Basic helix-loop-helix (bHLH) proteins, are important regulators of eukaryotic transcription and are particularly numerous in plants, playing roles in development and environmental responses (Pires and Dolan, 2010; Feller et al., 2011). Members of this family of transcription factors are generally characterized by the presence of the HLH signature motif required for homo- or heterodimer formation, and dimerization is essential for DNA binding when the HLH is accompanied by the adjacent basic region. Frequently, bHLH proteins harbor other conserved motifs that participate in protein-protein interactions central for their regulatory function.
Cui et al. (2016) recently reported the in planta interactions of the Arabidopsis thaliana bHLH factor DYT1 (DYSFUNCTIONAL TAPETUM1) with three other related members, bHLH010, bHLH089, and bHLH091, and the roles the three bHLH factors played in promoting the nuclear localization of DYT1 and transcriptional activation by DYT1 on target genes. DYT1 had been previously described as essential for male fertility in Arabidopsis by regulating key genes for anther and pollen development (Zhang et al., 2006). In addition to the bHLH domain necessary for DNA binding, DYT1 and bHLH010/089/091 share a domain conserved in topology, which Cui et al. named the BIF domain for bHLH protein interaction and function. The BIF domain is required for the dimerization, in vivo function, and transcriptional activity of DYT1. The authors proposed that this plant-specific domain is also present in 59/158 Arabidopsis bHLH proteins (Cui et al., 2016).
In this letter, we want to clarify that this purported novel BIF domain was previously described in studies of maize (Zea mays) and Arabidopsis bHLH proteins and was referred to as the ACT-like domain (Anantharaman et al., 2001; Feller et al., 2006; Kong et al., 2012) (Figure 1). The ACT-like domain in the maize bHLH transcription factor R (RED1) is important for the regulation of anthocyanin biosynthesis and behaves as a switch that permits distinct configurations of a regulatory complex to be tethered to different anthocyanin pathway gene promoters (Feller et al., 2006; Kong et al., 2012). The ACT-like domain mediates homodimerization and is essential for the transcriptional activity of R, similar to what was reported by Cui et al. for the DYT1 BIF domain. Using sequence-structure homology recognition analyses, ACT-like domains were identified over a decade ago in ∼30% of all the Arabidopsis bHLH transcription factors, including DYT1, but not in bHLH010/089/091 (Feller et al., 2006). Our inability to detect ACT-like domains in bHLH group II (to which bHLH010/089/091 belong) back then could have been a consequence of a significantly smaller structure database or limitations in the algorithm used. Below, we provide a brief historical perspective of ACT and ACT-like domains and how sequence/structural diversity can result in the misclassification of related motifs.
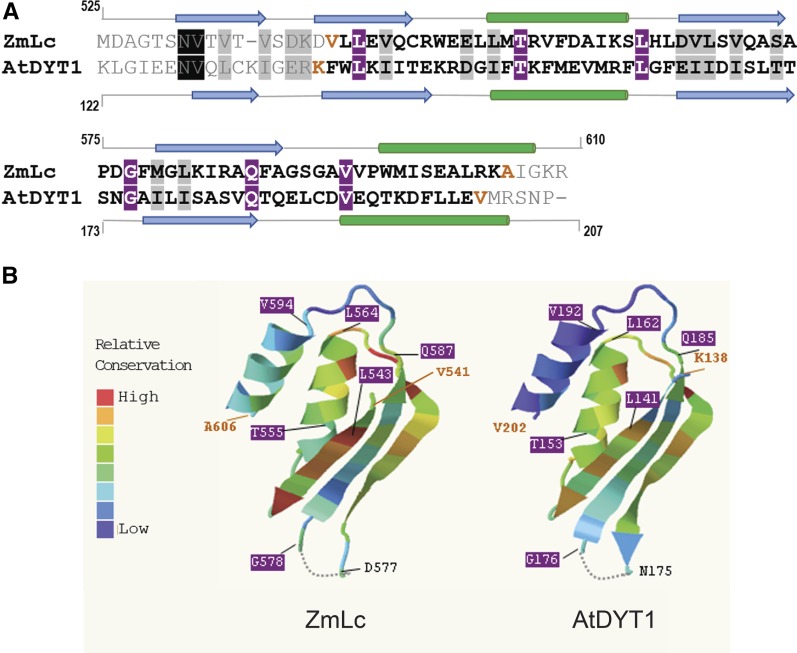
Figure 1.
Sequence and Structure Comparison of Maize R (represented by ZmLc) and AtDYT1.
(A) Alignment of amino acids 525 to 610 of ZmLc (R family member) and amino acids 122 to 207 of AtDYT1 was created by the ClustalW Sequence Alignment Program v1.83 (http://www.genome.jp/tools/clustalw/) and displayed with Expasy Boxshade (http://embnet.vital-it.ch/software/BOX_form.html). Highly conserved amino acids are indicated with black or purple boxes, while gray boxes indicate less conserved ones. Orange-colored amino acids indicate the beginning and end of the structure in (B). The secondary structure was analyzed with the Predictprotein secondary structure prediction program (www.predictprotein.org); blue arrows indicate β-sheets and green cylinders α-helices.
(B) The 3D structures of the protein sequences depicted in (A) for ZmLc (left) and AtDYT1 (right) were predicted using the Phyre2 Protein Homology/Analogy Recognition Engine V2.0, followed by in-depth analysis of model quality using Phyre Investigator (Kelley et al., 2015). For both proteins, the library entry d1u8sa2, which corresponds to the ACT-like superfamily ferredoxin-like fold of the glycine-cleavage system transcriptional repressor, was used by Phyre2 to model the 3D structures. Phyre2 was used to perform conservation analyses of each amino acid; those shown in red correspond to high and those in blue to low conservation with respect to d1u8sa2. The dashed line at the bottom of both structures represents regions with undetermined conformation.
The ACT domain was originally recognized by iterative BLAST searches seeded with the amino acid sequence of the small subunit of the acetolactate synthase and named after the three enzymes in which it was initially found, aspartate kinase-chorismate mutase-TyrA (prephenate dehydrogenase) (Aravind and Koonin, 1999). The original suggestion that the ACT domain corresponded to a binding domain for regulatory small molecules was subsequently confirmed by a number of functional and structural studies (Chipman and Shaanan, 2001; Curien et al., 2008). ACT domains are widely present in amino acid biosynthetic enzymes where they participate in allosteric regulation often involving the formation of homodimers. They are generally 60 to 80 amino acids long and adopt a βαββαβ secondary structure, although significant variants in this topological arrangement have been described (Curien et al., 2008). Amino acid sequence identities between ACT domains are very low, possibly because of the variety of ligands to which they can bind. For example, despite adopting similar βαββαβ folds, the ACT domains in threonine deaminase (TD) and 3-phosphoglycerate dehydrogenase have no recognizable amino acid sequence similarity, resulting in Chipman and Shaanan (2001) suggesting the use of “ACT-like” to describe the structurally related domain in TD. Some proteins involved in the regulation of amino acid biosynthesis contain an ACT-like domain with the βαββαβ fold (named RAM for regulator of amino acid metabolism), which may differ from ACT domains in the ligand binding region (Ettema et al., 2002). Besides their presence in bHLH factors, ACT-like domains have been found in additional proteins in Arabidopsis and other plants, for example, as part of the ACR (ACT domain repeats) protein family (Hsieh and Goodman, 2002; Liu, 2006; Sung et al., 2011). More recently, four tandem ACT domains were identified in the human Cellular Arginine Sensor for mTORC1 (CASTOR) proteins and shown to participate in arginine sensing (Chantranupong et al., 2016; Saxton et al., 2016). To date, the protein family database (Pfam PF01842, http://pfam.xfam.org/family/ACT) shows 15,117 sequences containing ACT domains from 4123 species with 133 different domain architectures and 159 diverse structures. The ACT-like domains in plant bHLH proteins, including those in R and DYT1, show a ββαββα topology (Feller et al., 2006; Cui et al., 2016) (Figure 1), compared with the more characteristic βαββαβ topology of ACT domains (Curien et al., 2008). However, three independent studies conducted on the crystal structure of arginine-bound CASTOR1 proposed somewhat different ACT domain topologies (βαββαβ, ββαββα, and other variants) (Gai et al., 2016; Saxton et al., 2016; Xia et al., 2016), suggesting some flexibility in the structure of the domain.
It is clear from these studies that the BIF domain is an ACT-like domain. While ultimately it is just semantics whether this domain is named BIF or ACT-like, we feel that it is important for the research community to be aware that they correspond to the same structural motif with diverse functions.
Acknowledgments
Research on ACT-like domains is supported by National Science Foundation Grant MCB-1513807 in the E.G. lab and by the National Science Foundation under Cooperative Agreement 1355438 to L.Y.
AUTHOR CONTRIBUTIONS
A.F., L.Y., and E.G. wrote the manuscript.
Footnotes
Articles can be viewed without a subscription.
References
- Anantharaman V., Koonin E.V., Aravind L. (2001). Regulatory potential, phyletic distribution and evolution of ancient, intracellular small-molecule-binding domains. J. Mol. Biol. 307: 1271–1292. [DOI] [PubMed] [Google Scholar]
- Aravind L., Koonin E.V. (1999). Gleaning non-trivial structural, functional and evolutionary information about proteins by iterative database searches. J. Mol. Biol. 287: 1023–1040. [DOI] [PubMed] [Google Scholar]
- Chantranupong L., Scaria S.M., Saxton R.A., Gygi M.P., Shen K., Wyant G.A., Wang T., Harper J.W., Gygi S.P., Sabatini D.M. (2016). The CASTOR proteins are arginine sensors for the mTORC1 pathway. Cell 165: 153–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chipman D.M., Shaanan B. (2001). The ACT domain family. Curr. Opin. Struct. Biol. 11: 694–700. [DOI] [PubMed] [Google Scholar]
- Cui J., You C., Zhu E., Huang Q., Ma H., Chang F. (2016). Feedback regulation of DYT1 by interactions with downstream bHLH factors promotes DYT1 nuclear localization and anther development. Plant Cell 28: 1078–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curien G., Biou V., Mas-Droux C., Robert-Genthon M., Ferrer J.L., Dumas R. (2008). Amino acid biosynthesis: new architectures in allosteric enzymes. Plant Physiol. Biochem. 46: 325–339. [DOI] [PubMed] [Google Scholar]
- Ettema T.J., Brinkman A.B., Tani T.H., Rafferty J.B., Van Der Oost J. (2002). A novel ligand-binding domain involved in regulation of amino acid metabolism in prokaryotes. J. Biol. Chem. 277: 37464–37468. [DOI] [PubMed] [Google Scholar]
- Feller A., Hernandez J.M., Grotewold E. (2006). An ACT-like domain participates in the dimerization of several plant basic-helix-loop-helix transcription factors. J. Biol. Chem. 281: 28964–28974. [DOI] [PubMed] [Google Scholar]
- Feller A., Machemer K., Braun E.L., Grotewold E. (2011). Evolutionary and comparative analysis of MYB and bHLH plant transcription factors. Plant J. 66: 94–116. [DOI] [PubMed] [Google Scholar]
- Gai Z., Wang Q., Yang C., Wang L., Deng W., Wu G. (2016). Structural mechanism for the arginine sensing and regulation of CASTOR1 in the mTORC1 signaling pathway. Cell Discov. 2: 16051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh M.-H., Goodman H.M. (2002). Molecular characterization of a novel gene family encoding ACT domain repeat proteins in Arabidopsis. Plant Physiol. 130: 1797–1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley L.A., Mezulis S., Yates C.M., Wass M.N., Sternberg M.J.E. (2015). The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10: 845–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong Q., Pattanaik S., Feller A., Werkman J.R., Chai C., Wang Y., Grotewold E., Yuan L. (2012). Regulatory switch enforced by basic helix-loop-helix and ACT-domain mediated dimerizations of the maize transcription factor R. Proc. Natl. Acad. Sci. USA 109: E2091–E2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q. (2006). Computational identification and systematic analysis of the ACR gene family in Oryza sativa. J. Plant Physiol. 163: 445–451. [DOI] [PubMed] [Google Scholar]
- Pires N., Dolan L. (2010). Origin and diversification of basic-helix-loop-helix proteins in plants. Mol. Biol. Evol. 27: 862–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxton R.A., Chantranupong L., Knockenhauer K.E., Schwartz T.U., Sabatini D.M. (2016). Mechanism of arginine sensing by CASTOR1 upstream of mTORC1. Nature 536: 229–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung T.Y., Chung T.Y., Hsu C.P., Hsieh M.H. (2011). The ACR11 encodes a novel type of chloroplastic ACT domain repeat protein that is coordinately expressed with GLN2 in Arabidopsis. BMC Plant Biol. 11: 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia J., Wang R., Zhang T., Ding J. (2016). Structural insight into the arginine-binding specificity of CASTOR1 in amino acid-dependent mTORC1 signaling. Cell Discov. 2: 16035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Sun Y., Timofejeva L., Chen C., Grossniklaus U., Ma H. (2006). Regulation of Arabidopsis tapetum development and function by DYSFUNCTIONAL TAPETUM1 (DYT1) encoding a putative bHLH transcription factor. Development 133: 3085–3095. [DOI] [PubMed] [Google Scholar]