The histone deacetylase HDA6 interacts with the histone H3K9 methyltransferases SUVH4/5/6 to coordinately regulate transposon silencing.
Abstract
Histone deacetylases (HDACs) play important roles in regulating gene expression. In yeast and animals, HDACs act as components of multiprotein complexes that modulate transcription during various biological processes. However, little is known about the interacting proteins of plant HDACs. To identify the plant HDAC complexes and interacting proteins, we developed an optimized workflow using immunopurification coupled to mass spectrometry-based proteomics in Arabidopsis thaliana. We found that the histone deacetylase HDA6 can interact with the histone methyltransferases SUVH4, SUVH5, and SUVH6 (SUVH4/5/6). Domain analysis revealed that the C-terminal regions of HDA6 and SUVH5 are important for their interaction. Furthermore, HDA6 interacts with SUVH4/5/6 and coregulates a subset of transposons through histone H3K9 methylation and H3 deacetylation. In addition, two phosphorylated serine residues, S427 and S429, were unambiguously identified in the C-terminal region of HDA6. Phosphomimetics (amino acid substitutions that mimic a phosphorylated protein) of HDA6 resulted in increased enzymatic activity, whereas the mutation of S427 to alanine in HDA6 abolished its interaction with SUVH5 and SUVH6, suggesting that the phosphorylation of HDA6 is important for its activity and function.
INTRODUCTION
Reversible histone acetylation and deacetylation at the N termini of histone tails play a crucial role in regulating gene activity. Hyperacetylation of histones relaxes chromatin structure and is associated with transcriptional activation, whereas hypoacetylation of histones induces chromatin compaction and gene repression. Histone acetylation and deacetylation are dynamically regulated by histone acetyltransferases and histone deacetylase (HDACs), respectively. Arabidopsis thaliana contains 18 HDACs, which can be divided into three families, namely, the RPD3/HDA1-like family, the Sirtuin 2-like family, and the HD2 family (Pandey et al., 2002). Members of the RPD3/HDA1-like family can be further divided into three classes (Pandey et al., 2002; Hollender and Liu, 2008; Alinsug et al., 2009; Liu et al., 2016). Class I includes HDA6, HDA7, HDA9, HDA10, HDA17, and HDA19. Class II has five members, HDA5, HDA8, HDA14, HDA15, and HDA18. HDA2 is the sole plant HDAC under the class III group (Hollender and Liu, 2008; Liu et al., 2016).
HDA6 was initially identified as an important factor required for the silencing of transgenes and transposable elements (TEs), since mutations in HDA6 result in the loss of transcriptional silencing from several repetitive transgenic and endogenous templates (Murfett et al., 2001; Lippman et al., 2003; Lawrence et al., 2004; Probst et al., 2004; Aufsatz et al., 2007). HDA6 regulates various plant development processes such as embryo development (Tanaka et al., 2008), seed maturation (Perrella et al., 2013), and the vegetative-to-reproductive phase transition in the meristem (Yu et al., 2011). The Arabidopsis hda6 mutant hda6-6 (also named axe1-5) displays a late flowering phenotype, and HDA6 interacts with FLOWERING LOCUS D (FLD) and the histone binding protein FVE to repress FLOWERING LOCUS C (FLC) expression (Yu et al., 2011). Moreover, HDA6 influences leaf development, since hda6 mutants display serrated leaf and short petiole phenotypes (Luo et al., 2012b). In addition, hda6 mutants showed lower germination rates under abscisic acid (ABA) and salt stress compared with the wild type, suggesting that HDA6 may play an important role in plant ABA and salt stress responses (Chen et al., 2010; Chen and Wu, 2010; Luo et al., 2012a).
More recent studies indicate that HDA6 functions by interacting with other proteins involved in transcriptional regulation (Liu et al., 2014). HDA6 physically associates with the lysine-specific LSD1-type histone demethylase FLD to regulate flowering time (Yu et al., 2011), suggesting that histone deacetylases and demethylases may interact to regulate gene expression. HDA6 also associates with MSI5/FVE, a homolog of Retinoblastoma-Associated Protein 46/48 (RbAp46/48), to repress FLC expression in the control of flowering time (Gu et al., 2011). In addition, HDA6 controls leaf development by interacting with ASYMMETRIC LEAVES1 (AS1) and AS2 (Luo et al., 2012b). Furthermore, HDA6 directly interacts with the DNA methyltransferase MET1, which coregulates a large subset of transposons and repeat sequences (Liu et al., 2012). Both HDA6 and MET1 are required for silencing multiple classes of TEs, suggesting that HDA6-dependent epigenetic regulation might be involved in the maintenance of DNA methylation by MET1 (Lippman et al., 2003; To et al., 2011; Liu et al., 2012; Blevins et al., 2014).
SUVH4, SUVH5, and SUVH6 (SUVH4/5/6) are histone H3 lysine 9 methyltransferases belonging to the SUV(R) group of SET domain proteins (Ebbs and Bender, 2006; Zhang et al., 2002). The SRA domain from SUVH4 and SRA-Ring proteins binds to methylated DNA and prefers methyltyrosines in different sequence contexts (Johnson et al., 2007). SUVH4 and SUVH6 are responsible for maintaining H3K9 methylation on inverted repeats undergoing transcription (Ebbs et al., 2005). Moreover, the functional SUVH5 SRA domain is required for both DNA methylation and the accumulation of the H3K9 dimethylation modification (Rajakumara et al., 2011). In Arabidopsis, TE reactivation was observed in suvh4 suvh5 suvh6 triple mutants (Ebbs and Bender, 2006), suggesting that SUVH4/5/6 are important for maintaining TE silencing.
Histone modifications can act alone or in concert in a context-dependent manner to promote or repress chromatin-mediated processes (Berger, 2007; Bannister and Kouzarides, 2011). Hyperacetylated histones are primarily associated with activated genomic regions at both the local and global levels. By contrast, deacetylation mainly results in repression and silencing (Grunstein, 1997; Turner, 2000). In human cells, HDAC1 and HDAC2 interact with SUV39H1, a histone K9 methyltransferase, forming a complex to maintain histone methylation and deacetylation, resulting in transcriptional repression (Vaute et al., 2002). In yeast and animal systems, members of RPD3/HDA1 HDACs were identified as components of several multiprotein HDAC complexes, such as Sin3, Mi2/NuRD, and CoREST (Seto and Yoshida, 2014). However, little is known about plant HDAC protein complexes and their interaction partners.
In this study, we used immunopurification coupled to mass spectrometry-based proteomics to identify the interacting proteins of HDA6 in Arabidopsis. Our results reveal that HDA6 can interact with the SET domain histone H3K9 methyltransferases SUVH4/5/6, which function coordinately to regulate transposon silencing. Furthermore, the phosphorylation of HDA6 is important for its activity and function. The results of this study shed light on the role of HDAC protein complexes in transposon silencing in plants.
RESULTS
HDA6 Interacts with the Histone Lysine 9 Methyltransferases SUVH4, SUVH5, and SUVH6
To identify the HDA6 complex and interacting proteins, we developed an optimized workflow using immunopurification coupled to mass spectrometry-based proteomics (Miteva et al., 2013). We used Arabidopsis lines expressing GFP-tagged HDA6 (GFP-HDA6) driven by the HDA6 native promoter (proHDA6:HDA6-GFP) or the 35S promoter (pro35S:HDA6-GFP) in the hda6-6 mutant (Yu et al., 2011) and immunoaffinity-purified HDA6 complexes. HDA6-GFP can complement the late flowering phenotype of hda6-6, suggesting that the GFP-HDA6 fusion protein is functional in vivo (Yu et al., 2011). Protein complexes were isolated by immunoprecipitation using GFP-Trap magnetic beads followed by liquid chromatography-tandem mass spectrometry (LC-MS/MS) and Mascot analysis. Proteins that only appeared in the HDA6-GFP immunoprecipitation samples but not in the control (hda6-6) samples were considered to be the potential interaction proteins of the HDA6 complex. Our results confirmed the previous findings that HDA6 can interact with HDC1, MET1, FVE/MSI4, HD2A, and HD2B (Table 1) (Liu et al., 2012; Luo et al., 2012a). In addition, we also identified other HDA6 interacting proteins involved in chromatin remodeling and histone modifications (Table 1).
Table 1. HDA6 Interacting Proteins Involved in Chromatin Remodeling and Histone Modifications Identified by LC-MS/MS.
| AGI Code | Coverage | Unique Peptides | Description |
|---|---|---|---|
| AT5G63110ab | 47% | 16 | HDA6 |
| AT5G22650a | 30.07% | 5 | HD2B |
| AT5G08450ab | 26% | 8 | HDC1, histone deacetylase complex 1 |
| AT1G64490a | 8.87% | 1 | DEK, chromatin associated protein |
| AT1G20696a | 8.84% | 1 | HMGB3, high mobility group B3 protein |
| AT3G44750a | 8.16% | 2 | HD2A |
| AT2G35160ab | 5.16% | 3 | SUVH5, histone methyltransferase (MTase) |
| AT5G05330a | 5% | 1 | HMG-box (high mobility group) DNA binding family protein |
| AT2G19520b | 3.75% | 1 | MSI4/FVE |
| AT3G22590a | 3.37% | 1 | PHP, a homolog of human Paf1 Complex subunit Parafibromin |
| AT1G24190b | 3.08% | 2 | SNL3 |
| AT5G67430a | 2.59% | 1 | GCN5-related N-acetyltransferase (GNAT) family protein |
| AT5G35600a | 2.2% | 1 | HDA7 |
| AT3G01320b | 2.11% | 2 | SNL1 |
| AT5G67320a | 1.96% | 1 | HOS15, histone deacetylation-related WD-40 repeat protein |
| AT5G58230a | 1.89% | 1 | MSI1, histone binding protein |
| AT5G53430a | 0.86% | 1 | SDG29/ATX5, histone-lysine N-methyltransferase |
| AT5G49160a | 0.85% | 2 | MET1 |
| AT4G34430a | 0.71% | 1 | SWI/SNF complex subunit SWI3D |
| AT4G15180a | 0.3% | 1 | SDG2/ATXR3, putative histone-lysine N-methyltransferase |
Unique peptides coverage (%) is indicated in the “Coverage” column.
Identified from pro35S:HDA6-GFP transgenic plants.
Identified from proHDA6:HDA6-GFP transgenic plants.
Identified from both pro35S:HDA6-GFP and proHDA6:HDA6-GFP transgenic plants.
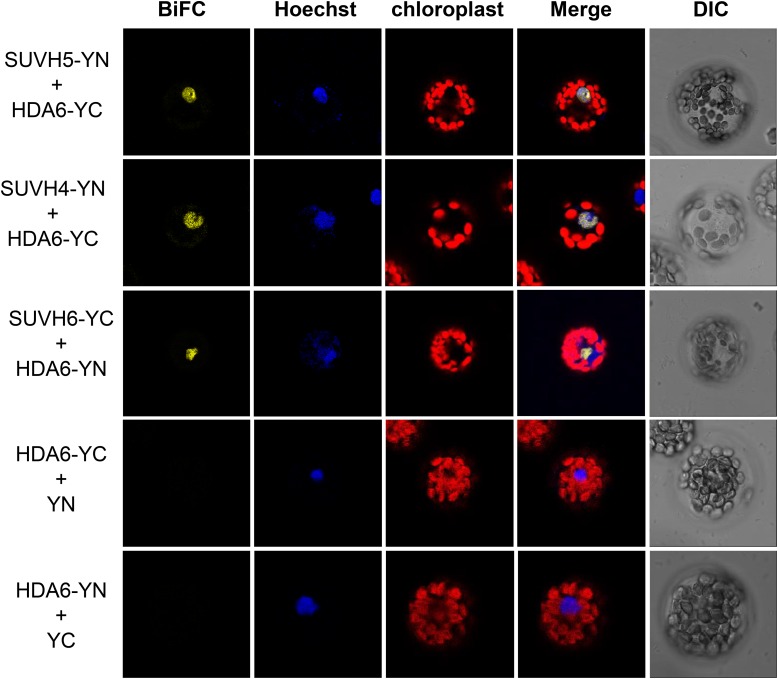
We found that the histone lysine methyltransferase (HKMT) SUVH5 is the potential interacting protein of HDA6 (Table 1). SUVH5 was identified from the LC-MS/MS results using both proHDA6:HDA6-GFP and pro35S:HDA6-GFP plants, supporting the notion that HDA6 and SUVH5 interact under physiological conditions. The interaction between HDA6 and SUVH5 was confirmed by bimolecular fluorescence complementation (BiFC) assays, in vitro pull-down assays, coimmunoprecipitation (Co-IP) assays, and yeast two-hybrid assays (Figures 1 and 2; Supplemental Figure 1). In the BiFC assays, SUVH5 interacted with HDA6 in the nuclei of Arabidopsis protoplasts (Figure 1). Since SUVH5 functions redundantly with KYP (SUVH4) and SUVH6, we also analyzed the interaction of HDA6 with SUVH4 and SUVH6 by BiFC assays. As shown in Figure 1, SUVH4 and SUVH6 also interacted with HDA6.
Figure 1.
SUVH4, SUVH5, and SUVH6 Interact with HDA6.
BiFC assays of Arabidopsis protoplasts showing that SUVH4, SUVH5, and SUVH6 interact with HDA6. SUVH4/5/6 and HDA6 fused with the N terminus (YN) or the C terminus (YC) of YFP were cotransfected into protoplasts and visualized by confocal microscopy. Hoechst staining indicates the nucleus. DIC, differential interference contrast.
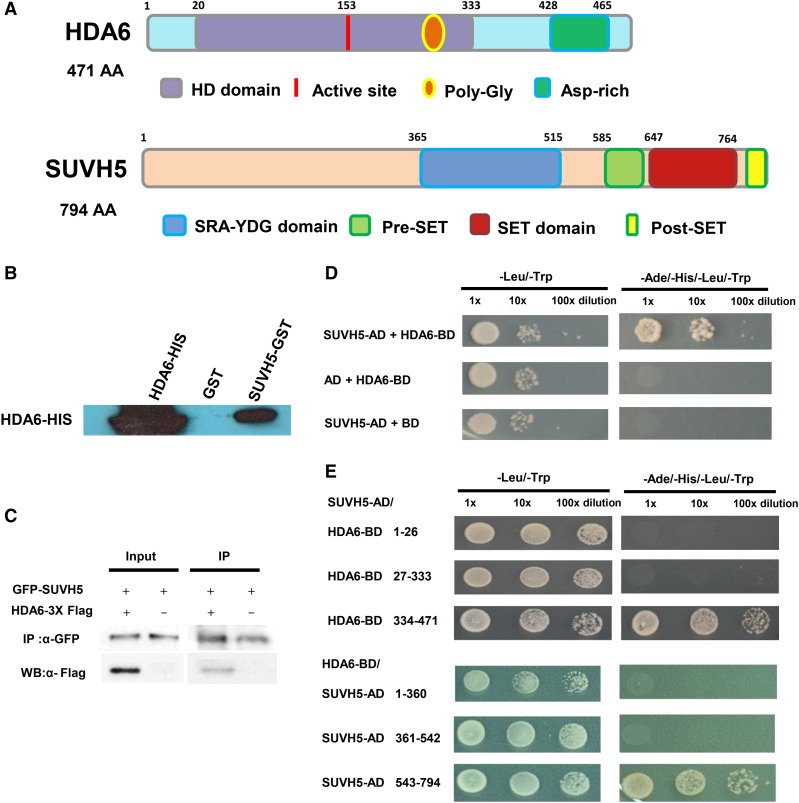
Figure 2.
Interaction of HDA6 and SUVH5 in in Vitro Pull-Down, Co-IP, and Yeast Two-Hybrid Assays.
(A) HDA6 and SUVH5 domain structures.
(B) SUVH5 interacts with HDA6 in in vitro pull-down assays. GST-SUVH5 or GST was incubated with HDA6-His and GST affinity resins, and the bound proteins were eluted from the resins and probed with the anti-His antibody.
(C) Interaction of HDA6 and SUVH5 in coimmunoprecipitation assays. Protein extracts coexpressing pro35S:GFP-SUVH5 and pro35S:HDA6-3X FLAG in N. benthamiana leaves were immunoprecipitated using anti-GFP antibody and analyzed by protein gel blot analysis.
(D) Yeast two-hybrid assays showing that full-length SUVH5 interacts with full-length HDA6.
(E) The C-terminal regions of HDA6 (334–471 amino acids) and SUVH5 (543–794 amino acids with Pre-SET and SET domains) are responsible for the protein-protein interactions.
For the in vitro pull-down assay, purified recombinant HDA6-His protein was incubated with glutathione S-transferase (GST)-SUVH5 protein. As shown in Figure 2B, HDA6-His was pulled down by GST-SUVH5. For the Co-IP assays, FLAG-tagged HDA6 (HDA6-3X FLAG) and GFP-tagged SUVH5 (SUVH5-GFP) were coexpressed in wild tobacco (Nicotiana benthamiana) leaves. HDA6-3X FLAG was coimmunoprecipitated by SUVH5-GFP (Figure 2C). These results indicate that HDA6 can interact with SUVH5 both in vitro and in vivo.
We also mapped the domain of HDA6 responsible for the interaction with SUVH5 using yeast two-hybrid assays. The C-terminal region of HDA6 (334–471 amino acids) is important for its interaction with SUVH5 (Figures 2A and 2E). We divided SUVH5 into three parts, the N-terminal region (1–360 amino acids), the SRA-YDG domain (361–542 amino acids), and the C-terminal region including the pre-SET and SET domains (543–794 amino acids). As shown in Figure 2E, the C-terminal region of HDA6 interacted with the C-terminal region of SUVH5. These results indicate that the C-terminal regions of HDA6 and SUVH5 are important for their interaction.
SUVH4/5/6 and HDA6 Coregulate Transposable Element Silencing
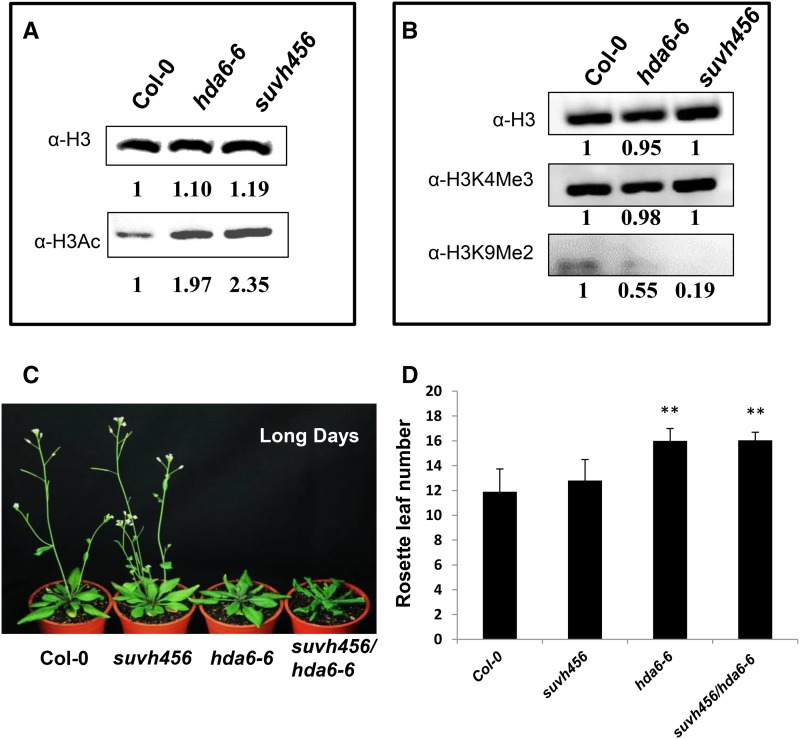
The interaction of HDA6 with SUVH4/5/6 may mediate the crosstalk between histone deacetylation and methylation in Arabidopsis. We analyzed the histone modification levels in hda6, suvh4 suvh5 suvh6 (suvh456) triple, and hda6-6/suvh456 quadruple mutants. As shown in Figure 3 and Supplemental Figure 2, compared with the wild type, the histone H3 acetylation level was higher in the hda6-6, suvh456 triple, and suvh456/hda6-6 quadruple mutants, whereas the histone H3K9me2 level was lower in hda6-6, suvh456 triple, and suvh456/hda6-6 quadruple mutants, suggesting that both HDA6 and SUVH4/5/6 regulate histone H3 acetylation and H3K9me2. Furthermore, the histone H3K9me2 level in the suvh456/hda6-6 quadruple mutant was much lower than that of the hda6-6 and suvh456 triple mutants (Supplemental Figure 2), suggesting that HDA6 and SUVH4/5/6 act synergistically to regulate H3K9me2 levels.
Figure 3.
Histone Modifications and Flowering Phenotypes of hda6 and suvh456.
(A) and (B) Total histone H3 acetylation (A) and histone H3K9 methylation (B) of hda6-6 and suvh456, as detected by protein gel blot analysis. Histone H3 antibody (a-H3) is shown as a loading control. The numbers shown on the gels represent the quantitative results (in arbitrary units). Proteins were extracted from the leaves of 20-d-old plants.
(C) The flowering phenotypes of suvh456, hda6-6, and suvh456/hda6-6.
(D) Rosette leaf number at flowering. Plants were grown under long-day conditions. At least 20 plants were scored for each line. Values are means ± sd from three independent plant batches. **P < 0.01, t test.
The hda6 mutants display a late flowering phenotype due to the derepression of FLC expression (Yu et al., 2011). We compared the flowering time of suvh456, hda6-6, and suvh456/hda6-6 mutant plants grown under long-day conditions. As shown in Figures 3B and 3C, the late flowering phenotype was only found in the hda6-6 single mutant but not in suvh456. The flowering phenotype of suvh456/ hda6-6 was similar to that of the hda6-6 single mutant, indicating that HDA6 regulates flowering independent of SUVH4/5/6.
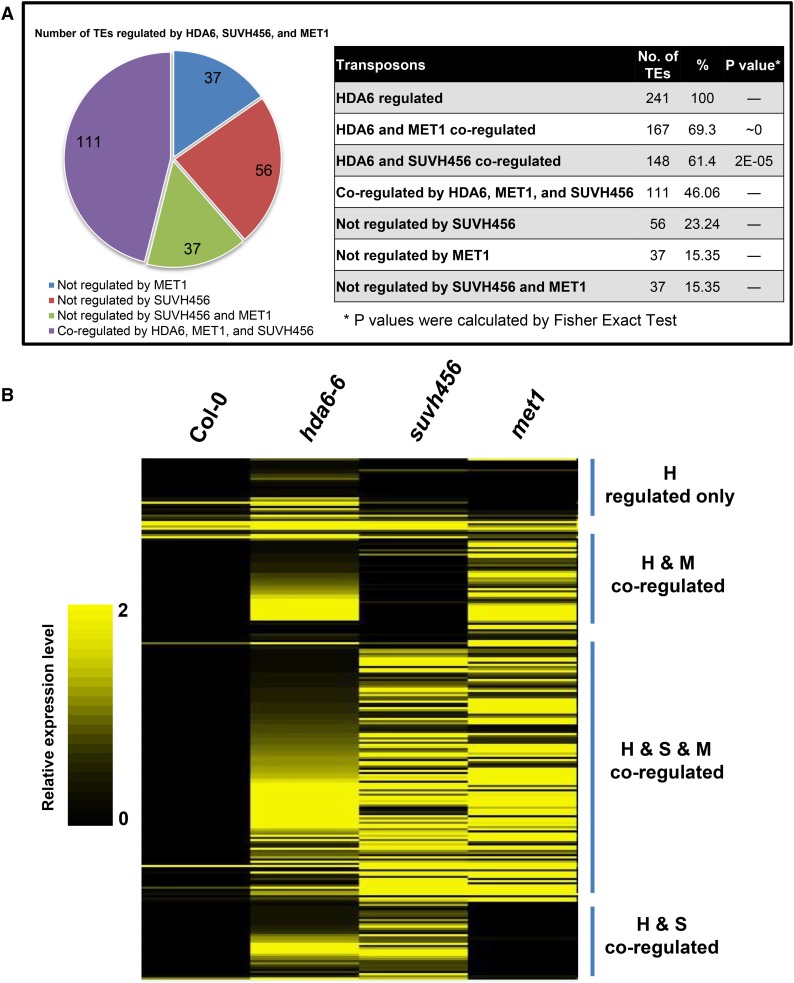
Our recent transcriptome analysis by RNA-seq revealed that 241 TEs are upregulated in the hda6-6 mutant compared with the wild type (Yu et al., 2016). Interestingly, among these 241 upregulated TEs in hda6, 148 (61.4%) and 167 (69.3%) TEs are also upregulated in the suvh456 triple mutant and met1 mutant, respectively (Figure 4A; Supplemental Figure 3) (Kuhn et al., 2009; Stroud et al., 2013). In addition, 111 (46.06%) TEs are coregulated by HDA6, SUVH4/5/6, and MET1 (Figure 4A; Supplemental Figure 3), suggesting that the functions of HDA6, SUVH4/5/6, and MET1 are highly correlated. Based on the results of heat map analysis, we further classified these TEs into four groups: coregulated by HDA6 and SUVH4/5/6 (H and S); coregulated by HDA6, SUVH4/5/6, and MET1 (H and S and M); coregulated by HDA6 and MET1 (H and M); and regulated by HDA6 (H) only (Figures 4A and 4B). Nine TEs (AT5G26345, AT2G20460, AT2G26630, AT3G43530, AT1G52850, AT5G27925, AT1G42705, AT5G59620, and AT5G46645) that were coregulated by HDA6 and SUVH4/5/6 were further analyzed by qRT-PCR. As shown in Supplemental Figure 4, all of these TEs were highly induced in hda6, suvh456, and suvh456/hda6 quadruple mutants.
Figure 4.
HDA6 and SUVH4/5/6 Coregulate Transposable Element Expression.
(A) Number of TEs regulated by HDA6, MET1, and SUVH4/5/6.
(B) Heat map analysis of TEs coregulated by HDA6, SUVH4/5/6, and MET1. H, HDA6; S, SUVH4/5/6; M, MET1. The color scale indicates the relative expression levels of TEs in the respective mutants compared with Col-0 wild type.
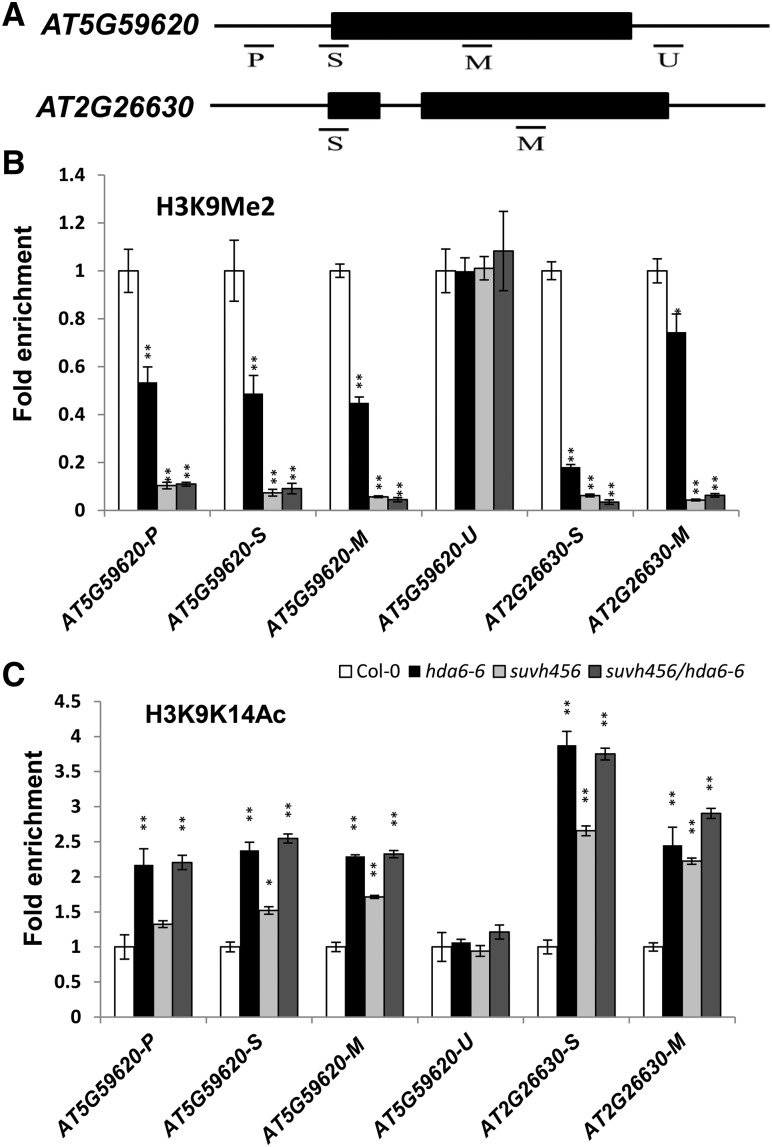
We selected two TEs (AT5G59620 and AT2G26630) for further histone modification analysis via chromatin immunoprecipitation (ChIP) assays. The histone H3K9me2 level of the promoter and gene body regions was dramatically reduced in both hda6-6 and suvh456 mutant plants compared with the wild type (Figures 5A and 5B). In addition, the H3K9K14Ac level was also significantly higher in the hda6-6, suvh456, and suvh456/hda6 quadruple mutants than in the wild type (Figure 5C).
Figure 5.
Histone Modification Levels in the Two Transposons.
(A) Schematic structures of the genomic sequences of AT5G59620 and AT2G26630 and the regions examined by ChIP-qPCR.
(B) and (C) Relative levels of H3K9Me2 (B) and H3Ac (C) in AT5G59620 and AT2G26630 in Col-0, hda6-6, suvh456, and suvh456/hda6-6. Leaves of 14-d-old plants were harvested for ChIP assays. The amounts of DNA were quantified and normalized to that of input DNA. The fold enrichment of histone modification levels in hda6-6 and suvh456 versus Col-0 at the indicated regions is shown. Values are means ± sd of three independent measurements. *P < 0.05, **P < 0.01, t test.
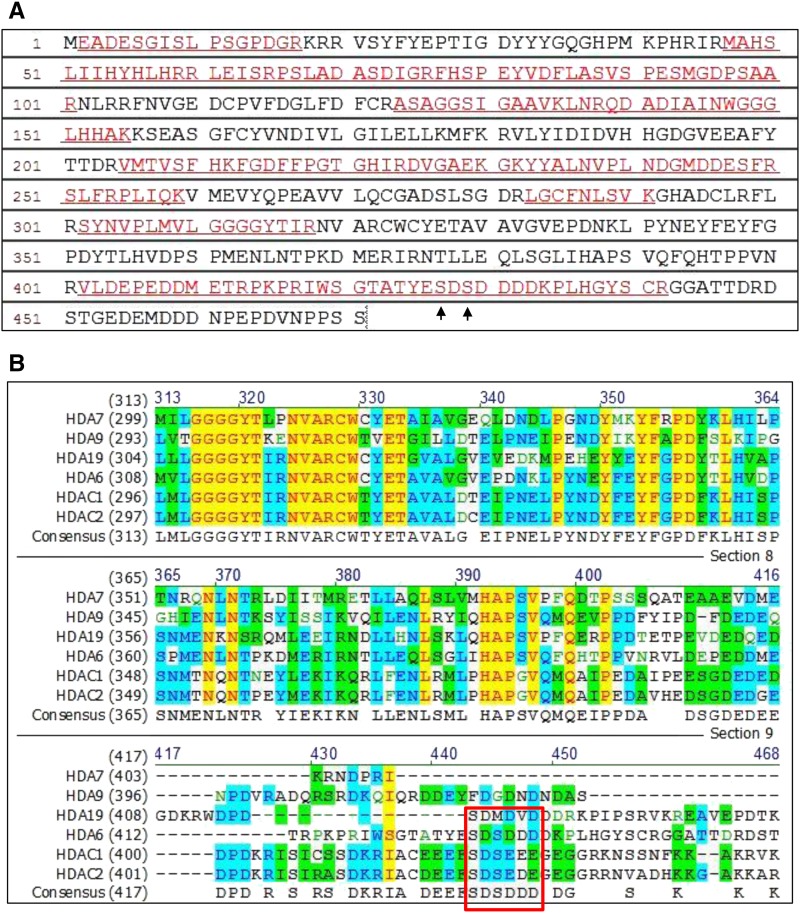
Identification of the Phosphorylation Sites of HDA6
In our LC-MS/MS assays, the sequence coverage matched 47% of the amino acids of HDA6, and two phosphorylated serine were unambiguously identified at S427 and S429 (Figure 6A). We compared the amino acid sequences of Arabidopsis HDA6, HDA7, HDA9, and HDA19 with those of human HDAC1 and HDAC2 (Figure 6B). The conserved motif of SDS(D/E) (D/E) (D/E) in human HDAC1 and HDAC2 (Pflum et al., 2001) was also found in Arabidopsis HDA6 but not in HDA7, HDA9, or HDA19 (Figure 6B).
Figure 6.
The Conserved Motif in Phosphorylation Sites among HDA6, HDAC1, and HDAC2.
(A) Peptide sequence of HDA6 detected by LC-MS/MS. The peptides detected by LC-MS/MS are indicated by underlined red letters. The matched peptides exhibit 47% amino acid sequence coverage. Arrows indicate the phosphorylation sites (S427 and S429) reveal by LC-MS/MS.
(B) Amino acid sequence alignment of Arabidopsis HDA6, HDA7, HDA9, and HDA19 with human HDAC1 and HDAC2. HDA6, HDAC1, and HDAC2 contain the conserved motif SDS(E/D)(E/D)(E/D) (boxed in red). Conservative, identical, and blocks of similar amino acid residues are shaded in blue, yellow, and green, respectively.
Since the phosphorylation of human HDAC1 can promote HDAC1 enzymatic activity and complex formation (Pflum et al., 2001), we were interested in determining whether the phosphorylation of S427 and S429 would affect the functioning and enzymatic activity of HDA6. We first examined the subcellular localization of transiently expressed wild-type HDA6-GFP and mutated HDA6-GFP (S427A, S429A, and S427/429A) in Arabidopsis protoplasts. Like HDA6-GFP, GFP fluorescence was observed in both the nucleus and cytosol using mutated HDA6-GFP (Supplemental Figure 5). Similar results were also obtained with transgenic plants expressing wild-type HDA6-GFP and mutated HDA6-GFP (S427A, S429A, and S427/429A) driven by the 35S promoter. These results indicate that S427A and S429A phosphorylation does not alter the subcellular localization of HDA6.
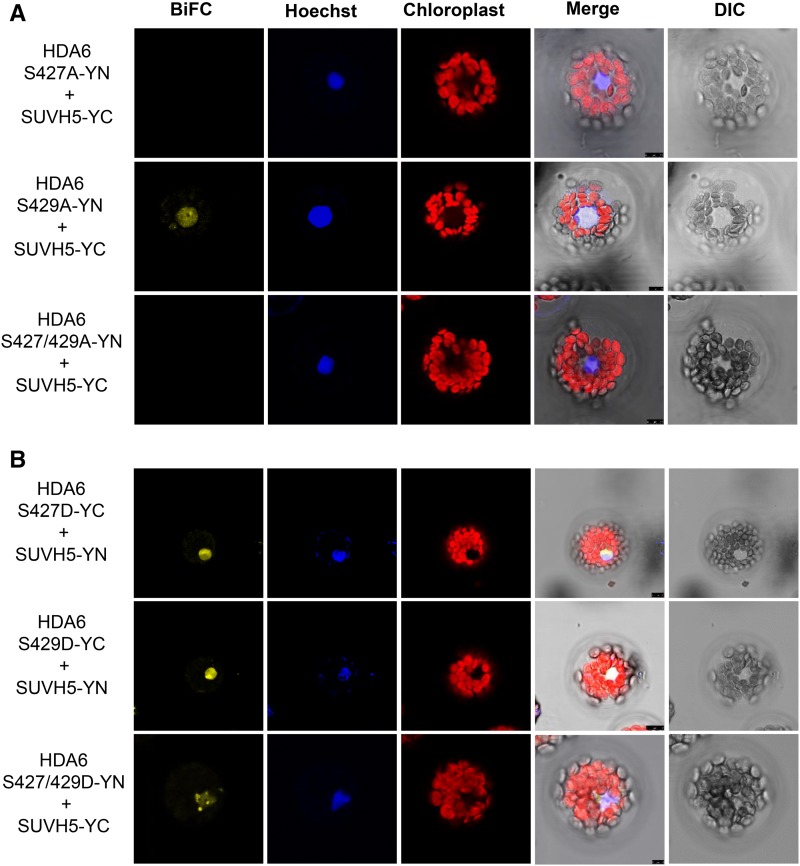
Next, we investigated whether phosphorylation of S427 and S429 affects the protein-protein interaction between HDA6 and SUVH5. As shown in Figure 7A, SUVH5 did not interact with HDA6-S427A or S427/429A. We also generated S427D, S429D, and S427/429D mutants to mimic the phosphorylation at serine. BiFC assays revealed that SUVH5 interacted with S427D, S429D, and S427/429D (Figure 7B). Similar results were also obtained with SUVH6 (Supplemental Figure 6). HDA6 can also interact with FLD and MET1 (Yu et al., 2011; Liu et al., 2012). BiFC assays revealed that FLD and MET1 did not interact with HDA6-S427A (Supplemental Figure 7). Together, these results indicate that the phosphorylation on S427 plays an important role in the interaction of HDA6 with its interacting proteins such as SUVH5, SUVH6, FLD, and MET1.
Figure 7.
Interaction of SUVH5 with HDA6 Phosphorylation Mutant Proteins.
(A) SUVH5 does not interact with HDA6 S427A in BiFC assays using Arabidopsis protoplasts.
(B) HDA6 S427/429D interact with SUVH5 in BiFC assays. DIC, differential interference contrast. Bars = 7.5 μm.
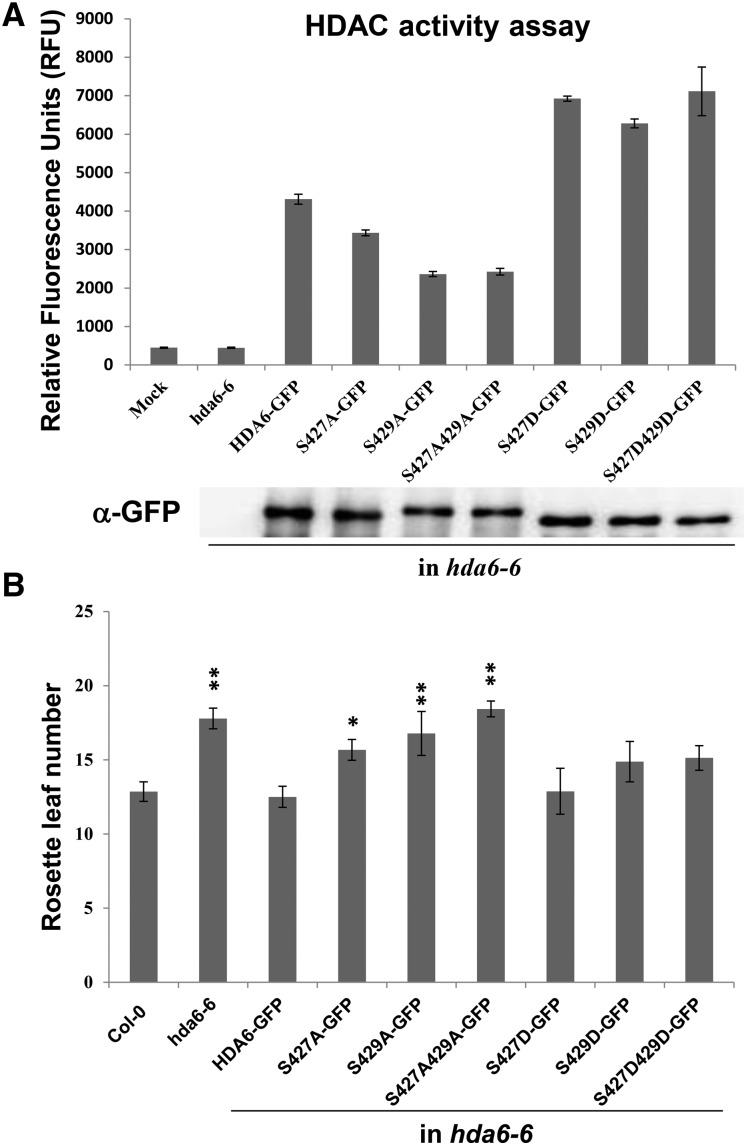
HDA6 S427 and S429 Phosphorylation Increases HDA6 Enzymatic Activity
To determine whether the phosphorylation of HDA6 influences its enzymatic activity, we expressed HDA6 phosphorylation mutant proteins fused to a GFP tag in hda6-6 plants. The HDA6 fusion proteins were immunoprecipitated with GFP trap beads, and enzymatic activity was determined in vitro. S427A, S429A, and S427/429A mutant proteins displayed diminished deacetylase activity compared with wild-type HDA6 (Figure 8A). In contrast, the S427D, S429D, and S427/S429D phosphorylation mimics showed increased enzymatic activity compared with wild-type and alanine mutant proteins.
Figure 8.
HDAC Activity Assay of HDA6 Phosphorylation Mutant Proteins.
(A) HDA6 and phosphorylation mutant proteins were expressed in hda6-6. Total protein extracts were immunoprecipitated by GFP-Trap magnetic beads. The expression of GFP-tagged proteins was analyzed by protein gel blot analysis with an anti-GFP antibody. Enzymatic activity is shown as relative fluorescence units (RFU). Values are means ± sd of three independent measurements.
(B) Rosette leaf number at flowering. Plants were grown under long-day conditions (16 h light/8 h dark). At least 20 plants were scored for each line. Values are means ± sd from three independent plant batches. *P < 0.05, **P < 0.01, t test.
We also analyzed the flowering phenotypes of hda6-6 plants expressing mutated HDA6. S427A, S429A, and S427/429A failed to restore the late flowering phenotype of hda6-6 (Figure 8B; Supplemental Figure 8). In contrast, S427D and S429D restored the late flowering phenotype of hda6-6 (Figure 8B; Supplemental Figure 8). Finally, we analyzed the expression of the flowering repressor genes FLC, MAF4, and MAF5. Consistent with the flowering phenotype, only S427D and S429D decreased the expression of FLC, MAF4, and MAF5 (Supplemental Figure 9). Furthermore, the expression of the TEs was reduced in hda6-6 overexpressing HDA6 and HDA6-S427D/429D but not HDA6-S427A/S429A (Supplemental Figure 10). Taken together, these results demonstrate that the phosphorylation of S427 and S429 is essential for both the function and enzymatic activity of HDA6.
DISCUSSION
HDA6 Interacts with Proteins Involved in Chromatin Remodeling and Histone Modification
In yeast and mammals, various RPD3/HDA1 HDACs were shown to function as components of several multiprotein complexes (Seto and Yoshida, 2014). However, little is known about plant HDAC protein complexes and their interaction partners. Here, we found that several histone modification enzymes can be coimmunoprecipitated by HDA6-GFP. Three histone methyltransferases (SUVH5, SDG2, and SDG29) as well as three histone deacetylases (HDA7, HD2A, and HD2B) were found to be associated with HDA6, suggesting that HDA6 might mediate crosstalk between histone deacetylation and methylation by interacting with other proteins involved in histone modification.
The cytosine methylation inhibitor 5-azadeoxycytidine and the HDAC inhibitor TSA showed similar efficacy in reactivating silenced rRNA genes (Chen and Pikaard, 1997), indicating that both DNA methylation and histone deacetylation play critical roles in regulating chromatin remodeling. We found that the DNA methyltransferase MET1 is associated with HDA6, confirming the previous finding that HDA6 directly interacts with MET1 to coregulate gene expression (Liu et al., 2012). Furthermore, HDA6 was also copurified with MSI1 and SWI3D. MSI1-like proteins bind histones and tether various protein complexes to histones or chromatin (Hennig et al., 2005). More recent studies indicated that MSI1 forms a complex with HDA19 to fine-tune ABA signaling by binding to the chromatin of ABA receptor genes and maintaining low histone H3K9ac levels (Mehdi et al., 2015). The SWI3D subunit is the core component of SWITCH/SUCROSE NONFERMENTING (SWI/SNF) chromatin-remodeling complexes, which are involved in controlling reproductive organ and embryonic development (Sarnowski et al., 2005). Taken together, these findings indicate that HDA6 functions in association with other proteins involved in chromatin remodeling to regulate gene expression.
The Phosphorylation of HDA6 Is Important for Its Activity and Function
Protein phosphorylation plays a crucial role in regulating many cellular processes in eukaryotes (Hunter, 1995). Sequence alignment comparing the Arabidopsis class I HDACs (HDA6, HDA7, HDA9, and HDA19) with human HDAC1/HDAC2 identified the conserved motif (SDS E/D E/D E/D) in HDA6, HDAC1, and HDAC2. Phosphorylation of human HDAC1 at two phosphorylation sites, S421 and S423, promotes its enzymatic activity and complex formation (Pflum et al., 2001). In contrast, mutations of S421 and S423 to alanine in human HDAC1 reduce its deacetylase activity and disrupt its interaction with its associated proteins (Pflum et al., 2001; Walters et al., 2009). In addition, inhibiting the phosphorylation of human HDAC2 also disrupts its interaction with other proteins (Galasinski et al., 2002; Walters et al., 2009). In this study, we found that mutations at HDA6 phosphorylation sites (S427A, S429A, and S427/429A) reduce its enzymatic activity, whereas phophomimics (S427D, S429D, and S427/429D) increase its enzymatic activity. Furthermore, the interaction between HDA6 and SUVH5 was abolished when S427 was changed to alanine. Interestingly, the S429 mutation affected the enzymatic activity of HDA6 but not its interaction with SUVH5, suggesting that the phosphorylation of S429 may affect the substrate binding of HDA6 but not its interaction with its partner proteins.
HDA6-S427A, S429A, and S427A/429A failed to restore the late flowering phenotype of hda6-6. In contrast, HDA6-S427D and S429D restored its late flowering phenotype. Furthermore, the expression of TEs was reduced in hda6-6 overexpressing HDA6-GFP and HDA6-S427D/429D but not HDA6-S427A/S429A. Taken together, our results support the notion that the phosphorylation of HDA6 promotes its enzyme activity and is required for its function and interaction with interacting proteins.
HDA6 and SUVH4/5/6 Act Coordinately to Regulate Transposon Silencing
Histone modification of proteins can promote or repress chromatin-mediated processes and alter transcription by acting alone or in concert in a context-dependent manner (Berger, 2007; Kouzarides, 2007; Bannister and Kouzarides, 2011). The crosstalk between histone modifications can occur via multiple mechanisms, and one modification may be dependent upon another (Bannister and Kouzarides, 2011). There is evidence for H3K4me3-dependent recruitment of histone acetyltransferases (Barski et al., 2007). In mammals, H3 acetylation and H3K4me signals outside of promoter regions have been correlated with functional enhancers in various cell types (Barski et al., 2007). Different histone modifications may also act cooperatively in order to efficiently recruit specific factors. For example, the human histone demethylase PHF8 specifically binds to H3K4me3 via its PHD finger, and this interaction is stronger when H3K9 and H3K14 are also acetylated on the same tail of H3 (Vermeulen et al., 2010). In mammals, transcriptional repression by the histone methyltransferase SUV39H1 is largely abolished in the presence of the HDAC inhibitor TSA, indicating that the repressive activity of SUV39H1 is dependent on HDAC activity (Vaute et al., 2002). Moreover, SUV39H1 can interact with HDAC1 and HDAC2 (Vaute et al., 2002).
In this study, we found that H3 acetylation levels are higher in the suvh456 mutant than in the wild type, supporting the notion that the activity of HDACs such as HDA6 is dependent on the functioning of SUVH4/5/6. In addition, the hda6-6 mutant has reduced H3K9me2 levels compared with the wild type, indicating that the activity of the H3K9 methyltransferases SUVH4/5/6 may also be dependent on HDA6. These findings indicate that both HDA6 and SUVH4/5/6 can regulate histone H3 acetylation and H3K9me2. The expression of transposons was higher in hda6-6 and suvh456 than in the wild type, indicating that both HDA6 and SUVH4/5/6 are required to maintain the stability of TEs. Furthermore, HDA6 and SUVH5 interact both in vivo and in vitro, supporting the notion that they function together to regulate transposon expression.
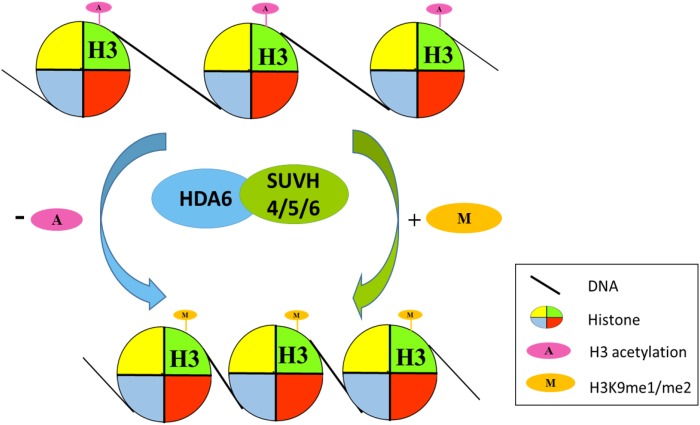
In conclusion, we constructed a model describing how SUVH4/5/6 and HDA6 are involved in transposon silencing by mediating the crosstalk between histone deacetylation and methylation (Figure 9). HDA6 and SUVH4/5/6 act collaboratively by removing the acetyl group from histone H3 and adding the methyl group to histone H3K9, resulting in chromatin condensation and transcriptional silencing.
Figure 9.
Working Model of the Coregulation of Transposons by SUVH4/5/6 and HDA6.
HDA6 and SUVH4/5/6 act collaboratively by removing the acetyl group from histone H3 and adding the methyl group to histone H3K9, resulting in chromatin condensation and transcriptional silencing.
METHODS
Plant Materials
Arabidopsis thaliana plants were grown under long-day (16 h light/8 h dark) conditions. hda6-6 (axe1-5) is a hda6 mutant carrying a point mutation on an HDA6 splicing site. suvh5 (GABI_263C05) and suvh456 (suvh4 suvh5 suvh6) were described previously (Johnson et al., 2008; Zheng et al., 2012). All mutant plants are in the Columbia (Col-0) background.
Plant Transformation
Amplified fragments of the full-length coding sequence of HDA6 were cloned into the Gateway pENTR SD/D Topo entry vector (Invitrogen) and transferred to a destination vector containing an N-terminal GFP tag (pK7WGF2) by LR recombination. HDA6 fragments containing point mutations (S427A, S429A, and S427/429A) were PCR amplified and subcloned into the pK7WGF2 vector. To add phosphomimetics (amino acid substitutions that mimic a phosphorylated protein) of the phosphorylation on serine, HDA6 fragments containing point mutations (S427D, S429D, and S427/429D) were PCR amplified and subcloned into the C-terminal GFP tag (pEarleyGate 103) vector. The primers used for produce these constructs are listed in Supplemental Table 1. The recombinant plasmids were introduced into hda6-6 mutant plants by the floral dip method using Agrobacterium tumefaciens strain GV3101.
RNA Extraction and Quantitative RT-PCR Analysis
Total RNA was extracted from 2-week-old plants using the TRIZOL reagent (Invitrogen) according to the manufacturer’s protocol. To synthesize cDNA, 2 μg of total RNA was used to synthesize cDNA with MMLV reverse transcriptase (Promega). qRT-PCR was performed using iQ SYBR Green Supermix solution (Bio-Rad). The gene-specific primers used for qRT-PCR are listed in Supplemental Table 1. Each sample was quantified at least in triplicate and normalized using Ubiquitin10 (UBQ) as an internal control.
Immuno-Isolation and LC-MS/MS Analysis
Approximately 25 g of leaf tissues from 20-d-old hda6-6 plants (control), hda6-6 plants expressing HDA6-GFP driven by the 35S promoter (pro35S: HDA6-GFP), or the 2-kb HDA6 native promoter (proHDA6: HDA6-GFP) was harvested and ground in liquid nitrogen. Immunoprecipitation buffer (100 mM Tris-HCl, pH 7.4, 75 mM NaCl, 0.05% SDS, 0.01% Triton X-100, 10% glycerol, and Protease inhibitor cocktail [Roche]) was used to extract the total cellular lysates. Thirty microliters of GFP-Trap_M beads (Chromotek) was used to perform the immunoprecipitation of GFP-HDA6 and control plants overnight with rotation at 4°C. The GFP-Trap bead-bound complex was washed five times with immunoprecipitation buffer at 4°C for 5 min each time. Then, on-bead trypsin digestion was performed as previously described (Turriziani et al., 2014). To further identify the HDA6-interacting proteins in the proHDA6:HDA6-GFP line, in-gel trypsin digestion was also performed for the bands from SDS-PAGE with relative molecular masses ranging from 75 to 100 kD, followed by analysis on a Thermo LTQ-Orbitrap XL mass spectrometer. Data were analyzed using a MASCOT MS/MS ions search.
Yeast Two-Hybrid Assays
Yeast two-hybrid assays were performed according to the instructions for the Matchmaker GAL4-based two-hybrid system 3 (Clontech). Different regions of HDA6 and SUVH5 cDNA fragments were subcloned into the pGADT7 (AD) and pGBKT7 (BD) vectors. All constructs were transformed into yeast strain AH109 by the lithium acetate method, and yeast cells were grown on a minimal medium/-Leu/-Trp according to the manufacturer’s instructions (Clontech). Transformed colonies were cultured overnight and diluted 10-fold and 100-fold before dropping onto minimal medium (-Ade/-Leu/-Trp/-His) plates.
Co-IP Assays
Co-IP assays were performed as previous described (Yu et al., 2011). Agrobacterium cultures carrying pro35S:GFP-SUVH5 and pro35S:HDA6-3X FLAG were coinfiltrated into wild tobacco (Nicotiana benthamiana) leaves. Two days after infiltration, the N. benthamiana leaves were harvested and ground in liquid nitrogen. Proteins were extracted in extraction buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 2 mM MgCl2, 1 mM DTT, 20% glycerol, and 1% Igepal CA-630 [Sigma-Aldrich]) containing protease inhibitor cocktail (Roche). The cell debris was pelleted by centrifugation at 14,000g for 20 min. Crude extracts (input) were immunoprecipitated with an anti-GFP antibody (catalog number ab290; Abcam). After 1 h of incubation at 4°C, 50 μL of protein G Mag Sepharose beads (GE) was added, followed by an additional hour of incubation. The beads were then washed with washing buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 2 mM MgCl2, 1 mM DTT, 10% glycerol, and 1% CA-630). The eluted protein samples were analyzed by protein gel blot analysis using anti-GFP (1:5000 dilution; catalog number ab290; Abcam) and anti-FLAG (1:3000 dilution; catalog number F3165; Sigma-Aldrich).
ChIP Assays
ChIP assays were performed as described (Yu et al., 2011). The chromatin was sheared to an average length of 500 bp by sonication prior to immunoprecipitation. The following antibodies were used: anti-acetylated histone H3K9K14 (catalog no. 06-599; Millipore) and anti-dimethylated histone H3K9 (catalog no. ab1220; Abcam). The cross-linking of DNA to immunoprecipitated proteins was reversed and the DNA was recovered by phenol:chloroform:isoamyl alcohol (25:24:1) purification. The purified DNA was analyzed by qRT-PCR using gene-specific primers (Supplemental Table 1).
BiFC Assays
To generate the constructs for BiFC assays, full-length cDNA fragments of HDA6, SUVH4, SUVH5, and SUVH6 were PCR amplified and subcloned into the pCR8/GW/TOPO (Invitrogen) vector and then recombined into the YN (pEarleyGate201-YN) and YC (pEarleyGate202-YC) vectors (Lu et al., 2010). The constructed vectors were transiently transformed into Arabidopsis protoplasts by PEG-mediated transfection. Transfected cells were then examined using a TCS SP5 confocal spectral microscope imaging system (Leica).
HDAC Activity Assays
Total proteins were extracted from 2-week-old transgenic plants expressing either wild-type or phosphorylation mutant HDA6 proteins (S427A, S429A, S427/429A, S427D, S429D, and S427/429D) fused with GFP using protein extraction buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl,1 mM PMSF, 10% glycerol, and 1% CA-630) containing protease inhibitor cocktail (Roche). The protein extracts were immunoprecipitated by GFP-Trap_MA (Chromotek) overnight at 4°C with rotation and washed twice with wash buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 10% glycerol, and 0.1% CA-630). HDAC enzymatic activity assays were performed using a Fluorometric HDAC activity assay kit (BioVision) following the manufacturer’s instructions. HDAC activity was measured in a fluorescence plate reader (excitation/emission = 360/460 nm).
Heat Map Analysis of Relative Expression Pattern of Transposons
The RPKM values of 241 transposons upregulated in the hda6-6 mutant were described previously (Yu et al., 2016). The average expression values of these transposons in suvh456 and met1 were obtained from the University of California Santa Cruz Genome Browser (genome.ucsc.edu) (Kuhn et al., 2009; Stroud et al., 2013). Heat map analysis was performed to compare the expression patterns of the transposons in different mutants using NetWalker (Komurov et al., 2012).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL libraries under the following accession numbers: HDA6, AT5G63110, SUVH4, AT5G13960; SUVH5, AT2G35160; SUVH6, AT2G22740; FLD, AT3G10390; and MET1, AT5G49160.
Supplemental Data
Supplemental Figure 1. An original image from an in vitro pull-down assay.
Supplemental Figure 2. Histone modification levels of hda6-6 and suvh456.
Supplemental Figure 3. Venn diagram showing the number of HDA6-, MET1-, and SUVH456-regulated transposons.
Supplemental Figure 4. Validation of transposon expression by qRT-PCR.
Supplemental Figure 5. Subcellular localization of HDA6 phosphorylation mutant proteins.
Supplemental Figure 6. Interaction of SUVH6 with HDA6 phosphorylation mutant proteins.
Supplemental Figure 7. Interaction of MET1 and FLD with HDA6 phosphorylation mutant proteins.
Supplemental Figure 8.Flowering phenotypes of transgenic plants expressing the HDA6 phosphorylation mutant proteins in hda6-6.
Supplemental Figure 9. Expression of the flowering repressor genes FLC, MAF4, and MAF5.
Supplemental Figure 10. Expression of transposon genes in transgenic plants expressing HDA6 phosphorylation mutant proteins.
Supplemental Table 1. Primers used in this study.
Acknowledgments
We thank Technology Commons, College of Life Science, National Taiwan University for the convenient use of the Bio-Rad real-time PCR system and the confocal spectral microscope imaging system. This work was supported by the Ministry of Science and Technology, Taiwan (105-2311-B-002-012-MY3, 105-2313-B-002-005, and 106-2313-B-002 -003). This work was also supported by the National Natural Science Foundation of China (31371308).
AUTHOR CONTRIBUTIONS
C.-W.Y., Y.-S.C., and K.W. designed research. C.-W.Y., R.T., S.-C.W., P.Y., M.L., S.Y., K.C., and W.-C.W. performed research. C.-W.Y., R.T, S.-C.W., Y.-S.C., and K.W. analyzed data. C.-W.Y., S.-C.W., and K.W. wrote the article.
References
- Alinsug M.V., Yu C.-W., Wu K. (2009). Phylogenetic analysis, subcellular localization, and expression patterns of RPD3/HDA1 family histone deacetylases in plants. BMC Plant Biol. 9: 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aufsatz W., Stoiber T., Rakic B., Naumann K. (2007). Arabidopsis histone deacetylase 6: a green link to RNA silencing. Oncogene 26: 5477–5488. [DOI] [PubMed] [Google Scholar]
- Bannister A.J., Kouzarides T. (2011). Regulation of chromatin by histone modifications. Cell Res. 21: 381–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barski A., Cuddapah S., Cui K., Roh T.Y., Schones D.E., Wang Z., Wei G., Chepelev I., Zhao K. (2007). High-resolution profiling of histone methylations in the human genome. Cell 129: 823–837. [DOI] [PubMed] [Google Scholar]
- Berger S.L. (2007). The complex language of chromatin regulation during transcription. Nature 447: 407–412. [DOI] [PubMed] [Google Scholar]
- Blevins T., Pontvianne F., Cocklin R., Podicheti R., Chandrasekhara C., Yerneni S., Braun C., Lee B., Rusch D., Mockaitis K., Tang H., Pikaard C.S. (2014). A two-step process for epigenetic inheritance in Arabidopsis. Mol. Cell 54: 30–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L.-T., Wu K. (2010). Role of histone deacetylases HDA6 and HDA19 in ABA and abiotic stress response. Plant Signal. Behav. 5: 1318–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L.-T., Luo M., Wang Y.-Y., Wu K. (2010). Involvement of Arabidopsis histone deacetylase HDA6 in ABA and salt stress response. J. Exp. Bot. 61: 3345–3353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z.J., Pikaard C.S. (1997). Epigenetic silencing of RNA polymerase I transcription: a role for DNA methylation and histone modification in nucleolar dominance. Genes Dev. 11: 2124–2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebbs M.L., Bender J. (2006). Locus-specific control of DNA methylation by the Arabidopsis SUVH5 histone methyltransferase. Plant Cell 18: 1166–1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebbs M.L., Bartee L., Bender J. (2005). H3 lysine 9 methylation is maintained on a transcribed inverted repeat by combined action of SUVH6 and SUVH4 methyltransferases. Mol. Cell. Biol. 25: 10507–10515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galasinski S.C., Resing K.A., Goodrich J.A., Ahn N.G. (2002). Phosphatase inhibition leads to histone deacetylases 1 and 2 phosphorylation and disruption of corepressor interactions. J. Biol. Chem. 277: 19618–19626. [DOI] [PubMed] [Google Scholar]
- Grunstein M. (1997). Histone acetylation in chromatin structure and transcription. Nature 389: 349–352. [DOI] [PubMed] [Google Scholar]
- Gu X., Jiang D., Yang W., Jacob Y., Michaels S.D., He Y. (2011). Arabidopsis homologs of retinoblastoma-associated protein 46/48 associate with a histone deacetylase to act redundantly in chromatin silencing. PLoS Genet. 7: e1002366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennig L., Bouveret R., Gruissem W. (2005). MSI1-like proteins: an escort service for chromatin assembly and remodeling complexes. Trends Cell Biol. 15: 295–302. [DOI] [PubMed] [Google Scholar]
- Hollender C., Liu Z. (2008). Histone deacetylase genes in Arabidopsis development. J. Integr. Plant Biol. 50: 875–885. [DOI] [PubMed] [Google Scholar]
- Hunter T. (1995). Protein kinases and phosphatases: the yin and yang of protein phosphorylation and signaling. Cell 80: 225–236. [DOI] [PubMed] [Google Scholar]
- Johnson L.M., Law J.A., Khattar A., Henderson I.R., Jacobsen S.E. (2008). SRA-domain proteins required for DRM2-mediated de novo DNA methylation. PLoS Genet. 4: e1000280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson L.M., Bostick M., Zhang X., Kraft E., Henderson I., Callis J., Jacobsen S.E. (2007). The SRA methyl-cytosine-binding domain links DNA and histone methylation. Curr. Biol. 17: 379–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komurov K., Dursun S., Erdin S., Ram P.T. (2012). NetWalker: a contextual network analysis tool for functional genomics. BMC Genomics 13: 282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T. (2007). Chromatin modifications and their function. Cell 128: 693–705. [DOI] [PubMed] [Google Scholar]
- Kuhn R.M., et al. (2009). The UCSC genome browser database: update 2009. Nucleic Acids Res. 37: D755–D761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence R.J., Earley K., Pontes O., Silva M., Chen Z.J., Neves N., Viegas W., Pikaard C.S. (2004). A concerted DNA methylation/histone methylation switch regulates rRNA gene dosage control and nucleolar dominance. Mol. Cell 13: 599–609. [DOI] [PubMed] [Google Scholar]
- Lippman Z., May B., Yordan C., Singer T., Martienssen R. (2003). Distinct mechanisms determine transposon inheritance and methylation via small interfering RNA and histone modification. PLoS Biol. 1: E67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Yu C.-W., Duan J., Luo M., Wang K., Tian G., Cui Y., Wu K. (2012). HDA6 directly interacts with DNA methyltransferase MET1 and maintains transposable element silencing in Arabidopsis. Plant Physiol. 158: 119–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Yang S., Zhao M., Luo M., Yu C.-W., Chen C.-Y., Tai R., Wu K. (2014). Transcriptional repression by histone deacetylases in plants. Mol. Plant 7: 764–772. [DOI] [PubMed] [Google Scholar]
- Liu X., Yang S., Yu C.W., Chen C., Wu K. (2016). Histone acetylation and plant development. In Developmental Signaling in Plants: The Enzymes, Vol. 40, C. Lin and S. Luan, eds (Burlington: Academic Press), pp. 173–199. [DOI] [PubMed] [Google Scholar]
- Lu Q., Tang X., Tian G., Wang F., Liu K., Nguyen V., Kohalmi S.E., Keller W.A., Tsang E.W., Harada J.J., Rothstein S.J., Cui Y. (2010). Arabidopsis homolog of the yeast TREX-2 mRNA export complex: components and anchoring nucleoporin. Plant J. 61: 259–270. [DOI] [PubMed] [Google Scholar]
- Luo M., Wang Y.-Y., Liu X., Yang S., Lu Q., Cui Y., Wu K. (2012a). HD2C interacts with HDA6 and is involved in ABA and salt stress response in Arabidopsis. J. Exp. Bot. 63: 3297–3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M., Yu C.-W., Chen F.-F., Zhao L., Tian G., Liu X., Cui Y., Yang J.-Y., Wu K. (2012b). Histone deacetylase HDA6 is functionally associated with AS1 in repression of KNOX genes in arabidopsis. PLoS Genet. 8: e1003114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehdi S., Derkacheva M., Ramström M., Kralemann L., Bergquist J., Hennig L. (2015). MSI1 functions in a HDAC complex to fine-tune ABA signaling. Plant Cell 28: 42–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miteva Y.V., Budayeva H.G., Cristea I.M. (2013). Proteomics-based methods for discovery, quantification, and validation of protein-protein interactions. Anal. Chem. 85: 749–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murfett J., Wang X.-J., Hagen G., Guilfoyle T.J. (2001). Identification of Arabidopsis histone deacetylase HDA6 mutants that affect transgene expression. Plant Cell 13: 1047–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey R., Müller A., Napoli C.A., Selinger D.A., Pikaard C.S., Richards E.J., Bender J., Mount D.W., Jorgensen R.A. (2002). Analysis of histone acetyltransferase and histone deacetylase families of Arabidopsis thaliana suggests functional diversification of chromatin modification among multicellular eukaryotes. Nucleic Acids Res. 30: 5036–5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrella G., Lopez-Vernaza M.A., Carr C., Sani E., Gosselé V., Verduyn C., Kellermeier F., Hannah M.A., Amtmann A. (2013). Histone deacetylase complex1 expression level titrates plant growth and abscisic acid sensitivity in Arabidopsis. Plant Cell 25: 3491–3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pflum M.K.H., Tong J.K., Lane W.S., Schreiber S.L. (2001). Histone deacetylase 1 phosphorylation promotes enzymatic activity and complex formation. J. Biol. Chem. 276: 47733–47741. [DOI] [PubMed] [Google Scholar]
- Probst A.V., Fagard M., Proux F., Mourrain P., Boutet S., Earley K., Lawrence R.J., Pikaard C.S., Murfett J., Furner I., Vaucheret H., Mittelsten Scheid O. (2004). Arabidopsis histone deacetylase HDA6 is required for maintenance of transcriptional gene silencing and determines nuclear organization of rDNA repeats. Plant Cell 16: 1021–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajakumara E., Law J.A., Simanshu D.K., Voigt P., Johnson L.M., Reinberg D., Patel D.J., Jacobsen S.E. (2011). A dual flip-out mechanism for 5mC recognition by the Arabidopsis SUVH5 SRA domain and its impact on DNA methylation and H3K9 dimethylation in vivo. Genes Dev. 25: 137–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarnowski T.J., Ríos G., Jásik J., Świeżewski S., Kaczanowski S., Li Y., Kwiatkowska A., Pawlikowska K., Koźbiał M., Koźbiał P., Koncz C., Jerzmanowski A. (2005). SWI3 subunits of putative SWI/SNF chromatin-remodeling complexes play distinct roles during Arabidopsis development. Plant Cell 17: 2454–2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seto E., Yoshida M. (2014). Erasers of histone acetylation: the histone deacetylase enzymes. Cold Spring Harb. Perspect. Biol. 6: a018713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroud H., Greenberg M.V., Feng S., Bernatavichute Y.V., Jacobsen S.E. (2013). Comprehensive analysis of silencing mutants reveals complex regulation of the Arabidopsis methylome. Cell 152: 352–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M., Kikuchi A., Kamada H. (2008). The Arabidopsis histone deacetylases HDA6 and HDA19 contribute to the repression of embryonic properties after germination. Plant Physiol. 146: 149–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- To T.K., et al. (2011). Arabidopsis HDA6 regulates locus-directed heterochromatin silencing in cooperation with MET1. PLoS Genet. 7: e1002055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner B.M. (2000). Histone acetylation and an epigenetic code. BioEssays 22: 836–845. [DOI] [PubMed] [Google Scholar]
- Turriziani B., Garcia-Munoz A., Pilkington R., Raso C., Kolch W., von Kriegsheim A. (2014). On-beads digestion in conjunction with data-dependent mass spectrometry: a shortcut to quantitative and dynamic interaction proteomics. Biology (Basel) 3: 320–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaute O., Nicolas E., Vandel L., Trouche D. (2002). Functional and physical interaction between the histone methyl transferase Suv39H1 and histone deacetylases. Nucleic Acids Res. 30: 475–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeulen M., Eberl H.C., Matarese F., Marks H., Denissov S., Butter F., Lee K.K., Olsen J.V., Hyman A.A., Stunnenberg H.G., Mann M. (2010). Quantitative interaction proteomics and genome-wide profiling of epigenetic histone marks and their readers. Cell 142: 967–980. [DOI] [PubMed] [Google Scholar]
- Walters M.S., Erazo A., Kinchington P.R., Silverstein S. (2009). Histone deacetylases 1 and 2 are phosphorylated at novel sites during varicella-zoster virus infection. J. Virol. 83: 11502–11513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C.-W., Chang K.-Y., Wu K. (2016). Genome-wide analysis of gene regulatory networks of the FVE-HDA6-FLD complex in Arabidopsis. Front. Plant Sci. 7: 555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C.-W., Liu X., Luo M., Chen C., Lin X., Tian G., Lu Q., Cui Y., Wu K. (2011). HISTONE DEACETYLASE6 interacts with FLOWERING LOCUS D and regulates flowering in Arabidopsis. Plant Physiol. 156: 173–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Tamaru H., Khan S.I., Horton J.R., Keefe L.J., Selker E.U., Cheng X. (2002). Structure of the Neurospora SET domain protein DIM-5, a histone H3 lysine methyltransferase. Cell 111: 117–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J., Chen F., Wang Z., Cao H., Li X., Deng X., Soppe W.J., Li Y., Liu Y. (2012). A novel role for histone methyltransferase KYP/SUVH4 in the control of Arabidopsis primary seed dormancy. New Phytol. 193: 605–616. [DOI] [PubMed] [Google Scholar]