Mutants in the translation initiation factor eIF5B reveal the importance of translation for recovery from heat stress and a possible role for this conserved factor in mRNA-specific translation.
Abstract
The conserved eukaryotic translation initiation factor 5B, eIF5B, is a GTPase that acts late in translation initiation. We found that an Arabidopsis thaliana mutant sensitive to hot temperatures 3 (hot3-1), which behaves as the wild type in the absence of stress but is unable to acclimate to high temperature, carries a missense mutation in the eIF5B1 gene (At1g76810), producing a temperature sensitive protein. A more severe, T-DNA insertion allele (hot3-2) causes pleiotropic developmental phenotypes. Surprisingly, Arabidopsis has three other eIF5B genes that do not substitute for eIF5B1; two of these appear to be in the process of pseudogenization. Polysome profiling and RNA-seq analysis of hot3-1 plants show delayed recovery of polysomes after heat stress and reduced translational efficiency (TE) of a subset of stress protective proteins, demonstrating the critical role of translational control early in heat acclimation. Plants carrying the severe hot3-2 allele show decreased TE of auxin-regulated, ribosome-related, and electron transport genes, even under optimal growth conditions. The hot3-2 data suggest that disrupting specific eIF5B interactions on the ribosome can, directly or indirectly, differentially affect translation. Thus, modulating eIF5B interactions could be another mechanism of gene-specific translational control.
INTRODUCTION
Eukaryotic protein synthesis requires numerous accessory factors that facilitate translation initiation, elongation, and termination. Translational control has been documented primarily at translation initiation, which involves multiple steps and protein factors (Sonenberg and Hinnebusch, 2009). In the canonical model of cap-dependent initiation, the small ribosomal subunit containing bound initiator methionyl-tRNA (Met-tRNAi) is recruited to the capped 5′ end of mRNA and scans to the AUG start codon to form the 48S initiation complex. Subsequently, the large ribosomal subunit joins to yield elongation-competent 80S ribosomes. The potential for regulatory interactions at initiation are obvious from the fact that there are at least 12 distinct eukaryotic translation initiation factors (eIFs), many with multiple subunits (Jackson et al., 2010).
It is becoming apparent that translation initiation factors play various important roles in plants (Browning and Bailey-Serres, 2015; Dutt et al., 2015), as they provide focal points for control of gene expression in response to intrinsic and external cues. Several factors have been demonstrated to modulate vegetative and reproductive growth processes. For example, eIF3h and eIF3e are essential during pollen germination (Roy et al., 2011), mutation in eIF4A confers a dwarfing phenotype (Vain et al., 2011), eIF4E is involved in root development (Martínez-Silva et al., 2012), and eIF5A plays a role in specification of cytokinin-mediated root protoxylem (Ren et al., 2013). In addition, there are reports regarding involvement of eIFs in abiotic stresses. Overexpression of eIF1A improved tolerance to salt stress (Rausell et al., 2003), while overexpression of eIF5A increased resistance to osmotic, nutrient, oxidative, and high temperature stresses (Ma et al., 2010; Xu et al., 2011; Wang et al., 2012). eIF4G plays significant roles against tungro spherical virus (Lee et al., 2010), and eIF4E is involved in resistance against plum pox virus (Wang et al., 2013), potato virus Y (Duan et al., 2012), and maize rough dwarf disease (Shi et al., 2013). Thus, specific manipulation of initiation factors may afford an approach to improve plant survival under stress or pathogen attack. However, a better understanding of the roles, regulation, and network interactions of these factors is required.
Only two IFs are universally conserved in prokaryotes, archaea, and eukaryotes, IF1/eIF1A and IF2/eIF5B (Wei et al., 1995; Choi et al., 1998; Kyrpides and Woese, 1998; Lee et al., 1999). eIF1A stimulates binding of eIF2-GTP-Met-tRNAi to the 40S ribosomal subunit and cooperates with eIF1 in promoting ribosome scanning and initiation codon selection. eIF5B is a ribosome-dependent GTPase that interacts with the Met-tRNAi, facilitates the final cleavage of the 20S pre-rRNA, and recruits the 60S ribosomal subunit to form the 80S ribosome in the final steps of translation initiation (Lebaron et al., 2012). On the ribosome, eIF1A and eIF5B interact with other initiation factors and GTP to position Met-tRNAi on the start codon to initiate translation accurately (Choi et al., 2000; Marintchev et al., 2003; Fringer et al., 2007; Zheng et al., 2014). However, the role of these factors in plants has not been well characterized. Here, we focus on translational control of gene expression mediated by eIF5B, for which only limited data are available from any organism. The eIF5B gene was first described in yeast (Choi et al., 1998), and strains deleted for this gene are viable but show a severe slow growth phenotype due to impaired translation initiation. Subsequently, eIF5B was described in Homo sapiens (Lee et al., 1999; Wilson et al., 1999) and Drosophila melanogaster (Carrera et al., 2000). Mutations in the GTP binding domain of human eIF5B affected its ability to promote translation (Wilson et al., 1999), and in Drosophila, homozygous dIF2 mutants are lethal in larval stages (Carrera et al., 2000). The only report of eIF5B in plants is of PeIF5B from pea (Pisum sativum), which in addition to its role in translation initiation was reported to have high thermostability (up to 95°C) and the ability to prevent thermal aggregation and promote refolding of heat-labile proteins (Rasheedi et al., 2007, 2010; Suragani et al., 2011).
We previously identified an ethyl methanesulfonate (EMS) mutant of Arabidopsis thaliana, hot3-1, that was unable to acquire tolerance to high temperature (Hong and Vierling, 2000). Further studies indicated that the hot3-1 mutant had reduced heat shock protein (HSP) accumulation and an inability to recover the activity of introduced firefly luciferase after heat stress (Hong et al., 2003). Here, we report that hot3-1 has a single point mutation in gene At1g76810 encoding a translation initiation factor 5B (eIF5B1), and investigation of additional, more severe alleles shows that eIF5B1 is essential for plant growth and development. Three other eIF5B homologs in the Arabidopsis genome cannot substitute for eIF5B1 function, and evidence indicates at least two are in the process of pseudogenization. Furthermore, the temperature sensitivity of the protein produced by the hot3-1 allele allows us to demonstrate the critical role of translation very early in the process of acclimation to high temperature. Using RNA-seq, we identify subsets of genes with altered translational efficiency in the hot3 mutants, providing insights into the importance of specific initiation factors in posttranscriptional gene regulation.
RESULTS
The Thermotolerance-Defective Mutant hot3-1 Encodes a Translation Initiation Factor
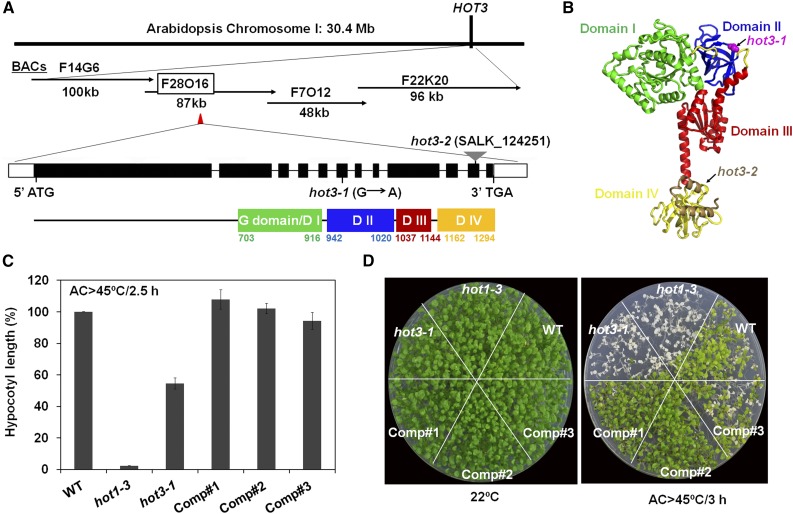
In previous research, we identified EMS-generated, recessive mutations in several loci in Arabidopsis that impaired the ability of seedlings to acclimate to severe high temperatures (Hong and Vierling, 2000). One of these mutants, hot3-1, showed reduced levels of HSPs and apparently reduced ability to recover translation after heat stress (Hong et al., 2003), but growth was like wild type in the absence of stress. We used map-based cloning to identify the HOT3 gene. A point mutation (G-to-A) that changes Gly-952 to Ser was found in gene AT1G76810, encoding an eIF5B translation initiation factor (Figure 1A). For clarity, we renamed HOT3 as eIF5B1 (AT1G76810). The 1294-amino acid eIF5B1 protein contains four conserved domains in the C-terminal half of the protein, and an ∼700-amino acid, N-terminal intrinsically disordered region (Figure 1A; Supplemental Figure 1). A structural model of eIF5B indicates that Gly-952 is a conserved residue located at the beginning of a β-strand in Domain II (Figure 1B) and could affect eIF5B interactions with the 40S ribosomal subunit.
Figure 1.
Identification and Characterization of HOT3.
(A) Diagram of the region identified by map-based cloning of HOT3. A G-to-A mutation changes Gly-952 to Ser in eIF5B (At1g76810; eIF5B1) in the hot3-1 mutant. Position of a T-DNA insertion in SALK_124251 (hot3-2) is also shown. Four conserved domains are found in the C-terminal half of eIF5B1 as analyzed at http://pfam.xfam.org/: EF-Tu GTP binding domain/G domain/Domain I, green; EF-Tu domain/Domain II, blue; IF-2 domain/Domain III, red; and Domain IV, orange.
(B) Structural model of the conserved domains of eIF5B1 based on eIF5B from the thermophilic yeast Chaetomium thermophilum (PDB: 4N3N) (the N-terminal ∼700 amino acids of HOT3 are not modeled as they are part of a disordered region not present in any crystal structure). Position of the hot3-1 mutation is shown in magenta as a space-filled residue. Conserved domains are colored as in (A), Domain IV is in yellow, with the exception of the final two helices (brown), which are deleted in hot3-2.
(C) Hypocotyl elongation after heat stress for the wild type, hot3-1, and three complemented lines (Comp#1, #2, #3) shows that eIF5B1 restores heat acclimation ability to hot3-1 seedlings. A T-DNA protein null allele of HSP101 (hot1-3) was used as a control for heat sensitivity. Hypocotyl elongation 2.5 d after heat treatment (AC >45°C /2.5 h: acclimation at 38°C for 1.5 h and 22°C for 2 h, followed by 45°C for 2.5 h) was measured and expressed as a percentage of growth during the same time period for seedlings that had not been heat treated. Error bars indicate se; n = 12.
(D) Complementation of hot3-1 with eIF5B1 rescues the heat sensitivity of 6-d-old light-grown hot3-1 seedlings. Seedlings were maintained at 22°C or subjected to heat stress: acclimation at 38°C for 1.5 h and 22°C for 2 h, followed by 45°C for 3 h (AC >45°C/3 h). Plates were photographed 5 d later.
To confirm that this mutation in eIF5B1 is responsible for the hot3-1 thermotolerance defect, we introduced the wild-type genomic sequence of eIF5B1 into the hot3-1 mutant. Three independent transgenic lines were generated (Supplemental Figure 2A) and tested for complementation. As expected, the hot3-1 mutant and the complemented lines showed a wild-type phenotype during all phases of growth (Supplemental Figure 2B). Heat stress tolerance assays demonstrated that the transgenic lines were complemented for the hot3-1 thermotolerance defects (Figures 1C and 1D). Additionally, the complemented lines showed a wild-type pattern of HSP expression after 2 h of recovery following heat stress (Supplemental Figure 2C). These data confirm that the missense mutation in eIF5B1 is responsible for the hot3-1 thermotolerance defects.
A Second eIF5B1 Allele Has Strong Effects on Plant Growth and Development
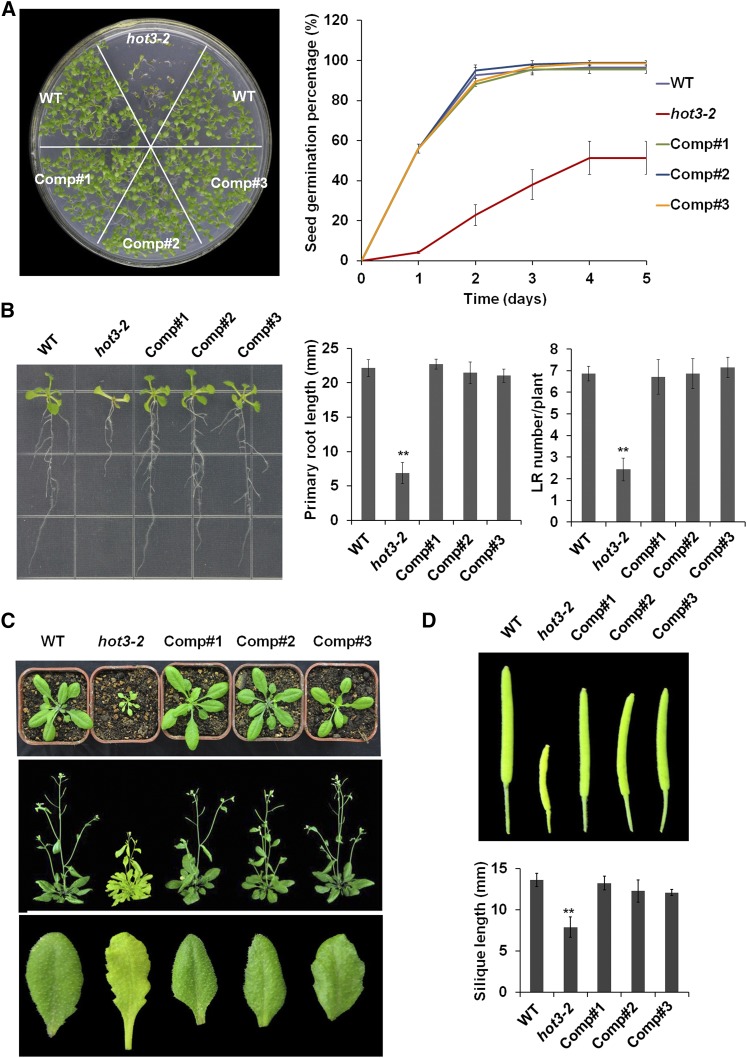
To better understand the function of eIF5B1, we isolated a T-DNA insertion mutation, hot3-2 (SALK_124251), which truncates the eIF5B1 protein by 30 amino acids (Figures 1A and 1B; Supplemental Figure 3A). The hot3-2 mutant produced a truncated transcript, but not a full-length transcript (Supplemental Figures 3B and 3C), and transcript sequence analysis revealed an in-frame stop codon in the T-DNA. Therefore, the hot3-2 mutant could potentially produce a partially functional protein.
Unlike hot3-1, the hot3-2 mutant shows pleiotropic developmental phenotypes; seed germination was significantly retarded, and total germination was decreased by 50% (Figure 2A). Seedlings of hot3-2 show reduced growth compared with the wild type, with shorter primary roots and arrested lateral root growth (Figure 2B), and an overall delay throughout vegetative growth and flowering, requiring >4 months to complete the life cycle (Figure 2C). Rosettes have an increased number of shallowly lobed and yellow-green leaves, and siliques are short (Figures 2C and 2D). To confirm these diverse phenotypes result from the hot3-2 defect, a wild-type genomic fragment of eIF5B1 was transferred into the hot3-2 mutant. Analysis of these transgenic plants showed complementation for all the hot3-2 phenotypes (Figure 2; Supplemental Figure 3). Therefore, eIF5B1 function is essential for multiple developmental and growth processes, consistent with the role of this factor in translation initiation. We also tested the heat tolerance phenotype of hot3-2, and results indicate that hot3-2 is heat sensitive (Figures 3B and 3C). However, the poor growth of hot3-2 makes it difficult to assess the degree of heat sensitivity compared with the wild type, as wild-type plants are always more vigorous even in the absence of stress.
Figure 2.
Phenotypes of the hot3-2 Mutant and Three Complemented Lines.
(A) Growth of hot3-2 seedlings compared with the wild type after 7 d (left). Seed germination assay in the wild type, hot3-2, and three complemented lines (right). Imbibed seeds were transferred to MS medium, and seed germination was recorded at the indicated time points. Each time point represents the means ± se of three replicates with >100 seeds per replicate.
(B) Phenotype of 10-d-old seedlings growing on vertically placed agar plates. Primary root length and the number of lateral roots were measured. Data are means ± se of three replicates (n = 15), **P < 0.01, compared with wild-type plants (two-tail Student t test).
(C) Morphology of the wild type, hot3-2, and three complemented lines. Two-week-old (top) and 6-week-old (middle) plants growing in soil and the shape of rosette leaves (bottom).
(D) Full-grown siliques. From left to right: the wild type, hot3-2, and three complemented lines (Comp #1, #2, #3). Silique length was measured and data are means ± se (n = 15), **P < 0.01, compared with wild-type plants (two-tail Student’s t test).
Figure 3.
Phenotypes of T-DNA Insertion Lines of Multiple eIF5B Homologs.
(A) Growth and development of 4-week-old mutants under normal conditions. hot3-1, hot3-2, eif5b2 (SALK_148816), eif5b3 (SALK_143304), and eif5b4 (SALK_056578) are shown.
(B) The 2.5-d-old dark-gown seedlings were treated by heat stress (A>45°C/1 h: acclimation at 38°C for 1.5 h and 22°C for 2 h, followed by 45°C for 1 h). Hypocotyl elongation was measured before (blue bars) and 2.5 d after (red bars) heat treatment. Data are means ± se (n = 12).
(C) Six-day-old seedlings were subjected to heat stress (AC>45°C/3 h: acclimation at 38°C for 1.5 h and 22°C for 2 h, followed by 45°C for 3 h) and photographed 5 d later.
Other eIF5B Homologs Do Not Substitute for the Function of eIF5B1
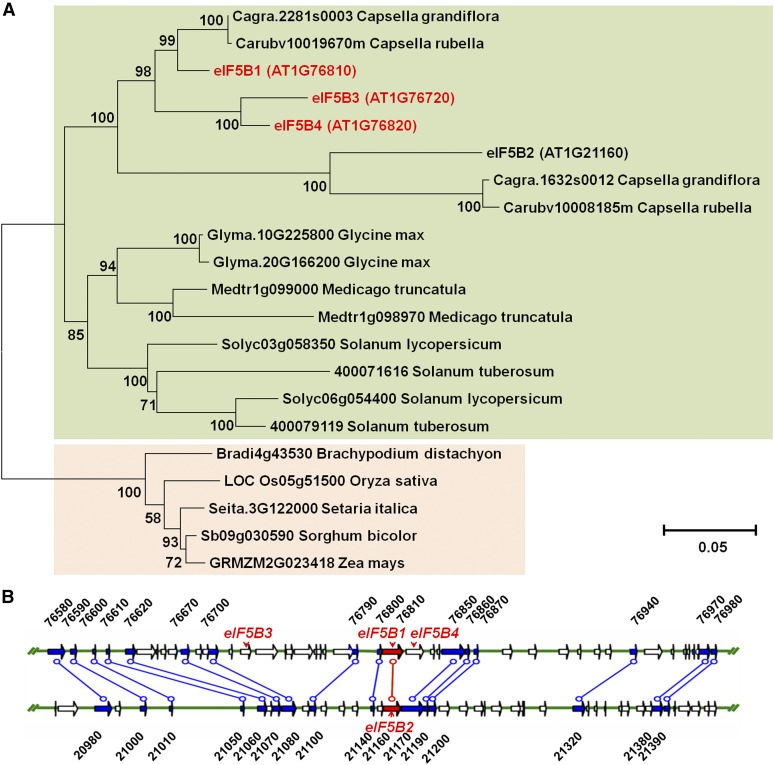
The phenotypes of the hot3 mutants was surprising, as there are three other homologous eIF5B proteins encoded in the Arabidopsis genome, which we designated eIF5B2 (AT1G21160), eIF5B3 (AT1G76720), and eIF5B4 (AT1G76820). These eIF5B homologs in the Col-0 accession share extensive conservation, including at the position of the hot3-1 mutation (Supplemental Figure 4). A phylogenetic tree of eIF5B from higher plant species shows two eIF5B homologs in most eudicots, compared with the four in Arabidopsis, while a single ortholog is found in monocots. Most eIF5B homologs from eudicots clustered together or with close relatives (Figure 4A; Supplemental Data Set 1), implying that multiple eIF5B genes emerged from recent duplications. Indeed, the eIF5B1 and eIF5B2 genes in Arabidopsis are contained in a large duplicated block of chromosome 1 (Figure 4B). Thus, specialized functions of the eIF5B genes would have evolved relatively recently.
Figure 4.
Phylogenetic Tree of eIF5B Genes from Various Species and the Chromosomal Positions of the Four eIF5B Homologs in Arabidopsis.
(A) The full-length amino acid sequences were aligned and the tree was generated using the neighbor-joining method in MEGA6. Bootstrap values (as a percentage of 1000 replicates) are provided at the branches. The scale bar represents two amino acid replacements per 100 positions.
(B) Data from the Plant Genome Duplication Database (http://chibba.agtec.uga.edu/duplication/) indicate that the four eIF5B genes in Arabidopsis are contained in a large duplicated block on chromosome 1. The four eIF5B genes are in red font.
The fact that the eIF5B1 mutant phenotypes are expressed in the background of the wild type, eIF5B2, 3, and 4 genes might be explained in different ways. These duplicated genes could have undergone subfunctionalization such that each eIF5B has a specialized role in translation initiation, or the genes could have different spatial and temporal patterns of expression. Alternatively, one or more of these genes might be nonfunctional or in the process of being eliminated or becoming pseudogenes. To distinguish these possibilities, we first tested if the three other eIF5B homologs are also required for plant development and thermotolerance by examining homozygous T-DNA insertion lines for each homolog (Supplemental Figure 5). Unlike hot3-1 and hot3-2, all these mutants showed wild-type growth and were not heat sensitive (Figure 3). Therefore, eIF5B1 clearly serves a unique role in Arabidopsis translation. We then evaluated the expression patterns of all four eIF5B genes. Consistent with publicly available data (http://jsp.weigelworld.org/expviz/expviz.jsp), the eIF5B homologs are differentially expressed, with eIF5B1 having the highest mRNA levels (Figures 5A and 5B). Analysis of transgenic plants carrying promoter-GUS fusions parallels the transcript analysis; eIF5B1 shows strong constitutive expression throughout the plant, while eIF5B2 shows much weaker expression and very limited expression in the roots. Virtually no GUS staining was detected for eIF5B3, and eIF5B4 had minimal staining (Figure 5C). Thus, the eIF5B1 homologs are not expected to compensate for the loss of eIF5B1 due to their much lower levels and different patterns of expression.
Figure 5.
Expression Patterns of Four eIF5B Genes from Arabidopsis.
(A) In silico analysis of the expression of four eIF5B genes. Data were obtained and organized from public microarray data (http://jsp.weigelworld.org/expviz/expviz.jsp) of transcript levels of the four eIF5B genes under heat stress. The black bar underneath the x axis indicates heat treatment period at 38°C, while the gray bar indicates the recovery period at 25°C following a 3-h heat treatment.
(B) Relative expression levels of eIF5B genes under heat stress. RNA from 5-d-old seedlings treated at 38°C in the dark for the indicated times was used for qRT-PCR. The relative levels of eIF5B mRNA were normalized with eIF5B1 set as 1.0. Actin 2 (At3g18780) was used as the internal standard. Data are means ± se from three technical replicates.
(C) Analysis of eIF5Bpro::GUS expression. Different tissue-specific expression patterns of the four eIF5B genes were observed. Bars = 1 mm.
Further analysis suggests that eIF5B3 and eIF5B4 are on the evolutionary road to elimination. These two genes appear as the most recent duplications, and their expression levels are very low. Consistent with eIF5B1 being the major functional copy, sequences from 80 Arabidopsis accessions (http://1001genomes.org/) show that eIF5B1 has <250 single nucleotide polymorphisms, 10 highly diverged regions and no major deletions, while eIF5B3 and eIF5B4 have >950 single nucleotide polymorphisms, at least 7 major deletions, and >200 highly diverged regions (Supplemental Figure 6). Furthermore, prediction and review of the sequence of eIF5B3 indicates the transcript might be cleaved by ath-MIR2936 (Grant-Downton et al., 2009), accounting in part for the low RNA levels (Supplemental Figure 7A). We also found that the eIF5B4 sequence in the database is incorrect; the correct coding region is deleted for a single “A” nucleotide, resulting in a premature stop codon (Supplemental Figure 7B). Therefore, eIF5B3 and eIF5B4 are very likely not contributing any significant functional eIF5B protein and are gradually being eliminated from the Arabidopsis genome or converted to pseudogenes.
hot3-1 Inhibits Translation Initiation during Recovery from Heat Stress
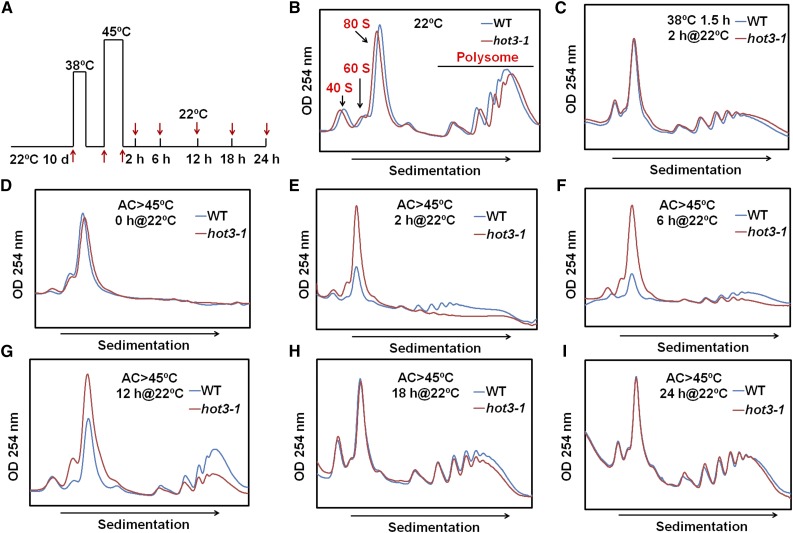
The reduced heat tolerance phenotypes of the hot3-1 mutant suggested that eIF5B1 may be involved in controlling translation of genes required for heat acclimation. To test this, we isolated polysome-bound, translationally active mRNAs on sucrose gradients (Juntawong et al., 2014) and identified changes in the translating mRNA profiles caused by mutation of eIF5B1 using genome-wide RNA sequencing (RNA-seq) (Supplemental Figure 8). First, we investigated the dynamic changes in polysome profiles during heat stress and recovery in wild type and hot3-1 mutant plants. Ten-day-old Arabidopsis seedlings were acclimated at 38°C for 1.5 h and 22°C for 2 h, subjected to heat stress at 45°C for 2 h, and then allowed to recover at 22°C for different times as indicated in Figure 6A. In accordance with the phenotype, essentially identical polysome profiles were obtained in the wild type and hot3-1 under control conditions (Figure 6B). Heat acclimation (treatment at 38°C plus 2 h recovery at 22°C) did not lead to a severe reduction in the polysome fraction in wild-type or mutant plants (Figure 6C). However, as reported for other plant species (Key et al., 1981; Porankiewicz and Gwozdz, 1995), severe heat stress, even after acclimation (45°C), caused essentially a complete loss of polysomes (Figure 6D), indicating a dramatic inhibition of translation in both the wild type and hot3-1. During recovery, the polysome fraction gradually increased. However, in the wild type, recovery of polysomes was observed even after 2 h, and recovery was mostly complete after 12 h, while in hot3-1 recovery lagged behind and required over 18 h to return to wild-type levels (Figures 6E to 6I). Thus, the hot3-1 mutation confers heat sensitivity on translation initiation.
Figure 6.
Time Course of Recovery of Polysome Accumulation after Heat Stress in 10-d-Old Seedlings of the Wild Type and hot3-1.
(A) Temperature treatment protocol. Materials for polysome profiles were obtained at the times indicated (AC>45°C: acclimation at 38°C for 1.5 h and 22°C for 2 h, followed by 45°C for 2 h).
(B) to (I) Polysome profiles were analyzed by sucrose gradient sedimentation, and OD254 (arbitrary units) was measured for the wild type (blue) and hot3-1 (red) at the times indicated. Samples represent equal amounts of RNA for the wild type and hot3-1 at each time point analyzed. The experiment was repeated more than three times with comparable results.
Differences in Translational Regulation in hot3-1 Emphasize the Importance of Translation in the Development of Heat Tolerance
To determine how the hot3-1 mutation might alter translating mRNA profiles, we used RNA-seq to examine total and polysome-bound (PB) mRNA from control plants (CK), from plants subjected to heat acclimation (AC) (as in Figure 6C) and from plants allowed to recover for 12 h after 45°C heat stress (HS) (as in Figure 6G). Although there is no obvious difference in polysome profiles between the wild type and hot3-1 after heat acclimation (Figure 6C), we hypothesized alterations during acclimation could contribute to hot3-1 heat sensitivity. We chose the 12-h recovery time point because, relative to the polysome fraction, the monosome fraction (80S) of hot3-1 was greater than that of the wild type, and polysomes were restored to a lesser extent in hot3-1 than in the wild type (Figure 6G). We obtained an average of ∼15 million reads that mapped to 20,611 annotated mRNA transcripts (Supplemental Data Set 2), and all were sufficiently represented to provide measurements of their translational status. The expression of each gene was normalized as counts per million reads mapped using edgeR software (Robinson et al., 2010) (Supplemental Data Set 3). There was high data reproducibility for all genes in both total and PB mRNA samples, with an r2 value of 0.92 to 0.98, with an average value of 0.96. Additionally, the data were validated by RT-qPCR of 10 randomly selected genes (Supplemental Figures 9A to 9C).
Quantifying total and PB mRNA levels allowed us to identify changes in translation efficiency (TE; calculated as PB mRNA/Total mRNA), which provides a better predictor of protein synthesis than measurements of mRNA levels (Ingolia et al., 2009). The correlation between the changes of control and heat acclimated or heat stressed samples in total and PB mRNA had an r2 value of 0.818 to 0.853 (Supplemental Figure 10), significantly lower than expected for identical samples, indicating a layer of regulation at the translational level during acclimation and recovery from heat stress.
To evaluate the extent of translational control, we calculated the fold change in TE using Xtail software with a false discovery rate (FDR) < 0.01. First, we compared TE between the wild type and hot3-1 under both control and heat acclimation conditions (CKTE:hot3-1/WT and ACTE:hot3-1/WT, as diagrammed in Supplemental Figure 8; Supplemental Data Set 4). No significant differences were observed for CKTE:hot3-1/WT (Figures 7A and 7B, lane 4), which was expected, as hot3-1 shows no significant phenotype under optimal growth conditions. By contrast, 233 genes (88 increased/145 decreased) were identified to have altered TE in hot3-1 compared with the wild type after heat acclimation (Figures 7A and 7B, lane 3). Gene Ontology (GO) analysis of translationally upregulated genes in hot3-1 after heat acclimation revealed enrichment in photosynthesis (GO:0015979), glucose catabolic process (GO:0006007), hexose metabolic process (GO:0019318), and monosaccharide catabolic process (GO:0046365), while there was no enrichment in any specific biological process for the downregulated genes (Supplemental Data Set 5, “GO-AC”; Figure 7C, lanes 1 and 2). To examine how translational differences during acclimation might further contribute to the hot3-1 heat-sensitive phenotype, we also compared TE between heat acclimation and control conditions for both the wild type and hot3-1. In the wild type, Xtail identified 1124 translationally regulated genes (286 upregulated/845 downregulated) of the 18,680 genes measured (Figures 7A and 7B, lane 1). Translationally upregulated genes included translation (GO:0006412), ribosome biogenesis (GO:0042254), chromatin assembly or disassembly (GO:0006333), response to stress (GO:0006950), and response to heat (GO:0009408), while developmental process (GO:0032502), plant-type cell wall biogenesis (GO:0009832), and organelle development (GO:0048513) gene categories were downregulated (Supplemental Data Set 5, “GO-AC-WT”; Figure 7C, lanes 3 and 4). In hot3-1, 1406 genes (587 downregulated/819 upregulated) translationally regulated genes were identified (Figures 7A and 7B, lane 2). Translation (GO:0006412), ribosome biogenesis (GO:0042254), and photosynthesis (GO:0015979) were enriched in the upregulated genes, and cellular component organization (GO:0016043), developmental process (GO:0032502), and plant-type cell wall biogenesis (GO:0009832) were enriched in the downregulated genes (Supplemental Data Set 5, “GO-AC-hot3-1”; Figure 7C, lanes 5 and 6). Additionally, genes responsive to stress and heat were translationally upregulated in the wild type, but not in hot3-1, suggesting that the loss of eIF5B1 activity in hot3-1 may affect heat tolerance due to a defect in translation initiation during acclimation that propagates effects to the later time points of recovery from heat stress. Thus, despite similar polysome profiles, the hot3-1 defect has already resulted in altered TE of specific genes during acclimation to heat stress.
Figure 7.
Identity of Genes with Altered TE in hot3-1 and after AC or during Recovery from 45°C HS.
(A) Volcano plots of TE changes generated from Xtail. Log2 of TE-fold change is shown on the horizontal axis, and –log10 of the adjusted P value is shown on the vertical axis. Red and blue dots represent significantly up- and downregulated genes (FDR < 0.01). Green dots are genes with a nonsignificant TE change. Total gene number used for the analysis is shown in parentheses.
(B) Hierarchical clustering analysis of the log2-transformed TE-fold change values. Each column represents the TE variation between the two samples indicated. Red and green indicate a relative increase or decrease of TE in the comparison. Black indicates that there is no significant difference in TE. The log2-transformed TE-fold changes are color coded according to the scale shown and plotted using the R heatmap.2 function from the R “gplots” library. The 3986 genes that changed TE in one comparison at least were used.
(C) GO functional categories of genes with translationally up- and downregulated levels for the indicated comparisons. The color in each cell indicates –log10 (P values) of the GO enrichment according to the scale shown, and blank cells indicate not significant. U, upregulation; D, downregulation.
The changes in TE of specific genes in hot3-1 compared with the wild type are even more evident following 12 h of recovery from the 45°C heat stress. The TE of 595 mRNAs is increased and 389 mRNAs decreased in hot3-1 compared with the wild type (Figures 7A and 7B, lane 6). The most abundant gene categories preferentially translated in hot3-1 were translation (GO:0006412), ribosome biogenesis (GO:0042254), gene expression (GO:0010467), and chromatin assembly or disassembly (GO:0006333), while translationally downregulated categories were stimulus response (GO:0050896), stress response (GO:0006950), and heat response genes (GO:0009408) (Supplemental Data Set 5, “GO-HS”; Figure 7C, lanes 7 and 8). In the wild type, 498 translationally regulated genes (204 upregulated/294 downregulated) were altered compared with the wild type control (Figures 7A and 7B, lane 5). Upregulated genes included gene expression (GO:0010467), response to stress (GO:0006950), and photosynthesis (GO:0015979), while biosynthetic process (GO:0009058), ATP synthesis-coupled proton transport (GO:0015986), and ion transmembrane transport (GO:0034220) gene categories were enriched in translationally downregulated genes (Supplemental Data Set 5, “GO-HS-WT”; Figure 7C, lanes 9 and 10), which is consistent with previous research (Yángüez et al., 2013). Notably, many more (total 2098; 878 downregulated/1220 upregulated) translationally regulated genes were identified in hot3-1 (Figures 7A and 7B, lane 7). Translation (GO:0006412), ribosome biogenesis (GO:0042254), and chromatin assembly or disassembly (GO:0006333) were enriched in the upregulated genes, and response to stress (GO:0006950), response to stimulus (GO:0050896), and response to heat (GO:0009408) were downregulated genes (Supplemental Data Set 5, “GO-HS-hot3-1”; Figure 7C, lanes 11 and 12). This misregulated translation during recovery from heat stress is likely causally related to the heat sensitivity phenotype of the hot3-1 mutant. Therefore, it was of particular interest to examine genes whose TE is downregulated during heat stress in hot3-1 relative to the wild type. GO analysis of the 389 translationally downregulated genes indicated that 69 genes are classified in the “response to stress” functional category (Supplemental Data Set 5, “Stress-related genes”) and 13 genes are classified in the “heat response” functional category (Supplemental Figure 11). Among the 13 heat-related genes, MULTIPROTIN BRIDGING FACTOR 1C (MBF1C) (Suzuki et al., 2005), HEAT SHOCK FACTOR 7A (HSFA7A) (Larkindale and Vierling, 2008), HEAT SHOCK PROTEIN 18.5 (HSP18.5) (Spano et al., 2004), HEAT SHOCK PROTEIN 17.6A (HSP17.6A) (Sun et al., 2001), CHITINASE-LIKE PROTEIN1 (CTL1) (Kotak et al., 2007), CASEIN LYTIC PROTEINASE B3 (CLPB3), and CLPB4 (Lee et al., 2007) have been shown to be positively correlated with heat tolerance in previous studies, likely contributing to the hot3-1 stress phenotype.
The hierarchical clustering of TE fold changes between the wild type and hot3-1 and between treatments (Figure 7B) provides a genome-wide picture of the differences in translational profiles. The wild type and hot3-1 after heat stress (HSTE:hot3-1/WT) were clearly separated from the control (CKTE:hot3-1/WT) and heat acclimation (ACTE:hot3-1/WT) samples. For the wild type and hot3-1, genes detected under the acclimation treatment were clustered together (WTTE:AC/CK with hot3-1TE:AC/CK), whereas after heat stress (WTTE:HS/CK with hot3-1TE:HS/CK), they were separated. This suggests that TE changes during acclimation are more dependent on treatment, while those after heat stress are more dependent on genotype. In total, a major change in TE between the wild type and hot3-1 occurred after heat stress compared with control and heat acclimation.
The fact that the hot3-1 phenotype is limited to temperature stress suggested the mutation makes eIF5B1 temperature sensitive. To address this possibility, we examined the time course of loss of polysomes after plants were shifted to 45°C and found that polysomes disappear more rapidly in the hot3-1 mutant than in the wild type (Supplemental Figure 12). These results support the conclusion that the hot3-1 mutant protein is rapidly inactivated by high temperature, leading to a global defect in translation initiation. Therefore, the slow recovery of polysomes is likely due to the need to refold or resynthesize eIF5B1 to fully restore translation. Altogether, the phenotypic results from this rapid, temperature-sensitive inactivation of eIF5B1 emphasize the significance of eIF5B1 activity and the critical role of translation in heat stress tolerance.
hot3-2 Decreases Translation Efficiency of Ribosome, Electron Transport, and Auxin-Related Genes
The multiple growth defects of the hot3-2 mutant indicate that this more severe allele of eIF5B1 has constitutive defects in translation. To better understand how the hot3-2 mutation impacts the phenotype, we analyzed polysome profiles from 10-d-old wild-type and hot3-2 seedlings in the absence of stress. Compared with the wild type, the hot3-2 mutant has decreased levels of 40S and 60S ribosomal subunits and of 80S monosomes relative to polysomes (Figure 8A). In addition, hot3-2 showed more small polysomes than large polysomes, which is indicative of a defect in translation, potentially at the initiation step.
Figure 8.
Identification of Genes with Altered TE in the hot3-2 Mutant.
(A) Polysome profiles of 10-d-old seedlings from the wild type and hot3-2. Positions of the 40S, 60S, 80S ribosomes, and polysomes are indicated. SP, small polysomes; LP, large polysomes. The experiment was repeated three times with similar results.
(B) Volcano plots for the differences in TE between the wild type and hot3-1 as generated by Xtail. Log2 of TE-fold change is shown on the horizontal axis, and –log10 of the adjusted P value is shown on the vertical axis. Red and blue dots represent significantly up- and downregulated genes in hot3-2 compared with the wild type (FDR < 0.01). Green dots represent genes with no significant change in TE. Total gene number used for analysis is shown in parentheses.
(C) Biological process GO category of genes with translationally up- (U) and downregulated (D) levels in hot3-2 compared with the wild type. Other details as in Figure 7.
To identify cohorts of mRNAs that are altered by impairing eIF5B1 function in the hot3-2 mutant, we again used RNA-seq to compare the relative abundance of PB and total mRNA in the wild type and hot3-2 under optimal growth conditions (Supplemental Data Sets 2 and 3). High data reproducibility of both total and PB mRNA levels, with an average r2 value of 0.96, was observed, and the RNA-seq data were validated by RT-qPCR (Supplemental Figure 9D). To identify genes with altered translation in hot3-2 compared with the wild type, we calculated TE for each gene as described for experiments with hot3-1 above. Results identified 2589 genes (1111 increased/1478 decreased) with altered TE in hot3-2 compared with the wild type (Figure 8B; Supplemental Data Set 4). The most abundant GO categories with decreased TE in hot3-2 were translation (GO:0006412), ribosome biogenesis (GO:0042254), nucleosome assembly (GO:0006334), electron transport chain (GO:0022900), response to hormone stimulus (GO:0009725), and response to auxin stimulus (GO:0009733). Downregulated translation of these genes might explain the phenotypic growth defects of hot3-2. In addition, the most abundant preferentially translated mRNAs in hot3-2 were in photosynthesis (GO:0015979), cellular nitrogen compound metabolic process (GO:0034641), carboxylic acid metabolic process (GO:0019752), transport (GO:0006810), and response to nutrient levels (GO:0031667) (Figure 8C; Supplemental Data Set 5, “GO-hot3-2”). These results suggest that specific mRNAs have a differential requirement for optimal eIF5B function. Further analysis found that the 178 translation-related genes (GO:0006412) with downregulated TE in hot3-2 include 16 translation initiation factors and 153 ribosomal proteins (Supplemental Data Set 5, “Translation-related genes”).
To examine whether mRNAs with altered TE in hot3-2 have any specific structure or sequence features, we calculated characteristics of the 5′UTR (untranslated region) and coding sequence (CDS) for each gene with altered TE and averaged the values for each up- or downregulated gene group. No significant differences were found in characteristics of the length, %GC content or normalized mean free energy of the 5′UTR (Supplemental Figure 13B). However, the 1478 translationally downregulated mRNAs in hot3-2 have, on average, a significantly shorter CDS length than that of the control (all Arabidopsis mRNAs or mRNAs randomly selected from the unregulated pools) or upregulated mRNAs (Supplemental Figure 13A). These results suggest that eIF5B activity directly or indirectly impacts efficiency of translation of genes with shorter CDS.
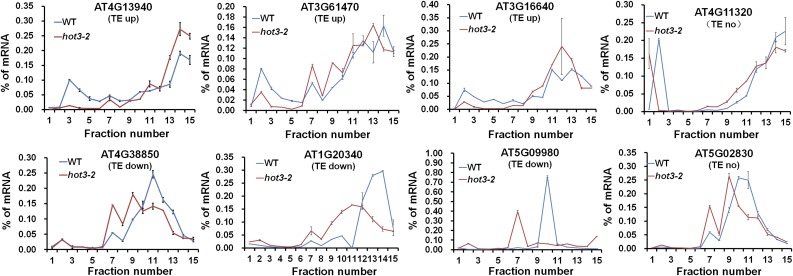
We considered that reduced translation of mRNAs with shorter CDS in hot3-2 could result from a general decrease in translation initiation, which could shift all mRNAs to smaller polysomes, resulting in loss of shorter mRNAs from the PB fraction. To test this possibility, we monitored the distribution of mRNA across the polysome fractions of eight selected genes, three that showed upregulated, three that showed downregulated, and two that showed unchanged TE in hot3-2. Polysome gradients from wild-type and hot3-2 seedlings were fractionated, and the resulting fractions were analyzed by RT-qPCR, using added luciferase RNA in each fraction to normalize the data. mRNAs identified as having an upregulated TE in hot3-2 (AT4G13940, AT3G61470, and AT3G16640) show shifts into larger polysomes, while three mRNAs with downregulated TE in hot3-2 (AT4G38850, AT1g02340, and AT5G09980) are shifted to smaller polysomes (Figure 9). Two of these mRNAs, AT3G61470 (LHCA2) involved in photosynthesis and At4g38850 (SAUX15) involved in response to auxin, were identified as relevant to the hot3-2 phenotype. We also assayed two genes (AT4G11320 and AT5G02830) whose TE appeared not to change. These data confirm that TE values obtained by the polysome profiling are consistent with changes in ribosome occupancy on specific transcripts and importantly indicate that reduced eIF5B function does not just shift all mRNAs to smaller polysomes as could be expected from a global decrease in translation initiation.
Figure 9.
Distribution of Specific mRNAs in Polysome Gradients.
The relative mRNA levels of three TE upregulated genes (AT4G13940, AT3G61470, and AT3G16640), three TE downregulated genes (AT4G38850, AT1g20340, and AT5G09980), and two genes (AT4G11320 and AT5G02830) whose TE appeared not to change were determined by RT-qPCR performed from the top (fraction 1) to the bottom (fraction 15) of the gradient. Expression values were normalized based on the addition of equal luciferase RNA to each fraction prior to RNA extraction, wild type (blue) and hot3-2 (red). Standard deviations were calculated from technical repeats.
DISCUSSION
Gene-specific regulation of translation initiation is widespread, phylogenetically conserved, and occurs under a variety of environmental and developmental conditions (Branco-Price et al., 2005; Liu et al., 2012; Ueda et al., 2012; Lin et al., 2014; Li et al., 2015). Previous work also indicates that mutations in individual eukaryotic translation initiation factors can disrupt gene-specific pathways of translational control (Kim et al., 2007; Martínez-Silva et al., 2012; Rubio et al., 2014), but there is no comprehensive information about the extent to which translational regulation may involve eIF5B. In Saccharomyces cerevisiae, eIF5B is required for rapid growth, but not for viability (Choi et al., 1998), and eIF5B from Drosophila has a role in germplasm function (Carrera et al., 2000). Upregulation of eIF5B controls cell cycle arrest and specific developmental stages in immature Xenopus laevis oocytes and mouse embryonic stem cells and has effects during growth factor deprivation of mammalian cells (Lee et al., 2014). Mammalian eIF5B and eIF5 together stimulate 48S initiation complex formation, influencing initiation codon selection during ribosomal scanning (Pisareva and Pisarev, 2014). Biochemical studies of a putative eIF5B from pea reported that it bound GTP as expected, was highly heat stable, and had properties of a molecular chaperone (Rasheedi et al., 2010; Suragani et al., 2011). However, the possibility that eIF5B could differentially impact translation of specific mRNAs has not been investigated, and there are no data on the requirement for eIF5B in plants. Through studies in Arabidopsis of a temperature-sensitive allele of eIF5B1 (hot3-1), we demonstrate that restoring translation immediately following heat stress is critical to stress recovery. In addition, translational profiling of a more severe allele (hot3-2) under optimal growth temperatures demonstrates that eIF5B1 is essential for normal growth and development in plants and further suggests that eIF5B may differentially affect translation of specific mRNAs.
Structural Basis of eIF5B Mutant Defects
We earlier isolated the Arabidopsis hot3-1 mutant based on the impaired ability of seedlings to acclimate to severe high temperature (Hong and Vierling, 2000; Hong et al., 2003). Here, we determined the hot3-1 phenotype resulted from a point mutation in an eIF5B gene that we designated eIF5B1. The mutated residue, Gly-952, is conserved in eIF5B from plants and other organisms, including bacteria and humans, but is not associated with any specific function. The wild-type phenotype of hot3-1 plants in the absence of heat stress indicates this mutation has little impact on eIF5B1 function at normal growth temperatures. We conclude that the hot3-1 mutation is temperature sensitive, as polysome collapse occurs more rapidly in the mutant than the wild type after heat stress. Delayed recovery of polysomes after heat stress could then be due to the need to refold heat-aggregated eIF5B or to resynthesize it. Detailed analysis of the stability and solubility of the mutant protein will be required to confirm this conclusion.
To better understand the function of eIF5B, we isolated a second mutant allele, hot3-2, that truncates eIF5B1 by 30 amino acids and has pleiotropic developmental phenotypes, including shorter primary roots, arrested lateral root growth, delayed flowering, and short siliques. A structural model reveals that the hot3-2 mutation removes the final two C-terminal helices of eIF5B, which are reported to bind to the C-terminal tail of eIF1A (Nag et al., 2016), an interaction required for efficient subunit joining (Acker et al., 2006), the final step in translation initiation. We do not think that hot3-2 is a null allele, as we recently obtained two other homozygous, insertional mutant alleles of eIF5B1, hot3-3 (SAIL_1147_G06) and hot3-4 (SALK_135561), with more severe phenotypes. Although the mutant plants are still viable, hot3-3 and hot3-4 are smaller than hot3-2 and produce very few seeds (Supplemental Figure 14). We suggest that expression of eIF5B2 may provide enough eIF5B activity to allow survival of the hot3-2 and hot3-4 mutants, but to date the poor fertility of the latter mutants has prevented us from creating double mutants to test this hypothesis.
The Four Arabidopsis eIF5B Homologs Show Divergent Fates
Arabidopsis has three other eIF5B homologs in addition to eIF5B1: eIF5B2 (At1g21160), eIF5B3 (At1g76720), and eIF5B4 (At1g76820) (Figure 3). We conclude that eIF5B1 and eIF5B2 arose from recent genome duplications and that eIF5B3 and eIF5B4 arose from local duplications. Furthermore, eIF5B1 and eIF5B3 are segmentally duplicated, while eIF5B1 and eIF5B4 are tandem duplications. Insertional mutations in eIF5B2, 3, and 4 have wild-type phenotypes under both control and heat stress conditions, supporting the major functional role of eIF5B1. Duplicate genes that are preserved usually have divergent functions (He and Zhang, 2005), while duplicated genes destined to “die” usually do so within a few million years after duplication, having acquired mutations that result in a nonfunctional gene (Lynch and Conery, 2000). We provide evidence that eIF5B3 and eIF5B4 are nonfunctional and on their way to pseudogenization.
Heat Sensitivity of hot3-1 Correlates with Reduced TE of a Subset of Protective Proteins Including HSPs
The hot3-1 phenotype emphasizes the importance of translation for acclimation to high-temperature stress. hot3-1 seedlings exhibit significant, gene-specific differences in TE after the heat acclimation treatment, even though their polysome profile overlaps with the wild type. Compared with control conditions, wild-type seedlings subjected to heat acclimation treatment already show translational upregulation of stress responsive genes, which is not seen in hot3-1. This translational difference does not simply reflect failure to induce stress responsive genes, as shown by analysis of the total mRNA (Supplemental Figure 15). TE differences between the wild type and hot3-1 are even more dramatic during recovery from 45°C stress, when hot3-1 lags behind the wild type on the order of 6 to 12 h in reestablishing polysome profiles to prestress levels. There were significant differences in TE of >900 mRNAs in the mutant compared with the wild type at the 12-h recovery time point examined. Notably, in hot3-1 translation is disproportionately decreased for a subset of proteins associated with protection from heat stress, including the molecular chaperones ClpB3, ClpB4, CR88/HSP90.5, and HSP17.6A (Wang et al., 2004; Waters, 2013), and the heat shock transcription factor HSF7A (Nishizawa-Yokoi et al., 2011). We do not conclude that these TE changes result from eIF5B1-dependent translation of specific mRNAs in the hot3-1 mutant. Rather, we suggest that the reduced amount of eIF5B1 during recovery results in a delay in recovery of overall translation initiation, which likely leads to an imbalance of multiple factors required for restoring normal translation. However, reduced TE of many newly transcribed genes linked to stress tolerance no doubt contributes to the heat sensitivity of hot3-1 plants. Importantly, these results demonstrate that rapid recovery of translation after heat stress is critical to optimal survival and that as little as a 6 to 12 h delay completely alters transcriptional and translational activity and has significant consequences for plant phenotype.
The eIF5B1 Allele hot3-2 Shows Defects in Certain Developmental Processes
Plants carrying the 30-amino acid truncation allele of eIF5B1, hot3-2, show pleiotropic phenotypes and major differences in translational regulation of gene expression under optimal growth conditions. The mutant exhibits auxin-related phenotypes, including stunted primary root growth and delayed lateral root emergence that may reflect decreased TE of certain auxin-related or auxin-regulated genes (GO:0009733; GO:0009725), such as SAUR15, SAUR24, SAUR66, IAA19, IAA26, and IAA27 (Supplemental Data Set 4, “Auxin-related genes”). The decrease in TE of many ribosomal proteins could also contribute to auxin-related phenotypes, as similar phenotypes have been reported in ribosome mutants. In addition, the phenotypes of hot3-2 are similar to some mutants of plant ribosome biogenesis factors (Weis et al., 2015), and the TE of mRNAs related to ribosome biogenesis is significantly decreased in hot3-2 (Supplemental Data Set 4, “Translation-related genes”).
In addition to functioning in ribosomal subunit joining, eIF5B is reported to enhance the final nuclease step in maturation of the 18S rRNA (Lebaron et al., 2012). Total RNA profiles of hot3-2 have slightly lower ratios of 18S to 28S rRNA, compared with the wild type, which could reflect decreased efficiency of 18S rRNA and 40S ribosome maturation. This apparent defect in 18S rRNA is not seen in the hot3-1 allele under either optimal growth conditions or after heat stress (Supplemental Figure 16). Decreased TE of many ribosomal proteins in hot3-2, which is not seen in hot3-1, could point to a mechanism linking ribosomal protein synthesis to the rate of mature ribosome production.
In addition, hot3-2 showed inhibited TE of 15 genes involved in the electron transport chain pathway (Supplemental Data Set 4, “ETC-related genes”), including NADH dehydrogenase, SDH2, ATPD, FdC1, and FdC2. Reduced TE of these genes might result in a decrease in respiration and photosynthesis explaining the slow growth of hot3-2.
mRNAs with Shorter CDSs Are Poorly Translated in the hot3-2 Mutant
Previous research indicates that the 5′UTR has the greatest effect on ribosome loading based on nucleotide composition, length, predicted secondary structure, and the presence of uAUGs (Kawaguchi and Bailey-Serres, 2005). We evaluated specific properties of mRNAs that were preferentially increased or decreased in TE in hot3-2 plants. The only significant mRNA feature associated with TE was mRNAs with short CDSs that showed a decrease in TE. As eIF5B performs a general initiation function in promoting ribosomal subunit joining, it is not clear how the hot3-2 mutation would specifically impact translation of mRNAs with short open reading frames (ORFs). As mRNA association with polysomes is dependent on the time to initiate and to elongate (Weinberg et al.,2016), whether initiation or elongation is rate limiting for translation of a specific mRNA will determine the polysome association of that mRNA. If elongation takes a long time, for example, on long ORFs, due to their length, then mRNAs will remain polysome-associated even when initiation is partially impaired. Alternatively, if elongation is completed relatively fast (as on short ORFs), mRNAs will be depleted from polysomes when initiation is impaired. It is possible that reduced TE of short ORFs in hot3-2 may simply reflect the shorter time required for their translation elongation. However, we did not see a general shift of mRNAs to smaller polysomes, suggesting there is not an overall reduction in initiation of all mRNAs. Importantly, we do not see reduced translation of mRNAs with short ORFs when eIF5B1 is limiting, but functional, as it is in hot3-1 when the plants are returned to room temperature after heat stress. Thus, we suggest that removing the C-terminal helix of eIF5B could alter an interaction with the ribosome that leads to impaired initiation of specific mRNAs with a bias against those containing a short CDS. Further studies of the interactions of eIF5B on the ribosome, including an understanding of the function of the >600-amino acid, intrinsically disordered amino terminal domain, will be required to better understand the full role of eIF5B in both global and, potentially, gene-specific translation. It will also be of interest to determine the extent to which providing plants with increased levels or a more heat-resistant eIF5B could increase plant heat tolerance.
METHODS
Plant Materials and Growth Conditions
All Arabidopsis thaliana mutants were in the Col-0 background. The missense mutant hot3-1 was obtained from a previous screen conducted in our laboratory (Hong and Vierling, 2000). The T-DNA insertion lines, SALK_124251 (eIF5B1, AT1G76810, hot3-2), SAIL_1147_G06 (eIF5B1, AT1G76810, hot3-3), SALK_135561 (eIF5B1, AT1G76810, hot3-4), SALK_148816 (eIF5B2, At1g21160, eif5b2), SALK_143304C (eIF5B3, At1g76720, eif5b3), and SALK_056878 (eIF5B4, At1g76820, eif5b4), were obtained from the Arabidopsis stock center. The mutant seeds were propagated to homozygosity and the T3 generation was used for all experiments. For seed germination, sterilized seeds were kept at 4°C in darkness for 3 d for stratification; seeds were then sown on MS medium plates containing 1% sucrose and 0.8% agar. Seedlings were grown in a lighted greenhouse (white light; ∼100 µmol m−2 s−1) on a 22°C/18°C, 16-h/8-h day/night cycle. For the seed germination assay, the germination percentage was evaluated daily. Germination was defined as emergence of the radical through the seed coat.
Thermotolerance Assays
Thermotolerance assays were performed according to Hong and Vierling (2000). All heat treatments were performed in the dark. For the hypocotyl elongation assay, 2.5-d-old dark-grown seedlings were heat treated at 38°C for 1.5 h, followed by 2 h at 22°C and then 45°C for 2 h, and measured after an additional 2.5 d in the dark. Hypocotyl elongation was expressed as a percentage of growth during the same time period for seedlings that had not been heat treated. For the seedling survival assay, 6-d-old light/dark-grown seedlings were heat treated at 38°C for 1.5 h, followed by 2 h at 22°C and then 45°C for 3 h, and photographs were taken after an additional 5 d.
Identification of the hot3-1 Mutant Allele
As described previously, we generated a mapping population by crossing the hot3-1 mutant with wild-type plants of the Landsberg erecta ecotype. In the segregating F2 population, homozygous hot3-1 mutants were selected on the basis of their phenotype in the hypocotyl thermotolerance assay. Simple sequence length polymorphism markers were developed to locate the hot3-1 mutation to the bottom of Chromosome 1 in a 150-kb interval. Thirty-two ORFs (plus 300 bp upstream and 150 bp downstream) were sequenced, including all genes from BAC F28O16 and F7O12 and two from F14G6. For complementation, the eIF5B1 genomic sequence (∼7 kb, from –1198 bp to +5833 bp containing the ∼1400-bp promoter; where +1 is the transcription start site from the TAIR database) was cloned into SalI and SacI sites of pCAMBIA1300 binary vector using two pairs of primers (eIF5B1pro-F and eIF5B1pro-R; eIF5B1CDS-F and eIF5B1CDS-R). The construct was introduced into hot3-1 and hot3-2 mutants using Agrobacterium tumefaciens GV3101. The transgenic plants were screened with 25 mg/L hygromycin. Primers used for the construct are listed in Supplemental Table 1.
Isolation of Total RNA and Polysome-Bound RNA
Ten-day-old Arabidopsis seedlings were acclimated at 38°C for 1.5 h and 22°C for 2 h, subjected to heat stress at 45°C for 2 h, and then allowed to recover at 22°C in light for different times. For the control conditions, seedlings taken directly from the growth chamber were used. All the seedlings were planted at the same time and divided into three parts as biological replicates. Whole plants were quickly frozen in liquid nitrogen, ground to powder, and stored at –80°C. Polysome isolation by differential centrifugation was performed as previously described (Juntawong et al., 2014). Two grams of frozen tissue was thawed in 5 mL of polysome extraction buffer (200 mM Tris-HCl, pH 9.0, 200 mM KCl, 35 mM MgCl2, 2 mM EGTA, 1% Triton X-100, 1% Tween 20, 2% polyoxyethylene, 0.5 mg/mL heparin, 5 mM DTT, 100 μg/mL chloramphenicol, and 25 μg/mL cycloheximide). Four milliliters of crude cell extract was clarified by centrifugation at 13,200g for 15 min at 4°C. The supernatants were layered on top of a 1.7 M sucrose cushion and centrifuged at 45,000 rpm (SW55Ti rotor in a Beckman L-100XP ultracentrifuge) for 3 h at 4°C. The ribosome pellet was resuspended in 300 μL resuspension buffer (200 mM Tris-HCl, pH 9.0, 200 mM KCl, 35 mM MgCl2, 2 mM EGTA, 100 µg/mL chloramphenicol, and 25 µg/mL cycloheximide). The solution was then layered over a 4.5 mL 20 to 60% sucrose density gradient and centrifuged at 41,000 rpm (SW55Ti rotor) for 2 h at 4°C. The polysome profile was recorded with a chart recorder using a gradient fractionator connected to a UA-5 detector (Biocomp). Fourteen fractions of 350 μL were automatically collected for polysome-bound RNA extraction. Three biological replications were performed and the results showed similar trends; thus, one replication is shown in the figures. For polysome-RNA isolation, fractions 8 to 14 were pooled and treated with 5% SDS/0.2 M EDTA, then extracted twice with an equal volume of phenol/chloroform/isoamyl alcohol [25:24:1] (v/v/v) by vortexing. The mixture was centrifuged at 12,000 rpm for 5 min at 10°C. The RNA was precipitated by 100% ethanol for 1 h at room temperature and centrifuged at 12,000 rpm for 15 min at 10°C. Total RNA extraction was performed using TRIzol reagent (Takara) according to the manufacturer’s protocol. Polysome gradient fractions to be analyzed by RT-qPCR for different mRNAs were prepared using a spiked-in luciferase mRNA control for normalization. To each sample (single or pooled fractions), 50 pg of luciferase RNA was added prior to mRNA extraction to allow normalization independent of the original RNA content of the sample.
Library Preparation and RNA-Seq
RNA concentration was measured using a NanoDrop 2000 spectrophotometer (ND-2000; Thermo Fisher Scientific). RNA integrity was assessed on an Agilent 2100 Bioanalyzer (Agilent Technologies). Paired-end sequencing libraries with an average insert size of 200 bp were prepared with the TruSeq RNA Sample Preparation Kit version 2 (Illumina) and sequenced on a HiSeq 2500 (Illumina) according to the manufacturer’s standard protocols. Raw data obtained from Illumina sequencing were processed and filtered using the Illumina pipeline (http://www.illumina.com) to generate FastQ files. Finally, ∼58 GB high-quality, 125-bp, paired-end reads were generated from 29 libraries. All data were provided by BerryGenomics.
Identification of Differentially Transcriptionally and Translationally Regulated Genes
FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) was initially run to assess the overall quality of all sample reads. Poor quality bases were filtered out using Sickle with parameters pe- mode; -t sanger –q 20 –l 50 (https://github.com/najoshi/sickle). The trimmed and quality filtered reads were mapped to the Arabidopsis Information Resource (TAIR10) version of the Arabidopsis genome using the splice junction aware short-read alignment suite TopHat v2.09 (Kim et al., 2013) with default settings and allowing only unique alignments (i.e., each read was allowed to map to one location only) and not more than two nucleotide mismatches. Reads located in the noncoding RNA regions were filtered out using the intersect function of BEDTools v2.17.0 (Quinlan and Hall, 2010). Only reads located in exons and CDSs were kept for further analyses for total and PB mRNA samples respectively. Cufflinks v2.1.1 (Trapnell et al., 2012) with default parameters was used for normalization and estimation of gene transcriptional and translational expression levels. Only genes with fragments per kilobases per million fragments ≥1 in total and PB mRNA were used to calculate the Pearson correlation for replicates of each sample. The Bioconductor package “edgeR” was used for differential expression analysis (Robinson et al., 2010). Only the genes with FDR < 0.05 and absolute value of log2 (fold change; mutants/WT, AC/CK, or HS/CK) ≥ 1.0 were considered as differentially expressed genes in total and PB mRNA samples.
Xtail software (https://github.com/xryanglab/xtail/releases) was used to identify genes with different TE in pairwise comparisons. Two parallel pipelines were implemented to compare the log2 fold changes (log2FC) of PB mRNA and total mRNA or the log2 ratios (log2R) of PB mRNA to Total mRNA between two conditions. First, Xtail adapts the strategy of DESeq2 (Love et al., 2014) to normalize read counts of mRNA and PB in all samples, and the negative binomial distribution is used to model the variation of mRNA data and PB data. Then, the probability distributions for fold changes of total mRNA or PB mRNA, or for PB mRNA-to-total mRNA ratios were established using the generalized linear model. Finally, the statistical significance and magnitude of differential translation for each gene were evaluated using the probability distributions. Specifically, a joint probability matrix of the outer product of the two probability density distributions for log2FC of total mRNA and log2FC of PB mRNA were generated, and the approximate P values were estimated to evaluate the degree of difference in changes of total mRNA and PB mRNA. In the other parallel pipeline, Xtail established the probability distributions for PB mRNA-to-total mRNA ratios in the two conditions and derived another distribution for differential translation. From these two parallel pipelines, the more conservative set of results (more significant P value) were derived as the final result. With this strategy, Xtail assesses discoordination between the change in PB mRNA abundance and total mRNA levels, theoretically equally evaluating the discoordination of a gene’s translational rate in any two conditions to be compared (Xiao et al., 2016). Differentially translated genes were defined here as those with a FDR < 0.01. Identification of significantly (hypergeometric test; FDR < 0.01) enriched GO categories was done using AgriGO (Du et al., 2010), a web-based tool and database for GO analysis (http://bioinfo.cau.edu.cn/agriGO/).
mRNA Feature Analyses
Data sets for 5′UTR, CDS, and 3′UTR regions and sequences were downloaded from TAIR. The representative gene model was used as the reference gene model for the analysis. Only genes with complete information for the different features analyzed were considered. Normalized minimal free energy was used to define the sequence stability of the secondary structure, which was calculated by RNAfold and normalized by corresponding length. Basic and advanced statistical analyses were performed using custom-made PERL scripts and R version 3.3.0.
Quantitative Real-Time RT-PCR
Quantitative real-time PCR was performed with SYBR Premix Ex TaqII (Takara) using a Bio-Rad CFX96 real-time PCR detection system. To determine the relative mRNA abundance, Ct values were calculated as Ct(sample) = Ct(gene)/Ct (control). In the final results, the relative expression level of the sample was determined by the 2−ΔCt method. All the RT-qPCR primers are listed in Supplemental Table 1.
Phylogenetic Analysis
Sequences of eIF5B homologs were found using the conserved C-terminal half of Arabidopsis eIF5B1 as a query sequence in a TBLASTP search of the database Phytozome (https://phytozome.jgi.gov/pz/portal.html). The full-length of amino acid sequences were aligned in ClustalW in MEGA6 (Tamura et al., 2013). The aligned sequences were then used to generate a phylogenetic tree by the neighbor-joining method. Bootstrap values (as a percentage of 1000 replicates) are provided at the branches.
GUS Histochemical Assays
The promoter fragments from eIF5B1 (1365 bp, from –1198 to +167 bp; where +1 is the transcription start site from the TAIR database), eIF5B2 (1489 bp, from –1491 to −2 bp), eIF5B3 (1475 bp, from –1475 to –1 bp), and eIF5B4 (1365 bp, from –1153 to +212 bp) were amplified and cloned into the pBGwFS7.0 vector using Gateway cloning (Invitrogen). The constructs were introduced into Col-0 via Agrobacterium-mediated transformation. The transgenic plants were screened on 100 mg/L spectinomycin. T4 homozygous transgenic plants were then used for GUS staining. Plant samples were vacuum-infiltrated with staining buffer [0.1 M phosphate buffer, pH 7.0, 5 mM K4Fe(CN)6, 5 mM K3Fe(CN)6, 0.1% Triton X-100, and 0.5 mg/mL X-Gluc] incubated at 37°C overnight, and cleared in 75% ethanol. Photographs were taken with an Olympus SEX16 microscope. Three independent lines were tested, and representative pictures are shown.
SDS-PAGE and Immunoblots
Total protein from 0.1 g seedlings was extracted in 0.5 mL SDS sample buffer (50 mM Tris-HCl, pH 6.8, 2% [w/v] SDS, 10% glycerol, 1% β-mercaptoethanol, 12.5 mM EDTA, and 0.02% bromophenol blue) in a ground glass homogenizer. All protein samples were examined by SDS-PAGE and Coomassie Brilliant Blue staining. For immunoblots, proteins were transferred to polyvinylidene difluoride membrane (Millipore), and the membrane was then saturated for 1 h at room temperature using 5% fat-free milk in Tris-buffered saline with 0.1% Tween 20 (TBST). Protein blots were probed with rabbit antiserum against Arabidopsis HSP101 (Agrisera; AS08287), cytosolic HSP70 (Agrisera; AS08371), and chloroplast HSP21 (Agrisera; AS08285) at a dilution of 1:1000. As a loading control, blots were probed with mouse antiserum against α-Actin antibody (Sigma-Aldrich; A0480). Blots were incubated with goat anti-rabbit or anti-mouse horseradish peroxidase. Specific protein bands were visualized by enhanced chemiluminescence (Amersham).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: eIF5B1, AT1G76810; eIF5B2, AT1G21160; eIF5B3, AT1G76720; eIF5B4, AT1G76820; HSP101, At1g74310; HSP70, AT3G12580; HSP21, AT4G27670; and ACTIN2, AT3G18780. All total mRNA and PB mRNA sequencing data generated in this study have been submitted to the NCBI Sequence Read Archive (http://www.ncbi.nlm.nih.gov/sra/) under accession number SRP082597.
Supplemental Data
Supplemental Figure 1. Amino acid sequence alignment of the conserved C-terminal half of eIF5B from S. cerevisiae (NP_009365), A. thaliana (NP_177807), P. sativum (AAN32916), O. sativa (NP_001056497), H. sapiens (NP_056988), and the prokaryotic homolog IF2 from E. coli (WP_001723945).
Supplemental Figure 2. Identification and characterization of three hot3-1 complemented lines.
Supplemental Figure 3. Identification and characterization of the hot3-2 mutant.
Supplemental Figure 4. Amino acid sequence alignment of eIF5B1 homologs in Arabidopsis.
Supplemental Figure 5. Identification of T-DNA insertion alleles for the three additional eIF5B homologs.
Supplemental Figure 6. Polymorphic analysis of the four eIF5B homologs.
Supplemental Figure 7. Predicted microRNA cleavage site of eIF5B3 and position of the stop codon in eIF5B4.
Supplemental Figure 8. Schematic illustration of the experimental design for estimation of translation efficiency.
Supplemental Figure 9. Ten genes were selected to examine the validity of the RNA-seq data using RT-qPCR.
Supplemental Figure 10. Scatterplot of the fold changes between control and heat-acclimated or heat-stressed samples for total and PB mRNA in the wild type or hot3-1.
Supplemental Figure 11. Analysis of the log2-transformed TE-fold change values for the genes in the heat responsive GO category.
Supplemental Figure 12. Time course of loss of polysomes when plants are shifted to 45°C after the acclimation treatment.
Supplemental Figure 13. Violin plots showing the distribution of different mRNA properties among differently translationally regulated genes.
Supplemental Figure 14. Characterization of two additional T-DNA insertion alleles for eIF5B1.
Supplemental Figure 15. Comparisons of Total mRNA profiles between control, acclimated, and heat stressed wild-type and hot3-1 seedlings.
Supplemental Figure 16. A representative analysis of total RNA samples from the wild type, hot3-1, and hot3-2.
Supplemental Table 1. Sequence of primers for cloning, identifying mutations and RT-qPCR.
Supplemental Data Set 1. Alignments used to generate the phylogeny presented in Figure 4.
Supplemental Data Set 2. RNA-seq data: sample reads.
Supplemental Data Set 3. RNA-seq data used for calculating total and PB mRNA.
Supplemental Data Set 4. RNA-seq data used for calculating TE.
Supplemental Data Set 5. Gene Ontology terms and TE among translationally regulated gene sets.
Acknowledgments
We thank Xuerui Yang and Zhengtao Xiao (Tsinghua University) for providing the Xtail software, Lijia Jia (Chinese Academy of Sciences) for helping with analysis of the 3D structure, Runlai Hang and Xiaofeng Cao (Chinese Academy of Sciences) for providing good suggestions for the polysome profiling assay, and three anonymous reviewers for giving us good ideas for analyzing and interpreting our data. E.V. also thanks Ung Lee and Joseph T. Carroll for the initial mapping data leading to identification of HOT3. This work was supported by grants from the National Science Foundation (NSF DBI-0820047 to E.V.), from the USDA National Institute of Food and Agriculture (USDA-NRICGP 3510014857 to E.V.), and from a new faculty award from the Massachusetts Life Sciences Center (to E.V.). This work was also supported by the National Natural Science Foundation of China (31171549 and 31571747 to H.P.).
AUTHOR CONTRIBUTIONS
E.V., H.P., Q.S., and Z.N. conceived this project, designed experiments, and analyzed data. L.Z., K.G., X.L., F.W., X.T., and M.X. performed experiments and analyzed data. L.Z. wrote the article. X.L. did the bioinformatics. E.V. and H.P. contributed to manuscript revision.
Glossary
- PB
polysome-bound
- TE
translation efficiency
- FDR
false discovery rate
- UTR
untranslated region
- CDS
coding sequence
- ORFs
open reading frames
Footnotes
Articles can be viewed without a subscription.
References
- Acker M.G., Shin B.S., Dever T.E., Lorsch J.R. (2006). Interaction between eukaryotic initiation factors 1A and 5B is required for efficient ribosomal subunit joining. J. Biol. Chem. 281: 8469–8475. [DOI] [PubMed] [Google Scholar]
- Branco-Price C., Kawaguchi R., Ferreira R.B., Bailey-Serres J. (2005). Genome-wide analysis of transcript abundance and translation in Arabidopsis seedlings subjected to oxygen deprivation. Ann. Bot. (Lond.) 96: 647–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning K.S., Bailey-Serres J. (2015). Mechanism of cytoplasmic mRNA translation. Arabidopsis Book 13: e0176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrera P., Johnstone O., Nakamura A., Casanova J., Jäckle H., Lasko P. (2000). VASA mediates translation through interaction with a Drosophila yIF2 homolog. Mol. Cell 5: 181–187. [DOI] [PubMed] [Google Scholar]
- Choi S.K., Lee J.H., Zoll W.L., Merrick W.C., Dever T.E. (1998). Promotion of met-tRNAiMet binding to ribosomes by yIF2, a bacterial IF2 homolog in yeast. Science 280: 1757–1760. [DOI] [PubMed] [Google Scholar]
- Choi S.K., Olsen D.S., Roll-Mecak A., Martung A., Remo K.L., Burley S.K., Hinnebusch A.G., Dever T.E. (2000). Physical and functional interaction between the eukaryotic orthologs of prokaryotic translation initiation factors IF1 and IF2. Mol. Cell. Biol. 20: 7183–7191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z., Zhou X., Ling Y., Zhang Z., Su Z. (2010). agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Res. 38: W64–W70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan H., Richael C., Rommens C.M. (2012). Overexpression of the wild potato eIF4E-1 variant Eva1 elicits Potato virus Y resistance in plants silenced for native eIF4E-1. Transgenic Res. 21: 929–938. [DOI] [PubMed] [Google Scholar]
- Dutt S., Parkash J., Mehra R., Sharma N., Singh B., Raigond P., Joshi A., Chopra S., Singh B.P. (2015). Translation initiation in plants: roles and implications beyond protein synthesis. Biol. Plant. 59: 401–412. [Google Scholar]
- Fringer J.M., Acker M.G., Fekete C.A., Lorsch J.R., Dever T.E. (2007). Coupled release of eukaryotic translation initiation factors 5B and 1A from 80S ribosomes following subunit joining. Mol. Cell. Biol. 27: 2384–2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant-Downton R., Le Trionnaire G., Schmid R., Rodriguez-Enriquez J., Hafidh S., Mehdi S., Twell D., Dickinson H. (2009). MicroRNA and tasiRNA diversity in mature pollen of Arabidopsis thaliana. BMC Genomics 10: 643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X., Zhang J. (2005). Rapid subfunctionalization accompanied by prolonged and substantial neofunctionalization in duplicate gene evolution. Genetics 169: 1157–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S.W., Vierling E. (2000). Mutants of Arabidopsis thaliana defective in the acquisition of tolerance to high temperature stress. Proc. Natl. Acad. Sci. USA 97: 4392–4397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S.W., Lee U., Vierling E. (2003). Arabidopsis hot mutants define multiple functions required for acclimation to high temperatures. Plant Physiol. 132: 757–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia N.T., Ghaemmaghami S., Newman J.R.S., Weissman J.S. (2009). Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324: 218–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson R.J., Hellen C.U., Pestova T.V. (2010). The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell Biol. 11: 113–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juntawong P., Girke T., Bazin J., Bailey-Serres J. (2014). Translational dynamics revealed by genome-wide profiling of ribosome footprints in Arabidopsis. Proc. Natl. Acad. Sci. USA 111: E203–E212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi R., Bailey-Serres J. (2005). mRNA sequence features that contribute to translational regulation in Arabidopsis. Nucleic Acids Res. 33: 955–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Key J.L., Lin C.Y., Chen Y.M. (1981). Heat shock proteins of higher plants. Proc. Natl. Acad. Sci. USA 78: 3526–3530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim B.H., Cai X., Vaughn J.N., von Arnim A.G. (2007). On the functions of the h subunit of eukaryotic initiation factor 3 in late stages of translation initiation. Genome Biol. 8: R60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S.L. (2013). TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotak S., Larkindale J., Lee U., von Koskull-Döring P., Vierling E., Scharf K.-D. (2007). Complexity of the heat stress response in plants. Curr. Opin. Plant Biol. 10: 310–316. [DOI] [PubMed] [Google Scholar]
- Kyrpides N.C., Woese C.R. (1998). Universally conserved translation initiation factors. Proc. Natl. Acad. Sci. USA 95: 224–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkindale J., Vierling E. (2008). Core genome responses involved in acclimation to high temperature. Plant Physiol. 146: 748–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebaron S., Schneider C., van Nues R.W., Swiatkowska A., Walsh D., Böttcher B., Granneman S., Watkins N.J., Tollervey D. (2012). Proofreading of pre-40S ribosome maturation by a translation initiation factor and 60S subunits. Nat. Struct. Mol. Biol. 19: 744–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.-H., et al. (2010). Single nucleotide polymorphisms in a gene for translation initiation factor (eIF4G) of rice (Oryza sativa) associated with resistance to Rice tungro spherical virus. Mol. Plant Microbe Interact. 23: 29–38. [DOI] [PubMed] [Google Scholar]
- Lee J.H., Choi S.K., Roll-Mecak A., Burley S.K., Dever T.E. (1999). Universal conservation in translation initiation revealed by human and archaeal homologs of bacterial translation initiation factor IF2. Proc. Natl. Acad. Sci. USA 96: 4342–4347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Truesdell S.S., Bukhari S.I., Lee J.H., LeTonqueze O., Vasudevan S. (2014). Upregulation of eIF5B controls cell-cycle arrest and specific developmental stages. Proc. Natl. Acad. Sci. USA 111: E4315–E4322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee U., Rioflorido I., Hong S.W., Larkindale J., Waters E.R., Vierling E. (2007). The Arabidopsis ClpB/Hsp100 family of proteins: chaperones for stress and chloroplast development. Plant J. 49: 115–127. [DOI] [PubMed] [Google Scholar]
- Li R., Sun R., Hicks G.R., Raikhel N.V. (2015). Arabidopsis ribosomal proteins control vacuole trafficking and developmental programs through the regulation of lipid metabolism. Proc. Natl. Acad. Sci. USA 112: E89–E98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S.Y., Chen P.W., Chuang M.H., Juntawong P., Bailey-Serres J., Jauh G.Y. (2014). Profiling of translatomes of in vivo-grown pollen tubes reveals genes with roles in micropylar guidance during pollination in Arabidopsis. Plant Cell 26: 602–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M.J., Wu S.H., Chen H.M., Wu S.H. (2012). Widespread translational control contributes to the regulation of Arabidopsis photomorphogenesis. Mol. Syst. Biol. 8: 566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M., Conery J.S. (2000). The evolutionary fate and consequences of duplicate genes. Science 290: 1151–1155. [DOI] [PubMed] [Google Scholar]
- Ma F., Liu Z., Wang T.W., Hopkins M.T., Peterson C.A., Thompson J.E. (2010). Arabidopsis eIF5A3 influences growth and the response to osmotic and nutrient stress. Plant Cell Environ. 33: 1682–1696. [DOI] [PubMed] [Google Scholar]
- Marintchev A., Kolupaeva V.G., Pestova T.V., Wagner G. (2003). Mapping the binding interface between human eukaryotic initiation factors 1A and 5B: a new interaction between old partners. Proc. Natl. Acad. Sci. USA 100: 1535–1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Silva A.V., Aguirre-Martínez C., Flores-Tinoco C.E., Alejandri-Ramírez N.D., Dinkova T.D. (2012). Translation initiation factor AteIF(iso)4E is involved in selective mRNA translation in Arabidopsis thaliana seedlings. PLoS One 7: e31606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nag N., Lin K.Y., Edmonds K.A., Yu J., Nadkarni D., Marintcheva B., Marintchev A. (2016). eIF1A/eIF5B interaction network and its functions in translation initiation complex assembly and remodeling. Nucleic Acids Res. 44: 7441–7456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishizawa-Yokoi A., Nosaka R., Hayashi H., Tainaka H., Maruta T., Tamoi M., Ikeda M., Ohme-Takagi M., Yoshimura K., Yabuta Y., Shigeoka S. (2011). HsfA1d and HsfA1e involved in the transcriptional regulation of HsfA2 function as key regulators for the Hsf signaling network in response to environmental stress. Plant Cell Physiol. 52: 933–945. [DOI] [PubMed] [Google Scholar]
- Pisareva V.P., Pisarev A.V. (2014). eIF5 and eIF5B together stimulate 48S initiation complex formation during ribosomal scanning. Nucleic Acids Res. 42: 12052–12069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porankiewicz J., Gwozdz E.A. (1995). Polysomes in lupin roots exposed to heat-shock. Acta Physiol. Plant. 17: 55–60. [Google Scholar]
- Quinlan A.R., Hall I.M. (2010). BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasheedi S., Ghosh S., Suragani M., Tuteja N., Sopory S.K., Hasnain S.E., Ehtesham N.Z. (2007). Pisum sativum contains a factor with strong homology to eIF5B. Gene 399: 144–151. [DOI] [PubMed] [Google Scholar]
- Rasheedi S., Suragani M., Haq S.K., Sachchidanand, Bhardwaj R., Hasnain S.E., Ehtesham N.Z. (2010). Expression, purification and ligand binding properties of the recombinant translation initiation factor (PeIF5B) from Pisum sativum. Mol. Cell. Biochem. 344: 33–41. [DOI] [PubMed] [Google Scholar]
- Rausell A., Kanhonou R., Yenush L., Serrano R., Ros R. (2003). The translation initiation factor eIF1A is an important determinant in the tolerance to NaCl stress in yeast and plants. Plant J. 34: 257–267. [DOI] [PubMed] [Google Scholar]
- Ren B., Chen Q., Hong S., Zhao W., Feng J., Feng H., Zuo J. (2013). The Arabidopsis eukaryotic translation initiation factor eIF5A-2 regulates root protoxylem development by modulating cytokinin signaling. Plant Cell 25: 3841–3857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M.D., McCarthy D.J., Smyth G.K. (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy B., Copenhaver G.P., von Arnim A.G. (2011). Fluorescence-tagged transgenic lines reveal genetic defects in pollen growth--application to the eIF3 complex. PLoS One 6: e17640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio C.A., Weisburd B., Holderfield M., Arias C., Fang E., DeRisi J.L., Fanidi A. (2014). Transcriptome-wide characterization of the eIF4A signature highlights plasticity in translation regulation. Genome Biol. 15: 476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L., et al. (2013). Identification of promoter motifs regulating ZmeIF4E expression level involved in maize rough dwarf disease resistance in maize (Zea Mays L.). Mol. Genet. Genomics 288: 89–99. [DOI] [PubMed] [Google Scholar]
- Sonenberg N., Hinnebusch A.G. (2009). Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell 136: 731–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spano G., Capozzi V., Vernile A., Massa S. (2004). Cloning, molecular characterization and expression analysis of two small heat shock genes isolated from wine Lactobacillus plantarum. J. Appl. Microbiol. 97: 774–782. [DOI] [PubMed] [Google Scholar]
- Sun W., Bernard C., van de Cotte B., Van Montagu M., Verbruggen N. (2001). At-HSP17.6A, encoding a small heat-shock protein in Arabidopsis, can enhance osmotolerance upon overexpression. Plant J. 27: 407–415. [DOI] [PubMed] [Google Scholar]
- Suragani M., Rasheedi S., Hasnain S.E., Ehtesham N.Z. (2011). The translation initiation factor, PeIF5B, from Pisum sativum displays chaperone activity. Biochem. Biophys. Res. Commun. 414: 390–396. [DOI] [PubMed] [Google Scholar]
- Suzuki N., Rizhsky L., Liang H., Shuman J., Shulaev V., Mittler R. (2005). Enhanced tolerance to environmental stress in transgenic plants expressing the transcriptional coactivator multiprotein bridging factor 1c. Plant Physiol. 139: 1313–1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. (2013). MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 30: 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D.R., Pimentel H., Salzberg S.L., Rinn J.L., Pachter L. (2012). Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7: 562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda K., Matsuura H., Yamaguchi M., Demura T., Kato K. (2012). Genome-wide analyses of changes in translation state caused by elevated temperature in Oryza sativa. Plant Cell Physiol. 53: 1481–1491. [DOI] [PubMed] [Google Scholar]
- Vain P., Thole V., Worland B., Opanowicz M., Bush M.S., Doonan J.H. (2011). A T-DNA mutation in the RNA helicase eIF4A confers a dose-dependent dwarfing phenotype in Brachypodium distachyon. Plant J. 66: 929–940. [DOI] [PubMed] [Google Scholar]
- Wang L., Xu C., Wang C., Wang Y. (2012). Characterization of a eukaryotic translation initiation factor 5A homolog from Tamarix androssowii involved in plant abiotic stress tolerance. BMC Plant Biol. 12: 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Vinocur B., Shoseyov O., Altman A. (2004). Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response. Trends Plant Sci. 9: 244–252. [DOI] [PubMed] [Google Scholar]
- Wang X., Kohalmi S.E., Svircev A., Wang A., Sanfaçon H., Tian L. (2013). Silencing of the host factor eIF(iso)4E gene confers plum pox virus resistance in plum. PLoS One 8: e50627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters E.R. (2013). The evolution, function, structure, and expression of the plant sHSPs. J. Exp. Bot. 64: 391–403. [DOI] [PubMed] [Google Scholar]
- Wei C.L., Kainuma M., Hershey J.W.B. (1995). Characterization of yeast translation initiation factor 1A and cloning of its essential gene. J. Biol. Chem. 270: 22788–22794. [DOI] [PubMed] [Google Scholar]
- Weinberg D.E., Shah P., Eichhorn S.W., Hussmann J.A., Plotkin J.B., Bartel D.P. (2016). Improved ribosome-footprint and mRNA measurements provide insights into dynamics and regulation of yeast translation. Cell Reports 14: 1787–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weis B.L., Kovacevic J., Missbach S., Schleiff E. (2015). Plant-specific features of ribosome biogenesis. Trends Plant Sci. 20: 729–740. [DOI] [PubMed] [Google Scholar]
- Wilson S.A., Sieiro-Vazquez C., Edwards N.J., Iourin O., Byles E.D., Kotsopoulou E., Adamson C.S., Kingsman S.M., Kingsman A.J., Martin-Rendon E. (1999). Cloning and characterization of hIF2, a human homologue of bacterial translation initiation factor 2, and its interaction with HIV-1 matrix. Biochem. J. 342: 97–103. [PMC free article] [PubMed] [Google Scholar]
- Xiao Z., Zou Q., Liu Y., Yang X. (2016). Genome-wide assessment of differential translations with ribosome profiling data. Nat. Commun. 7: 11194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Zhang B., Jiang C., Ming F. (2011). RceIF5A, encoding an eukaryotic translation initiation factor 5A in Rosa chinensis, can enhance thermotolerance, oxidative and osmotic stress resistance of Arabidopsis thaliana. Plant Mol. Biol. 75: 167–178. [DOI] [PubMed] [Google Scholar]
- Yángüez E., Castro-Sanz A.B., Fernández-Bautista N., Oliveros J.C., Castellano M.M. (2013). Analysis of genome-wide changes in the translatome of Arabidopsis seedlings subjected to heat stress. PLoS One 8: e71425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng A., Yu J., Yamamoto R., Ose T., Tanaka I., Yao M. (2014). X-ray structures of eIF5B and the eIF5B-eIF1A complex: the conformational flexibility of eIF5B is restricted on the ribosome by interaction with eIF1A. Acta Crystallogr. D Biol. Crystallogr. 70: 3090–3098. [DOI] [PubMed] [Google Scholar]