Abstract
U1snRNA, U3snRNA, 28 S ribosomal RNA, poly(A) RNA and a specific messenger RNA were visualized in living cells with microinjected fluorochrome-labeled 2′ O-Methyl oligoribonucleotides (2′ OMe RNA). Antisense 2′ OMe RNA probes showed fast hybridization kinetics, whereas conventional oligodeoxyribonucleotide (DNA) probes did not. The nuclear distributions of the signals in living cells were similar to those found in fixed cells, indicating specific hybridization. Cytoplasmic ribosomal RNA, poly(A) RNA and mRNA could hardly be visualized, mainly due to a rapid entrapment of the injected probes in the nucleus. The performance of linear probes was compared with that of molecular beacons, which due to their structure should theoretically fluoresce only upon hybridization. No improvements were achieved however with the molecular beacons used in this study, suggesting opening of the beacons by mechanisms other than hybridization. The results show that linear 2′ OMe RNA probes are well suited for RNA detection in living cells, and that these probes can be applied for dynamic studies of highly abundant nuclear RNA. Furthermore, it proved feasible to combine RNA detection with that of green fluorescent protein-labeled proteins in living cells. This was applied to show co-localization of RNA with proteins and should enable RNA–protein interaction studies.
INTRODUCTION
The eukaryotic cell nucleus is highly organized to tightly control and coordinate gene expression. This picture has emerged from fluorescence in situ hybridization and immunocytochemical studies, showing that chromosomes occupy distinct territories within the nucleus and that many of the nuclear factors that play a role in transcription and RNA processing are compartmentalized in nuclear bodies, as reviewed by Lamond and Earnshaw (1). The chromosome territories contain regions in which the degree of chromatin condensation varies. Transcriptionally active genes have been shown to be located at the borders of the condensed chromatin regions to have access to the transcription, RNA processing and transport machinery (2,3). Nuclear bodies, like coiled bodies, PML bodies and speckles may then serve as pre-assembly sites of the components of this machinery or as storage sites from which factors are recruited to active genes (4,5). Alternatively, or additionally, transcription and RNA processing complexes may be functional at these sites (6).
To relate the spatial organization of nuclear components to functional activities the dynamic behaviors of DNA, RNA and proteins need to be studied in living cells. Various strategies for in vivo labeling of DNA, RNA and proteins have been proposed. DNA can be labeled by the transient incorporation of fluorescent nucleotides during replication, which results in a partial labeling of chromatin in successive cell generations (7). Proteins can be tagged with the spectral variants of the green fluorescent protein (GFP) and their dynamic behavior can be analyzed by time-lapse experiments and by fluorescence redistribution after photo bleaching (FRAP) (8,9). More recently, Tsukamoto et al. (6) described a system to visualize a gene and its protein product in living cells. To detect the various RNA classes in living cells, several approaches have been developed (reviewed in 10–13). Ainger et al. (14) injected in vitro transcribed fluorescent RNAs, which act as duplicates of endogenous RNAs, and studied their localization upon delivery into cells. As an alternative, mRNA was visualized using GFP as a fluorescent tag on the RNA-loop binding protein MS2 (15–17). In addition to these two methods, in vivo hybridization methods were used that are based on the annealing of short antisense fluorescent probes to specific cellular RNAs. In most studies, short linear DNA oligonucleotides (20–40 bases) were used (18–21). As an alternative, Carmo-Fonseca et al. (22) described the use of 2′ O-Methyl (2′ OMe) RNA and 2′ O-allyl RNA probes for the detection of small nuclear RNAs. 2′ OMe RNA probes were considered to perform better then DNA oligonucleotides because they are not only nuclease resistant, but also possess a higher affinity, increased specificity, faster hybridization kinetics and a superior ability to bind to structured targets compared to DNA oligonucleotides (23,24). Recently, so-called molecular beacons were introduced for RNA detection in living cells (19,25). Molecular beacons are hairpin-shaped oligonucleotide probes that fluoresce only upon hybridization (26,27). The rationale for using molecular beacons to detect RNAs in living cells was to improve signal-to-noise ratios by eliminating fluorescence signals derived from non-hybridized probe sequences. In order to come to a generally applicable technique for the detection of RNAs in living cells, we systematically evaluated the performance of linear probes and molecular beacons synthesized both from DNA and 2′ OMe RNA monomers. The RNA targets detected were two small nuclear RNAs, U1 snRNA and U3 snRNA, ribosomal RNA of the 28S ribosomal subunit, poly(A) RNA and a specific mRNA, the mRNA encoding the human cytomegalovirus immediate early antigen (HCMV IE) in transformed rat fibroblasts. We demonstrate that of the four probe types tested, linear 2′ OMe RNA probes are the most suitable for specific detection of these RNAs, representing different classes of RNA, in the nuclei of living cells. Under the conditions used, molecular beacons did not result in images with improved signal to noise ratios, thereby leading to better detection sensitivity.
MATERIALS AND METHODS
Probes
For the various RNA targets described, probes were selected on the basis of sequences in the NCBI genome database. Single-stranded regions or partly single-stranded regions in the secondary RNA structure for U1 snRNA and U3 snRNA (28) were targeted. They were varied in length, structure and nucleotide modification. Sequences and GenBank IDs are listed in Table 1. DNA oligonucleotides were synthesized using standard phosphoramidite chemistry and purified by HPLC. 2′ OMe RNA probes were synthesized using standard 2′ OMe phosphoramidite monomers. For linear probes fluorescent dyes were covalently linked to the 5′-end via a succinimidyl ester derivative (Molecular Probes, Eugene, OR). Molecular beacons were synthesized automatically with the 3′-4-dimethylaminoazobenzene-4′-sulfonyl (dabcyl)-quencher linked to the solid phase support (DABCYL-CPG, Glen Research, Sterling, VA). Oligonucleotide chain elongation was performed via standard cyanoethyl-phosphoramidite chemistry and fluorophores were covalently linked to the 5′-end via iodoacetamide (Molecular Probes) or phosphoramidite derivatives (Glen Research) following release from the solid support. All 2′ OMe RNA probes were purified twice by reverse phase HPLC with a Waters 600E instrument, equipped with a Waters 996 Photodiode Array Detector for simultaneous detection at three different wavelengths. Ion-molecular weights of purified probes were determined by mass spectrometry using a Dynamo time-of-flight instrument. The initial molecular beacon probes were designed following the guidelines described by Tyagi and Kramer (26) (http:\\www.molecular-beacons.org). The intramolecular configuration of molecular beacons and their target sequences was modeled using the DNA mfold program (www.ibc.wustl.edu/~zuker/dna/form1.shtml), which uses thermodynamic parameters established by SantaLucia (26). The molecular beacons did not contain self-complementary sequences in the loop. Beacons were equipped with stem sequences of varying length.
Table 1. Probe sequences.
| RNA target |
Design |
Sequence |
GenBank ID |
Nucleotide positions |
| U1 snRNA |
linear probea |
(c)cugccagguaaguau(g) |
340085 |
1–17 |
| |
MB probe |
gcgac cugccagguaaguau gucgc |
|
|
| U3 snRNA |
Linear probea |
(c)ggcuucacgcucagg(a) |
2736114 |
205–221 |
| |
MB probe |
(c/g)cgacggcuucacgcucagggucg(c/g) |
|
|
| 28S rRNA |
Linear probe 17 bp |
uaccaccaagaucugca |
337381 |
2321–2337 |
| |
Linear probe 35 bpb |
gaatatttgctactaccaccaagatctgcacctgc |
|
2316–2350 |
| |
MB probe |
gcgacuaccaccaagaucugcagucgc |
|
|
| Poly(U) |
Linear probes |
(u)18(u)22(t)40b |
|
|
| |
MB probes |
gucgac(u)18gucgac;cgucgac(u)22gucgacg |
|
|
| Poly(A) |
Linear probe |
(a)18 |
|
|
| |
MB probe |
gucgac(a)18gucgac |
|
|
| CMV1 |
Linear probe |
aaacauccucccauca |
330617 |
119–134 |
| |
MB probe |
cgcacaaacauccucccaucagucag |
(exon 4) |
|
| CMV2 |
Linear probe |
caucuccucgaaaggcu |
|
169–186 |
| |
MB probe |
cgcacaucuccucgaaaggcugugcg |
|
|
| CMV3 |
Linear probe |
acaucaugcagcuccuu |
|
271–287 |
| |
MB probe |
ccagacaucaugcagcuccuucugg |
|
|
| CMV4 |
Linear probe |
gagcacugaggcaaguuc |
|
465–482 |
| |
MB probe |
ccagggagcacugaggcaaguucccugg |
|
|
| CMV5 |
Linear probe |
ugugaucaaugugcguga |
|
551–568 |
| MB probe | ccagugugaucaaugugcgugcugg |
aAdditional bases in longer probes for the same target are indicated within brackets.
bLonger linear probes.
All sequences were synthesized as DNA and as 2′ OMe RNA oligonucleotides, except for the longer linear probes that were only used as DNA probes.
All probes were labeled with TAMRA; molecular beacons were labeled additionally with DABCYL as quencher.
Cell culture and microinjection
For microinjection experiments and life cell observation, U2OS (human osteosarcoma) cells and R9G cells were cultured on coverslips in 3.5 cm Petri dishes (Mattek, Ashland, MA) in RPMI 1640, without phenol red supplemented with 5% fetal calf serum and buffered with 25 mM HEPES buffer to pH 7.2 (Life Technologies, Breda, The Netherlands). R9G is a rat fibroblast cell line harboring a tandem repeat of 50 copies of the HCMV IE gene in one of the rat chromosomes (29–31). To induce the S-phase dependent HCMV IE gene expression, 50 µg/ml cycloheximide (Sigma, St Louis, MO) was added to the culture medium, 3–4 h prior to injection. The temperature of the culture medium was kept at 37°C using a heated ring surrounding the culture chamber and a microscope objective heater (Bioptechs, Butler, PA). For microinjection, a Narishige IM 300 microinjector (Narishige, Tokyo, Japan) was used. Needles with a tip inner diameter of ∼0.5 µm were pulled from glass capillaries (no. MTW100F-3 WPI, Aston, UK) on a Sutter micropipette puller (p97, Sutter instrument company, Novato, CA). Probes were dissolved in a buffer containing 80 mM KCl, 10 mM K2PO4, 4 mM NaCl pH 7.2 to a final concentration of 5 µM and microinjected in the cytoplasm of cells. The optimal concentration and the amount of probe molecules injected was determined on the basis of a series of initial experiments taking the intensity of the hybridization signals and cell viability into account. Typically, for each probe about 50 cells were injected and monitored over time. Representative cells were selected and analyzed by digital fluorescence microscopy.
RNA-FISH on fixed cells
To detect the various RNAs in fixed cells, in situ hybridization was performed essentially as described previously (32). In brief, U2OS cells were fixed in 3.7% formaldehyde, 5% acetic acid in PBS for 15 min at room temperature. The cells were pretreated for 1 min with 0.1% pepsin pH 2.0 at 37°C before hybridization. The 2′ OMe RNA probes were dissolved in 40% deionized formamide/2× SSC pH 7.4 and the DNA probes were dissolved in 10% deionized formamide/2× SSC. Hybridization mixture (10 µl) containing 50 nM probe was applied on cells grown on glass slides and covered with an 18 × 18 mm coverslip. Slides were put on an 80°C hot plate for 2 min to denature nucleic acids and then incubated for 2 h at 37°C for hybridization. To remove excess of probe, slides were rinsed in 2× SSC for 5 min, 2× SSC/10% formamide for 10 min, 2× SSC for 5 min. Cells were then mounted in VectaShield (Vector Laboratories, Burlingame, CA).
Construction and expression of GFP-fusion proteins
The cDNAs encoding ASF/SF2 and fibrillarin were generated by RT–PCR and cloned into the pEGFP-C1 vector (Clontech, Palo Alto, CA) as described previously (9). The resulting plasmid constructs were microinjected into cells (50 ng/µl H2O), and cells showing GFP-expression were injected 24 h after transfection with antisense probes.
Life cell imaging
Microinjection and life cell imaging was performed on a Zeiss Axiovert 135 TV microscope (Zeiss, Jena, Germany), equipped with a 100 W mercury arc lamp. The objectives used were a 40×/NA 1.30 Plan Neofluar phase and a 100×/NA 1.30 Plan Neofluar phase. Tetramethylrhodamine (TAMRA) labeled probes were detected using a filter set consisting of a 546/10 nm BP filter, a 580 nm DM and a 590 LP barrier filter. Images were acquired using a cooled charged coupled device camera (type PXL, Photometrix, Tucson, AZ). At various time points after microinjection, images were acquired and fluorescence distribution patterns were analyzed. Typical exposure times were 1–2 s.
RESULTS
U1 snRNA and U3 snRNA, 28 S rRNA and poly(A) are present in high amounts in a cell, and specific nuclear compartments, such as nucleoli, speckles and Cajal bodies are enriched for some of these RNA species. U1 snRNA is a component of ‘spliceosomes’, structures that are responsible for mRNA splicing (33); U3 snRNA has a function in rRNA processing, and is mainly localized in nucleoli (34). Poly(A) RNA, representing mRNA or pre-mRNA, is present throughout the nucleoplasm and concentrated in speckles and also in the cytoplasm (35,36), and 28S rRNA is localized in nucleoli and cytoplasm. The high expression from the HCMV IE gene integration sites generates a local nuclear track consisting of transcript (31). For each of these targets four different types of probes were synthesized: linear 2′ OMe RNA probes, molecular beacon 2′ OMe RNA probes, linear DNA probes and molecular beacon DNA probes, and used for hybridization in fixed and living cells. Probe sequences and the fluorescent labels that were used are listed in Table 1.
Linear 2′ OMe RNA probes
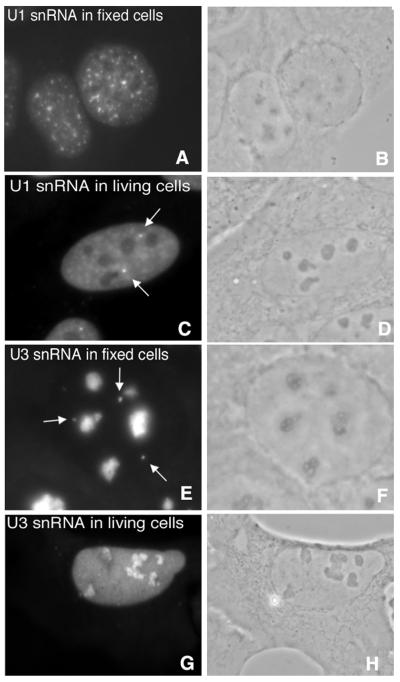
Probes were injected in the cytoplasm of U2OS cells, causing less damage to the cells compared to nuclear injections. Within 60 s of injection, all probes accumulated in the nucleus, an observation that is in agreement with earlier studies (37,38). Microinjected fluorescent linear 2′ OMe RNA probes to U1 snRNA were visible in speckles, Cajal bodies and throughout the nucleoplasm (Fig. 1C), excluding nucleoli, in agreement with earlier studies (22,39,40). The U3 snRNA 2′ OMe RNA probe was found to localize in nucleoli and in the nucleoplasm (Fig. 1G). Nucleoli were identified in phase contrast images (Fig. 1B, D, F and H). Similar staining patterns were observed when these probes were hybridized on fixed cells (Fig. 1A and E), indicating that they specifically hybridized to their respective RNA targets in living cells. In general the signal intensity of hybridized probe was higher in fixed cells compared to that in living cells. The pepsine-treatment and the denaturation step that are applied in FISH on fixed cells make the RNA more accessible for the probe, causing a much higher hybridization efficiency compared to that in living cells. Post-hybridization washing steps removed the unbound probe from fixed cells. The homogeneous nucleoplasmic staining observed with the U3 snRNA probe in living cells, that obscured the detection of U3 snRNA in Cajal bodies, was not observed in fixed cells (Fig. 1E). The microinjected cells were followed for several days and underwent mitosis. Similar localization patterns were observed in daughter cells (not shown). This indicated that the probes did not interfere with cell vitality. When non-fluorescent probes for U1 snRNA and U3 snRNA were injected in a 5-fold higher concentration prior to injection of fluorescent probes, the specific hybridization pattern was not obtained (data not shown), indicating blocking of the hybridization sites.
Figure 1.
Similar localization patterns of 2′ OMe RNA oligonucleotides hybridized to U1 snRNA and U3 snRNA in fixed and living U2OS cells (A, C, E and G). The distinct localization patterns observed for U1 snRNA (C) and U3 snRNA (G) detected in living cells after microinjection of the antisense oligonucleotides are similar to those observed in fixed cells (A and E), indicating a specific hybridization to RNA in living cells. Corresponding phase contrast images (B, D, F and H). The intense spots in (C) and (E), indicated by arrows, are Cajal bodies, the faint structures in the nucleoplasm in (C) are speckles.
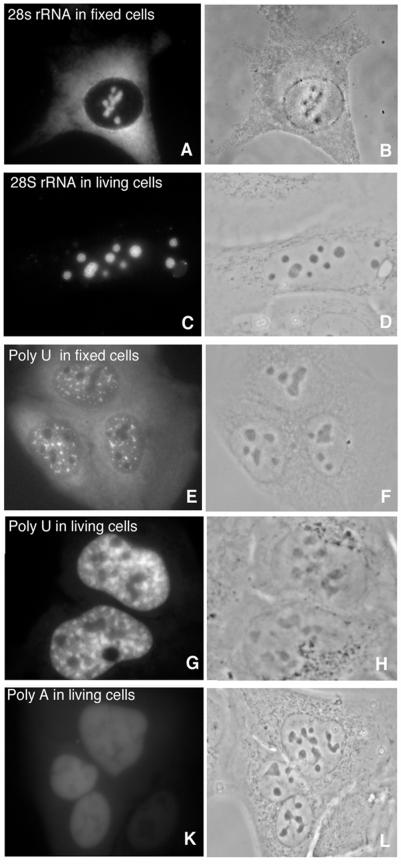
Following microinjection in U2OS cells, fluorescent linear 2′ OMe RNA probes designed to hybridize to 28S rRNA or poly(A) RNA localized at the expected nuclear sites. Ribosomal RNA was detected in nucleoli (Fig. 2C) and poly(A) RNA was detected in speckles and throughout the nucleoplasm (Fig. 2G). Similar nuclear hybridization patterns were obtained when these probes were hybridized to fixed cells (Fig. 2A). However, in fixed cells, the nucleoplasmic staining was less pronounced and the contrast between speckles and nucleoplasm was more apparent (Fig. 2E). The cytoplasmic staining of 28S rRNA and poly(A) RNA observed in fixed cells (Fig. 2A and E) was not observed in living cells (Fig. 2C and G), due to a rapid accumulation of the probes in the nucleus. Only a very low concentration of all injected probes could be observed in the cytoplasm.
Figure 2.
Hybridization of linear 2′ OMe RNA probes to abundant RNA targets in living cells. Probes for 28S rRNA (A and C) and poly(A) RNA (E and G) hybridized to fixed and living cells show a similar nuclear hybridization pattern but a different cytoplasmic localization pattern. Probes to 28S rRNA and poly(A) RNA accumulate in the nucleus within minutes of injection and the level of hybridization to the RNA present in the cytoplasm is negligible. The (A)18 probe shows no specific hybridization pattern (K). Corresponding phase contrast images (B, D, F, H and L).
To further prove the specificity of hybridization in live cells, cells were microinjected with control probes without a specific target sequence. Microinjection of the poly(A)18 2′ OMe RNA probe resulted in a diffuse staining throughout the nucleoplasm without revealing any specific hybridization pattern (Fig. 2K). A similar diffuse staining was observed when non-induced rat 9G cells or U2OS cells were microinjected with a 2′ OMe RNA probe specific for HCMV IE RNA (not shown).
Detection of the transcription site of CMV RNA in living R9G cells
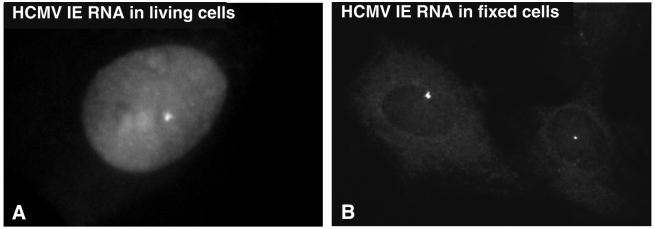
As 2′ OMe RNA oligonucleotides were shown to hybridize to high abundant RNAs in living cells these probes were subsequently used to detect CMV IE mRNA in a transformed rat fibroblast cell line. Induced R9G cells were microinjected with up to five TAMRA-labeled 2′ OMe RNA oligonucleotides specific for IE mRNA. Injection of single probes did not result in specific hybridization signals. Nevertheless, when a mixture of three 2′ OMe RNA oligonucleotides (I, II and IV in Table 1) was injected, a fluorescent dot representing IE RNA at the integrated IE gene repeat was visible (Fig. 3A). This nuclear signal was similar to the one observed in fixed cells after FISH with these probes (Fig. 3B), and as observed in previous studies (31,41). In addition to the fluorescent dot, live cells revealed a homogeneous fluorescent staining throughout the nucleus including nucleoli. This can be ascribed to non-hybridized probes and non-specifically hybridized probes. Cytoplasmic IE mRNA could not be detected in living cells.
Figure 3.
Detection of the transcription site of CMV RNA, transcribed from the integrated HCMV IE 1 gene in R9G cells. (A) Transcription sites and cytoplasmic mRNA are visible. Detection of transcription sites in living cells. The small bright spot shows transcription from the integration sites. A high background was observed in the nucleus and nucleoli. The signal in the cytoplasm was very weak. (B) Representative example of an in situ hybridization with three linear 2′ OMe RNA oligonucleotides on fixed R9G cells.
2′ OMe RNA molecular beacons
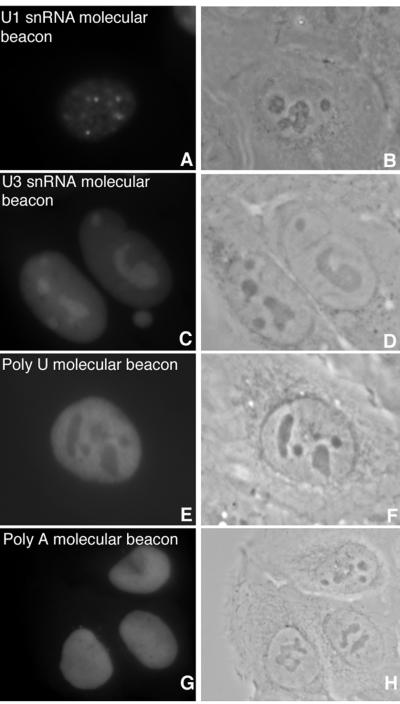
To test their performance in living cells, 2′ OMe RNA molecular beacon probes specific for U1 and U3 snRNA, for 28S rRNA and for poly(A) RNA were designed. As control probes, a poly(A) beacon and an HCMV IE mRNA specific 2′ OMe RNA beacon were used (Table 1). Since not much is known about the hybridization kinetics of these types of probes in living cells, all molecular beacons were tested in solution for specificity for their target sequences and their ability to fluoresce upon hybridization (not shown). 2′ OMe RNA molecular beacons specific for U1 snRNA, U3 snRNA, 28S rRNA and poly(A) RNA microinjected in U2OS cells showed initially a very low signal intensity because they are in a closed and thus quenched form. However, after 5–10 min, the signal intensity increased steadily throughout the nucleus following injection and similar localization patterns were observed as obtained with linear 2′ OMe RNA probes (Fig. 4). Overall, the maximal signal intensity achieved with the molecular beacons was 2–3-fold lower compared to the linear probes. This difference in intensity was also observed when the comparison was performed on fixed cells (data not shown). In addition to nuclear compartments a diffuse background staining of the nucleoplasm was observed following microinjection of the beacon probes in living cells. This background staining was independent from specific hybridization to an RNA target, since opening of the molecular beacon also occurred when 2′ OMe RNA beacons were injected with no RNA target in U2OS cells. This was observed after injection of a beacon with a loop containing only 18 adenines or a beacon specific for HCMV IE RNA (not shown).
Figure 4.
Hybridization of molecular beacons in living cells. U1 snRNA (A) and U3 snRNA (C) molecular beacons show similar hybridization patterns as the linear probes (Fig. 1), but have a lower signal-to-noise ratio. A molecular beacon targeted to poly(A) RNA consisting of a 22 base loop and a 7 bp stem showed only a very vague specific staining (E), compared to the linear probe (Fig. 2G). The overall intensity was lower for molecular beacons. For none of the RNAs that were targeted, a molecular beacon showed hybridization with an improved contrast. The control poly(A) molecular beacon probe does not show a specific hybridization pattern (G). Corresponding phase contrast images (B, D, F and H).
To find the optimal beacon probe structure for hybridization in living cells, probes for U3 snRNA and poly(A) RNA with different stem–loop lengths were tested (Table 1). The altered probe structures did not result in improved signal-to-noise ratios. For example, a poly(U) molecular beacon with a length of 28 bases (loop, 18 bases; stem, 5 bp) and a molecular beacon with a length of 36 bases (loop, 22 bases; stem, 7 bp) were used to detect poly(A) tails in live cells. Microinjection of the 28 base probe resulted in the specific ‘speckled’ pattern that was obtained with the linear probe, although the intensity of the signals was 2–3-fold lower (not shown). Injection of the 36 base probe resulted in a diffuse nuclear staining (Fig. 4E), similar to that obtained with the non-specific poly(A) beacon (Fig. 4G).
The performance of linear and molecular beacon DNA probes
To test the theoretical advantage of 2′ OMe RNA probes compared to unmodified oligodeoxynucleotides for detecting RNA species in vivo, we directly compared the two types of probes. Linear fluorescent DNA probes with the same sequences as the 2′ OMe RNA probes (Table 1) were synthesized. Upon microinjection, all DNA probes rapidly accumulated in the cell nucleus which resulted in a diffuse staining, without revealing any specific hybridization pattern (Fig. 5). Hybridization on fixed cells indicated that the probes were specific for their RNA target (data not shown). These results suggest that in living cells, linear DNA probes have an insufficient affinity for the RNA target sequences to allow specific and stable hybridization. The inability to show specific hybridization was not caused by degradation of the probes, since the observations were done within minutes of injection of the probes, and the half-life of DNA oligonucleotides injected in living cells was shown to be 15 min (42). It is feasible that a low percentage of the injected DNA oligonucleotides did in fact hybridize to the target RNA, but that the relatively dim specific signal was very much disturbed by the high amount of the non-specific staining. Even 35 and 40 base long DNA probes for 28S rRNA (Fig. 5A) and poly(A) RNA (Fig. 5B), possessing higher affinities compared to the 15–22 base probes, did not result in distinct localization patterns in living cells. Also molecular beacon DNA probes for the same targets did not reveal a specific hybridization pattern in living cells upon injection.
Figure 5.
No specific hybridization visible after microinjection of DNA oligonucleotides. Two examples of microinjected cells where RNA was targeted with DNA oligonucleotides. Thirty-five and 40 base antisense DNA probes for 28S ribosomal RNA (A) and the poly(A) tail of mRNA (C) were microinjected into living U2OS cells. The probes accumulated in the nucleus with time as was expected but did not show any further specific localization as was observed with the 2′ OMe RNA probes (as in Figs 1 and 2). (B and D) Corresponding phase contrast images.
Combined RNA and protein detection in living cells
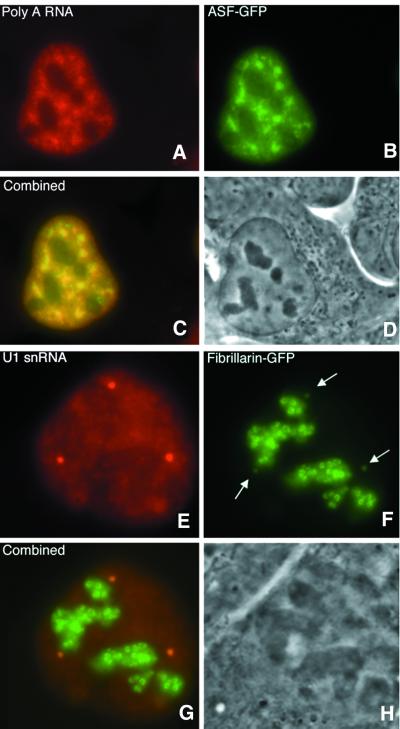
Having established that linear 2′ OMe RNA probes are suited for detection of RNA species in living cells, we examined the possibility to simultaneously detect RNA and protein molecules in living cells. U2OS cells were transiently transfected with GFP expression vectors containing the coding sequences for alternative splicing factor (ASF) and fibrillarin and expressing cells were microinjected with a 2′ OMe RNA probe. Figure 6A–D shows the simultaneous detection of ASF–GFP together with poly(A) RNA in a living cell. As expected from previous localization studies on fixed cells, ASF and poly(A) RNA were shown to co-localize in a speckled pattern. Fibrillarin–GFP, which localizes to nucleoli and Cajal bodies (9), was shown to co-localize with U1 snRNA in Cajal Bodies (Fig. 6E–H). These results indicate that proteins and RNA species can be visualized simultaneously in living cells without disturbing their integrity. The simultaneous labeling of RNA and proteins in living cells not only allows microscopic co-localization studies as was shown here, but could also provide the opportunity to study their interaction, by applying RNA–Protein Fluorescence Resonance Energy Transfer (FRET).
Figure 6.
Combined detection of RNA and expression of GFP in living cells. (A–D) Co-localization of poly(A) RNA and ASF–GFP in living U2OS cells in speckles and nucleoplasm. (E–H) Co-localization of U1 snRNA and fibrillarin–GFP in coiled bodies. Arrows in (F) indicate Cajal Bodies containing fibrillarin–GFP. Cells expressing ASF–GFP (B) were microinjected with a probe for poly(A) RNA (A) and cells expressing fibrillarin–GFP (F) with a probe for U1 snRNA (E).
DISCUSSION
In this study we evaluated the performance of different probe types, for the detection of representatives of the major RNA classes, rRNA, snRNA, poly(A) RNA and a specific mRNA, in living cells. Our results show that 2′ OMe RNA probes can be designed such that they hybridize with high specificity to the corresponding target sequences in living cells without affecting cell vitality, and that they are superior to DNA oligonucleotides. Cells microinjected with the 2′ OMe RNA probes could be monitored for several days and underwent mitosis, showing that processes like transcription and proliferation were not irreversibly disturbed. Long-term observation however was only found possible when low light level illumination was applied, in order to prevent fluorescence fading and phototoxic effects. As was observed before (37,38,43), oligonucleotide probes rapidly entered the cell nucleus after microinjection in the cytoplasm, and specific hybridization patterns were observed within 1–2 min irrespective of the target RNA. This suggests that oligonucleotide probes can move freely throughout the cell nucleus.
RNA targets in the cytoplasm could not be labeled presumably because rapid entrapment of the probes in the nucleus caused depletion of the cytoplasm and prevented successful hybridization to cytoplasmic RNA targets. One possibility to overcome this nuclear entry of probes when injected in the cytoplasm, is to couple a high molecular weight tag to the oligonucleotide. Tsuji et al. (18) used streptavidin to tag oligonucleotide probes to detect mRNA in the cytoplasm. Alternatively, oligo-peptide conjugates, where the peptide sequence is a nuclear export signal, could be tested (44). Interestingly, 24 h after injection of a probe to poly(A) tails, an increase in cytoplasmic signal was observed, suggestive for the export of mRNA from the nucleus to the cytoplasm. This signal, however, could also have been derived from degradation products.
Using linear 2′ OMe RNA probes, U1 and U3 snRNA, 28S rRNA and poly(A) RNA were detected with high signal-to-noise ratios, indicating that the majority of the injected probe was hybridized specifically to the RNA of interest. This was particularly true when the injected amount of probe was estimated to be lower or equal to the number of target RNA molecules. The probes for U1 and U3 snRNA and poly(A) RNA that localized to specific nuclear compartments as speckles, Cajal bodies and nucleoli, were also found present throughout the nucleoplasm. Since this diffuse staining was much weaker after FISH on fixed cells, it is possible that part of this signal is derived from non-hybridized fractions of probes.
HCMV IE mRNA was detected as a single confined spot in the nucleus. This shows that the high concentration of HCMV IE mRNA that is present at the transcribing HCMV gene cluster can be detected in living cells, and suggests that this method provides a useful additional tool for the visualization of gene activity in vivo. The IE mRNAs emanating from the transcription site towards the cytoplasm, as revealed by FISH on fixed cells (31), could however not be detected. This could be attributed to a low signal-to-noise ratio. Interestingly, Sei-Iida et al. (45) observed that DNA probes hybridize efficiently to RNA molecules in vitro during or just after synthesis and less efficiently after folding. Thus, also in vivo, the nascent RNA transcripts are probably better accessible for probes compared to the folded mRNA–protein complexes in transport through the nucleus. The efficiency of hybridization of antisense probes to mRNA varies along the target sequence, depending on secondary RNA structures and proteins occupying the probe binding sites (18). Testing a series of probes along the sequence may identify the most optimal probes for a particular mRNA. In this study 2′ OMe RNA oligonucleotides were shown to be well suited for detection of highly abundant nuclear RNA targets in living cells. The fact that 2′ OMe RNA probes and RNA form a hybrid that is not degraded by RNase H, whereas DNA/RNA and RNA/RNA hybrids do form substrates for this enzyme, is an additional argument to use the 2′ OMe modification. DNA analogs that were shown to have a higher affinity or specificity for RNA, such as 2′ allyl RNA (46), 2′ methoxy-ethoxy (2′ Moe) RNA (47,48), peptide nucleic acids (PNAs) (49) and several others, could possibly be used to further optimize hybridization in living cells.
Reducing or eliminating signals from unbound or non-specifically bound probe
The key question is whether the signals obtained by in vivo hybridization are actually derived from probes specifically hybridized to RNA. To reduce or circumvent the contribution of unwanted signals from unbound or non-specifically bound probes, a method may be applied that enables discrimination between signals derived from probes bound specifically to the RNA target and signals from unbound or non-specifically bound probes.
Therefore, we tested a series of molecular beacons for RNA detection in living cells. In vitro experiments demonstrated the improved specificity of 2′ OMe RNA molecular beacons compared to 2′ OMe RNA linear probes in vitro (data not shown). We could not confirm the beneficial properties of molecular beacons for detecting RNA targets in living cells. Although 2′ OMe RNA molecular beacons showed specific hybridization in our study and were applied before in live cells by Matsuo et al. (25) and Sokol et al. (19), for all RNAs in this study the linear probes showed better signal-to-noise ratios than molecular beacons. In general, the molecular beacons used by us showed lower affinity for RNA targets in fixed and living cells. Specific as well as control beacons were shown to open up shortly after entering the cell nucleus, indicating that their non-specific interaction with nuclear proteins or nucleic acids imposes a conformational change leading to fluorescence. Recently, it has been shown that a beacon type structure can be recognized by lactate dehydrogenase (50), and since many nuclear proteins can interact with structural motives present in RNA molecules, it is well possible that such interactions occur with molecular beacons, resulting in fluorescence regardless of specific hybridization.
Sixou et al. (51) proposed an alternative method to discriminate between specific and non-specific hybridization based on FRET. Recently Tsuji et al. (18) applied this method by using adjacently hybridizing probes, labeled 3′ and 5′ with donor and acceptor molecules, respectively, thereby facilitating FRET. This approach was used to detect cFos mRNA in transfected COS cells using Bodipy 493/503 and Cy5 labeled DNA oligonucleotides. For the detection of FRET it is required that both probes are simultaneously hybridized on the target molecule, demanding very efficient probe hybridization. For this reason, high affinity probes should improve FRET efficiency.
In conclusion, linear 2′ OMe RNA antisense probes promise to be a general applicable probe type allowing the detection of various endogenous nuclear RNAs in living cells, without interfering with cell vitality. In addition, we demonstrate that simultaneous detection of RNA species with GFP-tagged proteins is very feasible, opening a way to study the molecular interactions involved in RNA transcription, processing and transport in living cells.
Acknowledgments
ACKNOWLEDGEMENTS
We would like to thank S. Snaar, S. Khazen, H. Albus and E. Manders for their excellent technical assistance, J. Bonnet and H. Vrolijk for their expertise in digital imaging microscopy and C. Jost for her helpful comments on the manuscript. We would like to thank Michel Dechamps and Dario Largana (Eurogentec) for technical assistance in the synthesis of the probes used in this study. C. Molenaar is supported by the Dutch Scientific Organization NWO program ‘4D imaging of living cells and tissues’, grant no. 901-34-144.
References
- 1.Lamond A.I. and Earnshaw,W.C. (1998) Structure and function in the nucleus. Science, 280, 547–553. [DOI] [PubMed] [Google Scholar]
- 2.Verschure P.J., van Der Kraan,I., Manders,E.M. and van Driel,R. (1999) Spatial relationship between transcription sites and chromosome territories. J. Cell Biol., 147, 13–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Leonhardt H., Rahn,H.P., Weinzierl,P., Sporbert,A., Cremer,T., Zink,D. and Cardoso,M.C. (2000) Dynamics of DNA replication factories in living cells. J. Cell Biol., 149, 271–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dirks R.W., Hattinger,C.M., Molenaar,C. and Snaar,S.P. (1999) Synthesis, processing, and transport of RNA within the three-dimensional context of the cell nucleus. Crit. Rev. Eukaryot. Gene Expr., 9, 191–201. [DOI] [PubMed] [Google Scholar]
- 5.Misteli T. (2000) Cell biology of transcription and pre-mRNA splicing: nuclear architecture meets nuclear function. J. Cell Sci., 113, 1841–1849. [DOI] [PubMed] [Google Scholar]
- 6.Tsukamoto T., Hashiguchi,N., Janicki,S.M., Tumbar,T., Belmont,A.S. and Spector,D.L. (2000) Visualization of gene activity in living cells. Nature Cell Biol., 2, 871–878. [DOI] [PubMed] [Google Scholar]
- 7.Zink D., Cremer,T., Saffrich,R., Fischer,R., Trendelenburg,M.F., Ansorge,W. and Stelzer,E.H. (1998) Structure and dynamics of human interphase chromosome territories in vivo. Hum. Genet., 102, 241–251. [DOI] [PubMed] [Google Scholar]
- 8.Phair R.D. and Misteli,T. (2000) High mobility of proteins in the mammalian cell nucleus. Nature, 404, 604–609. [DOI] [PubMed] [Google Scholar]
- 9.Snaar S., Wiesmeijer,K., Jochemsen,A.G., Tanke,H.J. and Dirks,R.W. (2000) Mutational analysis of fibrillarin and its mobility in living human cells. J. Cell Biol., 151, 653–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pederson T. (1999) Movement and localization of RNA in the cell nucleus. FASEB J., 13 (supp. 2), S238–S242. [DOI] [PubMed] [Google Scholar]
- 11.Pederson T. (2001) Fluorescent RNA cytochemistry: tracking gene transcripts in living cells. Nucleic Acids Res., 29, 1013–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bassell G.J., Oleynikov,Y. and Singer,R.H. (1999) The travels of mRNAs through all cells large and small. FASEB J., 13, 447–454. [DOI] [PubMed] [Google Scholar]
- 13.Dirks R.W., Molenaar,C. and Tanke,H.J. (2001) Methods for visualizing RNA processing and transport pathways in living cells. Histochem. Cell Biol., 115, 3–11. [DOI] [PubMed] [Google Scholar]
- 14.Ainger K., Avossa,D., Diana,A.S., Barry,C., Barbarese,E. and Carson,J.H. (1997) Transport and localization elements in myelin basic protein mRNA. J. Cell Biol., 138, 1077–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rook M.S., Lu,M. and Kosik,K.S. (2000) CaMKIIalpha 3′ untranslated region-directed mRNA translocation in living neurons: visualization by GFP linkage. J. Neurosci., 20, 6385–6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beach D.L., Salmon,E.D. and Bloom,K. (1999) Localization and anchoring of mRNA in budding yeast. Curr. Biol., 9, 569–578. [DOI] [PubMed] [Google Scholar]
- 17.Bertrand E., Chartrand,P., Schaefer,M., Shenoy,S.M., Singer,R.H. and Long,R.M. (1998) Localization of ASH1 mRNA particles in living yeast. Mol. Cell, 2, 437–445. [DOI] [PubMed] [Google Scholar]
- 18.Tsuji A., Koshimoto,H., Sato,Y., Hirano,M., Sei-Iida,Y., Kondo,S. and Ishibashi,K. (2000) Direct observation of specific messenger RNA in a single living cell under a fluorescence microscope. Biophys. J., 78, 3260–3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sokol D.L., Zhang,X., Lu,P. and Gewirtz,A.M. (1998) Real time detection of DNA.RNA hybridization in living cells. Proc. Natl Acad. Sci. USA, 95, 11538–11543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Politz J.C., Tuft,R.A., Pederson,T. and Singer,R.H. (1999) Movement of nuclear poly(A) RNA throughout the interchromatin space in living cells. Curr. Biol., 9, 285–291. [DOI] [PubMed] [Google Scholar]
- 21.Paillasson S., Robert-Nicoud,M. and Ronot,X. (1996) Specific detection of RNA molecules by fluorescent in situ hybridization in living cells. Cell Biol. Toxicol., 12, 359–361. [DOI] [PubMed] [Google Scholar]
- 22.Carmo-Fonseca M., Pepperkok,R., Sproat,B.S., Ansorge,W., Swanson,M.S. and Lamond,A.I. (1991) In vivo detection of snRNP-rich organelles in the nuclei of mammalian cells. EMBO J., 10, 1863–1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pitts A.E. and Corey,D.R. (1998) Inhibition of human telomerase by 2′-O-methyl-RNA. Proc. Natl Acad. Sci. USA, 95, 11549–11554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Majlessi M., Nelson,N.C. and Becker,M.M. (1998) Advantages of 2′-O-methyl oligoribonucleotide probes for detecting RNA targets. Nucleic Acids Res., 26, 2224–2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matsuo T. (1998) In situ visualization of messenger RNA for basic fibroblast growth factor in living cells. Biochim. Biophys. Acta, 1379, 178–184. [DOI] [PubMed] [Google Scholar]
- 26.Tyagi S. and Kramer,F.R. (1996) Molecular beacons: probes that fluoresce upon hybridization. Nat. Biotechnol., 14, 303–308. [DOI] [PubMed] [Google Scholar]
- 27.Tyagi S., Bratu,D.P. and Kramer,F.R. (1998) Multicolor molecular beacons for allele discrimination. Nat. Biotechnol., 16, 49–53. [DOI] [PubMed] [Google Scholar]
- 28.Luhrmann R., Kastner,B. and Bach,M. (1990) Structure of spliceosomal snRNPs and their role in pre-mRNA splicing. Biochim. Biophys. Acta, 1087, 265–292. [DOI] [PubMed] [Google Scholar]
- 29.Boom R., Geelen,J.L., Sol,C.J., Raap,A.K., Minnaar,R.P., Klaver,B.P. and van der Noordaa,J. (1986) Establishment of a rat cell line inducible for the expression of human cytomegalovirus immediate-early gene products by protein synthesis inhibition. J. Virol., 58, 851–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van de Corput M.P., Dirks,R.W., van Gijlswijk,R.P., van de Rijke,F.M. and Raap,A.K. (1998) Fluorescence in situ hybridization using horseradish peroxidase-labeled oligodeoxynucleotides and tyramide signal amplification for sensitive DNA and mRNA detection. Histochem. Cell Biol., 110, 431–437. [DOI] [PubMed] [Google Scholar]
- 31.Dirks R.W., de Pauw,E.S. and Raap,A.K. (1997) Splicing factors associate with nuclear HCMV-IE transcripts after transcriptional activation of the gene, but dissociate upon transcription inhibition: evidence for a dynamic organization of splicing factors. J. Cell Sci., 110, 515–522. [DOI] [PubMed] [Google Scholar]
- 32.Dirks R.W., van de Rijke,F.M., Fujishita,S., van der,P.M. and Raap,A.K. (1993) Methodologies for specific intron and exon RNA localization in cultured cells by haptenized and fluorochromized probes. J. Cell Sci., 104, 1187–1197. [DOI] [PubMed] [Google Scholar]
- 33.Sleeman J.E. and Lamond,A.I. (1999) Nuclear organization of pre-mRNA splicing factors. Curr. Opin. Cell Biol., 11, 372–377. [DOI] [PubMed] [Google Scholar]
- 34.Tollervey D. and Kiss,T. (1997) Function and synthesis of small nucleolar RNAs. Curr. Opin. Cell Biol., 9, 337–342. [DOI] [PubMed] [Google Scholar]
- 35.Huang S., Deerinck,T.J., Ellisman,M.H. and Spector,D.L. (1994) In vivo analysis of the stability and transport of nuclear poly(A)+ RNA. J. Cell Biol., 126, 877–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fay F.S., Taneja,K.L., Shenoy,S., Lifshitz,L. and Singer,R.H. (1997) Quantitative digital analysis of diffuse and concentrated nuclear distributions of nascent transcripts, SC35 and poly(A). Exp. Cell Res., 231, 27–37. [DOI] [PubMed] [Google Scholar]
- 37.Chin D.J., Green,G.A., Zon,G., Szoka,F.C.,Jr and Straubinger,R.M. (1990) Rapid nuclear accumulation of injected oligodeoxyribonucleotides. New Biol., 2, 1091–1100. [PubMed] [Google Scholar]
- 38.Leonetti J.P., Mechti,N., Degols,G., Gagnor,C. and Lebleu,B. (1991) Intracellular distribution of microinjected antisense oligonucleotides. Proc. Natl Acad. Sci.USA, 88, 2702–2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lamond A.I. and Carmo-Fonseca,M. (1993) Localisation of splicing snRNPs in mammalian cells. Mol. Biol. Rep., 18, 127–133. [DOI] [PubMed] [Google Scholar]
- 40.Carmo-Fonseca M., Pepperkok,R., Carvalho,M.T. and Lamond,A.I. (1992) Transcription-dependent colocalization of the U1, U2, U4/U6, and U5 snRNPs in coiled bodies. J. Cell Biol., 117, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lawrence J.B., Singer,R.H. and Marselle,L.M. (1989) Highly localized tracks of specific transcripts within interphase nuclei visualized by in situ hybridization. Cell, 57, 493–502. [DOI] [PubMed] [Google Scholar]
- 42.Sixou S., Szoka,F.C.,Jr, Green,G.A., Giusti,B., Zon,G. and Chin,D.J. (1994) Intracellular oligonucleotide hybridization detected by fluorescence resonance energy transfer (FRET). Nucleic Acids Res., 22, 662–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fisher T.L., Terhorst,T., Cao,X. and Wagner,R.W. (1993) Intracellular disposition and metabolism of fluorescently-labeled unmodified and modified oligonucleotides microinjected into mammalian cells. Nucleic Acids Res., 21, 3857–3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Meunier L., Mayer,R., Monsigny,M. and Roche,A.C. (1999) The nuclear export signal-dependent localization of oligonucleopeptides enhances the inhibition of the protein expression from a gene transcribed in cytosol. Nucleic Acids Res., 27, 2730–2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sei-Iida Y., Koshimoto,H., Kondo,S. and Tsuji,A. (2000) Real-time monitoring of in vitro transcriptional RNA synthesis using fluorescence resonance energy transfer. Nucleic Acids Res., 28, e59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Iribarren A.M., Sproat,B.S., Neuner,P., Sulston,I., Ryder,U. and Lamond,A.I. (1990) 2′-O-alkyl oligoribonucleotides as antisense probes. Proc. Natl Acad. Sci. USA, 87, 7747–7751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McKay R.A., Miraglia,L.J., Cummins,L.L., Owens,S.R., Sasmor,H. and Dean,N.M. (1999) Characterization of a potent and specific class of antisense oligonucleotide inhibitor of human protein kinase C-alpha expression. J. Biol. Chem., 274, 1715–1722. [DOI] [PubMed] [Google Scholar]
- 48.Elayadi A.N., Demieville,A., Wancewicz,E.V., Monia,B.P. and Corey,D.R. (2001) Inhibition of telomerase by 2′-O-(2-methoxyethyl) RNA oligomers: effect of length, phosphorothioate substitution and time inside cells. Nucleic Acids Res., 29, 1683–1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nielsen P.E., Egholm,M. and Buchardt,O. (1994) Peptide nucleic acid (PNA). A DNA mimic with a peptide backbone. Bioconjug. Chem., 5, 3–7. [DOI] [PubMed] [Google Scholar]
- 50.Fang X., Li,J.J. and Tan,W. (2000) Using molecular beacons to probe molecular interactions between lactate dehydrogenase and single-stranded DNA. Anal. Chem., 72, 3280–3285. [DOI] [PubMed] [Google Scholar]
- 51.Sixou S., Szoka,F.C.J., Green,G.A., Giusti,B., Zon,G. and Chin,D.J. (1994) Intracellular oligonucleotide hybridization detected by fluorescence resonance energy transfer (FRET). Nucleic Acids Res., 22, 662–668. [DOI] [PMC free article] [PubMed] [Google Scholar]