Abstract
MicroRNAs (miRNAs) are known to suppress gene expression by binding to messenger RNAs and in turn regulate different pathophysiological processes. Transforming growth factor-β, mitogen-activated protein kinase signaling, and Wnt signaling-like major pathways associated with miRNAs are involved with kidney diseases. The discovery of miRNAs has provided new insights into kidney pathologies and may provide effective therapeutic strategies. Research has demonstrated the role of miRNAs in a variety of kidney diseases including diabetic nephropathy, lupus nephritis, hypertension, nephritic syndrome, acute kidney injury, renal cell carcinoma, and renal fibrosis. miRNAs are implicated as playing a role in these diseases due to their role in apoptosis, cell proliferation, differentiation, and development. As miRNAs have been detected in a stable condition in different biological fluids, they have the potential to be tools to study the pathogenesis of human diseases with a great potential to be used in disease diagnosis and prognosis. The purpose of this review is to examine the role of miRNA in kidney disease.
Keywords: Basic kidney diseases, end-stage renal disease, epigenetics, microRNA
Introduction
Chronic kidney disease (CKD) involves permanent loss of functioning of kidney ultimately leading to end-stage renal disease (ESRD). Nearly 10% of the Americans[1] and 7.5% of Indians[2] are reportedly suffering from ESRD, and this number is on constant rise. Pathogenesis of multifactorial ESRD is affected by environmental and genetic factors. Genetic causes such as multiple targeted genes, posttranscriptional and posttranslational modifications, interactions among RNA-DNA, RNA-RNA, RNA-protein, and microRNA (miRNA)-messenger RNA (mRNA) as well as epigenetic modifications in the genome are involved with ESRD.[3] Posttranscriptional events account to nearly 97% of altered kidney function involving the activities of noncoding RNA (ribosomal RNA, transfer tNA, introns, transposable elements, and intergenic regions), long noncoding RNAs, and miRNA.
According to miRBase 39,000 miRNAs have been identified in humans[4] of which nearly 365 miRNAs are present on the renal cortex. Emerging evidence suggest that miRNAs can reduce kidney fibrosis through regulation of targets associated with collagen and extracellular matrix accumulation. Unfortunately, due to loss in targeted and sustainable methods involved in miRNA delivery, the processes of developing miRNA therapies are hindered.[5]
miRNAs are short RNAs and are conventionally constituted of 22–25 nucleotides. These are well known to regulate gene expression at posttranscriptional level.[6] Through the regulation of gene expression, miRNAs play a very critical role in variety of cellular functions and physiological activities. Usually, miRNAs get attached to 3´ untranslated regions (UTRs) or to the 5´ UTR in seed region of the target mRNAs. Among prominent features, the miRNAs mimic mRNA translation and induce mRNA degradation, thus preventing the protein synthesis. miRNAs are produced by gene transcription of noncoding region which is catalyzed by polymerase II enzyme followed by editing or sequential processing. However, to understand how miRNAs are transcribed, it is important to know about miRNA biogenesis.
MicroRNA Biogenesis
miRNAs can be generated either by canonical or noncanonical pathways.[7] The miRNA genes first get transcribed by RNA polymerase II in the canonical pathway. This result in forming primary transcripts (pri-miRNAs) featured with hairpin structures. Subsequently, the pri-miRNAs get cleaved by the multiprotein complex called microprocessor complex in the nucleus. Drosha and DGCR8 are the two major proteins that comprise the multiprocessor complex. DGCR8 recruits Drosha after recognizing the hairpin structure in pri-miRNA. The Drosha then cleaves the pri-miRNAs at stem-loop structure giving rise to the pre-miRNAs (secondary precursor RNAs of seventy nucleotides).[6]
Subsequently, exportin-5 transfers the pre-miRNAs to cytoplasm. Exportin-5, which is a nuclear transport receptor family member specifically recognizes the stem-loop structure of pri-miRNAs. The pre-miRNAs are furthers cleaved, resulting in mature miRNA production.[6]
Mode of Action of MicroRNAs
It has been suggested that miRNAs inhibit protein synthesis. However, it is believed that inhibition of protein synthesis by miRNAs is due to mRNA destabilization. miRNAs may be expressed in a ubiquitous or specific manner to repress hundreds of genes. Microarray data further helped to detect the mRNA abundance and translational rates of approximately 8000 genes. miRNAs inhibit translation at an early step, probably at translation initiation, which is followed by deadenylation of target mRNAs ultimately leading to mRNA decay.
MicroRNAs in Kidney
miRNAs affect the cellular metabolism, differentiation, and proliferation, which directly or indirectly have a role in affecting the pathophysiology of an array of diseases.[8] In kidney diseases, the role of miRNAs may be initiated from the time of regulation of the nephron progenitors. For example, congenital anomalies of the kidney and urinary tract have been found to be associated with the deletion of miRNA 17–92 cluster ultimately resulting in delayed growth and improper functioning of the heart.[9] Further, renal hypoplasia is also associated with miRNA 17–92 cluster leading to fewer developing nephron structures.[10] The miRNAs present in the renal cortex are associated with basic kidney diseases such as Type 1[11] and Type 2[12] diabetes mellitus, diabetic nephropathy,[13] glomerulosclerosis,[14] lupus nephritis,[15] hypertension,[16] kidney neoplasm,[17] acute kidney injury (AKI),[18] autosomal dominant, and recessive polycystic kidney disease (PKD).[19]
MicroRNA Function in Mature Nephron
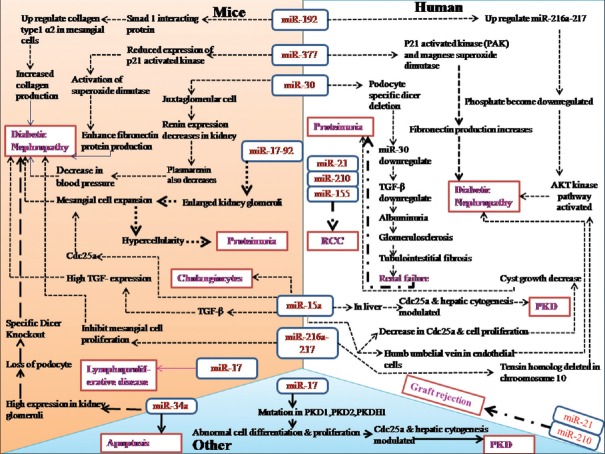
The role of conditionally knocking out Dicer has been associated with irregularities in proximal tubules[6] and podocytes,[7] decreased juxtaglomerular cells, low levels of renin concentration in plasma, and lowered blood pressure.[20] miRNAs reportedly regulate the tubular and glomerular damage and proteinuria due to podocyte-specific deletion of Dicer.[21] Down regulation of miRNA-30 family has been correlated with renal disease progression featured with albuminuria, glomerulosclerosis, and tubule-interstitial fibrosis. Further miRNA-30 family was observed to target connective tissue growth factor.[22] This review focuses on discussing different biological phenomenas involving miRNAs with lupus nephritis, hypertension, PKD, diabetic nephropathy, and AKI [Figure 1].
Figure 1.
Association of microRNAs with renal pathophysiology. MicroRNA reported in (i) human and mice; (ii) cell line and mice; (iii) in human; (iv) cell line; (v) mice
Lupus Nephritis
miRNAs act as epigenetic regulators toward genetic susceptibility of lupus nephritis in a heritable and subtle manner. An array of miRNAs such as miR-371-5p, miR-423-5p, miR-638, and miR-663 is known to be upregulated while certain miRNAs such as miR-31, miR-95, miR-99a, miR-130b, miR-10a, miR-134, and miR-146a are under expressed among the lupus nephritis cases.[23]
Lupus nephritis is a complication of systemic lupus erythematosus (SLE). In urine, the exosomal miRNAs otherwise found in SLE cases with or without nephritis are found to be increased.[24] More so, if an individual is affected with active lupus nephritis, overexpression of urinary exosomal miRNAs has been observed. Specifically, miR-146a discriminates the presence of active lupus nephritis.[24]
Hypertension
Apart from the genetic and environmental factors, involvements of miRNAs play a pivotal role in hypertension. miR-126 like endothelial-specific miRNAs is known to maintain vascular integrity. Another miRNA, i.e., miR-21 inhibits angiogenesis in endothelial cells which in turn causes hypertension and different cardiovascular abnormalities.[25] Further miR-217 and miR-122 are known to be involved in endothelial senescence among hypertensive cases.[25]
Importantly, the expressions of miR-200a, miR-200b, miR-141, miR-429, miR-205, and miR-192 have been found to be heightened among hypertension cases. The level of expression of these miRNAs is directly associated with severity of hypertensive nephrosclerosis.[26] It is well established that increases in β-myosin heavy chain (β-MHC) and decreases in α-myosin heavy chain (α-MHC) results in hypertension and progression to renal disease. Interestingly, miR-208a, miR-208b, and miR-499 are observed to regulate the β-MHC and α-MHC expression. Several other miRNAs such as miR-16, miR-19b, miR-20b, miR-93, miR-106b, miR-223, and miR-423-5p are also reportedly associated with hypertension.[27]
Polycystic Kidney Disease
The PKD is not ethnicity-specific and has been classified broadly into autosomal dominant and autosomal recessive. Potential role of certain miRNAs has been explored in controlling PKD associated gene expression. miR-17 targets UTR of PKD2 gene and suppresses the PKD2 expression in a posttranscriptional manner which promulgates cell proliferation. It is evident that PKD associated miRNAs are associated with Wnt, mTOR, mutagen-activated protein kinase, and transforming growth factor (TGF-β) like signaling pathways that play a role in cyst formation among the cases with renal defects.[28]
Another miRNA of relevance among PKD cases is miR-15a. Activity of Cdc25A, a cell cycle regulator, is altered by miR-15a. This in turn leads to progression of hepatic cystogenesis.[29] Such a stage is featured with altercation in G1 to S phase transition, proliferation of cell, and also a significant cyst growth in vitro when studied in mice model.
Diabetic Nephropathy
Cutting edge research on miRNAs role in diabetes has suggested some alarming findings, which may open up new prospects in modern medicine. miRNAs are reported to regulate glucose homeostasis. They are also associated with insulin secretion, pancreatic islet development, differentiation of β-cell, and controlling lipid and glucose metabolisms in context to diabetes.[30] miR-192 down regulation is correlated with tubule-interstitial fibrosis and decreased eGFR. miR-192 activates the Akt kinase which in turn causes fibrosis. It has been evidenced that the Akt activation in mice model inhibits apoptosis and hypertrophy. Further in response to TGF-β1, expression of miR-192 gets induced. Mesangial cells treated with high glucose and kidneys in diabetic mice models have reflected up regulation of miR-377 and down regulation of miR200a and miR-141. Another miRNA of interest, i.e., miR-29 which is induced by high glucose level and gets inhibited by TGF-β1 increases the risk of collagen deposition. These reports support the fact that TGF-β promotes mature miRNA synthesis from primary miRNAs by Drosha complex among diabetic cases. Further miR-93 directly targets and regulates the vascular endothelial growth factor (VEGF) expression.[6] It is noteworthy that the level of VEGF is normally heightened in diabetic kidney and is associated with angiogenesis among diabetic nephropathy cases.
Acute Kidney Injury
Genome expression profiling of mice models having ischemia-reperfusion injury showed differential expression of miR-20a, miR-21, miR-146a, miR-187, miR-192, miR-194, miR-199a, miR-214, and miR-805 in the kidney.[31] In context to human urine sample, collected from AKI cases, miR-21 and miR-155 are found to be upregulated.[32] Further the expression of miR-21 is observed to be induced by TGF-β signaling and suppressed by Smad7.[33] Promisingly 60-fold risk have been observed for miR-494 in individuals with AKI.[31] miR-494 targets activating transcription factor-3 and promotes nuclear factor-κB dependent immune response and some interleukin-6, monocyte chemoattractant protein-1 like inflammatory mediators in AKI cases.
TGF-β pathway also inhibits miR-29, miR-200 and induces miR-192, miR-491 in a unilateral ureteral obstruction model designed to study their importance in kidney injury.[33] More so miR-155, miR-210, and miR-21 which are involved in apoptosis and cell proliferation have been suggested as candidate biomarkers for AKI.
MicroRNAs in Urine
An array of miRNAs in urine has been reported by Melkonyan et al. of which interestingly none are observed to be specific to the kidney.[34] However, significant incidence of miR-152, miR-182, and miR-126 is reflected in urine. Notably, the presence of miR-152 and miR-126 heightens to nearly 9.6-fold among bladder cancer cases in comparison to normal individuals. Using such miRNAs of importance in a noninvasive manner, diagnosis and progression of kidney diseases may be followed. In urinary exosomes, which are often released from nephron, the levels of miR-192 and miR-27b have been proposed as biomarkers to differentiate among lupus and nonlupus nephritis cases.[35]
MicroRNAs in Therapeutics
Antagomirs (antisense oligonucleotide targeting specific miRNAs) are employed to systematically silence endogenous miRNAs in the kidney.[36] Single intravenous antagomirs dose lasts up to 21 days in the kidney.[37] It downplays activities of miR-21 like miRNAs, which alter extracellular signal-regulated kinase-mitogen-activated protein kinase pathway causing imbalance in growth factor secretion and fibroblast survival among CKD cases.[38] Similarly, anti-miR-192 treatment restructures glomerular fibrosis in diabetic nephropathy mice models by undermining mesangial fibronectin and collagen levels.[39] However, power for predicting disease susceptibility using this therapeutics is still under debate.
Epigenetic Factors and Kidney Disease
Epigenetic studies associated with CKD are reported for RNA interference (RNAi) along with chromatic modifications and DNA methylation. RNAi plays an important role in kidney development, kidney fibrosis, and homeostasis; for example, when Dicer is inactivated in mouse podocytes, it leads to renal failure and death. Skewing of female X-chromosome inactivation has been associated with kidney fibrosis, progressive renal disease, and kidney transplant outcomes. As with histone modifications, many kidney-miRNA studies have focused on fibrosis through the TGF-β pathway and limiting or reversing fibrotic damage through this mechanism. For example, miRNA-192 has been identified as a key regulator of collagen formation in diabetic kidney disease mouse models, whereas in human renal biopsies from patients with diabetic kidney disease, TGF-β upregulated miRNA-192 expression in proximal tubule cells, correlating with fibrosis and reduction in eGFR. Inhibition of kidney miRNA-192 was associated with decreased renal fibrosis and reduced proteinuria in a diabetic kidney disease mouse model. The defined influence of miRNAs in kidney damage and their ready accessibility in blood and urine makes them attractive biomarkers or targets for therapeutic intervention.
Future Challenges
There exist several major challenges in exploring the miRNAs role in kidney diseases. Regulatory mechanisms for miRNAs production are not clear. Many miRNAs are present in introns of host genes, and their expressions often do not correlate with the host genes suggesting subsequent posttranscriptional regulation.[40] However, specific targets for most miRNAs remain unclear till date. Large numbers of miRNA targets are predicted using various bioinformatics tools, but very less percentage of these have been experimentally validated and hence it faces considerable challenges in becoming a therapeutic agent. Amidst these challenges, the most instant clinical benefits are likely to appear from the identification of miRNAs that can be used as reliable biomarkers for diagnosis, prognosis, and response to therapy in context to both kidney and allograft outcome.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
References
- 1.Lerma EV, Nissenson AR. Nephrology Secrets. 3rd ed. Maryland Heights, Missouri, United States; 2012. [Google Scholar]
- 2.Agarwal SK, Srivastava RK. Chronic kidney disease in India: Challenges and solutions. Nephron Clin Pract. 2009;111:C197–203. doi: 10.1159/000199460. [DOI] [PubMed] [Google Scholar]
- 3.Mas VR, Dumur CI, Scian MJ, Gehrau RC, Maluf DG. MicroRNAs as biomarkers in solid organ transplantation. Am J Transplant. 2013;13:11–9. doi: 10.1111/j.1600-6143.2012.04313.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: Tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–8. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yao K, Ricardo SD. Mesenchymal stem cells as novel micro-ribonucleic acid delivery vehicles in kidney disease. Nephrology (Carlton) 2016;21:363–71. doi: 10.1111/nep.12643. [DOI] [PubMed] [Google Scholar]
- 6.Wei Q, Mi QS, Dong Z. The regulation and function of microRNAs in kidney diseases. IUBMB Life. 2013;65:602–14. doi: 10.1002/iub.1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bhatt K, Mi QS, Dong Z. microRNAs in kidneys: Biogenesis, regulation, and pathophysiological roles. Am J Physiol Renal Physiol. 2011;300:F602–10. doi: 10.1152/ajprenal.00727.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chandrasekaran K, Karolina DS, Sepramaniam S, Armugam A, Wintour EM, Bertram JF, et al. Role of microRNAs in kidney homeostasis and disease. Kidney Int. 2012;81:617–27. doi: 10.1038/ki.2011.448. [DOI] [PubMed] [Google Scholar]
- 9.Ventura A, Young AG, Winslow MM, Lintault L, Meissner A, Erkeland SJ, et al. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell. 2008;132:875–86. doi: 10.1016/j.cell.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marrone AK, Stolz DB, Bastacky SI, Kostka D, Bodnar AJ, Ho J. MicroRNA-17~92 is required for nephrogenesis and renal function. J Am Soc Nephrol. 2014;25:1440–52. doi: 10.1681/ASN.2013040390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hezova R, Slaby O, Faltejskova P, Mikulkova Z, Buresova I, Raja KR, et al. microRNA-342, microRNA-191 and microRNA-510 are differentially expressed in T regulatory cells of type 1 diabetic patients. Cell Immunol. 2010;260:70–4. doi: 10.1016/j.cellimm.2009.10.012. [DOI] [PubMed] [Google Scholar]
- 12.Bang-Berthelsen CH, Pedersen L, Fløyel T, Hagedorn PH, Gylvin T, Pociot F. Independent component and pathway-based analysis of miRNA-regulated gene expression in a model of type 1 diabetes. BMC Genomics. 2011;12:97. doi: 10.1186/1471-2164-12-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saal S, Harvey SJ. MicroRNAs and the kidney: Coming of age. Curr Opin Nephrol Hypertens. 2009;18:317–23. doi: 10.1097/MNH.0b013e32832c9da2. [DOI] [PubMed] [Google Scholar]
- 14.Gebeshuber CA, Kornauth C, Dong L, Sierig R, Seibler J, Reiss M, et al. Focal segmental glomerulosclerosis is induced by microRNA-193a and its downregulation of WT1. Nat Med. 2013;19:481–7. doi: 10.1038/nm.3142. [DOI] [PubMed] [Google Scholar]
- 15.Lu MC, Lai NS, Chen HC, Yu HC, Huang KY, Tung CH, et al. Decreased microRNA (miR)-145 and increased miR-224 expression in T cells from patients with systemic lupus erythematosus involved in lupus immunopathogenesis. Clin Exp Immunol. 2013;171:91–9. doi: 10.1111/j.1365-2249.2012.04676.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sen CK, Gordillo GM, Khanna S, Roy S. Micromanaging vascular biology: Tiny microRNAs play big band. J Vasc Res. 2009;46:527–40. doi: 10.1159/000226221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khella HW, White NM, Faragalla H, Gabril M, Boazak M, Dorian D, et al. Exploring the role of miRNAs in renal cell carcinoma progression and metastasis through bioinformatic and experimental analyses. Tumour Biol. 2012;33:131–40. doi: 10.1007/s13277-011-0255-5. [DOI] [PubMed] [Google Scholar]
- 18.Cantaluppi V, Gatti S, Medica D, Figliolini F, Bruno S, Deregibus MC, et al. Microvesicles derived from endothelial progenitor cells protect the kidney from ischemia-reperfusion injury by microRNA-dependent reprogramming of resident renal cells. Kidney Int. 2012;82:412–27. doi: 10.1038/ki.2012.105. [DOI] [PubMed] [Google Scholar]
- 19.Chu AS, Friedman JR. A role for microRNA in cystic liver and kidney diseases. J Clin Invest. 2008;118:3585–7. doi: 10.1172/JCI36870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sequeira-Lopez ML, Weatherford ET, Borges GR, Monteagudo MC, Pentz ES, Harfe BD, et al. The microRNA-processing enzyme dicer maintains juxtaglomerular cells. J Am Soc Nephrol. 2010;21:460–7. doi: 10.1681/ASN.2009090964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harvey SJ, Jarad G, Cunningham J, Goldberg S, Schermer B, Harfe BD, et al. Podocyte-specific deletion of dicer alters cytoskeletal dynamics and causes glomerular disease. J Am Soc Nephrol. 2008;19:2150–8. doi: 10.1681/ASN.2008020233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Duisters RF, Tijsen AJ, Schroen B, Leenders JJ, Lentink V, van der Made I, et al. miR-133 and miR-30 regulate connective tissue growth factor: Implications for a role of microRNAs in myocardial matrix remodeling. Circ Res. 2009;104:170–8. doi: 10.1161/CIRCRESAHA.108.182535. [DOI] [PubMed] [Google Scholar]
- 23.Dai Y, Sui W, Lan H, Yan Q, Huang H, Huang Y. Comprehensive analysis of microRNA expression patterns in renal biopsies of lupus nephritis patients. Rheumatol Int. 2009;29:749–54. doi: 10.1007/s00296-008-0758-6. [DOI] [PubMed] [Google Scholar]
- 24.Perez-Hernandez J, Forner MJ, Pinto C, Chaves FJ, Cortes R, Redon J. Increased urinary exosomal microRNAs in patients with systemic lupus erythematosus. PLoS One. 2015;10:e0138618. doi: 10.1371/journal.pone.0138618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bátkai S, Thum T. MicroRNAs in hypertension: Mechanisms and therapeutic targets. Curr Hypertens Rep. 2012;14:79–87. doi: 10.1007/s11906-011-0235-6. [DOI] [PubMed] [Google Scholar]
- 26.Wang G, Kwan BC, Lai FM, Choi PC, Chow KM, Li PK, et al. Intrarenal expression of miRNAs in patients with hypertensive nephrosclerosis. Am J Hypertens. 2010;23:78–84. doi: 10.1038/ajh.2009.208. [DOI] [PubMed] [Google Scholar]
- 27.Dickinson BA, Semus HM, Montgomery RL, Stack C, Latimer PA, Lewton SM, et al. Plasma microRNAs serve as biomarkers of therapeutic efficacy and disease progression in hypertension-induced heart failure. Eur J Heart Fail. 2013;15:650–9. doi: 10.1093/eurjhf/hft018. [DOI] [PubMed] [Google Scholar]
- 28.Pandey P, Brors B, Srivastava PK, Bott A, Boehn SN, Groene HJ, Gretz N. Microarray-based approach identifies microRNAs and their target functional patterns in polycystic kidney disease. BMC Genomics. 2008;9:624. doi: 10.1186/1471-2164-9-624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen J, Kam-Tao P, Kwan BC, Chow KM, Lai KB, Luk CC, et al. Relation between microRNA expression in peritoneal dialysis effluent and peritoneal transport characteristics. Dis Markers. 2012;33:35–42. doi: 10.3233/DMA-2012-0901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li R, Chung AC, Yu X, Lan HY. MicroRNAs in diabetic kidney disease. Int J Endocrinol. 2014;2014:593956. doi: 10.1155/2014/593956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Godwin JG, Ge X, Stephan K, Jurisch A, Tullius SG, Iacomini J. Identification of a microRNA signature of renal ischemia reperfusion injury. Proc Natl Acad Sci U S A. 2010;107:14339–44. doi: 10.1073/pnas.0912701107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lorenzen JM, Kielstein JT, Hafer C, Gupta SK, Kümpers P, Faulhaber-Walter R, et al. Circulating miR-210 predicts survival in critically ill patients with acute kidney injury. Clin J Am Soc Nephrol. 2011;6:1540–6. doi: 10.2215/CJN.00430111. [DOI] [PubMed] [Google Scholar]
- 33.Chung AC, Dong Y, Yang W, Zhong X, Li R, Lan HY. Smad7 suppresses renal fibrosis via altering expression of TGF-ß/Smad3-regulated microRNAs. Mol Ther. 2013;21:388–98. doi: 10.1038/mt.2012.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Melkonyan HS, Feaver WJ, Meyer E, Scheinker V, Shekhtman EM, Xin Z, et al. Transrenal nucleic acids: From proof of principle to clinical tests. Ann N Y Acad Sci. 2008;1137:73–81. doi: 10.1196/annals.1448.015. [DOI] [PubMed] [Google Scholar]
- 35.Zhou H, Lin K. Excess of microRNAs in large and very 5' biased introns. Biochem Biophys Res Commun. 2008;368:709–15. doi: 10.1016/j.bbrc.2008.01.117. [DOI] [PubMed] [Google Scholar]
- 36.Krützfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, et al. Silencing of microRNAs in vivo with 'antagomirs'. Nature. 2005;438:685–9. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 37.Obad S, dos Santos CO, Petri A, Heidenblad M, Broom O, Ruse C, et al. Silencing of microRNA families by seed-targeting tiny LNAs. Nat Genet. 2011;43:371–8. doi: 10.1038/ng.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thum T, Gross C, Fiedler J, Fischer T, Kissler S, Bussen M, et al. MicroRNA-21 contributes to myocardial disease by stimulating MAP kinase signalling in fibroblasts. Nature. 2008;456:980–4. doi: 10.1038/nature07511. [DOI] [PubMed] [Google Scholar]
- 39.Putta S, Lanting L, Sun G, Lawson G, Kato M, Natarajan R. Inhibiting microRNA-192 ameliorates renal fibrosis in diabetic nephropathy. J Am Soc Nephrol. 2012;23:458–69. doi: 10.1681/ASN.2011050485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lanford RE, Hildebrandt-Eriksen ES, Petri A, Persson R, Lindow M, Munk ME, et al. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science. 2010;327:198–201. doi: 10.1126/science.1178178. [DOI] [PMC free article] [PubMed] [Google Scholar]