activation of autophagy in hypoxic GBM cells.
ATG9A is involved in the hypoxia-dependent autophagic response
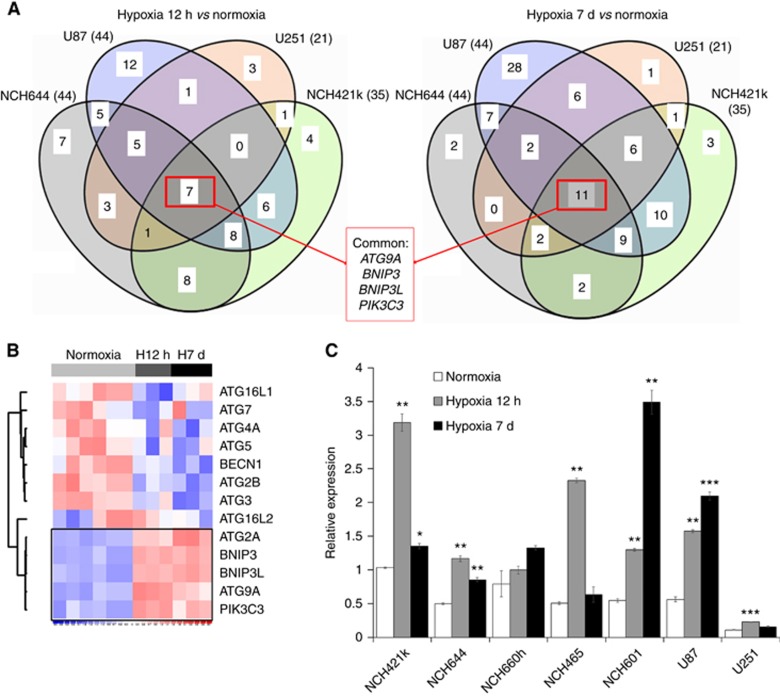
To further explore the GBM-specific response to hypoxia we focused on 98 specific regulators of autophagy (71-positive and 27-negative regulators, Supplementary Table S3). Although the number of deregulated genes and the extend of deregulation was variable, we found four commonly deregulated genes shared between short-term and long-term hypoxia (ATG9A, BNIP3, BNIP3L and PIK3C3; Figure 3A; Supplementary Table S3), showing increased levels upon hypoxia. BNIP3 and BNIP3L were previously associated with the autophagic response in hypoxic conditions (Mazure and Pouyssegur, 2010), whereas PIK3C3 is a well-known partner in the autophagy onset mechanism (Munson and Ganley, 2015). Of note, MTOR, a negative regulator of autophagy and of PIK3C3, was significantly downregulated in 3 out of 4 GBM cultures (Supplementary Table S3). Interestingly, within the ATG family, only ATG9A was upregulated in all GBM cells (Figure 3B; Supplementary Table S3), ATG2A was high only in 5 out of 8 conditions (Supplementary Table S3). The upregulation of ATG9A was confirmed by qPCR in GBM stem-like cells (NCH644, NCH421k, NCH660h, NCH601, NCH465) and adherent cultures (U87, U251) (Figure 3C).
Figure 3.
ATG9A is specifically activated upon autophagic response to hypoxia. (A) Genes directly related to autophagy (knowledge-driven selection) were extracted from DEG lists between hypoxia 12 h vs normoxia and hypoxia 7 days vs normoxia (FDR<0.01; any FC) for each culture (n=3–6). Venn diagrams reveal commonly deregulated genes. (B) Heatmap shows expression levels for selected genes in NCH421k in normoxia, 12 h and 7 days hypoxia. See Supplementary Table S3 for more autophagy-related genes. (C) QPCR confirmed increased ATG9A expression in hypoxia. EZRIN was used as a reference (mean±s.e.m.; n=3; *P<0.05, **P<0.01, ***P<0.001). NCH421k cells were used as an internal calibration (value=‘1’).
Interestingly, analysis of the ATG9A gene promoter revealed the presence of five hypoxia response elements (HREs) in close proximity to the canonical transcription start site, confirmed to be functional according to the TRANSFAC database (Matys et al, 2006; Mole et al, 2009) (Supplementary Table S4). This was true also for BNIP3, BNIP3L and PIK3C3 promoters, and is in line with the HIF-dependent regulation reported for the BNIP3 and BNIP3L (Kothari et al, 2003; Mole et al, 2009; Slemc and Kunej, 2016). In summary, we show for the first time that ATG9A expression is strongly induced in hypoxic conditions, implicating ATG9A as a new player of hypoxia-dependent autophagic response in GBM.
Silencing of ATG9A affects GBM cell proliferation and tumour growth in vivo
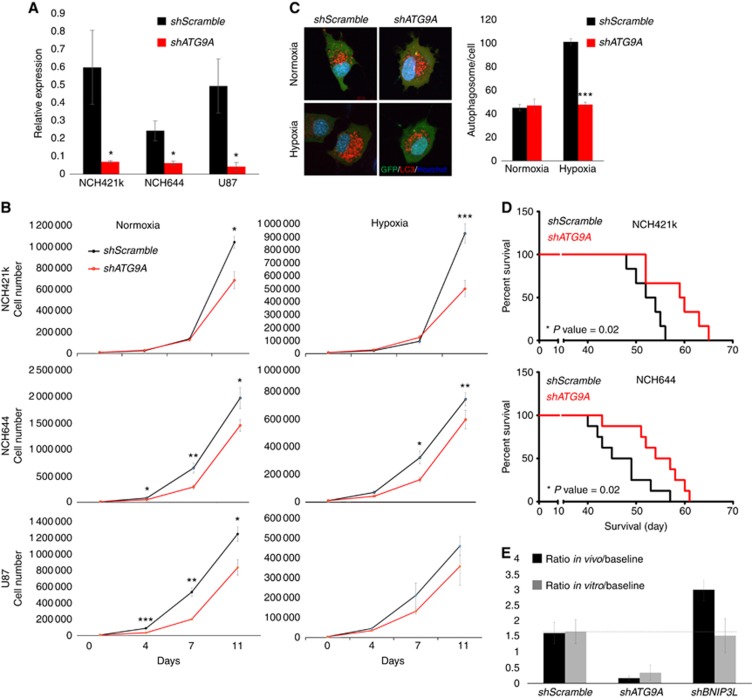
To study the involvement of ATG9 in autophagy, we generated an efficient long-term ATG9A knockdown (75–98%, Figure 4A) in three GBM cultures, resulting in reduced proliferation both in normoxia and hypoxia (Figure 4B). Contrary to the control, ATG9A-depleted U87 cells did not increase the number of LC3-positive vesicles upon hypoxia (Figure 4C), suggesting inefficient activation of autophagy. To examine the effect of ATG9A silencing on tumour growth in vivo, we implanted shATG9A NCH421k and NCH644 cells into the brain of immunodeficient mice. ATG9A knockdown led to a significant increase in mouse survival (+12–18% Figure 4D). Of note, two of the autopagy-associated genes, ATG9A and BNIP3L, were included in our previously reported targeted shRNA screen, where we examined the essentiality of 55 genes for survival of GBM cells in vitro and in vivo (Sanzey et al, 2015). Interestingly, ATG9A but not BNIP3L was also depleted both in vitro and in vivo (Figure 4E), indicating that ATG9A is essential for general GBM cell survival. Taken together, our data show that ATG9A is important for GBM growth both in normoxic and hypoxic conditions, and regulates activation of autophagy upon hypoxia. Interfering with ATG9A expression efficiently blocks tumour growth in vivo.
Figure 4.
ATG9A knockdown decreases GBM cell proliferation and increases mouse survival. (A) QPCR confirmation of shATG9A knockdown (mean±s.e.m.; n=3; *P<0.05). (B) Proliferation of shATG9A cells was decreased significantly in normoxic and hypoxic conditions (mean±s.e.m.; n=3; *P<0.05, **P<0.01, ***P<0.001). (C) Representative images show lack of increased autophagosome formation upon hypoxia in shATG9 U87 cells. Cells were exposed to hypoxia for 16 h in the presence of chloroquine. Autophagosomes were counted as LC3B–Tomato-positive vacuoles 24 h after transfection; GFP positivity confirms shRNA expression (mean±s.e.m.; n=34; ***P value <0.001). (D) ATG9A depletion in NCH421k and NCH644 prolonged the survival of tumour-bearing mice (n=6–8). (E) Targeted in vivo shRNA screen in NCH421k cells. shRNA targeting ATG9A but not BNIP3L was depleted after in vivo (n=5) and in vitro (normoxia, n=3) growth. Relative representation of respective shRNAs after selection pressure is presented as ratios compared with the original shRNA pool before selection (baseline). For detailed experimental setup, see (Sanzey et al, 2015).
Discussion
Hypoxia is a characteristic feature of malignant gliomas and drives tumour progression by adaptive cellular responses including angiogenesis, changes in tumour metabolism, motility and survival (Bertout et al, 2008). Increased hypoxia is also one of the escape mechanisms driving resistance to anti-angiogenic treatment in GBM. Here we find that the autophagy pathway is strongly induced in GBM under hypoxia, and we identify ATG9A as a novel regulator of autophagy induction. Inhibiting autophagy was shown to potentiate various anti-cancer therapies in vitro, including gliomas (Kanzawa et al, 2004; Shingu et al, 2009), where cells were subjected to external stress. Although, there are currently over 20 clinical studies involving the use of chloroquine and hydroxychloroquine in cancer treatment, in GBM both agents showed limited effect in non-toxic doses (Sotelo et al, 2006; Rosenfeld et al, 2014). Here we show a significant increase in survival of GBM PDXs when chloroquine was administered as a single agent, although with different effective dose. This is in accordance with the recent clinical trial showing dose-dependent inhibition of autophagy by hydroxychloroquine in GBM patients (Rosenfeld et al, 2014) and suggests that dosing needs to be adapted to the specific patient tumour. Of note, we show that certain tumour cells were sensitive to autophagy inhibitors also at normal oxygen, indicating a strong dependence on autophagy without additional environmental stress in a subgroup of GBM. This heterogeneous response suggests that the genetic background, for example, PTEN deletion, p53 mutation or EGFR amplification, may differentially affect the extent of basal level of autophagy and of treatment response in GBM and that appropriate biomarkers may be required to efficiently stratify patients. EGFR is known to negatively regulate autophagy (Chen et al, 2016) through multiple signalling pathways, thus EGFR overexpression may partly explain the lower sensitivity of T16 tumours to anti-autophagic treatment.
Importantly, we find that bevacizumab treatment sensitised GBM cells to autophagy inhibition allowing to reach survival benefit at lower dose. This was confirmed in vitro, where hypoxia increased sensitivity of GBM cells to autophagy inhibitors. The synergistic effect of bevacizumab was visible only when the anti-autophagy effect alone was mild or moderate at normal oxygen levels, but was masked if the autophagy inhibitor alone showed a strong effect. Interestingly, we have previously shown that bevacizumab leads to a lower number of mitochondria in tumour cells (Keunen et al, 2011), suggesting that mitophagy might be involved in the survival under hypoxia. A previous study has shown efficacy of chloroquine in combination with bevacizumab in subcutaneous U87 tumours, but failed to observe a tumour suppressive effect with chloroquine used as a single agent (Hu et al, 2012). This discrepancy may be due to the different tumour localisation and the heterogeneity in the GBM response to chloroquine described here. Although non-specific effects of chloroquine cannot be excluded (Maycotte et al, 2012; Maes et al, 2014), we did not observe vessel normalisation upon chloroquine treatment. In line with a previous study (Chen et al, 2008), normal astrocytes remained unaffected at the lowest chloroquine concentration affecting GBM cells, confirming more substantial dependence of tumour cells on autophagy. Although more potent inhibitors are warranted, our data suggest the existence of a ‘therapeutic window’ for autophagy inhibitors in GBM, and that co-treatment with anti-angiogenic agents allows to significantly lower effective doses.
We found that activation of autophagy in hypoxia was linked to transcriptional changes of numerous genes associated with autophagy, among which BNIP3, BNIP3L, ATG9A and PIK3C3 were upregulated in all GBM cells. BNIP3 and BNIP3L, while activated by HIF1α, mediate autophagy by releasing Beclin1 from complexes with Bcl-2 and Bcl-XL (Zhang et al, 2008; Bellot et al, 2009). Interestingly, within the ATG family, only ATG9A was transcriptionally activated in all GBM cells. Contrary to other ATG family members such as ATG5 and ATG7, but similarly to BNIP3 and BNIP3L, we identify ATG9A as potentially HIF1α responsive gene. These transcriptional changes were observed also in GBM cells that exhibit high basal autophagy at normal oxygen levels, suggesting that specific upstream molecules such as FOXO3A are involved in the regulation autophagy pathway at different oxygen levels. Pro-autophagic genes, such as Beclin1, ATG5, ATG7, BNIP3 and BNIP3L were previously found to be essential for autophagy in cancer cells (Zhang et al, 2008; Mazure and Pouyssegur, 2009). Here we show that ATG9A also represents an important pro-survival molecule, with ATG9A depletion leading to a strong reduction of tumour growth, thus confirming the relevance of autophagy as a promising target for GBM treatment. Of note, ATG7 knockdown displayed a therapeutic outcome only during anti-angiogenic treatment (Hu et al, 2012).
ATG9A was shown to be essential for autophagosome biogenesis and membrane maturation; however, its mode of action remains enigmatic. Recent data suggest that the Pho–Rpd3 complex regulates expression of ATG9A and other ATG genes upon induction of autophagy (Jin and Klionsky, 2014) and that ATG9A-containing vesicles are generated de novo upon starvation (Yamamoto et al, 2012). Here we show that upon ATG9A depletion, GBM cells were not able to activate autophagy upon hypoxia. We propose that the lack of autophagic activation upon hypoxia may be due to inhibition of de novo autophagosome synthesis. This is in accordance with a recent report, where ATG9A was shown to have a key role in autophagosome formation during hypoxic stress (Weerasekara et al, 2014). Thus, ATG9A may become essential upon autophagy induction and an increased demand for new autophagosome membranes (Orsi et al, 2012).
In conclusion, our data support the notion that inhibiting autophagy represents an effective therapy in primary GBM, although it may be concentration and patient dependent. Anti-autophagy treatment using genetic and pharmacological intervention was effective as a single treatment. However, currently available drugs, including chloroquine and hydroxychloroquine are non-curative in non-toxic doses and novel more potent agents will be necessary for GBM patients. Drugs directly targeting essential proteins such as ATG9A may be of particular interest and a combination with anti-angiogenic therapy may be beneficial. Finally, the hypoxic microenvironment also contributes to immunoresistance and hypoxia-induced autophagy impairs cytotoxic T-lymphocyte-mediated cell lysis of tumour cells (Noman et al, 2011, 2012) and NK-mediated target cell apoptosis (Baginska et al, 2013; Viry et al, 2014). Therefore, targeting autophagy in tumour cells may not only lead to increased tumour cell death but also sensitise tumours to immunotherapies.
Acknowledgments
We thank Morgane Sanzey, Virginie Baus, and Amandine Bernard for technical assistance. We thank Dr Christel Herold-Mende (Department of Neurosurgery, University of Heidelberg, Germany) and Dr Uros Rajcevic (National Institute of Biology, Ljubljana, Slovenia) for providing cell lines. This work was supported by the Luxembourg Institute of Health (LIH), the Fonds National de la Recherche (FNR) of Luxembourg (ESCAPE 784322 BM, AFR grand to SAAR and AD) and the Fondation Cancer Luxembourg (INVGBM).
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc)
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 4.0 Unported License.
The authors declare no conflict of interest.
Supplementary Material
References
- Baginska J, Viry E, Berchem G, Poli A, Noman MZ, van Moer K, Medves S, Zimmer J, Oudin A, Niclou SP, Bleackley RC, Goping IS, Chouaib S, Janji B (2013) Granzyme B degradation by autophagy decreases tumor cell susceptibility to natural killer-mediated lysis under hypoxia. Proc Natl Acad Sci USA 110: 17450–17455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellot G, Garcia-Medina R, Gounon P, Chiche J, Roux D, Pouyssegur J, Mazure NM (2009) Hypoxia-induced autophagy is mediated through hypoxia-inducible factor induction of BNIP3 and BNIP3L via their BH3 domains. Mol Cell Biol 29: 2570–2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertout JA, Patel SA, Simon MC (2008) The impact of O2 availability on human cancer. Nat Rev Cancer 8: 967–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bougnaud S, Golebiewska A, Oudin A, Keunen O, Harter PN, Mader L, Azuaje F, Fritah S, Stieber D, Kaoma T, Vallar L, Brons NH, Daubon T, Miletic H, Sundstrøm T, Herold-Mende C, Mittelbronn M, Bjerkvig R, Niclou SP (2016) Molecular crosstalk between tumour and brain parenchyma instructs histopathological features in glioblastoma. Oncotarget. [DOI] [PMC free article] [PubMed]
- Chen Y, Henson ES, Xiao W, Huang D, McMillan-Ward EM, Israels SJ, Gibson SB (2016) Tyrosine kinase receptor EGFR regulates the switch in cancer cells between cell survival and cell death induced by autophagy in hypoxia. Autophagy 12: 1029–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, McMillan-Ward E, Kong J, Israels SJ, Gibson SB (2008) Oxidative stress induces autophagic cell death independent of apoptosis in transformed and cancer cells. Cell Death Differ 15: 171–182. [DOI] [PubMed] [Google Scholar]
- Ciechomska IA, Gabrusiewicz K, Szczepankiewicz AA, Kaminska B (2013) Endoplasmic reticulum stress triggers autophagy in malignant glioma cells undergoing cyclosporine a-induced cell death. Oncogene 32: 1518–1529. [DOI] [PubMed] [Google Scholar]
- Corcelle-Termeau E, Vindelov SD, Hamalisto S, Mograbi B, Keldsbo A, Brasen JH, Favaro E, Adam D, Szyniarowski P, Hofman P, Krautwald S, Farkas T, Petersen NH, Rohde M, Linkermann A, Jäättelä M (2016) Excess sphingomyelin disturbs ATG9A trafficking and autophagosome closure. Autophagy 12: 833–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Witt Hamer PC, Van Tilborg AA, Eijk PP, Sminia P, Troost D, Van Noorden CJ, Ylstra B, Leenstra S (2008) The genomic profile of human malignant glioma is altered early in primary cell culture and preserved in spheroids. Oncogene 27: 2091–2096. [DOI] [PubMed] [Google Scholar]
- Demeure K, Fack F, Duriez E, Tiemann K, Bernard A, Golebiewska A, Bougnaud S, Bjerkvig R, Domon B, Niclou SP (2015) Targeted proteomics to assess the response to anti-angiogenic treatment in human glioblastoma. Mol Cell Proteomics 15: 481–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans SM, Judy KD, Dunphy I, Jenkins WT, Nelson PT, Collins R, Wileyto EP, Jenkins K, Hahn SM, Stevens CW, Judkins AR, Phillips P, Geoerger B, Koch CJ (2004) Comparative measurements of hypoxia in human brain tumors using needle electrodes and EF5 binding. Cancer Res 64: 1886–1892. [DOI] [PubMed] [Google Scholar]
- Fack F, Espedal H, Keunen O, Golebiewska A, Obad N, Harter PN, Mittelbronn M, Bahr O, Weyerbrock A, Stuhr L, Miletic H, Sakariassen PØ, Stieber D, Rygh CB, Lund-Johansen M, Zheng L, Gottlieb E, Niclou SP, Bjerkvig R (2015) Bevacizumab treatment induces metabolic adaptation toward anaerobic metabolism in glioblastomas. Acta Neuropathol 129: 115–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firat E, Weyerbrock A, Gaedicke S, Grosu AL, Niedermann G (2012) Chloroquine or chloroquine-PI3K/Akt pathway inhibitor combinations strongly promote gamma-irradiation-induced cell death in primary stem-like glioma cells. PloS ONE 7: e47357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, Botstein D, Brown PO (2000) Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell 11: 4241–4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert MR (2016) Antiangiogenic therapy for glioblastoma: complex biology and complicated results. J Clin Oncol 34: 1567–1569. [DOI] [PubMed] [Google Scholar]
- Glick D, Barth S, Macleod KF (2010) Autophagy: cellular and molecular mechanisms. J Pathol 221: 3–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golebiewska A, Bougnaud S, Stieber D, Brons NH, Vallar L, Hertel F, Klink B, Schrock E, Bjerkvig R, Niclou SP (2013) Side population in human glioblastoma is non-tumorigenic and characterizes brain endothelial cells. Brain 136: 1462–1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschhaeuser F, Menne H, Dittfeld C, West J, Mueller-Klieser W, Kunz-Schughart LA (2010) Multicellular tumor spheroids: an underestimated tool is catching up again. J Biotechnol 148: 3–15. [DOI] [PubMed] [Google Scholar]
- Hu YL, DeLay M, Jahangiri A, Molinaro AM, Rose SD, Carbonell WS, Aghi MK (2012) Hypoxia-induced autophagy promotes tumor cell survival and adaptation to antiangiogenic treatment in glioblastoma. Cancer Res 72: 1773–1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin M, Klionsky DJ (2014) Transcriptional regulation of ATG9 by the Pho23-Rpd3 complex modulates the frequency of autophagosome formation. Autophagy 10: 1681–1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanzawa T, Germano IM, Komata T, Ito H, Kondo Y, Kondo S (2004) Role of autophagy in temozolomide-induced cytotoxicity for malignant glioma cells. Cell Death Differ 11: 448–457. [DOI] [PubMed] [Google Scholar]
- Keunen O, Johansson M, Oudin A, Sanzey M, Abdul Rahim SA, Fack F, Thorsen F, Taxt T, Bartos M, Jirik R, Miletic H, Wang J, Stieber D, Stuhr L, Moen I, Rygh CB, Bjerkvig R, Niclou SP (2011) Anti-VEGF treatment reduces blood supply and increases tumor cell invasion in glioblastoma. Proc Natl Acad Sci USA 108: 3749–3754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, Abdelmohsen K, Abe A, Abedin MJ, Abeliovich H, Acevedo Arozena A, Adachi H, Adams CM, Adams PD, Adeli K, Adhihetty PJ, Adler SG, Agam G, Agarwal R, Aghi MK, Agnello M, Agostinis P, Aguilar PV, Aguirre-Ghiso J, Airoldi EM, Ait-Si-Ali S, Akematsu T, Akporiaye ET, Al-Rubeai M, Albaiceta GM, Albanese C, Albani D, Albert ML, Aldudo J, Algül H, Alirezaei M, Alloza I, Almasan A, Almonte-Beceril M, Alnemri ES, Alonso C, Altan-Bonnet N, Altieri DC, Alvarez S, Alvarez-Erviti L, Alves S, Amadoro G, Amano A, Amantini C, Ambrosio S, Amelio I, Amer AO, Amessou M, Amon A, An Z, Anania FA, Andersen SU, Andley UP, Andreadi CK, Andrieu-Abadie N, Anel A, Ann DK, Anoopkumar-Dukie S, Antonioli M, Aoki H, Apostolova N, Aquila S, Aquilano K, Araki K, Arama E, Aranda A, Araya J, Arcaro A, Arias E, Arimoto H, Ariosa AR, Armstrong JL, Arnould T, Arsov I, Asanuma K, Askanas V, Asselin E, Atarashi R, Atherton SS, Atkin JD, Attardi LD, Auberger P, Auburger G, Aurelian L, Autelli R, Avagliano L, Avantaggiati ML, Avrahami L, Awale S, Azad N, Bachetti T, Backer JM, Bae DH, Bae JS, Bae ON, Bae SH, Baehrecke EH, Baek SH, Baghdiguian S, Bagniewska-Zadworna A, Bai H, Bai J, Bai XY, Bailly Y, Balaji KN, Balduini W, Ballabio A, Balzan R, Banerjee R, Bánhegyi G, Bao H, Barbeau B, Barrachina MD, Barreiro E, Bartel B, Bartolomé A, Bassham DC, Bassi MT, Bast RC Jr, Basu A, Batista MT, Batoko H, Battino M, Bauckman K, Baumgarner BL, Bayer KU, Beale R, Beaulieu JF, Beck GR Jr, Becker C, Beckham JD, Bédard PA, Bednarski PJ, Begley TJ, Behl C, Behrends C, Behrens GM, Behrns KE, Bejarano E, Belaid A, Belleudi F, Bénard G, Berchem G, Bergamaschi D, Bergami M, Berkhout B, Berliocchi L, Bernard A, Bernard M, Bernassola F, Bertolotti A, Bess AS, Besteiro S, Bettuzzi S, Bhalla S, Bhattacharyya S, Bhutia SK, Biagosch C, Bianchi MW, Biard-Piechaczyk M, Billes V, Bincoletto C, Bingol B, Bird SW, Bitoun M, Bjedov I, Blackstone C, Blanc L, Blanco GA, Blomhoff HK, Boada-Romero E, Böckler S, Boes M, Boesze-Battaglia K, Boise LH, Bolino A, Boman A, Bonaldo P, Bordi M, Bosch J, Botana LM, Botti J, Bou G, Bouché M, Bouchecareilh M, Boucher MJ, Boulton ME, Bouret SG, Boya P, Boyer-Guittaut M, Bozhkov PV, Brady N, Braga VM, Brancolini C, Braus GH, Bravo-San Pedro JM, Brennan LA, Bresnick EH, Brest P, Bridges D, Bringer MA, Brini M, Brito GC, Brodin B, Brookes PS, Brown EJ, Brown K, Broxmeyer HE, Bruhat A, Brum PC, Brumell JH, Brunetti-Pierri N, Bryson-Richardson RJ, Buch S, Buchan AM, Budak H, Bulavin DV, Bultman SJ, Bultynck G, Bumbasirevic V, Burelle Y, Burke RE, Burmeister M, Bütikofer P, Caberlotto L, Cadwell K, Cahova M, Cai D, Cai J, Cai Q, Calatayud S, Camougrand N, Campanella M, Campbell GR, Campbell M, Campello S, Candau R, Caniggia I, Cantoni L, Cao L, Caplan AB, Caraglia M, Cardinali C, Cardoso SM, Carew JS, Carleton LA, Carlin CR, Carloni S, Carlsson SR, Carmona-Gutierrez D, Carneiro LA, Carnevali O, Carra S, Carrier A, Carroll B, Casas C, Casas J, Cassinelli G, Castets P, Castro-Obregon S, Cavallini G, Ceccherini I, Cecconi F, Cederbaum AI, Ceña V, Cenci S, Cerella C, Cervia D, Cetrullo S, Chaachouay H, Chae HJ, Chagin AS, Chai CY, Chakrabarti G, Chamilos G, Chan EY, Chan MT, Chandra D, Chandra P, Chang CP, Chang RC, Chang TY, Chatham JC, Chatterjee S, Chauhan S, Che Y, Cheetham ME, Cheluvappa R, Chen CJ, Chen G, Chen GC, Chen H, Chen JW, Chen JK, Chen M, Chen M, Chen P, Chen Q, Chen SD, Chen S, Chen SS, Chen W, Chen WJ, Chen WQ, Chen W, Chen X, Chen YH, Chen YG, Chen Y, Chen YJ, Chen YQ, Chen Z, Cheng A, Cheng CH, Cheng H, Cheong H, Cherry S, Chesney J, Cheung CH, Chevet E, Chi HC, Chi SG, Chiacchiera F, Chiang HL, Chiarelli R, Chiariello M, Chieppa M, Chin LS, Chiong M, Chiu GN, Cho DH, Cho SG, Cho WC, Cho YY, Cho YS, Choi AM, Choi EJ, Choi EK, Choi J, Choi ME, Choi SI, Chou TF, Chouaib S, Choubey D, Choubey V, Chow KC, Chowdhury K, Chu CT, Chuang TH, Chun T, Chung H, Chung T, Chung YL, Chwae YJ, Cianfanelli V, Ciarcia R, Ciechomska IA, Ciriolo MR, Cirone M, Claerhout S, Clague MJ, Clària J, Clarke PG, Clarke R, Clementi E, Cleyrat C, Cnop M, Coccia EM, Cocco T, Codogno P, Coers J, Cohen EE, Colecchia D, Coletto L, Coll NS, Colucci-Guyon E, Comincini S, Condello M, Cook KL, Coombs GH, Cooper CD, Cooper JM, Coppens I, Corasaniti MT, Corazzari M, Corbalan R, Corcelle-Termeau E, Cordero MD, Corral-Ramos C, Corti O, Cossarizza A, Costelli P, Costes S, Cotman SL, Coto-Montes A, Cottet S, Couve E, Covey LR, Cowart LA, Cox JS, Coxon FP, Coyne CB, Cragg MS, Craven RJ, Crepaldi T, Crespo JL, Criollo A, Crippa V, Cruz MT, Cuervo AM, Cuezva JM, Cui T, Cutillas PR, Czaja MJ, Czyzyk-Krzeska MF, Dagda RK, Dahmen U, Dai C, Dai W, Dai Y, Dalby KN, Dalla Valle L, Dalmasso G, D’Amelio M, Damme M, Darfeuille-Michaud A, Dargemont C, Darley-Usmar VM, Dasarathy S, Dasgupta B, Dash S, Dass CR, Davey HM, Davids LM, Dávila D, Davis RJ, Dawson TM, Dawson VL, Daza P, de Belleroche J, de Figueiredo P, de Figueiredo RC, de la Fuente J, De Martino L, De Matteis A, De Meyer GR, De Milito A, De Santi M, de Souza W, De Tata V, De Zio D, Debnath J, Dechant R, Decuypere JP, Deegan S, Dehay B, Del Bello B, Del Re DP, Delage-Mourroux R, Delbridge LM, Deldicque L, Delorme-Axford E, Deng Y, Dengjel J, Denizot M, Dent P, Der CJ, Deretic V, Derrien B, Deutsch E, Devarenne TP, Devenish RJ, Di Bartolomeo S, Di Daniele N, Di Domenico F, Di Nardo A, Di Paola S, Di Pietro A, Di Renzo L, DiAntonio A, Díaz-Araya G, Díaz-Laviada I, Diaz-Meco MT, Diaz-Nido J, Dickey CA, Dickson RC, Diederich M, Digard P, Dikic I, Dinesh-Kumar SP, Ding C, Ding WX, Ding Z, Dini L, Distler JH, Diwan A, Djavaheri-Mergny M, Dmytruk K, Dobson RC, Doetsch V, Dokladny K, Dokudovskaya S, Donadelli M, Dong XC, Dong X, Dong Z, Donohue TM Jr, Doran KS, D’Orazi G, Dorn GW 2nd, Dosenko V, Dridi S, Drucker L, Du J, Du LL, Du L, du Toit A, Dua P, Duan L, Duann P, Dubey VK, Duchen MR, Duchosal MA, Duez H, Dugail I, Dumit VI, Duncan MC, Dunlop EA, Dunn WA Jr, Dupont N, Dupuis L, Durán RV, Durcan TM, Duvezin-Caubet S, Duvvuri U, Eapen V, Ebrahimi-Fakhari D, Echard A, Eckhart L, Edelstein CL, Edinger AL, Eichinger L, Eisenberg T, Eisenberg-Lerner A, Eissa NT, El-Deiry WS, El-Khoury V, Elazar Z, Eldar-Finkelman H, Elliott CJ, Emanuele E, Emmenegger U, Engedal N, Engelbrecht AM, Engelender S, Enserink JM, Erdmann R, Erenpreisa J, Eri R, Eriksen JL, Erman A, Escalante R, Eskelinen EL, Espert L, Esteban-Martínez L, Evans TJ, Fabri M, Fabrias G, Fabrizi C, Facchiano A, Færgeman NJ, Faggioni A, Fairlie WD, Fan C, Fan D, Fan J, Fang S, Fanto M, Fanzani A, Farkas T, Faure M, Favier FB, Fearnhead H, Federici M, Fei E, Felizardo TC, Feng H, Feng Y, Ferguson TA, Fernández ÁF, Fernandez-Barrena MG, Fernandez-Checa JC, Fernández-López A, Fernandez-Zapico ME, Feron O, Ferraro E, Ferreira-Halder CV, Fesus L, Feuer R, Fiesel FC, Filippi-Chiela EC, Filomeni G, Fimia GM, Fingert JH, Finkbeiner S, Finkel T, Fiorito F, Fisher PB, Flajolet M, Flamigni F, Florey O, Florio S, Floto RA, Folini M, Follo C, Fon EA, Fornai F, Fortunato F, Fraldi A, Franco R, Francois A, François A, Frankel LB, Fraser ID, Frey N, Freyssenet DG, Frezza C, Friedman SL, Frigo DE, Fu D, Fuentes JM, Fueyo J, Fujitani Y, Fujiwara Y, Fujiya M, Fukuda M, Fulda S, Fusco C, Gabryel B, Gaestel M, Gailly P, Gajewska M, Galadari S, Galili G, Galindo I, Galindo MF, Galliciotti G, Galluzzi L, Galy V, Gammoh N, Gandy S, Ganesan AK, Ganesan S, Ganley IG, Gannagé M, Gao FB, Gao F, Gao JX, García Nannig L, García Véscovi E, Garcia-Macía M, Garcia-Ruiz C, Garg AD, Garg PK, Gargini R, Gassen NC, Gatica D, Gatti E, Gavard J, Gavathiotis E, Ge L, Ge P, Ge S, Gean PW, Gelmetti V, Genazzani AA, Geng J, Genschik P, Gerner L, Gestwicki JE, Gewirtz DA, Ghavami S, Ghigo E, Ghosh D, Giammarioli AM, Giampieri F, Giampietri C, Giatromanolaki A, Gibbings DJ, Gibellini L, Gibson SB, Ginet V, Giordano A, Giorgini F, Giovannetti E, Girardin SE, Gispert S, Giuliano S, Gladson CL, Glavic A, Gleave M, Godefroy N, Gogal RM Jr, Gokulan K, Goldman GH, Goletti D, Goligorsky MS, Gomes AV, Gomes LC, Gomez H, Gomez-Manzano C, Gómez-Sánchez R, Gonçalves DA, Goncu E, Gong Q, Gongora C, Gonzalez CB, Gonzalez-Alegre P, Gonzalez-Cabo P, González-Polo RA, Goping IS, Gorbea C, Gorbunov NV, Goring DR, Gorman AM, Gorski SM, Goruppi S, Goto-Yamada S, Gotor C, Gottlieb RA, Gozes I, Gozuacik D, Graba Y, Graef M, Granato GE, Grant GD, Grant S, Gravina GL, Green DR, Greenhough A, Greenwood MT, Grimaldi B, Gros F, Grose C, Groulx JF, Gruber F, Grumati P, Grune T, Guan JL, Guan KL, Guerra B, Guillen C, Gulshan K, Gunst J, Guo C, Guo L, Guo M, Guo W, Guo XG, Gust AA, Gustafsson ÅB, Gutierrez E, Gutierrez MG, Gwak HS, Haas A, Haber JE, Hadano S, Hagedorn M, Hahn DR, Halayko AJ, Hamacher-Brady A, Hamada K, Hamai A, Hamann A, Hamasaki M, Hamer I, Hamid Q, Hammond EM, Han F, Han W, Handa JT, Hanover JA, Hansen M, Harada M, Harhaji-Trajkovic L, Harper JW, Harrath AH, Harris AL, Harris J, Hasler U, Hasselblatt P, Hasui K, Hawley RG, Hawley TS, He C, He CY, He F, He G, He RR, He XH, He YW, He YY, Heath JK, Hébert MJ, Heinzen RA, Helgason GV, Hensel M, Henske EP, Her C, Herman PK, Hernández A, Hernandez C, Hernández-Tiedra S, Hetz C, Hiesinger PR, Higaki K, Hilfiker S, Hill BG, Hill JA, Hill WD, Hino K, Hofius D, Hofman P, Höglinger GU, Höhfeld J, Holz MK, Hong Y, Hood DA, Hoozemans JJ, Hoppe T, Hsu C, Hsu CY, Hsu LC, Hu D, Hu G, Hu HM, Hu H, Hu MC, Hu YC, Hu ZW, Hua F, Hua Y, Huang C, Huang HL, Huang KH, Huang KY, Huang S, Huang S, Huang WP, Huang YR, Huang Y, Huber TB, Huebbe P, Huh WK, Hulmi JJ, Hur GM, Hurley JH, Husak Z, Hussain SN, Hussain S, Hwang JJ, Hwang S, Hwang TI, Ichihara A, Imai Y, Imbriano C, Inomata M, Into T, Iovane V, Iovanna JL, Iozzo RV, Ip NY, Irazoqui JE, Iribarren P, Isaka Y, Isakovic AJ, Ischiropoulos H, Isenberg JS, Ishaq M, Ishida H, Ishii I, Ishmael JE, Isidoro C, Isobe K, Isono E, Issazadeh-Navikas S, Itahana K, Itakura E, Ivanov AI, Iyer AK, Izquierdo JM, Izumi Y, Izzo V, Jäättelä M, Jaber N, Jackson DJ, Jackson WT, Jacob TG, Jacques TS, Jagannath C, Jain A, Jana NR, Jang BK, Jani A, Janji B, Jannig PR, Jansson PJ, Jean S, Jendrach M, Jeon JH, Jessen N, Jeung EB, Jia K, Jia L, Jiang H, Jiang L, Jiang T, Jiang X, Jiang Y, Jiménez A, Jin C, Jin H, Jin L, Jin M, Jin S, Jinwal UK, Jo EK, Johansen T, Johnson DE, Johnson GV, Johnson JD, Jonasch E, Jones C, Joosten LA, Jordan J, Joseph AM, Joseph B, Joubert AM, Ju D, Ju J, Juan HF, Juenemann K, Juhász G, Jung HS, Jung JU, Jung YK, Jungbluth H, Justice MJ, Jutten B, Kaakoush NO, Kaarniranta K, Kaasik A, Kabuta T, Kaeffer B, Kågedal K, Kahana A, Kajimura S, Kakhlon O, Kalia M, Kalvakolanu DV, Kamada Y, Kambas K, Kaminskyy VO, Kampinga HH, Kandouz M, Kang C, Kang R, Kang TC, Kanki T, Kanneganti TD, Kanno H, Kanthasamy AG, Kantorow M, Kaparakis-Liaskos M, Kapuy O, Karantza V, Karim MR, Karmakar P, Kaser A, Kaushik S, Kawula T, Kaynar AM, Ke PY, Ke ZJ, Kehrl JH, Keller KE, Kemper JK, Kenworthy AK, Kepp O, Kern A, Kesari S, Kessel D, Ketteler R, Kettelhut Ido C, Khambu B, Khan MM, Khandelwal VK, Khare S, Kiang JG, Kiger AA, Kihara A, Kim AL, Kim CH, Kim DR, Kim DH, Kim EK, Kim HY, Kim HR, Kim JS, Kim JH, Kim JC, Kim JH, Kim KW, Kim MD, Kim MM, Kim PK, Kim SW, Kim SY, Kim YS, Kim Y, Kimchi A, Kimmelman AC, Kimura T, King JS, Kirkegaard K, Kirkin V, Kirshenbaum LA, Kishi S, Kitajima Y, Kitamoto K, Kitaoka Y, Kitazato K, Kley RA, Klimecki WT, Klinkenberg M, Klucken J, Knævelsrud H, Knecht E, Knuppertz L, Ko JL, Kobayashi S, Koch JC, Koechlin-Ramonatxo C, Koenig U, Koh YH, Köhler K, Kohlwein SD, Koike M, Komatsu M, Kominami E, Kong D, Kong HJ, Konstantakou EG, Kopp BT, Korcsmaros T, Korhonen L, Korolchuk VI, Koshkina NV, Kou Y, Koukourakis MI, Koumenis C, Kovács AL, Kovács T, Kovacs WJ, Koya D, Kraft C, Krainc D, Kramer H, Kravic-Stevovic T, Krek W, Kretz-Remy C, Krick R, Krishnamurthy M, Kriston-Vizi J, Kroemer G, Kruer MC, Kruger R, Ktistakis NT, Kuchitsu K, Kuhn C, Kumar AP, Kumar A, Kumar D, Kumar D, Kumar R, Kumar S, Kundu M, Kung HJ, Kuno A, Kuo SH, Kuret J, Kurz T, Kwok T, Kwon TK, Kwon YT, Kyrmizi I, La Spada AR, Lafont F, Lahm T, Lakkaraju A, Lam T, Lamark T, Lancel S, Landowski TH, Lane DJ, Lane JD, Lanzi C, Lapaquette P, Lapierre LR, Laporte J, Laukkarinen J, Laurie GW, Lavandero S, Lavie L, LaVoie MJ, Law BY, Law HK, Law KB, Layfield R, Lazo PA, Le Cam L, Le Roch KG, Le Stunff H, Leardkamolkarn V, Lecuit M, Lee BH, Lee CH, Lee EF, Lee GM, Lee HJ, Lee H, Lee JK, Lee J, Lee JH, Lee M, Lee MS, Lee PJ, Lee SW, Lee SJ, Lee SJ, Lee SY, Lee SH, Lee SS, Lee SJ, Lee S, Lee YR, Lee YJ, Lee YH, Leeuwenburgh C, Lefort S, Legouis R, Lei J, Lei QY, Leib DA, Leibowitz G, Lekli I, Lemaire SD, Lemasters JJ, Lemberg MK, Lemoine A, Leng S, Lenz G, Lenzi P, Lerman LO, Lettieri Barbato D, Leu JI, Leung HY, Levine B, Lewis PA, Lezoualc’h F, Li C, Li F, Li FJ, Li J, Li K, Li L, Li M, Li M, Li Q, Li R, Li S, Li W, Li X, Li Y, Lian J, Liang C, Liang Q, Liao Y, Liberal J, Liberski PP, Lie P, Lieberman AP, Lim HJ, Lim KL, Lim K, Lima RT, Lin CS, Lin CF, Lin F, Lin FC, Lin K, Lin KH, Lin PH, Lin T, Lin WW, Lin YS, Linden R, Lindholm D, Lindqvist LM, Lingor P, Linkermann A, Liotta LA, Lipinski MM, Lira VA, Lisanti MP, Liton PB, Liu B, Liu C, Liu CF, Liu F, Liu HJ, Liu J, Liu JJ, Liu JL, Liu K, Liu L, Liu Q, Liu RY, Liu S, Liu S, Liu W, Liu XD, Liu X, Liu XH, Liu Y, Liu Z, Liuzzi JP, Lizard G, Ljujic M, Lodhi IJ, Logue SE, Lokeshwar BL, Long YC, Lonial S, Loos B, López-Otín C, López-Vicario C, Lorente M, Lorenzi PL, Lõrincz P, Los M, Lotze MT, Lovat PE, Lu B, Lu J, Lu Q, Lu SM, Lu S, Lu Y, Luciano F, Luckhart S, Lucocq JM, Ludovico P, Lugea A, Lukacs NW, Lum JJ, Lund AH, Luo H, Luo J, Luo S, Luparello C, Lyons T, Ma J, Ma Y, Ma Y, Ma Z, Machado J, Machado-Santelli GM, Macian F, MacIntosh GC, MacKeigan JP, Macleod KF, MacMicking JD, MacMillan-Crow LA, Madeo F, Madesh M, Madrigal-Matute J, Maeda A, Maeda T, Maegawa G, Maellaro E, Maes H, Magariños M, Maiese K, Maiti TK, Maiuri L, Maiuri MC, Maki CG, Malli R, Malorni W, Maloyan A, Mami-Chouaib F, Man N, Mancias JD, Mandelkow EM, Mandell MA, Manfredi AA, Manié SN, Manzoni C, Mao K, Mao Z, Mao ZW, Marambaud P, Marconi AM, Marelja Z, Marfe G, Margeta M, Margittai E, Mari M, Mariani FV, Marin C, Marinelli S, Mariño G, Markovic I, Marquez R, Martelli AM, Martens S, Martin KR, Martin SJ, Martin S, Martin-Acebes MA, Martín-Sanz P, Martinand-Mari C, Martinet W, Martinez J, Martinez-Lopez N, Martinez-Outschoorn U, Martínez-Velázquez M, Martinez-Vicente M, Martins WK, Mashima H, Mastrianni JA, Matarese G, Matarrese P, Mateo R, Matoba S, Matsumoto N, Matsushita T, Matsuura A, Matsuzawa T, Mattson MP, Matus S, Maugeri N, Mauvezin C, Mayer A, Maysinger D, Mazzolini GD, McBrayer MK, McCall K, McCormick C, McInerney GM, McIver SC, McKenna S, McMahon JJ, McNeish IA, Mechta-Grigoriou F, Medema JP, Medina DL, Megyeri K, Mehrpour M, Mehta JL, Mei Y, Meier UC, Meijer AJ, Meléndez A, Melino G, Melino S, de Melo EJ, Mena MA, Meneghini MD, Menendez JA, Menezes R, Meng L, Meng LH, Meng S, Menghini R, Menko AS, Menna-Barreto RF, Menon MB, Meraz-Ríos MA, Merla G, Merlini L, Merlot AM, Meryk A, Meschini S, Meyer JN, Mi MT, Miao CY, Micale L, Michaeli S, Michiels C, Migliaccio AR, Mihailidou AS, Mijaljica D, Mikoshiba K, Milan E, Miller-Fleming L, Mills GB, Mills IG, Minakaki G, Minassian BA, Ming XF, Minibayeva F, Minina EA, Mintern JD, Minucci S, Miranda-Vizuete A, Mitchell CH, Miyamoto S, Miyazawa K, Mizushima N, Mnich K, Mograbi B, Mohseni S, Moita LF, Molinari M, Molinari M, Møller AB, Mollereau B, Mollinedo F, Mongillo M, Monick MM, Montagnaro S, Montell C, Moore DJ, Moore MN, Mora-Rodriguez R, Moreira PI, Morel E, Morelli MB, Moreno S, Morgan MJ, Moris A, Moriyasu Y, Morrison JL, Morrison LA, Morselli E, Moscat J, Moseley PL, Mostowy S, Motori E, Mottet D, Mottram JC, Moussa CE, Mpakou VE, Mukhtar H, Mulcahy Levy JM, Muller S, Muñoz-Moreno R, Muñoz-Pinedo C, Münz C, Murphy ME, Murray JT, Murthy A, Mysorekar IU, Nabi IR, Nabissi M, Nader GA, Nagahara Y, Nagai Y, Nagata K, Nagelkerke A, Nagy P, Naidu SR, Nair S, Nakano H, Nakatogawa H, Nanjundan M, Napolitano G, Naqvi NI, Nardacci R, Narendra DP, Narita M, Nascimbeni AC, Natarajan R, Navegantes LC, Nawrocki ST, Nazarko TY, Nazarko VY, Neill T, Neri LM, Netea MG, Netea-Maier RT, Neves BM, Ney PA, Nezis IP, Nguyen HT, Nguyen HP, Nicot AS, Nilsen H, Nilsson P, Nishimura M, Nishino I, Niso-Santano M, Niu H, Nixon RA, Njar VC, Noda T, Noegel AA, Nolte EM, Norberg E, Norga KK, Noureini SK, Notomi S, Notterpek L, Nowikovsky K, Nukina N, Nürnberger T, O’Donnell VB, O’Donovan T, O’Dwyer PJ, Oehme I, Oeste CL, Ogawa M, Ogretmen B, Ogura Y, Oh YJ, Ohmuraya M, Ohshima T, Ojha R, Okamoto K, Okazaki T, Oliver FJ, Ollinger K, Olsson S, Orban DP, Ordonez P, Orhon I, Orosz L, O’Rourke EJ, Orozco H, Ortega AL, Ortona E, Osellame LD, Oshima J, Oshima S, Osiewacz HD, Otomo T, Otsu K, Ou JH, Outeiro TF, Ouyang DY, Ouyang H, Overholtzer M, Ozbun MA, Ozdinler PH, Ozpolat B, Pacelli C, Paganetti P, Page G, Pages G, Pagnini U, Pajak B, Pak SC, Pakos-Zebrucka K, Pakpour N, Palková Z, Palladino F, Pallauf K, Pallet N, Palmieri M, Paludan SR, Palumbo C, Palumbo S, Pampliega O, Pan H, Pan W, Panaretakis T, Pandey A, Pantazopoulou A, Papackova Z, Papademetrio DL, Papassideri I, Papini A, Parajuli N, Pardo J, Parekh VV, Parenti G, Park JI, Park J, Park OK, Parker R, Parlato R, Parys JB, Parzych KR, Pasquet JM, Pasquier B, Pasumarthi KB, Patschan D, Patterson C, Pattingre S, Pattison S, Pause A, Pavenstädt H, Pavone F, Pedrozo Z, Peña FJ, Peñalva MA, Pende M, Peng J, Penna F, Penninger JM, Pensalfini A, Pepe S, Pereira GJ, Pereira PC, Pérez-de la Cruz V, Pérez-Pérez ME, Pérez-Rodríguez D, Pérez-Sala D, Perier C, Perl A, Perlmutter DH, Perrotta I, Pervaiz S, Pesonen M, Pessin JE, Peters GJ, Petersen M, Petrache I, Petrof BJ, Petrovski G, Phang JM, Piacentini M, Pierdominici M, Pierre P, Pierrefite-Carlevarle V, Pietrocola F, Pimentel-Muiños FX, Pinar M, Pineda B, Pinkas-Kramarski R, Pinti M, Pinton P, Piperdi B, Piret JM, Platanias LC, Platta HW, Plowey ED, Pöggeler S, Poirot M, Polčic P, Poletti A, Poon AH, Popelka H, Popova B, Poprawa I, Poulose SM, Poulton J, Powers SK, Powers T, Pozuelo-Rubio M, Prak K, Prange R, Prescott M, Priault M, Prince S, Proia RL, Proikas-Cezanne T, Prokisch H, Promponas VJ, Przyklenk K, Puertollano R, Pugazhenthi S, Puglielli L, Pujol A, Puyal J, Pyeon D, Qi X, Qian WB, Qin ZH, Qiu Y, Qu Z, Quadrilatero J, Quinn F, Raben N, Rabinowich H, Radogna F, Ragusa MJ, Rahmani M, Raina K, Ramanadham S, Ramesh R, Rami A, Randall-Demllo S, Randow F, Rao H, Rao VA, Rasmussen BB, Rasse TM, Ratovitski EA, Rautou PE, Ray SK, Razani B, Reed BH, Reggiori F, Rehm M, Reichert AS, Rein T, Reiner DJ, Reits E, Ren J, Ren X, Renna M, Reusch JE, Revuelta JL, Reyes L, Rezaie AR, Richards RI, Richardson DR, Richetta C, Riehle MA, Rihn BH, Rikihisa Y, Riley BE, Rimbach G, Rippo MR, Ritis K, Rizzi F, Rizzo E, Roach PJ, Robbins J, Roberge M, Roca G, Roccheri MC, Rocha S, Rodríguez CI, de Cordoba SR, Rodriguez-Muela N, Roelofs J, Rogov VV, Rohn TT, Rohrer B, Romanelli D, Romani L, Romano PS, Roncero MI, Rosa JL, Rosello A, Rosen KV, Rosenstiel P, Rost-Roszkowska M, Roth K, Rodrigues CMA, Roué G, Rouis M, Rouschop KM, Ruan DT, Ruano D, Rubinsztein DC, Rucker EB 3rd, Rudich A, Rudolf E, Rudolf R, Ruegg MA, Ruiz-Roldan C, Ruparelia AA, Rusmini P, Russ DW, Russo GL, Russo G, Russo R, Rusten TE, Ryabovol V, Ryan KM, Ryter SW, Sabatini DM, Sacher M, Sachse C, Sack MN, Sadoshima J, Saftig P, Sagi-Eisenberg R, Sahni S, Saikumar P, Saito T, Saitoh T, Sakakura K, Sakoh-Nakatogawa M, Sakuraba Y, Salazar-Roa M, Salomoni P, Saluja AK, Salvaterra PM, Salvioli R, Samali A, Sanchez AM, Sánchez-Alcázar JA, Sanchez-Prieto R, Sandri M, Sanjuan MA, Santaguida S, Santambrogio L, Santoni G, Dos Santos CN, Saran S, Sardiello M, Sargent G, Sarkar P, Sarkar S, Sarrias MR, Sarwal MM, Sasakawa C, Sasaki M, Sass M, Sato K, Sato M, Satriano J, Savaraj N, Saveljeva S, Schaefer L, Schaible UE, Scharl M, Schatzl HM, Schekman R, Scheper W, Schiavi A, Schipper HM, Schmeisser H, Schmidt J, Schmitz I, Schneider BE, Schneider EM, Schneider JL, Schon EA, Schönenberger MJ, Schönthal AH, Schorderet DF, Schröder B, Schuck S, Schulze RJ, Schwarten M, Schwarz TL, Sciarretta S, Scotto K, Scovassi AI, Screaton RA, Screen M, Seca H, Sedej S, Segatori L, Segev N, Seglen PO, Seguí-Simarro JM, Segura-Aguilar J, Seki E, Sell C, Seiliez I, Semenkovich CF, Semenza GL, Sen U, Serra AL, Serrano-Puebla A, Sesaki H, Setoguchi T, Settembre C, Shacka JJ, Shajahan-Haq AN, Shapiro IM, Sharma S, She H, Shen CK, Shen CC, Shen HM, Shen S, Shen W, Sheng R, Sheng X, Sheng ZH, Shepherd TG, Shi J, Shi Q, Shi Q, Shi Y, Shibutani S, Shibuya K, Shidoji Y, Shieh JJ, Shih CM, Shimada Y, Shimizu S, Shin DW, Shinohara ML, Shintani M, Shintani T, Shioi T, Shirabe K, Shiri-Sverdlov R, Shirihai O, Shore GC, Shu CW, Shukla D, Sibirny AA, Sica V, Sigurdson CJ, Sigurdsson EM, Sijwali PS, Sikorska B, Silveira WA, Silvente-Poirot S, Silverman GA, Simak J, Simmet T, Simon AK, Simon HU, Simone C, Simons M, Simonsen A, Singh R, Singh SV, Singh SK, Sinha D, Sinha S, Sinicrope FA, Sirko A, Sirohi K, Sishi BJ, Sittler A, Siu PM, Sivridis E, Skwarska A, Slack R, Slaninová I, Slavov N, Smaili SS, Smalley KS, Smith DR, Soenen SJ, Soleimanpour SA, Solhaug A, Somasundaram K, Son JH, Sonawane A, Song C, Song F, Song HK, Song JX, Song W, Soo KY, Sood AK, Soong TW, Soontornniyomkij V, Sorice M, Sotgia F, Soto-Pantoja DR, Sotthibundhu A, Sousa MJ, Spaink HP, Span PN, Spang A, Sparks JD, Speck PG, Spector SA, Spies CD, Springer W, Clair DS, Stacchiotti A, Staels B, Stang MT, Starczynowski DT, Starokadomskyy P, Steegborn C, Steele JW, Stefanis L, Steffan J, Stellrecht CM, Stenmark H, Stepkowski TM, Stern ST, Stevens C, Stockwell BR, Stoka V, Storchova Z, Stork B, Stratoulias V, Stravopodis DJ, Strnad P, Strohecker AM, Ström AL, Stromhaug P, Stulik J, Su YX, Su Z, Subauste CS, Subramaniam S, Sue CM, Suh SW, Sui X, Sukseree S, Sulzer D, Sun FL, Sun J, Sun SY, Sun Y, Sundaramoorthy V, Sung J, Suzuki H, Suzuki K, Suzuki N, Suzuki T, Suzuki YJ, Swanson MS, Swanton C, Swärd K, Swarup G, Sweeney ST, Sylvester PW, Szatmari Z, Szegezdi E, Szlosarek PW, Taegtmeyer H, Tafani M, Taillebourg E, Tait SW, Takacs-Vellai K, Takahashi Y, Takáts S, Takemura G, Takigawa N, Talbot NJ, Tamagno E, Tamburini J, Tan CP, Tan L, Tan ML, Tan M, Tan YJ, Tanaka K, Tanaka M, Tang D, Tang G, Tanida I, Tanji K, Tannous BA, Tapia JA, Tasset-Cuevas I, Tatar M, Tavassoly I, Tavernarakis N, Taylor A, Taylor GS, Taylor GA, Taylor JP, Taylor MJ, Tchetina EV, Tee AR, Teixeira-Clerc F, Telang S, Tencomnao T, Teng BB, Teng RJ, Terro F, Tettamanti G, Theiss AL, Theron AE, Thomas KJ, Thomé MP, Thomes PG, Thorburn A, Thorner J, Thum T, Thumm M, Thurston TL, Tian L, Till A, Ting JP, Titorenko VI, Toker L, Toldo S, Tooze SA, Topisirovic I, Torgersen ML, Torosantucci L, Torriglia A, Torrisi MR, Tournier C, Towns R, Trajkovic V, Travassos LH, Triola G, Tripathi DN, Trisciuoglio D, Troncoso R, Trougakos IP, Truttmann AC, Tsai KJ, Tschan MP, Tseng YH, Tsukuba T, Tsung A, Tsvetkov AS, Tu S, Tuan HY, Tucci M, Tumbarello DA, Turk B, Turk V, Turner RF, Tveita AA, Tyagi SC, Ubukata M, Uchiyama Y, Udelnow A, Ueno T, Umekawa M, Umemiya-Shirafuji R, Underwood BR, Ungermann C, Ureshino RP, Ushioda R, Uversky VN, Uzcátegui NL, Vaccari T, Vaccaro MI, Váchová L, Vakifahmetoglu-Norberg H, Valdor R, Valente EM, Vallette F, Valverde AM, Van den Berghe G, Van Den Bosch L, van den Brink GR, van der Goot FG, van der Klei IJ, van der Laan LJ, van Doorn WG, van Egmond M, van Golen KL, Van Kaer L, van Lookeren Campagne M, Vandenabeele P, Vandenberghe W, Vanhorebeek I, Varela-Nieto I, Vasconcelos MH, Vasko R, Vavvas DG, Vega-Naredo I, Velasco G, Velentzas AD, Velentzas PD, Vellai T, Vellenga E, Vendelbo MH, Venkatachalam K, Ventura N, Ventura S, Veras PS, Verdier M, Vertessy BG, Viale A, Vidal M, Vieira HL, Vierstra RD, Vigneswaran N, Vij N, Vila M, Villar M, Villar VH, Villarroya J, Vindis C, Viola G, Viscomi MT, Vitale G, Vogl DT, Voitsekhovskaja OV, von Haefen C, von Schwarzenberg K, Voth DE, Vouret-Craviari V, Vuori K, Vyas JM, Waeber C, Walker CL, Walker MJ, Walter J, Wan L, Wan X, Wang B, Wang C, Wang CY, Wang D, Wang F, Wang G, Wang HJ, Wang H, Wang HG, Wang HD, Wang J, Wang M, Wang MQ, Wang PY, Wang P, Wang RC, Wang S, Wang TF, Wang X, Wang XJ, Wang XW, Wang Y, Wang YJ, Wang YT, Wang ZN, Wappner P, Ward C, Ward DM, Warnes G, Watada H, Watanabe Y, Watase K, Weaver TE, Weekes CD, Wei J, Weide T, Weihl CC, Weindl G, Weis SN, Wen L, Wen X, Wen Y, Westermann B, Weyand CM, White AR, White E, Whitton JL, Whitworth AJ, Wiels J, Wild F, Wildenberg ME, Wileman T, Wilkinson DS, Wilkinson S, Willbold D, Williams C, Williams K, Williamson PR, Winklhofer KF, Witkin SS, Wohlgemuth SE, Wollert T, Wolvetang EJ, Wong E, Wong GW, Wong RW, Wong VK, Woodcock EA, Wright KL, Wu C, Wu D, Wu GS, Wu J, Wu J, Wu M, Wu M, Wu S, Wu WK, Wu Y, Wu Z, Xavier CP, Xavier RJ, Xia GX, Xia T, Xia W, Xia Y, Xiao H, Xiao J, Xiao S, Xiao W, Xie CM, Xie Z, Xilouri M, Xiong Y, Xu C, Xu F, Xu H, Xu J, Xu L, Xu X, Xu Y, Xu ZX, Xu Z, Xue Y, Yamada T, Yamamoto A, Yamanaka K, Yamashina S, Yamashiro S, Yan B, Yan X, Yan Z, Yanagi Y, Yang DS, Yang JM, Yang L, Yang M, Yang PM, Yang P, Yang Q, Yang W, Yang WY, Yang X, Yang Y, Yang Z, Yao MC, Yao PJ, Yao X, Yao Z, Yasui LS, Ye M, Yedvobnick B, Yeganeh B, Yeh ES, Yeyati PL, Yi F, Yi L, Yin XM, Yip CK, Yoo YM, Yoo YH, Yoon SY, Yoshida K, Yoshimori T, Young KH, Yu H, Yu JJ, Yu JT, Yu J, Yu L, Yu WH, Yu XF, Yu Z, Yuan J, Yuan ZM, Yue BY, Yue J, Yue Z, Zacks DN, Zacksenhaus E, Zaffaroni N, Zaglia T, Zakeri Z, Zecchini V, Zeng J, Zeng M, Zeng Q, Zervos AS, Zhang DD, Zhang F, Zhang G, Zhang GC, Zhang H, Zhang J, Zhang JP, Zhang L, Zhang MY, Zhang X, Zhang XD, Zhang Y, Zhao M, Zhao WL, Zhao X, Zhao YG, Zhao Y, Zhao YX, Zhao Z, Zhao ZJ, Zheng D, Zheng XL, Zheng X, Zhivotovsky B, Zhong Q, Zhou GZ, Zhou G, Zhou H, Zhou SF, Zhou XJ, Zhu H, Zhu WG, Zhu W, Zhu XF, Zhu Y, Zhuang SM, Zhuang X, Ziparo E, Zois CE, Zoladek T, Zong WX, Zorzano A, Zughaier SM (2016) Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy 12: 1–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kothari S, Cizeau J, McMillan-Ward E, Israels SJ, Bailes M, Ens K, Kirshenbaum LA, Gibson SB (2003) BNIP3 plays a role in hypoxic cell death in human epithelial cells that is inhibited by growth factors EGF and IGF. Oncogene 22: 4734–4744. [DOI] [PubMed] [Google Scholar]
- Lamb CA, Nuhlen S, Judith D, Frith D, Snijders AP, Behrends C, Tooze SA (2016) TBC1D14 regulates autophagy via the TRAPP complex and ATG9 traffic. EMBO J 35: 281–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maes H, Kuchnio A, Peric A, Moens S, Nys K, De Bock K, Quaegebeur A, Schoors S, Georgiadou M, Wouters J, Vinckier S, Vankelecom H, Garmyn M, Vion AC, Radtke F, Boulanger C, Gerhardt H, Dejana E, Dewerchin M, Ghesquière B, Annaert W, Agostinis P, Carmeliet P (2014) Tumor vessel normalization by chloroquine independent of autophagy. Cancer Cell 26: 190–206. [DOI] [PubMed] [Google Scholar]
- Matys V, Kel-Margoulis OV, Fricke E, Liebich I, Land S, Barre-Dirrie A, Reuter I, Chekmenev D, Krull M, Hornischer K, Voss N, Stegmaier P, Lewicki-Potapov B, Saxel H, Kel AE, Wingender E (2006) TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res 34: D108–D110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maycotte P, Aryal S, Cummings CT, Thorburn J, Morgan MJ, Thorburn A (2012) Chloroquine sensitizes breast cancer cells to chemotherapy independent of autophagy. Autophagy 8: 200–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazure NM, Pouyssegur J (2009) Atypical BH3-domains of BNIP3 and BNIP3L lead to autophagy in hypoxia. Autophagy 5: 868–869. [DOI] [PubMed] [Google Scholar]
- Mazure NM, Pouyssegur J (2010) Hypoxia-induced autophagy: cell death or cell survival? Curr Opin Cell Biol 22: 177–180. [DOI] [PubMed] [Google Scholar]
- Mole DR, Blancher C, Copley RR, Pollard PJ, Gleadle JM, Ragoussis J, Ratcliffe PJ (2009) Genome-wide association of hypoxia-inducible factor (HIF)-1alpha and HIF-2alpha DNA binding with expression profiling of hypoxia-inducible transcripts. J Biol Chem 284: 16767–16775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moussay E, Kaoma T, Baginska J, Muller A, Van Moer K, Nicot N, Nazarov PV, Vallar L, Chouaib S, Berchem G, Janji B (2011) The acquisition of resistance to TNFalpha in breast cancer cells is associated with constitutive activation of autophagy as revealed by a transcriptome analysis using a custom microarray. Autophagy 7: 760–770. [DOI] [PubMed] [Google Scholar]
- Munson MJ, Ganley IG (2015) MTOR, PIK3C3, and autophagy: signaling the beginning from the end. Autophagy 11: 2375–2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niclou SP, Danzeisen C, Eikesdal HP, Wiig H, Brons NH, Poli AM, Svendsen A, Torsvik A, Enger PO, Terzis JA, Bjerkvig R (2008) A novel eGFP-expressing immunodeficient mouse model to study tumor-host interactions. FASEB J 22: 3120–3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noman MZ, Janji B, Berchem G, Mami-Chouaib F, Chouaib S (2012) Hypoxia-induced autophagy: a new player in cancer immunotherapy? Autophagy 8: 704–706. [DOI] [PubMed] [Google Scholar]
- Noman MZ, Janji B, Kaminska B, Van Moer K, Pierson S, Przanowski P, Buart S, Berchem G, Romero P, Mami-Chouaib F, Chouaib S (2011) Blocking hypoxia-induced autophagy in tumors restores cytotoxic T-cell activity and promotes regression. Cancer Res 71: 5976–5986. [DOI] [PubMed] [Google Scholar]
- Orsi A, Razi M, Dooley HC, Robinson D, Weston AE, Collinson LM, Tooze SA (2012) Dynamic and transient interactions of Atg9 with autophagosomes, but not membrane integration, are required for autophagy. Mol Biol Cell 23: 1860–1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pouyssegur J, Dayan F, Mazure NM (2006) Hypoxia signalling in cancer and approaches to enforce tumour regression. Nature 441: 437–443. [DOI] [PubMed] [Google Scholar]
- Rabinowitz JD, White E (2010) Autophagy and metabolism. Science 330: 1344–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori F, Tooze SA (2012) Autophagy regulation through Atg9 traffic. J Cell Biol 198: 151–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeld MR, Ye X, Supko JG, Desideri S, Grossman SA, Brem S, Mikkelson T, Wang D, Chang YC, Hu J, McAfee Q, Fisher J, Troxel AB, Piao S, Heitjan DF, Tan KS, Pontiggia L, O’Dwyer PJ, Davis LE, Amaravadi RK (2014) A phase I/II trial of hydroxychloroquine in conjunction with radiation therapy and concurrent and adjuvant temozolomide in patients with newly diagnosed glioblastoma multiforme. Autophagy 10: 1359–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouschop KM, van den Beucken T, Dubois L, Niessen H, Bussink J, Savelkouls K, Keulers T, Mujcic H, Landuyt W, Voncken JW, Lambin P, van der Kogel AJ, Koritzinsky M, Wouters BG (2010) The unfolded protein response protects human tumor cells during hypoxia through regulation of the autophagy genes MAP1LC3B and ATG5. J Clin Invest 120: 127–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanzey M, Abdul Rahim SA, Oudin A, Dirkse A, Kaoma T, Vallar L, Herold-Mende C, Bjerkvig R, Golebiewska A, Niclou SP (2015) Comprehensive analysis of glycolytic enzymes as therapeutic targets in the treatment of glioblastoma. PLoS One 10: e0123544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shingu T, Fujiwara K, Bogler O, Akiyama Y, Moritake K, Shinojima N, Tamada Y, Yokoyama T, Kondo S (2009) Inhibition of autophagy at a late stage enhances imatinib-induced cytotoxicity in human malignant glioma cells. Int J Cancer 124: 1060–1071. [DOI] [PubMed] [Google Scholar]
- Shintani T, Klionsky DJ (2004) Autophagy in health and disease: a double-edged sword. Science 306: 990–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slemc L, Kunej T (2016) Transcription factor HIF1A: downstream targets, associated pathways, polymorphic hypoxia response element (HRE) sites, and initiative for standardization of reporting in scientific literature. Tumour Biol 37: 14851–14861. [DOI] [PubMed] [Google Scholar]
- Sotelo J, Briceno E, Lopez-Gonzalez MA (2006) Adding chloroquine to conventional treatment for glioblastoma multiforme: a randomized, double-blind, placebo-controlled trial. Ann Intern Med 144: 337–343. [DOI] [PubMed] [Google Scholar]
- Vaupel P, Mayer A (2007) Hypoxia in cancer: significance and impact on clinical outcome. Cancer Metastasis Rev 26: 225–239. [DOI] [PubMed] [Google Scholar]
- Viry E, Baginska J, Berchem G, Noman MZ, Medves S, Chouaib S, Janji B (2014) Autophagic degradation of GZMB/granzyme B: a new mechanism of hypoxic tumor cell escape from natural killer cell-mediated lysis. Autophagy 10: 173–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weerasekara VK, Panek DJ, Broadbent DG, Mortenson JB, Mathis AD, Logan GN, Prince JT, Thomson DM, Thompson JW, Andersen JL (2014) Metabolic-stress-induced rearrangement of the 14-3-3zeta interactome promotes autophagy via a ULK1- and AMPK-regulated 14-3-3zeta interaction with phosphorylated Atg9. Mol Cell Biol 34: 4379–4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto H, Kakuta S, Watanabe TM, Kitamura A, Sekito T, Kondo-Kakuta C, Ichikawa R, Kinjo M, Ohsumi Y (2012) Atg9 vesicles are an important membrane source during early steps of autophagosome formation. J Cell Biol 198: 219–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Lin C, Wang L, Guo H, Wang X (2012) Hypoxia and hypoxia-inducible factors in glioblastoma multiforme progression and therapeutic implications. Exp Cell Res 318: 2417–2426. [DOI] [PubMed] [Google Scholar]
- Yang ZJ, Chee CE, Huang S, Sinicrope FA (2011) The role of autophagy in cancer: therapeutic implications. Mol Cancer Ther 10: 1533–1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Bosch-Marce M, Shimoda LA, Tan YS, Baek JH, Wesley JB, Gonzalez FJ, Semenza GL (2008) Mitochondrial autophagy is an HIF-1-dependent adaptive metabolic response to hypoxia. J Biol Chem 283: 10892–10903. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.