Abstract
Context:
Oral squamous cell carcinoma (OSCC) is the sixth most common cancer in the world. As per previous studies, most patients who develop oral cancer are elderly males who are heavy users of tobacco and alcohol; however, the incidence is increasing in younger individuals and in those who neither smoke nor drink. Many of the genes that code for the detoxification enzymes are polymorphic with abnormal activity profiles.
Aims and Objectives:
The aim of this study was to evaluate the risk of development of oral leukoplakia (OLP) and OSCC in glutathione S-transferase polymorphisms genes in the east coast of Andhra Pradesh population with tobacco consumption habit and habit-free controls using polymerase chain reaction (PCR-restriction fragment length polymorphism).
Materials and Methods:
This study included 15 patients each with histologically proven epithelial dysplasia and OSCC and compared with age- and gender-related controls with no tobacco habits in any form. A volume of 2 ml of blood sample was collected into presterilized vials containing ethylenediaminetetracetic acid from each individual under aseptic conditions. DNA extraction was done from whole blood, and PCR was performed. Statistical analysis was performed using Chi-square test and odds ratio (OR) and 95% confidence intervals (CIs).
Results:
The results are suggestive that glutathione S-transferase mu1 (GSTM1) null was associated with increased risk of OLP (OR = 5.5, 95% CI = 1.14–26.41, P = 0.021) and OSCC (OR = 11, 95% CI = 1.99–60.5, P = 0.021). Glutathione S-transferase theta1 (GSTT1) null genotype was associated with increased risk of OLP (OR = 2.154, 95% CI = 0.74–26.672, P > 0.99) and OSCC (OR = 2.154, 95% CI = 0.74–26.672, P > 0.99). The glutathione S-transferase mu3 (GSTM3) AB + BB genotypes appear to be risk factors for OSCC (OR = 1.31, 95% CI = 0.31–5.58, P = 0.7) although statistically insignificant.
Conclusion:
Hence to conclude, because of small sample size in the present study, statistically insignificant results were found and this study failed to observe the relationship between GSTM3 and GSTT1 polymorphism and risk of developing OSCC and positive relationship was observed with GSTM1 polymorphism and risk of developing OSCC.
Keywords: Glutathione S-transferases, polymerase chain reaction, polymorphisms
Introduction
Oral potentially malignant disorders are OLP, erythroplakia, and oral submucous fibrosis of which OLP is the most common.[1] The incidence rates of OLP are increasing for many years as the commercially available tobacco products are freely available in many Asiatic countries including India.[2] The prevalence of tobacco use among men is high (>50%) from almost all parts of India, the prevalence is more in rural than in urban areas.[3] Incidence rates of OLP per 1000 individuals per year varied from 1.1 to 2.4 among men and 0.2–1.3 among women.[4] Malignant transformation rate of OLP worldwide is 3.6% and in India varies from 0.13% to 10%.[3,5] Globally, Oral cancer ranks sixth in the cancer burden.[6] In India, incidence rates of oral squamous cell carcinoma (OSCC) are 7–17 per 100,000 persons per year. Around 75,000–80,000 new cases develop in India every year.[7]
As an early sign of damage to oral mucosa by the carcinogens in tobacco smokers and chewers, visible alterations occur and appear usually as white patches with or without red patches. These changes are usually precancerous.[1] OLP, the common premalignant lesion among the smokers. In 2005, the World Health Organization defined it as “a white plaque of questionable risk having excluded (other) known diseases or disorders that carry no increased risk for cancer.”[8] This lesion is easily accessible to diagnosis and can be considered as indicators of oral cancer risk. The consequence of malignant transformation of OLP is OSCC. OSCC is defined as a malignant epithelial neoplasm exhibiting squamous differentiation as characterized by the formation of keratin and/or the presence of intercellular bridges.[7]
This cancer is a global burden to humanity, and lifestyle-related addictive habits such as tobacco smoking, chewing, and alcohol consumption are the main culprits. Chemical carcinogens from smoking and chewing of tobacco and betel-quid act synergistically in oral carcinogenesis and that persons with mixed habits form a substantially high-risk population.[9]
Polycyclic aromatic hydrocarbons (PAHs), aldehydes, ethylene oxide, aromatic amines, and nitrosamines are thought to be carcinogenic components present in tobacco smoke. In general, PAHs are carcinogens that act locally and other carcinogenic agents act systemically. Some PAHs, such as benzopyrene (BP), have powerful carcinogenic activity.[10] However, chewing tobacco with betel quid increases concentrations of carcinogenic tobacco-specific nitrosamines and reactive oxygen species in mouth.[11]
Chemicals that initiate carcinogenesis can act directly or indirectly. Direct carcinogens do not require metabolic conversion to become carcinogenic and are weak carcinogenic agents such as alkylating and acylating. Indirect carcinogens require metabolic conversion to become active carcinogen such as PAH, BP, and ethylene oxides.[12]
Cytochrome P450s are mostly the Phase I enzymes involved in the activation pathway in the liver.[13] These enzymes catalyze the introduction of a functional group such as an epoxide into PAH and BP, which offers electrophilic intermediates that can damage DNA.[7,13]
Elimination of these reactive electrophilic intermediate metabolites requires action by Phase II enzymes such as N-acetyltransferases,[13] glutathione S-transferases (GSTs)[4,12] that converts them into water-soluble excretable compounds. Human GSTs are divided into three main families: Cytosolic or soluble, mitochondrial, and membrane-bound microsomal. Sixteen members of GST superfamily have been identified in humans so far.[14] Of which four members of the GST genes – glutathione S-transferase mu3 (GSTM3), glutathione S-transferase theta1 (GSTT1), glutathione S-transferase mu1 (GSTM1), and glutathione S-transferase pi1 (GSTP1) display polymorphisms that have been associated with increased risk for cancers.[4,15,16] Among them, GSTM and GSTT have attracted most of the interest, mainly because they are involved in detoxification of reactive metabolites of carcinogenic substances from tobacco smoke.[17]
The GSTT1 gene is located at 22q11.23, and its polymorphism is characterized by a deletion of almost the entire gene, causing the absence of the protein GSTT1. This enzyme metabolizes various potential indirect carcinogens such as monohalomethanes and ethylene oxide present in tobacco smoke and some endogenously generated reactive products derived from lipid peroxides.[15]
The gene cluster of GSTM1-GSTM5 is localized on chromosome 1p13.[15] The GSTM1 catalyzes the conjugation of glutathione to a variety of electrophilic compounds, including carcinogens and cytotoxic drugs. The homozygous GSTM1 null genotype results in the loss of expression of the GSTM1 enzyme and decreased capacity to detoxify carcinogens. GSTM3 plays a role in the metabolism of harmful agents such as PAH and BP.[4] GSTM3 gene has two alleles identified so far: GSTM3*A and GSTM3*B.[18] GSTM3*B has been postulated to regulate gene expression for GSTM3 protein.[17]
Materials and Methods
This study was done on 45 consecutive patients with 15 cases each of OLP, OSCC were selected from the outpatient department of GITAM Dental College and Hospital and Lions Cancer Hospital, Visakhapatnam, Andhra Pradesh, India, and 15 normal individuals without deleterious habits were taken as controls after obtaining ethical clearance from the committee of the college. A volume of 2 ml of blood was collected from histopathologically confirmed individuals from antecubital vein of patients with OLP, OSCC, and controls using 2 ml disposable syringe under aseptic conditions after obtaining informed consent from the patients. The blood samples were stored in ethylenediaminetetracetic acid vials and were kept in container packed with ice and protected from light until the samples were transported and stored at −20°C until DNA isolation. The blood sample of 200 μl was taken into a fresh sterilized 1.5 ml Eppendorf Tube. The DNA was isolated from the peripheral white blood cells.
Quantification of DNA was done using spectrophotometer. The ratio of 260/280 nm for each sample was calculated for both quality check and quantification of DNA. The samples with an acceptable range from 1.8 to 2.0 were used for polymerase chain reaction (PCR) studies. Simultaneously, the quality of DNA was checked by standard 1% agarose weight/volume electrophoresis. The genomic DNA isolated from the control, OLP, and OSCC samples were used as the template for the amplification of internal control gene (β-actin) along with GSTT1 and GSTM3 and β-globin as an internal control for GSTM1. Amplification was performed using β-actin universal primers, GSTT1, GSTM3 gene-specific primers in a multiplex PCR and β-globin as an internal control for GSTM1 gene separately in a conventional PCR (Corbett Research Australia). The sequences of primers used for PCR amplification are mentioned in Table 1.
Table 1.
Primers of β-actin, Glutathione S-transferase theta1, Glutathione S-transferase Mu3, β-globin, Glutathione S-transferase Mu1
DNA samples were mixed with the appropriate amount of gel loading buffer. The electrodes were connected to a power pack; running of the gel was done until the marker dyes have migrated to the desired distance. The gels were stained with ethidium bromide, and the dried gels were exposed to phosphorimager screen.
Statistical analysis
The comparison of GSTT1, GSTM1 null genotype, and GSTM3 polymorphisms among controls without habits, OLP and OSCC, was done using Statistical Package for Social Sciences Software 17 version (SPSS, Chicago, Il, USA). Statistical comparison of GSTT1, GSTM1 null genotype, and GSTM3 polymorphisms with tobacco habits in controls with habits and OLP, OSCC individuals was done and was analyzed using Chi-square test and Fischer's exact t-test. Odds ratio (OR) and 95% confidence intervals (CIs) were calculated for GSTT1, GSTM1 null genotypes, and GSTM3 polymorphisms in controls with habits and OLP, OSCC individuals.
Results
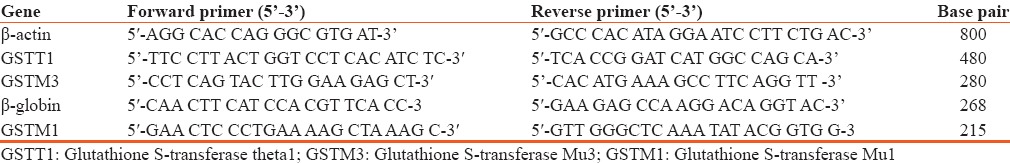
Distribution of GSTT1 genotype among controls, OLP and OSCC using Chi-square test, relative risk of GSTT1, OLP, OSCC, and controls with OR and 95% CI [Table 2].
Table 2.
Distribution of glutathione S-transferase theta1 genotype among controls, oral leukoplakia and oral squamous cell carcinoma using Chi-square test, relative risk of glutathione S-transferase theta1, oral leukoplakia and oral squamous cell carcinoma, and controls with odds ratio and 95% confidence interval
The PCR analysis showed that the GSTT1 null genotype was greater in OLP and OSCC (13.3%) in comparison to the controls (6.7%) indicating the absence of GSTT1 increases risk of developing OLP and OSCC (P value nonsignificant >0.99).
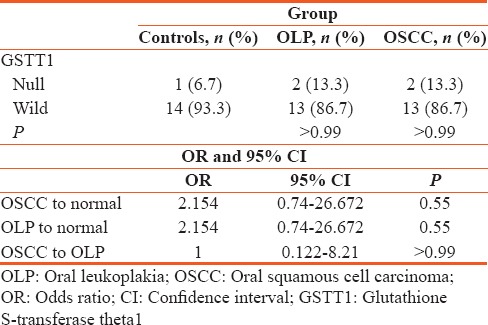
Distribution of GSTM1 genotype among controls, OLP, and OSCC using Chi-square test, relative risk of GSTM1, OLP, OSCC, and controls with OR and 95% CI [Table 3].
Table 3.
Distribution of glutathione S-transferase Mu1 genotype among controls, oral leukoplakia and oral squamous cell carcinoma using Chi-square test, relative risk of glutathione S-transferase Mu1, oral leukoplakia, oral squamous cell carcinoma, and controls with odds ratio and 95% confidence interval
The PCR analysis showed that the GSTM1 null genotype was greater in OLP (66.7%) and OSCC (80%) in comparison to the controls (26.7%) indicating null GSTM1 increases the risk of developing OLP and OSCC (P value: Statistically significant).
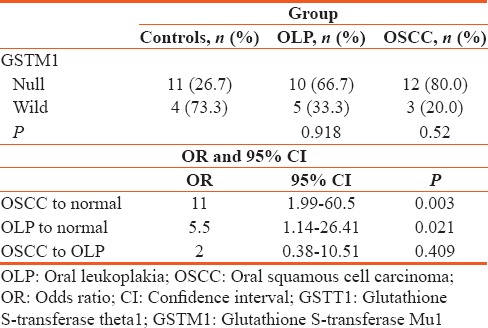
Distribution of GSTM3 genotype among controls, OLP, and OSCC using Chi-square test, relative risk of GSTM3 AB + BB, OLP, OSCC, and controls with OR, 95% CI [Table 4].
Table 4.
Distribution of glutathione S-transferase Mu3 genotype among controls, oral leukoplakia and oral squamous cell carcinoma using Chi-square test, relative risk of glutathione S-transferase Mu3 AB + BB, oral leukoplakia, oral squamous cell carcinoma and controls with odds ratio, 95% confidence interval
The PCR analysis showed that the GSTM3 AB + BB polymorphisms increase the risk of developing OLP and OSCC (P value nonsignificant).
Discussion
Some heavy smokers and chewers do not develop precancerous states and cancer even after prolonged periods of usage while such lesions are seen in early smokers and even in nonsmokers. Differences in carcinogen metabolism and DNA repair due to polymorphisms are suggested for this variance in susceptibility. Hence, individual's susceptibility to OLP and cancer is modulated by both environmental and genetic factors which are confirmed by molecular epidemiologic studies.[4]
In this study, we examined the association between polymorphisms in GSTs genes and the associated risk of OLP and OSCC among the east Coast of Andhra Pradesh tobacco users. The null genotype percentage varies in ethnic groups and also in geographical locations. In various previously conducted studies, the null genotype was reported to be deleted in 12%–27% of Brazilian population,[19,20] 48% of Taiwan population,[21] 18% of the US population.[13,22] Similarly in India, it was observed to be 16%–25% in Mumbai,[23,24,25] 8% in Thiruvananthapuram.[26] In the present study, GSTT1 null genotype was greater in OLP and OSCC (13.3%) in comparison to the controls (6.7%). Although the study is statistically insignificant, it suggests a positive association between GSTT1 null genotype and OLP and OSCC. One study conducted in 1999 by Cheng et al.,[22] in Houston, USA, found that 32.7% of cases and 17.5% of controls were null for GSTT1. These results are in relation to the present study and are in contrast to studies of Olshan et al.,[13] Buch et al.[24] on Mumbai population who found no difference of null genotype between cases and controls and interestingly Anantharaman et al.[25] even found that GSTT1 null genotype conferred reduced risk to OSCC. The OR associated with the GSTT1 null genotype was increased, (OR = 2.154; 95% CI = 0.74–26.672) although not significantly, the relative risk observed in the study was in relation to the studies conducted by Sreelekha et al.[26] (OR = 2.475; 95% CI = 0.2821–21.712), Cheng et al.[22] in Houston (USA) (OR = 2.27; 95% CI = 1.43–3.60). The same risk was also found in the studies of Hung et al.[21] showing an adjusted OR = 3.1; 95% CI = 0.9–10.9.
Studies conducted by Sikdar et al.,[4] Sreelekha et al.,[26] and Bathi et al.[27] in West Bengal, Kerala, and Karnataka, respectively, also confirmed that GSTT1 null had an increased risk for OLP and OSCC which are in correlation to the present study.
In the present study, GSTM1 null genotype, there was increased risk between the normal and OSCC subjects. The OR of null genotype in OSCC is two times greater than OLP and in OSCC is 11 times greater than the odds of null genotype in normal subjects, whereas odds of null genotype in OLP is 5.5 times greater than the odds of null genotype in normal subjects. These are in correlation with Duarte et al.[28] and Bathi et al.,[27] who found an association between the GSTM1 null genotype and an increased risk of development of OLP and OSCC.
In this study, we found a higher frequency of the GSTM3 AA genotype in OLP patients than in controls and a lower frequency of GSTM3 AB genotype in OLP patients than in controls. This was in correlation to the studies conducted by Sikdar et al.[4] and Majumder et al.[29] where higher frequency of the GSTM3 AA genotype was observed in cases than in controls. The OR associated with the GSTM3 AB + BB genotype was slightly increased, the relative risk observed in the study was in relation to the studies conducted by Majumder et al.,[29] Jain et al.[16] and Jourenkova-Mironova et al.[17] They found a 2-fold risk of laryngeal cancer associated with the GSTM3 (AB or BB) genotype in relation to the present study.
Conclusion
In spite of small sample size and certain other limitations, in this present study, we observed a modest association between polymorphisms of GSTT1, GSTM1, and GSTM3 genotypes. This study suggests that GSTT1, GSTM1 null genotype; GSTM3 AB + BB null genotypes apparently play a significant role in mediating susceptibility for developing OLP and OSCC.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
References
- 1.Garg KN, Raj V, Chandra S. Trends in frequency and duration of tobacco habit in relation to potentially malignant lesion: A 3 years retrospective study. J Oral Maxillofac Pathol. 2013;17:201–6. doi: 10.4103/0973-029X.119760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chadda R, Sengupta S. Tobacco use by Indian adolescents. Tob Induc Dis. 2002;1:111–9. doi: 10.1186/1617-9625-1-2-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reddy KS, Gupta PC. Report on Tobacco Control in India. Joint Report Supported by Ministry of Health and Family Welfare, Government of India, Centre of Disease Control and Prevention. USA: World Health Organization; 2004. Economic history of tobacco production: From colonial origins to contemporary trends; pp. 19–32. [Google Scholar]
- 4.Sikdar N, Paul RR, Roy B. Glutathione S-transferase M3 (A/A) genotype as a risk factor for oral cancer and leukoplakia among Indian tobacco smokers. Int J Cancer. 2004;109:95–101. doi: 10.1002/ijc.11610. [DOI] [PubMed] [Google Scholar]
- 5.Sridharan G. Epidemiology, control and prevention of tobacco induced oral mucosal lesions in India. Indian J Cancer. 2014;51:80–5. doi: 10.4103/0019-509X.134651. [DOI] [PubMed] [Google Scholar]
- 6.Warnakulasuriya S. Global epidemiology of oral and oropharyngeal cancer. Oral Oncol. 2009;45:309–16. doi: 10.1016/j.oraloncology.2008.06.002. [DOI] [PubMed] [Google Scholar]
- 7.Shafer WG, Hine MK, Levy BM. Benign and malignant tumors of oral cavity. In: Rajendran R, Sivapathasundaram B, editors. Shafer's Textbook of Oral Pathology. 6th ed. Livingstone: Elsevier Company; 2009. pp. 80–6. [Google Scholar]
- 8.Brouns ER, Baart JA, Bloemena E, Karagozoglu H, van der Waal I. The relevance of uniform reporting in oral leukoplakia: Definition, certainty factor and staging based on experience with 275 patients. Med Oral Patol Oral Cir Bucal. 2013;18:e19–26. doi: 10.4317/medoral.18756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Patil PB, Bathi R, Chaudhari S. Prevalence of oral mucosal lesions in dental patients with tobacco smoking, chewing, and mixed habits: A cross-sectional study in South India. J Family Community Med. 2013;20:130–5. doi: 10.4103/2230-8229.114777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ko YC, Huang YL, Lee CH, Chen MJ, Lin LM, Tsai CC. Betel quid chewing, cigarette smoking and alcohol consumption related to oral cancer in Taiwan. J Oral Pathol Med. 1995;24:450–3. doi: 10.1111/j.1600-0714.1995.tb01132.x. [DOI] [PubMed] [Google Scholar]
- 11.How Tobacco Smoke Causes Disease: The Biology and Behavioral Basis for Smoking-Attributable Disease. A Report of the Surgeon General. Atlanta, GA: U.S. Cancer; 2010. U.S. Department of Health and Human Services. [PubMed] [Google Scholar]
- 12.Kumar V, Abbas AK, Astar JC. Robbins Pathological Basis of Disease. 9th ed. Philadelphia: Elsevier, Saunders; 2013. Neoplasia; pp. 322–4. [Google Scholar]
- 13.Olshan AF, Weissler MC, Watson MA, Bell DA. GSTM1, GSTT1, GSTP1, CYP1A1, and NAT1 polymorphisms, tobacco use, and the risk of head and neck cancer. Cancer Epidemiol Biomarkers Prev. 2000;9:185–91. [PubMed] [Google Scholar]
- 14.Chatterjee S, Rudra T, Poddar S, Chakrabarti S, Sengupta B, Biswas D, et al. GST and CYP polymorphism related to tea drinking and oral pathology. Int J Hum Genet. 2008;8:295–9. [Google Scholar]
- 15.Sherratt PJ, Hayes JD. In: Glutathione S-transferases, in Enzyme Systems that Metabolise Drugs and Other Xenobiotics. Ioannides C., editor. Chichester, UK: John Wiley & Sons, Ltd; 2001. pp. 320–52. [Google Scholar]
- 16.Jain M, Kumar S, Lal P, Tiwari A, Ghoshal UC, Mittal B. Role of GSTM3 polymorphism in the risk of developing esophageal cancer. Cancer Epidemiol Biomarkers Prev. 2007;16:178–81. doi: 10.1158/1055-9965.EPI-06-0542. [DOI] [PubMed] [Google Scholar]
- 17.Jourenkova-Mironova N, Voho A, Bouchardy C, Wikman H, Dayer P, Benhamou S, et al. Glutathione S-transferase GSTM3 and GSTP1 genotypes and larynx cancer risk. Cancer Epidemiol Biomarkers Prev. 1999;8:185–8. [PubMed] [Google Scholar]
- 18.Inskip A, Elexperu-Camiruaga J, Buxton N, Dias PS, MacIntosh J, Campbell D, et al. Identification of polymorphism at the glutathione S-transferase, GSTM3 locus: Evidence for linkage with GSTM1*A. Biochem J. 1995;312(Pt 3):713–6. doi: 10.1042/bj3120713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rossini A, Rapozo DC, Soares Lima SC, Guimarães DP, Ferreira MA, Teixeira R, et al. Polymorphisms of GSTP1 and GSTT1, but not of CYP2A6, CYP2E1 or GSTM1, modify the risk for esophageal cancer in a Western population. Carcinogenesis. 2007;28:2537–42. doi: 10.1093/carcin/bgm222. [DOI] [PubMed] [Google Scholar]
- 20.Drummond SN, Gomez RS, Motta Noronha JC, Pordeus IA, Barbosa AA, De Marco L. Association between GSTT-1 gene deletion and the susceptibility to oral squamous cell carcinoma in cigarette-smoking subjects. Oral Oncol. 2005;41:515–9. doi: 10.1016/j.oraloncology.2004.12.014. [DOI] [PubMed] [Google Scholar]
- 21.Hung HC, Chuang J, Chien YC, Chern HD, Chiang CP, Kuo YS, et al. Genetic polymorphisms of CYP2E1, GSTM1, and GSTT1; environmental factors and risk of oral cancer. Cancer Epidemiol Biomarkers Prev. 1997;6:901–5. [PubMed] [Google Scholar]
- 22.Cheng L, Sturgis EM, Eicher SA, Char D, Spitz MR, Wei Q. Glutathione-S-transferase polymorphisms and risk of squamous-cell carcinoma of the head and neck. Int J Cancer. 1999;84:220–4. doi: 10.1002/(sici)1097-0215(19990621)84:3<220::aid-ijc4>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 23.Nair UJ, Nair J, Mathew B, Bartsch H. Glutathione S-transferase M1 and T1 null genotypes as risk factors for oral leukoplakia in ethnic Indian betel quid/tobacco chewers. Carcinogenesis. 1999;20:743–8. doi: 10.1093/carcin/20.5.743. [DOI] [PubMed] [Google Scholar]
- 24.Buch SC, Notani PN, Bhisey RA. Polymorphism at GSTM1, GSTM3 and GSTT1 gene loci and susceptibility to oral cancer in an Indian population. Carcinogenesis. 2002;23:803–7. doi: 10.1093/carcin/23.5.803. [DOI] [PubMed] [Google Scholar]
- 25.Anantharaman D, Chaubal PM, Kannan S, Bhisey RA, Mahimkar MB. Susceptibility to oral cancer by genetic polymorphisms at CYP1A1, GSTM1 and GSTT1 loci among Indians: Tobacco exposure as a risk modulator. Carcinogenesis. 2007;28:1455–62. doi: 10.1093/carcin/bgm038. [DOI] [PubMed] [Google Scholar]
- 26.Sreelekha TT, Ramadas K, Pandey M, Thomas G, Nalinakumari KR, Pillai MR. Genetic polymorphism of CYP1A1, GSTM1 and GSTT1 genes in Indian oral cancer. Oral Oncol. 2001;37:593–8. doi: 10.1016/s1368-8375(01)00028-8. [DOI] [PubMed] [Google Scholar]
- 27.Bathi RJ, Rao R, Mutalik S. GST null genotype and antioxidants: Risk indicators for oral pre-cancer and cancer. Indian J Dent Res. 2009;20:298–303. doi: 10.4103/0970-9290.57365. [DOI] [PubMed] [Google Scholar]
- 28.Duarte EC, Silva MS, Gomez MV, Gomez RS. GSTT1 polymorphism and oral OLP. Anticancer Res. 2006;26:427–30. [PubMed] [Google Scholar]
- 29.Majumder M, Sikdar N, Paul RR, Roy B. Increased risk of oral OLP and Cancer among mixed tobacco users carrying XRCC1 variant haplotypes and cancer among smokers carrying two risk genotypes: One on each of two loci, GSTM3 and XRCC1. Cancer Epidemiol Biomarkers Prev. 2005;14:2106–12. doi: 10.1158/1055-9965.EPI-05-0108. [DOI] [PubMed] [Google Scholar]


