As a proton-coupled H+/K+ antiporter, NRT1.5/NPF7.3 mediates K+ release from root parenchyma cells into the xylem and coordinates K+/NO3− distribution in Arabidopsis.
Abstract
Potassium and nitrogen are essential macronutrients for plant growth and have a positive impact on crop yield. Previous studies have indicated that the absorption and translocation of K+ and NO3− are correlated with each other in plants; however, the molecular mechanism that coordinates K+ and NO3− transport remains unknown. In this study, using a forward genetic approach, we isolated a low-K+-sensitive Arabidopsis thaliana mutant, lks2, that showed a leaf chlorosis phenotype under low-K+ conditions. LKS2 encodes the transporter NRT1.5/NPF7.3, a member of the NRT1/PTR (Nitrate Transporter 1/Peptide Transporter) family. The lks2/nrt1.5 mutants exhibit a remarkable defect in both K+ and NO3− translocation from root to shoot, especially under low-K+ conditions. This study demonstrates that LKS2 (NRT1.5) functions as a proton-coupled H+/K+ antiporter. Proton gradient can promote NRT1.5-mediated K+ release out of root parenchyma cells and facilitate K+ loading into the xylem. This study reveals that NRT1.5 plays a crucial role in K+ translocation from root to shoot and is also involved in the coordination of K+/NO3− distribution in plants.
INTRODUCTION
Potassium (K+) is an essential macronutrient for plant growth and development. K+ also contributes to the regulation of crop yield and quality in agricultural production (Clarkson and Hanson, 1980; Zörb et al., 2014). As the most abundant cation in plants, K+ in plants is an important source of K+ intake for animals. Consuming K+-rich plants is also beneficial for human health (Whelton and He, 2014). K+ absorption by plant roots and K+ transport inside plants are mediated through K+ channels and transporters (Wang and Wu, 2013; Véry et al., 2014). To date, many K+ channels and transporters involved in K+ absorption have been identified in plant root cells (Véry et al., 2014). After being absorbed into root hairs and epidermis, K+ is transported into root stelar tissues and then translocated from roots toward shoots via xylem vessels so that K+ can be used in plant shoots. So far, only the K+ channel SKOR has been implicated in root-to-shoot K+ translocation in Arabidopsis thaliana (Gaymard et al., 1998). Encoding an outwardly rectifying channel, SKOR is expressed in stelar tissues and is responsible for the K+ release from root parenchyma cells into the xylem (Gaymard et al., 1998). However, skor null mutants do not exhibit a particular K+-deficient phenotype, which suggests that other unidentified components may also have important functions in this process.
K+ is also necessary for the electroneutrality of anions during ion absorption and transport in plants. Nitrate (NO3−) is one of the most abundant anions in plants, whose absorption and transport are believed to be accompanied by K+ (White, 2012a). In plants, NO3− absorption and transport are mainly conducted by the nitrate transporters from two families NRT1/PTR (Nitrate Transporter 1/Peptide Transporter) and NRT2. Both families belong to the major facilitator superfamily that represents the largest secondary active transporters in all species (Pao et al., 1998; Law et al., 2008; Yan, 2015). The NRT1/PTR family is also known as the POT (proton-dependent oligopeptide transporter) family, which is present in all living organisms and prevalent in higher plants, referred to as the NPF (NRT1/PTR family) (Steiner et al., 1995; Daniel et al., 2006; Léran et al., 2014). In Arabidopsis, the NRT1/PTR family contains as many as 53 members, which implies that this family has diverse functions (Tsay et al., 2007). Some members of this family have been shown to participate in the transport of diverse substrates in plants, including nitrate, peptides, glucosinolate defense compounds, and plant hormones (Krouk et al., 2010; Nour-Eldin et al., 2012; Léran et al., 2014; Saito et al., 2015; Tal et al., 2016). Members of the NRT1/PTR family are thought to transport numerous unidentified substrates and have multiple unknown functions (Tsay et al., 2007; Léran et al., 2014).
In plants, it has been noticed for a long time that K+/NO3− absorption and transport are somehow coordinated (Blevins et al., 1978; Triplett et al., 1980; White, 2012a). Here, we report that NRT1.5/NPF7.3 from the NRT1/PTR family functions as a proton-coupled H+/K+ antiporter. NRT1.5 directly mediates K+ release from root parenchyma cells into the xylem. At the same time, NRT1.5 regulates NO3− loading into the xylem. This study reveals that K+ is a newly identified substrate of NRT1/PTR transporters. NRT1.5 plays a crucial role in K+ translocation from root to shoot and is also involved in the coordination of K+/NO3− distribution in plants.
RESULTS
Isolation of the Low-K+-Sensitive Mutant lks2
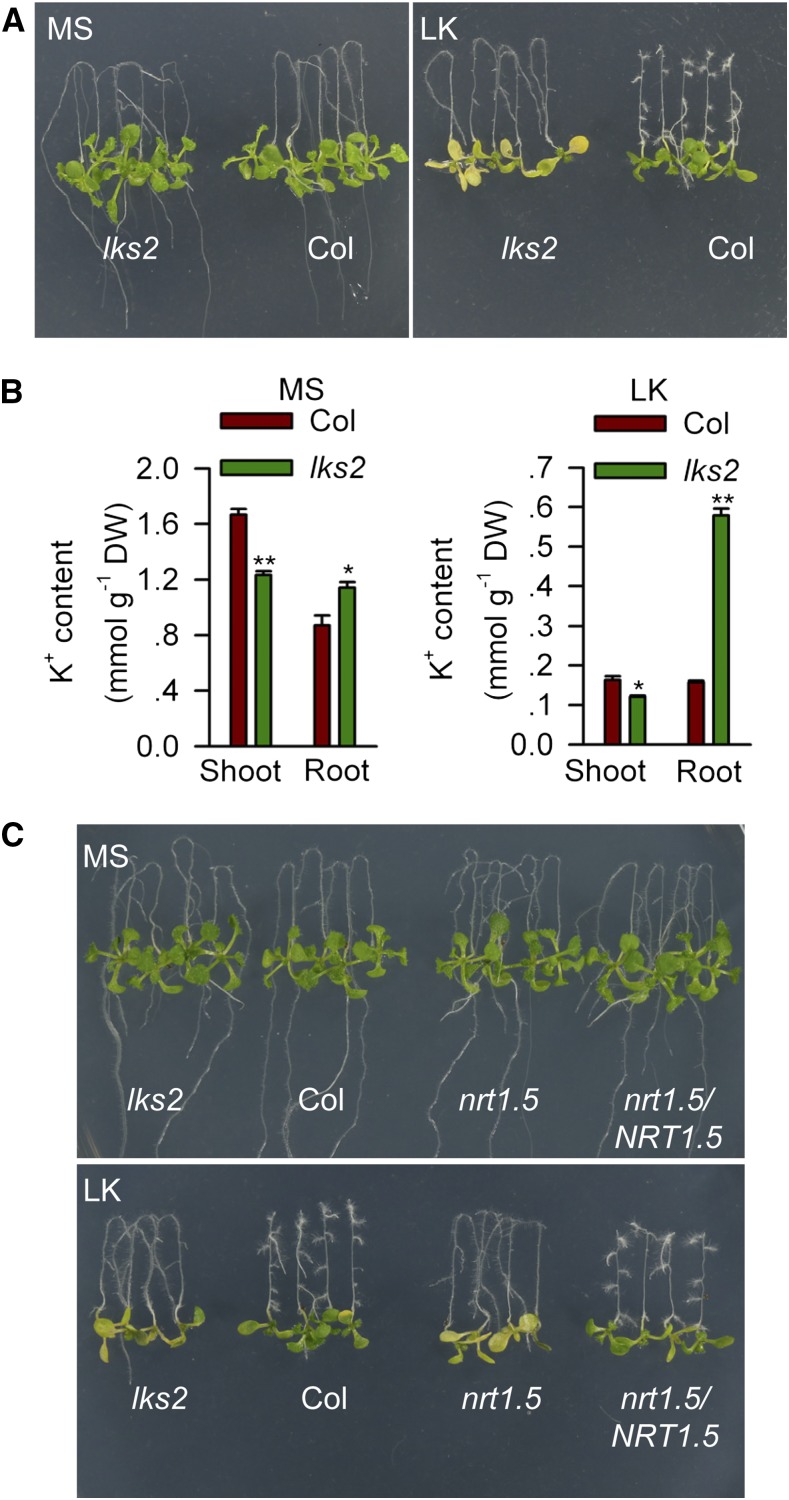
To identify the key components involved in K+ absorption or translocation in plants, we isolated low-K+-sensitive (lks) Arabidopsis mutants from EMS-mutagenized M2 seedlings. Among these mutants, lks2 showed a significantly sensitive phenotype on low K+ (LK; 0.1 mM K+) medium. After the plants were transferred to LK medium for 7 d, the mutant shoot became yellow, but the wild-type (Col-0) plants did not (Figure 1A). However, the primary root of lks2 continued growing on LK medium, while the root growth of wild-type plants stopped (Figure 1A). Under LK conditions, the K+ content of lks2 shoots was reduced, while the K+ content of lks2 roots was significantly increased (Figure 1B), which may explain the shoot chlorosis phenotype and the root growth phenotype of lks2 (Figure 1A). This K+ distribution feature suggests that the K+ translocation from root to shoot is disrupted in the lks2 mutants.
Figure 1.
lks2 and nrt1.5 Mutants Are Sensitive to LK Stress.
(A) Phenotype comparison between wild-type Arabidopsis (Col) and the lks2 mutant. Seeds were germinated on MS medium for 4 d, and seedlings were then transferred to MS or LK (0.1 mM K+) medium for 7 d.
(B) K+ content measurement of Col and lks2. Data are mean ± se (n = 3). Student’s t test (*P < 0.05 and **P < 0.01) was used to analyze statistical significance.
(C) Phenotype comparison of lks2 and nrt1.5 mutants as well as the complementation line.
Genetic analyses demonstrated that lks2 harbors a monogenic recessive mutation in a nuclear gene (Supplemental Figures 1A and 1E). Map-based cloning revealed that LKS2 encodes NRT1.5/NPF7.3, a member of the NRT1/PTR family. In the lks2 mutant, a single-nucleotide mutation (G3242A) was found in NRT1.5, which resulted in a change of Gly-209 to Glu (G209E) in NRT1.5 (Supplemental Figure 1B). A T-DNA insertion mutant of NRT1.5 (nrt1.5, a Salk line obtained from the ABRC; Supplemental Figure 1B) was used as a control of lks2. The nrt1.5 null mutant was sensitive to LK medium, similar to lks2 (Figure 1C; Supplemental Figures 1C and 2). The complementation line nrt1.5/NRT1.5 was able to rescue the low-K+-sensitive phenotype of nrt1.5 (Figure 1C; Supplemental Figure 2), which confirms that the low-K+-sensitive phenotype of lks2 is due to the loss of function of NRT1.5.
The lks2 mutant shows a similar low-K+-sensitive phenotype to the lks1/cipk23 mutant reported previously (Xu et al., 2006; Supplemental Figure 3A). However, they are different in K+ content. In lks2, K+ was accumulated in the root but reduced in the shoot (Figure 1B). In lks1, K+ content was reduced in both root and shoot (Xu et al., 2006). In addition, CIPK23 transcript was not significantly altered in the lks2 mutant (Supplemental Figure 3B). Therefore, the lks2 phenotype does not arise from defects in the CIPK23 signaling pathway.
K+ and NO3− Accumulate in the lks2 Root
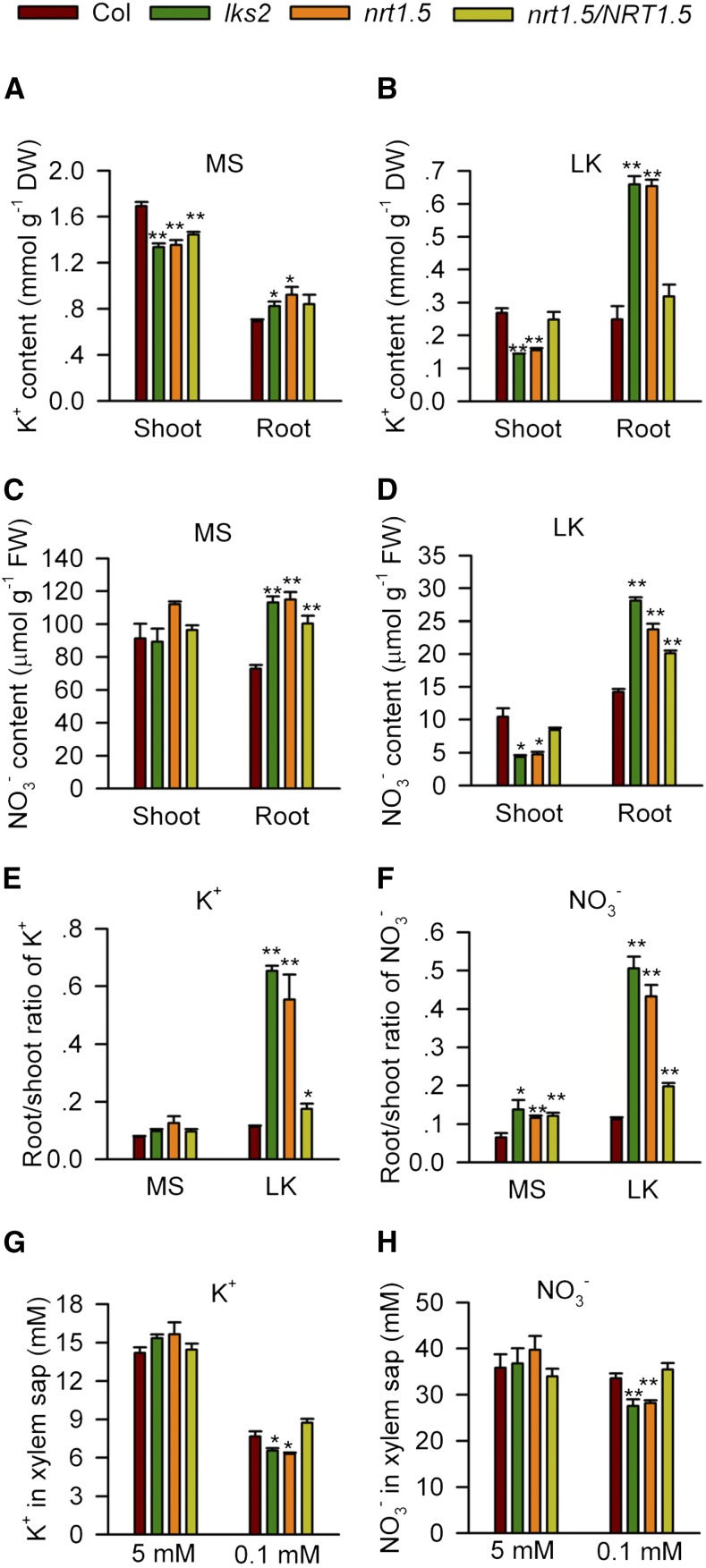
NRT1.5 was reported as a bidirectional, low-affinity nitrate transporter (Lin et al., 2008). NRT1.5 is expressed in root pericycle cells close to the xylem where NO3− loading into the xylem occurs (Lin et al., 2008). In this study, we measured both the K+ and NO3− contents in nrt1.5 and lks2 mutant plants. Under K+-sufficient conditions (on Murashige and Skoog [MS] medium), the K+ content in mutant shoots was lower than that in wild-type shoots, while the K+ content in mutant roots was greater (Figure 2A). After LK treatment, the K+ content in mutant roots was significantly greater than that in wild-type roots (Figure 2B). Similar to K+, the NO3− content was greater in mutant roots on MS medium, but there was no difference in shoot NO3− content between the mutant and wild-type plants (Figure 2C). Under LK conditions, the NO3− content was less in mutant shoots than in wild-type shoots but was significantly greater in mutant roots than in wild-type roots (Figure 2D). Both the K+ and NO3− contents in the complementation line nrt1.5/NRT1.5 were almost the same as those in the wild-type plants, especially on LK medium (Figures 2A to 2D), which may explain the restored phenotype of the complementation line (Figure 1C). The ion contents were also analyzed using hydroponically cultured plants. The results indicated that lks2 and nrt1.5 accumulated higher K+/NO3− in the root but lower K+/NO3− in the shoot under LK conditions (Supplemental Figure 4).
Figure 2.
lks2 and nrt1.5 Are Defective in the Root-to-Shoot Transport of K+ and NO3−.
(A) to (D) K+ ([A] and [B]) and NO3− ([C] and [D]) content measurement in various plant materials after being transferred to MS or LK medium for 7 d.
(E) and (F) The root/shoot ratios of K+ (E) and NO3− (F) in various plant materials.
(G) and (H) K+ (G) and NO3− (H) concentrations in xylem exudates from mutant and complemented plants after being cultured in solutions containing different K+ concentrations (5 and 0.1 mM) for 20 d.
Data in (A) to (F) are mean ± se (n = 3). Data in (G) and (H) are mean ± se (n = 4). Student’s t test (*P < 0.05 and **P<0.01) was used to analyze statistical significance.
Under LK conditions, the root/shoot ratios of K+ and NO3− in mutant plants were significantly higher than those in the wild-type plants, indicating the increased accumulation of K+ and NO3− in mutant roots (Figures 2E and 2F). Furthermore, both K+ and NO3− concentrations in the xylem exudates of mutant plants were significantly reduced under LK (0.1 mM K+) conditions, but did not changed under high K+ (5 mM) conditions (Figures 2G and 2H). These results demonstrate that the translocation of both K+ and NO3− from root to shoot was impaired in the lks2 and nrt1.5 mutant plants, especially under LK conditions, suggesting that NRT1.5 not only participates in NO3− translocation but also regulates K+ transport from root to shoot.
The Sensitive Phenotype of lks2 Depends on Low K+
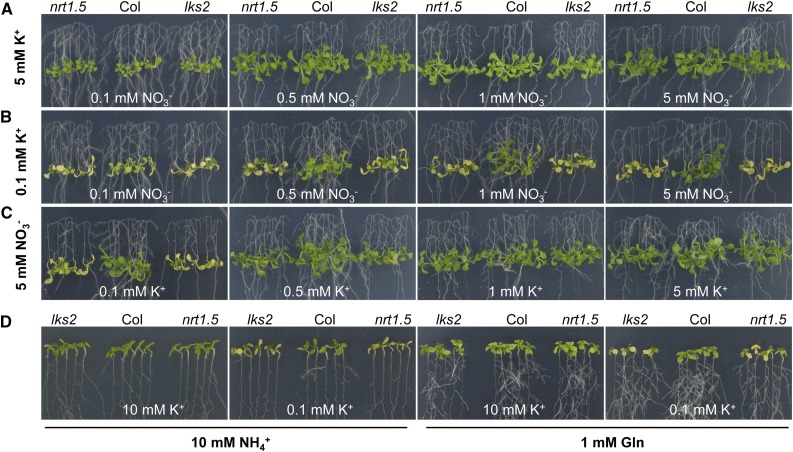
Previous reports have observed that K+ transport in the nrt1.5 mutant was reduced (Lin et al., 2008; Drechsler et al., 2015; Meng et al., 2016), and this reduction was dependent on NO3− (Lin et al., 2008). Our results here showed that there was no obvious phenotypic difference between wild-type and mutant plants when grown on high K+ (5 mM) medium, although the NO3− concentration in the medium was decreased from 5 to 0.1 mM (Figure 3A; Supplemental Figure 5C). However, when the K+ concentration in the medium was decreased to 0.1 mM, the mutant plants exhibited a typical leaf chlorosis phenotype and showed reduced dry weight, regardless of the external NO3− concentration (Figure 3B; Supplemental Figure 5C). Along with the increment of external K+ concentration, the sensitive phenotype of mutant plants disappeared (Figure 3C; Supplemental Figure 5C). To further confirm the possible effect of NO3− on the leaf chlorosis phenotype, NO3− in the medium was replaced with ammonium succinate (NH4+) or Gln as the sole nitrogen source. The results showed that the leaf chlorosis phenotype of mutant plants only appeared under LK conditions but not high K+ conditions, even after the NO3− was completely removed (Figure 3D; Supplemental Figure 6). These data demonstrate that the leaf chlorosis phenotype of lks2 specifically depends on low K+.
Figure 3.
The Sensitive Phenotype of nrt1.5 and lks2 Is Dependent on K+.
Phenotype comparison between the mutant and wild-type plants. Seeds were germinated on MS medium ([A] to [C]) or on the medium without NO3− (D) for 4 d and then seedlings were transferred to various media for 7 d.
(A) High K+ (5 mM) medium containing the indicated NO3− concentrations.
(B) Low K+ (0.1 mM) medium containing the indicated NO3− concentrations.
(C) High NO3− (5 mM) medium containing the indicated K+ concentrations.
(D) Medium without NO3−, which was replaced with 5 mM ammonium succinate (NH4+, 10 mM) or 1 mM Gln.
NRT1.5 Function Does Not Depend on SKOR
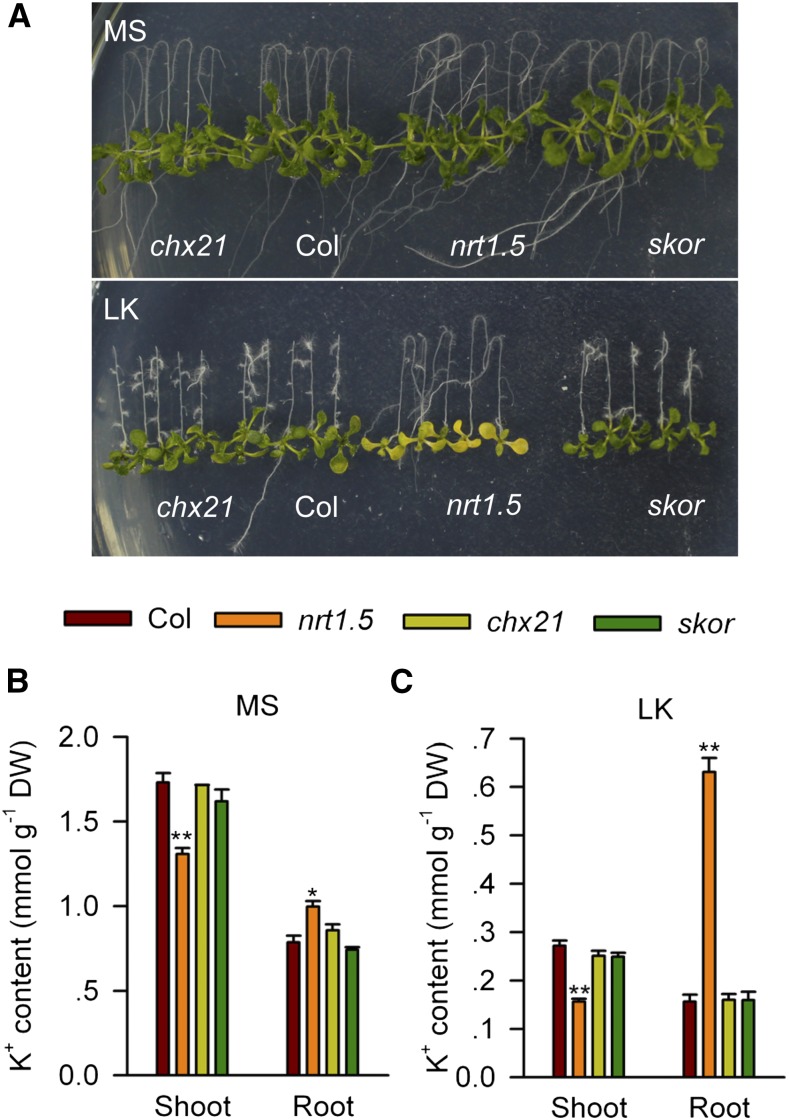
Previous studies suggested that NRT1.5 indirectly affects K+ transport, probably by influencing the K+ channel SKOR or cell membrane potential (Lin et al., 2008; Drechsler et al., 2015). To date, SKOR is the only identified component that participates in K+ loading into the xylem (Gaymard et al., 1998; Véry et al., 2014). The results presented here show that the skor null mutant did not exhibit the same sensitive phenotype as the nrt1.5 mutant under this LK condition (Figure 4A). The K+ content of the root or shoot of the skor mutant did not differ from that of wild-type plants (Figures 4B and 4C). We also tested another K+ (or Na+) transporter mutant chx21, in which the K+ content in the xylem may be affected (Hall et al., 2006). However, chx21 showed neither a sensitive phenotype nor an altered K+ content under LK conditions in our experiments (Figure 4). These results demonstrate that NRT1.5 does not rely on SKOR during K+ transport and suggest that NRT1.5 itself may play important roles in K+ loading into the xylem.
Figure 4.
Phenotype Comparison between nrt1.5 and skor Mutants.
(A) Phenotype comparison between nrt1.5, skor, and chx21 mutants and wild-type plants (Col). Seeds were germinated on MS medium for 4 d, and seedlings were then transferred to MS or LK medium for 7 d.
(B) and (C) K+ content measurement in various plant materials after 4-d-old seedlings were transferred to MS (B) or LK (C) medium for 7 d. Data are mean ± se (n = 3). Student’s t test (*P < 0.05 and **P < 0.01) was used to analyze statistical significance.
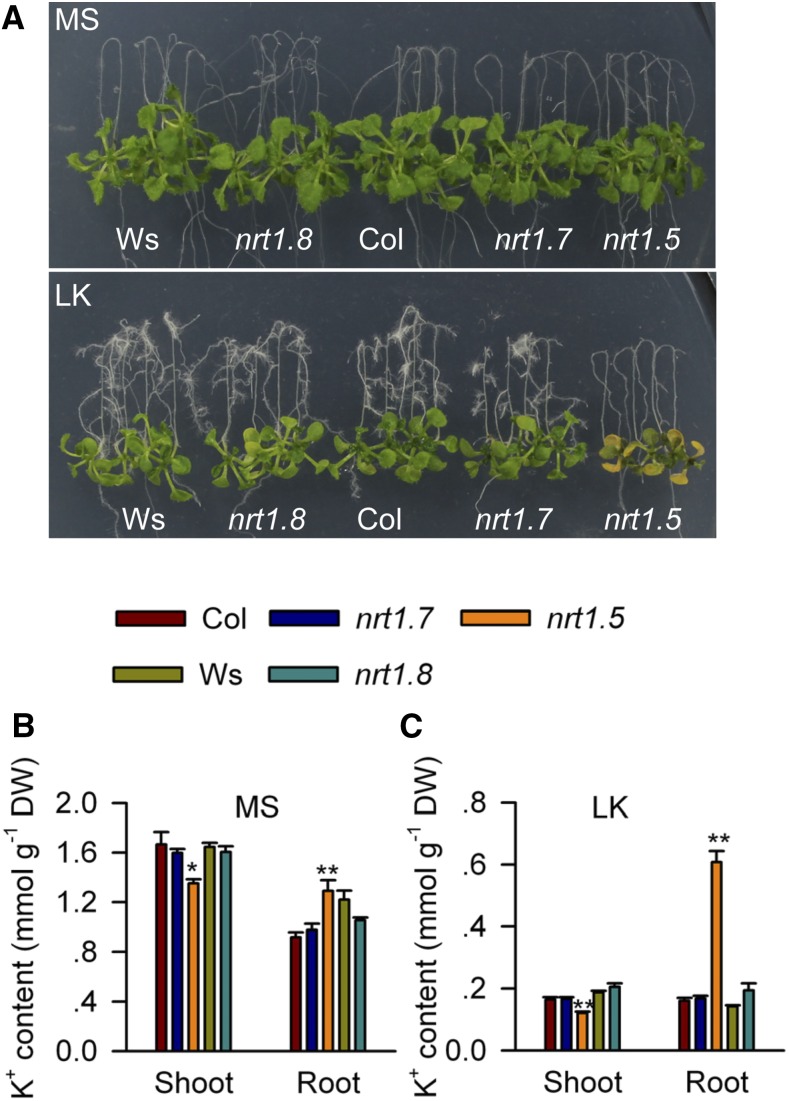
Previous reports have shown that NRT1.8/NPF7.2 and NRT1.7/NPF2.13 from the NRT1/PTR family play roles in NO3− removal from the xylem and NO3− loading into the phloem, respectively (Fan et al., 2009; Li et al., 2010). Thus, we examined the phenotypes of the nrt1.7 and nrt1.8 mutants under the LK condition. These results showed that neither nrt1.7 nor nrt1.8 was sensitive to LK treatment (Figure 5), suggesting that, unlike NRT1.5, NRT1.7, and NRT1.8 may not directly affect K+ translocation in Arabidopsis.
Figure 5.
Phenotype Test of nrt1.7 and nrt1.8 Mutants.
(A) Phenotype comparison between nrt1.5, nrt1.8, and nrt1.7 mutants and wild-type plants. Seeds were germinated on MS medium for 4 d, and seedlings were then transferred to MS or LK medium for 7 d.
(B) and (C) K+ content measurement in various plant materials after 4-d-old seedlings were transferred to MS (B) or LK (C) medium for 7 d. Data are mean ± se (n = 3). Student’s t test (*P < 0.05 and **P < 0.01) was used to analyze statistical significance.
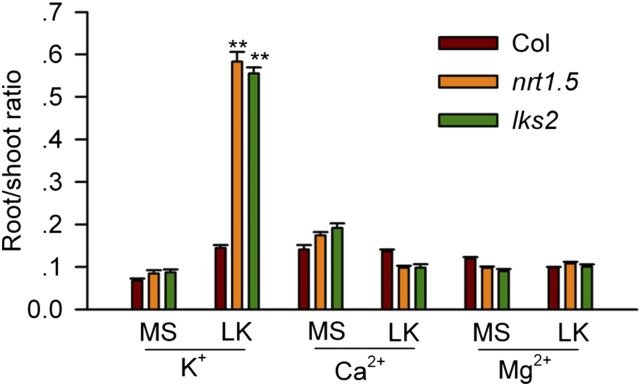
In addition, the possible involvement of other major cations (namely Ca2+ and Mg2+) in NO3− loading was investigated. The results showed that the root/shoot distribution of only K+, but not of Ca2+ or Mg2+, was significantly changed in the lks2 and nrt1.5 mutants (Figure 6), suggesting that NRT1.5 specifically regulates the transport of K+ rather than that of any other cation.
Figure 6.
The Root/Shoot Ratios of Cations in nrt1.5 and lks2.
Seeds were germinated on MS medium for 4 d and then transferred to MS or LK (0.1 mM K+) medium for 7 d. After this, the root/shoot ratios of different cations (K+, Ca2+, and Mg2+) were calculated. Data are mean ± se (n = 3). Student’s t test (**P < 0.01) was used to analyze statistical significance.
NRT1.5 Acts as a K+ Efflux Transporter
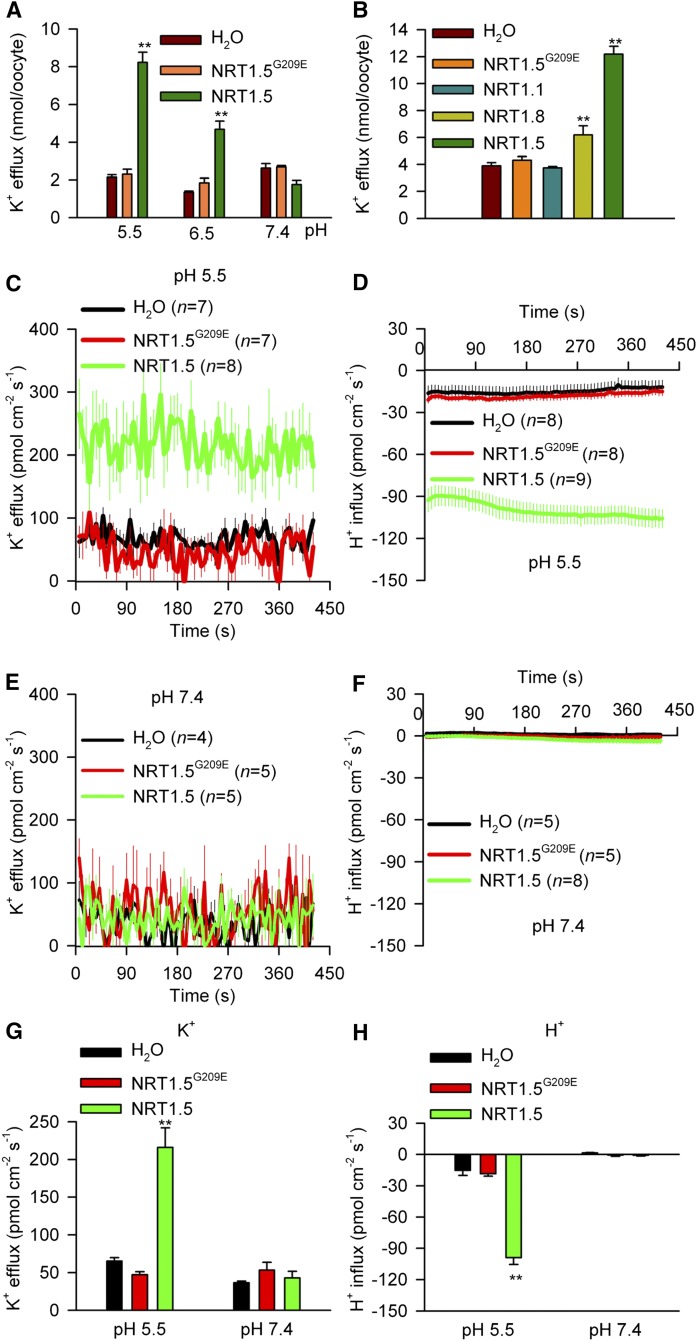
All the results described above imply that NRT1.5 directly regulates K+ transport; therefore, we validated the function of NRT1.5 in facilitating K+ transport in Xenopus laevis oocytes. As shown in Figure 7A, NRT1.5 was indeed able to mediate K+ release from the oocytes, while NRT1.5G209E (point mutation of NRT1.5 in lks2) entirely lost the ability to transport K+ (Figure 7A; Supplemental Figure 7C). G209 is located in the fifth transmembrane segment of NRT1.5 (Supplemental Figure 7A). NRT1.5G209E can still localize at the plasma membrane of oocytes, (Supplemental Figure 7B), suggesting that the G209E mutation may alter protein structure and abolish ion transport activity.
Figure 7.
NRT1.5 Mediates K+ Efflux in X. laevis Oocytes.
(A) K+ release activities of oocytes expressing NRT1.5 or NRT1.5G209E. Oocytes injected with water were used as the control. Oocytes were cultured in K+-free MBS solution at different pH for 3 h.
(B) K+ release activities of oocytes expressing NRT1.1 or NRT1.8 at pH 5.5.
(C) and (E) K+ flux rates of oocytes expressing NRT1.5 or NRT1.5G209E at pH 5.5 (C) or 7.4 (E) measured in NMT/MIFE assays.
(D) and (F) H+ flux rates of oocytes expressing NRT1.5 or NRT1.5G209E at pH 5.5 (D) or 7.4 (F) measured in NMT/MIFE assays.
(G) and (H) Histogram analyses of K+ (G) and H+ (H) flux rates.
All data are mean ± se. The data in (A) and (B) are from four samples (n = 4), and each sample contained five oocytes. Student’s t test (**P < 0.01) was used to analyze statistical significance.
It has been reported that the transport activity of the NRT1/PTR members depends on the proton electrochemical gradient across the membrane (Tsay et al., 2007; Newstead, 2015). Our results demonstrated that the K+ release activity of NRT1.5 relies on an external acidic pH (Figure 7A). An increase in the external pH from 5.5 to 7.4 dramatically reduced the K+ release activity (Figure 7A), suggesting that NRT1.5 may function as a proton-coupled H+/K+ antiporter. As the closest member of NRT1.5, NRT1.8 was also able to transport K+ in oocytes, but had a much lower activity than that of NRT1.5 (Figure 7B). Meanwhile, another important nitrate transporter NRT1.1/NPF6.3 did not exhibit any K+ release activity (Figure 7B). In addition, K+ content in oocytes was also measured. Before the K+ release assay, the K+ contents of oocytes expressing NRT1.5 or NRT1.5G209E were not significantly different from the K+ contents in control oocytes. After the K+ release assay, the K+ content in the oocytes expressing NRT1.5 was significantly reduced compared with the K+ contents in control oocytes (Supplemental Figure 8), suggesting that K+ was released from the NRT1.5 oocytes.
The K+ efflux rate of NRT1.5 was also measured using the NMT/MIFE (noninvasive microelectrode ion flux estimation; Shabala et al., 2013) technique in oocytes (Figures 7C and 7E). In the oocytes expressing NRT1.5, the K+ efflux rates across oocyte membrane ranged from 150 to 250 pmol cm−2 s−1 at pH 5.5 (Figures 7C and 7G). By contrast, the K+ efflux rates in control oocytes (water and NRT1.5G209E) were only ∼50 pmol cm−2 s−1 (Figures 7C and 7G). In addition, H+ influx was recorded in the oocytes expressing NRT1.5 at pH 5.5 (Figures 7D and 7H), suggesting that NRT1.5 is an H+/K+ antiporter. When the external pH was increased from 5.5 to 7.4, both K+ efflux and H+ influx in the oocytes expressing NRT1.5 were completely depressed to the control level (Figures 7E to 7H). These results indicate that the proton gradient across the plasma membrane can promote NRT1.5-mediated K+ efflux out of cells.
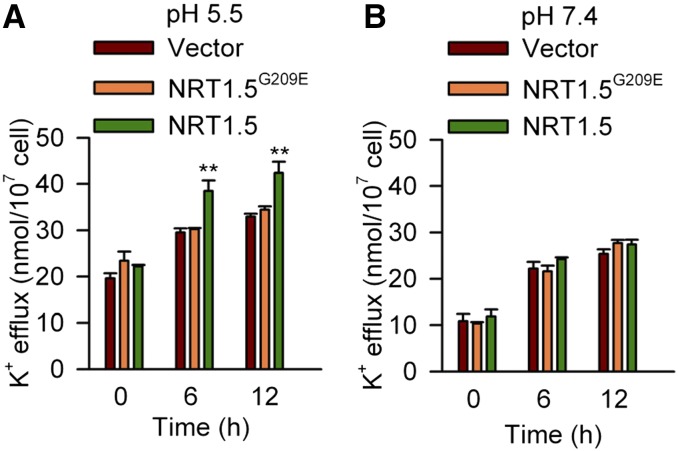
Furthermore, the K+ transport activity of NRT1.5 was tested in the yeast mutant strain ANT5 (ena1-4Δ and nha1Δ) that is defective in K+/Na+ efflux activity at the plasma membrane. The results showed that NRT1.5 can mediate K+ efflux from yeast cells into liquid medium at pH 5.5 (Figure 8A). However, NRT1.5-mediated K+ efflux was entirely inhibited at pH 7.4 (Figure 8B). NRT1.5G209E did not show any K+ efflux activity at both pH 5.5 and 7.4 (Figure 8). These results are consistent with the data from X. laevis oocytes, which suggests that NRT1.5 is a K+ efflux transporter.
Figure 8.
NRT1.5 Mediates K+ Efflux in Yeast.
K+ efflux activities of the yeast mutant strain ANT5 (ena1-4Δ and nha1Δ) expressing NRT1.5 or NRT1.5G209E. Yeast transformed with empty vector (p416GPD) was used as the control. Yeast cells were cultured in K+-free testing buffer at pH 5.5 (A) and 7.4 (B), respectively. All data are mean ± se (n = 4). Student’s t test (**P < 0.01) was used to analyze statistical significance.
To test whether the K+ efflux was voltage dependent, we used the two-electrode voltage clamp (TEVC) technique. However, we did not observe any difference in ionic currents between NRT1.5-expressing oocytes and control oocytes at different voltages (Supplemental Figures 9A and 9B), suggesting that NRT1.5 may mediate electroneutral transport for H+ and K+.
Since NRT1.5 was reported as a nitrate transporter, we investigated whether extracellular or intracellular NO3− would affect the K+ release activity of NRT1.5 in oocytes. In the presence of 10 mM NO3− in the bath solution, the K+ release activity of NRT1.5 was slightly increased; however, the K+ release in the negative control (water and NRT1.5G209E) also increased (Supplemental Figure 10A). These results were further confirmed by measuring K+ efflux rates in NMT/MIFE assays (Supplemental Figure 10B). In addition, when NO3− was injected into oocytes, the K+ release activity of NRT1.5 was not affected (Supplemental Figure 10C). These results demonstrate that neither extracellular nor intracellular NO3− can significantly influence the K+ release activity of NRT1.5 in oocytes.
DISCUSSION
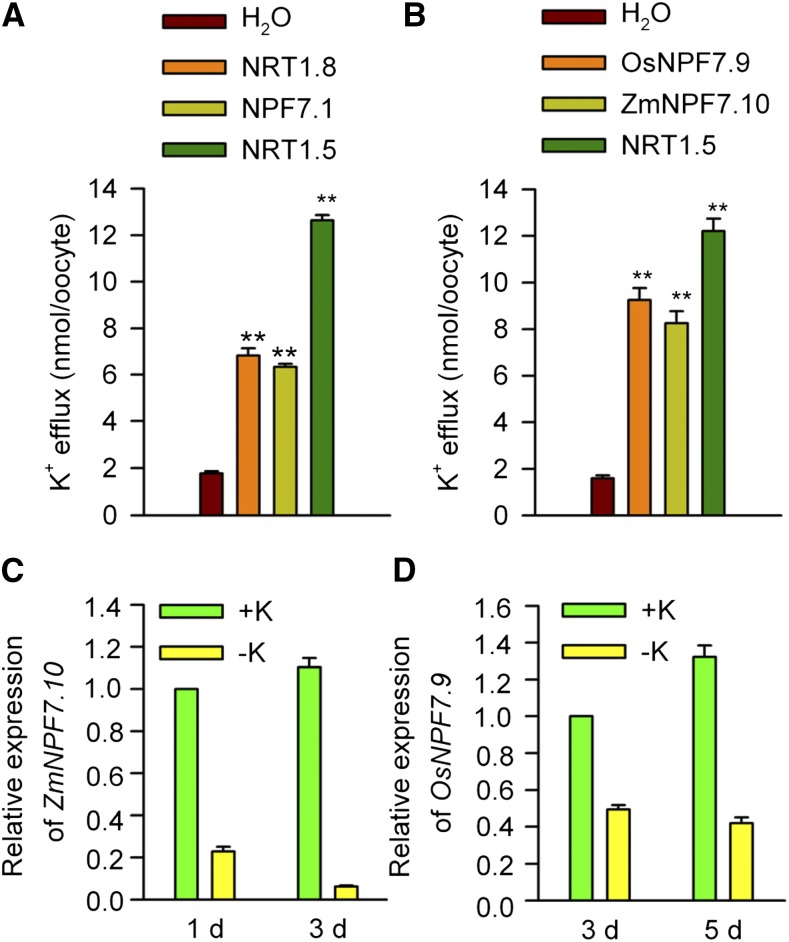
NRT1/PTR is a ubiquitous family that exists in all kingdoms of life (Steiner et al., 1995; Daniel et al., 2006). In contrast to the low number of family members in other organisms, such as in yeast (one), Drosophila melanogaster (three), Caenorhabditis elegans (four), and human (six), the NRT1/PTR family is prevalent in higher plants (Tsay et al., 2007). Arabidopsis and rice (Oryza sativa) contain 53 and 80 NRT1/PTR members, respectively (Tsay et al., 2007), which implies that these proteins have diverse functions in plants, not necessarily limited to nitrate and peptide transport. Most of the identified members in plants are nitrate transporters, but some transport different substrates, for example, indole-3-acetic acid, abscisic acid, gibberellic acid, jasmonoyl-isoleucine, glucosinolates, or amino acids (Krouk et al., 2010; Nour-Eldin et al., 2012; Léran et al., 2014; Saito et al., 2015; Tal et al., 2016). In this study, we identified K+ as a novel substrate of NRT1/PTR transporters. We have shown that NRT1.5 and NRT1.8 are able to directly transport K+. In addition to NRT1.5 and NRT1.8, another unexplored NRT1/PTR member (At5G19640/NPF7.1) similar to NRT1.5 also possesses the ability to transport K+ (Figure 9A; Supplemental Figure 11A and Supplemental Data Set 1). Furthermore, the putative orthologs of NRT1.5 from rice (Os02g0689900/OsNPF7.9) and maize (Zea may; GRMZM2G044851/ZmNPF7.10) can function as K+ efflux transporters as well (Figure 9B; Supplemental Figure 11B and Supplemental Data Set 2), suggesting that the K+ transport function of NRT1.5 and its orthologs are highly conserved in different plants. Similar to NRT1.5, its putative orthologs in rice and maize are both responsive to LK stress and their transcription are downregulated by LK stress (Lin et al., 2008; Ma et al., 2012) (also see Figures 9C and 9D). Under LK conditions, K+ uptake and K+ accumulation in roots are reduced. Less K+ needs to be translocated from the root to shoot and subsequently the transcript of NRT1.5 is downregulated.
Figure 9.
Functional Characterization of NRT1.5 Putative Orthologs.
(A) K+ efflux activities of the X. laevis oocytes expressing NRT1.5, NRT1.8, and NPF7.1.
(B) K+ efflux activities of the X. laevis oocytes expressing NRT1.5 putative orthologs from rice and maize. All the data in (A) and (B) are shown as mean ± se from five samples (n = 5), and each sample contained five oocytes. The Student’s t test (**P < 0.01) was used to analyze statistical significance.
(C) and (D) Real-time PCR analyses of ZmNPF7.10 (C) and OsNPF7.9 (D) in maize and rice responses to low K+ stress. Maize (B73-329) seedlings were cultured in solution containing different K+ concentrations (+K, 1.85 mM K+ and –K, 0 K+) for 1 and 3 d. UBQ was used as the internal control. Data are mean ± se (n = 3). Data in (D) were derived from those reported in a previous study (Ma et al., 2012).
The TEVC assay suggested that NRT1.5 may mediate electroneutral transport for H+ and K+ (Supplemental Figures 9A and 9B); however, the absolute values of H+ influx rate were smaller than those of K+ efflux rate in the NMT/MIFE assay (Figures 7C and 7D). The difference in values may be due to two reasons. First, different ionophores were used to measure K+ and H+ flux rates, respectively. They may have different affinities for K+ and H+. Second, MES/Tris in the recording solution can buffer and reduce H+ flux rate in the NMT/MIFE assay.
The findings of this study expand the substrates of plant NRT1/PTR transporters and suggest that NRT1.5 together with its orthologs is a novel type of K+ transporter in plants. The substrate specificity of NRT1/PTR transporters is mainly determined by the specific side chains that line the aqueous pocket at the center of the membrane (Parker and Newstead, 2014; Sun et al., 2014; Yan, 2015). However, how plant NRT1/PTR transporters are able to recognize and transport diverse substrates remains unknown. Therefore, it would be interesting to examine the crystal structures of these transporters in further investigations.
To date, most of the NRT1/PTR members have not been functionally characterized yet. Some members already identified in plants are H+/substrate symporters that utilize the inwardly direct proton gradient (ΔμH+) across the cell membrane and drive the uptake of the substrates (Krouk et al., 2010; Nour-Eldin et al., 2012; Léran et al., 2014; Saito et al., 2015; Tal et al., 2016). However, this study reveals that NRT1.5 functions as a proton-coupled H+/K+ antiporter, and external acidic pH promotes K+ release out of cells (Figures 7 and 8). In plants, xylem sap has an acidic pH from 5.5 to 6.5 (De Boer and Volkov, 2003; White, 2012b), which facilitates NRT1.5-mediated K+ release into the xylem. Since the cytoplasmic K+ concentrations in root parenchyma cells are over dozens of millimolars, NRT1.5 should be a low-affinity H+/K+ antiporter. According to the crystal structure of NRT1.1, the “EXXER” motif (containing three conserved residues) on TMH1 together with the conserved residue Lys-164 on TMH4 is responsible for proton coupling (Doki et al., 2013; Parker and Newstead, 2014; Sun et al., 2014). These four charged residues are highly conserved in most Arabidopsis NRT1 members. However, NRT1.5 and NRT1.8 have simultaneously evolved into noncharged residues in these four sites, suggesting that NRT1.5 and NRT1.8 must have an alternative proton-coupling mechanism compared with other NRT1 members (Sun et al., 2014). This independent evolution of NRT1.5 and NRT1.8 may endow them with the substrate specificity for K+ as well as the unique antiport function. However, the binding and transport mechanisms of NRT1.5 for H+ and K+ need further investigation.
A previous study reported that NRT1.5 is a bidirectional nitrate transporter in oocytes (Lin et al., 2008); thus, we reasoned that NRT1.5 might be a K+ and NO3− cotransporter. However, both the 15NO3− uptake and release assays (from over 10 independent experiments) conducted in our study indicated that NRT1.5 did not show an obvious NO3− transport activity in oocytes (Supplemental Figures 9E and 9F), even though the positive control NRT1.1 had a very strong NO3− uptake activity in each independent experiment (Supplemental Figure 9E). In addition, NRT1.5-mediated NO3− currents were not recorded in oocytes under different K+ and NO3− conditions (Supplemental Figures 9C and 9D). Nevertheless, NO3− transport is indeed impaired in the lks2 mutant, especially under LK conditions (Figure 2). We speculate that NRT1.5 mediates NO3− transport in planta but shows very weak transport activity for NO3− in oocytes. Possibly, NRT1.5 requires some special partner components or regulators for functional NO3− transport in oocytes. For example, the K+ transport activity of the AKT1 channel requires coexpression of protein kinase CIPK23 and calcium binding proteins CBL1/9 in oocytes (Xu et al., 2006). Nonetheless, NRT1.5 plays an essential role in coordinating K+/NO3− transport in xylem under K+-limited conditions. NRT1.5 may function as a dual-role transporter in Arabidopsis; however, crystal structure studies will need to be performed to decipher the mechanism by which NRT1.5 simultaneously transports NO3− and K+.
In addition, we observed that external NO3− supply affected the K+ concentrations in nrt1.5 xylem exudates (Supplemental Figure 9G). Low NO3− supply significantly depressed the K+ concentrations in nrt1.5 xylem exudates even under K+ replete conditions. Along with the increment of NO3− supply, the K+ concentrations were gradually restored to the wild-type level. This K+ increase in nrt1.5 may be caused by other K+ transporters such as SKOR. It has been reported that the expression of SKOR is upregulated by NO3− (Wang et al., 2004). Furthermore, the expression of NRT1.5 is also induced by NO3− supply (Lin et al., 2008). We propose that K+/NO3− transport may also be coordinated via regulation of K+ transporter genes at the transcriptional level.
A recent report showed that the K+ concentrations in skor xylem exudates were significantly reduced under both high and low K+ conditions (Han et al., 2016), while the decline in K+ concentrations in nrt1.5 only occurred under low K+ conditions (Figure 2G). These data demonstrate that SKOR and NRT1.5 operate under different K+ conditions. The LK phenotypes of the mutants suggest that NRT1.5 may be more important under K+-limited conditions (Figure 4). However, these two proteins coordinate and complement each other in K+ loading into the xylem.
METHODS
Plant Materials and Growth Conditions
The lks2 mutant was isolated from EMS-mutagenized M2 seeds of wild-type Arabidopsis thaliana (Columbia ecotype, Col-0) as described previously (Xu et al., 2006). Arabidopsis T-DNA insertion lines, including nrt1.5 (SALK_043036), nrt1.7 (SALK_022429), chx21 (SALK_061500), and skor (SALK_132944) were obtained from the ABRC. The nrt1.8 mutant was isolated from the ALPHA T-DNA insertion population (Krysan et al., 1999). To generate the complementation line of the nrt1.5 mutant, the construct was generated by cloning the genomic sequence of NRT1.5 into the pCAMBIA1300 vector driven by its own promoter (2192 bp). The construct was introduced into Agrobacterium tumefaciens GV3101 and transformed into nrt1.5 mutant plants by the floral dip method (Clough and Bent, 1998). Primer sequences are provided in Supplemental Table 1.
Map-Based Cloning
The lks2 (Col-0 ecotype) mutant was crossed with wild-type Arabidopsis (Landsberg erecta ecotype) to create mapping populations. Then, 943 individual F2 plants showing the lks2 phenotype were selected for chromosomal mapping of LKS2.
Phenotype Analyses
Plant growth, mutant isolation and low K+ phenotype analyses were performed as described previously (Xu et al., 2006). For the phenotype analyses using different concentrations of K+ and NO3−, seeds were germinated on MS medium at 22°C under constant illumination at 60 μmol m−2 s−1 (Philips; TLD 36W/865 Cool Daylight) for 4 d and then transferred to different media containing various concentrations of K+ and NO3− for 7 d. The LK (0.1 mM) media with different NO3− concentrations (0.1, 0.5, 1, and 5 mM) were made by omitting NH4NO3 (28.5, 28.1, 27.6, and 23.6 mM) from LK medium. For the medium with 5 mM K+ and serial NO3− concentrations, 5 mM K+ was obtained by adding 5 mM KCl to the medium, while the different NO3− concentrations were obtained using the same method as described above. For the medium containing different K+ concentrations, NH4NO3 (23.6 mM) was omitted from the LK medium described before, while the different concentrations of KCl were added to the medium. For the phenotype test on NO3−-free medium, the medium was made by modifying the MS medium, as follows: KNO3, NH4H2PO4, and NH4NO3 were replaced with 10 mM KCl, 1.25 mM NaH2PO4, and 5 mM ammonium succinate or 1 mM glutamine, respectively. Sucrose was reduced to 0.5% (w/v). Seeds were germinated on medium containing 10 mM KCl, 5 mM ammonium succinate, or 1 mM glutamine at 22°C under constant illumination at 60 μmol m−2 s−1 (Philips; TLD 36W/865 Cool Daylight) for 4 d, and the seedlings were then transferred to medium containing low (0.1 mM) or high (10 mM) K+ for 10 d.
Seedlings were transferred to different media for 7 or 10 d, and then photographs were taken. ImageJ software was used to measure primary root length. The 7-d-old seedlings grown on different media were harvested. The dry weight was measured as biomass. In addition, the fresh shoots of 7-d-old seedlings were harvested for chlorophyll content measurement. The chlorophyll was extracted in 80% acetone (v/v) in the dark for 2 d. Then, the absorbance of the extraction buffer at 645 and 663 nm was measured. Each biological replicate contained more than 15 individual plants.
Ion Content Measurement
After germination on MS medium at 22°C under constant illumination at 60 μmol m−2 s−1 for 4 d, seedlings were transferred to MS or LK medium for 7 d. Shoots and roots were harvested separately. Samples were dried at 80°C for 24 h and then treated in a muffle furnace at 575°C for 6 h and dissolved in 0.1 N HCl. K+, Mg2+, and Ca2+ contents were measured using the 4100-MP AES system (Agilent). A pool containing more than 60 individual plants represented one biological replicate. For NO3− content analyses, fresh weights of plant roots and shoots were measured. Plant tissues were boiled in distilled water for 20 min and then frozen at –80°C for 12 h, as described previously (Chiu et al., 2004). The NO3− content of the samples was measured using HPLC. Each biological replicate contained more than 20 individual plants. For measuring K+ and NO3− concentrations in xylem sap, seeds were germinated on MS medium at 22°C under constant illumination at 60 μmol m−2 s−1 (Philips; TLD 36W/865 Cool Daylight) for 10 d and then transferred to vermiculite irrigated with one-quarter-strength MS nutrient solution that contained different K+ (0.1 or 5 mM) or irrigated with nutrient solution containing 5 mM K+ and different concentrations of NO3− (0.5, 1, or 2 mM). After growth in nutrient solution for 20 d at 22°C with illumination at 120 μmol m−2 s−1 (Philips; TLD 36W/865 Cool Daylight) under a 14-h-light/10-h-dark photoperiod, the plant shoots were excised. Xylem sap was then collected using glass capillaries within 2 h. Finally, the K+ and NO3− concentrations in xylem sap were measured using the 4100-MP AES system or HPLC. Each biological replicate contained 20 to 30 individual plants.
For hydroponic culture, nrt1.5, lks2, and the complementation line nrt1.5/NRT1.5 were germinated on MS medium at 22°C under constant illumination at 60 μmol m−2 s−1 for 12 d. Then the plants were transferred to hydroponic solution (5 or 0.1 mM K+) and kept in a growth chamber at 22°C with illumination at 120 μmol m−2 s−1 (Philips; TLD 36W/865 Cool Daylight) for an 8-h daily light period for 3 weeks. Shoots and roots were harvested separately. Then, the K+ and NO3− contents were measured as described above.
Vector Construction and Oocyte Assay
Coding sequences of NRT1.5, NRT1.1, and NRT1.8 were codon optimized (Feng et al., 2013) and synthesized. These cDNAs were then cloned into the expression vector pT7TS. The NRT1.5G209E mutant was generated using the QuikChange Lightning site-directed mutagenesis kit (Agilent Technologies). After linearization of pT7TS plasmids with BamHI, RNA was transcribed in vitro using an mRNA synthesis kit (mMESSAGE mMACHINE T7 kit; Ambion). Xenopus laevis oocytes were isolated in 25 mL ND96 solution containing 43 mg collagenase and 12.5 mg trypsin inhibitor for 1.5 h and were then recovered in ND96 for 24 h. Oocytes were injected with 20 ng RNA after recovery and were incubated in NO3−-free MBS solution (in mM: 88 NaCl, 1 KCl, 2.4 NaHCO3, 0.71 CaCl2, 0.82 MgSO4, and 15 HEPES, pH 7.4) for 30 h. Oocytes injected with water were used as the control.
For K+ release assays, oocytes were washed five times with K+-free MBS solution and incubated in K+-free MBS solution (in mM: 89 NaCl, 2.4 NaHCO3, 0.71 CaCl2, 0.82 MgSO4, and 15 MES/HEPES) with 10 mM NO3− or without NO3− for 3 h. Five oocytes incubated in each Petri dish represented one sample. K+ content of the bath solution in each Petri dish was analyzed using the 4100-MP AES system. K+ efflux activities of the oocytes were then calculated.
For K+ measurement of oocytes, the oocytes before and after the K+ release assay were collected and washed five times with K+-free MBS solution. Oocytes were homogenized in 0.1 n HCl. The suspensions were filtered using Millipore filters (0.22 μm). Then, the K+ concentration was measured as described above.
For the localization assay, the coding sequences of NRT1.5 and NRT1.5G209E were fused with GFP. The oocytes expressing NRT1.5-GFP and NRT1.5G209E-GFP were cultured in ND96 solution for 30 h. Fluorescence was observed using a confocal scanning microscope (LSM510; Carl Zeiss).
For the TEVC assay, the TEVC technique was applied using a GeneClamp 500B amplifier (Molecular Devices) at room temperature (∼20°C). The microelectrodes were filled with 3 M KCl. The bath solution contained (in mM) 230 mannitol, 0.15 CaCl2, and 10 MES with different K+ and NO3− concentrations (0 KNO3, 10 KCl, 10 HNO3, or 10 KNO3) at pH 5.5. For the 15N assay in oocytes, the 15NO3− uptake and release assays in oocytes were modified according to the previous report (Lin et al., 2008). HNO3 was replaced with Na15NO3, and 15NO3− was measured using an isotope ratio mass spectrometer (IRMS; DELTAplus XP).
NMT/MIFE Assays
Oocytes were injected with water, NRT1.5G209E, or NRT1.5 RNA and incubated in NO3−-free MBS solution for 30 h. Prior to K+ and H+ flux rate measurement, oocytes were equilibrated in the measuring solution for 15 min. The measuring solution for K+ contained (in mM) 88.95 NaCl, 0.05 KCl, 2.4 NaHCO3, 0.71 CaCl2, 0.82 MgSO4.7H2O, and 15 MES/HEPES, pH 5.5/7.4. The measuring solution for H+ contained (in mM) 88.95 NaCl, 0.05 KCl, 0.71 CaCl2, 0.82 MgSO4.7H2O, and 0.03 MES/HEPES, pH 5.5/7.4. The K+ and H+ flux rates across the oocyte membrane were measured using the NMT/MIFE technique (Xuyue Beijing Science and Technology Company). Data were analyzed using Jcal V3.3 software (http://www.youngerusa.com).
K+ Efflux Assay in Yeast
The yeast mutant strain ANT5 (ena1::KanMX4::ena4, nha1::hisG) expressing NRT1.5, NRT1.5G209E and empty vector (p416GPD) were grown overnight in SC-Ura medium with 0.5 M KCl. After rinsing, the yeast cells were washed with fresh SC-Ura medium (half strength) three times and then washed with water three times and resuspended in K+-free testing buffer (20 mM Tris, 10 mM RbCl, 10 mM glucose, and 0.1 mM MgCl2, pH adjusted to 5.5/7.4 with citric acid). For the K+ efflux assay, yeast cells were added to 50 mL of testing buffer (starting OD600 = 0.9). The OD600 was measured after culture for 6 and 12 h. The testing buffer was collected by rinsing and filtering. The K+ concentration of the testing buffer was measured using the 4100-MP AES system and then the K+ efflux activity of yeast cells was calculated.
Phylogenetic Analyses of NRT1.5 Orthologs
The amino acid sequences for 10 NPF members in Arabidopsis were retrieved from TAIR (http://www.arabidopsis.org). The amino acid sequences of NRT1.5 orthologs from wheat (Triticum aestivum, Traes_6DL_0A59B823C), rice (Oryza sativa, Os02g0689900/OsNPF7.9), barley (Hordeum vulgare, MLOC_57296), and maize (Zea mays, GRMZM2G044851/ZmNPF7.10) were derived from EnsemblPlants (http://www.plants.ensembl.org). The phylogenetic tree was analyzed using ClustalX and MEGA4 software.
Accession Numbers
Sequence data for the genes described in this article can be found in the Arabidopsis TAIR database (https://www.arabidopsis.org) under the following accession numbers: At1g12110 for NRT1.1, At1g32450 for NRT1.5, At1g69870 for NRT1.7, At4g21680 for NRT1.8, At5g19640 for NPF7.1, At3g02850 for SKOR, and At2g31910 for CHX21.
Supplemental Data
Supplemental Figure 1. Genetic analyses of the lks2 mutant.
Supplemental Figure 2. Phenotype test of various plants on vertical plates.
Supplemental Figure 3. Phenotype comparison of lks1 and lks2 mutants.
Supplemental Figure 4. K+ and NO3− contents of various plants grown in hydroponic system.
Supplemental Figure 5. Biomass comparison of lks2 and nrt1.5 mutants under different conditions.
Supplemental Figure 6. The sensitive phenotype of nrt1.5 and lks2 is dependent on K+.
Supplemental Figure 7. Expression and localization analyses of NRT1.5 and NRT1.5G209E in X. laevis oocytes and yeast cells.
Supplemental Figure 8. K+ content of oocytes before and after K+ release assay.
Supplemental Figure 9. NO3− transport analyses in X. laevis oocytes.
Supplemental Figure 10. Effect of NO3− on NRT1.5-mediated K+ efflux in X. laevis oocytes.
Supplemental Figure 11. Phylogenetic analyses of NRT1.5 putative orthologs.
Supplemental Table 1. Primer sequences used in this study.
Supplemental Data Set 1. Alignments used to generate the phylogeny in Supplemental Figure 11A.
Supplemental Data Set 2. Alignments used to generate the phylogeny in Supplemental Figure 11B.
Acknowledgments
We thank Guohua Xu (Nanjing Agricultural University) and Jiming Gong (Institute of Plant Physiology and Ecology, Chinese Academy of Sciences) for providing us with the pT7TS vector and nrt1.8 mutant seeds, respectively. We also thank Xiaorong Fan (Nanjing Agricultural University), Lixing Yuan (China Agricultural University), Jörg Kudla (Universität Münster), and Yi-Fang Tsay (Academia Sinica) for discussions and help in general. This work was supported by grants from the National Key Research and Development Program of China (2016YFD0100700) and the National Natural Science Foundation of China (31622008 and 31270306 to Y.W.; 31421062 to W.-H.W.). F.J.Q. was supported by Spanish Ministry MINECO Grant BIO2015-70946R cofinanced by the European Regional Development Fund.
AUTHOR CONTRIBUTIONS
W.-H.W. and Y.W. designed the research. H.L., M.Y., X.-Q.D., and Z.-F.W. conducted the phenotype test, ion content measurement, and oocyte experiments. X.-H.J. performed the map-based cloning. H.-D.L. conducted the mutant isolation. F.J.Q. generated the yeast strain ANT5. H.L., W.-H.W., and Y.W. wrote and revised the article.
Glossary
- LK
low K+
- MS
Murashige and Skoog
- TEVC
two-electrode voltage clamp
Footnotes
Articles can be viewed without a subscription.
References
- Blevins D.G., Barnett N.M., Frost W.B. (1978). Role of potassium and malate in nitrate uptake and translocation by wheat seedlings. Plant Physiol. 62: 784–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu C.C., Lin C.S., Hsia A.P., Su R.C., Lin H.L., Tsay Y.F. (2004). Mutation of a nitrate transporter, AtNRT1:4, results in a reduced petiole nitrate content and altered leaf development. Plant Cell Physiol. 45: 1139–1148. [DOI] [PubMed] [Google Scholar]
- Clarkson D.T., Hanson J.B. (1980). The mineral nutrition of higher plants. Annu. Rev. Plant Physiol. 31: 239–298. [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Daniel H., Spanier B., Kottra G., Weitz D. (2006). From bacteria to man: archaic proton-dependent peptide transporters at work. Physiology (Bethesda) 21: 93–102. [DOI] [PubMed] [Google Scholar]
- De Boer A.H., Volkov V. (2003). Logistics of water and salt transport through the plant: structure and functioning of the xylem. Plant Cell Environ. 26: 87–101. [Google Scholar]
- Doki S., et al. (2013). Structural basis for dynamic mechanism of proton-coupled symport by the peptide transporter POT. Proc. Natl. Acad. Sci. USA 110: 11343–11348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drechsler N., Zheng Y., Bohner A., Nobmann B., von Wirén N., Kunze R., Rausch C. (2015). Nitrate-dependent control of shoot K homeostasis by the nitrate transporter1/peptide transporter family member NPF7.3/NRT1.5 and the stelar K+ outward rectifier SKOR in Arabidopsis. Plant Physiol. 169: 2832–2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan S.C., Lin C.S., Hsu P.K., Lin S.H., Tsay Y.F. (2009). The Arabidopsis nitrate transporter NRT1.7, expressed in phloem, is responsible for source-to-sink remobilization of nitrate. Plant Cell 21: 2750–2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng H., Xia X., Fan X., Xu G., Miller A.J. (2013). Optimizing plant transporter expression in Xenopus oocytes. Plant Methods 9: 48–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaymard F., Pilot G., Lacombe B., Bouchez D., Bruneau D., Boucherez J., Michaux-Ferrière N., Thibaud J.B., Sentenac H. (1998). Identification and disruption of a plant shaker-like outward channel involved in K+ release into the xylem sap. Cell 94: 647–655. [DOI] [PubMed] [Google Scholar]
- Hall D., Evans A.R., Newbury H.J., Pritchard J. (2006). Functional analysis of CHX21: a putative sodium transporter in Arabidopsis. J. Exp. Bot. 57: 1201–1210. [DOI] [PubMed] [Google Scholar]
- Han M., Wu W., Wu W.H., Wang Y. (2016). Potassium transporter KUP7 is involved in K+ acquisition and translocation in Arabidopsis root under K+-limited conditions. Mol. Plant 9: 437–446. [DOI] [PubMed] [Google Scholar]
- Krouk G., et al. (2010). Nitrate-regulated auxin transport by NRT1.1 defines a mechanism for nutrient sensing in plants. Dev. Cell 18: 927–937. [DOI] [PubMed] [Google Scholar]
- Krysan P.J., Young J.C., Sussman M.R. (1999). T-DNA as an insertional mutagen in Arabidopsis. Plant Cell 11: 2283–2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law C.J., Maloney P.C., Wang D.N. (2008). Ins and outs of major facilitator superfamily antiporters. Annu. Rev. Microbiol. 62: 289–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Léran S., et al. (2014). A unified nomenclature of NITRATE TRANSPORTER 1/PEPTIDE TRANSPORTER family members in plants. Trends Plant Sci. 19: 5–9. [DOI] [PubMed] [Google Scholar]
- Li J.Y., et al. (2010). The Arabidopsis nitrate transporter NRT1.8 functions in nitrate removal from the xylem sap and mediates cadmium tolerance. Plant Cell 22: 1633–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S.H., Kuo H.F., Canivenc G., Lin C.S., Lepetit M., Hsu P.K., Tillard P., Lin H.L., Wang Y.Y., Tsai C.B., Gojon A., Tsay Y.F. (2008). Mutation of the Arabidopsis NRT1.5 nitrate transporter causes defective root-to-shoot nitrate transport. Plant Cell 20: 2514–2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma T.L., Wu W.H., Wang Y. (2012). Transcriptome analysis of rice root responses to potassium deficiency. BMC Plant Biol. 12: 161–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng S., Peng J.S., He Y.N., Zhang G.B., Yi H.Y., Fu Y.L., Gong J.M. (2016). Arabidopsis NRT1.5 mediates the suppression of nitrate starvation-induced leaf senescence by modulating foliar potassium level. Mol. Plant 9: 461–470. [DOI] [PubMed] [Google Scholar]
- Newstead S. (2015). Molecular insights into proton coupled peptide transport in the PTR family of oligopeptide transporters. Biochim. Biophys. Acta 1850: 488–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nour-Eldin H.H., Andersen T.G., Burow M., Madsen S.R., Jørgensen M.E., Olsen C.E., Dreyer I., Hedrich R., Geiger D., Halkier B.A. (2012). NRT/PTR transporters are essential for translocation of glucosinolate defence compounds to seeds. Nature 488: 531–534. [DOI] [PubMed] [Google Scholar]
- Pao S.S., Paulsen I.T., Saier M.H. Jr (1998). Major facilitator superfamily. Microbiol. Mol. Biol. Rev. 62: 1–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker J.L., Newstead S. (2014). Molecular basis of nitrate uptake by the plant nitrate transporter NRT1.1. Nature 507: 68–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito H., et al. (2015). The jasmonate-responsive GTR1 transporter is required for gibberellin-mediated stamen development in Arabidopsis. Nat. Commun. 6: 6095–7005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabala S., Shabala L., Bose J., Cuin T., Newman I. (2013). Ion flux measurements using the MIFE technique. Methods Mol. Biol. 953: 171–183. [DOI] [PubMed] [Google Scholar]
- Steiner H.Y., Naider F., Becker J.M. (1995). The PTR family: a new group of peptide transporters. Mol. Microbiol. 16: 825–834. [DOI] [PubMed] [Google Scholar]
- Sun J., Bankston J.R., Payandeh J., Hinds T.R., Zagotta W.N., Zheng N. (2014). Crystal structure of the plant dual-affinity nitrate transporter NRT1.1. Nature 507: 73–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tal I., et al. (2016). The Arabidopsis NPF3 protein is a GA transporter. Nat. Commun. 7: 11486–11496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triplett E.W., Barnett N.M., Blevins D.G. (1980). Organic acids and ionic balance in xylem exudate of wheat during nitrate or sulfate absorption. Plant Physiol. 65: 610–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsay Y.F., Chiu C.C., Tsai C.B., Ho C.H., Hsu P.K. (2007). Nitrate transporters and peptide transporters. FEBS Lett. 581: 2290–2300. [DOI] [PubMed] [Google Scholar]
- Véry A.A., Nieves-Cordones M., Daly M., Khan I., Fizames C., Sentenac H. (2014). Molecular biology of K+ transport across the plant cell membrane: what do we learn from comparison between plant species? J. Plant Physiol. 171: 748–769. [DOI] [PubMed] [Google Scholar]
- Wang R., Tischner R., Gutiérrez R.A., Hoffman M., Xing X., Chen M., Coruzzi G., Crawford N.M. (2004). Genomic analysis of the nitrate response using a nitrate reductase-null mutant of Arabidopsis. Plant Physiol. 136: 2512–2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Wu W.H. (2013). Potassium transport and signaling in higher plants. Annu. Rev. Plant Biol. 64: 451–476. [DOI] [PubMed] [Google Scholar]
- Whelton P.K., He J. (2014). Health effects of sodium and potassium in humans. Curr. Opin. Lipidol. 25: 75–79. [DOI] [PubMed] [Google Scholar]
- White P.J. (2012a). Ion uptake mechanisms of individual cells and roots: short-distance transport. In Marschner’s Mineral Nutrition of Higher Plants, Marschner P., ed (Amsterdam: Elsevier; ), pp. 28–46. [Google Scholar]
- White P.J. (2012b). Long-distance transport in the xylem and phloem. Marschner’s Mineral Nutrition of Higher Plants, Marschner P., ed (Amsterdam: Elsevier; ), pp. 50–58. [Google Scholar]
- Xu J., Li H.D., Chen L.Q., Wang Y., Liu L.L., He L., Wu W.H. (2006). A protein kinase, interacting with two calcineurin B-like proteins, regulates K+ transporter AKT1 in Arabidopsis. Cell 125: 1347–1360. [DOI] [PubMed] [Google Scholar]
- Yan N. (2015). Structural biology of the major facilitator superfamily transporters. Annu. Rev. Biophys. 44: 257–283. [DOI] [PubMed] [Google Scholar]
- Zörb C., Senbayram M., Peiter E. (2014). Potassium in agriculture--status and perspectives. J. Plant Physiol. 171: 656–669. [DOI] [PubMed] [Google Scholar]