A ketoacyl CoA synthase (PotriKCS1) is key to the biosynthesis of alkenes in the cuticular waxes of poplar leaves.
Abstract
Alkenes are linear hydrocarbons with one or more double bonds. Despite their potential as biofuels and precursors for specialty chemicals, the underlying biochemistry and genetics of alkene biosynthesis in plants remain elusive. Here, we report on a screen of natural accessions of poplar (Populus trichocarpa), revealing that the leaf cuticular waxes are predominantly composed of alkanes and alkenes. Interestingly, the accumulation of alkenes increases with leaf development, is limited to the abaxial side of the leaf, and is impaired in a few accessions. Among other genes, a β-ketoacyl CoA synthase gene (PotriKCS1) was downregulated in leaves from non-alkene-producing accessions. We demonstrated biochemically that PotriKCS1 elongates monounsaturated fatty acids and is responsible for the recruitment of unsaturated substrates to the cuticular wax. Moreover, we found significant associations between the presence of alkenes and tree growth and resistance to leaf spot. These findings highlight the crucial role of cuticular waxes as the first point of contact with the environment, and they provide a foundation for engineering long-chain monounsaturated oils in other species.
INTRODUCTION
The cuticle is a thin film that covers the surfaces of all terrestrial plants (Riederer and Schreiber, 1995). As such, the cuticle serves important roles in nonstomatal water loss prevention, defense against pathogens and insects, and plant development (Bernard and Joubès, 2013; Bargel et al., 2006). The cuticle is made of cutin, a polymeric matrix of oxygenated lipids and waxes, a complex mixture predominated by fatty acid (FA)-derived metabolites, interspersed with triterpenoids, flavonoids, and phenolic lipids (Haslam and Kunst, 2013; von Wettstein‐Knowles, 2012). Cuticular wax composition varies among species, tissues, and developmental stages (Busta et al., 2017; Guo et al., 2017; Cameron et al., 2002).
The biosynthesis of the FA-derived metabolites starts in the plastids of epidermal cells from palmitic (C16:0) and stearic (C18:0) acyl carrier protein (ACP)-bound acyl chains. The free FAs are subsequently transported out of the plastid and esterified to CoA. A four-step process is then initiated by the fatty acid elongation (FAE) complex, a set of endoplasmic reticulum-bound enzymes that elongate the C16:0- and C18:0-CoAs into very-long-chain fatty acids (VLCFAs). Elongation is facilitated by repeated rounds of four reactions starting with condensation of malonyl-CoA and an acyl-CoA primer to produce a β-ketoacyl-CoA (catalized by β-ketoacyl CoA synthase [KCS]) (Haslam and Kunst, 2013). Different KCSs are required during elongation to synthesize the C20 to C34 VLCFAs, which ultimately act as precursors during wax biosynthesis (von Wettstein-Knowles, 1982).
Free VLCFAs can be transported directly to the cuticle after removal of the CoA, or they can enter one of the wax biosynthetic pathways. The pathway then branches to produce several compound classes differing in decorating functionality: one branch produces alkanes, secondary alcohols, and ketones (alkane-forming pathway), while another produces primary alcohols and wax esters (alcohol-forming pathway). The products of these pathways are widespread in plant cuticular waxes, and many of the enzymes involved have been characterized (Bernard and Joubès, 2013; Samuels et al., 2008; Yeats and Rose, 2013; Metz et al., 2000; Lardizabal et al., 2000; Rowland et al., 2006). In contrast, it is much less clear how alkenes, cyclopropanes, and internally branched alkanes are synthesized, but this is proposed to occur via an uncharacterized enoic pathway (von Wettstein-Knowles, 2007). Previous studies describing alkenes in maize (Zea mays) silk and barley (Hordeum vulgare) spike waxes eluded to this independent pathway, but it is currently unclear which genes or enzymes are involved in this process in planta (Perera et al., 2010; von Wettstein-Knowles, 2007). Thus, the biosynthesis of alkenes in plants has remained obscure.
Populus trichocarpa (black cottonwood) is an ecologically and economically important forest species that occupies a large climatic range, spanning the west coast of North America from California to Alaska. This widespread distribution has translated into large phenotypic variation to adapt to very different environments (McKown et al., 2013; Evans et al., 2014). Here, we report the chemical diversity of cuticular waxes found in a collection of unrelated, wild accessions of P. trichocarpa. We found alkenes as predominant components of the wax and exploited the inherent diversity in wax composition to identify an enzyme involved in the enoic pathway for the biosynthesis of alkenes.
RESULTS
Diversity in Cuticular Wax Composition in P. trichocarpa Leaves
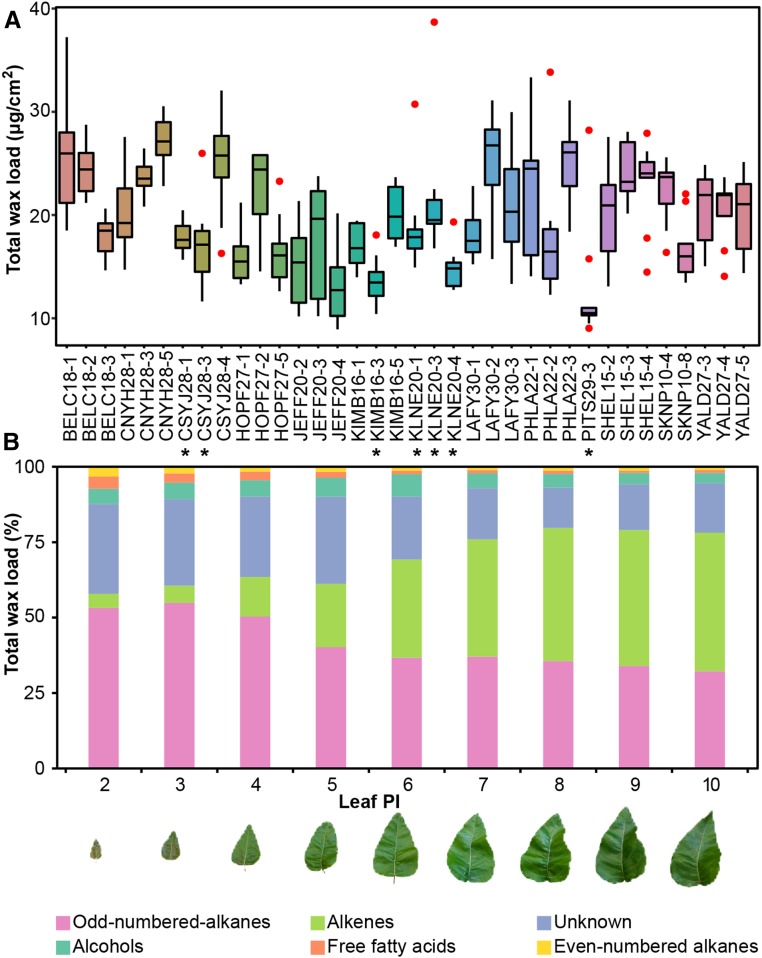
We investigated the natural variation in wax load and composition in the leaves of 36 accessions of P. trichocarpa collected from the natural range of distribution of the species in western North America and grown in a common garden (Supplemental Data Set 1) (McKown et al., 2013). From each accession, nine leaves spanning plastochron index (PI) 2 through 10 were sampled to investigate the underlying changes in wax accumulation that accompany leaf ontogeny (Lamoreaux et al., 1978). This developmental series therefore included both expanding (PI-2 to 6) and fully expanded leaves (PI-7 to 10). Using gas chromatography and mass spectroscopy (GC-MS), 45 analytical signals (peaks) were detected. Overall, a 4-fold variation in total wax load was observed among samples (Figure 1). Moreover, within an accession, large differences were observed in the contribution of each chemical class to the total wax load as leaves expanded (Figure 1B). These data indicate that, even when grown under the same environmental conditions, accessions of P. trichocarpa differ in the accumulation of cuticular waxes.
Figure 1.
Variation in Leaf Cuticular Wax in P. trichocarpa.
(A) Large variation in the dispersion of the data across the nine samples (tall box plot for PHLA22-1 and short in PITS29-1). Box plots show the lower and upper quartiles and the median of the total wax load collected from nine leaves sampled from 36 natural accessions. Outliers are shown as red dots. Stars point to accessions that fail to accumulate alkenes (AM).
(B) Changes in relative abundance of chemical classes with leaf ontology. Odd- and even-numbered alkanes, alkenes, alcohols, free fatty acids, and unidentified peaks in leaves derived from BELC18-1 spanning PI-2 through 10. Representative leaves from plastochron index 2 through 10 are shown below.
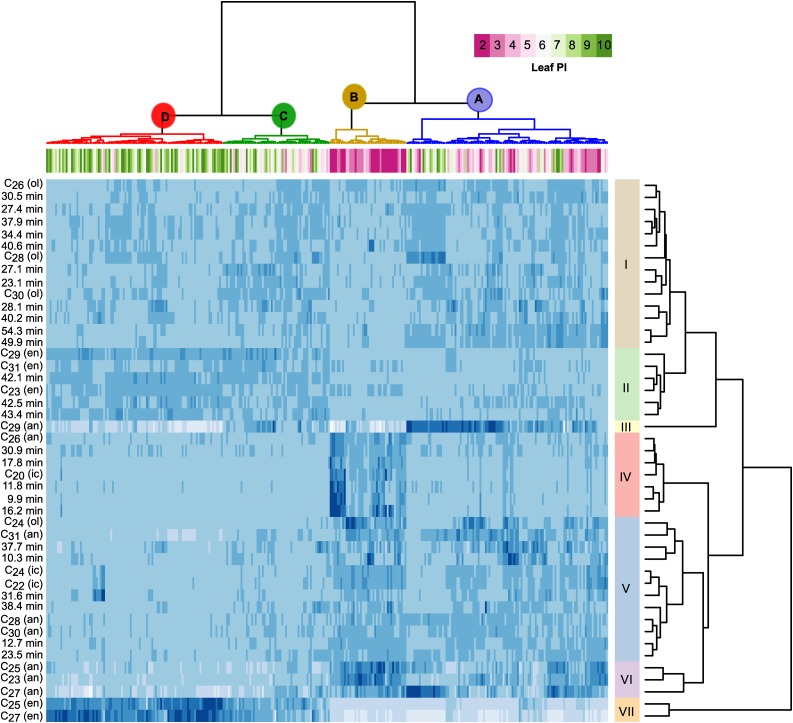
The high level of variation among accessions and within leaves from the same accession suggested that wax composition in P. trichocarpa is highly plastic. To identify patterns of variation, we performed hierarchical clustering analysis. Samples fell into four distinct clusters (A, B, C, and D; Figure 2). Clusters A and B were predominantly composed of young expanding leaves. PI-2 leaves differed from the other expanding leaves largely due to the presence of peaks from group IV and the higher amounts of alkanes and FAs found within them. Group IV consists of seven peaks that were found almost exclusively in PI-2 leaves and rapidly decreased below detection levels as leaves expanded (Supplemental Figure 1). Wax biosynthesis appears to be more active in the younger leaves, slowing as leaves expand, which manifests in an apparent reduction in most compounds per leaf area. In contrast, clusters C and D were enriched in fully expanded leaves (Figure 2). Five of the peaks that are more abundant in expanded leaves correspond to the monoalkene homologous series, ranging in chain lengths from C23 to C31 (Supplemental Figure 2).
Figure 2.
Clustering of Accessions by Developmental Stage.
Two-way cluster analysis of analytical signatures from P. trichocarpa cuticular waxes. Samples (columns), labeled by leaf plastochron index (Leaf PI), and grouped into four clusters. Peaks (rows) grouped into seven clusters. an, alkane; en, alkene; 01, alcohol; ic, fatty acid. Unidentified analytical signals are labeled with the retention time in minutes. In the heat map, darker shades correspond to higher concentration of the compound.
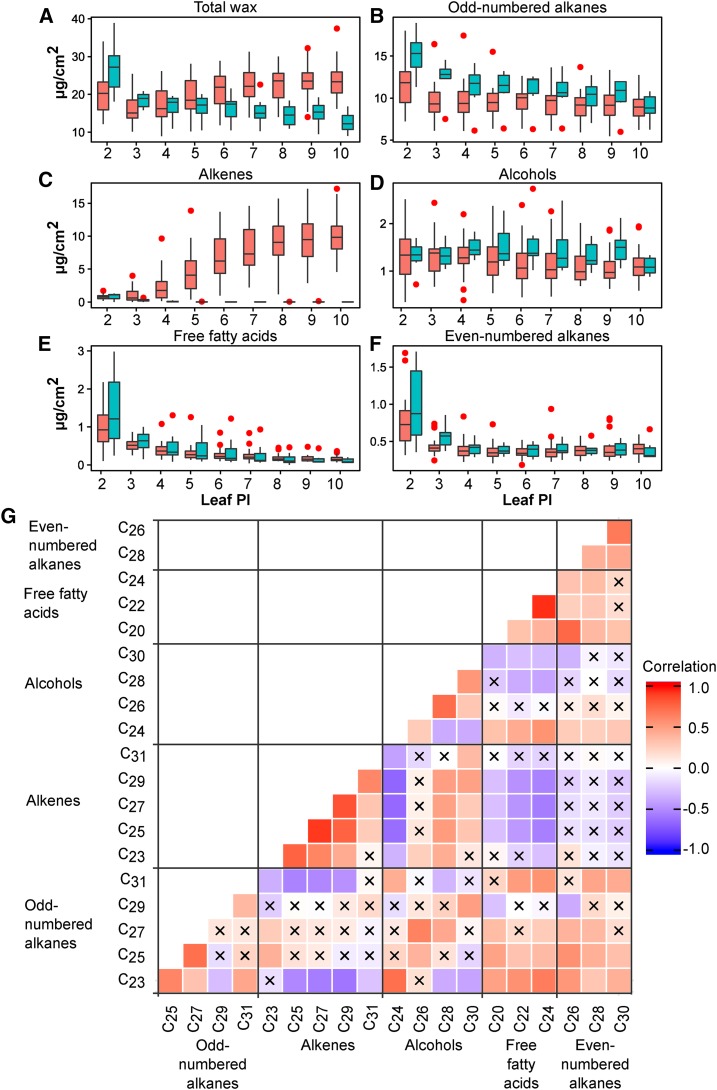
The influence of leaf ontology on wax profile was most evident in the alkene and the free FAs homologous series (Figure 3). Alkene levels exponentially increased as leaves expanded, from an average of 0.79 ± 0.07 μg/cm2 in PI-2 leaves (3.7% of the total wax load) to 8.36 ± 0.81 μg/cm2 in PI-10 leaves (34.9%; Figure 3C). In contrast, a reduction in the free saturated FAs detected was apparent (Figure 3E). However, the mass balance between FAs and alkenes did not support a precursor-product relationship between these compounds.
Figure 3.
Changes in Wax Quantity with Leaf Ontology.
(A) to (F) Box plots were used to summarize the data from 29 AP (pink) and 7 AM (blue) accessions. Outliers are shown as red dots. The total wax load (A), odd-numbered alkanes (B), alkenes (C), alcohols (D), free fatty acids (E), and even-numbered alkanes (F) are shown for leaves spanning PI-2 through 10.
(G) Pearson’s correlation heat map for wax classes. Wax components were grouped according to chemical classes. Positive correlations are shown in red and negative correlations in blue (see correlation color key). Coefficients with P values higher than 0.000125 (Bonferroni corrected P value) are marked with an X to represent that they are not significant. Samples from AM accessions were removed from analysis to focus on correlations in the presence of alkenes.
To gather information on the biochemical origin of alkenes, we used Pearson correlations to infer relationships among wax components (Figure 3G). With a few exceptions, compounds from the same chemical class had high correlation values, as would be expected from metabolites derived from the same biosynthetic pathway. Among compound classes, odd-numbered alkanes were positively correlated with even-numbered alkanes and FAs, suggesting that their accumulation was coordinated. In contrast, alkenes were negatively correlated with odd-numbered alkanes and FAs.
Thus, hierarchical analysis confirmed that leaf ontogeny has a strong influence on the cuticular wax profile of poplar, where young leaves of different accessions are more similar to each other than to older leaves from the same tree.
Natural Mutants in Alkene Accumulation
Surprisingly, the increase in alkene levels with leaf development was not observed across all accessions. Seven accessions failed to accumulate alkenes in fully expanded leaves. These accessions will hereafter be referred to as AM (alkene minus) to differentiate them from AP (alkene plus) accessions, which accumulate alkenes.
The absence of alkenes was most evident in the wax of fully expanded P. trichocarpa leaves. PI-10 leaves from AM accessions had a 47% reduction in the total wax load relative to AP leaves (12.58 ± 1.17 μg/cm2 in AM compared with 23.66 ± 0.86 μg/cm2 in AP; Figure 3A). This indicated that the absence of alkenes in expanded leaves was not fully compensated for by the synthesis of other compound classes.
Despite the fact that alkane and alkene chain lengths match (C23 to C31), their relative distribution was not the same. The alkane biosynthetic machinery has a preference for C30 precursors; therefore, nonacosane (C29) is the predominant alkane. In contrast, the alkene biosynthetic machinery displays a preference for C26 and C28 precursors. This observation and the negative correlation between alkanes and alkenes imply that different biochemical mechanisms are controlling their accumulation.
Leaf Surfaces Differ in Their Wax Profiles
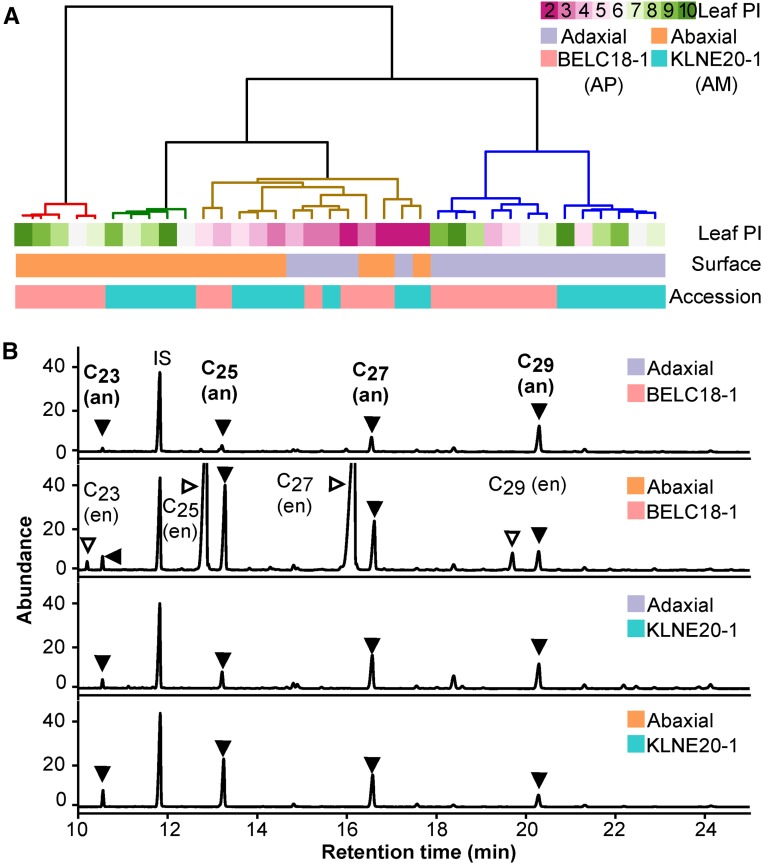
Alkenes show a developmentally controlled pattern of accumulation. Therefore, we investigated whether this pattern was present on both sides of the leaf. We determined the wax composition of the adaxial and abaxial surfaces by collecting waxes separately from either side of one AP and one AM accession. We then performed hierarchical clustering to assess how developmental stage, surface, and accession affect leaf wax composition (Figure 4). Wax profiles from young leaves clustered together, regardless of leaf surface or accession (Figure 4A). However, samples from PI-6 and higher fell into different clusters depending on whether they came from abaxial or adaxial surfaces. This indicated that early in leaf ontology, both surfaces had similar wax profiles, but as leaves reached maturity, there was a clear difference between the abaxial and adaxial surface.
Figure 4.
Leaf Surfaces Differ in Wax Composition as They Develop.
(A) Dendogram grouping samples collected from nine leaves, two leaf surfaces, and two accessions.
(B) Gas chromatogram traces of waxes collected from the adaxial and abaxial surfaces of leaf PI-10 from BELC18-1 (AP) and KLNE20-1 (AM). IS, internal standard tetracosane; an, alkane series marked with filled triangles; en, alkene alkane series marked with open triangles.
A major difference between leaf surfaces was the accumulation of alkenes (Figure 4B). Only the abaxial side of BELC18-1 showed a 200-fold increase in the accumulation of alkenes with leaf development, resulting in the abaxial surface containing 98 times more alkenes than the associated adaxial surface of the same leaf (Supplemental Figure 3). Notably, the alkene chain length distribution was similar on both surfaces, with pentacosene (C25) and heptacosene (C27) dominating. The lack of accumulation of alkenes in some genotypes (KLNE 20-1) did not affect the biosynthesis of other compound classes with the same chain length (Supplemental Figure 4). All these observations highlight the differential regulation of wax biosynthesis on the adaxial and abaxial surfaces, and they suggest that spatially regulated FAE systems exist in the same leaf.
KCS Is a Candidate Gene for the Biosynthesis of Alkenes
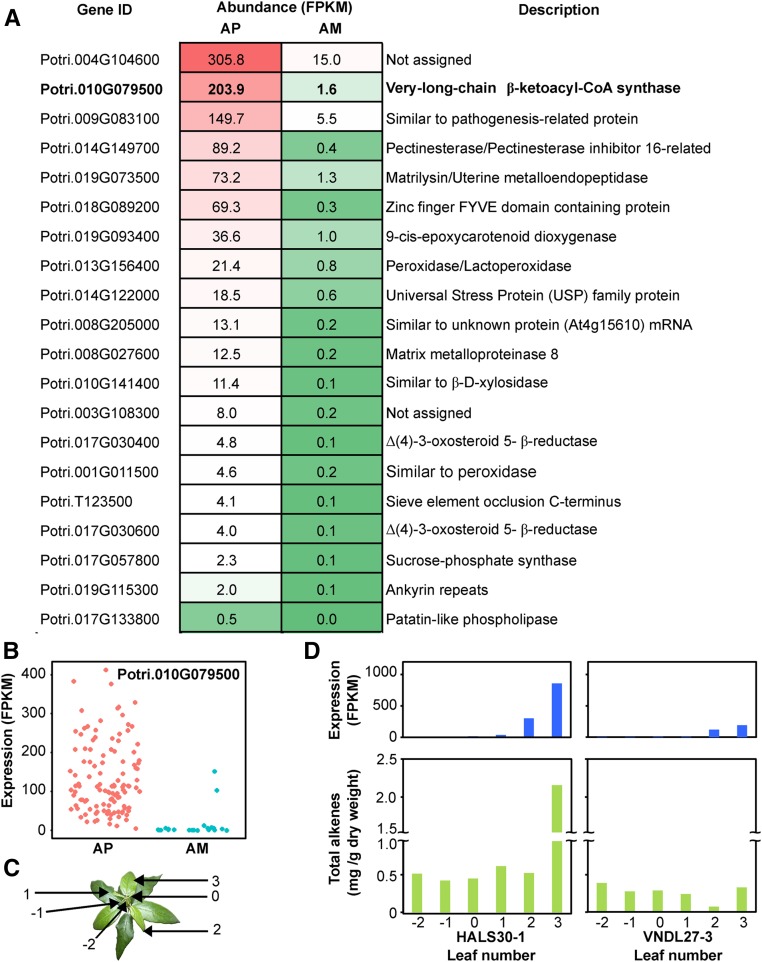
To identify putative genes involved in the biosynthesis or regulation of alkenes, we combined the whole leaf transcriptomes of 16 AP accessions and compared them to the transcriptomes of six AM accessions. A total of 258 genes were found to be differentially regulated, including genes involved in protein synthesis, hormone metabolism, signaling, response to biotic and abiotic stress, and transport (Supplemental Data Set 2). Notably, a putative KCS encoded by Potri.010G079500 (PotriKCS1) was identified as it displayed high levels of expression in AP accessions (Figure 5). KCSs, which are part of the FAE complex, take an acyl-CoA primer and condense it with malonyl-CoA, catalyzing the initial and rate limiting step in the elongation of VLCFAs (Haslam and Kunst, 2013). KCS activity is known to determine the chain length and the degree of saturation in VLCFAs (Blacklock and Jaworski, 2006). This led to the hypothesis that PotriKCS1 utilizes a monounsaturated acyl-CoA primer, culminating in the accumulation of alkenes in the cuticular waxes of P. trichocarpa.
Figure 5.
PotriKCS1 Expression Predicts Alkene Accumulation.
(A) The 20 most significantly upregulated genes in AP compared with AM accessions. Average read values in units of fragments per kilobase per million fragments mapped (FPKM) are shown.
(B) RNA abundance of Potri.010G079500 in an independent set of 136 unrelated accessions grouped by their alkene phenotype.
(C) Representative plant showing the leaves that were harvested to test the developmental expression.
(D) Developmental gene expression and accumulation of alkenes in leaves from early developmental stages. Top panel: expression of Potri.010G079500 with leaf ontogeny for accessions HALS30-6 (left) and VNDL27-3 (right). Bottom panel: total amount of alkenes (C25 to C31) per gram of dry weight.
To validate PotriKCS1 as a candidate gene, we screened the leaf wax of 139 additional accessions housed in the same common garden. During this process, 23 further accessions were identified as AM. The average expression levels for PotriKCS1 in all AM accessions were 13.4 ± 7.6 FPKM (mean ± se), compared with 134.2 ± 8.5 across all AP accessions (Figure 5B).
Alkene accumulation in P. trichocarpa leaves is delayed developmentally compared with the accumulation of other wax compounds. To test if PotriKCS1 expression is responsible for the developmental gradient, we analyzed the transcriptomes of six leaves at different developmental stages. A sharp increase in gene expression was indeed observed as the leaves transition from PI-1 to 2 preceding the drastic increase in alkene production seen in PI-2 and 3, suggesting that alkene accumulation is controlled at the transcriptional level by the expression of PotriKCS1 (Figure 5C). Similarly, in Arabidopsis thaliana, the increase in alkane accumulation parallels the upregulation of gene expression of the KCS CER6 (Busta et al., 2017).
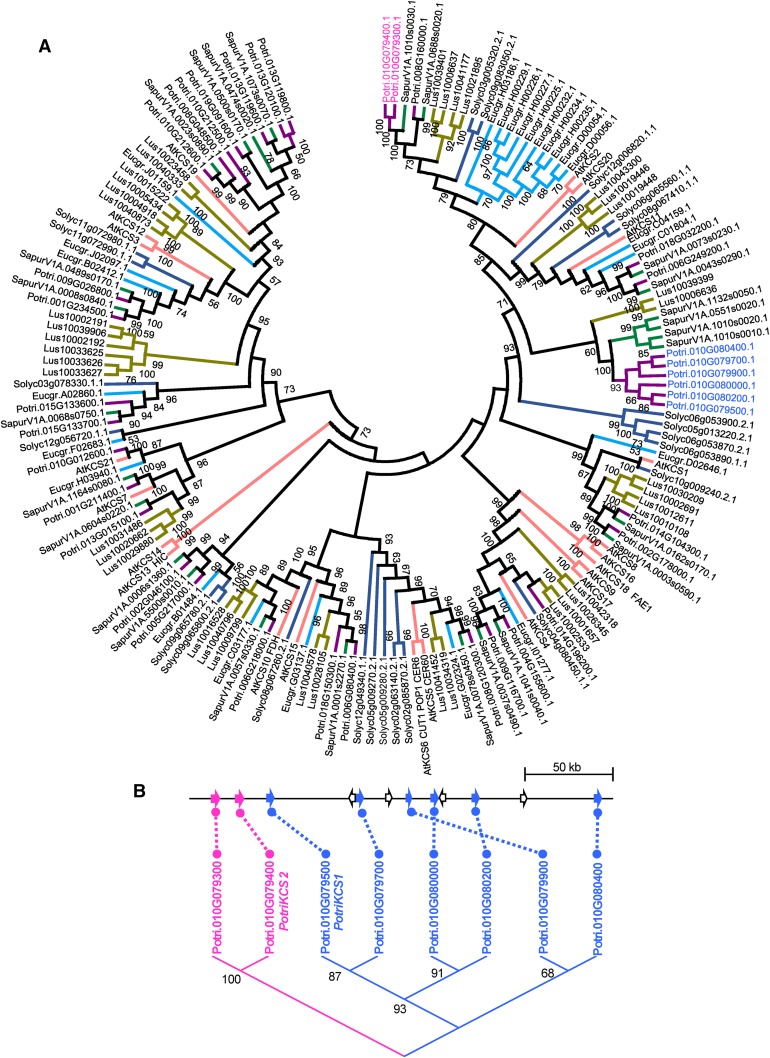
We reconstructed the phylogenetic relationships of KCSs in plants (Figure 6; Supplemental File 1). Several cases of lineage-specific expansions due to tandem duplications were detected (Guo et al., 2016; Joubès et al., 2008), indicating that KCS-encoding genes have a high probability of retention following tandem duplication (Supplemental Data Set 3; Figure 6A). Notably, in P. trichocarpa, 17 out of 37 KCS are the result of tandem duplication, and eight are located within a 210-kb region on chromosome 10, including PotriKCS1. The eight KCSs on chromosome 10 cluster into sequence-related pairs in two clades (Figure 6B). The first clade is composed of the most distant pair: Potri.010G079300 and Potri.010G079400 (PotriKCS2 herein), displaying 77% identity to PotriKCS1 in the coding sequence. The second clade is composed of six genes with more than 97% sequence identity. The low sequence divergence and gene family expansion after the split from Salix 52 million years ago (Dai et al., 2014) indicates that the KCS cluster containing PotriKCS1 has recently evolved.
Figure 6.
Phylogenetic Analysis of PotriKCS1.
(A) Minimum evolution phylogenetic tree for the KCS gene family. P. trichocarpa (purple), Salix purpurea (green), Linum usitatissimum (brown), Eucalyptus grandis (light blue), Arabidopsis (red), and Solanum lycopersicum (dark blue). Bootstrap values ≥ 50 are shown. Protein alignment was conducted using MUSCLE. The evolutionary distances were computed using the p-distance method. The analysis involved 169 amino acid sequences. Potri.010G012500 was removed from the analysis because there were no overlapping regions between this sequence and Potri.010G012600. Evolutionary analyses were conducted in MEGA6.
(B) KCS gene cluster on chromosome 10. Putative KCS-encoding genes are shown as filled arrows, and non-KCS encoding genes are shown as empty arrows. A maximum likelihood tree is shown below, with bootstrap values depicted above the branches.
Physiological Traits as a Function of Alkene Phenotype
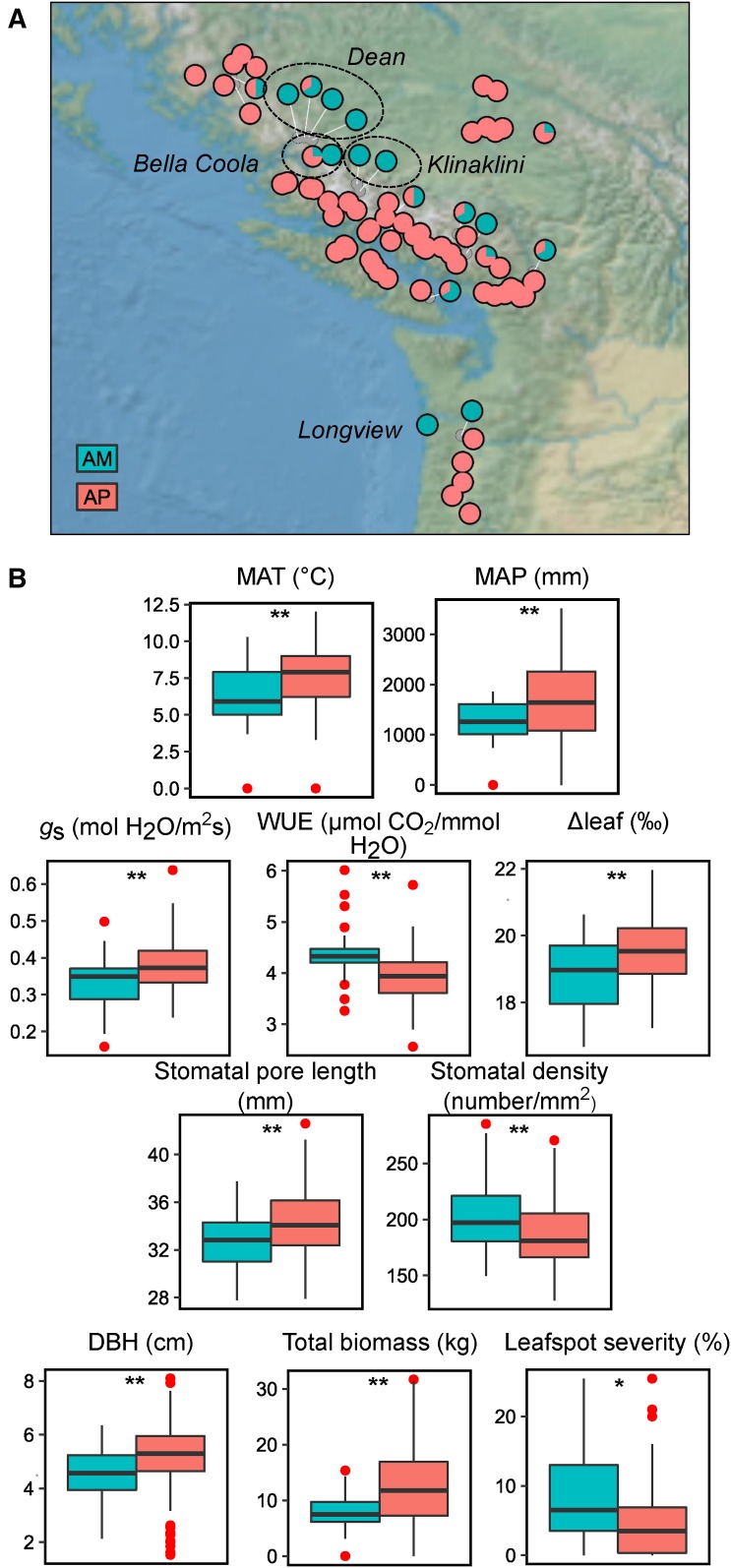
Eighty-three percent of the accessions employed in this study accumulated alkenes in mature leaves. To examine the geographical distribution of the alkene phenotypes, we mapped AM and AP accessions to their place of origin. Despite the widespread distribution of AM accessions without correlation to geographic origin, they do generally stem from regions with lower seasonal precipitation and temperature (Figure 7).
Figure 7.
Distribution and Effects of the Absence of Alkenes.
(A) Geographical distribution of P. trichocarpa accessions in British Columbia and Oregon classified according to their alkene phenotype. River drainages with clusters of AM accessions are shown.
(B) Climatic variables, biomass, ecophysiology, and disease resistance traits were compared as a function of accumulation of alkenes. AP is in pink, AM is in blue, and outliers are shown as red dots. MAT, mean annual temperature; MAP, mean annual precipitation; DBH, diameter at breast height; gs, stomatal conductance; WUE, instantaneous water-use efficiency (photosynthetic rate/transpiration); Δleaf, net leaf carbon isotope discrimination. Asterisks denote significant differences between groups at P value < 0.05 (*) and <0.01 (**) (Welsch’s t test).
As the first barrier against the environment, the cuticle can influence water relations and physiology (Agrawal et al., 2009). AM accessions have less wax per leaf area than AP accessions and a profile dominated by saturated lipids. These differences in quantity and composition could affect other tree traits. To test this, several previously published traits were compared as a function of the alkene phenotype (McKown et al., 2014a, 2014b; La Mantia et al., 2013). Alkene absence did not appear to affect photosynthetic rate, but reduced transpiration: AM trees showed lower stomatal conductance and higher instantaneous water-use efficiency (WUE; Figure 7B; Supplemental Data Set 4). It is possible that the reduced wax load in the AM trees increases cuticle permeability, which in turn leads to tighter control of water loss through the stomata. This could be achieved by smaller and more numerous stomata on the surfaces of AM trees (Figure 7B), which might be an adaptation to the drier, more arid regions from which AM trees commonly originate.
In contrast to the instantaneous WUE, the 13C discrimination rate is a measurement of the long-term WUE of the leaf. Δleaf was lower in AM trees versus AP trees, suggesting lower long-term WUE (Figure 7B). However, wax composition itself can affect carbon discrimination measurements, as wax is depleted in 13C relative to total plant carbon (Conte et al., 2003). As such, the higher Δleaf in AP could also be a consequence of the higher wax load per surface area.
Stomata are the point of entry for several common pathogens of poplar trees, such as the rust-causing Melampsora spp and the leaf spot pathogen Septoria musiva (Hacquard et al., 2011; Thompson, 1941). Consistent with the higher number of stomata per leaf surface, AM trees were more susceptible to leaf spot than their AP counterparts (Figure 7B). Remarkably, AP accessions displayed better overall growth than AM accessions, as indicated in their significantly higher diameters at breast height and stem and total biomass after 4 years of field growth in the common garden (Figure 7B). This suggests that the presence of alkenes in mature leaves has a large impact on tree physiology, growth, and biomass accrual.
PotriKCS1 Elongates Monounsaturated FAs
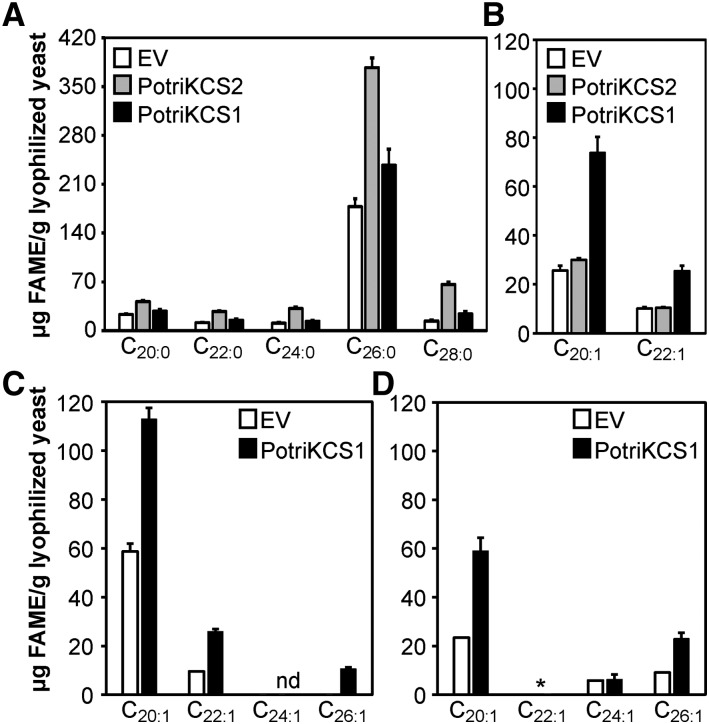
Gene duplication can provide the means for the evolution of new enzymatic functions by allowing selection to modify the activity of one of the gene copies (Hanada et al., 2008). To test this hypothesis, PotriKCS1 and the adjacent gene PotriKCS2 were expressed in yeast (Saccharomyces cerevisiae) and their biochemical activities compared. S. cerevisiae has two elongation proteins that are responsible for the biosynthesis of saturated and monounsaturated VLCFAs, with hexacosanoic acid being the predominant product (Oh et al., 1997). Upon induction of the cultures with galactose, strains containing either PotriKCS1 or PotriKCS2 independently displayed increased total VLCFA contents. The expression of PotriKCS2 led to a 2.3-fold increase in the amount of saturated products (307 μg fatty acid methyl ester [FAME]/g lyophilized yeast), ranging in size from eicosanoic to octacosanoic acid, relative to the empty vector control, and an increase of 13% (5 μg FAME/g lyophilized yeast) in the unsaturated products of eicosenoic and docosenoic acid (Figure 8). In contrast, the expression of PotriKCS1 increased the saturated VLCFAs content to 34% (80 μg FAME/g lyophilized yeast) and the monounsaturated by a 2.8-fold (63 μg FAME/g lyophilized yeast) (Figure 8B). This indicates that PotriKCS1 and PotriKCS2 are both able to elongate endogenous S. cerevisiae FAs but that they differ in their substrate preference. In yeast, PotriKCS1 elongates monounsaturated and saturated FAs, whereas PotriKCS2 preferentially elongates saturated FAs. This ability to elongate saturated and monounsaturated FAs is shared by other KCSs (Kunst et al., 1992; Mietkiewska et al., 2004; Tresch et al., 2012).
Figure 8.
Heterologous Expression of Poplar KCS Increases the Amount of VLCFAs in Yeast.
Changes in the total amount of saturated (A) and monounsaturated (B) FAMEs upon expression of PotriKCS1 and PotriKCS2 in comparison with an empty vector control (EV). Bars represent the average of four biological replicates (independent yeast transformants). Error bars indicate standard errors. Changes in monounsaturated fatty acids upon addition of C18:1Δ9 (C) and C22:1Δ13 (D). nd, not detected; *, supplemented to the medium.
Position of the Double Bond Suggests Oleic Acid Is a PotriKCS1 Substrate
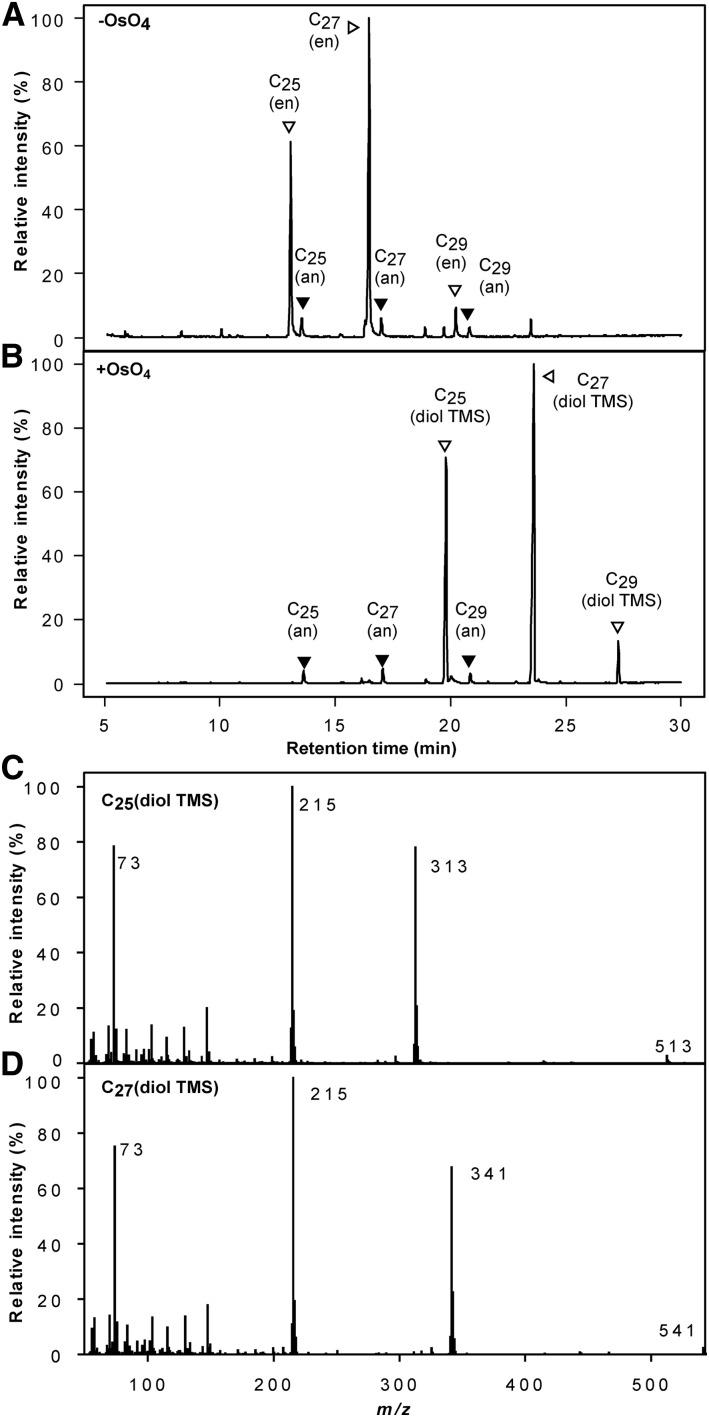
To determine the position of the double bond, the alkenes were oxidized with OsO4, producing α-diols with hydroxyl groups marking the position of the double bond (Capella and Zorzut, 1968). The fragment pairs 215/313 and 215/341 obtained from the C25- and C27-diol trimethylsilylated derivatives indicate cleavage of the carbon-carbon bond between carbon 9 and 10 (Figure 9). The ion fragments at 513 and 541 corresponding to M-15 ions were also observed. No positional isomers were identified from the mass spectra, suggesting that each peak corresponds to just one analytical signal.
Figure 9.
Determination of Double Bond Position in Alkenes from P. trichocarpa.
GC-MS chromatograms of alkene-enriched fraction before (A) and after (B) oxidation with osmium tetroxide and subsequent derivatization. Mass spectrum of the C25- (C) and C27-diol TMS derivative (D) showing the major ions resulting from cleavage at the original position of the double bond, as resolved on an HP-1 column run on a 6890 N Network GC. an, alkane; en, alkene; diol TMS, diol trimethylsilylated.
The double bond at carbon 9 on both alkenes suggests that the biosynthesis begins with a monounsaturated FA with the double bond at this position, as is the case of oleic acid (C18:1Δ9). To support this finding, we supplemented the yeast induction medium with two FAs with a double bond at carbon 9. The addition of oleic acid resulted in a 53% increase in C22:1 FAME levels (relative to medium not supplemented with oleic acid) and the appearance of C26:1 FAME (Figure 8C; Supplemental Figure 5). The levels of C26:1 were further increased when the longer C22:1 Δ13 (erucic acid) was supplied to the medium (Figure 8D). Therefore, C18:1Δ9 and C22:1Δ13 are metabolizable substrates that increase the production of monounsaturated FAs by PotriKCS1.
DISCUSSION
Several observations indicate that the production of alkenes is independent of the biosynthesis of other wax components. First, the absence of alkenes in AM accessions did not affect the accumulation of alkanes with the same carbon length. Second, the absence was not compensated for by the accumulation of other wax hydrocarbons. Third, alkene levels are negatively correlated with the levels of other wax chemical classes. Fourth, only alkenes are developmentally and spatially regulated. These observations are consistent with the enoic pathway branching out early on and utilizing a different substrate pool. In poplar leaves, we show that this was achieved through the recruitment of PotriKCS1, an elongation enzyme with the capacity to utilize substrates like oleic acid (C18:1Δ9).
Recruiting oleic acid for the biosynthesis of cuticular waxes has permitted mature P. trichocarpa leaves to double their wax load. In most vegetative tissues, oleic acid is used in glycerolipid biosynthesis; therefore, in epidermal cells, the enoic pathway would need to compete with glycerolipids for the substrate. In such a scenario, partitioning between both pathways may be determined by the supply of C18:1Δ9, which may explain why although monounsaturated FAs are widespread in plants, their use as substrates for the biosynthesis of cuticular waxes is limited. For example, oleic acid is abundant in Arabidopsis seeds and hence can be elongated into VLCFAs in this tissue (Post-Beittenmiller, 1996). Several cell compartments could supply the proposed C18:1Δ9 precursor used by PotriKCS1: It could come from C18:1Δ9-ACP synthesized in the plastid, from C18:0-CoA after desaturation in the endoplasmic reticulum, or be released from membranes by lipases. The involvement of a lipase in the biosynthesis of cuticular waxes has recently been proposed (Schneider et al., 2016; Kosma and Rowland, 2016), and in our study, 11 lipase genes were found among the list of differentially regulated genes between AM and AP accessions (Supplemental Data Set 2). Future experiments will look at the role of these and other lipid-related enzymes in the biosynthesis of alkenes. Additionally, a complete lipid profile would help determine if the downregulation of PotriKCS1 affects other lipid classes beyond waxes. For instance, downregulation of PotriKCS1 in AM trees could result in the accumulation of C18:1Δ9 in glycerolipids, or it could be redirected elsewhere, such as to sphingolipids or cutin.
Once monounsaturated FAs are elongated by PotriKCS1, they would still need to undergo terminal modification by CER1/CER3 to yield odd-numbered alkenes (Bernard et al., 2012). In theory, the mechanism for loss of carbon could be the same as that used in the biosynthesis of alkanes, if the enzymes of the alkane-forming pathway could also use monounsaturated substrates.
Gene duplication can provide the means for the evolution of new enzymatic functions by allowing selection to modify the activity of one of the gene copies (Des Marais and Rausher, 2008). PotriKCS1 is part of a recently evolved gene cluster that contains eight KCS-encoding genes. KCSs, in contrast to the other three enzymes of the FAE complex, have a narrow substrate specificity (Millar and Kunst, 1997). Therefore, it is not surprising that selection favors the maintenance of duplicates to increase metabolic diversity. This is especially true in long-lived organisms such at trees (i.e., Populus, in this case), as having multiple copies of KCS genes may provide the adaptive ability to optimize the utilization of monounsaturated fatty acids as substrates and cope with diverse geographies.
In vivo approaches using yeast as a heterologous expression system have proven very valuable in the characterization of ER-bound KCS enzymes (Joubès et al., 2008; Tresch et al., 2012; Trenkamp et al., 2004; Blacklock and Jaworski, 2006). Nevertheless, these approaches are limited by the availability of VLCFAs present in native yeast that can be used as substrates. The substrate availability issue has been proposed to be responsible for the differences in activity observed between yeast expression and plant mutant analysis (Lassner et al., 1996). This might explain why even though PotriKCS1 is able to elongate saturated FAs in yeast, alkanes and other saturated wax classes are not affected by the lower expression of PotriKCS1 in AM accessions.
The ability of PotriKCS1 and 2 to elongate monounsaturated and saturated FAs in yeast would indicate that the ancestral protein had activity on both, but after duplication, PotriKCS1 became specialized to use monounsaturated FAs. PotriKCS1 and 2 fall into the same clade as Arabidopsis KCS2 and 20, two condensing enzymes that have been reported to produce predominantly saturated VLCFAs up to 26 carbons in length and are implicated in suberin biosynthesis in roots and wax formation in stems (Tresch et al., 2012; Trenkamp et al., 2004; Lee et al., 2009; Franke et al., 2009; Blacklock and Jaworski, 2006). This suggests that KCSs are very plastic enzymes, whose substrate specificity can quickly change to increase wax diversity.
Even though heterologous expression of PotriKCS1 in yeast led to the production of monounsaturated FAs up to C26, it is likely that in planta more than one KCS is involved in elongating monounsaturated FAs up to the observed C32. Given the 97% sequence identity among the six KCSs in the clade, functional redundancy is expected and the other copies may work on unsaturated FAs of different chain lengths. Tandem duplication could have assisted in the evolution of alkene biosynthesis, as there is a high frequency of recombination between paralogs to generate sequence variation (Hanada et al., 2008).
In most P. trichocarpa trees, leaf wax is dominated by alkenes. The allocation of alkenes to leaf cuticular wax represents an important carbon sink (mature leaves of AP accessions accumulate 9.5 ± 0.4 μg/cm2 of alkenes). Nevertheless, doing so potentially provides an adaptive advantage to those plants, since in the common garden, AP trees accumulated 60% more biomass on average than AM trees. This growth advantage probably led to strong selection for AP accessions in most of the distribution range of the species. However, in the common garden, AP accessions had higher transpiration rates and lower WUE (as measured by gas exchange) than AM accessions. Although the increased gas exchange seems maladaptive, it is possible that it is required to support the improved growth of these accessions. In the drier and colder regions where AM accessions are usually found, a tighter control of water loss may be required. Interestingly, a mutation in a KCS gene that affects stomatal density and WUE has been reported in the C24 ecotype of Arabidopsis. Reduced expression of the HIC gene, encoding a KCS, resulted in increased stomatal density in response to elevated CO2 (Gray et al., 2000). Similarly, the wax mutants eceriferum-g in barley and cer1 and cer6 in Arabidopsis have reduced wax phenotypes accompanied by altered stomatal density compared with the wild type (Gray et al., 2000; Zeiger and Stebbins, 1972).
The fast growth of Populus tree species is hindered by several foliar and stem pathogens like the necrotroph fungus Septoria musiva. Chemical and biological control of Septoria leaf spot have proven ineffective, therefore entrusting genetic resistance as the best alternative for managing the disease (Liang et al., 2014). Traditionally, increased resistance comes with an associated cost in plant growth, which has proven to be the case in the Populus-Septoria interaction (Herms and Mattson, 1992; Liang et al., 2014). Despite this, AP trees were found to have better yield and increased resistance relative to AM accessions. Therefore, this study implicates cuticular waxes as an additional resistance factor against Septoria leaf spot, one that does not come at the expense of tree growth. Interestingly, resistance to leaf spot increases with leaf ontogeny (Dunnell and LeBoldus, 2016), matching the increase in the accumulation of alkenes. More detailed studies will be needed to test if the effect is direct and a function of the different composition and/or quantity of wax present in AP trees, or indirect through the reduced number of stomata associated with the presence of alkenes.
Although the association of alkenes with other traits does not indicate causation, we take it to indicate that the presence of alkenes and/or alteration of wax profiles can have a large impact on tree physiology. Further experiments would be required to determine if and how the accumulation of leaf alkenes leads to the observed effects on growth, resistance, and water use and if this trait is an adaptation to specific niches. Ultimately, this knowledge can be used to breed or engineer poplar for better growth and resilience to a changing climate.
METHODS
Plant Material
Leaf samples were taken from wild accessions of Populus trichocarpa originally collected by the British Columbia Ministry of Forests, Lands, and Natural Resource Operations and grown in a common garden at Totem Field in the University of British Columbia (McKown et al., 2013). The 36 accessions used in this study were selected to cover most of the distribution range of the collection sites. Accessions from Northern latitudes were not included because they had already entered dormancy by mid August, when the samples were harvested. From each tree, nine consecutive leaves were taken from the same branch. The leaf PI was used to identify leaves from similar morphological and physiological stages given the variability in leaf development across accessions (Lamoreaux et al., 1978).
Cuticular Wax Analysis
For cuticular wax extraction, two leaf discs of 13 mm in diameter were collected, avoiding mid veins and damaged areas. Waxes were extracted by dipping the discs for 30 s in 10 mL of HPLC-grade chloroform containing 10 μg of n-tetracosane (Sigma-Aldrich) as an internal standard. Samples were allowed to evaporate at room temperature, resuspended in 300 μL of chloroform, and transferred to 2 mL-insert-containing vials. Samples were dried under nitrogen before the addition of 10 μL of N,O-bis(trimethylsilyl) trifluoroacetamide and 10 μL of pyridine (Sigma-Aldrich) for derivatization for 60 min at 75°C. Derivatization reagents were then evaporated under nitrogen and the samples resuspended in 200 μL of chloroform for analysis with a GC-FID (gas chromatography-flame ionization detector). The amount of total wax was determined in comparison with the area of the internal standard and expressed per unit leaf area (cm2).
To collect waxes from separate leaf surfaces, a glass fiber swap method was used (Buschhaus and Jetter, 2011). The method was initially tested to confirm that waxes were only being extracted from one surface by spiking high quantities of tetracosane in the opposite surface. For sample collection, a 16-mm-wide leaf disc was carefully swiped with a chloroform-saturated swab 30 times in 1 min. In between swipes, the glass fiber was dipped in chloroform containing 10 μg of n-tetracosane. Three developmentally equivalent leaves from the same tree were used as replicates and showed comparable quantities of individual compounds, indicating that the method was reproducible. However, because this method cannot guarantee uniform removal of cuticular waxes per unit of surface area, the variation is larger compared with the disc-dipping method.
All samples were run on a 30 m × 320 μm × 1 μm HP-1 column on an HP 5890 Series II GC system with an FID detector (Agilent). The oven temperature was kept at 50°C for 2 min, increased at 40°C/min to 200°C, kept at 200°C for 1 min, raised at 3°C/min to 320°C, and maintained at 320°C for 15 min. For peak identification, samples were run as described previously on a 6890 N Network GC (Agilent) (Busta et al., 2016). Wax components were identified by comparing their mass spectra with those available in an in-house library, as well as authentic standards when available.
To validate PotriKCS1 as a candidate gene, frozen leaves from 139 accessions were used to collect cuticular waxes. Similarly, frozen samples from leaves of different developmental stages of HALS30-1 and VNDL27-3 were used to determine the amount of alkenes in the corresponding samples that were used for RNA-seq. In this case, the amount was determined in comparison with the area of tetracosane and expressed per gram of dry weight.
Analysis of metabolite data was conducted in R and METAGENassist (Arndt et al., 2012). For hierarchical clustering, samples were normalized (sum of 1) to make them comparable and peak concentrations normalized with a Pareto scaling. Euclidean distance and Ward clustering were used.
Leaf Transcriptome
The first fully uncurled leaf from field-grown trees was collected and used for RNA sequencing. The expression values for 31 samples coming from 16 AP trees (two replicates for most accessions) were averaged and compared with the average of 11 samples from six AM trees. Leaf PI-2 was chosen for the analysis based on the assumption that the upregulation of gene expression precedes the synthesis of enzymes and the accumulation of metabolites. Additionally, a developmental series set was available for accessions HALS30-6 and VNDL23-7. This consisted of leaves of PI-1 to 2 and the meristematic leaves above (labeled as 0, −1, and −2 from older to younger) (Figure 5C).
Total RNA was extracted with a Purelink kit (Invitrogen) and cleaned with an RNeasy plant mini kit (Qiagen). Quantity and quality were checked with a 2100 BioAnalizer (Agilent). Samples with an RNA integrity number ≥ 9 were used with a Nextera DNA library preparation kit (Illumina) to generate 75-nucleotide pair-end reads on the Illumina HiSeq 2000 platform. Short reads were trimmed for adapter sequences and quality using Trimmomatic version 0.32 (Bolger et al., 2014) (minimum quality 20 over 4-bp sliding window, minimum length of 50 bp) before alignment to the P. trichocarpa (version 3) genome using TopHat v2.0.8 (Trapnell et al., 2009) (mean inner distance 300 bp, 20 bp sd between reads). Reads mapping to multiple locations were given a map quality of 0. For FPKM calculations, reads mapping to more than one location were first excluded and then reassigned to the relevant transcript (or unique region). This information was then used to recalculate the FPKM. Using Cuffdiff v2.1.1, 258 genes were found to be differentially expressed between AP and AM samples at a P value of 0.05 (Trapnell et al., 2012).
Phylogenetic Analysis
A BLASTP search with a cutoff value of E-50 using Arabidopsis thaliana KCS protein sequences was performed in Phytozome to find KCS homologs in the genomes of P. trichocarpa (v3.0) (Tuskan et al., 2006), willow (Salix purpurea) (v1.0) (DOE-JGI, http://phytozome.jgi.doe.gov/pz/portal.html#!info?alias=Org_Spurpurea), flax (Linum usitatissimum) (v1.0) (Wang et al., 2012b), and eucalyptus (Eucalyptus grandis) v2.0 (Myburg et al., 2014), and the Sol Genomics Network for tomato (Solanum lycopersicum) (ITAG release 2.40) (Tomato Genome Consortium, 2012). This led to the identification of 37 putative KCS-encoding genes in poplar, a larger number than previously reported, likely due to the inclusion of sequences that lack one of the two protein domains (Guo et al., 2016). A total of 174 predicted proteins were aligned with MUSCLE (Edgar, 2004), and 169 amino acid sequences were used to determine phylogenetic relationships with a minimum evolution method and 10,000 bootstrap replications. Phylogenetic analysis was performed in MEGA6 (Tamura et al., 2013).
KCS-encoding genes were defined as originating from tandem duplications if the start codons were located within 120 kb from each other, even if they were separated by non-KCS genes (Supplemental Data Set 3). The selection of a 120-kb window was based on previous studies on tandem duplication in P. trichocarpa (Hanada et al., 2008).
Leaf Disc Bioassays
Leaf discs were taken from healthy greenhouse-grown vegetative shoot cuttings. Inoculations were performed as described previously (Ward and Ostry, 2005; Dowkiw et al., 2003). For Septoria musiva, discs were inoculated with 105 spores/mL of strain 6-1 obtained from Indian Head, Saskatchewan. Percent disease area was measured 14 d postinoculation using digital pixel analysis with WinFOLIA (Regent Instruments). Rust assays were performed with two species Melampsora larici-populina (MLP05Berth3729) and M. × columbiana (isolate collected in Vancouver, British Columbia). Leaf discs were inoculated with a solution of 8 mg of urediniospores in 100 mL of 0.01% agar in water. The total number of postules per leaf disc was counted 14 d postinoculation. Inoculated discs were incubated under controlled growth chamber conditions (18°C, 18-h photoperiod, mix of fluorescent and incandescent lights at an intensity of 400–500 µmoles/m2/s).
Yeast FAME Analysis
To determine the enzyme activity of PotriKCS1, Potri.010G079500 and the adjacent Potri.010G079400 were expressed in a galactose inducible system in Saccharomyces cerevisiae. The coding sequences of Potri.010G079400 and Potri.010G079500 were amplified from cDNA derived from leaves of P. trichocarpa accession HALS30-6 (AP) using primers designed from the reference genome sequence (PotriKCS1-F 5′-CACCATGGCAGATGAGAAGAAACAAAACC-3′, PotriKCS1-R stop 5′-TTAATAAACAAGAGGTGCCACTCTAGGC-3′, PotriKCS2-F 5′-CACCATGGCAGAGGATCAGAGCAAACAAAG-3′, and PotriKCS2-R stop 5′-TCAAGAGGCGATGGGCACCACTTTAG-3′). The coding sequences were inserted into the pENTR/D-TOPO vector (Invitrogen) and sequence verified. The sequence for Potri.010G079500 from HALS30-6 differed at two nucleotides from the reference sequence (370 T> C that results in a Ser-to-Pro mutation and a silent mutation 1128 A>G), whereas the sequence for Potri.010G079400 had two synonymous mutations (150 A>C and 542 G>A). The genes were transferred into the pYES-DEST52 Gateway (Invitrogen) vector though LR Clonase reaction and transformed into S. cerevisiae strain INVSc1 (Invitrogen). As a negative control for the KCS activity assay, pENTR-GUS containing the Arabidopsis β-glucuronidase (GUS) gene was recombined with pYES-DEST52 (referred to as empty vector control).
Overnight cultures in synthetic complete (SC) medium lacking uracil and supplied with 2% (w/v) glucose were used to inoculate 10 mL of SC lacking uracil supplied with 1% raffinose and 2% galactose. In experiments with fed substrates, FAs were dissolved in acetone at a concentration of 0.5 mM with 0.2% (w/v) tergitol and added 2 d after induction. Four days after induction, the cultures were centrifuged, washed twice with 2.5% sodium chloride, and freeze-dried.
FAs from 8 to 10 mg of freeze-dried cells were transmethylated with methanolic HCl containing 10 μg of triheptadecanoin and 0.2% butylated hydroxytoluene for 2 h at 80°C (Trenkamp et al., 2004). FAMEs were collected in pentane and analyzed by GC-FID on a 30 m × 0.25 mm × 0.5-μm DB-WAX column. Peaks were identified by comparison of retention times with authentic standards, and spectra were collected on a Polaris Q GC-MS (Thermo Finnigan) equipped with a 30 m × 320 μm × 0.1-μm Rtx-1 (Restek).
Alkene Chemistry
For alkene purification, waxes were collected with n-pentane from the abaxial surface of 20 fully expanded leaves from a pool of AP accessions to enrich for the presence of alkenes relative to alkanes. The wax mixture was concentrated in vacuo in a rotary evaporator. An aliquot was separated on a 21-cm SyliaFlash G60 (Silicycle) flash column eluted with n-pentane. One-milliliter fractions were collected and run on a GC-MS as described previously. The alkene-enriched fraction consisting of 79% alkenes and 8% alkanes (as determined by peak area) was used for the following steps.
To determine the position of the double bond, osmium tetroxide was used to hydroxylate alkenes (von Wettstein-Knowles, 2007; Capella and Zorzut, 1968). Roughly 1 mg of the alkene-enriched fraction was oxidized by the addition of 2 mg of osmium tetroxide in 200 μL of dioxane. The reaction was allowed to proceed for 90 min at room temperature before the addition of 3 mL of 16% Na2SO3 in water (w/v) and 3.3 volumes of methanol. The resulting α-diols were collected with 2 mL of pentane, followed by centrifugation to aid phase separation. The recovered pentane layer was washed with water. Dried α-diols were derivatized with of N,O-bis(trimethylsilyl) trifluoroacetamide and pyridine and examined by GC-MS as described for wax extractions.
Accession Numbers
The accession numbers of sequences used in this article can be found in Supplemental Data Set 3. Sequences for PotriKCS1 (Potri.010G079500) and PotriKCS2 (Potri.010G079400) can be found in Phytozome (https://phytozome.jgi.doe.gov/pz/portal.html). The raw reads from the RNA-seq experiment can be found at the NCBI Sequence Read Archive under accession number PRJNA300564 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA300564).
Supplemental Data
Supplemental Figure 1. GC profiles of waxes collected from expanding leaves of PHLA22-1.
Supplemental Figure 2. Identification of monoalkenes identified in P. trichocarpa leaves.
Supplemental Figure 3. Wax composition in BELC18-1.
Supplemental Figure 4. Wax composition in KLNE20-1.
Supplemental Figure 5. Identification of elongated products in yeast.
Supplemental Data Set 1. List of accessions used in this study.
Supplemental Data Set 2. List of genes differentially expressed between AM and AP accessions.
Supplemental Data Set 3. List of sequences used in the phylogenetic analysis.
Supplemental Data Set 4. Alkene accumulation as a proxy of other traits.
Supplemental File 1. Protein sequence alignment used to produce the phylogenetic tree shown in Figure 6.
Acknowledgments
We thank Lacey Samuels for discussions during the course of this research, Simon Turner and Erick Cardenas for providing feedback on this manuscript, and Lucas Busta and Reinhard Jetter for mass spectral interpretation. This work was funded by the Genome Canada Large Scale Applied Research Project (168BIO) and the U.S. Department of Energy, Great Lakes Bioenergy Research Center (DOE Office of Science BER DE-FC02-07ER64494) held by S.D.M. Carl Douglas, a co-principal investigator of the POPCAN project under which this research was conducted, passed away on July 25, 2016. We wish to honor his memory with this work.
AUTHOR CONTRIBUTIONS
E.G.-V. and S.D.M. were responsible for the conception and planning of the project. E.G.-V. performed research and analyzed data. C.A.H. analyzed transcriptome data. M.E.v.L. performed cuticular wax extractions. J.L.M. designed and performed leaf disc bioassays. The manuscript was written by E.G.-V. and S.D.M. with input from all other authors.
Glossary
- FA
fatty acid
- FAE
fatty acid elongation
- VLCFA
very-long-chain fatty acid
- KCS
β-ketoacyl CoA synthase
- GC-MS
gas chromatography and mass spectroscopy
- WUE
water-use efficiency
- FAME
fatty acid methyl ester
Footnotes
Articles can be viewed without a subscription.
References
- Agrawal A.A., Fishbein M., Jetter R., Salminen J.-P., Goldstein J.B., Freitag A.E., Sparks J.P. (2009). Phylogenetic ecology of leaf surface traits in the milkweeds (Asclepias spp.): chemistry, ecophysiology, and insect behavior. New Phytol. 183: 848–867. [DOI] [PubMed] [Google Scholar]
- Arndt D., Xia J., Liu Y., Zhou Y., Guo A.C., Cruz J.A., Sinelnikov I., Budwill K., Nesbø C.L., Wishart D.S. (2012). METAGENassist: a comprehensive web server for comparative metagenomics. Nucleic Acids Res. 40: W88–W95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bargel H., Koch K., Cerman Z., Neinhuis C. (2006). Evans Review No. 3: Structure–function relationships of the plant cuticle and cuticular waxes — a smart material? Funct. Plant Biol. 33: 893–910. [DOI] [PubMed] [Google Scholar]
- Bernard A., Domergue F., Pascal S., Jetter R., Renne C., Faure J.-D., Haslam R.P., Napier J.A., Lessire R., Joubès J. (2012). Reconstitution of plant alkane biosynthesis in yeast demonstrates that Arabidopsis ECERIFERUM1 and ECERIFERUM3 are core components of a very-long-chain alkane synthesis complex. Plant Cell 24: 3106–3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard A., Joubès J. (2013). Arabidopsis cuticular waxes: advances in synthesis, export and regulation. Prog. Lipid Res. 52: 110–129. [DOI] [PubMed] [Google Scholar]
- Blacklock B.J., Jaworski J.G. (2006). Substrate specificity of Arabidopsis 3-ketoacyl-CoA synthases. Biochem. Biophys. Res. Commun. 346: 583–590. [DOI] [PubMed] [Google Scholar]
- Bolger A.M., Lohse M., Usadel B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buschhaus C., Jetter R. (2011). Composition differences between epicuticular and intracuticular wax substructures: how do plants seal their epidermal surfaces? J. Exp. Bot. 62: 841–853. [DOI] [PubMed] [Google Scholar]
- Busta L., Budke J.M., Jetter R. (2016). Identification of β-hydroxy fatty acid esters and primary, secondary-alkanediol esters in cuticular waxes of the moss Funaria hygrometrica. Phytochemistry 121: 38–49. [DOI] [PubMed] [Google Scholar]
- Busta L., Hegebarth D., Kroc E., Jetter R. (2017). Changes in cuticular wax coverage and composition on developing Arabidopsis leaves are influenced by wax biosynthesis gene expression levels and trichome density. Planta 245: 297–311. [DOI] [PubMed] [Google Scholar]
- Cameron K.D., Teece M.A., Bevilacqua E., Smart L.B. (2002). Diversity of cuticular wax among Salix species and Populus species hybrids. Phytochemistry 60: 715–725. [DOI] [PubMed] [Google Scholar]
- Capella P., Zorzut C.M. (1968). Determination of double bond position in monounsaturated fatty acid esters by mass spectrometry of their trimethylsilyloxy derivatives. Anal. Chem. 40: 1458–1463. [Google Scholar]
- Conte M.H., Weber J.C., Carlson P.J., Flanagan L.B. (2003). Molecular and carbon isotopic composition of leaf wax in vegetation and aerosols in a northern prairie ecosystem. Oecologia 135: 67–77. [DOI] [PubMed] [Google Scholar]
- Dai X., et al. (2014). The willow genome and divergent evolution from poplar after the common genome duplication. Cell Res. 24: 1274–1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Des Marais D.L., Rausher M.D. (2008). Escape from adaptive conflict after duplication in an anthocyanin pathway gene. Nature 454: 762–765. [DOI] [PubMed] [Google Scholar]
- Dowkiw A., Husson C., Frey P., Pinon J., Bastien C. (2003). Partial resistance to Melampsora larici-populina leaf rust in hybrid poplars: Genetic variability in inoculated excised leaf disk bioassay and relationship with complete resistance. Phytopathology 93: 421–427. [DOI] [PubMed] [Google Scholar]
- Dunnell K.L., LeBoldus J.M. (2016). The correlation between Septoria leaf spot and stem canker resistance in hybrid poplar. Plant Dis. 101: 464–469. [DOI] [PubMed] [Google Scholar]
- Edgar R.C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32: 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans L.M., Slavov G.T., Rodgers-Melnick E., Martin J., Ranjan P., Muchero W., Brunner A.M., Schackwitz W., Gunter L., Chen J.-G., Tuskan G.A., DiFazio S.P. (2014). Population genomics of Populus trichocarpa identifies signatures of selection and adaptive trait associations. Nat. Genet. 46: 1089–1096. [DOI] [PubMed] [Google Scholar]
- Franke R., Höfer R., Briesen I., Emsermann M., Efremova N., Yephremov A., Schreiber L. (2009). The DAISY gene from Arabidopsis encodes a fatty acid elongase condensing enzyme involved in the biosynthesis of aliphatic suberin in roots and the chalaza-micropyle region of seeds. Plant J. 57: 80–95. [DOI] [PubMed] [Google Scholar]
- Gray J.E., Holroyd G.H., van der Lee F.M., Bahrami A.R., Sijmons P.C., Woodward F.I., Schuch W., Hetherington A.M. (2000). The HIC signalling pathway links CO2 perception to stomatal development. Nature 408: 713–716. [DOI] [PubMed] [Google Scholar]
- Guo H.-S., Zhang Y.-M., Sun X.-Q., Li M.-M., Hang Y.-Y., Xue J.-Y. (2016). Evolution of the KCS gene family in plants: the history of gene duplication, sub/neofunctionalization and redundancy. Mol. Genet. Genomics 291: 739–752. [DOI] [PubMed] [Google Scholar]
- Guo Y., Busta L., Jetter R. (2017). Cuticular wax coverage and composition differ among organs of Taraxacum officinale. Plant Physiol. Biochem. 115: 372–379. [DOI] [PubMed] [Google Scholar]
- Hacquard S., Petre B., Frey P., Hecker A., Rouhier N., Duplessis S. (2011). The poplar-poplar rust interaction: insights from genomics and transcriptomics. J. Pathogens 2011: 716041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada K., Zou C., Lehti-Shiu M.D., Shinozaki K., Shiu S.-H. (2008). Importance of lineage-specific expansion of plant tandem duplicates in the adaptive response to environmental stimuli. Plant Physiol. 148: 993–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haslam T.M., Kunst L. (2013). Extending the story of very-long-chain fatty acid elongation. Plant Sci. 210: 93–107. [DOI] [PubMed] [Google Scholar]
- Herms D.A., Mattson W.J. (1992). The dilemma of plants: to grow or defend. Q. Rev. Biol. 67: 283–335. [Google Scholar]
- Joubès J., Raffaele S., Bourdenx B., Garcia C., Laroche-Traineau J., Moreau P., Domergue F., Lessire R. (2008). The VLCFA elongase gene family in Arabidopsis thaliana: phylogenetic analysis, 3D modelling and expression profiling. Plant Mol. Biol. 67: 547–566. [DOI] [PubMed] [Google Scholar]
- Kosma D.K., Rowland O. (2016). Answering a four decade-old question on epicuticular wax biosynthesis. J. Exp. Bot. 67: 2538–2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunst L., Taylor D.C., Underhill E.W. (1992). Fatty acid elongation in developing seeds of Arabidopsis thaliana. Plant Physiol. Biochem. 30: 425–434. [Google Scholar]
- La Mantia J., Klápště J., El-Kassaby Y.A., Azam S., Guy R.D., Douglas C.J., Mansfield S.D., Hamelin R. (2013). Association analysis identifies Melampsora ×columbiana poplar leaf rust resistance SNPs. PLoS One 8: e78423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamoreaux R.J., Chaney W.R., Brown K.M. (1978). The plastochron index: A review after two decades of use. Am. J. Bot. 65: 586–593. [Google Scholar]
- Lardizabal K.D., Metz J.G., Sakamoto T., Hutton W.C., Pollard M.R., Lassner M.W. (2000). Purification of a jojoba embryo wax synthase, cloning of its cDNA, and production of high levels of wax in seeds of transgenic arabidopsis. Plant Physiol. 122: 645–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassner M.W., Lardizabal K., Metz J.G. (1996). A jojoba beta-Ketoacyl-CoA synthase cDNA complements the canola fatty acid elongation mutation in transgenic plants. Plant Cell 8: 281–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.-B., Jung S.-J., Go Y.-S., Kim H.-U., Kim J.-K., Cho H.-J., Park O.K., Suh M.-C. (2009). Two Arabidopsis 3-ketoacyl CoA synthase genes, KCS20 and KCS2/DAISY, are functionally redundant in cuticular wax and root suberin biosynthesis, but differentially controlled by osmotic stress. Plant J. 60: 462–475. [DOI] [PubMed] [Google Scholar]
- Liang H., Staton M., Xu Y., Xu T., Leboldus J. (2014). Comparative expression analysis of resistant and susceptible Populus clones inoculated with Septoria musiva. Plant Sci. 223: 69–78. [DOI] [PubMed] [Google Scholar]
- McKown A.D., Guy R.D., Azam M.S., Drewes E.C., Quamme L.K. (2013). Seasonality and phenology alter functional leaf traits. Oecologia 172: 653–665. [DOI] [PubMed] [Google Scholar]
- McKown A.D., Guy R.D., Klápště J., Geraldes A., Friedmann M., Cronk Q.C.B., El-Kassaby Y.A., Mansfield S.D., Douglas C.J. (2014a). Geographical and environmental gradients shape phenotypic trait variation and genetic structure in Populus trichocarpa. New Phytol. 201: 1263–1276. [DOI] [PubMed] [Google Scholar]
- McKown A.D., Guy R.D., Quamme L., Klápště J., La Mantia J., Constabel C.P., El-Kassaby Y.A., Hamelin R.C., Zifkin M., Azam M.S. (2014b). Association genetics, geography and ecophysiology link stomatal patterning in Populus trichocarpa with carbon gain and disease resistance trade-offs. Mol. Ecol. 23: 5771–5790. [DOI] [PubMed] [Google Scholar]
- Metz J.G., Pollard M.R., Anderson L., Hayes T.R., Lassner M.W. (2000). Purification of a jojoba embryo fatty acyl-coenzyme A reductase and expression of its cDNA in high erucic acid rapeseed. Plant Physiol. 122: 635–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mietkiewska E., Giblin E.M., Wang S., Barton D.L., Dirpaul J., Brost J.M., Katavic V., Taylor D.C. (2004). Seed-specific heterologous expression of a nasturtium FAE gene in Arabidopsis results in a dramatic increase in the proportion of erucic acid. Plant Physiol. 136: 2665–2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar A.A., Kunst L. (1997). Very-long-chain fatty acid biosynthesis is controlled through the expression and specificity of the condensing enzyme. Plant J. 12: 121–131. [DOI] [PubMed] [Google Scholar]
- Myburg A.A., et al. (2014). The genome of Eucalyptus grandis. Nature 510: 356–362. [DOI] [PubMed] [Google Scholar]
- Oh C.-S., Toke D.A., Mandala S., Martin C.E. (1997). ELO2 and ELO3, homologues of the Saccharomyces cerevisiae ELO1 gene, function in fatty acid elongation and are required for sphingolipid formation. J. Biol. Chem. 272: 17376–17384. [DOI] [PubMed] [Google Scholar]
- Perera M.A., Qin W., Yandeau-Nelson M., Fan L., Dixon P., Nikolau B.J. (2010). Biological origins of normal-chain hydrocarbons: a pathway model based on cuticular wax analyses of maize silks. Plant J. 64: 618–632. [DOI] [PubMed] [Google Scholar]
- Post-Beittenmiller D. (1996). Biochemistry and molecular biology of wax production in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47: 405–430. [DOI] [PubMed] [Google Scholar]
- Riederer M., Schreiber L. (1995). Waxes: The Transport Barriers of Plant Cuticles. (Dundee, Scotland: The Oily Press; ). [Google Scholar]
- Rowland O., Zheng H., Hepworth S.R., Lam P., Jetter R., Kunst L. (2006). CER4 encodes an alcohol-forming fatty acyl-coenzyme A reductase involved in cuticular wax production in Arabidopsis. Plant Physiol. 142: 866–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuels L., Kunst L., Jetter R. (2008). Sealing plant surfaces: cuticular wax formation by epidermal cells. Annu. Rev. Plant Biol. 59: 683–707. [DOI] [PubMed] [Google Scholar]
- Schneider L.M., Adamski N.M., Christensen C.E., Stuart D.B., Vautrin S., Hansson M., Uauy C., von Wettstein-Knowles P. (2016). The Cer-cqu gene cluster determines three key players in a β-diketone synthase polyketide pathway synthesizing aliphatics in epicuticular waxes. J. Exp. Bot. 67: 2715–2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. (2013). MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 30: 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson G.E. (1941). Leaf-spot diseases of poplars caused by Septoria musiva and S. populicola. Phytopathology 31: 241–254. [Google Scholar]
- Tomato Genome Consortium (2012). The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485: 635–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Pachter L., Salzberg S.L. (2009). TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D.R., Pimentel H., Salzberg S.L., Rinn J.L., Pachter L. (2012). Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7: 562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trenkamp S., Martin W., Tietjen K. (2004). Specific and differential inhibition of very-long-chain fatty acid elongases from Arabidopsis thaliana by different herbicides. Proc. Natl. Acad. Sci. USA 101: 11903–11908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tresch S., Heilmann M., Christiansen N., Looser R., Grossmann K. (2012). Inhibition of saturated very-long-chain fatty acid biosynthesis by mefluidide and perfluidone, selective inhibitors of 3-ketoacyl-CoA synthases. Phytochemistry 76: 162–171. [DOI] [PubMed] [Google Scholar]
- Tuskan G.A., et al. (2006). The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313: 1596–1604. [DOI] [PubMed] [Google Scholar]
- Wang Z., et al. (2012b). The genome of flax (Linum usitatissimum) assembled de novo from short shotgun sequence reads. Plant J. 72: 461–473. [DOI] [PubMed] [Google Scholar]
- Ward K.T., Ostry M.E. (2005). Variation in Septoria musiva and implications for disease resistance screening of poplars. Plant Dis. 89: 1077–1082. [DOI] [PubMed] [Google Scholar]
- von Wettstein-Knowles P. (2007). Analyses of barley spike mutant waxes identify alkenes, cyclopropanes and internally branched alkanes with dominating isomers at carbon 9. Plant J. 49: 250–264. [DOI] [PubMed] [Google Scholar]
- von Wettstein-Knowles P. (1982). Elongases and epicuticular wax biosynthesis. Physiol. Veg. 20: 797–809. [Google Scholar]
- von Wettstein‐Knowles P. (2012). Plant Waxes. eLS. doi/10.1002/9780470015902.a0001919.pub2. [Google Scholar]
- Yeats T.H., Rose J.K.C. (2013). The formation and function of plant cuticles. Plant Physiol. 163: 5–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeiger E., Stebbins G.L. (1972). Developmental genetics in barley: A mutant for stomatal development. Am. J. Bot. 59: 143–148. [Google Scholar]