Abstract
Background:
Chronic rhinosinusitis (CRS) is a heterogeneous inflammatory disorder in which many pathways contribute to end-organ disease. Small proline-rich proteins (SPRR) are polypeptides that have recently been shown to contribute to epithelial biomechanical properties relevant in T-helper type 2 inflammation. There is evidence that genetic polymorphism in SPRR genes may predict the development of asthma in children with atopy and, correlatively, that expression of SPRRs is increased under allergic conditions, which leads to epithelial barrier dysfunction in atopic disease.
Methods:
RNAs from uncinate tissue specimens from patients with CRS and control subjects were compared by RNA sequencing by using Ingenuity Pathway Analysis (n = 4 each), and quantitative polymerase chain reaction (PCR) (n = 15). A separate cohort of archived sinus tissue was examined by immunohistochemistry (n = 19).
Results:
A statistically significant increase of SPRR expression in CRS sinus tissue was identified that was not a result of atopic presence. SPRR1 and SPRR2A expressions were markedly increased in patients with CRS (p < 0.01) on RNA sequencing, with confirmation by using real-time PCR. Immunohistochemistry of archived surgical samples demonstrated staining of SPRR proteins within squamous epithelium of both groups. Pathway analysis indicated tumor necrosis factor (TNF) alpha as a master regulator of the SPRR gene products.
Conclusion:
Expression of SPRR1 and of SPRR2A is increased in mucosal samples from patients with CRS and appeared as a downstream result of TNF alpha modulation, which possibly resulted in epithelial barrier dysfunction.
Keywords: Chronic rhinosinusitis, sinonasal inflammation, sinusitis, SPRR, TNF-α
Chronic rhinosinusitis (CRS) is the second most common chronic condition reported in the United States and affects 31 million people (i.e., an estimated 12% of the population) and leads to a significantly decreased quality of life.1 Despite comparable quality-of-life alterations to diseases, such as congestive heart failure, chronic obstructive pulmonary disease, and chronic back pain, a thorough understanding of CRS etiology is lacking.2 Coupled with the current emphasis on medical cost reduction and a direct national health care cost of $8.6 billion per year, CRS is a disease that is in justifiable need of further understanding and novel evidenced-based management options.3 Although the pathogenesis of CRS remains poorly understood, the evidence to date points toward a multifactorial process instigated and/or maintained by a dysfunctional host-environment interaction. As such, it is also likely that CRS is an end point of disease manifested by one of many possible pathophysiologic processes. To date, many studies of CRS pathophysiology have focused on single genes or pathways, with little ability to describe complex interactions or to evaluate the relative importance of the numerous simultaneously occurring processes.
The contribution of various inflammatory pathways, remodeling processes, immunoglobulin E production, microorganisms, and epithelial barrier malfunctions have all been proposed as etiologic mechanisms in CRS.4,5 Given such heterogeneity found in CRS, the general current approach is to explore tissue dysfunction at the microscopic level according to subclassifications of disease.5,6 One such subclass is patients with CRS and with atopy. The normal mechanical barrier of the sinonasal mucosa consists of pseudostratified epithelium, motile cilia, respiratory epithelial cells linked by apical tight junctions, and a mucus blanket. Dysfunction of this barrier has been indicated to be a major factor in the etiology of inflammatory disorders of the respiratory epithelium, classically described in asthma.6 Although the exact link between atopic disease and CRS remains unknown, there is some indication that a common underlying mechanism could be at play, given that patients with CRS have at least a 20% prevalence of concurrent asthma and up to 90% of patients with asthma have abnormal findings on computed tomography (CT) of the sinuses.7
Small proline-rich proteins (SPRR) are a group of proteins found in epithelial cells that help make up the cornified layer in squamous epithelium. Previous studies in an allergen-induced murine asthma model found interleukin (IL) 13 dependent increases in SPRR expression.8 In addition, a single nucleotide polymorphism in the SPRR2b gene has been shown, from two independent populations, to be predictive for asthma among children with eczema.9 We hypothesize that, although many genes are dysregulated in CRS sinus tissue, including a number of those found in allergy and asthma, a few genes and pathways will demonstrate a higher relative importance. Here we aimed to identify the broader gene pathways that are relevant and illustrate how single genes or gene families can contribute to pathway dysfunction. To test this hypothesis, we evaluated human sinonasal mucosa for differences in gene expression of inflammatory targets in patients with CRS and allergic inflammation by using unbiased RNA sequencing (RNA-seq), followed by molecular and tissue examination to confirm the findings of sequencing.
METHODS
Patient Sampling
Four adults with CRS and allergic rhinitis (AR) who underwent endoscopic sinus surgery were compared with four adult control subjects who underwent endoscopic dacryocystorhinostomy for epiphora. Demographic and clinical data recorded included age, gender, presence of allergy or asthma, tobacco use, presence of polyps, Lund-Mackay CT score, Lund-Kennedy endoscopy score, and 22-item Sino-Nasal Outcome Test. CRS was defined according to 2007 Adult Sinusitis Guidelines,10 and the diagnosis of AR was decided by classic patient history and positive result of skin-prick testing to at least one antigen.11 Asthma was confirmed on spirometric response with bronchodilator and methacholine administration, and was ruled out in those without a wheeze, cough, shortness of breath, and/or bronchodilator response. Exclusion criteria used in identifying subjects included recurrent acute rhinosinusitis, suspected diagnosis of cystic fibrosis, or immunocompromised state. Mucosa from the uncinate was collected from each patient during surgery and immediately transported to the laboratory. Biopsy tissue was placed in RNAlater (Qiagen, Valencia, CA) and stored at 4°C until RNA extraction.
Study protocols were reviewed and approved by the University of Colorado Institutional Review Board (HS11–1134), and written informed consent was obtained from each patient. All the authors contributed to this study and approve of the final manuscript.
RNA Extraction.
RNA was extracted from tissue by using the RNeasy Mini kit from Qiagen (Hilden, Germany), according to standard manufacturer protocol, including a 30-minute DNase I treatment at room temperature to remove genomic DNA.
RNA-seq
Samples that contained 500 ng of total RNA were used to construct complementary DNA (cDNA) libraries, and deep sequencing was performed by using the Illumina HiSeq 2000 (Illumina, San Diego, CA) RNA-seq method and protocol (see Supplemental Material for further details on methods and analysis). Power analysis was performed and, in this study, was 77% to detect a twofold change at a p value of 0.05.
Quantitative Polymerase Chain Reaction
Quantitative polymerase chain reaction (qPCR) was used to evaluate expression levels of SPRR1 and SPRR2A in the uncinate tissue samples. Adequate total RNA from human biopsy specimens of the four patients with CRS and three of four control patients remained after the previous experiment and was reverse transcribed into cDNA by using the Biorad iScript kit (Biorad Laboratories, Inc., Hercules, CA). Similarly, total RNA was extracted and processed from uncinate tissue of a separate cohort of patients with CRS and with and without atopy (n = 4 each) to evaluate the potential effect of atopy on SPRR expression. Aliquots of 2 μL from the cDNA libraries were used in each PCR sample, performed in triplicate reactions by using the Applied Biosystems SYBR Green PCR Master Mix (Foster City, CA).
Primers (10 μM) were designed for SPRR1, SPRR2A, and the reference gene, GAPDH, and purchased from Integrated DNA Technologies (Coralville, IA). Primer sequences (5′ to 3′) for SPRR1 were as follows: sense CCA GCA GAA GAC CAA GCA GAA and antisense GCA AAT GGG ACT CAT ACG CAG AAT G. Primer sequences (5′ to 3′) for SPRR2A were as follows: sense AGT GCC AGC AGA AAT ATC CTC C and antisense TGC TCT TGG GTG GAT ACT TTG A. PCR was performed in the CFX Real-Time PCR Detection System (Biorad), with an initial 10-minute denaturation at 95°C, followed by 40 cycles of 15-second denaturation at 95°C and 60-second annealing and extension at 60°C. To validate the PCR, we included a no-template control (water). Quantitative analysis was performed by using the comparative cycle threshold (Ct) relative expression method (or ΔΔCt) in which the Ct values for our reference gene, GAPDH, were subtracted from the Ct of our target genes (SPRR1 and SPRR2A). The fold change was calculated by using the formula, fold change = 2−ΔΔCt.
Immunohistochemistry
Immunohistochemical staining was performed on archived sinus tissue from 6 control subjects who underwent endoscopic surgery for localized orbital or sellar pathology, and 13 patients with CRS and AR. Tissue was fixed in 10% buffered formalin and embedded into paraffin blocks. Tissue sections of 5 μm were prepared. After deparaffinization, antigen retrieval was performed by using heat-induced epitope retrieval in citrate buffer (pH 6). All subsequent steps were performed on the Dako automated immunostainer (Agilent, Santa Clara, CA). In brief, antibody staining was performed by using an antibody directed against SPRR2 (1:1000, catalog ALX-210–901-R100; Enzo Life Sciences, Farmingdale, NY) for 30 minutes, and antibody detection was performed by using the Bio SB (Santa Barbara, CA) rabbit polydetector horseradish peroxidase (HRP) (DAB catalog BSB0221). A no-primary antibody control was used to confirm specificity of secondary antibody detection. Descriptive analysis was performed by a board-certified surgical pathologist (C.D.C.), and semiquantitative scoring of the staining was attempted based on the tissue compartment (squamous epithelium, respiratory epithelium, inflammatory cells) that demonstrated the appropriate nuclear staining pattern.
Statistical Analysis
The qPCR data were evaluated for significance by using a two-tailed unpaired t-test. All tests of null hypotheses were evaluated at α = 0.05. After RNA-seq, derived sequences were analyzed by applying a custom computational pipeline that consisted of the open-source gSNAP, Cufflinks, and R for sequence alignment and ascertainment of differential gene expression.12–15 In short, the reads generated were mapped to the human genome (hg10) by gSNAP, expression (fragments per kilobase of transcript per million mapped reads; FPKM) derived by Cufflinks, and differential expression was analyzed with analysis of variance in R.16,17 Analyses of variance were performed to compare the expression levels of all four control subjects and all four subjects with CRS on a gene-by-gene basis. Once the gene list was formed, data were analyzed by using Ingenuity Pathway Analysis (Qiagen, Redwood City, CA).
RESULTS
Demographics are shown in Table 1. The subject groups used for RNA extraction had similar mean ages (51 versus 50 years); CRS-specific symptom scores by using the 22-item Sino-Nasal Outcome Test questionnaire were notably higher in the diseased group (median, 81.5 [range, 71–87]) compared with controls (median, 5.5 [range, 2–10]), as expected. Two of the four patients with CRS also had a history of comorbid asthma.
Table 1.
Subject demographics (n = 4, each group)
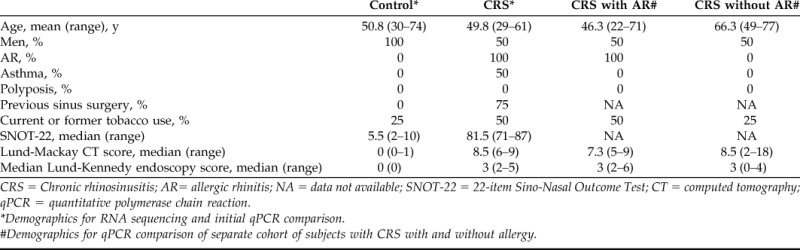
CRS = Chronic rhinosinusitis; AR= allergic rhinitis; NA = data not available; SNOT-22 = 22-item Sino-Nasal Outcome Test; CT = computed tomography; qPCR = quantitative polymerase chain reaction.
*Demographics for RNA sequencing and initial qPCR comparison.
#Demographics for qPCR comparison of separate cohort of subjects with CRS with and without allergy.
RNA-seq
RNA-seq produced a total of 151 million reads with an average of 20 million mapped reads per sample. The most significant of the 674 differentially expressed genes identified are presented in Table 2. A large and statistically significant increase in expression of SPPR genes was found in patients with CRS. Four of the top 10 upregulated genes in CRS were in the SPRR gene family; SPRR2A was the most discrepant gene product (a fold change of 50.06; p = 0.009), followed by SPRR1B (a fold change of 22.57; p = 0.002) and SPRR1A (a fold change of 15.30; p = 0.006). Transcription patterns associated with CRS tissue samples point to major signaling pathways associated with inflammation and cell proliferation that involve a number of well-characterized cytokines and transcription factors. An upstream regulator analysis within Ingenuity Pathway Analysis identified TNF-α as the most activated master regulator, with a relative activation z-score notably higher than other pathways (Table 3). These analyses indicated that the SPRR family of genes (SPRR1A, SPRR1B, SPRR2A, and SPRR3) were regulated by TNF-α and represent end points in such cascades, which indicated that they do not seem to have primary regulatory roles in CRS but rather may have structural importance to cells downstream in the response.
Table 2.
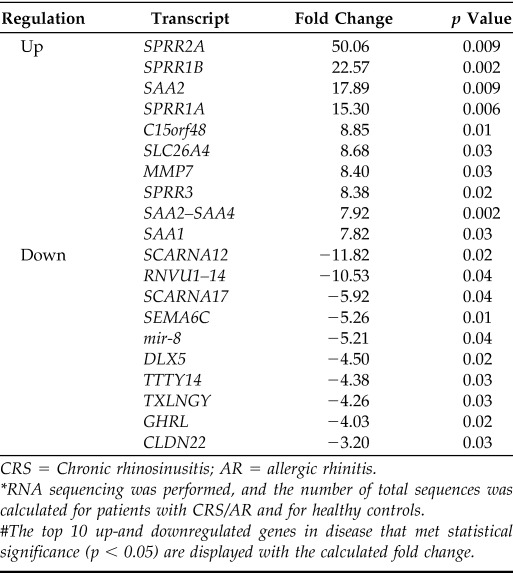
CRS = Chronic rhinosinusitis; AR = allergic rhinitis.
*RNA sequencing was performed, and the number of total sequences was calculated for patients with CRS/AR and for healthy controls.
The top 10 up-and downregulated genes in disease that met statistical significance (p < 0.05) are displayed with the calculated fold change.
Table 3.
Network pathway analysis of RNA sequencing of sinonasal mucosa from patients with CRS/AR and healthy controls*
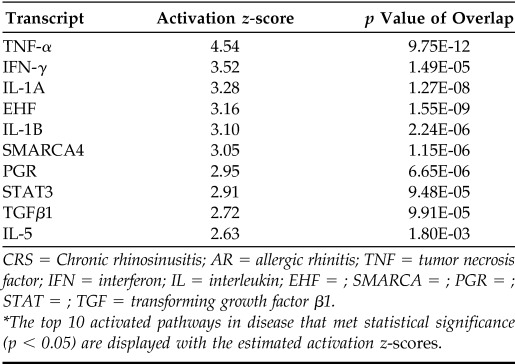
CRS = Chronic rhinosinusitis; AR = allergic rhinitis; TNF = tumor necrosis factor; IFN = interferon; IL = interleukin; EHF = ; SMARCA = ; PGR = ; STAT = ; TGF = transforming growth factor β1.
The top 10 activated pathways in disease that met statistical significance (p < 0.05) are displayed with the estimated activation z-scores.
qPCR
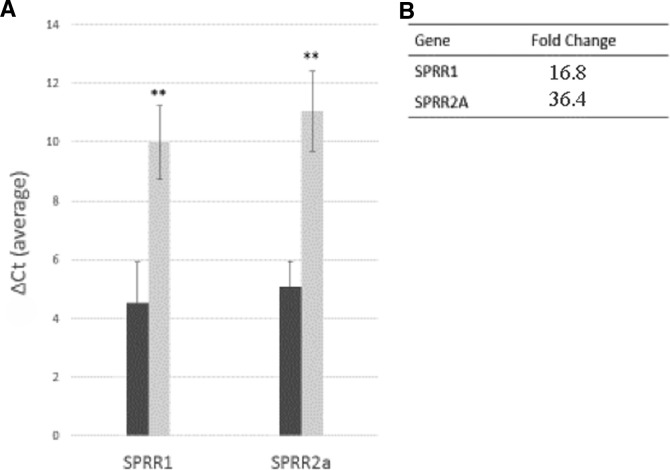
Given the significant dysregulation seen with the SPRR gene family, we performed qPCR to assess biologic variation in individual patients and to confirm the initial findings. This confirmed altered gene expression for SPRR1 and SPRR2A observed in our deep sequencing analysis (Fig. 1); the fold change in expression in this cohort was calculated to be 16.8 for SPRR1 and 36.4 for SPRR2A. To examine the potential effect of atopy in CRS on SPRR expression, we compared a separate group of patients with CRS and with and without allergy (n = 4 each), and found a higher fold-change expression in subject with CRS and without allergy (a fold change of 4.1 for SPRR1 and 7.2 for SPRR2a; p = 0.5 and 0.2, respectively).
Figure 1.
Quantitative analysis of dysregulated expression of small proline-rich proteins (SPRR) in sinonasal mucosa of patients with CRS/AR and of healthy controls. (A) Quantitative polymerase chain reaction (qPCR) analysis of SPRR1 and SPRR2A in uncinate mucosa from diseased versus the control patients. The ΔCt values (average of triplicates) from four diseased (dark gray bars) and three healthy controls (light gray bars) were averaged. All data are represented as mean ± SE. **p < 0.05. (B) Calculated fold change of SPRR1 and SPRR2A gene expression from qPCR data. Comparisons were made as diseased versus control fold change, calculated by using normalization to housekeeping gene (2−ΔΔCt).
Immunohistochemistry
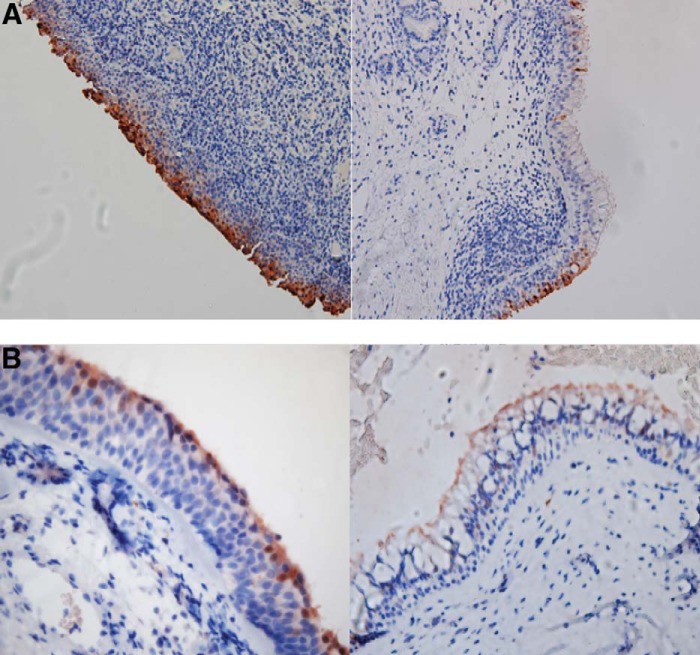
Given our observed increase in SPRR expression in patients with CRS at the mRNA level, we performed immunohistochemical staining of archived sinus tissue with an anti-SPRR2 antibody to localize the protein within the epithelial layer and assess for observable differences at the protein level. Consistent staining was seen in the epithelium of both patients with CRS and healthy controls however, there was no pattern of expression indicative of a specific compartment or cell type (Fig. 2). Staining was limited to areas of squamous epithelium, so direct quantitative assessment was not feasible due to potential sampling error of biopsied tissue.
Figure 2.
Immunohistochemical staining for small proline-rich protein 2 (SPRR2) expression in sinus tissue specimens. (A) Low-power view of SPRR2 antibody staining (dark brown) of archived sinus tissue from a subject with chronic rhinosinusitis and/or allergic rhinitis. A representative area of squamous epithelium (left) and ciliated respiratory epithelium (right) demonstrates intense and consistent staining of the squamous epithelium when compared with the respiratory epithelium. (B) High-power view of SPRR2 antibody staining (dark brown) of archived sinus specimens from a healthy control subject with similar findings. A representative area of squamous epithelium (left) and ciliated respiratory epithelium (right) is shown.
DISCUSSION
To identify relative molecular contributions in CRS, particularly in the presence of atopy, we used an RNA-seq analysis of uncinate mucosal tissues from healthy subjects and subjects with CRS. Our analysis revealed large magnitudes of dysregulation for SPRR1 and SPRR2A expression in disease, with extremely high statistical significance. On immunohistochemical evaluation, SPRR1a, SPRR2a, and SPRR2b, were expressed predominantly in squamous epithelium. These findings were consistent with a previous understanding of their roles in the formation of the insoluble cornified envelope, which provides structural integrity and limits epithelial permeability. We observed SPRR products consistently expressed in surgical samples from both control subjects and subjects with disease, and noted that expression was predominantly found in areas of squamous epithelium rather than respiratory epithelium. The lack of a distinctive visual SPRR expression pattern may indicate that it was only transiently expressed in the mucosa in response to specific stress and/or stimuli or, correlatively, that its expression was cell-cycle specific because this has been shown previously with SPRR1.18
The cyclic injury and remodeling of the sinonasal respiratory epithelium that occurs in CRS could also explain the upregulation of SPRRs because loss of healthy pseudostratified respiratory epithelium and continued barrier replacement occur in prolonged states of airway inflammation and repair, a process thoroughly reviewed earlier this year.19 Squamous metaplasia, the benign process of transformation from respiratory epithelium to squamous epithelium, is found in CRS specimens > 50% of the time; however, its presence does not seem to correlate with disease severity.20 A different study discovered squamous metaplasia in the septum and lateral nasal wall of patients who underwent septoplasty surgery and of a control group, and concluded that squamous metaplasia has a relationship to areas of complex and dynamic airflow,21 which indicated the possibility for squamous metaplasia found on the uncinate process accounted for some degree of SPRR expression in our tissue specimens.
At this point, relatively little is known about the functional role of SPRRs in squamous epithelium, and its role in CRS has not been evaluated. However, several studies8,22–25 indicate a role in atopic diseases. Zimmerman et al.8 showed that IL-13–associated allergic responses in the lung, which involved epithelial injury, and repair may be mediated by SPRR2, and proposed that the epithelial and goblet cell hyperplasia observed in asthma may be mediated by SPRR2. This is similar to the observations of de Koning et al.,22 who saw a strong upregulation of SPRR1 and SPRR2 in response to detergent-induced irritant contact dermatitis and tape stripping. SPRRs have also been linked to optimal tissue repair and wound healing via detoxification of reactive oxygen species and cell migration, mechanisms that are also at play in chronic sinonasal inflammation.23–25 The mechanisms behind these observations are poorly understood and warrant future investigation.
Most interesting are the results of analyses by using deep sequencing and pathway analysis that SPRR proteins are likely to be end products rather than mediators of the initial inflammatory response. They may be useful representative markers of CRS rather than major contributors to disease. However, the generally strong upregulation of the expression of these genes seemed to reflect their structural involvement during inflammation. These implications and the technology used to ascertain these data are potentially highly relevant. Many proinflammatory pathways are activated in CRS, which makes this disease challenging to study. Our finding that TNF-α was the most highly activated of the major gene pathways further supported its use in cell culture and animal model experimental research into the inflammatory mechanisms of disease in CRS. In addition to dysregulation of known gene pathways in CRS, our findings indicated relevance for other major pathways, which require further study, such as progesterone receptor and E-twenty six (ETS) transcription factor mechanisms (Table 3). In fact, e.g., these relatively unstudied pathways seemed more dysregulated in our cohort than the highly studied IL-5 and TGF-β pathways.
Here, we identified a strong association of the SPRR gene family with inflammation in patients with CRS and illustrated its broader context in the setting of TNF-α driven inflammation by using modern research and bioinformatics techniques. Given the clinical correlations between AR, asthma, and CRS, analysis of our data identified SPRR1 and SPRR2 as potential downstream targets of interest for further studies into tissue dysfunction in CRS. The most obvious limitation of the current study was the small sample size and potential heterogeneity of the subjects; as sequencing and informatics technology improves and costs decrease, experiments such as these can be repeated with larger sample sizes to test multiple CRS subgroups. Despite the small cohort, we identified extremely large fold changes with high statistical significance, which indicated that a larger N would not be necessary to confirm these findings but might uncover others that did not surpass statistical cutoffs in our analysis pipeline.
In the current exploratory study, we could not be certain that genes such as these would influence the disease process, but, instead, they may appear as a manifestation of the inflammatory response. Additional limitations included those associated with human tissue study in CRS, viz., the limited ability to select a well-defined cohort with a shared phenotype and endotype. Here, we attempted to use the clinical phenotype of nonpolypoid CRS and allergy, although there were other factors that were not controlled for, e.g., previous sinus surgery or nasal steroid medication use. In the immunohistochemical staining, representative sampling of the entire disease process or time in the disease development process was difficult to estimate. Finally, it is important to note that pathway analysis software is not always consistently reliable or reproducible because it depends on a network generation process that varies among providers.26 Future studies will focus on examining SPRR function at the protein level as well as elucidating the role of upstream inflammatory regulators in the creation of epithelium barrier dysfunction.
CONCLUSION
We observed a large and significant increase in SPRR gene expression in surgical specimens collected from patients with CRS and with atopy, and demonstrated its expression within squamous epithelium of the sinuses. These results raised the possibility of SPRRs having some role in sinonasal mucosal dysfunction in response to allergens and irritants in CRS, as has been noted in other atopic diseases. In addition, we described relative importances of potential disease-related pathways by using modern bioinformatics technology. Further study is warranted to determine the role of the SPRR gene family as a mediator in allergic sinonasal inflammation and its relation to upstream regulators.
ACKNOWLEDGMENTS
Author contributions included the following: (1) conception and design of study: V.R. Ramakrishnan, S.E. Cooper, H.P. Barham, C.B. Anderson, and S.C. Kinnamon; (2) data generation: V.R. Ramakrishnan, J.R. Gonzalez, S.E. Cooper, H.P. Barham, C.B. Anderson, C.D. Cool, J.D. Diller, and K. Jones; and (3) analysis and interpretation of data: all the authors; and (4) preparation and critical review of manuscript: V.R. Ramakrishnan, J.R. Gonzalez, C.B. Anderson, J.D. Diller, and S.C. Kinnamon.
Footnotes
This research was supported by an NIH/NIDCD training grant to the Department of Otolaryngology at the University of Colorado (T32DC012280), an NIH/NIDCD grant to V.R. Ramakrishnan (K23DC014747), and an American Rhinologic Society Resident Research Grant to H.P. Barham (235218)
The authors have no conflicts of interest to declare pertaining to this article
Supplemental data available at www.IngentaConnect.com
REFERENCES
- 1. Blackwell DL, Lucas JW, Clarke TC. Summary health statistics for U.S. adults: National Health Interview Survey, 2012. Vital Health Stat 10 (260):1–161, 2014. [PubMed] [Google Scholar]
- 2. Gliklich RE, Metson R. The health impact of chronic sinusitis in patients seeking otolaryngologic care. Otolaryngol Head Neck Surg 113:104–109, 1995. [DOI] [PubMed] [Google Scholar]
- 3. Bhattacharyya N. Incremental health care utilization and expenditures for chronic rhinosinusitis in the United States. Ann Otol Rhinol Laryngol 120:423–427, 2011. [DOI] [PubMed] [Google Scholar]
- 4. Van Crombruggen K, Zhang N, Gevaert P, et al. Pathogenesis of chronic rhinosinusitis: Inflammation. J Allergy Clin Immunol 128:728–732, 2011. [DOI] [PubMed] [Google Scholar]
- 5. Fokkens WJ, Lund VJ, Mullol, et al. European Position Paper on Rhinosinusitis and Nasal Polyps 2012. Rhinology 50(23), 2012. [PubMed] [Google Scholar]
- 6. Xiao C, Puddicombe SM, Field S, et al. Defective epithelial barrier function in asthma. J Allergy Clin Immunol 128:549–556.e1–e12, 2011. [DOI] [PubMed] [Google Scholar]
- 7. Pearlman AN, Chandra RK, Chang D, et al. Relationships between severity of chronic rhinosinusitis and nasal polyposis, asthma, and atopy. Am J Rhinol Allergy 23:145–148, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zimmermann N, Doepker MP, Witte DP, et al. Expression and regulation of small proline-rich protein 2 in allergic inflammation. Am J Respir Cell Mol Biol 2005:428–435, 2005. [DOI] [PubMed] [Google Scholar]
- 9. Epstein TG, LeMasters GK, Bernstein DI, et al. Genetic variation in small proline rich protein 2B as a predictor for asthma among children with eczema. Ann Allergy Asthma Immunol 108:145–150, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Rosenfeld RM, Andes D, Bhattacharyya N, et al. Clinical practice guideline: Adult sinusitis. Otolaryngol Head Neck Surg 137(suppl.):S1–S31, 2007. [DOI] [PubMed] [Google Scholar]
- 11. Boyd EL. Patient history. In Allergy and Immunology: An Otolaryngic Approach. Krouse JH, Chadwick SJ, Gordon BR, Dereberry JM. (Eds.). Philadelphia, PA: Lippincott Williams and Wilkins, 81–86, 2002. [Google Scholar]
- 12. Henderson HH, Timberlake KB, Austin ZA, et al. Occupancy of RNA polymerase II (S5P) and RNA polymerase II (S2P) on VZV genes 9, 51 and 66 is independent of transcript abundance and polymerase location within the gene. J Virol 90:1231–1243, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bradford AP, Jones KL, Kechris K, et al. Joint miRNA/mRNA expression profiling reveals changes consistent with development of dysfunctional corpus luteum after weight gain. PLoS One 10: e0135163, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Maycotte P, Jones KL, Goodall ML, et al. Autophagy supports breast cancer stem cell maintenance by regulating IL6 secretion. Mol Cancer Res 13:651–658, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Baird NL, Bowlin JL, Cohrs RJ, et al. Comparison of VZV RNA sequences in human neurons and fibroblasts. J Virol 88:5877–5880, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wu TD, Nacu S. Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics 26:873–881, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Trapnell C, Williams BA, Pertea G, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28:511–515, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tesfaigzi J, Carlson DM. Cell cycle-specific expression of G(0)SPR1 in Chinese hamster ovary cells. Exp Cell Res 228:277–282, 1996. [DOI] [PubMed] [Google Scholar]
- 19. Schleimer RP. Immunopathogenesis of chronic rhinosinusitis and nasal polyposis. Annu Rev Pathol 12:331–357, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Soler ZM, Sauer DA, Mace J, Smith TL. Relationship between clinical measures and histopathologic findings in chronic rhinosinusitis. Otolaryngol Head Neck Surg 141:454–461, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kamani T, Yilmaz T, Surucu S, et al. Histopathological changes in nasal mucosa with nasal septum deviation. Eur Arch Otorhinolaryngol 271:2969–2974, 2014. [DOI] [PubMed] [Google Scholar]
- 22. de Koning HD, van den Bogaard EH, Bergboer JG, et al. Expression profile of cornified envelope structural proteins and keratinocyte differentiation-regulating proteins during skin barrier repair. Br J Dermatol 166:1245–1254, 2012. [DOI] [PubMed] [Google Scholar]
- 23. Fordham MT, Mulligan JK, Casey SE, et al. Reactive oxygen species in chronic rhinosinusitis and secondhand smoke exposure. Otolaryngol Head Neck Surg 149:633–638, 2013. [DOI] [PubMed] [Google Scholar]
- 24. Yan Y, Gordon WM, Wang DY. Nasal epithelial repair and remodeling in physical injury, infection, and inflammation. Curr Opin Otolaryngol Head Neck Surg 21:263–270, 2013. [DOI] [PubMed] [Google Scholar]
- 25. Vermeij WP, Backendorf C. Skin cornification proteins provide global link between TOS detoxification and cell migration during wound healing. PLoS One 5:e11957, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Thomas S, Bonchev D. A survey of current software for network analysis in molecular biology. Hum Genomics 4:353–360, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.