The tomato transcription factor (TF) MYC2 and MYC2-targeted TFs form a hierarchical transcriptional cascade during jasmonate-mediated plant immunity that initiates and amplifies the transcriptional output.
Abstract
The hormone jasmonate (JA), which functions in plant immunity, regulates resistance to pathogen infection and insect attack through triggering genome-wide transcriptional reprogramming in plants. We show that the basic helix-loop-helix transcription factor (TF) MYC2 in tomato (Solanum lycopersicum) acts downstream of the JA receptor to orchestrate JA-mediated activation of both the wounding and pathogen responses. Using chromatin immunoprecipitation sequencing (ChIP-seq) coupled with RNA sequencing (RNA-seq) assays, we identified 655 MYC2-targeted JA-responsive genes. These genes are highly enriched in Gene Ontology categories related to TFs and the early response to JA, indicating that MYC2 functions at a high hierarchical level to regulate JA-mediated gene transcription. We also identified a group of MYC2-targeted TFs (MTFs) that may directly regulate the JA-induced transcription of late defense genes. Our findings suggest that MYC2 and its downstream MTFs form a hierarchical transcriptional cascade during JA-mediated plant immunity that initiates and amplifies transcriptional output. As proof of concept, we showed that during plant resistance to the necrotrophic pathogen Botrytis cinerea, MYC2 and the MTF JA2-Like form a transcription module that preferentially regulates wounding-responsive genes, whereas MYC2 and the MTF ETHYLENE RESPONSE FACTOR.C3 form a transcription module that preferentially regulates pathogen-responsive genes.
INTRODUCTION
Plants activate specific defense signaling pathways in response to pathogen infection or insect attack, and phytohormones play key regulatory roles in these pathways. Generally, salicylic acid signaling triggers plant immunity against biotrophic or hemibiotrophic pathogens and sucking insects, whereas jasmonic acid (JA) signaling activates plant resistance against necrotrophic pathogens and chewing insects. Accumulating evidence suggests that salicylic acid and JA have a mutually inhibitory effect on plant immunity (Glazebrook, 2005; Howe and Jander, 2008; Spoel and Dong, 2008; Pieterse et al., 2009; Mengiste, 2012; Dangl et al., 2013; Campos et al., 2014).
Tomato (Solanum lycopersicum) is a classic model plant used to investigate the mechanisms underlying JA-triggered plant immunity against insect attack and other forms of mechanical damage (Ryan, 2000; Schilmiller and Howe, 2005; Howe and Jander, 2008; Chen et al., 2011; Sun et al., 2011; Rosli and Martin, 2015). In this system, the 18-amino acid peptide systemin and JA act in a common signaling pathway to activate the systemic expression of numerous defense-related genes. Based on the timing of their expression in response to wounding and JA, these defense-related genes can be categorized as “early” or “late” genes (Ryan and Pearce, 1998; Ryan, 2000; Lee and Howe, 2003). Expression of the so-called “early” genes is induced rapidly and transiently in response to wounding. Genes whose expression reaches a maximum 12 to 24 h after wounding comprise the “late” genes. Whereas early JA-responsive genes mainly encode proteins involved in JA biosynthesis or signaling, late JA-responsive genes generally encode defense proteins that are directly involved in plant resistance against herbivores or pathogens (Ryan and Pearce, 1998; Ryan, 2000; Howe and Jander, 2008; Sun et al., 2011). Grafting experiments with tomato mutants defective in JA biosynthesis and signaling revealed that JA, rather than systemin, acts as a long-distance mobile signal in the systemic defense response (Li et al., 2002, 2004; Li et al., 2003).
Decades of studies, mainly in the model system Arabidopsis thaliana, have revealed a core JA signaling module consisting of the following: an F-box protein CORONATIN INSENSITIVE1 (COI1), which forms the functional Skp-Cullin-F-box (SCF) E3 ubiquitin ligase complex SCFCOI1 with Cullin1 and Skp1-like1 (Xie et al., 1998; Devoto et al., 2002; Xu et al., 2002); a group of jasmonate-ZIM domain (JAZ) proteins that function as transcriptional repressors (Chini et al., 2007; Thines et al., 2007; Yan et al., 2007); and the basic helix-loop-helix (bHLH) transcription factor (TF) MYC2, which differentially regulates diverse aspects of JA responses (Boter et al., 2004; Lorenzo et al., 2004; Dombrecht et al., 2007; Kazan and Manners, 2013; Zhai et al., 2013). A mechanistic understanding of the JA signaling pathway came from the findings that jasmonoyl-isoleucine (JA-Ile) is the receptor-active form of the hormone and that the SCFCOI1-JAZ complex acts as a coreceptor of JA-Ile (Yan et al., 2009; Pauwels et al., 2010; Sheard et al., 2010). In the steady state, JAZ proteins interact with and repress the transcriptional activity of MYC2. In response to internal or external stimuli, elevated JA-Ile levels promote SCFCOI1-dependent degradation of JAZ repressors, thereby activating (derepressing) MYC2-directed transcription of JA-responsive genes. These findings demonstrate that MYC2-orchestrated transcriptional reprogramming is a central theme of JA signaling (Kazan and Manners, 2013; Wasternack and Hause, 2013).
Botrytis cinerea, a fungal pathogen that infects many crops of economic importance, represents one of the most extensively studied necrotrophic pathogens (Elad et al., 2016; van Kan et al., 2017). Based on its scientific and economic importance, B. cinerea was ranked as the second most important plant-pathogenic fungus (Dean et al., 2012). This necrotrophic pathogen kills host cells at very early stages in the infection process and causes extensive tissue damage (Glazebrook, 2005; Mengiste, 2012). Therefore, plant resistance against B. cinerea involves the activation of both wounding- and pathogen-responsive genes (El Oirdi and Bouarab, 2007; El Oirdi et al., 2011; Yan et al., 2013).
The tomato-B. cinerea interaction is used as a model system to investigate the transcriptional mechanism underlying JA-mediated plant immunity. In this study, we showed that the tomato bHLH TF MYC2 (Sl-MYC2) plays a critical role in orchestrating JA-mediated defense gene expression during B. cinerea infection. We used genome-wide chromatin immunoprecipitation followed by sequencing (ChIP-seq) in combination with transcriptome profiling by RNA sequencing (RNA-seq) to identify MYC2-targeted JA-responsive (MTJA) genes. We found that MTJAs are enriched for MYC2-targeted TF (MTF) genes and early JA-responsive genes but are not enriched for the late JA-responsive genes, suggesting that MYC2 acts as a high-level regulator that initiates and controls the JA-elicited transcriptional cascade. Finally, we showed that during plant resistance to B. cinerea infection, tomato MYC2 positively regulates both wound-responsive and pathogen-responsive genes through intermediate MTFs.
RESULTS
B. cinerea Infection Activates Both Wounding-Responsive and Pathogen-Responsive Genes in a COI1-Dependent Manner
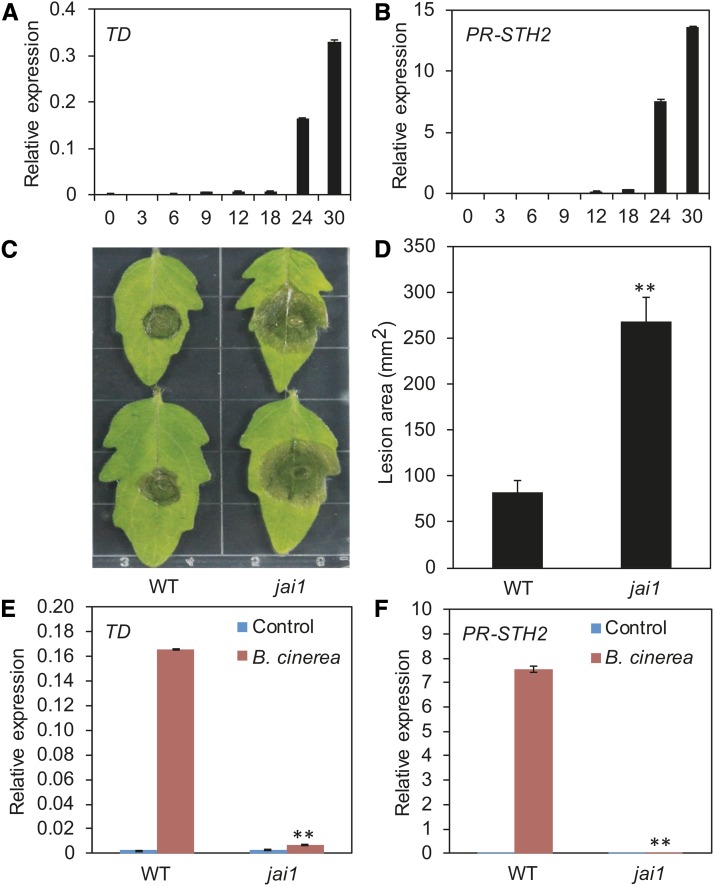
In response to insect attack and/or pathogen infection, plants activate the expression of two groups of defense genes. One group functions in wounding/insect resistance, and the other functions in the pathogen response. In tomato, THREONINE DEAMINASE (TD) (Chen et al., 2005) is a commonly used marker gene for the wounding/insect response, and PR-STH2 (Marineau et al., 1987; Matton and Brisson, 1989; Despres et al., 1995) is a marker gene for the pathogen response (Supplemental Figure 1). When wild-type tomato plants were inoculated with B. cinerea, the expression levels of TD and PR-STH2 significantly increased at 24 h after inoculation (HAI), which coincides with the time at which disease lesions began to expand (Figures 1A and 1B). These results indicate that resistance against B. cinerea infection in tomato involves the activation of both the wounding response and the pathogen response.
Figure 1.
COI1-Dependent JA Signaling Is Required for Resistance against B. cinerea Infection in Tomato.
(A) and (B) Time course of TD (A) and PR-STH2 (B) expression in response to B. cinerea inoculation.
(C) and (D) Response of wild-type and jai1 plants to B. cinerea infection. Detached leaves from 5-week-old tomato plants were spotted with a 5 μL spore suspension (106 spores/mL). Photographs were taken (C) and the lesion area analyzed (D) at 3 DAI. Error bars represent the sd from three independent experiments (n = 6). Asterisks indicate significant difference from the wild type according to Student’s t test at **P < 0.05.
(E) and (F) B. cinerea-induced expression of JA-mediated defense genes at 24 HAI in wild-type and jai1 plants.
For (A), (B), (E), and (F), 5-week-old plants were spotted with 10 μL spore suspension (106 spores/mL). Leaves were harvested at the indicated time points for RNA extraction and RT-qPCR analysis. Transcript levels of each gene were normalized to ACTIN2 expression. Error bars represent the sd of three biological replicates. Asterisks indicate significant difference from the wild type according to Student’s t test at **P < 0.05.
To assess the role of COI1-dependent JA signaling in resistance against B. cinerea infection in tomato, we compared the sizes of B. cinerea-triggered necrotic lesions on the leaves of wild-type plants with those of the jasmonic acid-insensitive1 (jai1) mutant, which harbors a mutation in the tomato homolog of Arabidopsis COI1 (Li et al., 2004). The lesion size on jai1 leaves was ∼3-fold that on wild-type leaves (Figures 1C and 1D), indicating that the disease severity of jai1 was much greater than that of the wild type. Consistent with this finding, the B. cinerea-induced expression of TD and PR-STH2 was much lower in jai1 plants than in the wild type (Figures 1E and 1F). Together, these results demonstrate that COI1-dependent JA signaling is required to regulate B. cinerea-induced activation of both wounding- and pathogen-responsive genes in tomato.
MYC2 Plays an Important Role in Plant Resistance against B. cinerea Infection
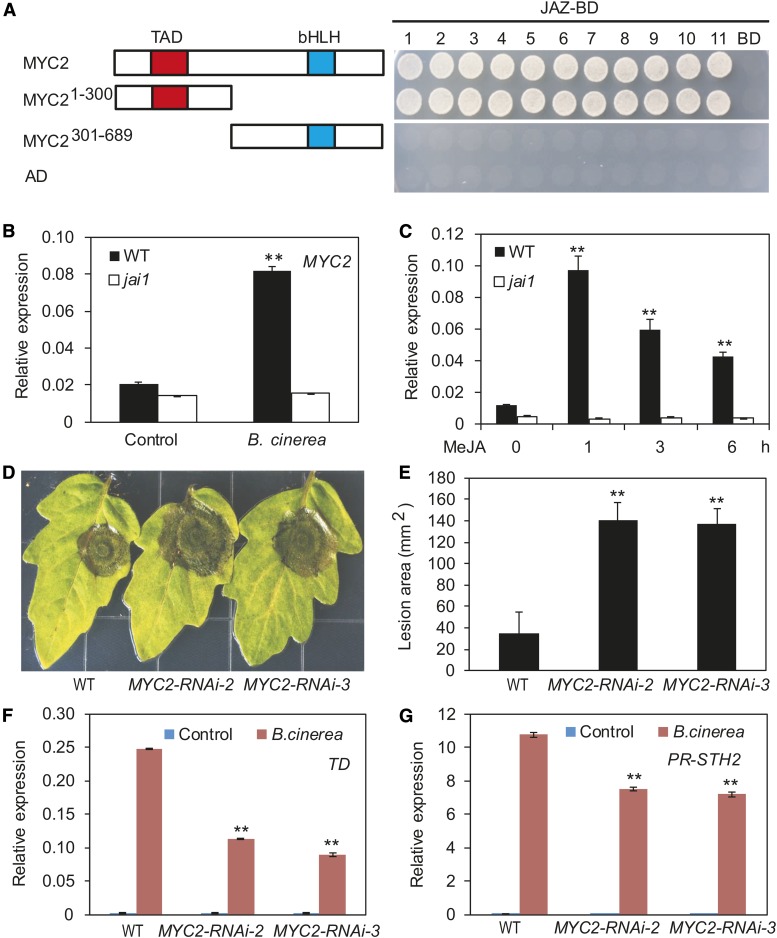
We attempted to identify TFs that act downstream of COI1 to regulate JA-mediated defense gene expression in tomato. The bHLH protein MYC2 was a strong candidate, based on several observations. First, tomato MYC2 shows high sequence similarity to At-MYC2 (Supplemental Figure 2), a master TF that regulates diverse aspects of JA responses in Arabidopsis (Boter et al., 2004; Lorenzo et al., 2004; Dombrecht et a., 2007; Kazan and Manners, 2013; Zhai et al., 2013). Second, in our yeast two-hybrid assays, MYC2 interacted with all 11 JAZ proteins in tomato, and the N-terminal fragment containing the transcriptional activation domain of MYC2 was involved in its interaction with JAZ proteins (Figure 2A). Consistent with this, in pull-down experiments using transgenic plants expressing a c-Myc-tagged MYC2 (MYC2-Myc) (Yan et al., 2013) and purified epitope-tagged JAZ proteins, we found that MYC2-Myc could be pulled down by maltose binding protein (MBP)-JAZ fusions or glutathione S-transferase (GST)-JAZ fusions (Supplemental Figure 3). Third, the expression of MYC2 was induced by methyl jasmonate (MeJA) treatment and B. cinerea infection, which depended on the function of Jai1/COI1 (Figures 2B and 2C). Finally, we recently found that the wounding-induced expression of a battery of JA-mediated defense genes is substantially reduced in MYC2-silenced plants compared with the wild type (Yan et al., 2013).
Figure 2.
Tomato MYC2 Plays a Key Role in Plant Resistance against B. cinerea Infection.
(A) Yeast two-hybrid assay to detect interactions of JAZs with MYC2. Tomato JAZ proteins were fused with the DNA binding domain (BD) in pGBKT7, and full-length MYC2 or different MYC2 domains were fused with the activation domain (AD) in pGADT7, respectively. Transformed yeast was grown on selective medium lacking Ade, His, Leu, and Trp to test protein interactions. The empty pGBKT7 vector was cotransformed with MYC2 and its derivatives in parallel as negative controls. The schematic diagram shows the MYC2 domain constructs. The conserved transcriptional activation domain (TAD) and bHLH domain are indicated with red and blue boxes, respectively.
(B) and (C) MYC2 transcripts were measured in the wild type and jai1 mutant treated with B. cinerea or MeJA. Asterisks indicate significant difference from the untreated control according to Student’s t test at **P < 0.05.
(D) and (E) The response of wild-type and MYC2-RNAi plants to B. cinerea infection. Detached leaves from 5-week-old tomato plants were spotted with 5 μL spore suspension. Photographs were taken (D) and the lesion area analyzed (E) at 3 DAI. Error bars represent the sd from three independent experiments (n = 6). Asterisks indicate significant difference from the wild type according to Student’s t test at **P < 0.05.
(F) and (G) B. cinerea-induced expression of JA-mediated defense genes (24 HAI) in wild-type and MYC2-RNAi plants. Asterisks indicate significant difference from wild type according to Student’s t test at **P < 0.05.
To verify the role of MYC2 in the B. cinerea-induced plant defense response, we generated MYC2-RNAi plants in which the expression of MYC2 was reduced by RNA interference (RNAi; Supplemental Figure 4A). As expected, B. cinerea infection led to significantly larger necrotic lesions in MYC2-RNAi plants than in the wild type (Figures 2D and 2E). To validate these results, we generated MYC2-SRDX lines (Supplemental Figure 4B) in which MYC2 was converted into a dominant-negative form by fusing it with the SUPERMAN repression domain X (SRDX) (Hiratsu et al., 2003; Du et al., 2014). B. cinerea-induced necrotic lesions on the leaves of these MYC2-SRDX plants were significantly larger than those on the leaves of wild-type plants (Supplemental Figures 4C and 4D), indicating that the MYC2-SRDX lines were more susceptible to B. cinerea infection than were wild-type plants. Together, these results demonstrate that tomato MYC2 positively regulates plant resistance to B. cinerea.
Tomato MYC2 Positively Regulates Both Wounding-Responsive and Pathogen-Responsive Genes
To investigate the mode of action of MYC2 in regulating the expression of wounding-responsive and pathogen-responsive genes, we compared the B. cinerea-induced expression of TD and PR-STH2 between MYC2-RNAi and wild type plants. At 24 HAI, the expression levels of TD and PR-STH2 were substantially lower in MYC2-RNAi plants than in wild-type plants (Figures 2F and 2G). Similarly, the B. cinerea-induced expression of TD and PR-STH2 was also reduced in the MYC2-SRDX lines compared with the wild type (Supplemental Figures 4E and 4F). These results indicate that MYC2 plays a positive role in regulating the B. cinerea-induced expression of both wounding- and pathogen-responsive genes in tomato.
Since Arabidopsis MYC2 positively regulates wounding-responsive genes while negatively regulating pathogen-responsive genes (Boter et al., 2004; Lorenzo et al., 2004; Nickstadt et al., 2004; Dombrecht et al., 2007; Zhai et al., 2013), a characteristic phenotype of the Arabidopsis myc2-2 mutant is reduced expression of the wounding-responsive marker gene VEGETATIVE STORAGE PROTEIN1 (VSP1) but elevated expression of the pathogen-responsive marker gene PLANT DEFENSIN 1.2 (PDF1.2) in response to JA. To determine whether the tomato MYC2 gene could rescue the JA response defects of the Arabidopsis myc2-2 mutant, we introduced Sl-MYC2 and At-MYC2 into the Arabidopsis myc2-2 mutant background and subjected the resulting transgenic plants (Supplemental Figure 5A) to standard JA response assays. Similar to At-MYC2, Sl-MYC2 could rescue the myc2-2 phenotype with respect to MeJA-induced root growth inhibition (Supplemental Figure 5B) and VSP1 expression to levels comparable to those of the wild type (Supplemental Figure 5C). Consistent with a previous observation (Boter et al., 2004), At-MYC2 only partially rescued the myc2-2 phenotype with respect to MeJA-induced PDF1.2 expression; MeJA-induced expression levels of PDF1.2 in Sl-MYC2/myc2-2 plants were more similar to those in myc2-2 mutants than to those in wild-type plants (Supplemental Figure 5D), indicating that Sl-MYC2 could not rescue the myc2-2 MeJA-induced PDF1.2 expression. Together, these findings substantiate the notion that tomato MYC2 regulates wounding-responsive and pathogen-responsive genes in a manner that is distinct from that of its Arabidopsis homolog.
Transcriptome Profiling of MYC2-Regulated Genes during JA Signaling
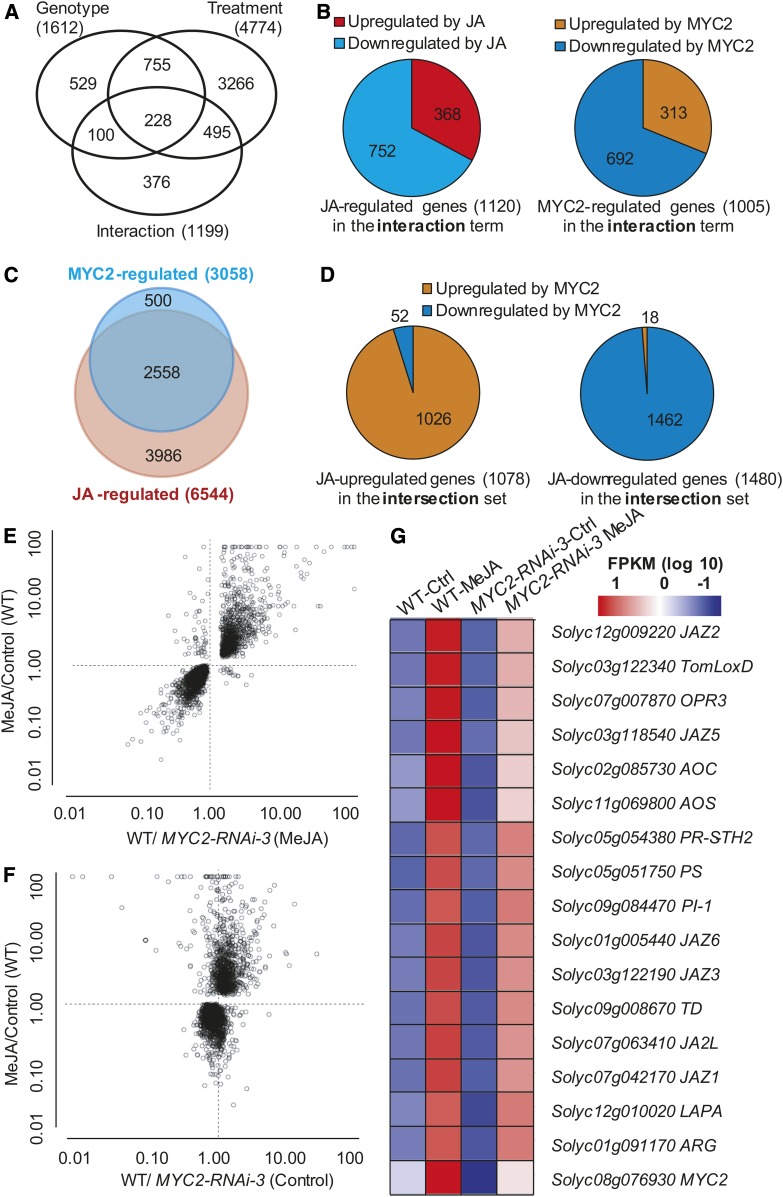
To identify genes regulated by MYC2 during JA signaling, we performed RNA-seq experiments with three biological replications and compared the transcriptome profiles of MYC2-RNAi-3 and wild-type seedlings treated with or without MeJA. The numbers of total reads, mapped reads, and the percentage of mapped reads in each replicate are shown in Supplemental Figure 6. Stringent statistical analysis of the RNA-seq data was performed by means of two-way ANOVA for the factors of genotype (wild type versus MYC2-RNAi-3) and treatment (control versus 50 μM MeJA). Based on this analysis, 4774 genes were significantly affected in their expression by treatment, 1612 genes were significantly affected by genotype, and 1199 genes were significantly affected by interaction between genotype and treatment (false discovery rate [FDR]-adjusted P value < 0.05; Figure 3A; Supplemental Data Set 1). These results are largely consistent with a similar ANOVA analysis to identify MYC2-mediated JA-responsive genes in Arabidopsis (Dombrecht et al., 2007). The 1199 genes in the interaction ANOVA term represent the “MYC2-mediated JA-responsive genes.” Gene Ontology (GO) analysis of these genes is shown in Supplemental Figure 7. Among these 1199 MYC2-mediated JA-responsive genes, 1120 were regulated by JA (i.e., had significant differential expression between MeJA-treated and untreated wild-type plants; FDR-adjusted P value < 0.05), and 1005 were regulated by MYC2 (i.e., had significant differential expression between MeJA-treated wild type and MeJA-treated MYC2-RNAi plants; FDR-adjusted P value < 0.05; Figure 3B; Supplemental Data Set 1). Surprisingly, most of the JAZ genes (i.e., JAZ3-JAZ11) were not among the 1199 genes of the interaction term data set. Considering that experimental evidence in Arabidopsis has demonstrated that JAZ genes are MYC2-mediated JA-responsive genes (Chini et al., 2007), it is likely that JAZ genes and possibly other MYC2-mediated JA-responsive genes might fail to cross the significance threshold of our stringent ANOVA analysis.
Figure 3.
Differentially Expressed Genes Identified by ANOVA Analysis and Pairwise Comparisons of the RNA-Seq Data.
(A) Statistical analysis of the effect on gene expression of the factors genotype (wild type versus MYC2-RNAi-3) and treatment (control versus 50 μM MeJA) by two-way ANOVA of the RNA-seq data. The numbers of genes showing a significant change (FDR-adjusted P value < 0.05) are shown in the Venn diagram.
(B) Distribution of genes upregulated or downregulated by JA or MYC2 among the 1199 genes in the interaction term.
(C) Venn diagrams of JA-regulated genes (differentially expressed between MeJA-treated and untreated wild-type plants; FDR-adjusted P value < 0.05) and MYC2-regulated genes (differentially expressed between MeJA-treated MYC2-RNAi-3 and the wild type, FDR-adjusted P value < 0.05). Genes coregulated by JA and MYC2 are shown in the overlapping region. The number of differentially expressed genes with significant differential expression (FDR-adjusted P value < 0.05) is shown.
(D) Distribution of genes upregulated or downregulated by MYC2 among the 2558 genes coregulated by JA and MYC2.
(E) and (F) Biplots of the ratios of expression values from the RNA-seq experiments. The JA- and MYC2-coregulated genes are indicated with white circles (2558 genes). Each data point is the ratio of the averages of three independent biological replicates. The y axes show the ratio of average expression levels of MeJA over control treatment in wild-type plants. The x axes show the ratio of average expression levels of wild-type plants over MYC2-RNAi-3 plants with (E) or without (F) MeJA treatment.
(G) Expression of representative JA-responsive genes in the RNA-seq experiments. The average FPKM (for fragments per kilobase of exon per million fragments mapped) (log10 scale) of each gene is shown.
By pairwise comparisons of the RNA-seq data, we identified 6544 genes that were differentially expressed between MeJA-treated and untreated wild-type plants (FDR-adjusted P value < 0.05) (Figure 3C; Supplemental Data Set 2); these genes were defined as JA-regulated genes. We identified 2567 genes showing significant expression differences between untreated MYC2-RNAi and wild-type seedlings and 3058 genes showing significant expression differences between MeJA-treated MYC2-RNAi and wild-type seedlings (FDR-adjusted P value < 0.05) (Figure 3C; Supplemental Data Set 2). As we are primarily interested in identifying MYC2-regulated genes during JA signaling, we defined the 3058 genes showing significant expression differences between MeJA-treated MYC2-RNAi and wild-type seedlings as MYC2-regulated genes. Analyses of these genes revealed that 40% (2558 of 6544) of the JA-regulated genes were also regulated by MYC2 (Figure 3C; Supplemental Data Set 2), confirming that MYC2 plays a key role in orchestrating the transcription of JA-regulated genes.
Among the 2558 genes coregulated by JA and MYC2, 1078 were upregulated and 1480 were downregulated by JA. Interestingly, 95% (1026 of 1078) of the genes upregulated by JA were also upregulated by MYC2 and were defined as differentially expressed genes with expression levels higher in MeJA-treated wild type versus MeJA-treated MYC2-RNAi plants (Figures 3D to 3F; Supplemental Data Set 2). Conversely, ∼98% (1462 of 1480) of the genes that were downregulated by JA were also downregulated by MYC2, and these were defined as differentially expressed genes with lower expression levels in MeJA-treated wild type versus MeJA-treated MYC2-RNAi plants (Figures 3D to 3F; Supplemental Data Set 2). These results suggest that MYC2 transduces JA signals by upregulating the expression of JA-induced genes and downregulating the expression of JA-repressed genes. GO analysis indicated that genes upregulated by JA and MYC2 are enriched in pathways related to JA biosynthesis and signaling, the oxidation-reduction process, lipid metabolism, and the wounding response. Genes downregulated by JA and MYC2 were significantly enriched in pathways related to the light response, cytoplasmic translation, photosynthesis, and other processes (Supplemental Figure 8).
Many well-characterized JA-inducible genes were identified as JA-regulated genes in our RNA-seq experiments (Figure 3G). This list includes several JA biosynthetic genes, such as ALLENE OXIDE CYCLASE (AOC), ALLENE OXIDE SYNTHASE (AOS), OXOPHYTODIENOATE-REDUCTASE3 (OPR3), and tomato LIPOXYGENASE D (TomLoxD), several JAZ genes, as well as several previously identified defense genes, including TD, LEUCINE AMINOPEPTIDASE A (LAPA), PROTEINASE INHIBITOR-1 (PI-1), and PR-STH2. Remarkably, MeJA-mediated upregulation of these genes was suppressed in MYC2-RNAi plants (Figure 3G), indicating that MYC2 plays a critical role in MeJA-mediated activation of these genes.
Genome-Wide Binding Profiles of MYC2 in Tomato
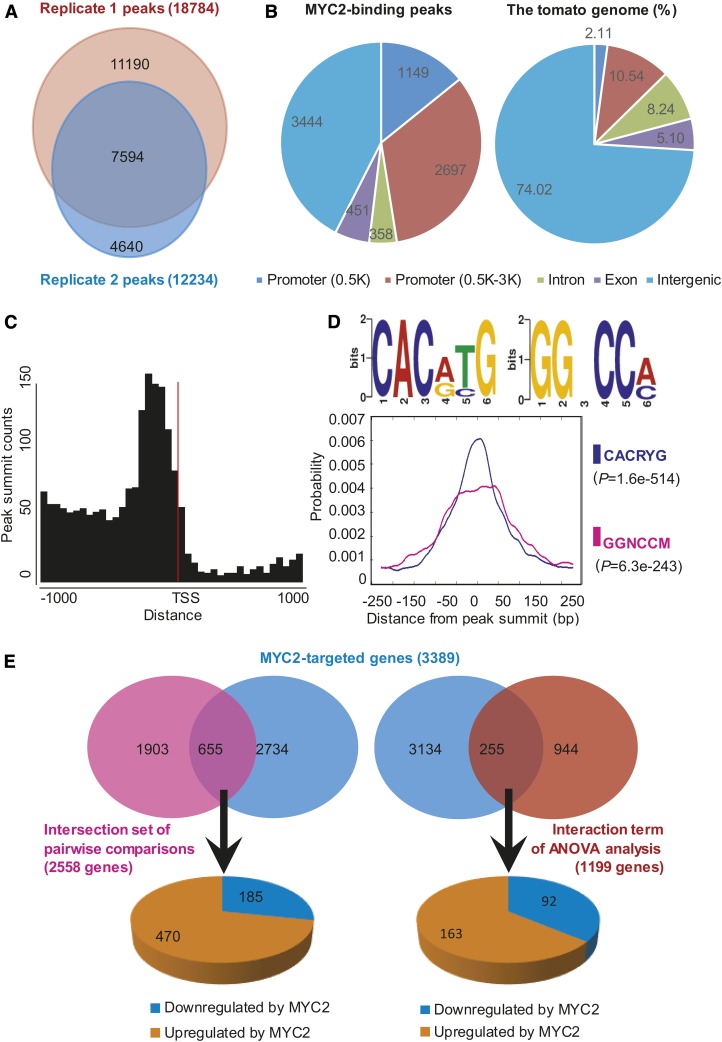
We then used ChIP-seq to investigate the genome-wide binding sites of MYC2 during JA signaling. ChIP-seq experiments were performed using MeJA-treated transgenic tomato seedlings expressing MYC2-GFP (Supplemental Figure 9) with two biological replicates (Supplemental Figure 10). Data quality assessment using the irreproducible discovery rate framework (Landt et al., 2012) indicated that the two replicates showed high reproducibility (Supplemental Figure 11). A saturation test indicated that the sequencing reads from the input libraries were sufficient to generate a suitable background for peak calling (Supplemental Figure 12). We then used model-based analysis of ChIP-seq to identify the MYC2 binding peaks (Zhang et al., 2008); genes that contain one or more binding sites within the 3-kb upstream putative promoter region are referred to as MYC2-targeted genes. We identified 18,784 and 12,234 putative MYC2 binding peaks from the two biological replicates, respectively. The two replicates shared 7594 peaks covering more than 62% of the smaller set (Figure 4A), confirming a good agreement between biological replicates. Using the 7594 overlapping peaks, we identified 3389 MYC2-targeted genes (Supplemental Data Set 3).
Figure 4.
Genome-Wide Identification of MYC2 Binding Sites and Motifs.
(A) Venn diagram depicting number of MYC2 binding peaks in two biological replicates of ChIP-seq analysis.
(B) Genome-wide distribution analysis of the overlapping MYC2 binding peaks. The distribution of individual gene regions in the tomato genome is shown as a control.
(C) MYC2 binding peaks are highly enriched in the 200-bp region immediately upstream of the TSS. The overlapping peaks were used for analysis.
(D) Binding motifs identified in the overlapping MYC2 binding peaks. DREME motif search identified two MYC2 binding motifs (CACRYG and GGNCCM). Distribution of different MYC2 binding motifs around the summits of overlapping peaks is shown by density plots. P value of one-tailed binomial test is shown in parentheses.
(E) Venn diagram showing the overlap of MYC2-targeted genes (3389, from ChIP-seq analysis) and 2558 JA- and MYC2-coregulated genes identified by pairwise comparisons of 1199 interaction genes identified by ANOVA analysis. Genes in the overlapping regions were identified as MYC2-targeted JA-responsive genes and were further divided into upregulated or downregulated sets based on RNA-seq analysis.
Genome-wide distribution analysis revealed that the MYC2 binding sites were highly enriched (Fisher’s exact test, P value < 2.20E-16) in the promoter region 3 kb upstream of the transcription start site (TSS), which accounted for 47% of all peaks (3389 genes) (Figure 4B; Supplemental Data Set 3). To investigate the detailed MYC2 binding profile in the promoter region, we calculated the distance between each peak summit and its nearest gene. A histogram of these distances at the ±1-kb region around TSSs revealed that the MYC2 binding sites were highly enriched in the proximal promoter region, peaking at ∼200 bp upstream of the TSSs (Figure 4C).
To identify the MYC2 binding motifs, the 500-bp flanking sequence around the peak summits was extracted and submitted to Multiple EM for Motif Elicitation (MEME)-ChIP to calculate the statistically overrepresented motifs (Machanick and Bailey, 2011). This analysis led to the identification of two motifs, CACRYG and GGNCCM (Figure 4D). Whereas the CACRYG motif was strongly enriched precisely at the peak summits, the GGNCCM motif was mainly enriched at ±50 bp around the peak summits (Figure 4D), suggesting that MYC2 might have a cofactor that targets the GGNCCM motif. Indeed, in our gel electrophoresis mobility shift assay (EMSA), MYC2 showed binding affinity to a DNA probe containing a CACATG (CACRYG) motif but did not show direct binding affinity to a DNA probe containing a GGACCA (GGNCCM) motif (Supplemental Figure 13). These results are consistent with our recent finding that MYC2 directly binds to the CACATG motif in vitro and in vivo (Yan et al., 2013).
MYC2-Targeted JA-Responsive Genes Are Enriched for Early JA-Responsive Genes
As described above, our ChIP-seq identified 3389 MYC2-targeted genes, and pairwise comparisons of the RNA-seq data identified 2558 genes that were coregulated by JA and MYC2. A comparison of these two data sets led to the identification of 655 overlapping genes (Figure 4E; Supplemental Data Set 4), which we refer to as genes. Among the 655 MTJA genes, 470 (71.8%) were upregulated by MYC2, while 185 (28.2%) were downregulated by MYC2 (Figure 4E; Supplemental Data Set 4), corroborating the notion that MYC2 mainly functions as a transcriptional activator in regulating the expression of JA-responsive genes. Similarly, comparison of 3389 MYC2-targeted genes and 1199 genes in the interaction term of ANOVA analysis of the RNA-seq data identified 255 MTJA genes, among which 163 were (63.9%) were upregulated by MYC2, while 92 (36.1%) were downregulated by MYC2 (Figure 4E; Supplemental Data Set 4). For further analyses, we mainly focused on the 655 MTJA genes identified by pairwise comparisons of the RNA-seq data. Notably, only ∼26% (655 out of 2558) of the JA- and MYC2-coregulated genes were MTJA genes, implying that MYC2 acts indirectly to regulate the remaining 74% (i.e., 1903 out of 2558) of the JA- and MYC2-coregulated genes.
We hypothesized that promoters of MTJA genes could contain cis-acting DNA elements that are involved in JA-stimulated MYC2 binding. In line with this hypothesis, it was recently shown that the promoters of early JA-induced genes in Arabidopsis contain a thymidine (T)-rich sequence immediately 3′ to the G-box motif (i.e., CACGTG-TTTT) that is required for full JA activation (Figueroa and Browse, 2012). Our analyses revealed that the frequency of CACRYG-TTTT motif-containing genes among MTJA genes was 30.23% (198 out of 655; Supplemental Data Set 4); This frequency is not only significantly higher (Fisher’s exact test, P value = 6.03E-11) than the frequency of CACRYG-TTTT motif-containing genes in the whole tomato genome (19.71%; 6843 out of 34,725; Tomato Genome Consortium, 2012), but also significantly higher (Fisher’s exact test, P value = 2.19E-07) than the frequency of CACRYG-TTTT motif-containing genes among the 2558 genes coregulated by JA and MYC2 (22.87%; 585 out of 2558). Thus, CACRYG-TTTT motif-containing genes are significantly enriched among MTJA genes.
For genome-wide evaluation of the timing of expression of the 2558 genes coregulated by JA and MYC2, we performed RNA-seq analyses with wild-type seedlings treated by mechanical wounding for different times (0, 1, 6, and 12 h) (Supplemental Figure 14). Genes with more than 1.5-fold change of reads count (FDR-adjusted P value < 0.05) between any time point after treatment (1, 6, or 12 h) and the untreated control (0 h) were selected for further investigation. Taking the definition of early or late genes by Ryan (2000) into consideration, we defined genes whose fold change reached a maximum at 1 h as early genes and defined genes whose fold change reached a maximum at 12 h as late genes (see Methods). Based on this criterion, 395 of the 2558 genes coregulated by JA and MYC2 could be classified as early genes, and 763 as late genes (Supplemental Data Set 5). Notably, the frequency of early genes among the MTJA genes (31.0%; 203 out of 655) was significantly higher (Fisher’s exact test, P value < 2.20E-16) than the frequency of early genes among the genes coregulated by JA and MYC2 (15.4%; 395 out of 2558). By contrast, the frequency of late genes among the MTJA genes (27.3.0%; 179 out of 655) was comparable (Fisher’s exact test, P value = 0.9946) to the frequency of late genes among the genes coregulated by JA and MYC2 (29.8%; 763 out of 2558). These statistics support the conclusion that early JA-responsive genes are enriched among MTJA genes but that late JA-responsive genes are not enriched among MTJA genes. However, since our ChIP-seq experiments were performed only at 1 h after MeJA treatment, we cannot rule out the possibility that MYC2 would target the late JA-responsive genes at later time points after treatment.
Many well-characterized early JA-responsive genes (Ryan, 2000; Li et al., 2004), including four JA biosynthetic genes (AOC, AOS, OPR3, and TomLoxD) and five JAZ genes (JAZ1, JAZ2, JAZ5, JAZ6, and JAZ9) were identified as MTJA genes (Figure 5A; Supplemental Table 1), suggesting that MYC2 directly regulates the transcription of these early JA-responsive genes. Surprisingly, most of the well-known late JA-responsive genes (Ryan, 2000; Li et al., 2004), such as the above-mentioned wounding-responsive marker gene TD and the pathogen-responsive marker gene PR-STH2, were not included in the list of MTJA genes (Supplemental Table 1), suggesting that MYC2 does not regulate the transcription of these late JA-responsive genes via direct promoter binding.
Figure 5.
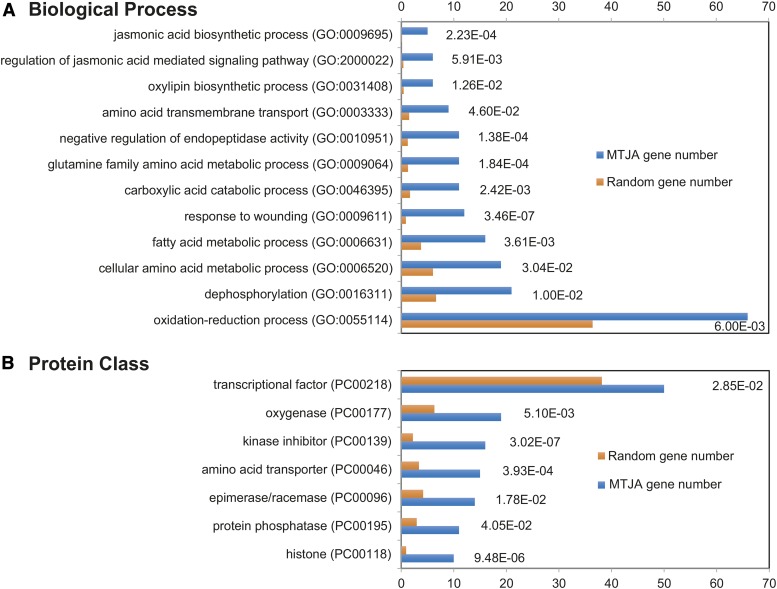
GO enrichment analysis was performed using the PANTHER (for protein annotation through evolutionary relationship) classification system (www.pantherdb.org) with default parameters. The 655 MTJA genes identified by pairwise comparisons of the RNA-seq data were used for analysis. Results of classification within the biological process (A) and protein class (B) categories with P values from the statistical overrepresentation test are shown. The P values are expressed in exponential notation, replacing part of the number with E+n, where E multiplies the preceding number by 10 to the nth power. For example, 1.00E-02 means 0.01. Categories with P values less are overrepresented among the MTJA genes.
MYC2-Targeted JA-Responsive Genes Are Enriched for Wounding- and Pathogen-Responsive TFs That Are Important for Plant Resistance against B. cinerea Infection
We reasoned that the role of MYC2 in regulating the transcription of the remaining 74% (i.e., 1903 out of 2558) of the JA- and MYC2-coregulated genes, including the well-known late JA-responsive genes (defense genes), is likely executed through intermediate JA-responsive TFs that act downstream of MYC2. Indeed, among the 655 MTJA genes, 50 were MTF genes, accounting for 7.63% of the MTJAs (Figure 5B; Supplemental Table 2). Among the 255 MTJA genes identified by ANOVA analysis of the RNA-seq data, 19 were MTF genes, accounting for 7.45% of the MTJAs (Supplemental Table 2). Notably, the frequency of MTF genes among the MTJA genes (7.63%; 50 out of 655) was significantly higher (Fisher’s exact test, P value = 0.0285) than that of TF genes in the whole tomato genome (5.83%; 2024 out of 34,725; Tomato Genome Consortium, 2012) (Figure 5B).
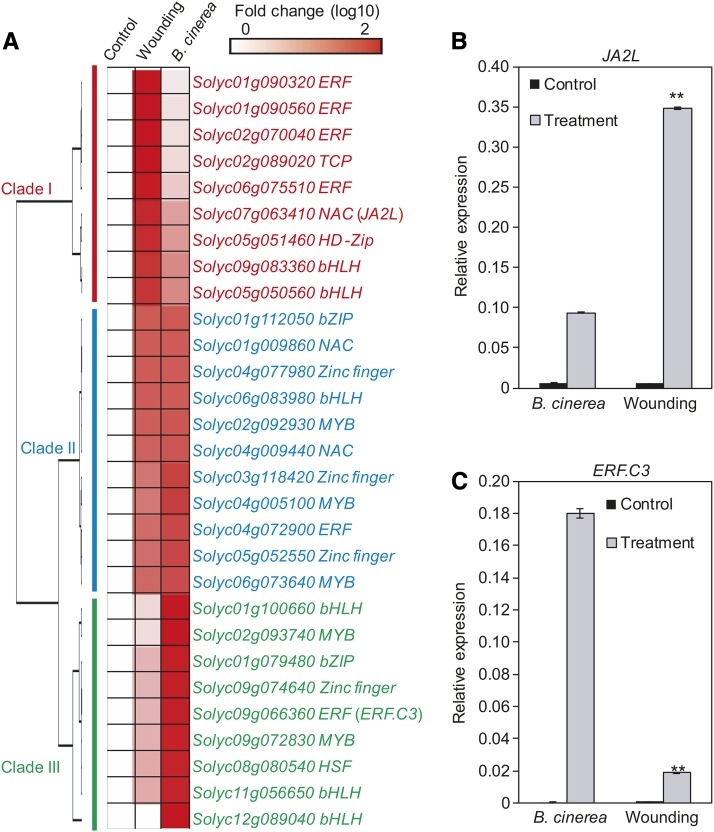
As a first step to understand the mode of action of these MTFs in regulating JA-responsive gene expression, we selected 29 MTF genes and examined their expression patterns in response to mechanical wounding and B. cinerea inoculation (Figure 6A). Based on their expression profiles in response to these two stimuli, the 29 MTFs were categorized into three clades (Figure 6A): MTFs in clade I (red) were strongly induced by mechanical wounding, but moderately induced by B. cinerea infection; MTFs in clade III (green) were strongly induced by B. cinerea, but moderately induced by mechanical wounding; and MTFs in clade II (blue) were induced by both mechanical wounding and B. cinerea infection (Figure 6A). These findings suggest that these MTFs might act downstream of MYC2 to differentially regulate the wounding response and/or pathogen response.
Figure 6.
Expression of MYC2-Targeted TF Genes in Response to Wounding and B. cinerea Infection.
(A) Expression of MTF genes in response to wounding and B. cinerea infection. Fold change of the average expression (log10 scale) of each gene is shown.
(B) and (C) Expression of JA2L (B) and ERF.C3 (C) in response to wounding and B. cinerea infection. For wounding-induced gene expression, 18-d-old wild-type seedlings were wounded with a hemostat as described in Methods. One hour after wounding, the wounded leaf tissues were harvested for RT-qPCR analysis. For pathogen-induced gene expression, leaves of 5-week-old plants were spotted with a 10 μL B. cinerea spore suspension (106 spores/mL) as described in Methods. At 24 HAI, infected leaf tissues were harvested for RT-qPCR analysis. Transcript levels of each gene were normalized to ACTIN2 expression. Error bars represent the sd of three biological replicates. Asterisks indicate significant difference between wounding and B. cinerea infection according to Student’s t test at **P < 0.05.
Taken together, the above results led us to hypothesize that MYC2, together with its downstream MTFs, orchestrates a hierarchical transcriptional cascade that regulates the JA-mediated expression of defense genes. To test this hypothesis and to validate the function of the identified MTFs in this transcriptional cascade, we selected the NAC (for NAM, ATAF1,2, and CUC2) TF gene JA2L (Du et al., 2014) from clade I and the ERF (for ETHYLENE RESPONSE FACTOR) TF gene ERF.C3 (Pirrello et al., 2012) from clade III (Figure 6A) for detailed analysis. Gene expression analysis confirmed that JA2L was strongly induced by mechanical wounding but only mildly induced by B. cinerea infection (Figure 6B). By contrast, ERF.C3 was strongly induced by B. cinerea infection, but only mildly induced by mechanical wounding (Figure 6C).
JA2L Is a Wounding-Responsive MTF That Plays an Important Role in Plant Resistance against B. cinerea Infection
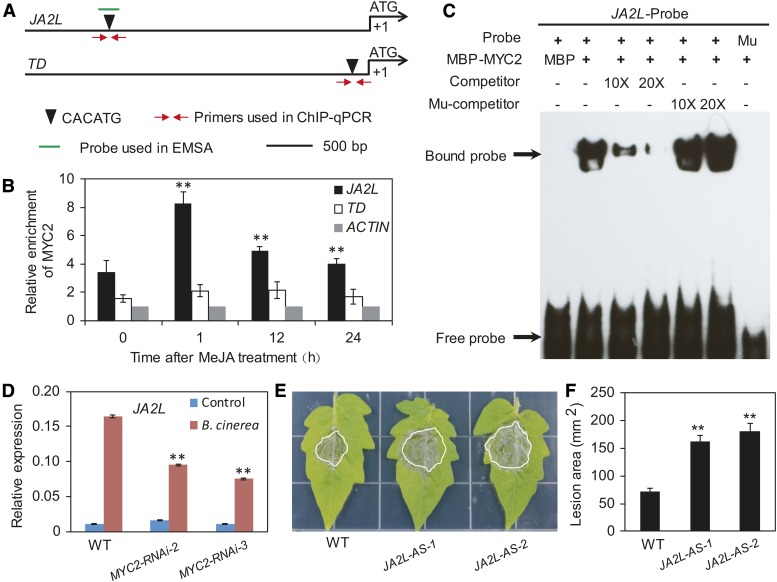
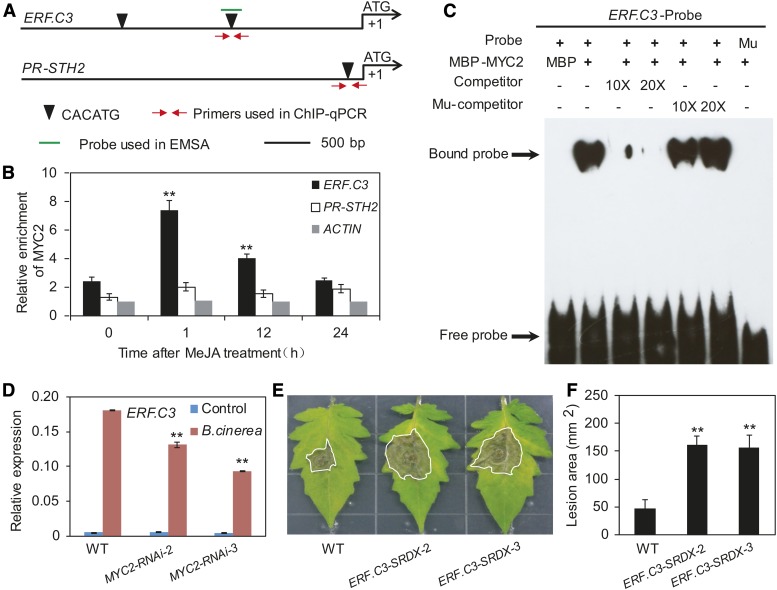
We conducted a series of experiments to verify that JA2L is a true MTF. ChIP-seq analysis, together with promoter sequence analysis, revealed that MYC2 binds to a CACATG motif located 2700 bp upstream of the TSS of JA2L (Supplemental Figure 15). ChIP-qPCR analysis using MYC2-GFP plants and anti-GFP antibodies confirmed binding enrichment of MYC2 to this motif at steady state (Figures 7A and 7B). In response to MeJA treatment, MYC2 binding enrichment to this motif was markedly increased at 1 h, then showed an obvious reduction at 12 and 24 h (Figures 7A and 7B). Parallel experiments revealed that, although a CACATG motif is present in the promoter of the wounding-responsive marker gene TD (Figure 7A), little or no MYC2 bound to this motif in the absence or presence of MeJA treatment (Figures 7A and 7B).
Figure 7.
The Wounding-Responsive MTF JA2L Is Involved in Plant Resistance against B. cinerea Infection.
(A) Schematic diagram of the promoters of the indicated genes. Black triangles indicate the CACATG motif. Red arrows indicate primers used for ChIP-qPCR assays, and short green lines indicate DNA probes used for EMSAs. The translational start site (ATG) is shown at position +1.
(B) ChIP-qPCR assays showing the fold enrichment of MYC2 at the promoters of the indicated genes. Ten-day-old MYC2-GFP-9 seedlings were treated with 50 μM MeJA for different times before chromatin was extracted for immunoprecipitation with an anti-GFP antibody. The immunoprecipitated chromatin was analyzed by qPCR using gene-specific primers as indicated in (A). The fold enrichment of MYC2 at each promoter was calculated against the ACTIN2 promoter. Error bars represent the sd of three biological replicates. Asterisks indicate significant difference from the enrichment at time 0 according to Student’s t test at **P < 0.05.
(C) EMSA showing that MBP-MYC2 could directly bind to the promoters of JA2L. The MBP protein was incubated with the labeled probe in the first lane to serve as a negative control; 10- and 20-fold excess of unlabeled or mutated probes were used for competition. Mu, mutated probe in which the CACATG motif was deleted.
(D) B. cinerea-induced expression of JA2L in wild-type and MYC2-RNAi plants. Transcript levels of JA2L were normalized to ACTIN2 expression. Error bars represent the sd of three biological replicates. Asterisks indicate significant difference from wild type according to Student’s t test at **P < 0.05.
(E) and (F) Response of JA2L-AS plants to B. cinerea. Photographs were taken (E) and the lesion area analyzed (F) at 3 DAI. Error bars represent the sd from three independent experiments (n = 6). Asterisks indicate significant difference from the wild type according to Student’s t test at **P < 0.05.
We then performed an EMSA to investigate whether the fusion protein MBP-MYC2 could directly bind to a 40-bp DNA probe containing the CACATG motif from the JA2L promoter. As shown in Figure 7C, MBP-MYC2 bound to the CACATG-containing DNA probe, which was outcompeted by the addition of unlabeled DNA probe. By contrast, MBP-MYC2 failed to bind to a mutant probe lacking the CACATG motif (Figure 7C), demonstrating that the binding of MYC2 to the JA2L promoter is dependent on the presence of the CACATG motif. Taken together, these results demonstrate that JA2L is a direct target of MYC2.
We hypothesized that MYC2 and JA2L form a transcriptional module, MYC2-JA2L, that plays an important role in plant resistance to B. cinerea infection. To test this, we first compared B. cinerea-induced expression of JA2L between wild-type and MYC2-RNAi plants. As revealed by RT-qPCR, B. cinerea-induced expression of JA2L was markedly reduced in MYC2-RNAi plants compared with wild-type plants (Figure 7D), implying that MYC2 activates JA2L expression during the plant response to B. cinerea infection. Next, we compared the performance of wild-type plants versus JA2L-AS plants in which the expression of JA2L was knocked down (Du et al., 2014). At 3 d after infection (DAI), B. cinerea-induced necrotic lesions on JA2L-AS leaves were larger than those on wild-type leaves (Figures 7E and 7F), indicating that JA2L-AS plants are more susceptible to B. cinerea than are wild-type plants.
These findings, together with the observation that JA2L did not interact with any member of the JAZ protein family in tomato (Supplemental Figure 16), support the notion that JA2L is a true MTF that acts downstream of MYC2 in the JA-mediated response to B. cinerea infection.
JA2L Directly Regulates the Late Wounding-Responsive Gene TD
Next, we explored the mechanism by which the MYC2-JA2L module regulates JA-mediated plant resistance to B. cinerea infection. Since JA2L is preferentially induced by mechanical wounding (Figures 6A6B), we hypothesized that the MYC2-JA2L module mainly regulates the group of wounding-responsive genes. Three lines of evidence support this hypothesis. First, as revealed by ChIP-qPCR assays, the binding of MYC2 to the JA2L promoter was rapidly and strongly induced by mechanical wounding (Figure 8A). Second, in line with our finding that JA2L-AS plants are more susceptible than wild-type plants to B. cinerea infection (Figures 7E and 7F), B. cinerea-induced expression of the wounding-responsive marker gene TD was significantly reduced in JA2L-AS plants compared with the wild type (Figure 8B), demonstrating a positive role for JA2L in regulating pathogen-induced expression of TD. Third, B. cinerea-induced expression levels of PR-STH2 were slightly higher in JA2L-AS lines than those in the wild-type plants (Supplemental Figure 17A), indicating that a reduction of JA2L causes a minor increase in B. cinerea-induced expression of the pathogen-responsive marker gene PR-STH2.
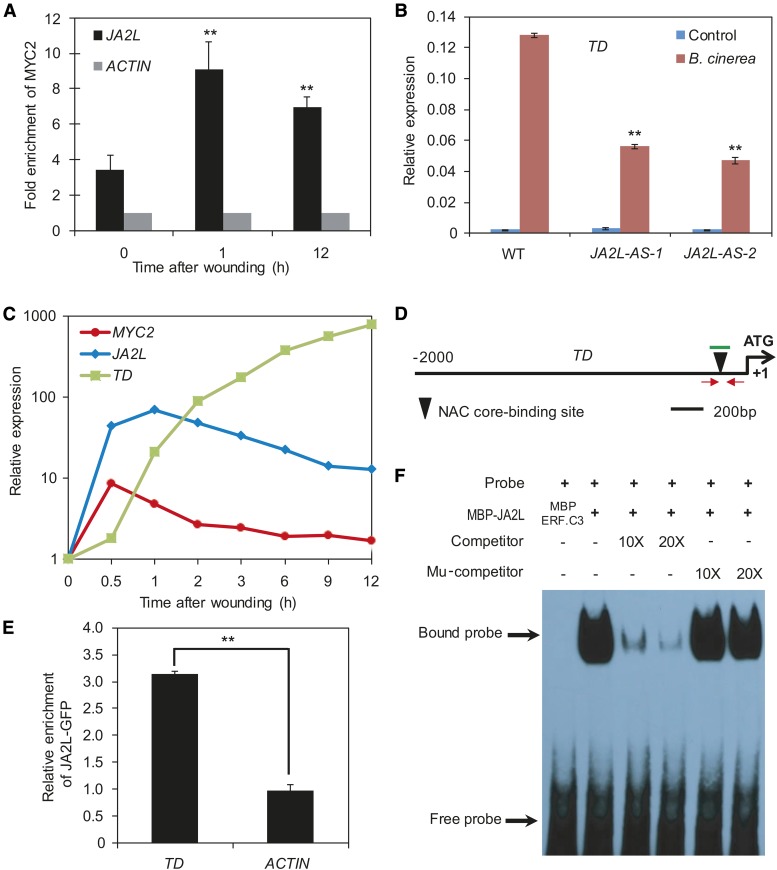
Figure 8.
The MTF JA2L Regulates the Expression of TD through Promoter Binding.
(A) Wounding-induced enrichment of MYC2 at the promoter of JA2L revealed by ChIP-qPCR assays. Immunoprecipitated chromatin was analyzed by qPCR using gene-specific primers as indicated in Figure 7A. The fold enrichment of MYC2 on each promoter was calculated against the ACTIN2 promoter. Error bars represent the sd of three biological replicates. Asterisks indicate significant difference from the enrichment at time 0 according to Student’s t test at **P < 0.05.
(B) B. cinerea-induced TD expression (24 HAI) in wild-type and JA2L-AS plants. Transcript levels of TD were normalized to ACTIN2 expression. Error bars represent the sd of three biological replicates. Asterisks indicate significant difference from the wild type according to Student’s t test at **P < 0.05.
(C) Wounding-induced expression pattern of MYC2, JA2L, and TD in wild-type seedlings. Transcript levels of each gene at different time points were normalized to that at time point 0. The experiment was repeated three times with similar results.
(D) Schematic diagram of the TD promoter. Black triangles represent the NAC core binding site (CACG). Red arrows indicate primers used for ChIP-qPCR assays, and short green lines indicate DNA probes used for EMSAs. Shown is the 2-kb upstream sequence of TD. The translational start site (ATG) is shown at position +1.
(E) ChIP-qPCR assays showing the fold enrichment of JA2L-GFP at the promoters of the indicated genes. Chromatin of JA2L-GFP seedlings was extracted for immunoprecipitation with an anti-GFP antibody. The immunoprecipitated chromatin was analyzed by qPCR using TD-specific primers as indicated in (D). The fold enrichment of JA2L-GFP was calculated against the ACTIN2 promoter. Error bars represent the sd of three biological replicates. Asterisks indicate significant difference according to Student’s t test at **P < 0.05.
(F) EMSA showing that the MBP-JA2L fusion protein binds to a DNA probe for the TD promoter in vitro. Biotin-labeled probes were incubated with MBP-JA2L protein, and free and bound DNA (arrows) was separated in an acrylamide gel. The MBP-ERF.C3 protein was incubated with the labeled probe in the first lane to serve as a negative control. Unlabeled probes were used as competitors, as indicated. Mu, mutated probe in which the NAC core binding site was deleted.
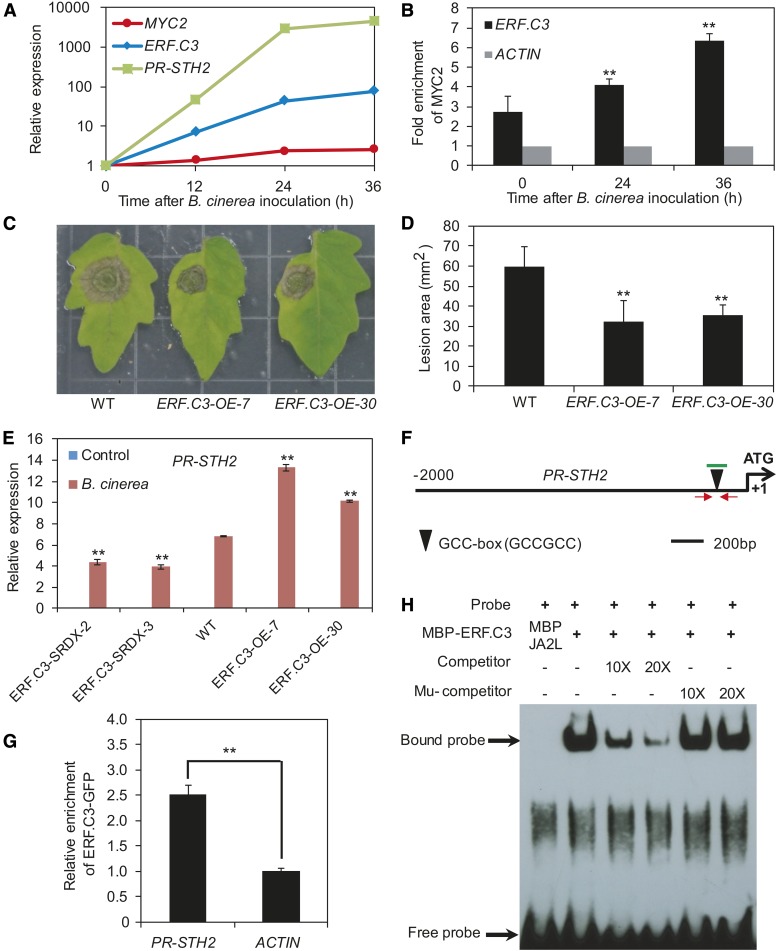
We also examined the wounding-inducible expression patterns of MYC2, JA2L, and TD and found that mechanical wounding induced the expression of these genes in a sequential manner. As shown in Figure 8C, MYC2 expression levels reached a maximum at 0.5 h after wounding and then decreased throughout the remainder of the experiment. JA2L expression levels peaked at 1 h after wounding before declining. Unlike MYC2 and JA2L, TD expression increased consistently throughout the experiment and reached a maximum at 12 h after wounding (Figure 8C). Notably, the maximum expression levels of MYC2, JA2L, and TD increased by ∼10-, 100-, and 1,000-fold, respectively (Figure 8C), suggesting a hierarchical effect of MYC2 and JA2L in amplifying the transcriptional output of mechanical wounding. Together, these results are consistent with our hypothesis that MYC2-mediated regulation of TD expression is achieved through the intermediate TF JA2L.
NAC family TFs preferentially bind to a CACG motif known as the NAC core binding site (Tran et al., 2004; Du et al., 2014). Indeed, our sequence analysis identified a CACG motif at 135 bp upstream of the TSS of TD (Figure 8D). ChIP-qPCR analysis with JA2L-GFP plants (Du et al., 2014) and anti-GFP antibodies confirmed binding enrichment of JA2L to this motif (Figure 8E). To investigate whether JA2L directly binds to this NAC core binding site, we performed EMSA with purified MBP-JA2L and a 38-bp DNA probe containing this CACG motif. As expected, MBP-JA2L bound to the DNA probe (Figure 8F). This binding was successfully outcompeted by unlabeled DNA probe, but not by a DNA probe in which the NAC core binding site was deleted (Figure 8F), demonstrating that JA2L directly binds to the TD promoter and that this binding is dependent on the presence of the CACG motif.
Taken together, our results support the hypothesis that, during the JA-mediated plant resistance response, MYC2 directly regulates the MTF JA2L, which in turn directly regulates the expression of the late wounding-responsive gene TD.
ERF.C3 Is a Pathogen-Responsive MTF That Plays an Important Role in Plant Resistance against B. cinerea Infection
In parallel with the JA2L experiments, we conducted a series of experiments to verify that ERF.C3 is a true MTF. ChIP-seq analysis, together with promoter sequence analysis, revealed that MYC2 binds to the CACATG motif at 1500 bp upstream of the ERF.C3 TSS (Supplemental Figure 15). ChIP-qPCR analysis confirmed binding enrichment of MYC2 to this motif at steady state (Figures 9A and 9B). In response to MeJA treatment, MYC2 binding enrichment to this motif was markedly increased at 1 h then showed an obvious reduction at 12 and 24 h (Figures 9A and 9B). Parallel experiments revealed that, although there is a CACATG motif in the promoter of the pathogen-responsive marker gene PR-STH2 (Figure 9A), MYC2 enrichment levels to this motif were very low both in the absence and presence of MeJA treatment (Figure 9B).
Figure 9.
The Pathogen-Responsive MTF ERF.C3 Is Involved in Plant Resistance against B. cinerea Infection.
(A) Schematic diagram of the promoters of the indicated genes. Black triangles indicate the CACATG motif. Red arrows indicate primers used for ChIP-qPCR assays, and short green lines indicate DNA probes used for EMSAs. The translational start site (ATG) is shown at position +1.
(B) ChIP-qPCR assays showing the fold enrichment of MYC2 at the promoters of the indicated genes. Ten-day-old MYC2-GFP-9 seedlings were treated with 50 μM MeJA for different times before chromatin was extracted for immunoprecipitation with an anti-GFP antibody. The immunoprecipitated chromatin was analyzed by qPCR using gene-specific primers as indicated in (A). The fold enrichment of MYC2 at each promoter was calculated against the ACTIN2 promoter. Error bars represent the sd of three biological replicates. Asterisks indicate significant difference from the enrichment at time 0 according to Student’s t test at **P < 0.05.
(C) EMSA showing that MBP-MYC2 could directly bind to the promoters of ERF.C3. The MBP protein was incubated with the labeled probe in the first lane to serve as a negative control; 10- and 20-fold excess of unlabeled or mutated probes were used for competition. Mu, mutated probe in which the CACATG motif was deleted.
(D) B. cinerea-induced expression of ERF.C3 in wild type and MYC2-RNAi plants. Transcript levels of ERF.C3 were normalized to ACTIN2 expression. Error bars represent the sd of three biological replicates. Asterisks indicate significant difference from wild type according to Student’s t test at **P < 0.05.
(E) and (F) Response of ERF.C3-SRDX plants to B. cinerea. Photographs were taken (E) and the lesion area analyzed (F) at 3 DAI. Error bars represent the sd from three independent experiments (n = 6). Asterisks indicate significant difference from the wild type according to Student’s t test at **P < 0.05.
We then performed an EMSA to test whether the MBP-MYC2 fusion protein could directly bind to a 40-bp DNA probe containing the CACATG motif from the ERF.C3 promoter. As shown in Figure 9C, MBP-MYC2 could indeed bind to the CACATG-containing DNA probe, which was successfully outcompeted by the addition of unlabeled DNA probe. By contrast, MBP-MYC2 failed to bind to a mutant probe in which the CACATG motif was deleted, and the addition of unlabeled mutant probe did not significantly affect the binding affinity of MBP-MYC2 (Figure 9C), demonstrating that this binding is dependent on the presence of the CACATG motif. Taken together, these results demonstrate that ERF.C3 is an MTF gene.
We reasoned that MYC2 and ERF.C3 might form a transcriptional module, MYC2-ERF.C3, that might play an important role in plant resistance to B. cinerea infection. To test this hypothesis, we first compared the B. cinerea-induced expression of ERF.C3 between wild-type and MYC2-RNAi plants. As revealed by RT-qPCR, B. cinerea-induced expression of ERF.C3 was reduced in MYC2-RNAi plants compared with the wild type (Figures 9D), implying that MYC2 activates ERF.C3 expression during the plant response to B. cinerea infection. Next, we compared the performance of wild-type plants versus ERF.C3-SRDX plants, in which ERF.C3 was expressed in dominant-negative form (Supplemental Figure 18). At 3 DAI, B. cinerea-induced necrotic lesions were larger on ERF.C3-SRDX leaves than on wild-type leaves (Figures 9E and 9F), indicating that ERF.C3-SRDX plants were more susceptible to B. cinerea than were wild-type plants.
These findings, together with the observation that ERF.C3 did not interact with any of the 11 JAZ protein family members in tomato (Supplemental Figure 16), support the notion that ERF.C3 is a true MTF that acts downstream of MYC2 in the JA-mediated plant response to B. cinerea infection.
ERF.C3 Directly Regulates the Late Pathogen-Responsive Gene PR-STH2
We explored the mechanism by which the MYC2–ERF.C3 module regulates JA-mediated plant resistance to B. cinerea infection. Since ERF.C3 is preferentially induced by pathogen infection (Figure 6C), we hypothesized that the MYC2-ERF.C3 module might be involved in regulating this group of pathogen-responsive genes. Supporting this hypothesis, we found that B. cinerea infection led to increased expression of MYC2, ERF.C3, and the pathogen-responsive marker gene PR-STH2 (Figure 10A). Moreover, as revealed by ChIP-qPCR, the binding enrichment of MYC2 to a CACATG motif of ERF.C3 promoter (Figure 9A) was induced by B. cinerea infection (Figure 10B). To confirm the role of ERF.C3 in pathogen resistance, we generated ERF.C3-OE plants in which ERF.C3 expression levels were elevated (Supplemental Figure 18). Pathogen infection assays indicated that, in contrast to ERF.C3-SRDX plants, which showed reduced resistance to B. cinerea infection (Figures 9E and 9F), ERF.C3-OE plants showed increased resistance to the pathogen, based on the size of the necrotic lesions (Figures 10C and 10D). Consistent with this pattern, the B. cinerea-induced expression levels of PR-STH2 were higher in ERF.C3-OE plants than in wild-type plants, but lower in ERF.C3-SRDX plants (Figure 10E) than in the wild type, demonstrating a positive role for ERF.C3 in regulating the pathogen-induced expression of PR-STH2. By contrast, B. cinerea-induced expression levels of TD were comparable or slightly higher in ERF.C3-SRDX lines than in wild-type plants, and B. cinerea-induced expression levels of TD were comparable in ERF.C3-OE lines compared with those in wild-type plants (Supplemental Figure 17), indicating that manipulation of ERF.C3 expression has a negligible or slightly positive effect on B. cinerea-induced expression of the wounding-responsive marker gene TD.
Figure 10.
The MTF ERF.C3 Regulates the Expression of PR-STH2 through Promoter Binding.
(A) B. cinerea-induced expression of MYC2, ERF.C3, and PR-STH2 in wild-type plants. Transcript levels of each gene at different time points were normalized to that at time point 0. The experiment was repeated three times with similar results.
(B) B. cinerea-induced enrichment of MYC2 to the promoter of ERF.C3 revealed by ChIP-qPCR. Immunoprecipitated chromatin was analyzed by qPCR using gene-specific primers as indicated in Figure 9A. The fold enrichment of MYC2 was calculated against the ACTIN2 promoter. Error bars represent the sd of three biological replicates. Asterisks indicate significant difference from the enrichment at time 0 according to Student’s t test at **P < 0.05.
(C) and (D) Response of wild-type and ERF.C3-OE plants to B. cinerea. Photographs were taken (C) and the lesion area analyzed (D) at 3 DAI. Error bars represent the sd from three independent experiments (n = 6). Asterisks indicate significant difference from the wild type according to Student’s t test at **P < 0.05.
(E) B. cinerea-induced expression of PR-STH2 (24 HAI) in wild-type, ERF.C3-OE, and ERF.C3-SRDX plants. Transcript levels were normalized to ACTIN2 expression. Error bars represent the sd of three biological replicates. Asterisks indicate significant difference from the wild type according to Student’s t test at **P < 0.05.
(F) Schematic diagram of the promoter of PR-STH2. Black triangles represent the GCC-box. Red arrows indicate primers used for ChIP-qPCR assays, and short green lines indicate DNA probes used for EMSAs. Shown is the 2-kb upstream sequence of PR-STH2. The translational start site (ATG) is shown at position +1.
(G) ChIP-qPCR assays showing the fold enrichment of ERF.C3 to the promoters of the indicated genes. Chromatin of ERF.C3-GFP-1 leaves was extracted for immunoprecipitation with an anti-GFP antibody. The immunoprecipitated chromatin was analyzed by qPCR using PR-STH2-specific primers as indicated in (F). The fold enrichment of ERF.C3-GFP was calculated against the ACTIN2 promoter. Error bars represent the sd of three biological replicates. Asterisks indicate significant difference according to Student’s t test at **P < 0.05.
(H) EMSA showing that the MBP-ERF.C3 fusion protein binds to the DNA probe for the PR-STH2 promoter in vitro. Biotin-labeled probes were incubated with MBP-ERF.C3 protein, and free and bound DNA (arrows) was separated in an acrylamide gel. The MBP-JA2L protein was incubated with the labeled probe in the first lane to serve as a negative control. Unlabeled probes were used as competitors, as indicated. Mu, mutated probe in which the GCC-box was deleted.
We then investigated whether ERF.C3 directly binds to the PR-STH2 promoter. Sequence analysis revealed that the PR-STH2 promoter contains a GCC-box (GCCGCC) (Figure 10F), which was previously shown to be an ERF.C3 binding motif (Pirrello et al., 2012). ChIP-qPCR analysis with ERF.C3-GFP-1 plants (Supplemental Figure 19) and anti-GFP antibodies confirmed binding enrichment of ERF.C3 to this motif (Figure 10G). In EMSA, MBP-ERF.C3 bound to a DNA probe containing the GCC-box from the PR-STH2 promoter (Figure 10F), which was successfully outcompeted using an unlabeled DNA probe, but not a DNA probe in which the GCC-box was deleted (Figure 10H), indicating that ERF.C3 binds to the PR-STH2 promoter in a GCC-box-dependent manner.
Taken together, our results support the hypothesis that, during the JA-mediated plant resistance response, MYC2 directly regulates the expression of the MTF gene ERF.C3, which in turn directly regulates the expression of the late pathogen-responsive gene PR-STH2.
DISCUSSION
As a major plant immunity hormone, JA orchestrates genome-wide transcriptional changes in response to versatile biotic and abiotic cues. Using the tomato-B. cinerea interaction as a model system, we illustrated here that the tomato master regulator MYC2 orchestrates a hierarchical transcriptional program that underlies JA-mediated plant immunity. Our results support a scenario in which, in response to JA elicitation, MYC2 and its downstream MTFs form various MYC2-MTF transcriptional modules, which initiate and amplify specific aspects of the transcriptional output. Considering that JA occurs in virtually all plants and controls plant responses to a wide range of stresses through transcriptional regulation, it is likely that the mechanism we described here also operates in other aspects of JA responses.
MYC2 and MTFs Form Transcriptional Modules That Regulate Diverse Aspects of the JA Response
Studies of the transcriptional regulation of hormone signaling in plants often focus on master regulators that act directly downstream of the hormone receptor and orchestrate hormone-mediated transcriptional reprogramming (Santner and Estelle, 2009; Kazan and Manners, 2013; Song et al., 2016). Here, we provide several lines of evidence that the tomato bHLH TF MYC2 acts as a master regulator in orchestrating JA-mediated plant immunity. First, MYC2 interacts with all 11 JAZ proteins, which are important components of the SCFCOI1-JAZ coreceptor complex. Second, our RNA-seq assays revealed that MYC2 regulates ∼40% (2558 of 6544) of the JA-responsive genes of the tomato genome. Third, pathogen infection assays using wild-type and MYC2-RNAi plants indicated that MYC2 plays a pivotal role in plant resistance to the necrotrophic pathogen B. cinerea.
To help uncover the mode of action of MYC2 in orchestrating JA-mediated transcriptional regulation, we combined RNA-seq and ChIP-seq data to identify JA-responsive genes that are directly bound by MYC2. Interestingly, among the 2558 JA-responsive genes that are regulated by MYC2, only ∼26% (655 out of 2558) are MTJA genes, indicating that MYC2 does not directly regulate the expression of the remaining 74% of the JA-responsive genes by binding to their promoters. Closer comparison of the promoter contexts in the 655 MTJA genes and the 2558 JA- and MYC2-coregulated genes revealed that a CACRYG-TTTT motif is enriched in promoters of MTJA genes. Considering that the CACRYG-TTTT motif is required for JA-mediated activation by MYC2 in Arabidopsis (Figueroa and Browse, 2012), it is reasonable to speculate that the CACRYG-TTTT motif is also important for JA-mediated binding and activation by tomato MYC2. Experimental evidence is required to support this hypothesis.
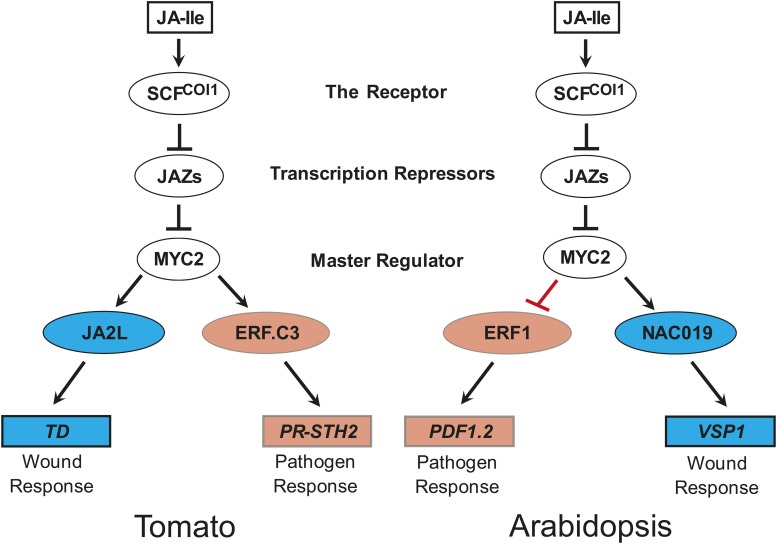
Further analyses revealed that MTJAs are enriched for MTFs and early JA-responsive genes, but not for late JA-responsive genes including the well-studied wounding-responsive marker gene TD and the pathogen-responsive marker gene PR-STH2, suggesting that MYC2 functions as a high-level regulator of the JA-mediated transcriptional program. Based on these observations, we propose a model in which MYC2 orchestrates a hierarchical transcriptional cascade that underlies JA-mediated plant immunity. According to this model, upon JA elicitation, MYC2 rapidly and directly regulates the transcription of downstream MTFs, which in turn regulate the expression of late wounding-responsive or pathogen-responsive genes (Figure 11).
Figure 11.
The Mode of Action of Tomato MYC2 in Regulating Wounding-Responsive and Pathogen-Responsive Genes Is Distinct from That of Its Arabidopsis Homolog.
In both tomato and Arabidopsis, the active hormone JA-Ile promotes COI1-dependent degradation of JAZ repressors and thereby activates (derepresses) the master TF MYC2. In tomato, MYC2 positively and directly regulates the transcription of its downstream MTFs, which in turn regulate the expression of the late wounding-responsive genes or pathogen-responsive genes. By contrast, in Arabidopsis, MYC2 positively regulates wounding-responsive genes, while negatively regulating pathogen-responsive genes.
If our model holds true, these MTFs should act differentially in regulating the late wounding-responsive versus pathogen-responsive genes. Indeed, based on their expression patterns, the MTFs can be generally categorized into three groups: those that are preferentially induced by wounding, those that are preferentially induced by pathogens, and those that are induced by both wounding and pathogens. Based on the notion that plant resistance against B. cinerea involves the JA-dependent activation of both the wounding response and the pathogen response (El Oirdi and Bouarab, 2007; Abuqamar et al., 2008, 2009; Chassot et al., 2008; El Oirdi et al., 2011; Yan et al., 2013), we conducted proof-of-concept experiments using the tomato-B. cinerea interaction system. We demonstrated that MYC2 and the wounding-inducible MTF JA2L form the MYC2-JA2L transcriptional module, which preferentially regulates the expression of wounding-responsive genes (Figures 7, 8, and 11). In parallel, we confirmed that MYC2 and the pathogen-inducible MTF ERF.C3 form the MYC2-ERF.C3 transcriptional module, which preferentially regulates pathogen-responsive genes (Figures 9 to 11). The frequencies of CACG (i.e., JA2L binding motif)-containing genes and GCCGCC (i.e., ERF.C3 binding motif)-containing genes are comparable among the 655 MTJA genes and in the 2558 JA- and MYC2-coregulated genes (Supplemental Figure 20), suggesting JA2L (or ERF.C3) may not directly regulate all the downstream wounding-responsive genes (or pathogen-responsive genes). Further functional study of the remaining MTFs will help to clarify this issue.
The Action Modes of the MYC2-JA2L and MYC2-ERF.C3 Modules Show Similarity and Difference
Two lines of evidence hint that both the MYC2-JA2L module and the MYC2-ERF.C3 module show a striking ability to amplify the JA-mediated transcriptional output. First, the binding enrichment of MYC2 to the promoters of JA2L and ERF.C3 are significantly enhanced by MeJA treatment. Since the MYC2-GFP plants used for ChIP assays are driven by the constitutive 35S promoter, it is unlikely that the MeJA-induced enhancement of MYC2 binding enrichment to these promoters is due to increased transcription of MYC2. We reasoned that the MeJA-induced enhancement of MYC2 binding enrichment is more likely due to the promoter context of its target promoters. Supporting to this hypothesis, we found that a CACRYG-TTTT motif, which is required for JA-mediated activation of Arabidopsis MYC2, is enriched in promoters of tomato MTJA genes. It is also possible that JA-induced posttranslational modification of MYC2, or JA-induced recruitment of MYC2 cofactors, contributes to the enhancement of MYC2 binding enrichment to its target promoters. Second, the expression levels of MYC2, MTFs (i.e., JA2L and ERF.C3), and the downstream defense genes (i.e., TD and PR-STH2) increased ∼10-, 100-, and 1000-fold, respectively, in response to wounding treatment or pathogen infection (Figures 8C and 10A), suggesting that the MYC2-MTF modules have a strong effect on amplifying the expression levels of the downstream defensive genes. In light of the observation that these downstream defensive genes actually determine the level of plant resistance to pests and pathogens (Despres et al., 1995; Ryan and Pearce, 1998; Ryan, 2000; Chen et al., 2005), the formation of MYC2-MTF transcription modules may reflect mechanisms by which plants optimize the strength of JA-mediated transcriptional output. It is of great importance in future studies to elucidate the molecular details by which the MYC2-MTF modules to amplify the JA-mediated transcriptional output.
Equally importantly, we found two aspects of difference between the MYC2-JA2L module and the MYC2-ERF.C3 module. First, whereas JA2L is preferentially induced by wounding, ERF.C3 is preferentially induced by pathogen infection. Since our results clearly demonstrate that MYC2 plays a similar role in activating these two MTFs, we reasoned that the differential induction of JA2L and ERF.C3 by wounding or pathogen infection may be achieved by regulators other than MYC2. These yet-to-be identified regulators could cooperate with MYC2 to specifically regulate wounding- or pathogen-induced expression of their downstream MTFs. Second, whereas the wounding-induced binding enrichment of MYC2 at the JA2L promoter and the temporal expression profiles of the MYC2-JA2L-TD module is transient (Figure 8), the pathogen-induced binding enrichment of MYC2 to the ERF.C3 promoter and the temporal expression profiles of the MYC2-ERF.C3-PR-STH2 module is relatively stable (Figure 10). This divergence could be caused by the fact that the wounding attack is usually very transient whereas pathogen infection is more durable. The distinct temporal expression profiles of the wounding-responsive MYC2-JA2L-TD module and the pathogen-responsive MYC2-ERF.C3-PR-STH2 module could reflect mechanisms by which plants optimize the JA-mediated transcriptional output in response to different attacking strategies of predators. It is of great interests in future studies to elucidate the mechanisms underlying the divergence of the two transcriptional modules.
The Mode of Action of Tomato MYC2 Is Distinct from That of Its Arabidopsis Homolog
Our understanding on the action mechanism of JA mainly comes from studies in the model systems of Arabidopsis and tomato (Wasternack and Hause, 2013). Molecular genetic studies indicated that, despite the conservation in Arabidopsis and tomato of the major molecular components (i.e., COI1, JAZs, and MYC2) that constitute the core JA signaling pathway, the action modes of specific molecular components from the two species show dramatic differences. For example, whereas Arabidopsis COI1 is required for anther and pollen development, its tomato homolog is required for maternal control of seed maturation and trichome development (Li et al., 2004), indicating that the JA-Ile receptor protein COI1 regulates distinct developmental processes in different plant species.
Here, we show that, as with its Arabidopsis homolog, the tomato MYC2 also plays a key role in regulating different aspects of JA responses. Furthermore, we reveal that the action mode of tomato MYC2 in regulating JA responses is distinct from its Arabidopsis homolog. An important feature of the role of Arabidopsis MYC2 in regulating plant immunity is that this protein positively regulates wounding-responsive genes, but negatively regulates pathogen-responsive genes. Thus, abolishing the functioning of MYC2 in Arabidopsis leads to reduced resistance to chewing insects, but increased resistance to B. cinerea (Boter et al., 2004; Lorenzo et al., 2004; Nickstadt et al., 2004; Dombrecht et al., 2007; Zhai et al., 2013). Here, we show that in tomato, MYC2 positively regulates both wounding- and pathogen-responsive genes and that this protein fails to rescue the increased disease resistance phenotype of the Arabidopsis myc2-2 mutant. Therefore, tomato MYC2 and its Arabidopsis homolog have distinct modes of action in regulating pathogen-responsive genes. Since tomato MYC2 and Arabidopsis MYC2 contain a highly conserved basic region (Supplemental Figure 2), which is thought to mediate binding to the JA-responsive G-boxes of their target promoters (Toledo-Ortiz et al., 2003; Kazan and Manners, 2013), we reasoned that the functional divergence of the two MYC2 TFs might not be due to their different binding abilities, but rather to their distinct transcriptional regulatory activities. Tomato MYC2 and Arabidopsis MYC2 likely recruit different partner proteins while regulating the transcription of pathogen-responsive genes. It would be interesting to further investigate this possibility. The finding that tomato MYC2 and its Arabidopsis homolog have distinct functional modes in regulating different JA responses highlights the striking capacity of plants to alter their defense strategies to adapt to the ever-changing environment.
METHODS
Plant Materials and Growth Conditions
Tomato (Solanum lycopersicum) cv M82 was used as the wild type in this study. Tomato seeds were germinated for 48 h on moistened filter paper. Subsequently, tomato seedlings were transferred to growth chambers and maintained under cycles of 16 h light (with a white light intensity of 200 μM photons m−2 s−1) at 25°C and 8 h darkness at 18°C. Homozygous jai1 plants were identified as described previously (Li et al., 2004).
Arabidopsis thaliana plants were grown in Murashige and Skoog (MS) medium at 22°C with a 16-h-light/8-h-dark photoperiod (with a white light intensity of 120 μM photons m−2 s−1), as previously described (Zhai et al., 2013). For JA-mediated root growth inhibition, seedlings were grown on MS medium with or without 20 µM JA (Sigma-Aldrich) for 10 d before seedlings were photographed.
DNA Constructs and Plant Transformation
DNA constructs for plant transformation were generated following standard molecular biology protocols and using Gateway (Invitrogen) technology (Nakagawa et al., 2007). To generate the MYC2-RNAi construct, a fragment of the MYC2 open read frame (1–400 bp) was selected. The sense sequence was cloned and inserted into the intron-containing intermediate vector pUCCRNAi in the positive orientation, thereby generating the vector pUCCRNAi-intron-F. The antisense sequence was cloned and inserted into pUCCRNAi-intron-F in the reverse orientation, generating the vector pUCCRNAi-R-intron-F. The target inverted repeat sequences (sense orientation fragment and antisense orientation fragment separated by an intron) were obtained by PstI digestion of pUCCRNAi-R-intron-F. Finally, the sequences obtained were inserted into pCAMBIA-1301 under the control of the CaMV 35S promoter, thereby generating the construct pCAMBIA-1301-MYC2-RNAi. For MYC2-SRDX or ERF.C3-SRDX plants, a 36-bp DNA sequence encoding the SRDX repression domain (LDLDLELRLGFA) was fused in-frame to the 3′ end of the MYC2 and ERF.C3 coding regions. These fusion sequences were then cloned into the pGWB2 vector to generate the Pro35S:MYC2-SRDX and Pro35S:ERF.C3-SRDX constructs, respectively. For ERF.C3-OE plants, the full-length coding sequence of ERF.C3 was amplified by PCR and cloned into the pGWB2 vector to generate the Pro35S:ERF.C3 construct. For MYC2-GFP tomato plants, the full-length coding sequence of MYC2 was amplified by PCR and cloned into the pGWB5 vector to generate the Pro35S:MYC2-GFP construct. For ERF.C3-GFP tomato plants, the full-length coding sequence of ERF.C3 was amplified by PCR and cloned into the pGWB5 vector to generate the Pro35S:ERF.C3-GFP construct. The JA2L-GFP plants were previously generated (Du et al., 2014). The primers used to generate these DNA constructs are listed in Supplemental Table 3.
The above constructs were introduced into tomato cv M82 by Agrobacterium tumefaciens-mediated transformation (Du et al., 2014). Transformants were selected based on their resistance to hygromycin B. Homozygous T2 or T3 transgenic plants were used for phenotypic and molecular characterization.
The tomato Pro35S:MYC2-GFP construct was also introduced into the Arabidopsis myc2-2 mutants as previously described (Zhai et al., 2013). Transformants were selected based on their resistance to hygromycin B. Homozygous T2 lines Sl-MYC2-GFP/myc2-2-6# and Sl-MYC2-GFP/myc2-2-8# were selected for further analysis. The At-MYC2-GFP/myc2-2 plants (6# and 13#) were previously generated (Zhai et al., 2013).
Plant Treatment and Gene Expression Analyses
Wounding treatment of 18-d-old tomato seedlings was performed as previously described (Yan et al., 2013). For MeJA treatment, 10-d-old tomato seedlings grown on 0.5× MS agar medium were transferred to liquid 0.5× MS medium supplemented with or without 50 μM MeJA for the indicated periods. Tomato leaves (wounding) or seedlings (MeJA) were harvested at various time points for RNA extraction and RT-qPCR analysis, which were performed as previously described (Du et al., 2014). The expression levels of target genes were normalized to that of the tomato ACTIN2 gene. Error bars represent the sd of three biological replicates. For wounding treatments, each biological replicate (sample) consisted of the pooled leaves of three soil-grown plants from one tray; for MeJA treatments, each biological replicate (sample) consisted of 10 pooled seedlings grown on 0.5× MS medium. Biological replicates (trays) were grown at different locations in growth chambers and treated separately. The primers used to quantify gene expression levels are listed in Supplemental Table 3.
For MeJA-induced defense gene expression of Arabidopsis plants, 10-d-old seedlings grown on plates were incubated in a 50 μM MeJA solution under continuous light. Seedlings were then harvested at 6 h to measure VSP1 expression and at 48 h to measure PDF1.2 expression.
Botrytis cinerea Inoculation Assays
B. cinerea isolate B05.10 was grown on 2xV8 agar (36% V8 juice, 0.2% CaCO3, and 2% Bacto-agar) for 14 d at 20°C under a 12-h photoperiod prior to spore collection. Spore suspensions were prepared by harvesting the spores in 1% Sabouraud Maltose Broth (SMB), filtering them through nylon mesh to remove hyphae, and adjusting the concentration to 106 spores/mL (Mengiste et al., 2003).
B. cinerea inoculation of tomato plants was performed as previously described (El Oirdi et al., 2011; Yan et al., 2013; Zhai et al., 2013), with minor modifications. For the pathogenicity test, detached leaves from 5-week-old tomato plants were placed in Petri dishes containing 0.8% agar medium (agar dissolved in sterile water), with the petiole embedded in the medium. Each leaflet was spotted with a single 5-μL droplet of B. cinerea spore suspension at a concentration of 106 spores/mL. The trays were covered with lids and kept under the same conditions used for plant growth. Photographs were taken after 3 d, and the mean lesion sizes of six leaves from various genotypes were recorded. Data from three independent experiments were collected, and error bars represent the sd from three independent experiments.
For RT-qPCR and ChIP-qPCR experiments, inoculations were performed in planta: Leaves of 5-week-old plants were spotted with a 10 μL B. cinerea spore suspension (106 spores/mL). The plants were then incubated in a growth chamber with high humidity. A similar experiment was performed using SMB-spotted plants as a control. Spotted leaves were harvested at various time points for RT-qPCR and ChIP-qPCR experiments. Error bars represent the sd of three biological replicates. Each biological replicate (sample) consisted of the pooled leaves of three spotted plants from one tray (different genotypes were grown together in a randomized design per tray). Biological replicates (trays) were grown at different locations in growth chambers and treated separately.
Yeast Two-Hybrid Assays
The full-length coding sequences of tomato JAZ genes were cloned into the pGBKT7 vector, and the full-length coding sequences of MYC2, JA2L, and ERF.C3 were cloned into pGADT7. Yeast two-hybrid assays were performed using the Matchmaker GAL4 Two-Hybrid System (Clontech). Constructs used to test interactions were cotransformed into yeast (Saccharomyces cerevisiae) strain AH109. The empty pGBKT7 vector was cotransformed in parallel as a negative control. To assess protein interactions, the transformed yeast cells were grown on SD/-4 (-Ade/-His/-Leu/-Trp) plates. The interactions were observed after 3 d of incubation at 30°C.
Pull-Down Experiments
To produce MBP-JAZ or GST-JAZ proteins, the full-length coding sequences of JAZs were amplified by PCR and cloned into the pMAL-c2X or the pGEX-4T-1 vector. The resulting constructs were transformed into Escherichia coli BL21 (DE3) and the recombinant proteins were abundantly expressed by adding 0.5 mM isopropyl-β-d-thiogalactoside. MBP-JAZ and GST-JAZ proteins were then purified using the amylose resin (NEB) and GST bind resin (Novagen), respectively.
Ten-day-old MYC2-Myc tomato seedlings were ground in liquid nitrogen and homogenized in extraction buffer containing 50 mM Tris-HCl, pH 7.4, 80 mM NaCl, 10% glycerol, 0.1% Tween 20, 1 mM DTT, 1 mM PMSF, 50 mM MG132 (Sigma-Aldrich), and protease inhibitor cocktail (Roche). After centrifugation (16,000g at 4°C), the supernatant was collected.
For in vivo pull-down experiments, 1 mg of total protein extract was incubated with resin-bound MBP-JAZ or GST-JAZ fusion proteins for 2 h at 4°C with rotation. After washing, samples were denaturalized in the SDS-PAGE loading buffer and detected by immunoblot analysis using an anti-c-Myc antibody (Roche 11667149001, lot 10653800). A 5-μL aliquot of MBP-JAZ or GST-JAZ fusion protein of each sample was run into SDS-PAGE gels and stained with Coomassie Brilliant Blue to serve as a loading control.
Construction of RNA-Seq Libraries
Ten-day-old wild-type and MYC2-RNAi-3 seedlings were treated with either mock (control; liquid 0.5× MS medium without MeJA) or MeJA (liquid 0.5× MS medium supplemented with 50 μM MeJA). At 1 h after treatment, seedlings were collected from three biological replicates (20 seedlings each) before total RNA extraction using a TRIzol kit (Invitrogen) according to the user manual. Total RNA was treated with DNase and purified with the RNA Clean and Concentrator-25 Kit (Zymo Research). The quality of the total RNA was assessed using a NanoDrop spectrophotometer and an Agilent 2100 Bioanalyzer. For each sample, 3 μg of total RNA was used to construct the Illumina sequencing libraries according to the manufacturer’s instructions. The libraries were sequenced using the Illumina HiSeq 2500 platform (Biomarker Technologies).
Analysis of RNA-Seq Data
About 4 Gb of high-quality 125-bp paired-end reads were generated from each library. RNA-seq reads were aligned to adaptor, rRNA, and tRNA sequences using Bowtie2 (-p 24 -x -U) (Langmead and Salzberg, 2012), and the remaining filtered reads were aligned to the tomato genome SL2.50 (https://solgenomics.net) using TopHat2 with default parameters (–read-realign-edit-dist 0–max-intron-length 10,000 -p 15–GTF–transcriptome-index–mate-inner-dist 50–mate-std-dev 70–max-segment-intron 10000–prefilter-multihits) (Kim et al., 2013). Finally, eXpress (Roberts and Pachter, 2013) was used to calculate gene expression levels in all biological replicates (–no-bias-correct). A two-way ANOVA analysis was performed to determine whether either factor (genotype or treatment) had a significant effect on the expression level of a certain gene (FDR-adjusted P value < 0.05). Differentially expressed genes between samples were identified using the DESeq2 package (Love et al., 2014) with standard parameters (FDR-adjusted P value < 0.05). GO enrichment analysis was performed using the PANTHER (for protein annotation through evolutionary relationship) classification system (www.pantherdb.org) with default parameters. A statistical overrepresentation test (P < 0.05, Bonferroni correction) was used to determine whether a particular class (e.g., a GO biological process or protein class) of genes was overrepresented or underrepresented in the test gene list (Mi et al., 2013). GO term enrichment is shown by the most specific subclass in the enrichment analysis. Since TFs are not all annotated in the PANTHER protein class database, the enrichment of TFs for the protein class was manually calculated using Fisher’s exact test in R3.0.
To evaluate, on a genome-wide scale, the expression timing of the 2558 genes coregulated by JA and MYC2, we performed three independent time-course RNA-seq experiments. For each experiment, 18-d-old wild-type seedlings were either not treated (control; 0 h) or treated by mechanical wounding. Seedlings were then harvested at 1, 6, or 12 h after wounding for RNA extraction and RNA-seq library construction as described above. The RNA-seq raw reads were mapped by HISAT2 (Kim et al., 2015), read counts were calculated by prepDE.py included in Stringtie2 program (Pertea et al., 2015), and differential gene expression analysis was conducted by DESeq2. Genes with more than 1.5-fold change in read count (FDR-adjusted P value < 0.05) between any time point after treatment (1, 6, or 12 h) and untreated control (0 h) were selected for further investigation. Genes with maximum fold change of read count at 1 h were defined as early genes, whereas those with maximum fold change of read count at 12 h were defined as the late genes.
ChIP Assays
ChIP assays were performed according to a published protocol (Zhu et al., 2012). Briefly, 5 g of seedling tissue was harvested and cross-linked with 1% (v/v) formaldehyde under a vacuum for 10 min and ground to a powder in liquid nitrogen. Subsequently, the chromatin complexes were isolated, sonicated, and immunoprecipitated with polyclonal anti-GFP antibodies (Abcam AB290, lot GR240324-1). The ChIP DNA and input DNA were recovered and dissolved in water for ChIP-seq or ChIP-qPCR analysis. For ChIP-seq, 10-d-old MYC2-GFP-9 seedlings treated with MeJA (liquid 0.5× MS medium supplemented with 50 μM MeJA) for 1 h were harvested for ChIP, and the recovered ChIP DNA and input DNA were used to construct Illumina sequencing libraries. For ChIP-qPCR, ChIP DNA was analyzed by qPCR using the respective primer pairs (Supplemental Table 3). Each ChIP value was normalized to its respective input DNA value. The fold enrichment on each promoter was calculated against the ACTIN2 promoter. Each PCR was repeated at least twice, and the mean value of technical replicates was recorded for each biological replicate. Data from three independent experiments were collected, and error bars represent the sd from three independent experiments.
For MeJA-induced enrichment of MYC2, 10-d-old MYC2-GFP-9 seedlings were treated with MeJA (liquid 0.5× MS medium supplemented with 50 μM MeJA) for indicated times, and 5 g of leaf tissue from each sample was harvested for the ChIP assay. For wounding-induced enrichment of MYC2, 18-d-old MYC2-GFP-9 seedlings were wounded with a hemostat. Wounded seedlings were incubated under continuous light for indicated times, and 5 g of leaf tissue from each sample was harvested for the ChIP assay. For B. cinerea-induced enrichment of MYC2, leaves of 5-week-old MYC2-GFP-9 plants were inoculated with B. cinerea for the indicated times, and 5 g of leaf tissue from each sample was harvested for the ChIP assay.
Construction of ChIP-Seq Libraries
Illumina sequencing libraries were constructed using the ChIP DNA and input DNA samples described above according to the manufacturer’s instructions, with minor modifications. The ends of the DNA fragments were repaired and ligated to an adaptor. DNA fragments of 270 to 330 bp in length were recovered from the gel and amplified by PCR for 18 cycles. The amplified DNA products were collected, ligated to the pEASY-Blunt vector for a quality test, and sequenced using the Illumina HiSeq 2500 platform.
Analysis of ChIP-Seq Data
The sequencing reads from two replicates were mapped to the tomato genome using Bowtie2 (version 2.2.9; parameters: -3 75 -p 20–met-stderr -t -phred33) (Langmead and Salzberg, 2012). The sequence alignment/map (SAM) file generated by Bowtie2 was transformed to a binary alignment/map (BAM) format file by SAMtools 0.1.19 (Li et al., 2009). To estimate the number of uniquely mapped reads, the SAMtools mapping score was used as a filter (SAMtools view -F 4 -Sb -q 10). Only uniquely mapped reads were used for peak identification. MYC2 binding peaks were obtained by model-based analysis of ChIP-seq with the genome size parameter (-g 7.6e+8 -B–keep-dup 1 -shift 73–SPMR) (Zhang et al., 2008). The q-value cutoff applied to calculate statistical significance was <0.05. Other parameters were set to default values. The WindowBed subcommand, contained in BEDTools with default parameters (-w 3000), was used to identify the peaks within the genic regions (including 3 kb up- and downstream of the gene).
Genes containing one or more MYC2 binding peaks within the 3-kb promoter region were considered to be MYC2-targeted genes. The MYC2 binding motifs were determined using MEME-ChIP with the default parameter (–meme-minw 8–meme-maxw 12) (MEME suite 4.11.2) by extracting 500-bp sequences around the peak summits (Machanick and Bailey, 2011). The graph of motif probability of the CACRYG and GGNCCM motifs was visualized using the density plot tool in R 3.0.
Identification of CACRYG-TTTT Motif in Promoters of JA-Responsive Genes
Based on the previous report (Figueroa and Browse, 2012), a thymidine (T)-rich sequence immediately 3′ to G-box (i.e., CACGTG-TTTT) was essential for expression of early JA-induced genes. The thymidine (T)-rich sequence sequences were considered as [A/T]TTT, T[A/T/G/C]TT, TT[A/T/G/C]T, or TTT[A/T/G/C] based on the report. We searched for the CACRYG motif and an extended CACRYG motif (CACRYG-TTTT) in the promoters (3 kb upstream of the TSS) of different gene sets, including the 655 MTJA genes and the 2558 JA- and MYC2-coregulated genes, using the script scanMotifGenomeWide.pl from homer2 package (Heinz et al., 2010). The Fisher’s exact test for enrichment analysis of CACRYG-TTTT motif-containing genes was conducted by fisher.test package in R3.0.
EMSAs
The full-length coding sequences of MYC2, JA2L, and ERF.C3 were amplified by PCR using the primers MBP-MYC2-F/R, MBP-JA2L-F/R, and MBP-ERF.C3-F/R and cloned into the pMAL-c2X vector. The recombinant MBP-fusion proteins were expressed in E. coli BL21 and purified to homogeneity using an amylase resin column. Oligonucleotide probes were synthesized and labeled with biotin at their 5′ ends (Invitrogen). EMSAs were performed as previously described (Chen et al., 2011; Du et al., 2014). Briefly, biotin-labeled probes were incubated with MBP-fusion proteins at room temperature for 20 min, and free and bound DNA was separated in an acrylamide gel. Unlabeled wild-type and mutated probes, in which the specific TF binding motif was deleted, were used as competitors. Probes used for EMSA are listed in Supplemental Table 3.
Accession Numbers
Sequence data from this article can be found in the Sol Genomics Network data libraries under the following accession numbers: MYC2, Solyc08g076930; JA2L, Solyc07g063410; TD, Solyc09g008670; ERF.C3, Solyc09g066360; PR-STH2, Solyc05g054380; JAZ1, Solyc07g042170; JAZ2, Solyc12g009220; JAZ3, Solyc03g122190; JAZ4, Solyc12g049400; JAZ5, Solyc03g118540; JAZ6, Solyc01g005440; JAZ7, Solyc11g011030; JAZ8, Solyc06g068930; JAZ9, Solyc08g036640; JAZ10, Solyc08g036620; JAZ11, Solyc08g036660; ACTIN2, Solyc11g005330. The raw sequence data reported in this article have been deposited in the Genome Sequence Archive (Wang et al., 2017) in the BIG Data Center (BIG Data Center Members, 2017), Beijing Institute of Genomics (BIG), Chinese Academy of Sciences, under accession numbers PRJCA000395, PRJCA000396, and PRJCA000440 that are publicly accessible at http://bigd.big.ac.cn/gsa.
Supplemental Data
Supplemental Figure 1. B. cinerea- and Wounding-Induced Expression Patterns of TD and PR-STH2.
Supplemental Figure 2. Sequence Alignment of MYC2 Proteins from Tomato and Arabidopsis.
Supplemental Figure 3. Association of MYC2 with JAZ Proteins Revealed by Pull-Down Experiments.
Supplemental Figure 4. MYC2-SRDX Plants Are More Susceptible to B. cinerea Infection.
Supplemental Figure 5. Sl-MYC2 Partially Rescues the JA Response Defects of the Arabidopsis myc2-2 Mutant.
Supplemental Figure 6. Overview of RNA-Seq Data from MeJA-Treated Wild-Type and MYC2-RNAi Plants.
Supplemental Figure 7. GO Analysis of the Genes in the Interaction Term of ANOVA Analysis of the RNA-Seq Data.
Supplemental Figure 8. GO Analysis of the JA- and MYC2-Coregulated Genes Identified from Pairwise Comparisons of the RNA-Seq Data.
Supplemental Figure 9. Characterization of MYC2-GFP Transgenic Plants.
Supplemental Figure 10. Overview of ChIP-Seq Data from Two Biological Replicates.
Supplemental Figure 11. The Irreproducible Discovery Rate Framework for Assessing the Reproducibility of ChIP-Seq Data Sets.
Supplemental Figure 12. Saturation Analysis of the ChIP-Seq Reads from the Input Libraries.
Supplemental Figure 13. MYC2 Binds to the CACATG Motif but Not the GGACCA/C Motif.
Supplemental Figure 14. Overview of the Time-Course RNA-Seq Data.
Supplemental Figure 15. MYC2 Binding Profile in the Promoters of JA2L and ERF.C3.
Supplemental Figure 16. Yeast Two-Hybrid Assay to Detect Interactions of JAZs with JA2L, ERF.C3, and MYC2.
Supplemental Figure 17. B. cinerea-induced Expression of TD and PR-STH2 in Wild-Type, JA2L-AS, ERF.C3-SRDX, and ERF.C3-OE Plants.
Supplemental Figure 18. Characterization of ERF.C3-SRDX and ERF.C3-OE Plants.
Supplemental Figure 19. Characterization of ERF.C3-GFP Transgenic Plants.
Supplemental Figure 20. Frequencies of GCCGCC- and CACG-Containing Genes.
Supplemental Table 1. Well-Known JA-Responsive Genes Targeted by MYC2.
Supplemental Table 2. MYC2-Targeted TFs Identified by ChIP-Seq and RNA-Seq Analysis.
Supplemental Table 3. DNA Primers Used in This Study.
Supplemental Data Set 1. ANOVA Analysis of the RNA-Seq Data of MeJA-Treated Wild-Type and MYC2-RNAi Plants.
Supplemental Data Set 2. Differentially Expressed Genes Identified by Pairwise Comparison Analysis of the RNA-Seq Data.
Supplemental Data Set 3. MYC2 binding Peaks and MYC2-Targeted Genes Identified from ChIP-Seq Analysis.
Supplemental Data Set 4. MYC2-Targeted JA-Responsive Genes.
Supplemental Data Set 5. Early and Late Genes Coregulated by JA and MYC2.
Acknowledgments
We thank Chang Li for the improvement of the English language. This work was supported by the Strategic Priority Research Program of the Chinese Academy of Sciences (Grant XDB11030200), by the National Key Research and Development Program of China (2016YFD0100603-10), by the Ministry of Agriculture of the People’s Republic of China (Chinese Ministry of Agriculture) (2016ZX08009-003-001), by the National Natural Science Foundation of China (Grants 31030006, 31672157, and 31390423), by the Tai-Shan Scholar Program from the Shandong Province, and by Hong Kong UGC (14104515/14119814).
AUTHOR CONTRIBUTIONS
M.D., J.Z., C.-B.L., and C.Y.L. designed the research strategy. C.Y.L. conceived and supervised the project. M.D., J.Z., and Y.L. performed most of the experiments. D.T.W.T., J.Z., Q.C., and S.Z. analyzed the sequencing data. L.D., T.Y., Q.Z., F.W., and Z.H. generated the constructs and transgenic plants. Q.W. and C.-B.L. helped grow the plants. M.D., J.Z., C.-B.L., and C.Y.L. analyzed the data. C.-B.L. and Q.W. contributed reagents/materials/analysis tools. M.D., J.Z., and C.Y.L. wrote the article.
Glossary
- JA
jasmonic acid
- ChIP-seq
chromatin immunoprecipitation followed by sequencing
- MTJA
MYC2-targeted JA-responsive
- MTF
MYC2-targeted TF
- JA-Ile
jasmonoyl-isoleucine
- TF
transcription factor
- HAI
hours after inoculation
- MeJA
methyl jasmonate
- FDR
false discovery rate
- GO
Gene Ontology
- TSS
transcription start site
- EMSA
electrophoresis mobility shift assay
- DAI
days after infection
- MS
Murashige and Skoog
Footnotes
Articles can be viewed without a subscription.
References
- Abuqamar S., Chai M.F., Luo H., Song F., Mengiste T. (2008). Tomato protein kinase 1b mediates signaling of plant responses to necrotrophic fungi and insect herbivory. Plant Cell 20: 1964–1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abuqamar S., Luo H., Laluk K., Mickelbart M.V., Mengiste T. (2009). Crosstalk between biotic and abiotic stress responses in tomato is mediated by the AIM1 transcription factor. Plant J. 58: 347–360. [DOI] [PubMed] [Google Scholar]
- BIG Data Center Members (2017). The BIG Data Center: from deposition to integration to translation. Nucleic Acids Res. 45: D18–D24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boter M., Ruíz-Rivero O., Abdeen A., Prat S. (2004). Conserved MYC transcription factors play a key role in jasmonate signaling both in tomato and Arabidopsis. Genes Dev. 18: 1577–1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos M.L., Kang J.H., Howe G.A. (2014). Jasmonate-triggered plant immunity. J. Chem. Ecol. 40: 657–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chassot C., Buchala A., Schoonbeek H.J., Métraux J.P., Lamotte O. (2008). Wounding of Arabidopsis leaves causes a powerful but transient protection against Botrytis infection. Plant J. 55: 555–567. [DOI] [PubMed] [Google Scholar]
- Chen H., Wilkerson C.G., Kuchar J.A., Phinney B.S., Howe G.A. (2005). Jasmonate-inducible plant enzymes degrade essential amino acids in the herbivore midgut. Proc. Natl. Acad. Sci. USA 102: 19237–19242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q., et al. (2011). The basic helix-loop-helix transcription factor MYC2 directly represses PLETHORA expression during jasmonate-mediated modulation of the root stem cell niche in Arabidopsis. Plant Cell 23: 3335–3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chini A., Fonseca S., Fernández G., Adie B., Chico J.M., Lorenzo O., García-Casado G., López-Vidriero I., Lozano F.M., Ponce M.R., Micol J.L., Solano R. (2007). The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448: 666–671. [DOI] [PubMed] [Google Scholar]
- Dangl J.L., Horvath D.M., Staskawicz B.J. (2013). Pivoting the plant immune system from dissection to deployment. Science 341: 746–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean R., Van Kan J.A., Pretorius Z.A., Hammond-Kosack K.E., Di Pietro A., Spanu P.D., Rudd J.J., Dickman M., Kahmann R., Ellis J., Foster G.D. (2012). The Top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 13: 414–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Despres C., Subramaniam R., Matton D.P., Brisson N. (1995). The activation of the potato PR-10a gene requires the phosphorylation of the nuclear factor PBF-1. Plant Cell 7: 589–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devoto A., Nieto-Rostro M., Xie D., Ellis C., Harmston R., Patrick E., Davis J., Sherratt L., Coleman M., Turner J.G. (2002). COI1 links jasmonate signalling and fertility to the SCF ubiquitin-ligase complex in Arabidopsis. Plant J. 32: 457–466. [DOI] [PubMed] [Google Scholar]
- Dombrecht B., Xue G.P., Sprague S.J., Kirkegaard J.A., Ross J.J., Reid J.B., Fitt G.P., Sewelam N., Schenk P.M., Manners J.M., Kazan K. (2007). MYC2 differentially modulates diverse jasmonate-dependent functions in Arabidopsis. Plant Cell 19: 2225–2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du M., et al. (2014). Closely related NAC transcription factors of tomato differentially regulate stomatal closure and reopening during pathogen attack. Plant Cell 26: 3167–3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Oirdi M., Bouarab K. (2007). Plant signalling components EDS1 and SGT1 enhance disease caused by the necrotrophic pathogen Botrytis cinerea. New Phytol. 175: 131–139. [DOI] [PubMed] [Google Scholar]
- El Oirdi M., El Rahman T.A., Rigano L., El Hadrami A., Rodriguez M.C., Daayf F., Vojnov A., Bouarab K. (2011). Botrytis cinerea manipulates the antagonistic effects between immune pathways to promote disease development in tomato. Plant Cell 23: 2405–2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elad Y., Vivier M., Fillinger S. (2016). Botrytis, the good, the bad and the ugly. In Botrytis: The Fungus, the Pathogen and Its Management in Agricultural Systems, Fillinger S., Elad Y., eds (Cham: Springer International Publishing; ), pp. 1–15. [Google Scholar]
- Figueroa P., Browse J. (2012). The Arabidopsis JAZ2 promoter contains a G-Box and thymidine-rich module that are necessary and sufficient for jasmonate-dependent activation by MYC transcription factors and repression by JAZ proteins. Plant Cell Physiol. 53: 330–343. [DOI] [PubMed] [Google Scholar]
- Glazebrook J. (2005). Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 43: 205–227. [DOI] [PubMed] [Google Scholar]
- Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., Glass C.K. (2010). Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38: 576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratsu K., Matsui K., Koyama T., Ohme-Takagi M. (2003). Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. Plant J. 34: 733–739. [DOI] [PubMed] [Google Scholar]
- Howe G.A., Jander G. (2008). Plant immunity to insect herbivores. Annu. Rev. Plant Biol. 59: 41–66. [DOI] [PubMed] [Google Scholar]
- Kazan K., Manners J.M. (2013). MYC2: the master in action. Mol. Plant 6: 686–703. [DOI] [PubMed] [Google Scholar]
- Kim D., Langmead B., Salzberg S.L. (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12: 357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S.L. (2013). TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landt S.G., et al. (2012). ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res. 22: 1813–1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Salzberg S.L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9: 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee G.I., Howe G.A. (2003). The tomato mutant spr1 is defective in systemin perception and the production of a systemic wound signal for defense gene expression. Plant J. 33: 567–576. [DOI] [PubMed] [Google Scholar]
- Li C., Liu G., Xu C., Lee G.I., Bauer P., Ling H.Q., Ganal M.W., Howe G.A. (2003). The tomato suppressor of prosystemin-mediated responses2 gene encodes a fatty acid desaturase required for the biosynthesis of jasmonic acid and the production of a systemic wound signal for defense gene expression. Plant Cell 15: 1646–1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R.; 1000 Genome Project Data Processing Subgroup (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Li C., Lee G.I., Howe G.A. (2002). Distinct roles for jasmonate synthesis and action in the systemic wound response of tomato. Proc. Natl. Acad. Sci. USA 99: 6416–6421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Zhao Y., McCaig B.C., Wingerd B.A., Wang J., Whalon M.E., Pichersky E., Howe G.A. (2004). The tomato homolog of CORONATINE-INSENSITIVE1 is required for the maternal control of seed maturation, jasmonate-signaled defense responses, and glandular trichome development. Plant Cell 16: 126–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo O., Chico J.M., Sánchez-Serrano J.J., Solano R. (2004). JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell 16: 1938–1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machanick P., Bailey T.L. (2011). MEME-ChIP: motif analysis of large DNA datasets. Bioinformatics 27: 1696–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marineau C., Matton D.P., Brisson N. (1987). Differential accumulation of potato tuber mRNAs during the hypersensitive response induced by arachidonic acid elicitor. Plant Mol. Biol. 9: 335–342. [DOI] [PubMed] [Google Scholar]
- Matton D.P., Brisson N. (1989). Cloning, expression, and sequence conservation of pathogenesis-related gene transcripts of potato. Mol. Plant Microbe Interact. 2: 325–331. [DOI] [PubMed] [Google Scholar]
- Mengiste T. (2012). Plant immunity to necrotrophs. Annu. Rev. Phytopathol. 50: 267–294. [DOI] [PubMed] [Google Scholar]
- Mengiste T., Chen X., Salmeron J., Dietrich R. (2003). The BOTRYTIS SUSCEPTIBLE1 gene encodes an R2R3MYB transcription factor protein that is required for biotic and abiotic stress responses in Arabidopsis. Plant Cell 15: 2551–2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H., Muruganujan A., Casagrande J.T., Thomas P.D. (2013). Large-scale gene function analysis with the PANTHER classification system. Nat. Protoc. 8: 1551–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T., Kurose T., Hino T., Tanaka K., Kawamukai M., Niwa Y., Toyooka K., Matsuoka K., Jinbo T., Kimura T. (2007). Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J. Biosci. Bioeng. 104: 34–41. [DOI] [PubMed] [Google Scholar]
- Nickstadt A., Thomma B.P., Feussner I., Kangasjärvi J., Zeier J., Loeffler C., Scheel D., Berger S. (2004). The jasmonate-insensitive mutant jin1 shows increased resistance to biotrophic as well as necrotrophic pathogens. Mol. Plant Pathol. 5: 425–434. [DOI] [PubMed] [Google Scholar]
- Pauwels L., et al. (2010). NINJA connects the co-repressor TOPLESS to jasmonate signalling. Nature 464: 788–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea M., Pertea G.M., Antonescu C.M., Chang T.C., Mendell J.T., Salzberg S.L. (2015). StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 33: 290–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieterse C.M., Leon-Reyes A., Van der Ent S., Van Wees S.C. (2009). Networking by small-molecule hormones in plant immunity. Nat. Chem. Biol. 5: 308–316. [DOI] [PubMed] [Google Scholar]
- Pirrello J., Prasad B.C., Zhang W., Chen K., Mila I., Zouine M., Latché A., Pech J.C., Ohme-Takagi M., Regad F., Bouzayen M. (2012). Functional analysis and binding affinity of tomato ethylene response factors provide insight on the molecular bases of plant differential responses to ethylene. BMC Plant Biol. 12: 190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts A., Pachter L. (2013). Streaming fragment assignment for real-time analysis of sequencing experiments. Nat. Methods 10: 71–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosli H.G., Martin G.B. (2015). Functional genomics of tomato for the study of plant immunity. Brief. Funct. Genomics 14: 291–301. [DOI] [PubMed] [Google Scholar]
- Ryan C.A. (2000). The systemin signaling pathway: differential activation of plant defensive genes. Biochim. Biophys. Acta 1477: 112–121. [DOI] [PubMed] [Google Scholar]
- Ryan C.A., Pearce G. (1998). Systemin: a polypeptide signal for plant defensive genes. Annu. Rev. Cell Dev. Biol. 14: 1–17. [DOI] [PubMed] [Google Scholar]
- Santner A., Estelle M. (2009). Recent advances and emerging trends in plant hormone signalling. Nature 459: 1071–1078. [DOI] [PubMed] [Google Scholar]
- Schilmiller A.L., Howe G.A. (2005). Systemic signaling in the wound response. Curr. Opin. Plant Biol. 8: 369–377. [DOI] [PubMed] [Google Scholar]
- Sheard L.B., et al. (2010). Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature 468: 400–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L., Huang S.C., Wise A., Castanon R., Nery J.R., Chen H., Watanabe M., Thomas J., Bar-Joseph Z., Ecker J.R. (2016). A transcription factor hierarchy defines an environmental stress response network. Science 354: 598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spoel S.H., Dong X. (2008). Making sense of hormone crosstalk during plant immune responses. Cell Host Microbe 3: 348–351. [DOI] [PubMed] [Google Scholar]
- Sun J.Q., Jiang H.L., Li C.Y. (2011). Systemin/Jasmonate-mediated systemic defense signaling in tomato. Mol. Plant 4: 607–615. [DOI] [PubMed] [Google Scholar]
- Tomato Genome Consortium (2012). The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485: 635–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thines B., Katsir L., Melotto M., Niu Y., Mandaokar A., Liu G., Nomura K., He S.Y., Howe G.A., Browse J. (2007). JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 448: 661–665. [DOI] [PubMed] [Google Scholar]
- Toledo-Ortiz G., Huq E., Quail P.H. (2003). The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell 15: 1749–1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran L.S., Nakashima K., Sakuma Y., Simpson S.D., Fujita Y., Maruyama K., Fujita M., Seki M., Shinozaki K., Yamaguchi-Shinozaki K. (2004). Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell 16: 2481–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Kan J.A., et al. (2017). A gapless genome sequence of the fungus Botrytis cinerea. Mol. Plant Pathol. 18: 75–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., et al. (2017). GSA: Genome Sequence Archive. Genomics Proteomics Bioinformatics 15: 14–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasternack C., Hause B. (2013). Jasmonates: biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann. Bot. (Lond.) 111: 1021–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie D.X., Feys B.F., James S., Nieto-Rostro M., Turner J.G. (1998). COI1: an Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280: 1091–1094. [DOI] [PubMed] [Google Scholar]
- Xu L., Liu F., Lechner E., Genschik P., Crosby W.L., Ma H., Peng W., Huang D., Xie D. (2002). The SCF(COI1) ubiquitin-ligase complexes are required for jasmonate response in Arabidopsis. Plant Cell 14: 1919–1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J., Zhang C., Gu M., Bai Z., Zhang W., Qi T., Cheng Z., Peng W., Luo H., Nan F., Wang Z., Xie D. (2009). The Arabidopsis CORONATINE INSENSITIVE1 protein is a jasmonate receptor. Plant Cell 21: 2220–2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L., Zhai Q., Wei J., Li S., Wang B., Huang T., Du M., Sun J., Kang L., Li C.B., Li C. (2013). Role of tomato lipoxygenase D in wound-induced jasmonate biosynthesis and plant immunity to insect herbivores. PLoS Genet. 9: e1003964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y., Stolz S., Chételat A., Reymond P., Pagni M., Dubugnon L., Farmer E.E. (2007). A downstream mediator in the growth repression limb of the jasmonate pathway. Plant Cell 19: 2470–2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai Q., Yan L., Tan D., Chen R., Sun J., Gao L., Dong M.Q., Wang Y., Li C. (2013). Phosphorylation-coupled proteolysis of the transcription factor MYC2 is important for jasmonate-signaled plant immunity. PLoS Genet. 9: e1003422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W., Liu X.S. (2008). Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9: R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J.Y., Sun Y., Wang Z.Y. (2012). Genome-wide identification of transcription factor-binding sites in plants using chromatin immunoprecipitation followed by microarray (ChIP-chip) or sequencing (ChIP-seq). Methods Mol. Biol. 876: 173–188. [DOI] [PubMed] [Google Scholar]