DNA binding specificity of floral MADS domain protein complexes depends on protein complex composition and provides clues to the mechanisms underlying target gene specificity.
Abstract
Floral organ identities in plants are specified by the combinatorial action of homeotic master regulatory transcription factors. However, how these factors achieve their regulatory specificities is still largely unclear. Genome-wide in vivo DNA binding data show that homeotic MADS domain proteins recognize partly distinct genomic regions, suggesting that DNA binding specificity contributes to functional differences of homeotic protein complexes. We used in vitro systematic evolution of ligands by exponential enrichment followed by high-throughput DNA sequencing (SELEX-seq) on several floral MADS domain protein homo- and heterodimers to measure their DNA binding specificities. We show that specification of reproductive organs is associated with distinct binding preferences of a complex formed by SEPALLATA3 and AGAMOUS. Binding specificity is further modulated by different binding site spacing preferences. Combination of SELEX-seq and genome-wide DNA binding data allows differentiation between targets in specification of reproductive versus perianth organs in the flower. We validate the importance of DNA binding specificity for organ-specific gene regulation by modulating promoter activity through targeted mutagenesis. Our study shows that intrafamily protein interactions affect DNA binding specificity of floral MADS domain proteins. Differential DNA binding of MADS domain protein complexes plays a role in the specificity of target gene regulation.
INTRODUCTION
The exact molecular mechanisms of how most transcription factors (TFs) achieve their DNA binding specificity are largely unknown. Most intriguing are the questions how closely related TFs control distinct biological processes and how heteromeric complex formation affects functional specificity. DNA binding specificity of proteins stems from primary DNA sequence and its structural properties (Rohs et al., 2009). Another aspect that contributes to functional specificity comes from the ability of TFs to form higher-order protein complexes. The protein interactions potentially modify the DNA binding affinity of individual members of the complex. For example, interactions of TFs from the same family with a common cofactor can evoke latent differences in DNA binding specificities (Slattery et al., 2011). Moreover, the formation of heterodimeric complexes between TFs of the same or different families results in the recognition of novel, composite DNA TF binding sites (TFBSs) (Jolma et al., 2013; Jolma et al., 2015; Bemer et al., 2017). The interplay of these molecular mechanisms could influence the DNA binding specificity of plant MADS domain TFs that are known to form a complex intrafamily protein interaction network.
MADS domain TFs are present in all major eukaryotic lineages. Especially in plants, they form a large family, e.g., of more than 100 members in the flowering plant Arabidopsis thaliana (Parenicová et al., 2003). MADS domain TFs have important roles in the regulation of many developmental processes (Smaczniak et al., 2012a). Remarkably, some MADS domain proteins acquired several distinct functions in different organs related with their ability to interact with other family members in a combinatorial manner. To explain the variety of regulatory functions of MADS domain proteins, we need to understand how the formation of heteromeric protein complexes affect their DNA binding specificity and target gene regulation. Previous studies showed that MADS domain proteins bind DNA sequence elements called CArG-boxes (consensus CC[A/T]6GG) (Pollock and Treisman, 1990; Schwarz-Sommer et al., 1992; Huang et al., 1993; Riechmann et al., 1996a, 1996b) by means of their highly conserved, 56-amino acid N-terminal DNA binding MADS domain (Schwarz-Sommer et al., 1990). Thousands of CArG-boxes are present in the genome of Arabidopsis, many of which seem not to be bound by MADS domain TFs (de Folter and Angenent, 2006; Kaufmann et al., 2009; Muiño et al., 2014). Besides that, a large fraction of genomic regions bound in vivo do not contain consensus CArG-box sequences (Kaufmann et al., 2009, 2010; Zheng et al., 2009; Deng et al., 2011). A good example how combinatorial protein interactions determine regulatory specificity of plant MADS domain proteins is flower development. According to the “floral quartet” model, each type of floral organ is specified by a distinct combination of MADS domain proteins that form quaternary protein complexes and bind two CArG-boxes (a protein dimer contacts a single TFBS in such tetrameric complex) in the regulatory regions of target genes (Theissen and Saedler, 2001). The interactions of Arabidopsis MADS domain proteins suggested in the floral quartet model as well as interactions with other TFs and chromatin-associated proteins were characterized in vitro and in vivo (Honma and Goto, 2001; Melzer and Theissen, 2009; Melzer et al., 2009; Smaczniak et al., 2012b). Therefore, part of the functional specificity may come from the ability of MADS domain TFs to form homo- and heteromeric protein complexes, which has been proposed before (Krizek and Meyerowitz, 1996; Riechmann et al., 1996b). The functional analysis of chimeric proteins, where MADS domain swaps between plant and human MADS domain TFs are able to rescue floral homeotic mutants, suggests, somewhat controversially, that their functional specificity is independent of DNA binding specificity (Riechmann and Meyerowitz, 1997; Krizek et al., 1999). However, recent genome-wide in vivo DNA binding for homeotic MADS domain proteins (Wuest et al., 2012; ÓMaoiléidigh et al., 2013; Pajoro et al., 2014) show between 30 and 60% nonoverlapping TFBSs (Yan et al., 2016). Also, the DNA looping may affect binding preferences, as shown in vitro for SEPALLATA homotetrameric complexes (Jetha et al., 2014). Together, these results indicate a complex recognition mechanism that depends on the DNA binding specificities of MADS domain TF dimers and their higher-order interactions.
Using systematic evolution of ligands by exponential enrichment followed by high-throughput sequencing (SELEX-seq), we determined DNA binding specificities of selected MADS domain protein complexes in vitro. Here, we show that MADS domain protein homodimers SEPALLATA3 (SEP3)-SEP3, AGAMOUS (AG)-AG, and APETALA1 (AP1)-AP1 and organ-specific heterodimers SEP3-AG (reproductive organs) and SEP3-AP1 (perianth) bind DNA sequences with different specificities and affinities. In particular, the identity specification of reproductive organs is linked with unique DNA binding preferences of SEP3-AG dimers, resulting in a set of target genes not bound by other MADS domain proteins. More generally, we show that differences in DNA binding specificities can discriminate between complex-specific in vivo DNA TFBSs (as identified by chromatin immunoprecipitation-sequencing [ChIP-seq] experiments). This approach allows us to modify the specificity of the native DNA TFBSs of MADS domain proteins. As a proof of concept, we modified TFBSs in the AP3 promoter to modulate organ-specific gene expression in vivo.
RESULTS
DNA Binding Specificities of Floral MADS Domain TF Complexes
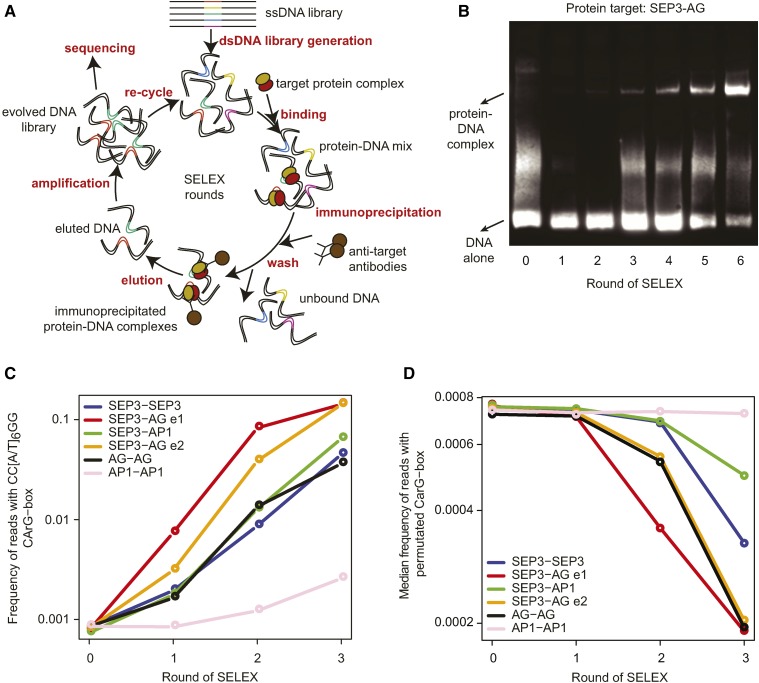
To determine DNA binding specificities of individual MADS domain TF complexes, we followed a SELEX-seq approach (Figure 1), in which we made use of in vitro transcribed/translated MADS domain protein complexes and double-stranded DNA (dsDNA) libraries containing a 20-bp region of randomized nucleotides (Jolma et al., 2010). Starting with this random pool of dsDNA, we isolated DNA sequences bound by MADS domain TF complexes by immunoprecipitation with immobilized, protein-specific antibodies (Figure 1A). For each MADS domain TF dimer combination, we performed at least three rounds of SELEX and characterized the evolved pools of sequences after each round by high-throughput DNA sequencing (Supplemental Table 1) and electrophoretic mobility shift assays (EMSAs) (Figure 1B). Heteromeric complexes were not separated from homomeric complexes during our SELEX experiment. However, our earlier data showed that SEP3 and AG or AP1 proteins, when incubated with the DNA, form predominantly heteromeric rather than homomeric complexes (Smaczniak et al., 2012b). This was confirmed by EMSA experiments using the SELEX-enriched DNA and AP1 or AG protein complexes with a truncated SEP3 protein (Supplemental Figure 1). The predominant formation of heteromeric complexes with specific DNA binding preferences is also confirmed here by high reproducibility of the SELEX-seq-derived affinities of the SEP3-AG complex when we used either the SEP3 or the AG antibody (Supplemental Figure 2). Furthermore, we observed a specific enrichment pattern of sequences for SEP3-AG heterodimers that is neither strong for the SEP3 homodimer nor for the AG homodimer (Figure 2).
Figure 1.
SELEX-Seq for MADS Domain Protein Complexes.
(A) Overview of the experimental setup for the SELEX-seq approach performed in this study.
(B) EMSA analysis of the DNA libraries obtained in different rounds of SELEX for the SEP3-AG complex.
(C) Enrichment of the putative CArG-box consensus sequence (CC[A/T]6GG) in the SELEX-seq rounds (log scale). Frequencies (in %) at Round 3 are: 14.2%, 14.1%, 6.4%, 4.5%, 3.6%, and 0.3% for SEP3-AG e1, SEP3-AG e2, SEP3-AP1, SEP3-SEP3, AG-AG, and AP1-AP1 respectively.
(D) Enrichment of randomly permutated CArG-box sequences in the SELEX-seq rounds (log scale). SEP3-AG e1 and SEP3-AG e2 indicate two independent experiments where different antibodies were used for immunoprecipitation, SEP3 antibodies (e1), and AG antibodies (e2).
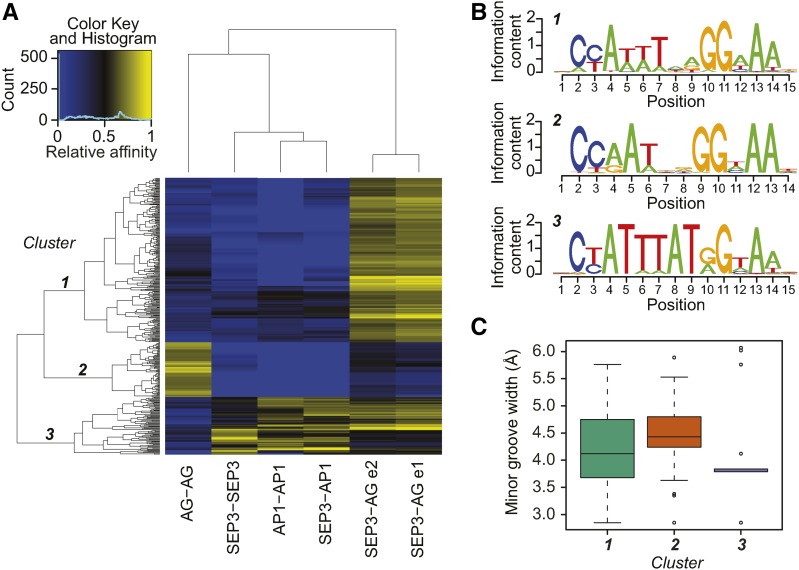
Figure 2.
DNA Binding Specificities of MADS Domain TF Complexes.
(A) Relative affinity heat map based on 12-mer sequences enriched in the 3rd round of SELEX for all studied MADS domain TF complexes. Each line in the heat map corresponds to a single 12-mer DNA fragment. High relative affinities for a particular sequence are marked in yellow and low relative affinities are in blue.
(B) Sequence logos corresponding to the three main clusters of sequences in the heat map built from the position weight matrices for all 20N sequences containing group specific 12-mers.
(C) Minimal DNA minor groove width predictions of the sequences from clusters 1 to 3. The mean value differences of the minor groove width are significant with P < 0.05 (t test) for all pairwise comparisons. Numbers of sequences used in predictions (sample size) are 1,298,340, 416,077, and 705,199 for clusters 1, 2, and 3, respectively.
To estimate affinities of MADS domain TF dimers to the DNA fragments, we used sequencing data of the initial randomized dsDNA libraries (Round 0 [R0]) and calculated the relative enrichments between R1-R3 of SELEX and R0 (Slattery et al., 2011). High-throughput sequencing of the first three rounds of SELEX (R1–R3) for the various MADS domain protein combinations showed enrichment of the generic MADS domain TFBS consensus in the evolved pools of sequences (Figure 1C). The enrichment of a generic CArG-box in the SELEX-seq for the AP1 homodimer was lower compared with any other studied MADS domain protein complex (Figure 1C). This suggests that either the AP1 homodimer binds more weakly to DNA compared with other MADS domain protein complexes, as was observed before in EMSA experiments on individual DNA probes (Smaczniak et al., 2012b), or AP1 homodimers are formed with lower efficiency. In comparison, we did not see any enrichment of randomly permutated CArG-box sequence fragments in the enriched DNA sequence pools, confirming DNA binding specificity of MADS domain proteins (Figure 1D).
SELEX-Seq Analyses Reveal DNA Binding Preferences of Floral Homeotic Protein Complexes
To determine the DNA binding preferences for MADS domain TF complexes independently of the presumed consensus CArG-box, we follow the methodology proposed by Slattery et al. (2011). We estimated the optimal length of the k-mer (subsequence of a length k) from which we can accurately predict the relative DNA binding affinities in the SELEX-seq data. We based our analysis on k-mers and not on the full-length 20-bp sequences because, in theory, there are more than 1012 possible combinations of unique 20-bp sequences, and our sequencing libraries contain on average 2.2 M reads (Supplemental Table 1). Depending on the library, the optimal length of the k-mer varied between 9 and 12 bp (Supplemental Figure 3). For further analyses, we chose k-mer length of 12 bp that had the highest information gain score in most of the libraries. Then, we calculated the relative DNA binding affinities using the ratio between frequencies of k-mers in R3 and R0 of SELEX. Owing to a large diversity of sequences in the randomized initial libraries (R0), their k-mer frequencies are low and, therefore, difficult to estimate directly from the sequencing results. For this reason, we used a Markov model to estimate the best frequency of R0. This workflow allowed us to calculate relative DNA binding affinities of MADS domain protein complexes to k-mers of length 12. We clustered those relative affinities in a heat map, highlighting diverse and overlapping binding preferences (Figure 2A). This analysis could differentiate the complexes based on their DNA binding preferences. Sequences in cluster 1 were specific for SEP3-AG, sequences in cluster 2 were specific for AG alone, while sequences in cluster 3 had mixed specificity mainly for SEP3, SEP3-AP1, and AP1 that could be further divided into three additional clusters 3a to 3c (Supplemental Figure 4). In detail, sequences in cluster 3a were not enriched for SEP3; sequences in cluster 3b were not enriched for SEP3-AG, while sequences in cluster 3c were specific to SEP3 alone. Remarkably, more k-mers were specifically enriched for SEP3-AG or for AG than for the other proteins and protein combinations. Also, the SEP3-AG complex seemed to bind a wide range of sequences including, but not limited to, the ones that were also bound by SEP3-AP1 or SEP3 complexes. This shows that MADS domain protein complexes bind overlapping sets of TFBSs and suggests a presence of active competition for those common sites when multiple proteins are expressed in the same tissues. However, SEP3-AG and AG complexes showed additional specific DNA binding capacities, which may suggest that reproductive organ specification by AG and SEP3 involves in vivo binding to specific target genes not bound by other MADS domain proteins.
To validate SELEX-seq-derived relative affinities for 12-mer sequences represented in the heat map, we performed quantitative multiple fluorescence relative affinity (quMFRA) assays (Man and Stormo, 2001). For these experiments, we used 20-bp fragments (plus barcodes and flanking regions with labeled primers) obtained from sequenced SELEX libraries that contained a particular 12-mer sequence (Supplemental Tables 2 and 3). Our SELEX and quMFRA approaches gave very similar results with a correlation coefficient (R2) between 0.6 and 0.8 (Supplemental Figure 5 and Supplemental Data Set 1). Additionally, for five selected 20-bp SELEX fragments and three protein complexes, we performed absolute protein-DNA binding affinity estimation by EMSA. We found that four out of five measured dissociation constants (Kd) were following relative affinity values observed in the SELEX-seq: High SELEX-seq relative affinity should relate to low Kd (Supplemental Figure 6 and Supplemental Data Set 2). Differences could be caused by the fact that SELEX-seq-derived relative affinities are based on the 12-mer sequences, while quMFRA and Kd estimations by EMSA were performed with the full-length 20-bp fragments containing a particular 12-mer and its flanking regions.
To identify consensus TFBSs of MADS domain protein complexes in each cluster, we extracted all full-length 20-bp fragments from the R3 sequencing data that contained specific 12-mer sequences and performed DNA motif discovery using the GADEM algorithm (Li, 2009). Comparison between different motifs revealed differences in nucleotide composition and number of consecutive nucleotides between different MADS domain TFBSs mainly in the central, A/T-rich region (Figure 2B). We observed that the motif for AG has the A/T-rich region length between 3 and 5 bp (cluster 2), while the A/T-rich region for SEP3, AP1, and SEP3-AP1 is longer, 6 to 8 bp (cluster 3). Especially long A/T-rich regions were found for sequences in the cluster 3c, which are specific for SEP3 alone (Supplemental Figure 4). The SEP3-AG dimer showed a binding preference (cluster 1) intermediate between SEP3-SEP3 and AG-AG dimers. The analysis of the sequence enrichment for all 64 possible variations of the consensus CArG-box allowing one nucleotide change each in the A/T-rich region of the CC[A/T]6GG sequence confirmed that different MADS domain protein complexes have different binding preferences, even in the case of the generic CArG box (Supplemental Figure 7).
Presence of additional nucleotides flanking the CArG-box region in the consensus motifs also constitutes important information for TFBS characterization. TTN- and -NAA nucleotide sequences were the prevalent flanking sites of the CArG-box motif. Moreover, the analysis of the 12-mer fragment enrichment revealed that MADS domain protein complexes bound non-CArG-box-like sequences. Examples of these sequences are listed in Supplemental Table 2 (confirmed by EMSA-based, quMFRA experiments provided in Supplemental Data Set 1) and detailed tables with relative affinities to all k-mers are available from the Gene Expression Omnibus database. Sequences with high relative affinity to AG complexes showed the strongest deviation from the generic CArG-box consensus, e.g., seq1 for AG, or seq5 for the SEP3-AG complex. On the other hand, sequences with high relative affinity to AP1 complexes, e.g., seq15 for SEP3-AP1 or seq12 for AP1-AP1, showed similarity to the generic CArG-box. This shows that TFBSs for AG and its heteromeric complexes differ strongly from the consensus CArG-box.
Because we found previously an association of DNA binding affinity of MADS domain TFs with structural parameters (e.g., narrower minor DNA groove width) (Muiño et al., 2014), we used the DNAshape tool to predict DNA structural features (Zhou et al., 2013) for our SELEX-seq data. For example, this allowed us to compare DNA minor groove width (MGW) between sequence clusters bound by different protein complexes (Figure 2C). We observed narrowing of the DNA minor groove for sequences more specific to SEP3 or SEP3-AP1 complexes (cluster 3), while the MGW for sequences specific for AG alone was the widest. Moreover, the roll and helix twist are the two DNA shape properties that mainly differed between studied sequences in clusters 1 to 3 (Supplemental Figure 8). Sequences with the longer A/T-rich region specific for SEP3 and SEP3-AP1 complexes have narrower minor groove and higher roll values compared to sequences with the shorter A/T-rich region specific for AG (P < 0.05; t test). These parameters determine intrinsic bending of the DNA (Dickerson, 1998; Rohs et al., 2009), which supports the idea that the bend level of the DNA might play a role in the DNA sequence selection by MADS domain TFs. This phenomenon was observed before for other types of TFs (Nelson and Laughon, 1990; Kneidl et al., 1995; Stella et al., 2010). The increased degree of bending of the prebent DNA sequence toward the minor groove was observed in the crystal structure of the mammalian MADS domain protein SRF bound to DNA (Pellegrini et al., 1995) and in in vitro experiments on Arabidopsis MADS domain TFs (Riechmann et al., 1996a; West et al., 1997, 1998). Our results suggest that these parameters not only influence DNA recognition by plant MADS domain TFs in general, but also contribute to DNA binding specificity.
Combining SELEX-Seq and ChIP-Seq to Predict Organ-Specific Targets of Floral Homeotic Protein Complexes
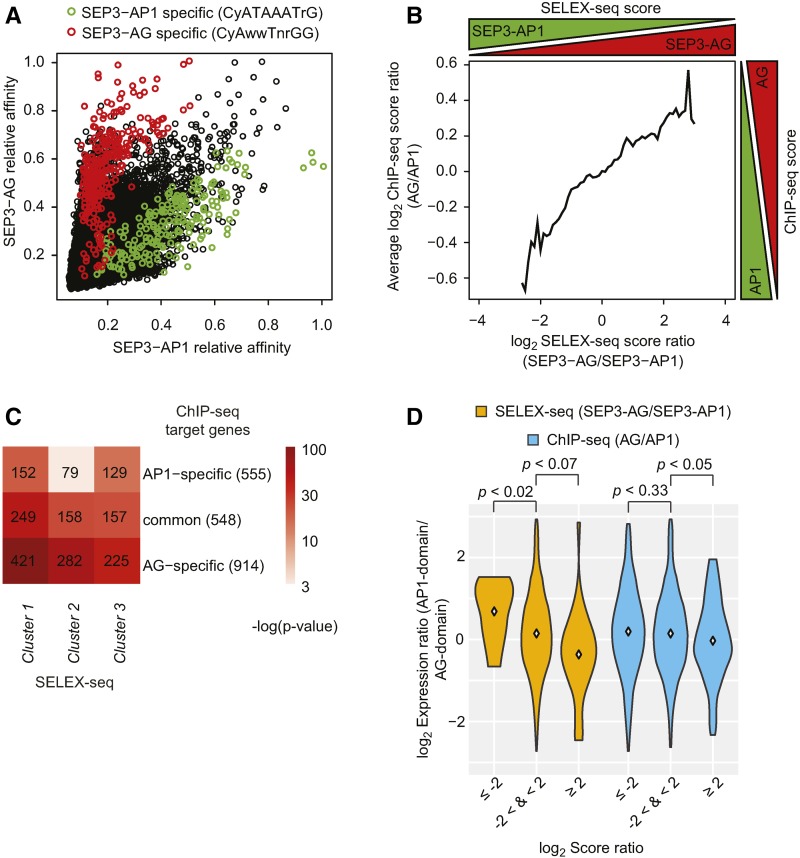
Since SELEX-seq data can separate the DNA binding specificities of different MADS domain protein complexes, we used this information to classify SEP3 binding events obtained by ChIP-seq (flower development stage 4-5 for SEP3, 4 d after induction) (Pajoro et al., 2014) depending on whether they were bound by SEP3-AP1 or SEP3-AG complexes (Figure 3). We chose these two dimers because they distinguish regulatory programs controlling the formation of perianth (sepals, and in combination with AP3/PI petals) and reproductive organs (carpels, and in combination with AP3/PI stamens), respectively. Among sequences bound by SEP3-AG and SEP3-AP1 complexes in SELEX-seq, we could distinguish the ones that are either more SEP3-AG or SEP3-AP1 specific (Figure 3A) based on the identified consensus sequences from Figure 2B. This prompted us to build a classifier to identify SEP3-AG or SEP3-AP1-specific SEP3 in vivo DNA TFBSs.
Figure 3.
Comparison between in Vitro SELEX-Seq and in Vivo ChIP-Seq.
(A) DNA specificity plots comparing the relative binding affinities of SELEX-seq sequences selected by SEP3-AG (y axis) and SEP3-AP1 (x axis). Each point represents a unique sequence that contains a color-coded motif. Black points represent all sequences.
(B) Association between SELEX-seq normalized score ratios (SEP3-AG/SEP3-AP1) and ChIP-seq normalized score ratios (AG/AP1) for TFBSs within the top 1500 SEP3 ChIP-seq peaks. The plot represents a moving average of overlapping windows of size 1 over the SELEX-seq log2 score ratio. Windows with less than five elements were not considered.
(C) Prediction of specific and common ChIP-seq target genes for AG and AP1 based on the SELEX-seq complex-specific motifs. The heat map shows significance (hypergeometric test) of the enrichment of SELEX-seq cluster motifs obtained in Figure 2C in specific and common ChIP-seq binding regions compared previously by Yan et al. (2016); numerical values represent gene numbers. For the raw data of this figure, see Supplemental Data Set 4.
(D) Comparison of the TRAP-seq expression data (Jiao and Meyerowitz, 2010) with the SELEX-seq and the ChIP-seq data (ÓMaoiléidigh et al., 2013; Pajoro et al., 2014). The violin plot visualizes the distribution of AP1-/AG-domain expression ratios of genes containing TFBSs with a certain SEP3-AG/SEP3-AP1 SELEX-seq score ratio (orange) or with a certain AG/AP1 ChIP-seq score ratio (blue).
To classify SEP3 binding events obtained by ChIP-seq depending on their specificity, we defined a score function (see Methods) to annotate positions in the Arabidopsis genome based on the SELEX-seq-inferred DNA binding specificities. Comparison of available ChIP-seq data of AP1 and AG (flower development stages 4–5 for AP1 and 5–6 for AG, 4 and 5 d after induction, respectively) (ÓMaoiléidigh et al., 2013; Pajoro et al., 2014) within 1500 most enriched SEP3 ChIP-seq peaks revealed that the score ratio for SELEX-seq derived peaks within those SEP3 binding regions predicts complex specific TFBSs (Figure 3B; Supplemental Data Set 3). In this manner, we were able to discriminate TFBSs that were either more SEP3-AP1 or more SEP3-AG specifically bound in the Arabidopsis genome. We also showed that the SEP3-AG-specific motif of cluster 1 (Figure 2B) overlaps very well with the AG-specific ChIP-seq regions (Figure 3C). In addition, SEP3-AP1 highly bound sequences from cluster 3 had good overlap with AP1-specific ChIP-seq bound regions. Moreover, the AG-specific motif from cluster 2 overlaps very well with the AG-specific ChIP-seq TFBSs. In general, the number of specifically bound TFBSs is higher for AG than for AP1, which is linked to the presence of specific DNA binding motifs (Figure 3C). This analysis showed that in vivo DNA binding specificities of reproductive organ-specific complexes can be derived from the SELEX-seq data.
Inferring protein complexes that bind to specific genomic regions is difficult based on ChIP-seq data alone, since standard ChIP pull-downs usually detect all genomic TFBSs of a TF, independent of its interaction partners. Combining ChIP-seq experiments of different TFs can identify which TFs bind to the same regions, but not necessarily together in a complex. However, SELEX-seq data allowed us to identify genomic regions that are directly bound by either SEP3-AG or by SEP3-AP1. To test whether protein complex-specific TFBSs drive expression in specific floral whorls, we integrated the information from SELEX-seq data for SEP3-AG and SEP3-AP1 with whorl-specific expression data (TRAP-seq) (Jiao and Meyerowitz, 2010). We used genome aligned SEP3-AP1 and SEP3-AG SELEX-seq data to classify the 1500 most enriched SEP3 ChIP-seq peaks. This analysis allowed us to predict genomic regions that are bound directly by a specific TF complex. As a result, the SELEX-based classification of SEP3-bound genomic regions correlated better with whorl-specific expression data, than a classification based on SEP3, AP1, and AG ChIP-seq data (Figure 3D; Supplemental Figure 9).
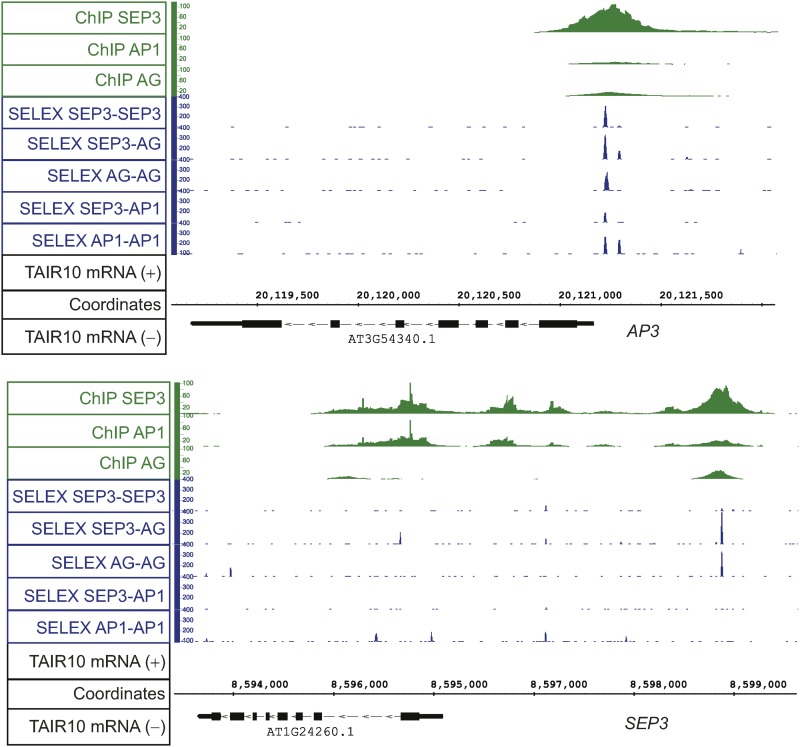
To validate TFBSs obtained with SELEX-seq, we focused on specific examples of regulatory regions that are bound by MADS domain proteins. The upstream promoter regions of SEP3 and AP3 were predicted by our SELEX-seq approach as binding regions of several MADS domain proteins (Figure 4). The position of the SELEX-seq peaks is in very good agreement with the position of ChIP-seq peaks for all MADS domain homeotic TFs within those two regulatory regions. One of these genomic regions, positioned 4.1 kb upstream in the promoter of SEP3, has two TFBSs in close proximity that show differential affinity (represented by the SELEX peak height) and binding specificity (not all heteromeric complexes bound to both TFBSs). These results are in agreement with our previous in vitro EMSA studies of the same regulatory fragment (Smaczniak et al., 2012b) where these two TFBSs showed variable binding efficiencies for different MADS domain protein complexes with one site being superior in importance to the other. Also in the AP3 promoter, two previously characterized TFBSs (Tilly et al., 1998; Jetha et al., 2014), positioned close to the transcriptional start site, showed differential binding affinity. In the same region, a third potential CArG-box has been identified (Tilly et al., 1998), but it is not predicted to be bound by MADS domain complexes in SELEX-seq, in agreement with other in vitro data (Tilly et al., 1998; Jetha et al., 2014).
Figure 4.
Examples of SELEX-Seq TFBSs and ChIP-Seq Profiles Mapped to the Genome of Arabidopsis.
Top: AP3 genomic locus. Bottom: SEP3 genomic locus.
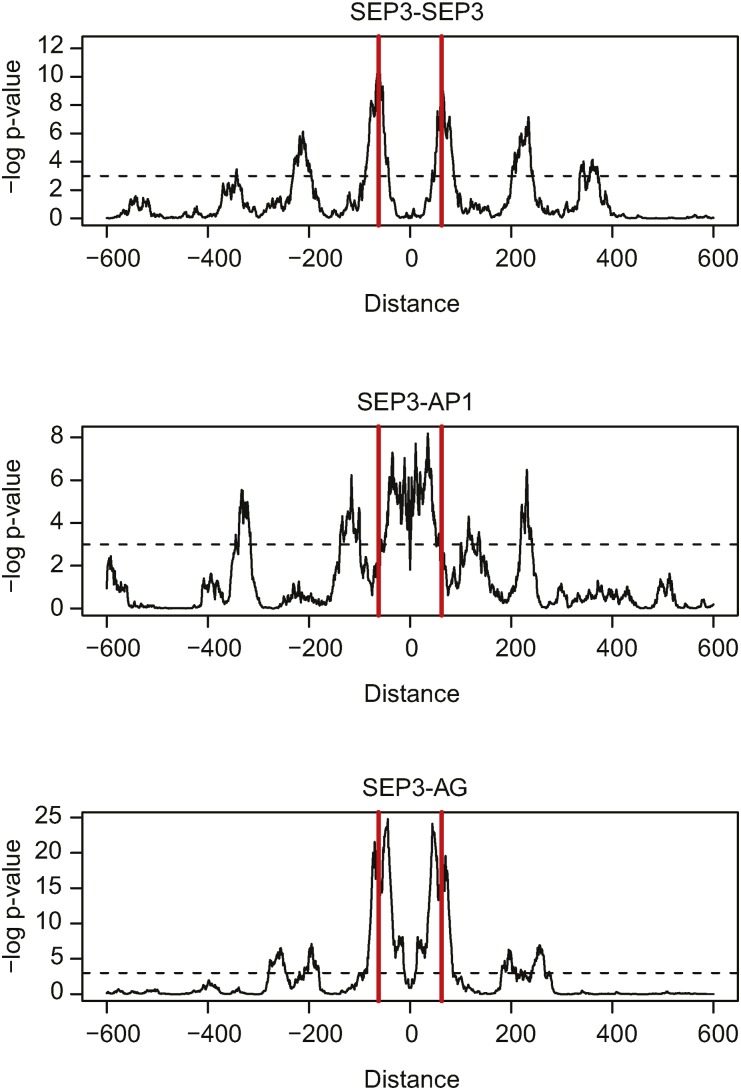
The SELEX-seq data mapped to the genome (Figure 4) allowed us to predict TFBSs at much higher resolution compared with the ChIP-seq data alone. Because of the high resolution, we estimated a preferred distance between located TFBSs. For all analyzed complexes, we showed that the preferred distance was around 62 bp (six helical turns, visualized as the first peak from the center and a red vertical line) (Figure 5). Moreover, we could see that for SEP3-AG, this was the only distance observed, while for SEP3-SEP3 complex TFBSs were also separated by a longer DNA stretch of around 210 bp. For SEP3-AP1, no strong preferences between more distal TFBSs were clearly defined. This suggests that SEP3-AG and SEP3-AP1 complexes prefer to act in different DNA loop size configurations, with one complex being stricter in the loop size than the other is.
Figure 5.
Characteristics of SELEX-Seq Peaks within ChIP-Seq Peaks.
Distribution of complex specific SELEX-seq TFBSs within ChIP-seq TFBSs of SEP3 (top), AP1 (middle), and AG (bottom). Shown is the frequency of distances normalized by the background frequency distances outside of the ChIP-seq peaks, plotted based on the calculated P values using the hypergeometric test. A distance of zero results when the position of compared TFBSs is the same.
Activity of the AP3 Promoter with Altered DNA TFBSs
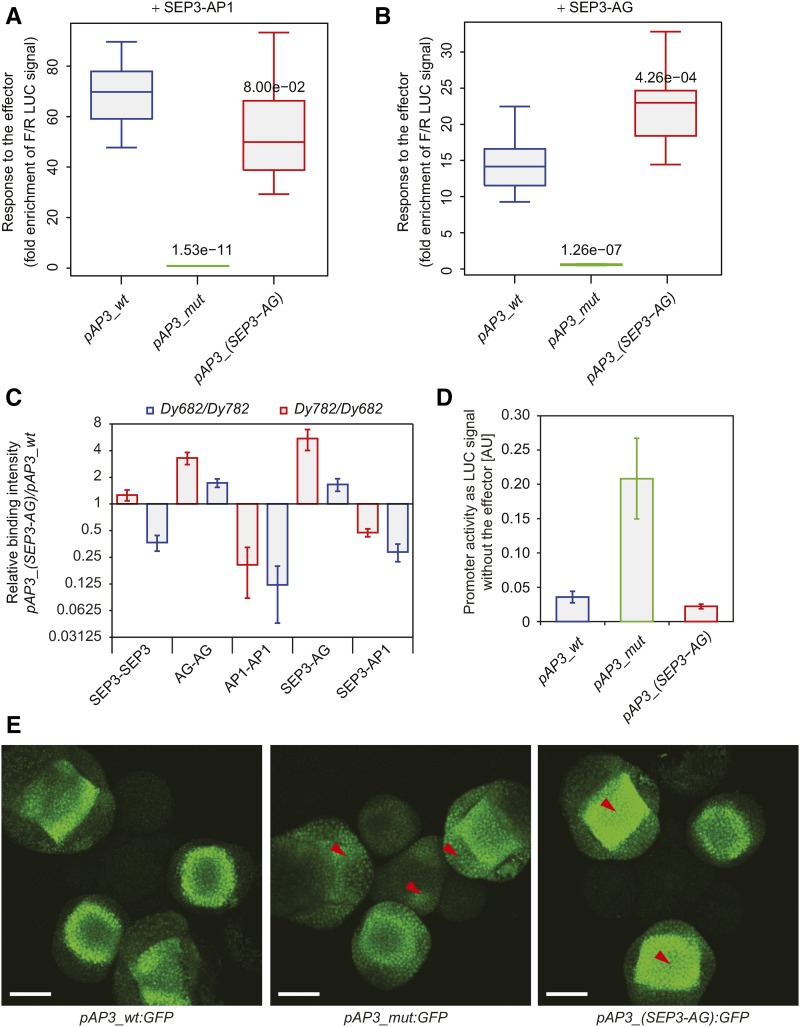
Combining information from SELEX-seq and ChIP-seq experiments to predict complex-specific TFBSs in the Arabidopsis genome suggested that TFBSs could be potentially engineered in vivo to modulate gene regulation by MADS domain factors. To test this hypothesis, we altered the AP3 promoter (pAP3_wt) in a reporter construct that contained two CArG-boxes (Figure 6). We chose the AP3 promoter for our in vivo and in vitro experiments as it is expressed in a floral whorl-specific manner (whorl 2 and whorl 3). The AP3 promoter is relatively short (0.9 kb) and well characterized (Tilly et al., 1998; Jetha et al., 2014), with two identified MADS TFBSs. The genomic region harboring the two functional CArG-boxes is bound by MADS domain proteins in vivo (Figure 4). These properties of the AP3 promoter make it ideal for mutagenesis studies because the compensation effects coming from other potential TFBSs would be minimized. The activity of the AP3 promoter, when the two CArG-boxes were completely mutated (pAP3_mut), did not respond to the presence of the effectors (SEP3-AP1 or SEP3-AG protein complex). Modifying the two CArG-boxes in the AP3 promoter based on the SELEX-seq data as having more affinity toward SEP3-AG [pAP3_(SEP3-AG)] protein complex moderately altered the activity of the AP3 promoter in response to the effectors (Figures 6A and 6B). The activity of the pAP3_(SEP3-AG) promoter comparing to the pAP3_wt promoter increased when SEP3-AG effector was used and decreased when SEP3-AP1 effector was present. The in vitro binding strength of corresponding MADS domain protein complexes to modified AP3 promoters was stronger comparing to wild-type AP3 promoter in case of the SEP3-AG complex and weaker in case of the SEP3-AP1 complex (Figure 6C).
Figure 6.
Comparison of Activities and Protein-DNA Binding between Various MADS Domain Protein Complexes to Modified AP3 Promoters.
(A) and (B) Promoter activity quantification in protoplasts using dual luciferase reporter assay with specific protein effector complexes SEP3-AP1 (A) and SEP3-AG (B). Error bars represent sd. Numbers above the boxes represent t test P value of the difference between wild-type and modified promoters.
(C) Protein-DNA relative binding intensities of various MADS domain protein complexes between modified and wild-type AP3 promoters studied by quMFRA. The quMFRA was performed using two sets of IR-labeled DNA sequences (Dy682 and Dy782) where IR fluorophores were reciprocally exchanged between AP3 promoter sequences. Error bars represent sd. Sequences used in EMSA are indicated in Supplemental Table 4.
(D) Basal promoter activity quantification in protoplasts using dual luciferase reporter assay. Error bars represent sd.
(E) Confocal pictures of the fluorescent reporter expression patterns of the wild-type and modified AP3 promoters. Red arrows indicate the most pronounced changes in spatial GFP expression in modified promoter signal compared with the wild-type signal. Bars = 50 µm.
The importance of these two TFBSs in the AP3 promoter was visualized by dual luciferase activity assays in protoplasts and in in planta fluorescent reporter assays. These two approaches showed that the expression of the mutated AP3 promoter without any functional CArG-box is higher (Figure 6D) and not restricted to whorl 2 and 3; it is detected throughout early stages of flower development and the inflorescence meristem (Figure 6E). This is in agreement with the previously reported GUS expression patterns (Tilly et al., 1998) and suggests that the CArG-boxes have also repressive roles, restricting AP3 expression to the developing flower. Thus, those CArG-boxes are likely bound by other MADS domain TF complexes as well, such as those that are expressed in the inflorescence meristem. Indeed, studies have shown that SOC1 and SVP (Gregis et al., 2009; Immink et al., 2012) directly repress AP3 expression in the inflorescence meristem and early floral meristem, providing an explanation of these results. Moreover, the activity of the promoter with TFBSs more specific to the SEP3-AG complex showed a spatial enhancement of the fluorescent signal in whorl 4 of the flower (Figure 6E). Taken together, these results indicate that MADS domain protein heteromeric complexes have partly different DNA binding specificities; consequently, this affects their target gene specificity.
DISCUSSION
The variety of functions of MADS domain TFs in the life cycle of Arabidopsis suggests that they may regulate different sets of target genes. Exactly how MADS domain TFs achieve their functional specificity is not yet fully understood. Here, we showed that part of the functional specificity can relate to DNA binding. By making use of the SELEX-seq approach, we were able to distinguish common and specific TFBSs for several key floral homeotic MADS domain TF complexes. Moreover, we presented here that differential binding of MADS domain protein complexes to their TFBSs plays a role in specificity of target gene regulation.
DNA Binding Specificities of Floral Homeotic Proteins Predict Organ-Specific Targets
Recent high-throughput approaches applied to large number of plant TFs aimed to identify their in vitro DNA binding specificities using protein binding microarrays (Franco-Zorrilla et al., 2014) or a modification of the SELEX, DNA affinity purification sequencing (O’Malley et al., 2016), did not focus on MADS domain proteins. Moreover, these studies did not take into consideration the impact of TF heterodimerization on DNA TFBS selection. SELEX-seq allows the systematic and sensitive characterization of DNA binding properties of TFs. Previously, “classical” SELEX followed by Sanger sequencing was used to study DNA binding properties of several plant MADS domain TFs (Huang et al., 1993, 1995, 1996; Tang and Perry, 2003). Some features of the consensus sequences found in those studies are common to our SELEX-seq derived consensus. For example, for most MADS domain TFs, the consensus motif resembles the putative CArG-box similar to our SELEX-seq motifs, and they also contain specific nucleotides in the flanking regions. Moreover, we identified additional TFBSs that strongly deviate from the consensus sequence. Although position weight matrices or consensus sequences give substantial information on the DNA sequence characteristics that determine TF binding, they fail, for example, to visualize the dependencies between nucleotide positions or, most importantly, the affinities to particular DNA structures. Our analysis shows that there are differences in DNA binding between selected MADS domain homo- and heterocomplexes and support the concept that different MADS domain protein complexes could bind overlapping sets of TFBSs, although with different affinities, which would allow for active competition between different MADS domain protein complexes for the same target genes in vivo. Floral homeotic complexes containing the AG protein, responsible for stamen and carpel specification, have distinct DNA binding preferences that result in the regulation of a specific set of target genes. On the other hand, other target genes are commonly bound by different MADS domain proteins but may be antagonistically regulated in perianth and reproductive organs in the flower (Yan et al., 2016).
DNA Structure Affects MADS TF Binding Specificity
MADS domain TFs bind DNA through interactions of the N-terminal part of the MADS domain with the CArG-box A/T-rich region of the minor groove (Pellegrini et al., 1995), causing substantial bending of the DNA. Although full crystal structures for plant MADS domains are not available, it was shown that plant MADS-domain proteins, similar to MADS proteins from animals and yeast, bend the DNA (Riechmann et al., 1996a; Melzer et al., 2009). Additionally, it was reported that DNA bending by MADS domain complexes could be DNA sequence specific (West et al., 1997, 1998), which supports the importance of the DNA sequence in the regulation of a protein-DNA complex structure. The determinants of this characteristic DNA binding are not well understood. Muiño et al. (2014) showed that there is an enrichment of a particular DNA structural pattern in regions bound by MADS domain TFs. Recently, large-scale DNA structure predictions for human and plant MADS domain ChIP-seq TF binding data showed that combining DNA sequence information with DNA shape features improves prediction of TF-bound genomic regions in vivo (Mathelier et al., 2016). In their analysis, authors captured the importance of the MGW and propeller twist in selection of TFBSs of MADS domain proteins. Our DNA-shape predictions of the SELEX-seq sequences show that two propeller twist maxima are present at the both ends of the CArG-box motif corresponding to CC and GG flanks.
Intrinsic shape properties of the DNA can influence DNA binding, especially of TFs that mainly recognize the DNA via interactions with the minor groove (Rohs et al., 2009). The A/T-rich region of the CArG-box can be considered as the “A-tract” (Muiño et al., 2014). It was previously reported that A-tracts facilitate MADS domain TF DNA binding when located inside the CArG-box motif and periodically distributed around it (Muiño et al., 2014). Based on our SELEX-seq sequence motifs, we can infer that differences in the length of the A/T-rich regions influence TF binding specificity. For example, AG alone usually prefers to bind sequences with shorter A/T-rich region, while the SEP3 alone binds sequences with a long A/T-region. The SEP3-AG heterodimer binds sequences with an intermediate A/T region. This, and the predicted structural characteristics of the DNA binding motifs of MADS domain TF dimers, suggests DNA structure, especially the width of the CArG-box minor groove, plays a role in determining DNA binding specificity. Since DNA bending (Haran and Mohanty, 2009) induced by the TF dynamically changes conformation of promoters and juxtaposition of regulatory regions, this can play a prominent role in MADS-DNA binding and gene regulation.
The Role of TF Multimerization in MADS DNA Binding Specificity
According to the floral quartet model (Theissen and Saedler, 2001), quaternary MADS domain TF complexes bend the DNA in order to bind simultaneously two different TFBSs. The exact nature of this binding and a possible regulation by formation of DNA regulatory loops by TFs is not fully explored. Recent in vitro studies on homotetrameric complexes by SEP1, SEP2, SEP3, or SEP4 proteins from Arabidopsis shows their ability to bind cooperatively to DNA as tetramers and loop the DNA, with different preferences for TFBS spacing (Jetha et al., 2014). The role of TFBS spacing in DNA binding specificity of MADS protein complexes is supported by the distribution of TFBSs predicted by SELEX-seq within genomic regions bound in vivo. We observed that spacing between SEP3/AG-specific TFBSs is more defined compared with that of SEP3-AP1 TFBSs. This might suggest that some of the functional specificity comes from the coordinated, simultaneous interaction of MADS domain proteins with more than one cis-regulatory element in the genome. Since MADS domain protein complexes can induce bending to a different degree (Pellegrini et al., 1995; Riechmann et al., 1996a; West et al., 1997; Huang et al., 2000), it also might be that stronger bending induced by SEP3-AG evokes interaction between two TFBSs at shorter distances when the floral quartet is formed, while a potentially mild bending induced by the SEP3-AP1 protein complex does not impose such a restriction. In summary, the commonly observed presence of more than one TFBS identified by SELEX-seq within a single ChIP-seq peak suggest a role of these TFBSs in recruitment of specific higher-order complexes and robust regulation of the target gene expression.
Toward Understanding the Specificity of Floral Homeotic Protein Complexes
Since different MADS domain proteins bind overlapping but not identical sets of genomic regions, so part of the functional specificity of MADS domain TFs is attributed to DNA binding specificity (Yan et al., 2016). High-throughput in vivo DNA binding experiments showed that MADS domain proteins bind the DNA in places that lack canonical CArG-box (or CArG-box-like) sequences (Kaufmann et al., 2009, 2010; Zheng et al., 2009; Deng et al., 2011). AG and SEP3-AG complexes bind DNA sequence elements that resemble, if at all, CArG-boxes with very short A/T-rich regions in the center. Comparing SELEX-seq with ChIP-seq binding profiles, we were able to unravel part of the cis-regulatory code for specification of perianth versus reproductive organs. We showed that AG and SEP3-AG complexes have unique DNA binding preferences that can be linked to a specific set of genomic regions bound in vivo, for example, HECATE2, which plays an important role in the gynoecium development (Gremski et al., 2007) or CAPRICE (Schellmann et al., 2002) and GLABRA3 (Payne et al., 2000) that regulate trichome development, a process that is repressed by AG in carpels (ÓMaoiléidigh et al., 2013). In line with these specific functions, only 12.4% of the AG-bound genomic regions are also bound by MADS domain TFs that act during vegetative development or floral transition (FLC, SVP, and SOC1), while ∼17% of the AP1-bound genomic regions show an overlap with these factors (D. Chen, personal communication).
Targeted in vivo immunoprecipitation experiments of several MADS domain TFs and other studies revealed that MADS domain proteins can form larger complexes with other transcriptional regulators (Brambilla et al., 2007; Simonini et al., 2012; Smaczniak et al., 2012b) and as such could bind the DNA. Whether the presence of other cofactors or other TFs that interact with the MADS domain TFs can modulate DNA binding specificity remains an important question to be resolved in future studies. Similarly, the chromatin status may have an impact on recruitment of specific complexes.
Our in vivo AP3 promoter studies suggest that the expression of MADS domain TF target genes could be modulated by single nucleotide changes in their regulatory regions. In agreement with these results, it was shown that differences between the spatio-temporal level of expression of AP1 and CAULIFLOWER, two recent Arabidopsis duplicated genes, were determined by the presence or absence of individual, functionally important TFBSs in regulatory regions (Ye et al., 2016). Together, the findings suggest that changes in individual MADS domain TFBSs contribute to regulating spatio-temporal levels of target gene expression but do not provide a strict on/off regulation of target gene expression.
METHODS
SELEX-Seq
The dsDNA libraries were made from the single-stranded DNA sequences by single-cycle PCR amplification with a complementary primer essentially as described before (Jolma et al., 2010). The dsDNA libraries contained 20 random nucleotide fragment flanked by specific barcodes that allowed for later characterization when multiplexed in high-throughput sequencing. The dsDNA libraries contained all necessary features required for direct sequencing with an Illumina platform (Jolma et al., 2010).
Proteins were produced using the TNT SP6 Quick Coupled Transcription/Translation System (Promega) following the manufacturer’s instructions in a total volume of 20 μL and equimolar expression plasmid concentrations (for protein dimers). The binding reaction mix was prepared essentially as described previously for EMSA experiments (Egea-Cortines et al., 1999; Smaczniak et al., 2012b) and contained 20 μL of in vitro-produced proteins and 50 to 100 ng of dsDNA library in a total volume of 120 µL. The binding reaction was incubated on ice for 1 h followed by 1 h immunoprecipitation with protein-specific antibodies (affinity-purified, polyclonal peptide antibodies; Eurogentec) coupled to magnetic beads (MyOne; Invitrogen) in a thermomixer at 4°C with constant mixing at 700 rpm. Antibodies were raised against the protein-specific C-terminal domain (the C-domain) or last part of the K-domain of MADS domain proteins. These domain parts are not responsible for protein-DNA (the M-domain is) or protein-protein interactions (the I- and first part of K-domain are); rather, they are assumed to stabilize higher-order protein complex formation (Kaufmann et al., 2005). Therefore, we expected no influence of antibody-protein interaction on studied protein-DNA interactions. The following synthetic peptides were used for immunizations: LNPNQEEVDHYGRHH for SEP3; EQWDQQNQGHNMPPPLPPQQ for AP1; and PPQTQSQPFDSRNYFC and QPNNHHYSSAGRQDQT for AG. The SEP3 antibodies were previously used in ChIP-seq experiments (Kaufmann et al., 2009; Pajoro et al., 2014). Magnetic beads with attached antibodies where prepared in advance according to the manufacturer’s instructions (MyOne) with purified antibodies resuspended in 1× PBS (∼1 mg/mL); 20 µg of antibodies and 0.5 mg of beads were used for a single binding reaction. After immunoprecipitation, beads were washed five times with 150 μL of binding buffer without salmon-sperm DNA and bound DNA was eluted with 50 μL 1× TE in a thermomixer at 90°C with full mixing speed. Afterwards, magnetic beads were immobilized and the supernatant was transferred to a 1.5-mL tube. DNA fragments were amplified with 10 to 15 cycles of PCR with SELEX round-specific primers (Jolma et al., 2010), and the total amplicon was used in the subsequent SELEX round. The amplification efficiency was checked on the agarose gel by comparing to a known concentration of a standard probe. Samples for sequencing, after amplification, were cut out from agarose gel and purified using MinElute gel extraction kit (Qiagen). Different libraries were multiplexed by mixing in equimolar amounts with 40% PhiX Control (Illumina) in elution buffer (Qiagen), and sequencing was performed on the HiSeq 2000 or GAII sequencers (Illumina).
Obtaining Relative Affinities from the SELEX-Seq Data
Sequence reads that did not pass the filter quality of CASAVA 1.8 or mapped with no mismatches to the phix174 genome were eliminated. The remaining sequences in fasta format were extracted and grouped according to library-specific barcodes, allowing no mismatches. Barcodes were removed leading to 20-bp sequence libraries used in the data analysis. The 20-bp sequences that were present in libraries in an unexpected high number (>1000) were eliminated, as well as 20-bp reads containing the sequence “TCGTATGCCG,” which is part of the Illumina adapter sequence used for sequencing.
Data analysis was essentially performed as described before (Slattery et al., 2011). Because the number of sequenced reads in each SELEX round (Supplemental Table 1) is much smaller than the 1012 possible different 20-mer sequences, we calculated which k-mer length should be used to obtain the maximum gain in information. For this, we computed the information gain (the Kullback-Leibler divergence of Round 3 relative to Round 0) for each k-mer length. For most libraries, the length 12 bp was the most optimal (Supplemental Figure 3); therefore, we based the further analysis on the 12-mer sequences.
Frequencies of 12-mer sequences in each round except Round 0 was calculated directly from the data using the function oligonucleotideFrequency from the Bioconductor R package: Biostrings. The 12-mer sequences that were present in libraries in an unexpected high number (>1000) were eliminated at this step.
Sequences in Round 0 represent a set of randomly synthesized oligonucleotides and their complexity did not allow for the direct calculation of 12-mer frequencies. Therefore, the sequence frequency in Round 0 was estimated by the sixth-order Monte Carlo model, as proposed before (Slattery et al., 2011). We chose the sixth-order Monte Carlo model because when the model was trained using 75% of the sequencing data, it resulted in the highest prediction value as measured by the Pearson correlation coefficient between the predicted and observed frequencies in the other 25% of the sequencing data.
Relative affinity for each possible 12-mer was calculated as the ratio between the frequencies of 12-mers in Round 3 to Round 0, and normalized to 1 by dividing for the highest affinity-predicted 12-mer. These affinities can be download in Excel file format from the GEO omnibus submission GSE95730.
Combining SEP3 ChIP-Seq and SELEX-Seq Data to Predict MADS Domain Dimer TFBSs
To in silico predict genomic regions bound by a given MADS domain dimer based on our SELEX-seq experiments, we obtained the affinity value for each k-mer of length 13 bp. For each studied MADS domain protein complex, we estimated the average affinity value to a particular 13-mer sequence and its reverse complementary sequence. We chose 13-mers instead of 12-mers for technical reasons: The mapping software soapv2 (Li et al., 2009) was only able to map sequences with a minimum length of 13 bp. We created fasta files for each library containing 13-mer sequences in a number equal to their estimated relative affinity multiplied by 100 and rounded up to the closest integer (e.g., a sequence with the relative affinity of 0.98 was present in 98 copies in the fasta file). Next, we mapped these fasta files to the TAIR10 genome with soapv2 (Li et al., 2009), allowing no mismatches. Later, the function mappedReads2Nhits from the peak caller CSAR (Muiño et al., 2011) was used to identify enriched regions and their associated score, extending the 13-bp reads and using no control since the relative affinities were already corrected by the Round 0 enrichment. This resulted in a genomic SELEX-seq score proportional to the affinity of the sum of 13-mers located on the particular genomic region.
To relate ChIP-seq scores with the SELEX-seq scores obtained by CSAR, we reanalyzed the SEP3 (4 d after induction; stage 4-5 of flower development) (Pajoro et al., 2014), AP1 (4 d after induction; stage 4-5 of flower development) (Pajoro et al., 2014), and AG (5 d after induction; stage 5-6 of flower development) (ÓMaoiléidigh et al., 2013) publically available ChIP-seq data using CSAR (default parameters expect the backg that was set to 3). Next, we linked the ChIP-seq and SELEX-seq peak to the SEP3 ChIP-seq when their distance was shorter than 500 bp. Only the top 1500 SEP3 ChIP-seq peaks were considered. Quantile normalization was used to normalize the SELEX-seq peaks scores and independently to normalize the ChIP-seq peak scores aligned to the SEP3 ChIP-seq peaks. When it was needed, a TFBS was associated to a gene when this was located 3 kb upstream or 1 kb downstream of a gene. The method to classify SEP3 TFBSs showed good precision/recall ratios compared with the random classifier (Supplemental Figure 10).
Other Bioinformatic Analyses
Position weight matrices were calculated based on the extracted 20N sequences containing 12-mers analyzed in the heat map with the GADEM algorithm (Li, 2009) and DNA sequence logos were built with the seqLogo R script.
We used the Find Individual Motif Occurrences tool (FIMO; version 4.10.2) (Grant et al., 2011) to scan the promoter region (TSS +3000 bp ∼ −1000 bp) of genes for TFBSs using the binding motifs generated from SELEX-seq data (with a P value threshold of 10e–5 and defaults for other parameters). A potential interaction was assigned if there was at least one TFBS in the promoter of a gene. Significant enrichment of specific and common ChIP-seq target genes for AP1 (Pajoro et al., 2014) and AG (ÓMaoiléidigh et al., 2013) that overlapped with the different SELEX-seq clusters was evaluated by a hypergeometric test (using the “phyper” function in R).
EMSA, Kd Estimation, and QuMFRA
SELEX-derived sequences for EMSAs were produced by PCR with biotin-labeled or IR-fluorophore-labeled primers and purified from 2% agarose gel. Electrophoretic mobility shift assays were performed essentially as described before (Smaczniak et al., 2012b). The detection was performed with the Odyssey scanner (Li-Cor) for IR-labeled fragments or with the Chemiluminescent Nucleic Acid Detection Module Kit (Pierce) for biotin-labeled fragments.
Absolute dissociation constants (Kd) for various protein complex-DNA interactions was performed with the EMSA-based method, essentially as described before (Riechmann et al., 1996a). The constant amount of the protein complex was used with the various, known concentrations of the IR-fluorophore-labeled DNA.
QuMFRA between selected 20-bp SELEX-seq-derived DNA fragments was performed as described (Muiño et al., 2014). DNA fragments were labeled with different fluorophores (Cy3/Cy5 or Dy682/Dy782) and binding quantification was recorded with a molecular imager (Bio-Rad) and Odyssey scanner.
Plant Materials and Growth Conditions
In our in vivo assays for AP3 promoter activity by fluorescent protein reporter, we used the 991-bp AP3 promoter region and we modified the known MADS domain TFBSs according to the SELEX-seq-inferred DNA sequences. Modifications were done following the highest relative affinity toward SEP3-AG protein complex [pAP3_(SEP3-AG)] and by mutating the CArG-boxes altogether (pAP3_mut). Modifications to the AP3_wt promoter were introduced by PCR and the promoter constructs were cloned into the Gateway entry vector pCR8/GW/TOPO (Invitrogen) followed by a subcloning via Gateway LR reaction into the destination vector pGREEN:GW:NLS-GFP (Horstman et al., 2015). The pAP3_x:GFP constructs were transformed into Col-0 plants using floral dip method (Clough and Bent, 1998). Plants were grown under long-day conditions (16 h of light/8 h of dark at 20–22°C) on soil/vermiculite mix (3:1; v/v) in a greenhouse. Plant inflorescences were studied by confocal microscopy (Zeiss LSM 510).
AP3 Promoter Activity Assays
In our in vivo AP3 promoter activity experiments by dual-LUC assays (Dual-Luciferase Reporter Assay System from Promega), the same promoter region of the AP3 gene was used and the same modifications were studied as in the fluorescent protein reporter assays. Promoters were inserted in the front of the firefly luciferase coding sequence (CDS) and together with the control Renilla luciferase CDS and corresponding effector protein complex CDS, transfected into Arabidopsis thaliana protoplasts cells (Díaz-Triviño et al., 2017). Protoplasts were isolated from young leaves essentially as reported before (Yoo et al., 2007). Relative value of the signal between firefly LUC and Renilla LUC was calculated and normalized to the relative signal of the sample without the effector protein complex. The resulting value corresponds to the activity of the promoter in the Arabidopsis protoplasts cells in arbitrary units.
Accession Numbers
Nucleotide sequence data are available from the NCBI/GenBank data library under the following accession numbers: NM_102272 (SEP3), NM_105581 (AP1), NM_118013 (AG), NM_115294 (AP3), and NM_122031 (PI). Accession numbers used in the supplemental data sets are from the TAIR10 annotation of the Arabidopsis genome (www.arabidopsis.org). SELEX-seq raw sequencing data are available from the Gene Expression Omnibus database under accession number GSE95730.
Supplemental Data
Supplemental Figure 1. EMSA analysis of homo- and heteromeric complexes bound to the SELEX DNA from round 5.
Supplemental Figure 2. SELEX-seq reproducibility.
Supplemental Figure 3. Optimal lengths of randomized fragments that sufficiently predict the specificity of bound MADS domain complexes.
Supplemental Figure 4. DNA binding specificities of MADS domain TF complexes in cluster 3.
Supplemental Figure 5. Comparison of relative binding affinities for MADS domain protein complexes obtained by different methods.
Supplemental Figure 6. Comparison of the absolute binding affinity (dissociation constants, Kd) by EMSA with the relative binding affinity by SELEX-seq.
Supplemental Figure 7. Enrichment of individual consensus CArG-boxes by MADS domain TF complexes.
Supplemental Figure 8. DNA structure predictions of the motifs from clusters 1 to 3 and subclusters 3a to 3c depicting an average value of the four DNA structural properties at each dinucleotide step in the sequences.
Supplemental Figure 9. Comparison of the SELEX-seq and ChIP-seq data with the whorl-specific expression data.
Supplemental Figure 10. Evaluation of the AP1/AG classifier.
Supplemental Table 1. Summary of the high-throughput sequencing analysis.
Supplemental Table 2. Sequences of selected 12-mers and their corresponding 20N most representative dsDNA SELEX library sequences.
Supplemental Table 3. The experimental DNA sequence setup and the number of observations for quMFRA experiments.
Supplemental Table 4. Wild-type and modified sequences of the AP3 promoter used in EMSA and quMFRA.
Supplemental Data Set 1. Results of the EMSA-based quMRFA experiments.
Supplemental Data Set 2. Measurement of DNA binding affinity of selected protein complexes for selected SELEX-seq DNA probes.
Supplemental Data Set 3. Comparison of the top 1500 SEP3 ChIP-seq TFBSs compared with the AG and AP1 ChIP-seq data and the SELEX-seq data for SEP3-AG versus SEP3-AP1 dimer.
Supplemental Data Set 4. The output of the FIMO and phyperanalysis for the prediction of specific and common ChIP-seq target genes for AG and AP1, based on the SELEX-seq complex-specific motifs.
Acknowledgments
We thank Arttu Jolma and Jussi Taipale for providing bar-coded single-stranded DNA libraries and sharing details on the dsDNA library preparation and amplification. We also thank Markus Schmid and Elio Schijlen for generating the Illumina GAII and HiSeq data, Anneke Horstman for providing pGREEN:GW:NLS-GFP reporter vector, and Ikram Blilou for providing dual-LUC reporter vectors. This work was supported by an NWO-VIDI grant to K.K. J.M.M. was supported by a DFG grant (TRR175).
AUTHOR CONTRIBUTIONS
C.S., J.M.M., G.C.A., and K.K. designed the research. C.S. performed the experiments. J.M.M. and D.C. analyzed the data. G.C.A. and K.K. supervised the project. C.S., J.M.M., and K.K. wrote the manuscript. All authors discussed the results and commented on the manuscript.
Glossary
- TF
transcription factor
- TFBS
TF binding site
- SELEX-seq
systematic evolution of ligands by exponential enrichment followed by high-throughput sequencing
- dsDNA
double-stranded DNA
- EMSA
electrophoretic mobility shift assay
- quMFRA
quantitative multiple fluorescence relative affinity
- MGW
minor groove width
- CDS
coding sequence
References
- Bemer M., van Dijk A.D., Immink R.G., Angenent G.C. (2017). Cross-family transcription factor interactions: an additional layer of gene regulation. Trends Plant Sci. 22: 66–80. [DOI] [PubMed] [Google Scholar]
- Brambilla V., Battaglia R., Colombo M., Masiero S., Bencivenga S., Kater M.M., Colombo L. (2007). Genetic and molecular interactions between BELL1 and MADS box factors support ovule development in Arabidopsis. Plant Cell 19: 2544–2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- de Folter S., Angenent G.C. (2006). trans meets cis in MADS science. Trends Plant Sci. 11: 224–231. [DOI] [PubMed] [Google Scholar]
- Deng W., Ying H., Helliwell C.A., Taylor J.M., Peacock W.J., Dennis E.S. (2011). FLOWERING LOCUS C (FLC) regulates development pathways throughout the life cycle of Arabidopsis. Proc. Natl. Acad. Sci. USA 108: 6680–6685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz-Triviño S., Long Y., Scheres B., Blilou I. (2017). Analysis of a plant transcriptional regulatory network using transient expression systems. Methods Mol. Biol. 1629: 83–103. [DOI] [PubMed] [Google Scholar]
- Dickerson R.E. (1998). DNA bending: the prevalence of kinkiness and the virtues of normality. Nucleic Acids Res. 26: 1906–1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egea-Cortines M., Saedler H., Sommer H. (1999). Ternary complex formation between the MADS-box proteins SQUAMOSA, DEFICIENS and GLOBOSA is involved in the control of floral architecture in Antirrhinum majus. EMBO J. 18: 5370–5379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco-Zorrilla J.M., López-Vidriero I., Carrasco J.L., Godoy M., Vera P., Solano R. (2014). DNA-binding specificities of plant transcription factors and their potential to define target genes. Proc. Natl. Acad. Sci. USA 111: 2367–2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant C.E., Bailey T.L., Noble W.S. (2011). FIMO: scanning for occurrences of a given motif. Bioinformatics 27: 1017–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregis V., Sessa A., Dorca-Fornell C., Kater M.M. (2009). The Arabidopsis floral meristem identity genes AP1, AGL24 and SVP directly repress class B and C floral homeotic genes. Plant J. 60: 626–637. [DOI] [PubMed] [Google Scholar]
- Gremski K., Ditta G., Yanofsky M.F. (2007). The HECATE genes regulate female reproductive tract development in Arabidopsis thaliana. Development 134: 3593–3601. [DOI] [PubMed] [Google Scholar]
- Haran T.E., Mohanty U. (2009). The unique structure of A-tracts and intrinsic DNA bending. Q. Rev. Biophys. 42: 41–81. [DOI] [PubMed] [Google Scholar]
- Honma T., Goto K. (2001). Complexes of MADS-box proteins are sufficient to convert leaves into floral organs. Nature 409: 525–529. [DOI] [PubMed] [Google Scholar]
- Horstman A., Fukuoka H., Muino J.M., Nitsch L., Guo C., Passarinho P., Sanchez-Perez G., Immink R., Angenent G., Boutilier K. (2015). AIL and HDG proteins act antagonistically to control cell proliferation. Development 142: 454–464. [DOI] [PubMed] [Google Scholar]
- Huang H., Mizukami Y., Hu Y., Ma H. (1993). Isolation and characterization of the binding sequences for the product of the Arabidopsis floral homeotic gene AGAMOUS. Nucleic Acids Res. 21: 4769–4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H., Tudor M., Weiss C.A., Hu Y., Ma H. (1995). The Arabidopsis MADS-box gene AGL3 is widely expressed and encodes a sequence-specific DNA-binding protein. Plant Mol. Biol. 28: 549–567. [DOI] [PubMed] [Google Scholar]
- Huang H., Tudor M., Su T., Zhang Y., Hu Y., Ma H. (1996). DNA binding properties of two Arabidopsis MADS domain proteins: binding consensus and dimer formation. Plant Cell 8: 81–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang K., Louis J.M., Donaldson L., Lim F.L., Sharrocks A.D., Clore G.M. (2000). Solution structure of the MEF2A-DNA complex: structural basis for the modulation of DNA bending and specificity by MADS-box transcription factors. EMBO J. 19: 2615–2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Immink R.G., Posé D., Ferrario S., Ott F., Kaufmann K., Valentim F.L., de Folter S., van der Wal F., van Dijk A.D., Schmid M., Angenent G.C. (2012). Characterization of SOC1's central role in flowering by the identification of its upstream and downstream regulators. Plant Physiol. 160: 433–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jetha K., Theißen G., Melzer R. (2014). Arabidopsis SEPALLATA proteins differ in cooperative DNA-binding during the formation of floral quartet-like complexes. Nucleic Acids Res. 42: 10927–10942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y., Meyerowitz E.M. (2010). Cell-type specific analysis of translating RNAs in developing flowers reveals new levels of control. Mol. Syst. Biol. 6: 419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolma A., Yin Y., Nitta K.R., Dave K., Popov A., Taipale M., Enge M., Kivioja T., Morgunova E., Taipale J. (2015). DNA-dependent formation of transcription factor pairs alters their binding specificity. Nature 527: 384–388. [DOI] [PubMed] [Google Scholar]
- Jolma A., et al. (2010). Multiplexed massively parallel SELEX for characterization of human transcription factor binding specificities. Genome Res. 20: 861–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolma A., et al. (2013). DNA-binding specificities of human transcription factors. Cell 152: 327–339. [DOI] [PubMed] [Google Scholar]
- Kaufmann K., Melzer R., Theissen G. (2005). MIKC-type MADS-domain proteins: structural modularity, protein interactions and network evolution in land plants. Gene 347: 183–198. [DOI] [PubMed] [Google Scholar]
- Kaufmann K., Muiño J.M., Jauregui R., Airoldi C.A., Smaczniak C., Krajewski P., Angenent G.C. (2009). Target genes of the MADS transcription factor SEPALLATA3: integration of developmental and hormonal pathways in the Arabidopsis flower. PLoS Biol. 7: e1000090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann K., Wellmer F., Muiño J.M., Ferrier T., Wuest S.E., Kumar V., Serrano-Mislata A., Madueño F., Krajewski P., Meyerowitz E.M., Angenent G.C., Riechmann J.L. (2010). Orchestration of floral initiation by APETALA1. Science 328: 85–89. [DOI] [PubMed] [Google Scholar]
- Kneidl C., Dinkl E., Grummt F. (1995). An intrinsically bent region upstream of the transcription start site of the rRNA genes of Arabidopsis thaliana interacts with an HMG-related protein. Plant Mol. Biol. 27: 705–713. [DOI] [PubMed] [Google Scholar]
- Krizek B.A., Meyerowitz E.M. (1996). Mapping the protein regions responsible for the functional specificities of the Arabidopsis MADS domain organ-identity proteins. Proc. Natl. Acad. Sci. USA 93: 4063–4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krizek B.A., Riechmann J.L., Meyerowitz E.M. (1999). Use of the APETALA1 promoter to assay the in vivo function of chimeric MADS box genes. Sex. Plant Reprod. 12: 14–26. [Google Scholar]
- Li L. (2009). GADEM: a genetic algorithm guided formation of spaced dyads coupled with an EM algorithm for motif discovery. J. Comput. Biol. 16: 317–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Yu C., Li Y., Lam T.W., Yiu S.M., Kristiansen K., Wang J. (2009). SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics 25: 1966–1967. [DOI] [PubMed] [Google Scholar]
- Man T.K., Stormo G.D. (2001). Non-independence of Mnt repressor-operator interaction determined by a new quantitative multiple fluorescence relative affinity (QuMFRA) assay. Nucleic Acids Res. 29: 2471–2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathelier A., Xin B., Chiu T.P., Yang L., Rohs R., Wasserman W.W. (2016). DNA shape features improve transcription factor binding site predictions in vivo. Cell Syst. 3: 278–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melzer R., Theissen G. (2009). Reconstitution of ‘floral quartets’ in vitro involving class B and class E floral homeotic proteins. Nucleic Acids Res. 37: 2723–2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melzer R., Verelst W., Theissen G. (2009). The class E floral homeotic protein SEPALLATA3 is sufficient to loop DNA in ‘floral quartet’-like complexes in vitro. Nucleic Acids Res. 37: 144–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muiño J.M., Kaufmann K., van Ham R.C.H.J., Angenent G.C., Krajewski P. (2011). ChIP-seq Analysis in R (CSAR): An R package for the statistical detection of protein-bound genomic regions. Plant Methods 7: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muiño J.M., Smaczniak C., Angenent G.C., Kaufmann K., van Dijk A.D. (2014). Structural determinants of DNA recognition by plant MADS-domain transcription factors. Nucleic Acids Res. 42: 2138–2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson H.B., Laughon A. (1990). The DNA binding specificity of the Drosophila fushi tarazu protein: a possible role for DNA bending in homeodomain recognition. New Biol. 2: 171–178. [PubMed] [Google Scholar]
- O’Malley R.C., Huang S.S., Song L., Lewsey M.G., Bartlett A., Nery J.R., Galli M., Gallavotti A., Ecker J.R. (2016). Cistrome and epicistrome features shape the regulatory DNA landscape. Cell 165: 1280–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ÓMaoiléidigh D.S., Wuest S.E., Rae L., Raganelli A., Ryan P.T., Kwasniewska K., Das P., Lohan A.J., Loftus B., Graciet E., Wellmer F. (2013). Control of reproductive floral organ identity specification in Arabidopsis by the C function regulator AGAMOUS. Plant Cell 25: 2482–2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pajoro A., et al. (2014). Dynamics of chromatin accessibility and gene regulation by MADS-domain transcription factors in flower development. Genome Biol. 15: R41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parenicová L., de Folter S., Kieffer M., Horner D.S., Favalli C., Busscher J., Cook H.E., Ingram R.M., Kater M.M., Davies B., Angenent G.C., Colombo L. (2003). Molecular and phylogenetic analyses of the complete MADS-box transcription factor family in Arabidopsis: new openings to the MADS world. Plant Cell 15: 1538–1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne C.T., Zhang F., Lloyd A.M. (2000). GL3 encodes a bHLH protein that regulates trichome development in Arabidopsis through interaction with GL1 and TTG1. Genetics 156: 1349–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrini L., Tan S., Richmond T.J. (1995). Structure of serum response factor core bound to DNA. Nature 376: 490–498. [DOI] [PubMed] [Google Scholar]
- Pollock R., Treisman R. (1990). A sensitive method for the determination of protein-DNA binding specificities. Nucleic Acids Res. 18: 6197–6204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann J.L., Meyerowitz E.M. (1997). Determination of floral organ identity by Arabidopsis MADS domain homeotic proteins AP1, AP3, PI, and AG is independent of their DNA-binding specificity. Mol. Biol. Cell 8: 1243–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann J.L., Wang M., Meyerowitz E.M. (1996a). DNA-binding properties of Arabidopsis MADS domain homeotic proteins APETALA1, APETALA3, PISTILLATA and AGAMOUS. Nucleic Acids Res. 24: 3134–3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann J.L., Krizek B.A., Meyerowitz E.M. (1996b). Dimerization specificity of Arabidopsis MADS domain homeotic proteins APETALA1, APETALA3, PISTILLATA, and AGAMOUS. Proc. Natl. Acad. Sci. USA 93: 4793–4798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohs R., West S.M., Sosinsky A., Liu P., Mann R.S., Honig B. (2009). The role of DNA shape in protein-DNA recognition. Nature 461: 1248–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schellmann S., Schnittger A., Kirik V., Wada T., Okada K., Beermann A., Thumfahrt J., Jürgens G., Hülskamp M. (2002). TRIPTYCHON and CAPRICE mediate lateral inhibition during trichome and root hair patterning in Arabidopsis. EMBO J. 21: 5036–5046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz-Sommer Z., Huijser P., Nacken W., Saedler H., Sommer H. (1990). Genetic control of flower development by homeotic genes in Antirrhinum majus. Science 250: 931–936. [DOI] [PubMed] [Google Scholar]
- Schwarz-Sommer Z., Hue I., Huijser P., Flor P.J., Hansen R., Tetens F., Lönnig W.E., Saedler H., Sommer H. (1992). Characterization of the Antirrhinum floral homeotic MADS-box gene deficiens: evidence for DNA binding and autoregulation of its persistent expression throughout flower development. EMBO J. 11: 251–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonini S., Roig-Villanova I., Gregis V., Colombo B., Colombo L., Kater M.M. (2012). Basic pentacysteine proteins mediate MADS domain complex binding to the DNA for tissue-specific expression of target genes in Arabidopsis. Plant Cell 24: 4163–4172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slattery M., Riley T., Liu P., Abe N., Gomez-Alcala P., Dror I., Zhou T., Rohs R., Honig B., Bussemaker H.J., Mann R.S. (2011). Cofactor binding evokes latent differences in DNA binding specificity between Hox proteins. Cell 147: 1270–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smaczniak C., Immink R.G., Angenent G.C., Kaufmann K. (2012a). Developmental and evolutionary diversity of plant MADS-domain factors: insights from recent studies. Development 139: 3081–3098. [DOI] [PubMed] [Google Scholar]
- Smaczniak C., et al. (2012b). Characterization of MADS-domain transcription factor complexes in Arabidopsis flower development. Proc. Natl. Acad. Sci. USA 109: 1560–1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stella S., Cascio D., Johnson R.C. (2010). The shape of the DNA minor groove directs binding by the DNA-bending protein Fis. Genes Dev. 24: 814–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W., Perry S.E. (2003). Binding site selection for the plant MADS domain protein AGL15: an in vitro and in vivo study. J. Biol. Chem. 278: 28154–28159. [DOI] [PubMed] [Google Scholar]
- Theissen G., Saedler H. (2001). Plant biology. Floral quartets. Nature 409: 469–471. [DOI] [PubMed] [Google Scholar]
- Tilly J.J., Allen D.W., Jack T. (1998). The CArG boxes in the promoter of the Arabidopsis floral organ identity gene APETALA3 mediate diverse regulatory effects. Development 125: 1647–1657. [DOI] [PubMed] [Google Scholar]
- West A.G., Shore P., Sharrocks A.D. (1997). DNA binding by MADS-box transcription factors: a molecular mechanism for differential DNA bending. Mol. Cell. Biol. 17: 2876–2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West A.G., Causier B.E., Davies B., Sharrocks A.D. (1998). DNA binding and dimerisation determinants of Antirrhinum majus MADS-box transcription factors. Nucleic Acids Res. 26: 5277–5287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuest S.E., O’Maoileidigh D.S., Rae L., Kwasniewska K., Raganelli A., Hanczaryk K., Lohan A.J., Loftus B., Graciet E., Wellmer F. (2012). Molecular basis for the specification of floral organs by APETALA3 and PISTILLATA. Proc. Natl. Acad. Sci. USA 109: 13452–13457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan W., Chen D., Kaufmann K. (2016). Molecular mechanisms of floral organ specification by MADS domain proteins. Curr. Opin. Plant Biol. 29: 154–162. [DOI] [PubMed] [Google Scholar]
- Ye L., Wang B., Zhang W., Shan H., Kong H. (2016). Gains and losses of cis-regulatory elements led to divergence of the Arabidopsis APETALA1 and CAULIFLOWER duplicate genes in the time, space, and level of expression and regulation of one paralog by the other. Plant Physiol. 171: 1055–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S.D., Cho Y.H., Sheen J. (2007). Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2: 1565–1572. [DOI] [PubMed] [Google Scholar]
- Zheng Y., Ren N., Wang H., Stromberg A.J., Perry S.E. (2009). Global identification of targets of the Arabidopsis MADS domain protein AGAMOUS-Like15. Plant Cell 21: 2563–2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T., Yang L., Lu Y., Dror I., Dantas Machado A.C., Ghane T., Di Felice R., Rohs R. (2013). DNAshape: a method for the high-throughput prediction of DNA structural features on a genomic scale. Nucleic Acids Res. 41: W56–W62. [DOI] [PMC free article] [PubMed] [Google Scholar]