Abstract
DNA helicase B is a major DNA helicase in mouse FM3A cells. A temperature-sensitive mutant defective in DNA replication, tsFT848, isolated from FM3A cells, has a heat-labile DNA helicase B. In this study, we purified DNA helicase B from mouse FM3A cells and determined partial amino acid sequences of the purified protein. By using a DNA probe synthesized according to one of the partial amino acid sequences, a cDNA was isolated, which encoded a 121.5 kDa protein containing seven conserved motifs for DNA/RNA helicase superfamily members. A database search revealed similarity between DNA helicase B and the α subunit of exodeoxyribonuclease V of a number of prokaryotes including Escherichia coli RecD protein, but no homologous protein was found in yeast. The cDNA encoding DNA helicase B from tsFT848 was sequenced and a mutation was found between DNA/RNA helicase motifs IV and V.
INTRODUCTION
The genetic information on a DNA strand is normally covered by a complementary strand and exposed only during DNA transaction processes such as DNA replication, repair, recombination and transcription. The unwinding of double-helical DNA is catalyzed by a class of enzyme designated DNA helicase. These enzymes accomplish the task by using the energy generated by ATP hydrolysis, so that they retain an intrinsic DNA-dependent ATPase activity (1).
A large number of mammalian DNA helicases have been identified, and the functions in cellular metabolism of some have been characterized. Two examples are the DNA helicases functioning in DNA excision repair, ERCC2 and ERCC3 (2,3). These enzymes are also known as subunits of the transcription factor, TFIIH (4,5). Recently, a class of mammalian DNA helicases that share similarity with Escherichia coli RecQ DNA helicase was identified. Five mammalian DNA helicases are classified into this group, and three of five genes encoding RecQ family helicases are found to be responsible for the genetic disorders Bloom syndrome, Werner syndrome and Rothmund–Thomson syndrome (6–9).
Mcm proteins were originally identified in a screen for mutations of budding yeast that were defective in replication of minichromosomes. Six Mcm proteins (Mcm2–7) are known to be involved in the initiation of DNA replication (10), and a complex of Mcm4, 6 and 7 possesses DNA helicase activity (11). Hence, the DNA helicase activity of the Mcm protein complex is thought to be involved in DNA replication in eukaryotic cells. Although analysis of human genetic disorders and yeast genetics has shed light on the physiological functions of some mammalian DNA helicases, the function of most of those originally identified by biochemical activity remains unknown.
We have isolated a temperature-sensitive mutant defective in DNA replication, tsFT848, from a mouse mammary carcinoma cell line, FM3A (12). Characterization of tsFT848 cells revealed that a major DNA-dependent ATPase activity in the mutant cells was diminished during incubation at the non-permissive temperature, 39°C. The DNA-dependent ATPase activity was found to be derived from a DNA helicase, DNA helicase B, whose biochemical properties we have characterized in detail (13–15). DNA helicase B is one of the mammalian proteins first identified as DNA helicases (14,15). The helicase stimulates primer RNA synthesis by DNA primase on M13 single-stranded DNA (16) and supports DNA replication in a model system which consists of plasmid DNA containing an autonomously replicating sequence from Saccharomyces cerevisiae, replication protein A, DNA polymerase α-primase complex and DNA gyrase (17,18).
In this study, we have purified DNA helicase B from FM3A cells, isolated its cDNA and identified a mutation in the DNA helicase B of tsFT848 cells.
MATERIALS AND METHODS
Purification of DNA helicase B from mouse FM3A cells
FM3A cells were grown in the abdominal cavity of ddY mice and harvested. The cells were washed twice with phosphate-buffered saline (PBS; 137 mM NaCl, 2.68 mM KCl, 8.04 mM Na2HPO4 and 1.47 mM KH2PO4) and once with 20 mM potassium phosphate buffer, pH 7.5, containing 0.1 mM EDTA, 1 mM 2-mercaptoethanol, 0.25 mM PMSF and 1% ethanol (buffer 1). The washed cells were stored at –80°C until use.
FM3A cells (1 × 1011) were suspended in 650 ml of buffer 1 supplemented with 10 mM NaHSO3 (buffer 1′) and homogenized by sonication. Extraction was performed with stirring for 30 min on ice after an addition of 1/10 vol of buffer 1′ containing 3.3 M KCl to adjust the salt concentration to 0.3 M KCl. The extract was cleared by centrifugation at 100 000 g for 60 min. DNA helicase B was purified by sequential column chromatography on first and second DEAE-cellulose (Brown), phosphocellulose (P11, Whatman), Bio-Gel HTP (Bio-Rad), single-stranded DNA cellulose, first Mono Q HR5/5 (Pharmacia), Mono S HR5/5 (Pharmacia) and second Mono Q HR5/5, as described previously (12,16).
The active fraction eluted from the second Mono Q column was dialyzed against buffer 2 (20 mM potassium phosphate buffer, pH 7.5, 0.1 mM EDTA, 1 mM 2-mercaptoethanol, 0.25 mM PMSF, 20% ethyleneglycol and 0.02% Triton X-100) containing 100 mM KCl and 50% glycerol, and stored at –80°C.
Determination of partial amino acid sequences of DNA helicase B
Purified DNA helicase B (50 µg) was subjected to 7.5% SDS–PAGE and stained with 0.2% Coomasie brilliant blue R-250. A gel piece containing a 130 kDa protein band was excised and immersed in a buffer containing 125 mM Tris–HCl, pH 6.8, 0.1% SDS, 2.5 mM dithiothreitol, 1 mM EDTA and 20% sucrose for 15 min, twice. Then, the gel piece was subjected to a second SDS–PAGE.
The separating gel for the second SDS–PAGE was 15% polyacrylamide (2 mm thick) containing 375 mM Tris–HCl, pH 8.8, 0.1% SDS and 1 mM EDTA, and the stacking gel was 5% polyacrylamide containing 125 mM Tris–HCl, pH 6.8, 0.1% SDS and 1 mM EDTA (second stacking buffer). Prior to loading of the gel piece, 2 µg of Achromobacter protease I (Lysyl Endopeptidase, Wako) was made to migrate into the stacking gel with a constant current at 65 mA for 10 min using a second stacking buffer for the cathode buffer. The gel piece was applied onto the polyacrylamide gel containing Achromobacter protease I and electrophoresed with a constant current at 30 mA until both the 130 kDa protein and the protease had migrated to the boundary between the stacking gel and the separating gel. The electrophoresis was then stopped for 30 min to allow the 130 kDa protein to be digested and resumed with a constant current at 65 mA for ∼1.5 h.
The digests in the second separating gel were electrically transferred onto Glassy Bond (Biometra) with a constant current at 0.3 mA for 2 h and stained with 0.2% Coomasie brilliant blue R-250. Stained bands on Glassy Bond were excised and analyzed by a Model 477A automated gas-phase protein sequencer (Applied Biosystems).
Screening for cDNA encoding DNA helicase B
The synthetic DNA (10 pmol) encoding one of the determined partial amino acid sequences of DNA helicase B was labeled with 50 pmol of [γ-32P]ATP using 10 U of T4 polynucleotide kinase, and separated from unused ATP by using Microspin S-300 HR (Pharmacia).
The labeled oligodeoxyribonucleotide was used as a probe for screening a poly(dT)-primed fetal mouse brain cDNA library in λgt10. More than 6 × 105 phage plaques transferred to nitrocellulose membranes were screened by plaque hybridization. Prehybridization proceeded at 68°C for 5 h in a solution containing 6× SSC (1× SSC; 150 mM NaCl, 15 mM sodium citrate), 1× Denhardt’s solution (0.02% Ficoll, 0.02% bovine serum albumin, 0.02% polyvinylpyrolidone) and 20 µg/ml of heat-denatured salmon sperm DNA. Hybridization was performed overnight at 60°C in a buffer containing 50 mM Tris–HCl, pH 8.0, 1× Denhardt’s solution, 1 M NaCl, 10 mM EDTA, 0.1% SDS, 50 µg/ml heat-denatured salmon sperm DNA and the 32P-labeled oligodeoxyribonucleotide probe. The membranes were rinsed twice with 2× SSC and 1× blot-wash buffer (1× SSC, 10 mM sodium phosphate and 0.025% SDS) for 10 min each at room temperature and at 68°C. The membranes were air-dried and subjected to autoradiography. Positive plaques were purified further.
Determination of a mutation point of DNA helicase B in tsFT848
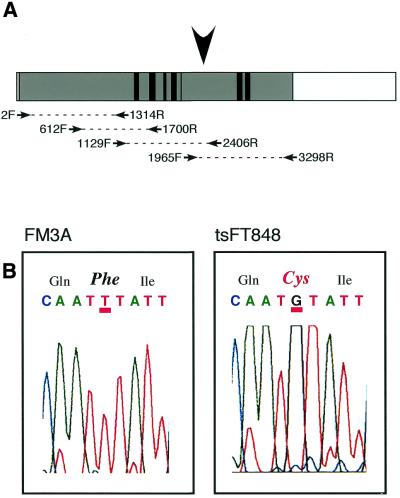
Total RNA was prepared from FM3A and tsFT848 cells with TRIZOL (Gibco). Poly(A)+-RNA derived from wild-type FM3A cells and tsFT848 cells was purified from total RNA with Oligotex-dT30 (Roche). cDNA was prepared from the poly(A)+-RNA by using oligo(dT) primer and First-Strand cDNA Synthesis Kit (Pharmacia). cDNAs encoding DNA helicase B were amplified by PCR with the primers indicated in Figure 3A. Pairs of primers used for the PCR were 2F–1314R, 612F–1700R, 1129F–2406R and 1965F–3298R. The PCR products were subjected to DNA sequencing to determine mutation sites in tsFT848 DNA helicase B.
Figure 3.
Determination of the mutation site of DNA helicase B in tsFT848 cells. (A) Schematic representation of RT–PCR for mapping of the mutation site in tsFT848 DNA helicase. The whole box represents the isolated DNA helicase B cDNA. An open reading frame and regions encoding seven conserved DNA/RNA helicase motifs are shown as shaded and black boxes, respectively. Primers for RT–PCR are indicated as arrows. The arrowhead indicates the position of the mutated nucleotide in tsFT848 DNA helicase B. (B) Analysis of DNA sequences of DNA helicase B cDNA from wild-type and tsFT848 cells. Charts from an automatic DNA sequencer corresponding to amino acids 723–725 of DNA helicase B are shown. Amino acid residues encoded by the nucleotide sequences are indicated on the top of the charts. Amino acid 724 in tsFT848 DNA helicase B is divergent from the wild-type residue and indicated in red.
Other methods
DNA-dependent ATPase activity was measured as described previously (13). One unit of ATPase activity was defined as the amount of enzyme needed to hydrolyze 1 nmol ATP/h at 37°C. Substrate for DNA helicase assay was prepared as described (15) with 21mer oligodeoxyribonucleotide complementary to M13mp19. The reaction and electrophoresis for the DNA helicase assay were carried out as described (15). SDS–PAGE was performed according to Laemmli’s method (19).
RESULTS
Purification of mouse DNA helicase B
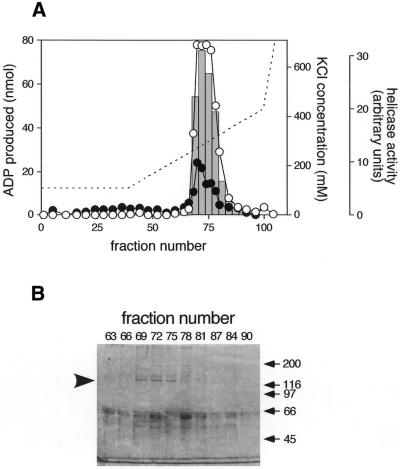
DNA helicase B was purified from 1 × 1011 mouse FM3A cells by sequential column chromatography using DNA-dependent ATPase activity as an index. The elution profile for ATPase activity on the second MonoQ column, the last column used in the purification, is shown in Figure 1A. The ATPase activity was eluted in fractions 68 to 78 with a peak at ∼280 mM KCl and was markedly stimulated by the addition of heat-denatured calf thymus DNA, indicating that the activity was DNA-dependent. The purified fraction contained 280 800 U of DNA-dependent ATPase activity and 331 µg of protein (848 441 U/mg). The specific activity of the DNA-dependent ATPase in the final fraction was increased 3270-fold as compared with that of the crude extract; however, the increase in the purity of DNA helicase B was not clear because of the existence of multiple ATPases in the crude extract. Bars in Figure 1A show DNA helicase activity in the fractions measured by release of 32P-labeled 21mer oligodeoxyribonucleotide from single-stranded circular M13mp19 DNA. A considerable amount of DNA helicase activity was also detected in fractions 69 to 78, suggesting that the protein in the eluted fraction possessed both DNA-dependent ATPase and DNA helicase activities.
Figure 1.
Purification of DNA helicase B. (A) The elution profile of ATPase and DNA helicase activities on the second Mono Q column. Open and closed circles represent ATPase activity in 5 µl of second Mono Q fractions with and without 5 µg of heat-denatured DNA, respectively. Bars in the graph indicate DNA helicase activity in 0.5 µl of the fractions. 32P-labeled 21mer oligodeoxyribonucleotides liberated from M13mp19 single-stranded circular DNA were separated in a 12% polyacrylamide gel. The radioactivity of the liberated oligodeoxyribonucleotides was measured by BAS2000 (Fuji Photo Film) and plotted. A broken line indicates the KCl concentration calculated from the conductivity of the fractions. (B) SDS–PAGE of the Mono Q fractions. The eluted fractions (8 µl) were loaded onto a 7.5% SDS–polyacrylamide gel, and proteins were stained with Coomasie brilliant blue R-250. The arrowhead on the left of the gel indicates the position of the 130 kDa protein. Positions of molecular weight markers are indicated on the right.
Proteins in the eluted fractions were analyzed by SDS–PAGE (Fig. 1B). Although several bands were detected on the gel, only one protein band at 130 kDa showed a good correlation to the activities of DNA-dependent ATPase and DNA helicase. In addition, SDS–PAGE of a fraction obtained by a similar purification process showed a clear 130 kDa band with fewer contaminating bands (16). We presumed that the 130 kDa protein was DNA helicase B, and proceeded to determine partial amino acid sequences of the protein as described in Materials and Methods. Three partial amino acid sequences were determined (Fig. 2, dots) and the longest one (Ile-Lys-Gln-Asp-Phe-Glu-Ser-Ser-Thr-Arg-Leu-X-Asn-Gly-Glu-Ile-Phe-Phe) was used to synthesize DNA probes for screening a cDNA library.
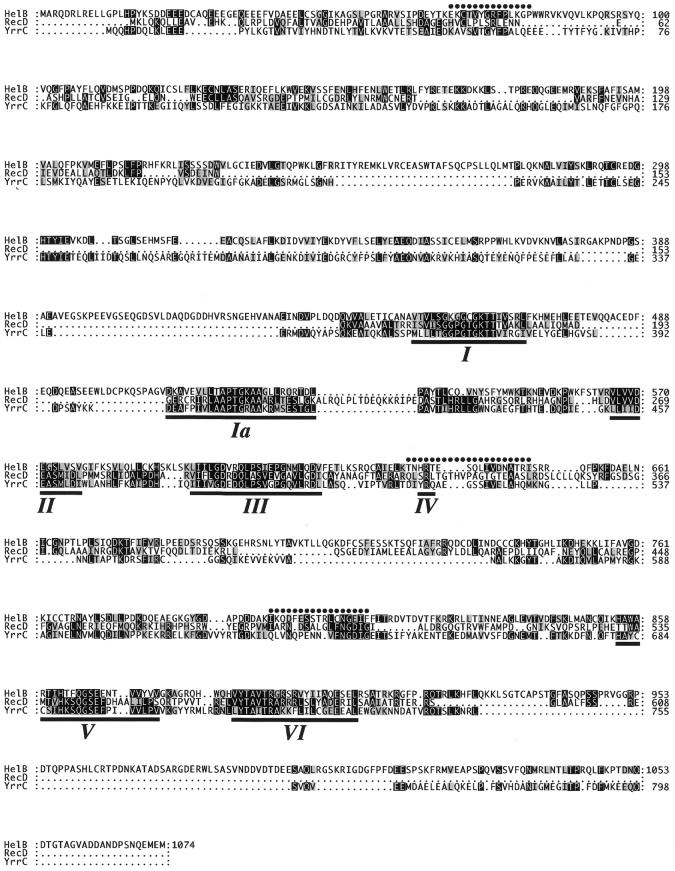
Figure 2.
Amino acid sequence of mouse DNA helicase B. The sequence was deduced from the longest open reading frame of a cDNA isolated from a fetal mouse brain cDNA library. The amino acid sequence of mouse DNA helicase B (HelB) was aligned with that of E.coli RecD protein (RecD) and B.subtilis YrrC protein (YrrC). Identical amino acids are boxed by black squares and similar amino acids are shaded. Partial amino acid sequences determined by using purified protein are indicated above the mouse DNA helicase B sequence by dots. The seven conserved DNA/RNA helicase motifs of mouse DNA helicase B, E.coli RecD protein and B.subtilis YrrC protein are underlined.
Molecular cloning of cDNA encoding mouse DNA helicase B
Possible nucleotide sequences corresponding to the middle of the determined amino acid sequence (Ser-Ser-Thr-Arg-Leu-X) were so diverse that we sequenced this region by RT–PCR using total RNA from FM3A cells. Oligodeoxyribonucleotides encoding six amino acids from the N-terminus (5′-ATI AAA/G CAA/G GAT/C TTT/C GA-3′, where I indicates inosine) and nucleotide sequences in the complementary strand encoding six amino acids from the C-terminus (5′-AAA/G AAI ATT/C TCI CCA/G TT-3′) were synthesized and used for PCR primers. The PCR product migrating at a position of 53 bp on a 12% polyacrylamide gel was extracted and subjected to DNA sequencing. A 53mer oligodeoxyribonucleotide (5′-ATI AAA/G CAA/G GAT/C TTT/C GAA AGC AGC ACC CGC CTG TGC AAT/C GGI GAA/G ATI TTT/C TT 3′) was synthesized according to the sequence and used for screening a fetal mouse brain cDNA library.
The screening of 6 × 105 plaques yielded two positive clones. The cDNA inserts of these clones were subcloned into pUC19 plasmid. Sequencing of these cDNA clones revealed that one clone was a part of the other clone. The longer clone (clone 1) consisted of 3232 bp, and the N-terminus of the protein was not encoded by this cDNA. To obtain cDNA encoding the N-terminus of the protein, DNA was amplified by PCR from the fetal mouse brain cDNA library using a primer corresponding to nucleotides 67 to 84 of clone 1 (5′-TCT GCA TTT GCG ACG TGT-3′) and a primer of 24 nt corresponding to the cloning site of λgt10 (5′-CTT ATG AGT ATT TCT TCC AGG GTA-3′). PCR was performed again using a primer corresponding to nucleotides 38 to 55 of clone 1 (5′-TCT AAC ATG GTC ATC GCC-3′) and the λgt10 primer used in the first PCR. The second PCR product was analyzed by Southern blotting with an oligodeoxyribonucleotide corresponding to nucleotides 1 to 37 of clone 1 as a probe. The largest positive band was subcloned into pUC19 and the DNA sequence was analyzed.
We finally cloned a 4464 bp cDNA with an open reading frame of 3222 bp. The result of DNA sequence analysis was confirmed more than three times with DNA products amplified by PCR from the cDNA library or by RT–PCR of FM3A mRNA. The amino acid sequence deduced from the open reading frame is shown in Figure 2. The open reading frame encodes a protein of 1074 amino acids with a predicted molecular weight of 121 483.5. All of the three determined partial amino acid sequences are contained in the amino acid sequence (Fig. 2, dots), indicating that the cDNA clone encodes the 130 kDa protein that we purified.
A search of amino acid sequence databases indicated that the protein had similarity to the YrrC protein of Bacillus subtilis and the α subunit of exodeoxyribonuclease V of several prokaryotes including E.coli RecD protein (Fig. 2), although no significant similarity was detected with known yeast proteins. The identity and similarity of amino acid sequences between mouse DNA helicase B and E.coli RecD protein are 11.1 and 16.9%, respectively, while between mouse DNA helicase B and B.subtilis YrrC protein they are 16.5 and 26.0%, respectively. The similarity revealed seven conserved helicase motifs, I, Ia, II, III, IV, V and VI (20), in the deduced amino acid sequence (Fig. 2, underlined), suggesting that the protein encoded by the isolated cDNA possessed DNA/RNA helicase activity. Accordingly, we concluded that the cDNA is the one encoding DNA helicase B. The motifs in mouse DNA helicase B are 46.8% identical and 66.1% similar to the motifs in E.coli RecD protein, and 42.7% identical and 71.0% similar to the motifs in B.subtilis YrrC protein.
Determination of a mutation site of DNA helicase B in tsFT848 cells
We previously reported that partially purified DNA helicase B from tsFT848 cells, a temperature-sensitive mutant of mouse FM3A cells, was more heat-labile than wild-type enzyme at the non-permissive temperature (12). Thus, we next investigated the mutation site(s) of DNA helicase B of the mutant cell. The four pairs of PCR primers indicated in Figure 3A were used and each segment of the cDNA was amplified by RT–PCR from poly(A)+-RNA prepared from FM3A and tsFT848 cells. Amplified DNAs were directly subjected to DNA sequence analysis. Every segment was amplified and analyzed using different preparations of poly(A)+-RNA more than three times to confirm the results.
The results revealed that only one nucleotide of mutant DNA helicase B cDNA was divergent from the wild-type. The charts obtained with the automated DNA sequencer are shown in Figure 3B. The thymine residue at 2199 in wild-type DNA helicase B cDNA was changed to guanine, resulting in the conversion of phenylalanine at 724 to cysteine. This position resides in between helicase motifs IV and V (Fig. 3A, arrowhead).
DISCUSSION
In this study, we have purified DNA helicase B, determined three partial amino acid sequences of the helicase, and cloned cDNAs encoding the enzyme by using a DNA probe prepared based on the longest of the partial amino acid sequences. The isolated cDNA was 4464 bp long and contained a 3222 bp open reading frame encoding a 1074 amino acid protein with a molecular weight of 121 483.5. The amino acid sequence deduced from the open reading frame contained the three partial sequences and seven motifs conserved in the DNA/RNA helicase superfamily, indicating that the cDNA actually encodes DNA helicase B.
A search of protein sequence databases revealed that the helicase motifs of DNA helicase B share remarkably high homology with those of the YrrC protein of B.subtilis and the α subunit of exodeoxyribonuclease V of various prokaryotes including E.coli. Although the homology is mainly observed in the helicase motifs, several homologous regions can be seen outside the motifs. Since helicase motifs share only limited homology between DNA/RNA helicases in general, it seems likely that DNA helicase B and the α subunit of exonuclease V in prokaryotes are structural homologs that have evolved from the same ancestor. On the other hand, it cannot plausibly be argued that mouse DNA helicase B is a functional homolog of E.coli RecD, because it acts as a DNA helicase without forming a hetero-complex with other proteins such as RecB or RecC. It is important to reveal the necessity of a DNA helicase containing a RecD-like structure in higher eukaryotes.
DNA helicase B in tsFT848 cells was inactivated by incubating cells at a non-permissive temperature, and a partially purified form from the mutant cells was inactivated more rapidly than that from wild-type cells upon incubation at 40°C. Thus, we assumed that DNA helicase B of the mutant cells carried a mutation and this mutation was responsible for the defect in DNA replication (12). Molecular cloning of the cDNA encoding the helicase allowed us to identify mutation points in tsFT848 cells. The mutation point of the DNA helicase B of tsFT848 cells was located in the sequence between motifs IV and V. Since this intervening sequence is very long compared with other intervening sequences, it seems likely that this region plays an important role in maintaining the spatial relationship between DNA helicase motifs I–IV and motifs V–VI, and at a non-permissive temperature, the spatial relationship is destroyed due to the mutation.
The physiological function of DNA helicase B seems to be related to DNA replication, but is still unknown. To obtain more information about the function of DNA helicase B, we analyzed the arrest point of the mutant cells in the cell cycle by immunostaining of chromatin-bound Mcm4, the staining patterns of which are gradually changed as S-phase proceeds. The result indicated that tsFT848 cells were arrested at early S-phase at a non-permissive temperature, suggesting that the helicase functions in the early stage of S-phase (Y.Kusa, T.Kobayashi, M.Seki, S.Tada and T.Enomoto, unpublished observation). One possible role of the enzyme is to function as a replicative helicase that unwinds double-stranded DNA ahead of the replication machinery. The MCM complex is known to be important in some aspects of DNA replication and to have DNA helicase activity (11,21,22), suggesting that the protein complex acts as a replicative DNA helicase. Nevertheless, it is possible that there are other DNA helicases that move ahead of the DNA replication forks, because the processivity of the helicase in the MCM complex is low. Our preliminary results, however, indicated that DNA helicase B was not necessarily localized to replication sites as revealed by staining with an anti-BrdU antibody (T.Kobayashi, M.Seki, S.Tada and T.Enomoto, unpublished observation).
Another possible function for the helicase is to remove barriers ahead of the DNA replication machinery, which may include nucleosome structures. Indeed, the RecBCD complex from E.coli was reported to be able to unwind DNA with a nucleosomal structure (23). T4 Dda, which is a DNA helicase with similarities to E.coli RecD (24), was able to overcome the inhibitory function of nucleosomes in the T4 bacteriophage DNA replication system (25). In addition, a T4 bacteriophage with a mutation in the dda gene is defective in DNA replication despite the fact that the replicative DNA helicase of the bacteriophage is not Dda but the gene41 product. Thus, because of the similarity of DNA helicase B to RecD, it is conceivable that DNA helicase B functions to remove barriers ahead of the DNA replication machinery.
There are a number of possible stages during S-phase in which the DNA helicase could be involved. It also cannot be ruled out that the enzyme is involved in the resolution of duplicated chromosomes together with DNA topoisomerases. We believe that elucidation of the function of DNA helicase B will uncover a new aspect of DNA metabolism that employs DNA helicase with significant structural similarity to prokaryotic proteins.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Dr J. Julian Blow and Dr Ellen Fanning for useful comments and suggestions in preparing this manuscript. This work was supported by Grants-in-Aid for Scientific Research and for Scientific Research on Priority Areas from the Ministry of Education, Science, Sports and Culture of Japan, and Health Sciences Research Grants from the Ministry of Health and Welfare of Japan.
DDBJ/EMBL/GenBank accession no. AB048542
References
- 1.Matson S.W. and Kaiser-Rogers,K.A. (1990) DNA helicases. Annu. Rev. Biochem., 59, 289–329. [DOI] [PubMed] [Google Scholar]
- 2.Weeda G., van Ham,R.C., Vermeulen,W., Bootsma,D., van der Eb,A.J. and Hoeijmakers,J.H. (1990) A presumed DNA helicase encoded by ERCC-3 is involved in the human repair disorders xeroderma pigmentosum and Cockayne’s syndrome. Cell, 62, 777–791. [DOI] [PubMed] [Google Scholar]
- 3.Weber C.A., Salazar,E.P., Stewart,S.A. and Thompson,L.H. (1990) ERCC2: cDNA cloning and molecular characterization of a human nucleotide excision repair gene with high homology to yeast RAD3. EMBO J., 9, 1437–1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schaeffer L., Roy,R., Humbert,S., Moncollin,V., Vermeulen,W., Hoeijmakers,J.H., Chambon,P. and Egly,J.M. (1993) DNA repair helicase: a component of BTF2 (TFIIH) basic transcription factor. Science, 260, 58–63. [DOI] [PubMed] [Google Scholar]
- 5.Schaeffer L., Moncollin,V., Roy,R., Staub,A., Mezzina,M., Sarasin,A., Weeda,G., Hoeijmakers,J.H. and Egly,J.M. (1994) The ERCC2/DNA repair protein is associated with the class II BTF2/TFIIH transcription factor. EMBO J., 13, 2388–2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Enomoto T. (2001) Function of RecQ family helicases: possible involvement of Bloom’s and Werner’s syndrome gene products in guarding genome integrity during DNA replication. J. Biochem., 129, 501–507. [DOI] [PubMed] [Google Scholar]
- 7.Karow J.K., Wu,L. and Hickson,I.D. (2000) RecQ family helicases: roles in cancer and aging. Curr. Opin. Genet. Dev., 10, 32–38. [DOI] [PubMed] [Google Scholar]
- 8.Chakraverty R.K. and Hickson,I.D. (1999) Defending genome integrity during DNA replication: a proposed role for RecQ family helicases. Bioessays, 21, 286–294. [DOI] [PubMed] [Google Scholar]
- 9.Kitao S., Shimamoto,A., Goto,M., Miller,R.W., Smithson,W.A., Lindor,N.M. and Furuichi,Y. (1999) Mutations in RECQL4 cause a subset of cases of Rothmund-Thomson syndrome. Nature Genet., 22, 82–84. [DOI] [PubMed] [Google Scholar]
- 10.Tye B.K. (1999) MCM proteins in DNA replication. Annu. Rev. Biochem., 68, 649–686. [DOI] [PubMed] [Google Scholar]
- 11.Ishimi Y. (1997) A DNA helicase activity is associated with an MCM4, -6, and -7 protein complex. J. Biol. Chem., 272, 24508–24513. [DOI] [PubMed] [Google Scholar]
- 12.Seki M., Kohda,T., Yano,T., Tada,S., Yanagisawa,J., Eki,T., Ui,M. and Enomoto,T. (1995) Characterization of DNA synthesis and DNA-dependent ATPase activity at a restrictive temperature in temperature-sensitive tsFT848 cells with thermolabile DNA helicase B. Mol. Cell. Biol., 15, 165–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Seki M., Enomoto,T., Watanabe,Y., Tawaragi,Y., Kawasaki,K., Hanaoka,F. and Yamada,M. (1986) Purification and characterization of a deoxyribonucleic acid dependent adenosinetriphosphatase from mouse FM3A cells: effects of ribonucleoside triphosphates on the interaction of the enzyme with single-stranded DNA. Biochemistry, 25, 3239–3245. [DOI] [PubMed] [Google Scholar]
- 14.Seki M., Enomoto,T., Hanaoka,F. and Yamada,M. (1987) DNA-dependent adenosinetriphosphatase B from mouse FM3A cells has DNA helicase activity. Biochemistry, 26, 2924–2928. [DOI] [PubMed] [Google Scholar]
- 15.Seki M., Enomoto,T., Yanagisawa,J., Hanaoka,F. and Ui,M. (1988) Further characterization of DNA helicase activity of mouse DNA-dependent adenosinetriphosphatase B (DNA helicase B). Biochemistry, 27, 1766–1771. [DOI] [PubMed] [Google Scholar]
- 16.Saitoh A., Tada,S., Katada,T. and Enomoto,T. (1995) Stimulation of mouse DNA primase-catalyzed oligoribonucleotide synthesis by mouse DNA helicase B. Nucleic Acids Res., 23, 2014–2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ishimi Y. and Matsumoto,K. (1993) Model system for DNA replication of a plasmid DNA containing the autonomously replicating sequence from Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA, 90, 5399–5403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matsumoto K., Seki,M., Masutani,C., Tada,S., Enomoto,T. and Ishimi,Y. (1995) Stimulation of DNA synthesis by mouse DNA helicase B in a DNA replication system containing eukaryotic replication origins. Biochemistry, 34, 7913–7922. [DOI] [PubMed] [Google Scholar]
- 19.Laemmli U.K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature, 227, 680–685. [DOI] [PubMed] [Google Scholar]
- 20.Gorbalenya A.E., Koonin,E.V., Donchenko,A.P. and Blinov,V.M. (1989) Two related superfamilies of putative helicases involved in replication, recombination, repair and expression of DNA and RNA genomes. Nucleic Acids Res., 17, 4713–4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ishimi Y., Komamura,Y., You,Z. and Kimura,H. (1998) Biochemical function of mouse minichromosome maintenance 2 protein. J. Biol. Chem., 273, 8369–8375. [DOI] [PubMed] [Google Scholar]
- 22.You Z., Komamura,Y. and Ishimi,Y. (1999) Biochemical analysis of the intrinsic Mcm4-Mcm6-mcm7 DNA helicase activity. Mol. Cell. Biol., 19, 8003–8015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eggleston A.K., O’Neill,T.E., Bradbury,E.M. and Kowalczykowski,S.C. (1995) Unwinding of nucleosomal DNA by a DNA helicase. J. Biol. Chem., 270, 2024–2031. [DOI] [PubMed] [Google Scholar]
- 24.Hacker K.J. and Alberts,B.M. (1992) Overexpression, purification, sequence analysis, and characterization of the T4 bacteriophage dda DNA helicase. J. Biol. Chem., 267, 20674–20681. [PubMed] [Google Scholar]
- 25.Bonne-Andrea C., Wong,M.L. and Alberts,B.M. (1990) In vitro replication through nucleosomes without histone displacement. Nature, 343, 719–726. [DOI] [PubMed] [Google Scholar]