Suppressing the expression of the putative sterol carrier protein gene GhSCP2D in cotton fibers leads to enhanced callose deposition, thereby reducing plasmodesmata permeability and fiber elongation.
Abstract
Plasmodesmata (PDs) play vital roles in cell-to-cell communication and plant development. Emerging evidence suggests that sterols are involved in PD activity during cytokinesis. However, whether sterols contribute to PD gating between established cells remains unknown. Here, we isolated GhSCP2D, a putative sterol carrier protein gene from elongating cotton (Gossypium hirsutum) fibers. In contrast to wild-type fiber PDs, which opened at 5 to 10 d postanthesis (DPA) and closed only at 15 to 25 DPA, plants with suppressed GhSCP2D expression had reduced sterol contents and closed PDs at 5 through 25 DPA. The GhSCP2D-suppressed fibers exhibited callose deposition at the PDs, likely due to reduced expression of GhPdBG3-2A/D, which encodes a PD-targeting β-1,3-glucanase. Both GhPdBG3-2A/D expression and callose deposition were sensitive to a sterol biosynthesis inhibitor. Moreover, suppressing GhSCP2D upregulated a cohort of SUT and SWEET sucrose transporter genes in fiber cells. Collectively, our results indicate that (1) GhSCP2D is required for GhPdBG3-2A/D expression to degrade callose at the PD, thereby contributing to the establishment of the symplasmic pathway; and (2) blocking the symplasmic pathway by downregulating GhSCP2D activates or increases the expression of SUTs and SWEETs, leading to the switch from symplasmic to apoplasmic pathways.
INTRODUCTION
Cotton (Gossypium hirsutum) fibers are single-celled trichomes derived from the ovule epidermis. These fibers represent the most important natural source of cellulose for the textile industry worldwide. Fiber development consists of four overlapping stages: initiation (2 d before to 2 d postanthesis [DPA]), rapid elongation (2–20 DPA), secondary wall thickening (16–35 DPA), and maturation (35–50 DPA) (Graves and Stewart, 1988; Kim and Triplett, 2001; Ji et al., 2003; Chen and Guan, 2011). Sucrose, the major carbon source imported into developing cotton fibers, is thought to be used directly as a substrate for the cellulose synthase complex (Amor et al., 1995) and for energy production (Ruan et al., 2003). Wang et al. (2010) suggested that sucrose, K+, and malate provide the major osmotically active solutes to cotton fiber cells to generate the turgor required to drive fiber elongation. Sucrose synthase, which degrades sucrose into UDP-glucose and fructose, plays important roles in fiber cell elongation and secondary cell wall synthesis (Ruan et al., 2003). Therefore, sucrose import and metabolism are essential for cotton fiber development.
Sucrose is imported into fiber cells from the underlying seed coat cells symplasmically through the plasmodesmata (PDs) or apoplasmically across the cell wall space and plasma membrane by SUCROSE TRANSPORT PROTEINS (SUTs), a process energized by plasma membrane H+-ATPases (Ruan et al., 2001; Wang and Ruan, 2010). In the apoplasmic pathway, SUTs function as sucrose/proton symporters to transport sucrose from the cell wall matrix into the cytosol (Ruan et al., 2000). Recently, a new family of sucrose transporters named SUGARS WILL EVENTUALLY BE EXPORTED TRANSPORTERs (SWEETs) was isolated from Arabidopsis thaliana, rice (Oryza sativa), and other species (Eom et al., 2015). SWEETs function in sugar uptake or efflux across the plasma membranes or tonoplast down a concentration gradient (Chen et al., 2012; Lin et al., 2014; Eom et al., 2015). In the symplasmic pathway, the movement of freely diffusing molecules strongly depends on the aperture size of the PD pores, which can be quantitatively described as the size exclusion limit (De Storme and Geelen, 2014). PD-mediated transport is a major mechanism for controlling cell-to-cell communication in plants (Levy et al., 2007; Guseman et al., 2010; Vatén et al., 2011). Compelling evidence suggests that the deposition of callose at the PD neck region reduces the size exclusion limit of the trans-PD cytosolic channel, thereby limiting symplasmic permeability between neighboring cells (Chen and Kim, 2009; Zavaliev et al., 2011). Callose deposition is controlled by GLUCAN SYNTHASE-LIKEs (GSLs) (Guseman et al., 2010) and β-1,3-glucanases (BGs) (Doxey et al., 2007; Levy et al., 2007), which synthesize and degrade the callose, respectively (Ruan et al., 2004). However, the molecular components regulating the expression of GSL and BG genes remain largely unknown.
Emerging evidence indicates that sterols play a role in PD activity. Sterols are integral components of the cell membrane. Over 100 types of sterols have been identified in plants, the most abundant being sitosterol, stigmasterol, and campesterol (Guo et al., 1995; Hartmann, 1998). The sterol-deficient mutants hyd1, fk/hyd2, and smt1/cph in Arabidopsis exhibit abnormal or ectopic accumulation of callose in their embryonic and vascular tissues (Diener et al., 2000; Souter et al., 2002; Schrick et al., 2004; Carland et al., 2010; Pullen et al., 2010). Modulating the overall sterol composition of young dividing cells in vitro reversibly impaired the PD localization of the glycosylphosphatidylinositol (GPI)-anchored protein, β-1,3-glucanase, and altered PD permeability (Grison et al., 2015). However, this study was limited to the analysis of primary PDs in dividing cells of young meristematic tissues. Whether PD gating among established cells is controlled by sterol, and the identity of the genes underlying the functioning of sterol in this process, remains to be investigated (Grison et al., 2015).
Cotton fibers are an excellent single-celled model for studying cell-to-cell communication in relation to cellular expansion and cellulose biosynthesis (Ruan et al., 2001, 2004). Here, we examined the role of a putative sterol carrier protein (SCP), GhSCP2D, in cotton fiber PD gating and cell expansion. GhSCP2D was predominantly expressed during the rapid elongation period in fibers of upland cotton. Transgenically suppressing GhSCP2D expression reduced the expression of GhPdBG3-2A/D, encoding a GPI-anchored, PD-targeted β-1,3-glucanase, leading to callose deposition at the PD and, consequently, PD closure during the 5- and 10-DPA elongation period. We substantiated the positive role of SCP2D in establishing PD permeability by investigating another cotton species, Gossypium barbadense, in which more extensive expression of a homologous SCP2D gene correlated with longer periods of PD opening and longer fibers. The GhSCP2D-suppressed transgenic fibers were characterized by reduced levels of sterol and soluble sugars but enhanced expression of a cohort of SUT and SWEET sucrose transporter genes. These findings provide insights into the GhSCP2D-mediated molecular control of PD permeability and the activation of sucrose transporters via the closure of the symplasmic pathway.
RESULTS
Isolation and Characterization of GhSCP2D in Cotton
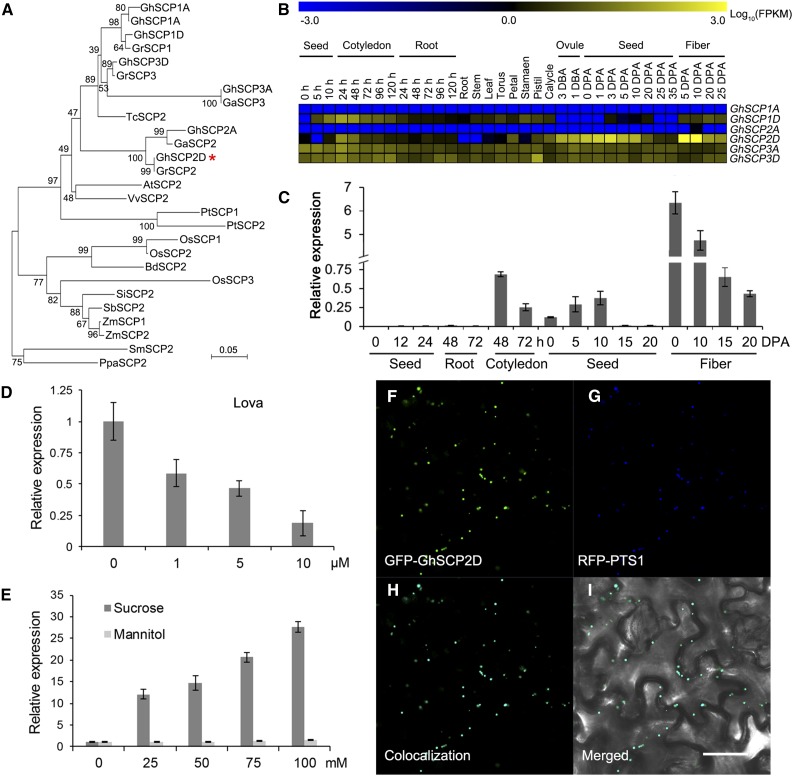
As the first step in our analysis, we isolated cDNA from the genetic standard upland cotton line TM-1 (G. hirsutum) encoding SCP2, a 121-amino acid protein with a conserved domain characteristic of a eukaryotic SCP (Figure 1A). Sequence analysis revealed that this putative SCP shares 77% sequence similarity with Arabidopsis AtSCP2 (Supplemental Figure 1A). The allotetraploid cotton was developed from an interspecific hybridization of the diploid ancestral species G. arboreum (A2) and G. raimondii (D5) (Cedroni et al., 2003; Wendel, 1989). The G. hirsutum (AD)1 genome usually contains two or more copies of each gene located on the A and D subgenome chromosomes, representing homoeologous or duplicate copies of the A and D genomes of the diploid ancestral species. Six putative SCP genes were previously found in the allotetraploid cotton genome (Zhang et al., 2015), each encoding a protein containing a complete SCP domain and peroxisome signal peptide 1 (PTS1) (Supplemental Table 1), which are characteristics of known SCPs (Edqvist et al., 2004; Edqvist and Blomqvist, 2006). Among these, GhSCP1D was found to have a longer amino acid sequence than its homologs due to a shift in the stop codon toward the 3′ coding sequence (Supplemental Figure 1A and Supplemental Table 1).
Figure 1.
Phylogenetic Analysis of the SCP2 Gene Family and Expression Pattern of GhSCP2D.
(A) Phylogenetic analysis of the deduced protein sequences of SCP2 family genes from G. hirsutum (Gh), G. arboreum (Ga), G. raimondii (Gr), Vitis vinifera (Vv), Populus trichocarpa (Pt), Carica papaya (Cp), Arabidopsis thaliana (At), Glycine max (Gm), Zea mays (Zm), Setaria italica (Si), Oryza sativa (Os), Brachypodium distachyon (Bd), Selaginella moellendorffii (Sm), and Physcomitrella patens (Pp). GhSCP2D is indicated by a red asterisk. The neighbor-joining tree was constructed using the MEGA 6.0 program (http://www.megasoftware.net/).
(B) RNA-seq analysis of GhSCPs in tissues and organs of G. hirsutum. Yellow and blue indicate up- and downregulated genes, respectively.
(C) qRT-PCR analysis of GhSCP2D expression in developing fibers and germinating seedlings of G. hirsutum. The “h” indicates hours of germination (after imbibition). RNA was extracted from germinating seeds at 0, 12, or 24 h and from roots and cotyledons at 48 or 72 h after imbibition. Three biological replicates were used for each reaction with two technical replicates each. Error bars represent the sd of triplicate experiments.
(D) The effect of lova treatment on GhSCP2D expression in wild-type cotton line W0. Seeds at 1 DPA were cultured in BT medium containing 0, 1, 5, or 10 µM lova for 2 d.
(E) qRT-PCR analysis of GhSCP2D expression in response to sucrose treatment in cv W0. Seeds at 1 DPA were cultured in BT medium containing different concentrations of sucrose or mannitol (instead of glucose, as used in the original BT medium) for 2 d.
(F) to (I) Transient expression of GhSCP2D fused to the C terminus of GFP (GFP-GhSCP2D) in epidermal cells of N. benthamiana leaves. Green and red fluorescent signals, representing GFP-GhSCP2D and the peroxisomal marker RFP-PTS1, respectively, were examined at 48 h after transformation. Bar = 50 μm.
RNA-seq and qRT-PCR analysis showed that the expression levels of the GhSCP genes are biased to the D subgenome and that these genes are predominantly expressed in fibers and the cotyledons of seedlings (Figure 1B; Supplemental Figure 2). Among the six homologs, GhSCP2D had the highest expression level in elongating cotton fibers (Figure 1B). GhSCP2D transcripts were detected in 3 d before anthesis ovules, reached the highest level in 5- and 10-DPA fibers, and decreased sharply thereafter. GhSCP2D was expressed at very low levels in other cotton tissues (Figure 1C; Supplemental Figure 1B). Importantly, GhSCP2D mRNA levels in cultured cotton seeds were reduced upon treatment with the sterol synthesis inhibitor lovastatin (lova) (Figure 1D) and increased in response to sucrose treatment (Figure 1E).
Since GhSCP2D has a peroxisome signal peptide (PTS1) at the C terminus (Figure 1B; Supplemental Table 1), a common feature of SCPs (Edqvist et al., 2004), we constructed two vectors with the GFP sequence fused with the N- and C-terminal coding sequences of GhSCP2D, respectively, and investigated which fusion protein would exhibit the expected localization after transformation. Green fluorescent signals distinct from chloroplasts were observed in protoplasts expressing GhSCP2D-N terminus fused to GFP, whereas fluorescent signals were evenly distributed in the cytosol in protoplasts expressing GhSCP2D-C terminus fused with GFP (Supplemental Figure 1C), indicating that the fusion of GFP to the C terminus of GhSCP2D interfered with PTS1-induced targeting of this protein. We then used a pumpkin (Cucurbita sp cv Kurokawa Amakuri Nankin) malate synthase gene as a positive control to verify the subcellular localization of GhSCP2D. This pumpkin protein contains a PTS1 signal peptide and is known to target peroxisomes when expressed in Arabidopsis (Hayashi et al., 1996). We fused the pumpkin cDNA encoding this protein with red fluorescent protein at the N terminus and cotransformed this construct with GFP-GhSCP2D. Green fluorescent signals from GFP (Figure 1F) and red fluorescent signals from the peroxisomal marker RFP-PTS1 (Figure 1G) completely overlapped in Nicotiana benthamiana epidermal cells (Figures 1H and 1I), indicating that GhSCP2D is localized to peroxisomes, as expected for SCPs (Edqvist et al., 2004).
Downregulation of GhSCP2D Shortens Fiber Length Due to the Reduced Sucrose and Hexose Levels
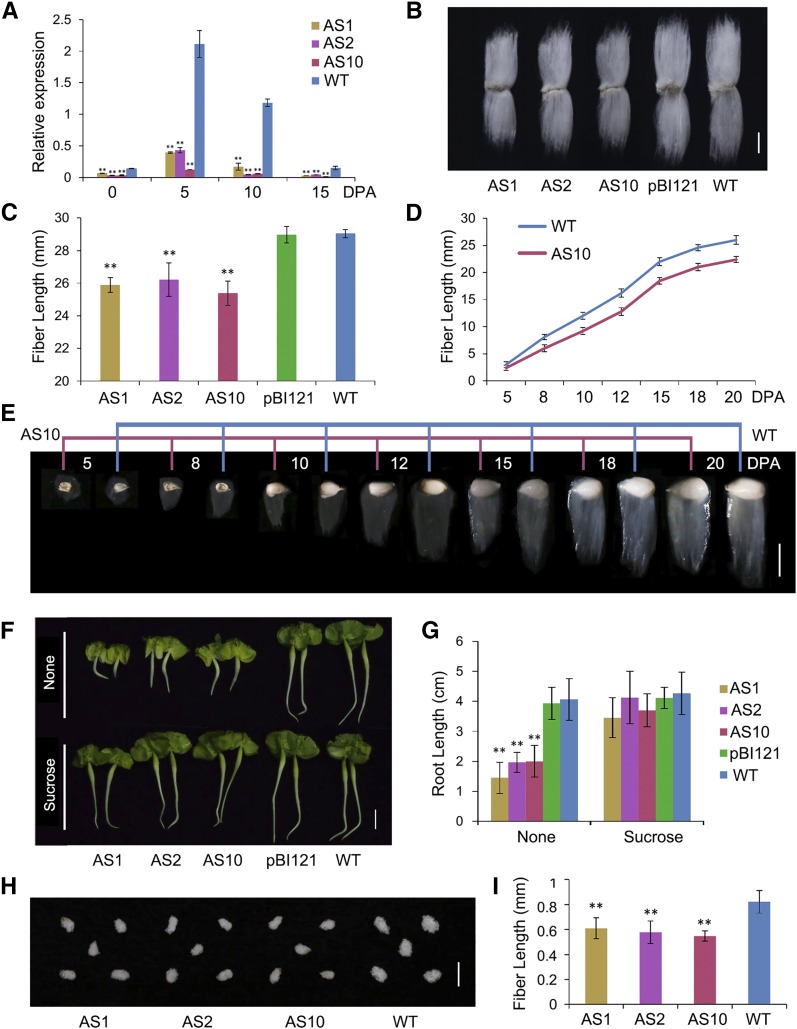
To explore the role of GhSCP2D in cotton fiber elongation, we produced GhSCP2D antisense and overexpression constructs using the full coding sequence of this gene (Supplemental Figures 3A and 3B), both driven by the constitutive 35S promoter. We introduced the constructs into cotton via Agrobacterium tumefaciens-mediated transformation. Comprehensive PCR-based genotyping and expression analysis of GhSCP2D mRNA identified eight antisense and two cosuppression primary transformants (Supplemental Figure 4A). The screening did not identify any overexpression lines (Supplemental Figure 4A), probably due to sequence homology-dependent cosuppression (Taylor, 1997). We selected three GhSCP2D-downregulated lines, designated AS1, AS2, and AS10, for further analysis based on the observation that GhSCP2D transcript levels were dramatically reduced in the fibers without affecting the expression levels of the other GhSCP homologs (Figure 2A; Supplemental Figures 4A to 4C). Compared with the wild type and empty vector transgenic control (pBI121), the GhSCP2D-downregulated lines showed reduced fiber length at maturity (Figures 2B and 2C), as well as smaller cotyledons and shorter hypocotyls and roots at the seedling stage (Figures 2F and 2G).
Figure 2.
Phenotypic Analysis of Cotton Fibers and Seedlings of GhSCP2D-Downregulated Plants.
(A) qRT-PCR analysis of the expression of GhSCP2D in 0-DPA seeds, 5- to 15-DPA fibers of the wild type, and 35S-antisense-GhSCP2D (35SAC) transgenic lines (AS1, AS2, and AS10). Error bars represent the sd of triplicate experiments (**P < 0.01, Student’s t test).
(B) Lint fibers, which become dehydrated and mature after 40 to 50 DPA, from 35SAC transgenic cotton lines (AS1, AS2, and AS10) appear shorter than those from the null control (pBI121) and wild-type fibers.
(C) The average length of mature fibers of the null control (pBI121), wild type, and 35SAC (lines AS1, AS2, and AS10). Error bars indicate the sd of triplicate experiments (**P < 0.01, Student’s t test).
(D) Reduced fiber elongation in transgenic line AS10 compared with the wild type. Each value is the mean ± sd of fiber length from at least ten seeds from three individual plants for transgenic line AS10 and the wild type.
(E) Phenotypes of fiber-bearing seeds in the wild type and transgenic line AS10. The stages are indicated above the graphs. Bar = 10 mm.
(F) Phenotypes of GhSCP2D-downregulated cotton seedling grown on Murashige and Skoog medium with and without 1% sucrose for 3 d after imbibition. Bar = 1 cm.
(G) Average root length of the null control (pBI121), wild-type, and 35SAC (lines AS1, AS2, and AS10) plants grown on MS medium with and without 1% sucrose. At least 30 seedlings were measured in each case. Error bars indicate the sd of triplicate experiments (**P < 0.01, Student’s t test).
(H) Phenotypes of GhSCP2D-downregulated and wild-type cotton seeds cultured in BT medium (in which 50 mM sucrose was added in place of glucose) for 10 d. Bar = 10 mm.
(I) Fiber length of wild-type and 35SAC (lines AS1, AS2, and AS10) fibers from seeds cultured in BT medium containing 50 mM sucrose for 10 d. At least 30 seeds were measured per genotype. Error bars indicate the sd of triplicate experiments (**P < 0.01, Student’s t test).
To investigate whether GhSCP2D is associated with fiber elongation, we examined the fiber growth rate in GhSCP2D-downregulated line AS10. In 5- to 10-DPA and 10- to 15-DPA wild-type fibers, in which GhSCP2D transcripts were enriched (Figure 1C), the fiber elongation rates were ∼1.80 and 2.00 mm/d, respectively, whereas in the GhSCP2D-downregulated line AS10, these rates were only 1.36 and 1.85 mm/d, respectively (Figures 2D and 2E). By 15 to 20 DPA, the fiber elongation rates in the wild type and AS10 were almost identical (Figures 2D and 2E). The sensitivity of fiber elongation to reduced GhSCP2D expression at 5 to 10 DPA matched the high expression level of GhSCP2D at this stage in the wild type (Figure 1C), suggesting that GhSCP2D expression is required for fiber elongation.
To explore the metabolic basis of reduced fiber elongation in the transgenic plants, we measured the contents of glucose, fructose, and sucrose, representing the major energy sources and osmotic solutes required for fiber elongation (Ruan, 2007). We detected a 10 to 15% reduction in hexose and sucrose levels in 5- and 10-DPA fibers of the GhSCP2D-downregulated lines compared with the wild type (Table 1). Perhaps sucrose and hexose levels were reduced to similar degrees because sucrose is transported into the cytoplasm of the fiber, where it is cleaved by invertase or sucrose synthase into hexoses (Ruan et al., 2003; Wang et al., 2010). Interestingly, adding sucrose to the germination medium complemented the defect in seedling growth in the GhSCP2D-downregulated lines (Figures 2F and 2G), which was also observed in the Atscp2-1 Arabidopsis mutants (Zheng et al., 2008). However, the reduced fiber growth of the GhSCP2D-downregulated lines was not restored by sucrose treatment, as demonstrated in in vitro-cultured cotton seeds (Figures 2H and 2I). These results suggest that the sucrose transport pathway may be blocked in the 5- and 10-DPA fibers of GhSCP2D-downregulated plants.
Table 1. Sugar Contents in Fibers of GhSCP2D-Downregulated and Wild-Type Plants.
| Fiber Stage | Line | Fructose (µg/mg FW) | Glucose (µg/mg FW) | Sucrose (µg/mg FW) | Total (µg/mg FW) |
|---|---|---|---|---|---|
| 5-DPA fiber | Wild type | 19.21 ± 3.67 | 21.69 ± 1.28 | 11.51 ± 1.05 | 52.57 ± 1.87 |
| AS1 | 16.65 ± 2.57* | 18.79 ± 0.56* | 8.62 ± 0.89** | 44.24 ± 2.49** | |
| AS2 | 17.08 ± 1.96* | 19.67 ± 1.45* | 8.51 ± 0.64** | 45.04 ± 1.36** | |
| AS10 | 16.59 ± 2.43* | 18.73 ± 1.75* | 8.07 ± 1.32** | 43.40 ± 3.23** | |
| 10-DPA fiber | Wild type | 18.74 ± 0.58 | 21.21 ± 2.64 | 11.30 ± 2.16 | 51.15 ± 2.17 |
| AS1 | 15.23 ± 1.10* | 17.19 ± 0.98* | 9.09 ± 1.45** | 45.25 ± 2.49** | |
| AS2 | 16.01 ± 2.24* | 18.32 ± 1.01* | 9.22 ± 1.05** | 47.05 ± 2.61** | |
| AS10 | 16.96 ± 2.16* | 18.61 ± 0.62* | 8.57 ± 2.05** | 44.24 ± 3.65** |
Mean values and standard errors were calculated from three biological replicates. The analysis of significance was calculated via comparison with data from the wild type. Significance of the phenotypic results was assessed using Student’s t tests (*P < 0.05 and **P < 0.01). FW, fresh weight.
Suppressing GhSCP2D Reduces Fiber PD Permeability
Sucrose is imported into fiber cells from the underlying seed coat cells through the PDs or by sucrose transporters (Ruan et al., 2001; Wang and Ruan, 2010). To evaluate the permeability of PDs in the fibers of GhSCP2D-suppressed lines, we used a phloem-mobile fluorescent probe, the membrane-impermeable fluorescent dye carboxyfluorescein (CF), since unloaded CF can only enter fiber cells from the phloem of the outer seed coat through the PD (Ruan et al., 2001, 2004). We monitored the movement of CF into fiber cells in situ via confocal laser scanning microscopy.
In the wild type, the fluorescent dye entered 5- and 10-DPA fibers from the vascular bundles of the seed coat (Figures 3A and 3B), indicating that the PDs connecting fiber cells with the adjacent cells at the fiber base were open at this stage. However, almost no fluorescent dye was observed in 15-, 20-, and 25-DPA wild-type fibers (Figures 3C to 3E), indicating that the PDs were closed during this period. In the three transgenic lines in which GhSCP2D expression was dramatically suppressed (Figure 2A), no or only trace amounts of fluorescent dye were observed in 5-DPA fibers compared with the wild type (Supplemental Figure 5). In line AS10, fluorescent dye signals were very weak or undetectable in 5- to 25-DPA fibers (Figures 3F to 3J), suggesting that fiber PDs are closed in the transgenic plants during the entire fiber elongation period. These results suggest that suppressing GhSCP2D expression results in PD closure and that GhSCP2D plays a key role in controlling PD gating in cotton fibers.
Figure 3.
Confocal Images of CF Transport to Fibers of the Wild Type and GhSCP2D-Downregulated Line AS10.
(A) and (B) Optical sections of 5- and 10-DPA seeds in the wild type, showing transport and accumulation of CF from vascular bundles to fibers and the outer seed coat but not to the inner seed coat.
(C) to (E) Optical sections of 15-, 20-, and 25-DPA seeds in the wild type, showing strong, widespread CF signals in the vascular region but few or no signals in the fibers.
(F) to (J) Optical sections of 5-, 10-, 15-, 20-, and 25-DPA seeds from transgenic lines AS10, showing strong, widespread CF signals in the vascular region but few or no signals in the fibers. The upper and lower panels show bright-field and merged images, respectively. For each line, imaging analysis was performed on five seeds from three plants with a total of at least 15 optical sections. F, fibers; VB, vascular bundle; OSC, outer seed coat; ISC, inner seed coat. Bars = 50 µm.
GhSCP2D-Downregulated Plants Exhibit Extensive Callose Deposition at PD in the Base of the Fiber
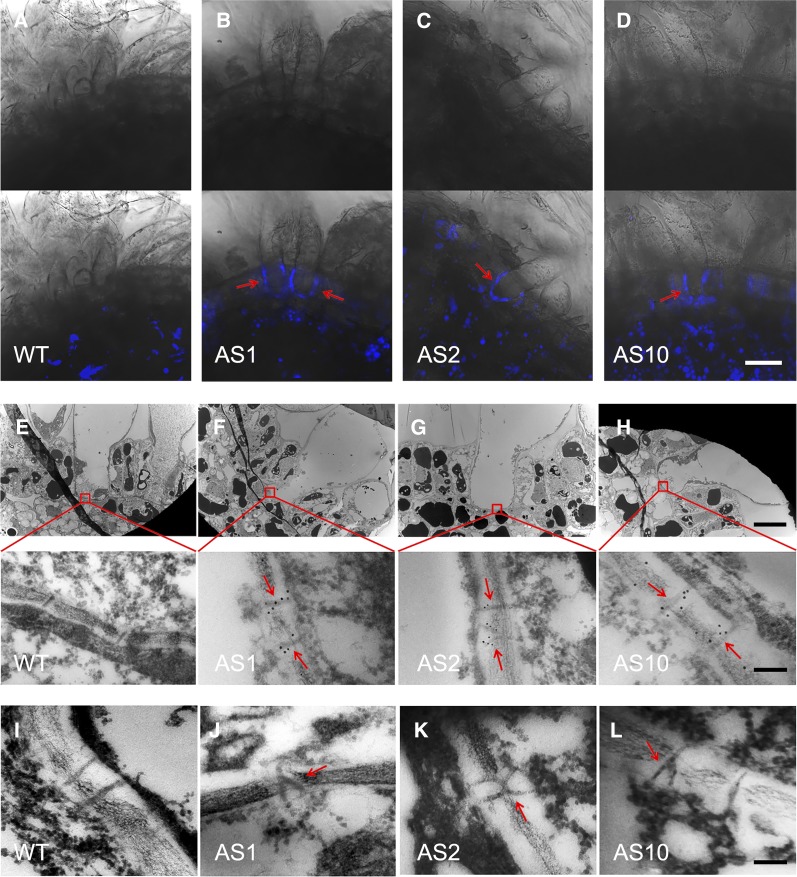
A major mechanism regulating symplasmic conductivity (PD permeability) is callose deposition at the PD (De Storme and Geelen, 2014; Guseman et al., 2010; Levy et al., 2007; Vatén et al., 2011). To determine whether callose deposition is altered in the fiber PDs of GhSCP2D-downregulated plants, we used aniline blue, a callose-specific dye, to detect callose in 5-DPA fibers. In the fibers of wild-type plants, no fluorescent signals from callose were detected at the bases of fibers (Figure 4A) when the PDs were open (Figure 3A). By contrast, in the three transgenic lines, fluorescent signals from callose were evident at the fiber bases located in the seed coat epidermis (Figures 4B to 4D), indicating that callose deposition was induced at the bases of fibers in GhSCP2D-downregulated plants.
Figure 4.
Detection of Callose at the Base of 5-DPA Fibers in GhSCP2D-Downregulated and Wild-Type Plants.
(A) to (D) Aniline blue staining for callose at the fiber base in GhSCP2D-downregulated and wild-type plants. No aniline blue fluorescent signals (indicating callose) were detected at the fiber bases of wild-type plants. By contrast, AS1, AS2, and AS10, representing the three transgenic lines of GhSCP2D-downregulated plants, exhibited clear fluorescent signals (callose) at the fiber base (red arrows). The upper and lower panels show bright-field and merged images, respectively. For each line, imaging analysis was performed from three individual plants with a total of at least 15 optical sections. Bar = 20 µm.
(E) to (H) Electron microscopy immunolocalization of callose at the fiber base of GhSCP2D-downregulated (AS1, AS2, and AS10) and wild-type plants. Immunogold labeling with a monoclonal antibody against callose was performed in cross sections of 5-DPA seeds. No immunogold particles (indicating callose) were detected at the fiber base (boxed area in the upper panel) of wild-type plants. By contrast, the three transgenic lines showed strong immunogold-labeling of callose at the fiber base (red arrows). Bars = 10 µm in the upper panel and 100 nm in the lower panel.
(I) to (L) Transmission electron microscopy analysis of fiber sections showing simple PDs at the fiber base of wild-type plants but branched PDs at the fiber base of GhSCP2D-downregulated plants (red arrows). Bar = 100 nm.
We also performed electron microscopic immunogold labeling experiments to detect callose deposition at PDs at the bases of fibers. Gold particles (labeling callose) were localized in or around the PDs of 5-DPA fibers in the three transgenic lines (Figures 4F to 4H), but not in the wild-type fiber PDs (Figure 4E). We detected 1.8 to 2.9 gold particles per PD in the three transgenic lines compared with only 0.5 per PD in wild-type plants (Table 2). These data confirm the notion that suppressing GhSCP2D expression resulted in callose deposition at PDs at the bases of cotton fibers, which connect the underlying or adjacent seed coat cells in GhSCP2D-downregulated plants.
Table 2. Number of Immunogold Particles Representing Callose at the PD in Fibers from GhSCP2D-Downregulated and Wild-Type Plants.
| Genotype | Total Cell Wall Length at Fiber Base (μm) | No. of PD Observed | PD Frequency (PD/μm) | Total No. of Gold Particles at PD Relative to the Cell Wall | Gold Particles per PD |
|---|---|---|---|---|---|
| Wild type | 749 | 102 | 0.14 ± 0.03 | 41/54 | 0.5 ± 0.2 |
| AS1 | 849 | 127 | 0.15 ± 0.04 | 278/75 | 2.3 ± 0.3** |
| AS2 | 805 | 112 | 0.14 ± 0.04 | 197/49 | 1.8 ± 0.3** |
| AS10 | 768 | 97 | 0.13 ± 0.03 | 295/77 | 2.9 ± 0.5** |
All branches of a branched PD were counted as one PD. For PD frequency and gold particles per PD, each value was derived from at least 10 fiber cells. Total number of gold particles at PD relative to the cell wall was counted across the specified total cell wall length at the fiber base. Statistical significance was assessed using Student’s t tests (**P < 0.01).
During fiber development, some PDs become branched when CF import into fibers is restricted (Ruan et al., 2001). We therefore compared the PD ultrastructure in 5-DPA fibers of GhSCP2D-downregulated lines versus the wild type. While the frequency of PDs per unit of cell wall length at the fiber base was similar in 5-DPA fibers between transgenic and wild-type plants (Table 2), more branched PDs were observed in the GhSCP2D-downregulated lines than in the wild type (Table 3). In wild-type fibers, almost all PDs were unbranched, as reported by Ruan et al. (2001) (Figure 4I). By contrast, two types of PD structures, simple PDs and branched PDs, were observed in the GhSCP2D-downregulated lines, as revealed by transmission electron microscopy (Figures 4J to 4L). The three GhSCP2D-downregulated lines had a higher frequency of branched PDs (9–18%) than the wild type (∼2%; Table 3).
Table 3. PD Morphology in Fibers from GhSCP2D-Downregulated and Wild-Type Plants.
| Genotype | Total | Simple | Branched |
|---|---|---|---|
| Wild type | 111 | 109 | 2 (1.8%) |
| AS1 | 132 | 117 | 15 (11.4%)* |
| AS2 | 102 | 84 | 18 (17.6%)* |
| AS10 | 117 | 106 | 11 (9.4%)* |
PDs from at least 10 cell wall interfaces between fiber bases and adjacent cells (neighboring nontrichomes and underlying seed coat cells) were examined from 5 to 10 nonserial sections derived from three seeds per genotype (*t test versus the wild type at P < 0.05).
Downregulation of GhSCP2D and the Application of a Sterol Biosynthesis Inhibitor Suppress the Expression of the β-1,3 Glucanase Gene GhPdBG3-2A/D in Cotton Fibers
Callose accumulation is controlled by the joint action of GSLs and BGs, which synthesize and degrade callose, respectively (Bucher et al., 2001; Ruan et al., 2004). We identified 16 and 26 GSLs in the G. raimondii and G. hirsutum genomes, respectively (Supplemental Figure 6A and Supplemental Table 2). RNA-seq analysis showed that some genes are expressed in G. hirsutum fibers (Supplemental Figure 6B) and that this expression is unaffected in the fibers of GhSCP2D-downregulated lines (Supplemental Figure 6C).
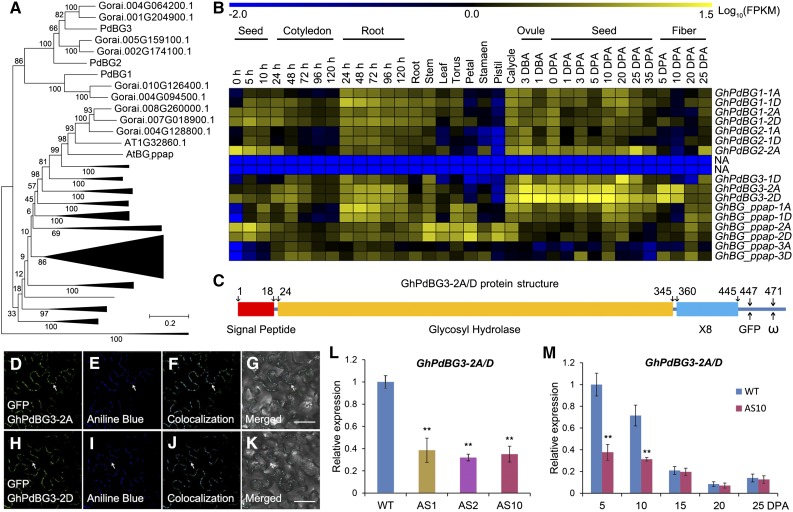
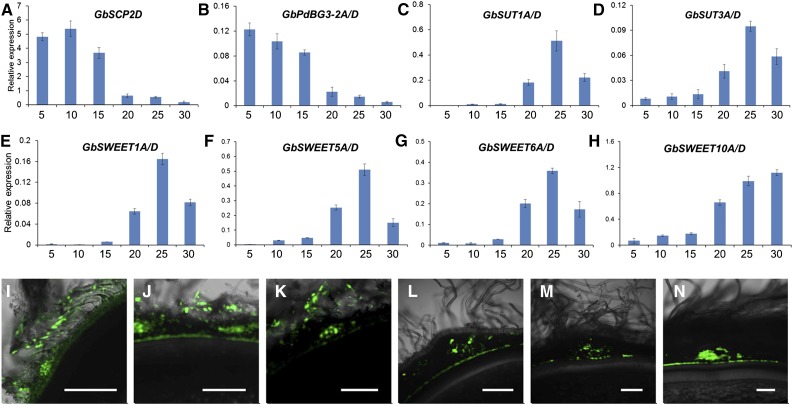
On the other hand, 51 and 66 BG genes were identified in the Arabidopsis and G. raimondii genomes, respectively (Figure 5A). In Arabidopsis, several GPI-anchored and plasmodesmal-localized β-1,3 glucanases, including the putative PD-associated protein (AtBG_ppap), as well as PdBG1, PdBG2, and PdBG3, which localize to the PD (hence their name, PdBGs), are thought to play key roles in callose degradation at the PD neck region (Levy et al., 2007; Benitez-Alfonso et al., 2013). Sixteen genes in G. hirsutum are phylogenetically homologous to these PdBGs (Figure 5A; Supplemental Table 3). RNA-seq and qRT-PCR analysis revealed that transcripts from a pair of homoeologous genes, GhPdBG3-2A/D, were abundant in 5- and 10-DPA versus 15-, 20-, and 25-DPA fibers in wild-type plants (Figures 5B and 5M), which corresponds with the opening and closure of the PDs in wild-type fibers, respectively (Figures 3A to 3E).
Figure 5.
GhPdBG3-2A and GhPdBG3-2D Expression Is Suppressed in GhSCP2D-Downregulated Lines (AS1, AS2, and AS10) Compared with Wild-Type Plants.
(A) Phylogenetic analysis of BGs in G. raimondii and Arabidopsis. A total of 66 BGs in G. raimondii and 51 BGs in Arabidopsis were used to construct the neighbor-joining tree using MEGA 6.0 software. The names are given for the genes encoding PD-localized β-1,3 glucanase proteins.
(B) RNA-seq analysis of GhPdBGs in different tissues and organs of G. hirsutum. Yellow and blue indicate up- and downregulated genes, respectively.
(C) Schematic model of GhPdBG3-2A and GhPdBG3-2D protein structure. GhPdBG3-2A/D contain a signal peptide, a typical glycosyl hydrolase domain, and a X8 domain involved in carbohydrate binding. The insertion site for GFP and the predicted ω site are at positions 447 and 471, respectively.
(D) to (K) GhPdBG3-2A- and GhPdBG3-2D-GFP (GFP was fused in frame to the structural protein at position 447) green fluorescent signals colocalized with callose deposits, as revealed by aniline blue staining at the PD. Bar = 50 µm.
(L) Transcript levels of GhPdBG3-2A/D in 5-DPA fibers of GhSCP2D-downregulated lines (AS1, AS2, and AS10) and the wild type. Error bars represent the sd of triplicate experiments (**P < 0.01, Student’s t test).
(M) Transcript levels of GhPdBG3-2A/D in fibers of AS10 and wild-type plants. Error bars represent the sd of triplicate experiments (**P < 0.01, Student’s t test).
Analysis using Big PI plant predictor and GPI-SOM suggested that GhPdBG3-2A and GhPdBG3-2D are GPI-anchored proteins (Figure 5C). To assess the cellular localization of these proteins, we produced GFP internal fusion constructs for GhPdBG3-2A/D and transiently expressed them in N. benthamiana leaves. Strong GFP signals were detected in punctate spots at the cell periphery, a pattern reminiscent of PD localization (Figures 5D and 5H), as indicated by aniline blue staining for callose at PDs (Figures 5E and 5I). Indeed, GhPdBG3-2A- and GhPdBG3-2D-GFP signals colocalized with aniline blue-stained callose (Figures 5F and 5J). The findings suggest that GhPdBG3-2A and GhPdBG3-2D localize to PDs and might control callose turnover by degrading callose at the PDs during fiber development.
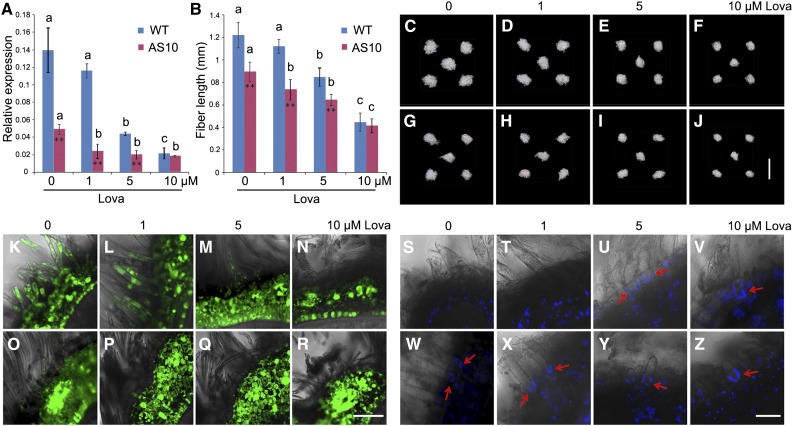
Importantly, GhPdBG3-2A/D expression was dramatically suppressed in the transgenic fibers (Figures 5L and 5M), indicating that transgenic suppression of GhSCP2D reduced GhPdBG3-2A/D expression, allowing callose to be deposited at the PD. Interestingly, Grison et al. (2015) reported that altering sterol composition in young, dividing cells of Arabidopsis seedlings impaired the PD localization of the GPI-anchored proteins PdBG2 and altered callose-mediated PD permeability. To investigate whether the expression of GhPdBG3-2A and GhPdBG3-2D is affected by sterol, we cultured wild-type and transgenic seeds at 1 DPA in medium containing various concentrations of the sterol biosynthesis inhibitor lova (Grison et al., 2015) for 10 d using an in vitro cotton ovule culture system described by Beasley (1971). As shown in Figure 6A, GhPdBG3-2A and GhPdBG3-2D mRNA levels were reduced in the fibers of both GhSCP2D-downregulated and wild-type plants treated with lova at concentrations of 5 µM or higher (Figure 6A; Supplemental File 6). The fiber length was also reduced by lova treatment in a similar manner (Figures 6B to 6J).
Figure 6.
Inhibiting Sterol Biosynthesis Alters PD Permeability (Controlled by Callose) in Cotton Fibers.
(A) The effect of lova treatment on GhPdBG3-2A/D expression in fibers of the wild type and the GhSCP2D-downregulated line AS10. 1 DPA seeds were cultured in BT medium containing 0, 1, 5, or 10 µM lova for 10 d. For each condition, the seeds removed from the independent boll were used as the biological replicate. Error bars indicate the sd of three biological replicates. Asterisks indicate significant difference at *P < 0.05 and **P < 0.01 based on Student’s t test for pairwise comparisons of AS10 and wild-type plants at a given concentration of lova. Different letters indicate significant differences among different concentrations of lova within AS10 or wild-type plants at P < 0.01 according to a randomization one-way ANOVA test (Supplemental File 6).
(B) Lova treatment reduces fiber length in wild-type and GhSCP2D-downregulated AS10 plants following in vitro culture in BT medium containing lova for 10 d starting from 1 DPA. At least 30 seeds were measured in each case. Error bars indicate the sd of three biological replicates. Asterisks indicate significant difference at *P < 0.05 and **P < 0.01 based on Student’s t test for pairwise comparisons between AS10 and wild-type plants at a given concentration of lova. Different letters indicate significant difference among different concentrations of lova within AS10 or wild-type plants at P < 0.01 according to a randomization one-way ANOVA test (Supplemental File 6).
(C) to (J) Phenotypes of wild-type and GhSCP2D-downregulated cotton seeds cultured in BT medium containing 0, 1, 5, or 10 µM lova for 10 d. (C) to (F) represent wild-type seeds exposed to lova treatment at the specified concentration. (G) to (J) represent AS10 seeds subjected to lova treatment at the specified concentration. Bar = 10 mm.
(K) to (N) Optical sections of wild-type seeds treated with 0, 1, 5, or 10 µM lova for 10 d starting from 1 DPA, showing transport and accumulation of CF from vascular bundles to fibers and the outer seed coat but not to the inner seed coat. Confocal images showing strong CF signals in (K) and (L), but few or no signals in (M) and (N). Bar = 50 µm.
(O) to (R) Optical sections of 0, 1, 5, or 10 µM lova-treated AS10 seeds, showing transport and accumulation of CF from vascular bundles to fibers and the outer seed coat but not to the inner seed coat. Confocal images show few or no signals. At least 30 sections produced from ten seeds from three cotton plants were examined for each growth condition. Bar = 50 µm.
(S) to (V) Confocal images of aniline blue staining for callose at the fiber base at 10 DPA in wild-type and GhSCP2D-downregulated plants cultured in medium containing 0, 1, 5, or 10 µM lova for 10 d. Wild-type fibers from seeds treated with 0 and 1 µM lova showed no aniline blue signals ([S] and [T]), but strong signals were observed in seeds treated with 5 and 10 µM lova ([U] and [V]).
(W) to (Z) Confocal images of 0, 1, 5, or 10 µM lova-treated fibers from line AS10, showing stronger callose signals at the fiber base upon treatment with lova compared with the control. At least 30 sections from 10 seeds were analyzed in each case. Bar = 20 µm.
The permeability of PDs in wild-type fibers, as judged by the movement of CF, was not affected in 10-d fibers treated with 1 µM lova compared with the control (Figures 6L versus 6K). Consistently, no fluorescent signals from aniline blue (indicating callose) were observed at the bases of fibers (Figures 6S and 6T). However, treatment of wild-type fibers with 5 or 10 µM lova dramatically reduced the permeability of PDs (Figures 6M and 6N), owing to callose deposition at the fiber base (Figures 6U and 6V). As observed in vivo (Figure 3F), some weak CF signals were detected in fibers from cultured seeds of GhSCP2D-downregulated plants (Figure 6O), and these transgenic fibers exhibited fluorescent signals from callose at their bases (Figure 6W). Treatment with 1, 5, or 10 µM lova elicited the production of callose, as revealed by intense fluorescent signals (Figures 6X to 6Z), with no fluorescent signals from CF were detected in fibers (Figures 6P to 6R). These results indicate that the defect in callose-mediated PD permeability in fibers of the GhSCP2D-downregulated plants intensified when sterol biosynthesis was inhibited.
Suppression of GhSCP2D Reduces Sterol Contents in Cotton Fibers
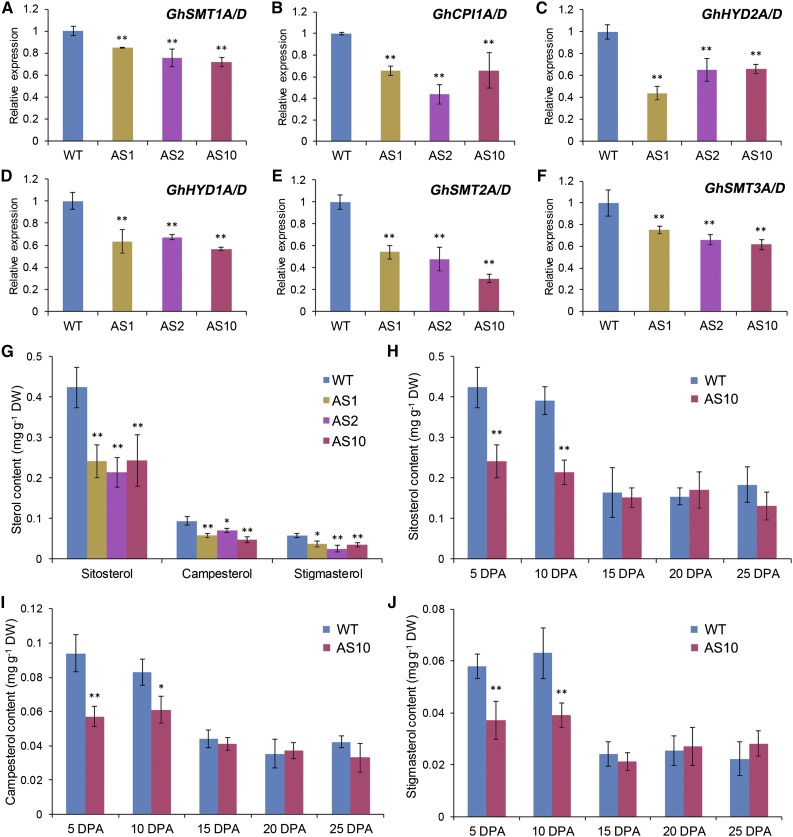
We then investigated whether suppressing GhSCP2D expression reduces sterol contents and alters the expression of sterol biosynthesis genes. Sterols are synthesized from cycloartenol and converted into a wide variety of sterol variants, including sitosterol, campesterol, and stigmasterol (Edwards and Ericsson, 1999). RNA-seq analysis revealed the expression of several sterol synthesis genes in elongating cotton fibers, including SMT1, CPI1, HYD2, HYD1, SMT2, and SMT3 (Supplemental Figure 7), all of which exhibited significantly lower mRNA levels in GhSCP2D-suppressed fibers than in the wild type (Figures 7A to 7F). Measurement of sterol contents using gas chromatography-mass spectrometry (GC-MS) showed that sitosterol, campesterol, and stigmasterol levels were enriched in 5- and 10-DPA fibers compared with 15-, 20-, and 25-DPA fibers in wild-type plants but were significantly reduced in 5- and 10-DPA fibers in the GhSCP2D-downregulated lines (Figures 7G to 7J). These results indicate that GhSCP2D is required for maintaining sterol levels and the expression of sterol biosynthesis genes in elongating cotton fibers.
Figure 7.
The Sterol Content Is Lower in 5- and 10-DPA Fibers of GhSCP2D-Downregulated Plants Than in the Wild Type.
(A) to (F) Transcript levels of the genes involved in sterol biosynthesis in 5-DPA fibers of GhSCP2D-downregulated (AS1, AS2, and AS10) and wild-type plants. Error bars represent the sd of triplicate experiments (**P < 0.01, by Student’s t test).
(G) GC-MS quantification of the sitosterol, campesterol, and stigmasterol contents in 5-DPA fibers of GhSCP2D-downregulated (AS1, AS2, and AS10) and wild-type plants. Error bars indicate sd from six replicate samples. The data were evaluated by Student’s t test; *P < 0.05 and **P < 0.01. DW, dry weight.
(H) to (J) Quantification by GC-MS of the sitosterol (H), campesterol (I), and stigmasterol (J) content in 5- to 25-DPA fibers of transgenic line AS10 and wild-type plants. Error bars indicate sd from six replicate samples. The data were evaluated by Student’s t test; *P < 0.05 and **P < 0.01. DW, dry weight.
Suppression of GhSCP2D Activates the Expression of Sucrose Transporter Genes in Elongating Fibers
The developmentally controlled closure of the symplasmic pathway may switch on the apoplasmic route for nutrient uptake (Ruan et al., 2001). We therefore investigated whether the closure of PDs in 5 to 10-DPA fibers observed in GhSCP2D-suppressed plants would activate or increase the expression of sugar transporter genes to compensate for the reduced symplasmic nutrient transport.
Plants contain two distinct types of sucrose transporters: SUT and clade III SWEET proteins (Eom et al., 2015). SUTs fall into the major facilitator superfamily (MFS) (Eom et al., 2015; Tao et al., 2015). There are 77 and 93 genes belonging to the MFS gene family in Arabidopsis and G. raimondii, respectively. Three SUTs in G. raimondii (Supplemental Figure 8A) corresponding to six SUTs in G. hirsutum (Supplemental Table 4) are phylogenetically clustered with Arabidopsis SUTs (AtSUC1, AtSUC2, AtSUC5, AtSUC6, AtSUC7, AtSUC8, and AtSUC9). These SUTs encode proteins responsible for sucrose transport across the plasma membrane, a process energized by H+-ATPases (Ruan et al., 2001; Baud et al., 2005; Dasgupta et al., 2014; Pommerrenig et al., 2013; Sivitz et al., 2007; Wippel and Sauer, 2012). RNA-seq and qRT-PCR analysis revealed that the transcripts of GhSUT1A/D, including GhSUT1 (Ruan et al., 2001), GhSUT2A/D, and GhSUT3A/D, were highly expressed in 15- to 25-DPA fibers, but only weakly expressed (<10%) in the early stage of fiber development (5 to 10 DPA) in wild-type plants (Figures 8A and 8B; Supplemental Figure 9B). Notably, GhSUT1 has been immunologically localized to the plasma membranes at the bases of cotton fibers (Ruan et al., 2001). However, the suppression of GhSCP2D drastically activated the expression of GhSUT1A/D and GhSUT3A/D in 5- and 10-DPA fibers but had no obvious effect on the expression of these genes at stage 15-DPA onwards (Figures 8A and 8B; Supplemental Figure 8C).
Figure 8.
Suppression of GhSCP2D Activates the Expression of SUT and Clade III SWEET Sucrose Transporter Genes in 5- and 10-DPA Fibers.
qRT-PCR analysis of genes encoding sucrose transporter (SUTs and SWEETs) in 5- to 25-DPA fibers of the wild type and GhSCP2D-downregulated line AS10. Three biological replicates were performed per reaction, each with two technical replicates (using the same sample). Each value represents mean ± se (**P < 0.01, Student’s t test).
We further investigated the effect of suppressing GhSCP2D expression on SWEET gene expression in cotton fibers. There are 12 and 21 SWEETs in G. raimondii and G. hirsutum, respectively, which are clustered with clade III SWEETs (SWEET 9-15) in Arabidopsis (Supplemental Figure 9A and Supplemental Table 5). RNA-seq showed that clade III SWEET genes, including GhSWEET1A/D, GhSWEET5A/D, GhSWEET6A/D, and GhSWEET10A/D, were expressed at extremely low levels in 5- and 10-DPA wild-type fibers (Supplemental Figure 9B). Most obviously, in the GhSCP2D-downregulated lines, the transcript levels of these SWEETs dramatically increased in 5- and 10-DPA fibers but remained unchanged in 15-, 20-, and 25-DPA fibers compared with the wild type (Figures 8C to 8F; Supplemental Figure 9C). These results indicate that suppressing GhSCP2D expression in cotton activates or enhances the expression of SUT and clade III SWEET genes during early fiber elongation.
Prolonged Expression of GbSCP2D, a GhSCP2D Homolog, Correlates with Extended PD Opening and Delayed Expression of SUTs and SWEETs during G. barbadense Fiber Development
Finally, we investigated whether overexpressing GhSCP2D prolongs PD opening by increasing the expression of GhPdBG3-2A/D. As mentioned, we failed to obtain transgenic lines overexpressing GhSCP2D from cotton plants transformed with the overexpression construct (Supplemental Figure 4), probably due to sequence homology-dependent cosuppression (Taylor, 1997). To circumvent this problem, we isolated GbSCP2D and GbPdBG3-2A/D, which are the homologous genes of GhSCP2D and GhPdBG3-2A/D, respectively, from another cultivated tetraploid cotton species, G. barbadense cultivar H7124, which has much longer fibers than G. hirsutum accession W0 (wild type) (Wendel and Cronn, 2003). The transcript levels of GbSCP2D and GbPdBG3-2A/D in H7124 fibers were enriched not only at 5 and 10 DPA, as observed for GhSCP2D and GhPdBG2-2A/D (Figures 1C and 5M), but also at 15-DPA, and they became undetectable at 20 to 30 DPA (Figures 9A and 9B). Accordingly, this prolonged expression of GbSCP2D and GbPdBG3-2A/D correlated with the detection of CF fluorescent signals in 5-, 10-, and 15-DPA H7124 fibers (Figures 9I to 9K), whereas these signals were undetectable in 20-, 25-, and 30-DPA fibers (Figures 9L to 9N). The CF data indicate that an extensive period of PD opening occurred at 5 to 15 DPA in H7124, whereas in the wild type, fiber PDs were open only at 5 to 10 DPA (Figures 3A and 3B). In conjunction with the prolonged PD opening (Figures 9I to 9K), little or no GbSUT and GbSWEET expression was detected during this period (Figures 9C to 9H). These findings help confirm the notion that SCP2D helps keep the PD open by positively regulating PdBG expression.
Figure 9.
GbSCP2D Is Expressed in 5- to 15-DPA Fibers of G. barbadense, Which Is Accompanied by the Prolonged Opening of PDs in G. barbadense.
(A) to (H) qRT-PCR analysis of the SCP2D, PdBG3-2A/D, SUTs, and SWEETs in 5 to 30 fibers of G. barbadense. SCP2D and PdBG3-2A/D transcripts accumulate in 5- to 15-DPA fibers of G. barbadense. By contrast, SUT and SWEET transcripts accumulate in 20- to 30-DPA fibers of G. barbadense.
(I) to (K) Optical sections of 5-, 10-, and 15-DPA G. barbadense seeds, showing the transport and accumulation of CF from vascular bundles to fibers and the outer seed coat but not to the inner seed coat.
(L) to (N) Optical sections of 20-, 25-, and 30-DPA G. barbadense seeds, showing strong, widespread CF signals in the vascular region but few or no signals in fibers. Bars = 50 µm.
DISCUSSION
GhSCP2D Is Required for Maintaining Symplasmic Permeability by Positively Regulating GhPdBG3-2A/D Expression to Degrade Callose at the PD during Cotton Fiber Elongation
Callose turnover represents a major mechanism regulating symplasmic conductivity at the PD (De Storme and Geelen, 2014; Guseman et al., 2010; Levy et al., 2007; Ruan et al., 2004; Vatén et al., 2011). Callose deposition at the PD generally correlates with reduced PD permeability and PD branching (Benitez-Alfonso et al., 2009; Ruan et al., 2001). In this study, we found that suppressing the expression of GhSCP2D, encoding a putative SCP, resulted in intensive callose deposition at the PD region at the base of the cotton fiber during early elongation as well as increased PD branching, leading to the closure of PDs throughout fiber elongation, as revealed by the movement of the symplasmic dye CF. By contrast, in wild-type plants, little or no callose was observed at the fiber base, and the PDs remained open at 5 and 10 DPA (Figures 3 and 4). Furthermore, prolonged expression of GbSCP2D, a homolog of GhSCP2D in G. barbadense, correlated with a longer period of PD opening (Figure 9). Collectively, these findings suggest that GhSCP2D plays a major role in controlling symplasmic transport, likely via regulating callose turnover in the PD.
Callose accumulation is controlled by the joint action of GSLs and BGs, which synthesize and degrade callose, respectively (Bucher et al., 2001; Ruan et al., 2004). GhGSL expression was not affected in the fibers of GhSCP2D-downregulated plants. However, the expression of GhPdBG3-2A/D, a putative β-1,3 glucanase gene that is predominantly expressed in 5- and 10-DPA fibers in wild-type plants, was dramatically reduced in the fibers of GhSCP2D-suppressed plants (Figure 5). These results suggest that GhSCP2D positively regulates GhPdBG3-2A/D expression to degrade the callose deposited at the PD during cotton fiber elongation.
In wild-type cotton, a cohort of genes involved in sterol biosynthesis are highly expressed during early fiber elongation (Supplemental Figure 7). Consistently, sterol is enriched in fibers at the elongation stage compared with the late secondary wall thickening period (Figure 7; Deng et al., 2016). Suppressing GhSCP2D expression downregulated sterol biosynthesis genes and reduced the sterol contents in 5- and 10-DPA fibers compared with wild-type plants (Figure 7), demonstrating a role for GhSCP2D in maintaining sterol homeostasis within the elongating cotton fiber cell.
We found that pharmacologically inhibiting sterol biosynthesis using lova significantly reduced the expression of GhPdBG3-2A/D, encoding GPI-anchored and PD-targeted proteins, in cotton fibers (Figures 5 and 6). Interestingly, Grison et al. (2015) used a similar approach to reduce the sterol contents in Arabidopsis root tips, leading to the mislocalization of AtPdBG2 in sterol-enriched PDs in the plasma membrane and the accumulation of callose in this region. Further studies are required to elucidate exactly how GhSCP2D regulates the expression of GhPdBG3-2A/D. Nevertheless, given the reduced sterol content and the expression of sterol synthesizing genes observed in the fibers of GhSCP2D-suppressed plants, it is plausible that GhSCP2D regulates GhPdBG3-2A/D expression through modulating sterol contents or composition. Altering sterol composition might affect the expression of GhPdBG3-2A/D through a feedback mechanism, as well as the targeting of GhPdBG3-2A/D to PDs in the plasma membranes of cotton fiber cells.
Closure of PD Induced by the Suppression of GhSCP2D Activates the Expression of Sucrose Transporter Genes SUT and SWEET during Fiber Elongation
Sucrose is the major organic nutrient and energy source imported into cotton fiber cells (Ruan et al., 2001, 2003). We found that fiber and seed growth were positively correlated with sucrose concentration in the culture medium (Supplemental Figure 10), indicating the importance of sucrose import to cotton fiber development. The GhSCP2D-downregulated cotton plants had reduced sucrose and hexose levels and shortened fiber length (Table 1, Figure 2). Unlike what was observed in seedlings, the addition of sucrose did not compensate for the defects in fiber growth where GhSCP2D transcripts were enriched (Figure 2). These findings, together with the observation that GhSCP2D is required for PD opening during fiber elongation, indicate that GhSCP2D plays a role in symplasmic sucrose transport to support fiber development.
SUTs and clade III SWEETs are the two classes of sucrose transporters responsible for sucrose transport across the plasma membrane (Eom et al., 2015; Chen et al., 2012; Gottwald et al., 2000). In Arabidopsis, SUC2, SWEET11, and SWEET12 are expressed in the vascular tissues of mature leaves, and their encoded proteins are localized to the plasma membranes of phloem parenchyma (Truernit and Sauer, 1995; Chen et al., 2012). The growth of Arabidopsis mutants suc2 and sweet11 sweet12 is retarded, likely due to reduced sucrose efflux from leaves to sink organs such as roots (Chen et al., 2012). Interestingly, the Arabidopsis scp2 mutant shows reduced seedling growth, a phenotype complemented by the addition of 1% sucrose to the medium, which is similar to that observed for the suc2 and sweet11 sweet12 mutants and for GhSCP2D-suppressed cotton seedlings in this study (Figure 2). These observations point to a link between SCP2 and sucrose transport.
Notably, in parallel to the strong expression of GhSCP2D and GhPdBG3-2A/D in 5- and 10-DPA fibers when PDs were open and the weak expression of these genes during the late stage of fiber development (Figures 2 and 5), we detected little or no GhSUT and clade III GhSWEET transcripts during the early stage of elongation, which significantly increased during late elongation when the PD were closed (Figure 9). Such an inverse relationship between PD gating and the expression of SUTs and clade III SWEETs was further demonstrated by suppressing GhSCP2D expression. The transgenic fibers exhibited PD closure during the early stage of elongation and, hence, the blockage of PD permeability throughout the elongation period (Figures 3, 4, and 10). Interestingly, the expression of SUTs and clade III SWEETs increased dramatically in 5- and 10-DPA fibers of GhSCP2D-downregulated plants compared with the wild type (Figure 8). Genotypic evidence from G. barbadense further supports the absence or activation of sugar transporter gene expression via the opening or closure of PDs in cotton fibers, respectively. In this genotype, fiber PDs were open at 5, 10, and 15 DPA but closed at 20, 25, and 30 DPA. Accordingly, GbSUT and clade III GbSWEET mRNA was undetectable in 5-, 10-, and 15-DPA fibers but enriched in late-stage fibers (Figure 9). These results indicate that sucrose import into fiber cells occurs through the symplasmic pathway controlled by SCP2D and PdBG3-2A/D during early fiber elongation but switches to a SUT- and SWEET-mediated apoplasmic route once SCP2D and PdBG3-2A/D expression is reduced or suppressed. These findings provide an example of the developmental versatility of cell-to-cell communication linking SCP2D with callose turnover, PD gating, and the expression of sugar transporter genes.
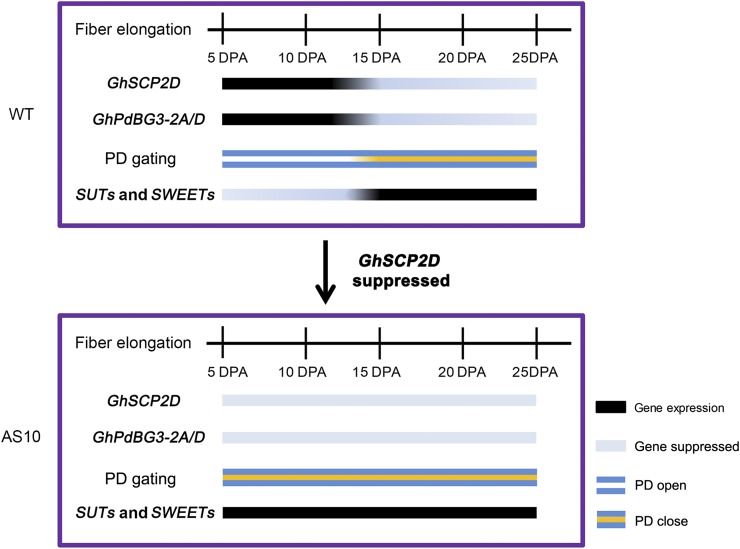
Figure 10.
Model Describing the Roles of GhSCP2D in PD Gating for Fiber Cell Elongation.
GhSCP2D modulates the expression of GhPdBG3-2A/D, encoding a β-1,3-glucanase, as well as the expression of sugar transporter genes SUTs and clade III SWEETs.
We found that SUTs and clade III SWEETs were coexpressed in cotton fibers (Figure 8). How this coordination is achieved remains to be determined. We hypothesize that this coordination might contribute to cytosolic sugar homeostasis, which is vital for metabolism and cellular function (Ruan, 2014). The joint action of energy-coupled sucrose influx mediated by the SUTs could lead to increased sucrose or hexose levels in the fiber cytosol, whereas the clade III SWEETs might transport sucrose from the cytosol back to the apoplasm once the intracellular sucrose concentration is high enough, since SWEETs are uniporters that mediate bidirectional sugar transport depending on the concentration difference (Chen et al., 2012). It is also possible that both the SUTs and clade III SWEETs might function in the uptake of phloem-unloaded sucrose into fiber cells from the apoplasm at the interface between the fiber base and seed coat. Under this scenario, the GhSUTs might function as energy-independent sucrose facilitators to transport sucrose from the apoplasm into the fiber cytosol in a manner similar to that predicted for the uniporters, GhSWEETs. Indeed, such a function in facilitating sucrose diffusion has been demonstrated for a class of SUTs in the legume seed coat (Zhou et al., 2007).
Overall, our study shows that suppressing GhSCP2D expression in cotton fibers reduced the expression of GhPdBG3-2A/D, likely by disrupting sterol homeostasis, leading to enhanced callose deposition and reduced PD permeability as well as fiber elongation. Moreover, we provided evidence that the symplasmic pathway prevails during early cotton fiber elongation and that the apoplasmic route for sucrose transport is switched on during later development to compensate for the loss of the symplasmic pathway (Figure 10). Further studies are needed to elucidate the biochemical pathway by which peroxisome-localized GhSCP2D regulates sterol content. In this context, several key enzymes for the formation of sterol precursors are also localized to the peroxisome (Simkin et al., 2011). Thus, it will be interesting to establish whether GhSCP2D acts alone or with sterol-synthesizing enzymes to transfer sterol or its precursors into or out of peroxisomes.
METHODS
Plant Materials and Growth Conditions
Cotton plants (Gossypium hirsutum accession W0 and G. barbadense cultivar H7124) were cultivated in the field at the experimental station of Nanjing Agricultural University and Zhejiang University, China. Cotton boll age was determined by tagging each pedicel on the day of flowering. The bolls were harvested in the afternoon. Fiber cells at different developmental stages were carefully removed from the seeds and immediately snap-frozen in liquid nitrogen for DNA and RNA extraction and sugar and sterol content determinations.
Phylogenic Analysis and Gene Cloning
The genome and annotation database used in this study (except for cotton) was obtained from Phytozome (https://phytozome.jgi.doe.gov/pz/portal.html). The genome and annotation database of cotton was obtained from the laboratory website (http://mascotton.njau.edu.cn) and in the CottonGen database (http://www.cottongen.org). Profile generation and searches were performed using the HMMER 3.0 software package (Finn et al., 2014). The conserved domains of SCP, BG, GSL, SUT, and SWEET genes were used as queries to search the genome database by tBLASTn using the local BLAST program version 2.2.28. Prediction of gene structure from genomic contigs was performed using FGENESH software in the Softberry server (http://www.softberry.com/berry.phtml). Sequences were edited and analyzed using DNAMAN software version 6.0 (Lynnon Biosoft). Phylogenetic and molecular evolutionary analyses were conducted using MEGA version 6.0 (Tamura et al., 2013). The protein sequences were aligned using ClustalX (Larkin et al., 2007). The results of multiple sequence alignments are provided in Supplemental Files 1 to 5. The neighbor-joining method was used to construct different phylogenetic trees with the following parameters: Poisson model, pairwise deletion, and bootstrap method (1000 replicates). The tree nodes were evaluated by bootstrap analysis with 1000 replicates and are shown in the phylogenetic trees. Genome-wide transcriptome data from G. hirsutum at different developmental stages were downloaded from the database (SRA: PRJNA248163) as described by Zhang et al. (2015). The method for calculating gene expression levels was performed according to Zhang et al. (2015). MeV software (http://www.tm4.org/mev.html) was used to display the data.
The GhSCP and GhPdBG3-2A/D sequences were obtained from the prediction described above. Pairs of gene-specific primers (Supplemental Table 6) were designed to amplify full-length cDNA from 5-DPA fibers of G. hirsutum accession TM-1. After purification (Qiagen), the PCR products were cloned into the pMD18-T plasmid (Takara) and sequenced.
qRT-PCR
Total RNA from various cotton tissues was isolated using a Biospin Plant Total RNA Extraction kit (BioFlux) and reverse transcribed to cDNA with SuperScript III reverse transcriptase (Invitrogen). Primers based on the SNP site between the exons of GhSCP genes in the At and Dt subgenomes from WebSNAPER (http://pga.mgh.harvard.edu/cgi-bin/snap3/websnaper3.cgi) were used to detect the expression of GhSCP genes in these subgenomes. EF-1α (Supplemental Table 7) from cotton was used as an internal control for normalization of the cDNA samples. qRT-PCR was performed using a LightCycler FastStart DNA Master SYBR Green I kit (Roche) in a CFX96 Touch real-time PCR detection system according to the manufacturer’s protocol (Bio-Rad). The data were evaluated using the comparative cycle threshold method described by Livak and Schmittgen (2001). Three biological replicates (three samples harvested from three plants, one from each) were performed per reaction, each with two technical replicates (using the same sample). Mean values and standard errors were calculated according the data from three replicates. The primers used for qRT-PCR are listed in Supplemental Table 7.
Vector Construction and Plant Transformation
To produce the overexpression/suppression constructs, two pairs of primers (OE-GhSCP2D-F/R and AS-GhSCP2D-F/R) with added BamHI and SacI, SacI, and BamHI restriction sites, respectively, were used to amplify the open reading frame of GhSCP2D, which was then cloned into the pBI121 vector under the control of the constitutive Cauliflower mosaic virus (CaMV) 35S promoter. GhSCP2D overexpression (35SSC) and GhSCP2D-downregulated (35SAC) constructs (Supplemental Figure 3A) were introduced into G. hirsutum accession W0 via Agrobacterium tumefaciens-mediated transformation as described previously (Wu et al., 2008). The homozygosity of the transgenic plants was determined using the kanamycin selection marker coupled with PCR-based genotyping. The primers used for vector construction and PCR-based screening are listed in Supplemental Table 6.
Subcellular Location in Cotton Protoplasts and Nicotiana benthamiana Leaves
For subcellular location of GhSCP2D in cotton protoplasts, the full-length cDNA of GhSCP2D without a stop codon was introduced into pJIT166-GFP upstream of the GFP sequence through a recombination reaction. In addition, the full-length cDNA of GhSCP2D with a stop codon was introduced into pJIT166-GFP downstream of the GFP sequence through overlap extension PCR (Bryksin and Matsumura, 2010). The resulting constructs consisted of GhSCP2D fused to the N terminus of GFP and GhSCP2D fused to the C terminus of GFP under the control of the CaMV 35S promoter. These constructs were introduced into cotton protoplasts via PEG-mediated transformation as described previously (Li et al., 2014).
For subcellular location of GhSCP2D in N. benthamiana leaves, the coding sequence of GhSCP2D was cloned into vector pBINGFP4 (GFP) to form GFP-GhSCP2D. GPI-anchor domains of GhPdBG3-2A/D were predicted using two programs, GPI-modified: Big PI plant predictor (http://mendel.imp.ac.at/gpi/plant_server.html) and GPI-SOM (http://gpi.unibe.ch; Fankhauser and Mäser, 2005). The coding sequence of GFP was inserted within the GhPdBG3-2A/D coding sequence immediately after amino acid 447, which is located 22 amino acids before the ω site, using published protocols (Tian et al., 2004). This region into which GFP was inserted does not affect the X8 domain, which may be involved in carbohydrate binding, as predicted by the SMART program (http://smart.embl-heidelberg.de; Letunic et al., 2015). The genes for GFP-tagged proteins were subcloned into pBINGFP4 (GFP). Agrobacterium strain GV3101 carrying the construct was used together with the p19 strain (Lombardi et al., 2009) and the peroxisomal marker, RFP-PTS1 (Hayashi et al., 1996), to infiltrate 5- to 6-week-old N. benthamiana leaves. Analysis was performed on a Zeiss LSM780 confocal microscope using a 488-nm excitation laser for GFP, a 405-nm laser for aniline blue fluorochrome, and a 561-nm laser for mCherry.
Cotton Ovule Culture
Bolls at 1 DPA were harvested and surface-sterilized in 75% ethanol for 5 min, followed by washing with sterile water. The seeds were peeled from the bolls and placed in liquid BT medium (Beasley, 1971) supplemented with the following compounds at various concentrations: sucrose (0, 25, 50, or 75 mM), or lova (stock solution in DMSO) (0, 1, 5, or 10 μM) (Sigma-Aldrich). For lova treatment, the control contained an equal amount of 0.1% DMSO solvent.
Soluble Carbohydrate and Sterol Analysis
Carbohydrate analysis was performed according to Jiang et al. (2012). Fibers (∼200 mg) were ground in liquid nitrogen and extracted with 80% ethanol, followed by 50% ethanol and water. The aqueous phase of the samples was collected, dried in a vacuum, and redissolved in 1 mL water. An anion-exchange HPLC system (Agilent 1100 series; Agilent Technologies) was used to measure the sucrose, glucose, and fructose contents of the samples. Quantification of each sugar was accomplished by comparing the peak areas of the samples with standard samples of sucrose, glucose, and fructose (Sigma-Aldrich).
Sterol extraction was performed according to Henry et al. (2015) with some modifications: 100 mg of dried cotton fibers was ground in liquid nitrogen and extracted with 4 mL chloroform:methanol (2:1) (v/v) containing 1.25 mg/L 5-α-cholestan-3-α-ol as an internal standard. After incubation at 70°C for 1 h, the samples were dried under nitrogen at 25°C and saponified with 2 mL 6% (w/v) KOH in methanol for 3 h at 90°C to release the sterol moiety of steryl esters. Sterols were extracted three times with 2 mL hexane:water (1:1) (v/v) and dried under nitrogen at 25°C. The dried residues were derived using 100 μL of BSTFA-TMCS (1:1) (v/v). Sterol levels were analyzed on a Thermo-Fish GC-MS (DSQ II mass spectrometer combined with focus GC). Separation was performed on a HP-5MS column (30 m × 0.25 mm × 0.25 μm film; Agilent Technologies) with helium as the carrier gas. The temperature program was 170°C for 1.5 min, ramp to 280°C at 37°C/min, 300°C at 1.5°C/min, and hold for 5 min.
Loading of CF and Confocal Laser Scanning Microscopy
The loading of CF and confocal imaging of the movement of CF into fibers were conducted according to Ruan et al. (2001). After excision, the detached fruit-bearing shoots were immediately recut under water, with or without 100 μg/mL of the nonfluorescent dye 5(6)-carboxyfluorescein diacetate (Sigma-Aldrich), and illuminated using light emitting diode bulb at 6500K color temperature and the photon flux density was 500 μmol m−2 s−1 at 25°C. After 24 h, intact seeds were removed from the fruits. Free-hand sections (∼1 mm) of seeds were produced, and CF was viewed in the seed coat and interconnecting fibers via confocal laser scanning microscopy (LSM780; Zeiss) at an excitation wavelength of 488 nm and emission wavelength range of 493 to 598 nm.
Aniline Blue Staining, Electron Microscopy of PD Structure, and Immunolocalization of Callose
Aniline blue was prepared in water at 0.05% (w/v) and stored in the dark at 4°C. Free-hand sections (∼1 mm) of seeds were produced and stained with aniline blue for 10 min in the dark, followed by a 30-s wash with water (Shang et al., 2015). Fluorescent signals from aniline blue were observed under a Zeiss LSM780 confocal microscope using a 405-nm excitation laser (Benitez-Alfonso et al., 2013). Sections stained with water only were used as a control.
Seeds fixation and embedding were performed as described by Shang et al. (2015). Cotton seeds (5 DPA) were fixed in 2% (v/v) paraformaldehyde and 0.1% (v/v) glutaraldehyde in phosphate buffer (25 mM, pH 7.2) for 2 h. The seeds were dehydrated in a step-graded ethanol series and embedded in LR White resin (medium grade; Alltech) through a step-graded series after washing in buffer. Infiltrated seeds were polymerized in gelatin capsules at 70°C for ∼2 h under nitrogen gas. Sections (90 nm thick) were cut with an ultramicrotome (EMUC7; Leica), collected on nickel mesh, and stained with uranyl acetate for 10 min and lead citrate for 5 min, followed by washing with 0.01 M PBS (six times, 3 min each) and water (four times, 3 min each). The sections were viewed under a Hitachi H-7650 transmission electron microscope at 80 kV to observe PD structure. Five to ten nonserial sections per genotype from three seeds were examined per line. At least 10 cell wall interfaces between the fiber and fiber base and adjacent seed coat cells from three seeds per genotype were examined.
For immunolocalization of callose, the sections were preincubated in 0.01 M PBS (1% BSA, 0.05 Triton X-100, and 0.05% Tween 20, pH 7.4) for 5 min, followed by incubation in 1:300 diluted monoclonal antibody to (1→3)-β-glucan (Biosupplies Australia) for 4 h at room temperature. After washing in PBS, the sections were incubated in 1:500 diluted goat anti-mouse IgG-Gold antibody (Bellancom Chemistry) for 1 h. The sections were stained with uranyl acetate for 5 min and lead citrate for 3 min, followed by washing with 0.01 M PBS (six times, 3 min each) and water (four times, 3 min each). The sections were viewed under a Hitachi H-7650 transmission electron microscope at 80 kV. Negative control samples were produced by omitting the incubation with primary antibody or by incubating with normal mouse serum only. Five to ten nonserial sections per genotype from three seeds were examined per line. At least 10 cell wall interfaces between the fiber and fiber base and the adjacent seed coat cells from three seeds per genotype were examined.
Accession Numbers
Sequence data for Arabidopsis from this article can be found in TAIR10 under accession number AT5G42890 (AtSCP2). Sequence data for cotton can be found on the laboratory website (http://mascotton.njau.edu.cn) and in the CottonGen database (http://www.cottongen.org). Accessions numbers are listed in Supplemental Tables 1 to 5.
Supplemental Data
Supplemental Figure 1. Expression and sequence analyses of GhSCP2D in cotton.
Supplemental Figure 2. qRT-PCR analysis of GhSCP1D, GhSCP3A, and GhSCP3D expression during fiber and seed germination in G. hirsutum.
Supplemental Figure 3. GhSCP2D constructs used for genetic transformation.
Supplemental Figure 4. Expression analysis of GhSCPs in 5-DPA fibers of wild-type, GhSCP2D-overexpressing, and GhSCP2D-downregulated cotton plants.
Supplemental Figure 5. Confocal images of CF transport to fibers in GhSCP2D-downregulated lines (AS1, AS2, and AS10) and wild-type plants.
Supplemental Figure 6. Expression analysis of GSL in GhSCP2D-downregulated lines (AS1, AS2, and AS10) and wild-type cotton plants.
Supplemental Figure 7. RNA-seq analysis of genes involved in sterol synthesis in various tissues and organs of G. hirsutum.
Supplemental Figure 8. Expression analysis of GhSUT in GhSCP2D-downregulated lines (AS1, AS2, and AS10) and wild-type cotton plants.
Supplemental Figure 9. Expression analysis of GhSWEET in GhSCP2D-downregulated lines (AS1, AS2, and AS10) and wild-type cotton plants.
Supplemental Figure 10. Sucrose is positively correlated with fiber development.
Supplemental Table 1. Distribution of SCP genes in cotton.
Supplemental Table 2. Distribution of GSL in the genomes of diploid and tetraploid cotton.
Supplemental Table 3. Distribution of PdBG in the genomes of tetraploid and diploid cotton.
Supplemental Table 4. Distribution of GhSUT in the genomes of tetraploid and diploid cotton.
Supplemental Table 5. Distribution of GhSWEET in the genomes of tetraploid and diploid cotton.
Supplemental Table 6. Oligonucleotides used for gene cloning and vector construction in this study.
Supplemental Table 7. Oligonucleotides used for qRT-PCR in this study
Supplemental File 1. Alignment used to construct the phylogenetic tree shown in Figure 1A.
Supplemental File 2. Alignment used to construct the phylogenetic tree shown in Figure 5A.
Supplemental File 3. Alignment used to construct the phylogenetic tree shown in Supplemental Figure 6A.
Supplemental File 4. Alignment used to construct the phylogenetic tree shown in Supplemental Figure 8A.
Supplemental File 5. Alignment used to construct the phylogenetic tree shown in Supplemental Figure 9A.
Supplemental File 6. One-way ANOVA tables.
Acknowledgments
This study was supported in part by grants from the NSFC (31330058), The Distinguished Discipline Support Program of Zhejiang University, National Research and Development Project of Transgenic Crops of China (2011ZX08009-003), the Australian Research Council (DP110104931 and DP120104148), the Priority Academic Program Development of Jiangsu Higher Education Institutions, and the JCIC-MCP Project. We thank Tiegang Lu (Biotechnology Research Institute, Chinese Academy of Agricultural Sciences) for the monoclonal antibody for (1→3)-β-glucan.
AUTHOR CONTRIBUTIONS
T.Z., Z.Z., Y.-L.R., X.G., W.G., and X.S. designed the research. Z.Z., N.Z., and F.W. performed research. Z.Z., Y.-L.R., and L.F. analyzed data. T.Z., Z.Z., and Y.-L.R. wrote the article.
Glossary
- DPA
days postanthesis
- PD
plasmodesmata
- GPI
glycosylphosphatidylinositol
- CF
carboxyfluorescein
- GC-MS
gas chromatography-mass spectrometry
References
- Amor Y., Haigler C.H., Johnson S., Wainscott M., Delmer D.P. (1995). A membrane-associated form of sucrose synthase and its potential role in synthesis of cellulose and callose in plants. Proc. Natl. Acad. Sci. USA 92: 9353–9357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baud S., Wuillème S., Lemoine R., Kronenberger J., Caboche M., Lepiniec L., Rochat C. (2005). The AtSUC5 sucrose transporter specifically expressed in the endosperm is involved in early seed development in Arabidopsis. Plant J. 43: 824–836. [DOI] [PubMed] [Google Scholar]
- Beasley C.A. (1971). In vitro culture of fertilized cotton ovules. Bioscience 21: 906–907. [Google Scholar]
- Benitez-Alfonso Y., Cilia M., San Roman A., Thomas C., Maule A., Hearn S., Jackson D. (2009). Control of Arabidopsis meristem development by thioredoxin-dependent regulation of intercellular transport. Proc. Natl. Acad. Sci. USA 106: 3615–3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benitez-Alfonso Y., Faulkner C., Pendle A., Miyashima S., Helariutta Y., Maule A. (2013). Symplastic intercellular connectivity regulates lateral root patterning. Dev. Cell 26: 136–147. [DOI] [PubMed] [Google Scholar]
- Bryksin A.V., Matsumura I. (2010). Overlap extension PCR cloning: a simple and reliable way to create recombinant plasmids. Biotechniques 48: 463–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucher G.L., Tarina C., Heinlein M., Di Serio F., Meins F. Jr., Iglesias V.A. (2001). Local expression of enzymatically active class I β-1,3-glucanase enhances symptoms of TMV infection in tobacco. Plant J. 28: 361–369. [DOI] [PubMed] [Google Scholar]
- Carland F., Fujioka S., Nelson T. (2010). The sterol methyltransferases SMT1, SMT2, and SMT3 influence Arabidopsis development through nonbrassinosteroid products. Plant Physiol. 153: 741–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cedroni M.L., Cronn R.C., Adams K.L., Wilkins T.A., Wendel J.F. (2003). Evolution and expression of MYB genes in diploid and polyploid cotton. Plant Mol. Biol. 51: 313–325. [DOI] [PubMed] [Google Scholar]
- Chen L.Q., Qu X.Q., Hou B.H., Sosso D., Osorio S., Fernie A.R., Frommer W.B. (2012). Sucrose efflux mediated by SWEET proteins as a key step for phloem transport. Science 335: 207–211. [DOI] [PubMed] [Google Scholar]
- Chen X.Y., Kim J.Y. (2009). Callose synthesis in higher plants. Plant Signal. Behav. 4: 489–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z.J., Guan X. (2011). Auxin boost for cotton. Nat. Biotechnol. 29: 407–409. [DOI] [PubMed] [Google Scholar]
- Dasgupta K., Khadilkar A.S., Sulpice R., Pant B., Scheible W.R., Fisahn J., Stitt M., Ayre B.G. (2014). Expression of sucrose transporter cDNAs specifically in companion cells enhances phloem loading and long-distance transport of sucrose but leads to an inhibition of growth and the perception of a phosphate limitation. Plant Physiol. 165: 715–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng S., Wei T., Tan K., Hu M., Li F., Zhai Y., Ye S., Xiao Y., Hou L., Pei Y., Luo M. (2016). Phytosterol content and the campesterol:sitosterol ratio influence cotton fiber development: role of phytosterols in cell elongation. Sci. China Life Sci. 59: 183–193. [DOI] [PubMed] [Google Scholar]
- De Storme N., Geelen D. (2014). Callose homeostasis at plasmodesmata: molecular regulators and developmental relevance. Front. Plant Sci. 5: 138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diener A.C., Li H., Zhou W., Whoriskey W.J., Nes W.D., Fink G.R. (2000). Sterol methyltransferase 1 controls the level of cholesterol in plants. Plant Cell 12: 853–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doxey A.C., Yaish M.W.F., Moffatt B.A., Griffith M., McConkey B.J. (2007). Functional divergence in the Arabidopsis beta-1,3-glucanase gene family inferred by phylogenetic reconstruction of expression states. Mol. Biol. Evol. 24: 1045–1055. [DOI] [PubMed] [Google Scholar]
- Edqvist J., Blomqvist K. (2006). Fusion and fission, the evolution of sterol carrier protein-2. J. Mol. Evol. 62: 292–306. [DOI] [PubMed] [Google Scholar]
- Edqvist J., Rönnberg E., Rosenquist S., Blomqvist K., Viitanen L., Salminen T.A., Nylund M., Tuuf J., Mattjus P. (2004). Plants express a lipid transfer protein with high similarity to mammalian sterol carrier protein-2. J. Biol. Chem. 279: 53544–53553. [DOI] [PubMed] [Google Scholar]
- Edwards P.A., Ericsson J. (1999). Sterols and isoprenoids: signaling molecules derived from the cholesterol biosynthetic pathway. Annu. Rev. Biochem. 68: 157–185. [DOI] [PubMed] [Google Scholar]
- Eom J.S., Chen L.Q., Sosso D., Julius B.T., Lin I.W., Qu X.Q., Braun D.M., Frommer W.B. (2015). SWEETs, transporters for intracellular and intercellular sugar translocation. Curr. Opin. Plant Biol. 25: 53–62. [DOI] [PubMed] [Google Scholar]
- Fankhauser N., Mäser P. (2005). Identification of GPI anchor attachment signals by a Kohonen self-organizing map. Bioinformatics 21: 1846–1852. [DOI] [PubMed] [Google Scholar]
- Finn R.D., et al. (2014). Pfam: the protein families database. Nucleic Acids Res. 42: D222–D230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottwald J.R., Krysan P.J., Young J.C., Evert R.F., Sussman M.R. (2000). Genetic evidence for the in planta role of phloem-specific plasma membrane sucrose transporters. Proc. Natl. Acad. Sci. USA 97: 13979–13984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves D.A., Stewart J.M. (1988). Analysis of the protein constituency of developing cotton fibers. J. Exp. Bot. 39: 59–69. [Google Scholar]
- Grison M.S., et al. (2015). Specific membrane lipid composition is important for plasmodesmata function in Arabidopsis. Plant Cell 27: 1228–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo D.A., Venkatramesh M., Nes W.D. (1995). Developmental regulation of sterol biosynthesis in Zea mays. Lipids 30: 203–219. [DOI] [PubMed] [Google Scholar]
- Guseman J.M., Lee J.S., Bogenschutz N.L., Peterson K.M., Virata R.E., Xie B., Kanaoka M.M., Hong Z., Torii K.U. (2010). Dysregulation of cell-to-cell connectivity and stomatal patterning by loss-of-function mutation in Arabidopsis chorus (glucan synthase-like 8). Development 137: 1731–1741. [DOI] [PubMed] [Google Scholar]
- Hartmann M.A. (1998). Plant sterols and the membrane environment. Trends Plant Sci. 3: 170–175. [Google Scholar]
- Hayashi M., Aoki M., Kato A., Kondo M., Nishimura M. (1996). Transport of chimeric proteins that contain a carboxy-terminal targeting signal into plant microbodies. Plant J. 10: 225–234. [DOI] [PubMed] [Google Scholar]
- Henry L.K., Gutensohn M., Thomas S.T., Noel J.P., Dudareva N. (2015). Orthologs of the archaeal isopentenyl phosphate kinase regulate terpenoid production in plants. Proc. Natl. Acad. Sci. USA 112: 10050–10055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji S.J., Lu Y.C., Feng J.X., Wei G., Li J., Shi Y.H., Fu Q., Liu D., Luo J.C., Zhu Y.X. (2003). Isolation and analyses of genes preferentially expressed during early cotton fiber development by subtractive PCR and cDNA array. Nucleic Acids Res. 31: 2534–2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y., Guo W., Zhu H., Ruan Y.L., Zhang T. (2012). Overexpression of GhSusA1 increases plant biomass and improves cotton fiber yield and quality. Plant Biotechnol. J. 10: 301–312. [DOI] [PubMed] [Google Scholar]
- Kim H.J., Triplett B.A. (2001). Cotton fiber growth in planta and in vitro. Models for plant cell elongation and cell wall biogenesis. Plant Physiol. 127: 1361–1366. [PMC free article] [PubMed] [Google Scholar]
- Larkin M.A., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948. [DOI] [PubMed] [Google Scholar]
- Letunic I., Doerks T., Bork P. (2015). SMART: recent updates, new developments and status in 2015. Nucleic Acids Res. 43: 257–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy A., Guenoune-Gelbart D., Epel B.L. (2007). Beta-1,3-Glucanases: Plasmodesmal gate keepers for intercellular communication. Plant Signal. Behav. 2: 404–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N., Ding L., Zhang Z., Guo W. (2014). Isolation of mesophyll protoplast and establishment of gene transient expression system in cotton. Acta Agron. Sin. 40: 231–239. [Google Scholar]
- Lin I.W., et al. (2014). Nectar secretion requires sucrose phosphate synthases and the sugar transporter SWEET9. Nature 508: 546–549. [DOI] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- Lombardi R., Circelli P., Villani M.E., Buriani G., Nardi L., Coppola V., Bianco L., Benvenuto E., Donini M., Marusic C. (2009). High-level HIV-1 Nef transient expression in Nicotiana benthamiana using the P19 gene silencing suppressor protein of Artichoke Mottled Crinckle Virus. BMC Biotechnol. 9: 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pommerrenig B., Popko J., Heilmann M., Schulmeister S., Dietel K., Schmitt B., Stadler R., Feussner I., Sauer N. (2013). SUCROSE TRANSPORTER 5 supplies Arabidopsis embryos with biotin and affects triacylglycerol accumulation. Plant J. 73: 392–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pullen M., Clark N., Zarinkamar F., Topping J., Lindsey K. (2010). Analysis of vascular development in the hydra sterol biosynthetic mutants of Arabidopsis. PLoS One 5: e12227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan Y.L. (2007). Rapid cell expansion and cellulose synthesis regulated by plasmodesmata and sugar: insights from the single-celled cotton fibre. Funct. Plant Biol. 34: 1–10. [DOI] [PubMed] [Google Scholar]
- Ruan Y.L. (2014). Sucrose metabolism: gateway to diverse carbon use and sugar signaling. Annu. Rev. Plant Biol. 65: 33–67. [DOI] [PubMed] [Google Scholar]
- Ruan Y.L., Llewellyn D.J., Furbank R.T. (2000). Pathway and control of sucrose import into initiating cotton fibre cells. Funct. Plant Biol. 27: 795–800. [Google Scholar]
- Ruan Y.L., Llewellyn D.J., Furbank R.T. (2001). The control of single-celled cotton fiber elongation by developmentally reversible gating of plasmodesmata and coordinated expression of sucrose and K+ transporters and expansin. Plant Cell 13: 47–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan Y.L., Llewellyn D.J., Furbank R.T. (2003). Suppression of sucrose synthase gene expression represses cotton fiber cell initiation, elongation, and seed development. Plant Cell 15: 952–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan Y.L., Xu S.M., White R., Furbank R.T. (2004). Genotypic and developmental evidence for the role of plasmodesmatal regulation in cotton fiber elongation mediated by callose turnover. Plant Physiol. 136: 4104–4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrick K., Fujioka S., Takatsuto S., Stierhof Y.D., Stransky H., Yoshida S., Jürgens G. (2004). A link between sterol biosynthesis, the cell wall, and cellulose in Arabidopsis. Plant J. 38: 227–243. [DOI] [PubMed] [Google Scholar]
- Shang X., Chai Q., Zhang Q., Jiang J., Zhang T., Guo W., Ruan Y.L. (2015). Down-regulation of the cotton endo-1,4-β-glucanase gene KOR1 disrupts endosperm cellularization, delays embryo development, and reduces early seedling vigour. J. Exp. Bot. 66: 3071–3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simkin A.J., Guirimand G., Papon N., Courdavault V., Thabet I., Ginis O., Bouzid S., Giglioli-Guivarc’h N., Clastre M. (2011). Peroxisomal localisation of the final steps of the mevalonic acid pathway in planta. Planta 234: 903–914. [DOI] [PubMed] [Google Scholar]
- Sivitz A.B., Reinders A., Johnson M.E., Krentz A.D., Grof C.P.L., Perroux J.M., Ward J.M. (2007). Arabidopsis sucrose transporter AtSUC9. High-affinity transport activity, intragenic control of expression, and early flowering mutant phenotype. Plant Physiol. 143: 188–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souter M., Topping J., Pullen M., Friml J., Palme K., Hackett R., Grierson D., Lindsey K. (2002). hydra Mutants of Arabidopsis are defective in sterol profiles and auxin and ethylene signaling. Plant Cell 14: 1017–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30: 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Y., Cheung L.S., Li S., Eom J.S., Chen L.Q., Xu Y., Perry K., Frommer W.B., Feng L. (2015). Structure of a eukaryotic SWEET transporter in a homotrimeric complex. Nature 527: 259–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor C.B. (1997). Comprehending cosuppression. Plant Cell 9: 1245–1249. [Google Scholar]
- Tian G.W., et al. (2004). High-throughput fluorescent tagging of full-length Arabidopsis gene products in planta. Plant Physiol. 135: 25–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truernit E., Sauer N. (1995). The promoter of the Arabidopsis thaliana SUC2 sucrose-H+ symporter gene directs expression of beta-glucuronidase to the phloem: evidence for phloem loading and unloading by SUC2. Planta 196: 564–570. [DOI] [PubMed] [Google Scholar]
- Vatén A., et al. (2011). Callose biosynthesis regulates symplastic trafficking during root development. Dev. Cell 21: 1144–1155. [DOI] [PubMed] [Google Scholar]
- Wang L., Ruan Y.L. (2010). Unraveling mechanisms of cell expansion linking solute transport, metabolism, plasmodesmtal gating and cell wall dynamics. Plant Signal. Behav. 5: 1561–1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Li X.R., Lian H., Ni D.A., He Y.K., Chen X.Y., Ruan Y.L. (2010). Evidence that high activity of vacuolar invertase is required for cotton fiber and Arabidopsis root elongation through osmotic dependent and independent pathways, respectively. Plant Physiol. 154: 744–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendel J.F. (1989). New World tetraploid cottons contain Old World cytoplasm. Proc. Natl. Acad. Sci. USA 86: 4132–4136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendel J.F., Cronn R.C. (2003). Polyploidy and the evolutionary history of cotton. Adv. Agron. 78: 139–186. [Google Scholar]
- Wippel K., Sauer N. (2012). Arabidopsis SUC1 loads the phloem in suc2 mutants when expressed from the SUC2 promoter. J. Exp. Bot. 63: 669–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S., Wang H., Li F., Chen T., Zhang J., Jiang Y., Ding Y., Guo W., Zhang T. (2008). Enhanced Agrobacterium-mediated transformation of embryogenic calli of upland cotton via efficient selection and timely subculture of somatic embryos. Plant Mol. Biol. Report. 26: 174–185. [Google Scholar]
- Zavaliev R., Ueki S., Epel B.L., Citovsky V. (2011). Biology of callose (β-1,3-glucan) turnover at plasmodesmata. Protoplasma 248: 117–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T., et al. (2015). Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nat. Biotechnol. 33: 531–537. [DOI] [PubMed] [Google Scholar]
- Zheng B.S., Rönnberg E., Viitanen L., Salminen T.A., Lundgren K., Moritz T., Edqvist J. (2008). Arabidopsis sterol carrier protein-2 is required for normal development of seeds and seedlings. J. Exp. Bot. 59: 3485–3499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Qu H., Dibley K.E., Offler C.E., Patrick J.W. (2007). A suite of sucrose transporters expressed in coats of developing legume seeds includes novel pH-independent facilitators. Plant J. 49: 750–764. [DOI] [PubMed] [Google Scholar]