ETTIN translates changes in auxin levels into diverse transcriptional outcomes to control specific processes during Arabidopsis plant development.
Abstract
The phytohormone auxin governs crucial developmental decisions throughout the plant life cycle. Auxin signaling is effectuated by auxin response factors (ARFs) whose activity is repressed by Aux/IAA proteins under low auxin levels, but relieved from repression when cellular auxin concentrations increase. ARF3/ETTIN (ETT) is a conserved noncanonical Arabidopsis thaliana ARF that adopts an alternative auxin-sensing mode of translating auxin levels into multiple transcriptional outcomes. However, a mechanistic model for how this auxin-dependent modulation of ETT activity regulates gene expression has not yet been elucidated. Here, we take a genome-wide approach to show how ETT controls developmental processes in the Arabidopsis shoot through its auxin-sensing property. Moreover, analysis of direct ETT targets suggests that ETT functions as a central node in coordinating auxin dynamics and plant development and reveals tight feedback regulation at both the transcriptional and protein-interaction levels. Finally, we present an example to demonstrate how auxin sensitivity of ETT-protein interactions can shape the composition of downstream transcriptomes to ensure specific developmental outcomes. These results show that direct effects of auxin on protein factors, such as ETT-TF complexes, comprise an important part of auxin biology and likely contribute to the vast number of biological processes affected by this simple molecule.
INTRODUCTION
The word hormone (from the ancient Greek hormo: “that which sets in motion”) is widely used to describe mobile signaling molecules able to generate specific developmental responses at a site far from their origin of synthesis. Auxins comprise a class of plant hormones including the predominant auxin, indole-3-acetic acid (IAA), which is involved in an exceptional range of developmental processes during organ growth and differentiation (Benjamins and Scheres, 2008; Vanneste and Friml, 2009). In the nucleus, high auxin levels cause degradation of the Aux/IAA repressors, allowing auxin response factors (ARFs) to regulate transcription of their target genes in a signaling cascade that culminates in a myriad of different pathways (Vernoux et al., 2011; Rademacher et al., 2012; Calderón-Villalobos et al., 2012; Weijers and Wagner, 2016). The Arabidopsis thaliana genome encodes 23 ARFs and the N-terminal region of each ARF possesses a conserved DNA binding domain (DBD) able to mediate ARF dimerization and recognize auxin response elements (AuxREs) with the general sequence motif TGTCNN. Individual ARFs have certain degrees of specificity and preference for both sequence and motif spacing (Boer et al., 2014). In the C-terminal region, all ARFs, except ARF3, ARF13, and ARF17, contain the conserved PB1 (Phox/Bem1p) domain through which they interact with Aux/IAA proteins (Boer et al., 2014; Guilfoyle, 2015; Korasick et al., 2015; Salehin et al., 2015; Chandler, 2016).
ARF3/ETTIN (ETT) is a master regulator of development and morphogenesis of the female reproductive structure, the gynoecium (Sessions et al., 1997; Nemhauser et al., 2000; Simonini et al., 2016). A wild-type gynoecium (Figure 1A) is patterned into highly distinct tissues with specific function and identity: The basal ovary is divided into two symmetrical valves that are separated by the medial region where replum and valve margins will form. At the apex, the gynoecium develops a radially symmetric style topped with stigmatic papillae (Moubayidin and Østergaard, 2017). In ett mutants, the polarity, patterning, and identity of these different tissues are severely affected (Figure 1B), ultimately making fertilization of the ett gynoecium highly inefficient (Sessions et al., 1997).
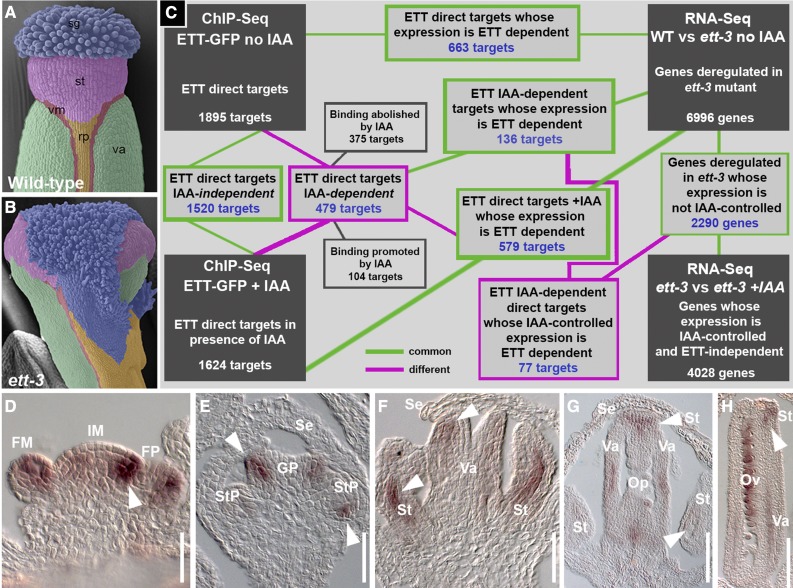
Figure 1.
Strategy of the Analyses.
(A) and (B) Scanning electron microscopy images of wild-type (A) and ett-3 (B) gynoecium apices. Tissues are false-colored: green, valves; pink, style; red, valve margin; orange, replum; blue, stigma.
(C) Schematic representation of strategy adopted to compare the data sets obtained through ChIP-seq and RNA-seq analyses.
(D) to (H) In situ hybridization with an ETT antisense probe in developing inflorescences. ETT expression can be observed in inflorescence and floral meristem (D), gynoecium and stamen primordia ([E] and [F]), and in developing ovules ([G] and [H]).
FM, floral meristem; FP, floral primordia; Gp, gynoecium primordia, IM, inflorescence meristem; Op, ovule primordia; Ov, ovules; rp, replum; SE, sepal; st, style; StP, stamen primordia; sg, stigma; va, valves; vm, valve margin. Bars = 100 µm.
In agreement with its lack of a PB1 domain, ETT was recently shown to mediate auxin signaling via an alternative Aux/IAA-independent pathway (Simonini et al., 2016). This signaling mechanism involves auxin (specifically IAA) modulating the interaction between ETT and its protein partners, thereby facilitating spatial and temporal changes in transcriptional responses during organ morphogenesis (Simonini et al., 2016).
Here, we take a genomic approach toward understanding how this ETT-unique IAA-sensing mechanism functions during plant development to modulate the expression of target genes. We show that ETT directly regulates several hundred genes but that a subset of these targets is regulated by ETT in an IAA-sensitive manner. Among the genes in this subset are some that also appear as targets of ETT-interacting transcription factors (TFs). A reverse genetics approach verified the biological relevance of the regulation of such targets by an ETT-TF module to control developmental processes. While the genomic data sets allowed us to test specific target genes, this genome-wide analysis demonstrates the significant involvement of IAA-sensitive ETT-controlled gene expression in Arabidopsis plant growth and development.
RESULTS
ETT Directly Controls Expression of Specific Gene Classes
As a first approach, we combined chromatin immunoprecipitation coupled with deep sequencing (ChIP-seq) of a pETT:ETT-GFP line (in ett-3; Simonini et al., 2016) normalized against a wild-type background (Supplemental Data Set 1) with transcriptome analysis (RNA-seq) of the ett-3 loss-of-function mutant versus the wild type (Supplemental Data Set 2). To explain the rationale of the analyses described in this work, Figure 1C graphically summarizes the experimental strategy adopted and the different data set subgroups that emerged from the comparison between the main data sets.
Both ChIP-seq and RNA-seq analyses were conducted by pooling in triplicate tissue from three independent harvests of both pETT:ETT-GFP line and wild-type plants to compose a biological triplicate for each genotype (i.e., nine tissue samples in total). Whole-inflorescence tissues, excluding mature flowers and developing siliques, were collected for both ChIP-seq and RNA-seq analyses. Given the material considered and the known ETT expression pattern in the Arabidopsis shoot (Sessions et al., 1997; Nemhauser et al., 2000; Simonini et al., 2016), this study was designed to identify ETT targets involved in aspects of inflorescence and floral meristem (Figure 1D), gynoecium (Figures 1E to 1H), stamen (Figures 1E and 1F), and ovule (Figures 1G and 1H) development and differentiation.
Out of the 1895 loci identified as being directly bound by ETT (P < 0.01), 663 showed differential expression of the cognate genes in the ett-3 mutant background (Figure 1C). This set is therefore expected to contain the genes whose expression is directly controlled by ETT during development of shoot structures. In agreement with published roles of ETT during plant development, the 663 ETT-target candidates were classified under Gene Ontology (GO) term clusters involved in processes such as transcriptional regulation, hormone dynamics, and plant organ development (in particular floral organ development) (Supplemental Data Set 1). Families of transcriptional regulators included homeobox, WRKY DNA binding proteins, basic HELIX-LOOP-HELIX (bHLH), MADS box, and TEOSINTE BRANCHED1/CYCLOIDEA/PCF.
Enrichment was also observed for factors involved in the signaling for responses to several plant hormones, predominantly auxin, ethylene, jasmonic acid (JA), and cytokinin (Table 1; Supplemental Data Set 1). Notably, while ETT appears to repress most of its auxin signaling-related targets (targets upregulated in ett-3), ETT emerged as a positive regulator of most of its ethylene- and JA-related targets (reduced in ett-3). These data agree with the proposed role of auxin in promoting both ethylene and JA synthesis (Muday et al., 2012; Chandler, 2009).
Table 1. ETT Targets Differentially Expressed in the ett-3 Mutant.
| Process | Gene | Name | Expression Level in ett-3 |
|---|---|---|---|
| Auxin | |||
| Synthesis/metabolism | At3g02875 | ILR1 | −0.332787 |
| At4g03400 | GH3.10 | −0.30953 | |
| At4g27260 | GH3.5 | −0.207892 | |
| At5g11320 | YUC4 | 0.739941 | |
| At5g55250 | IAMT1 | −0.434828 | |
| Transport | At1g73590 | PIN1 | −0.316707 |
| At1g70940 | PIN3 | 0.519409 | |
| At1g23080 | PIN7 | 0.319203 | |
| At2g38120 | AUX1 | −0.29821 | |
| At3g28860 | ABCB19 | −0.227607 | |
| At5g01240 | LAX1 | −0.729782 | |
| Signaling | At1g77850 | ARF17 | −0.360739 |
| At4g30080 | ARF16 | 0.295047 | |
| At2g33310 | IAA13 | 0.266635 | |
| At3g04630 | IAA16 | 0.21003 | |
| At3g16500 | IAA26 | 0.385515 | |
| At3g23030 | IAA2 | −0.192655 | |
| At3g23050 | IAA7 | 0.427759 | |
| At5g43700 | IAA4 | 0.309409 | |
| Response | At1g21210 | SAUR6 | 0.560865 |
| At4g34760 | SAUR50 | 0.69973 | |
| At1g75580 | SAUR51 | 0.23934 | |
| At4g38860 | SAUR16 | 0.331208 | |
| Ethylene | |||
| Synthesis/metabolism | At1g73500 | MKK9 | −0.872741 |
| At3g59060 | PIF5 | −0.252658 | |
| Signaling | At1g28360 | ERF12 | −0.787087 |
| At1g28730 | ERF11 | −0.780156 | |
| At1g43160 | RAP2.6 | −1.99865 | |
| At1g44830 | ERF14 | 1.03618 | |
| At2g27050 | EIL1 | −0.301253 | |
| At3g14230 | RAP2.2 | −0.434167 | |
| At4g17490 | ERF6 | −0.967468 | |
| At4g17500 | ERF-1 | −0.87404 | |
| At4g34410 | ERF109 | −1.72887 | |
| At5g07580 | ERF106 | −0.672901 | |
| At5g47220 | ERF2 | −1.2461 | |
| Atg547230 | ERF5 | −0.491374 | |
| Jasmonic acid | |||
| Synthesis/metabolism | At1g17420 | LOX3 | −0.900455 |
| At1g72520 | LOX4 | −0.900221 | |
| At3g25770 | AOC2 | −1.65739 | |
| At3g25780 | AOC3 | −1.05354 | |
| Signaling | At1g17380 | JAZ5 | −0.492825 |
| At1g19180 | JAZ1 | −0.22666 | |
| At3g27810 | MYB21 | 3.10916 | |
| At5g67300 | MYB44 | −0.425472 | |
Expression level indicated is the log2 value compared to the wild type.
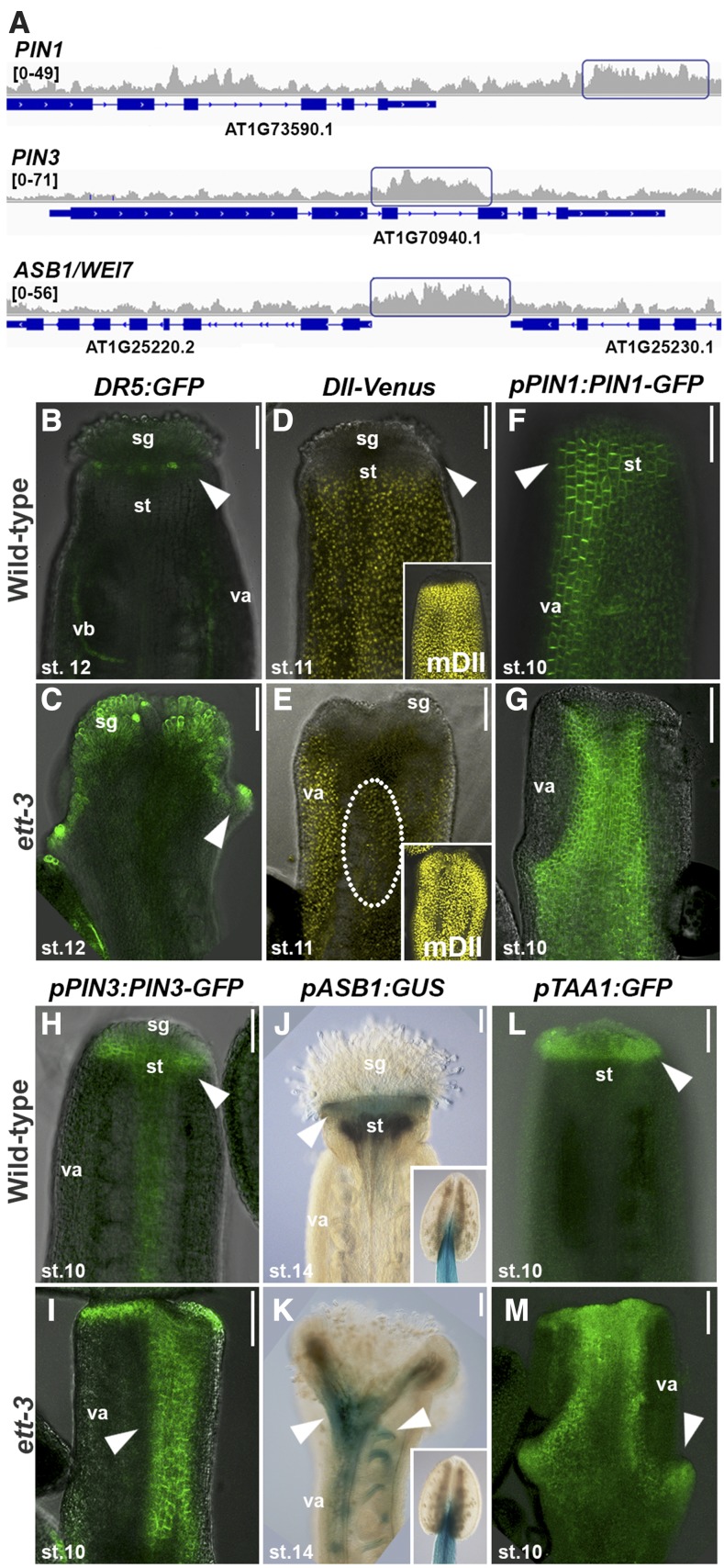
While ETT activity is affected by auxin, our genome-wide analysis also implicates ETT in regulating auxin dynamics itself. Notably, auxin efflux as well as auxin influx transporters were among the ETT targets (PIN-FORMED1/3/7, AUXIN RESISTANT1, LIKE AUXIN RESISTANT1, ARABIDOPSIS THALIANA ATP BINDING CASSETTE B19; Table 1, Figure 2A; Supplemental Figure 1).
Figure 2.
ETT Regulates Auxin Dynamics in Developing Gynoecium.
(A) Representative raw ChIP-seq peaks (only one replicate shown) for PIN1, PIN3, and ASB1/WEI7 target genes and their expression levels in the ett-3 mutant (on the right). Blue rectangles mark the peak regions. Each gene is represented below with blue bars (exons) and lines (introns).
(B) and (C) Confocal images of DR5:GFP in wild type (B) and ett-3 mutant (C) at stage 12 of gynoecium development.
(D) and (E) Confocal images of DII-VENUS and mDII-VENUS (insets) in the wild type (D) and ett-3 mutant (E) at stage 11 of gynoecium development.
(F) and (G) Confocal analyses of PIN1-GFP fusion protein in wild-type (F) and ett-3 (G) stage-10 gynoecia.
(H) and (I) Confocal analyses of PIN3-GFP fusion protein in wild-type (H) and ett-3 (I) stage-10 gynoecia.
(J) and (K) GUS staining of pASB1:GUS marker line in wild-type (J) and ett-3 (K) stage-14 gynoecia and anthers (inset).
(L) and (M) Confocal analyses of TAA1-GFP fusion protein in wild-type (L) and ett-3 (M) stage-10 gynoecia.
st, style; sg, stigma; va, valves; vb, vascular bundle. Bars = 100 µm.
These observations are in agreement with ett mutants having gynoecial defects that are reminiscent of those observed in plants carrying mutations in for example auxin transport and auxin biosynthesis genes (Benjamins et al., 2001; Cheng et al., 2006). Indeed, reporter lines of auxin signaling such as DR5:GFP (Benková et al., 2003) and DII-VENUS (Brunoud et al., 2012) showed abnormal distribution in ett-3 mutant gynoecia compared with the wild type, thus demonstrating that auxin dynamics is affected in ett mutant gynoecia (Figures 2B to 2E). Moreover, confocal analysis of GFP marker lines of the ETT-direct targets PIN1 (Benková et al., 2003) and PIN3 (Zádníková et al., 2010) revealed abnormal expression for both efflux carrier genes in the ett-3 mutant background during carpel development (Figures 2F to 2I). Auxin biosynthesis also appears to be a target of ETT control with direct targets including YUCCA4 (YUC4; Cheng et al., 2006), ANTHRANILATE SYNTHASE BETA SUBUNIT1/WEI7 (ASB1; Stepanova et al., 2005), and TRYPTOPHAN AMINOTRANSFERASE OF ARABIDOPSIS1/WEI8 (TAA1; Stepanova et al., 2008) (Figure 2A, Table 1; Supplemental Data Set 1 and Supplemental Figure 1). Compared with the wild type, expression of both ASB1 and TAA1 expanded throughout the carpel body at all developmental stages (stages according to Smyth et al., 1990) of the ett-3 mutant (Figures 2J to 2M).
Together, these data strongly suggest that ETT regulates gynoecium development at least in part by modulating auxin dynamics at all levels and by acting as a node in connecting hormonal pathways and floral tissues identity.
ETT Regulates a Subset of Its Targets in an IAA-Sensitive Manner
To explore the role of ETT in translating auxin cellular levels into diverse transcriptional outcomes, inflorescence tissue from the pETT:ETT-GFP line, wild type, and the ett-3 mutant was treated with IAA. ChIP-seq and RNA-seq were performed in triplicate for each genotype, and each individual sample derived from the pooling of five independent harvests to minimize the effects of any variability during the IAA treatment. Subsequently, the identified direct targets and transcriptomes were analyzed and compared (Figure 3A; Supplemental Data Sets 1 and 2). A total of 1624 loci were bound by ETT in ChIP-seq +IAA analysis (P < 0.01), and 579 showed differential expression of the cognate genes in the ett-3 mutant background (Supplemental Data Set 1).
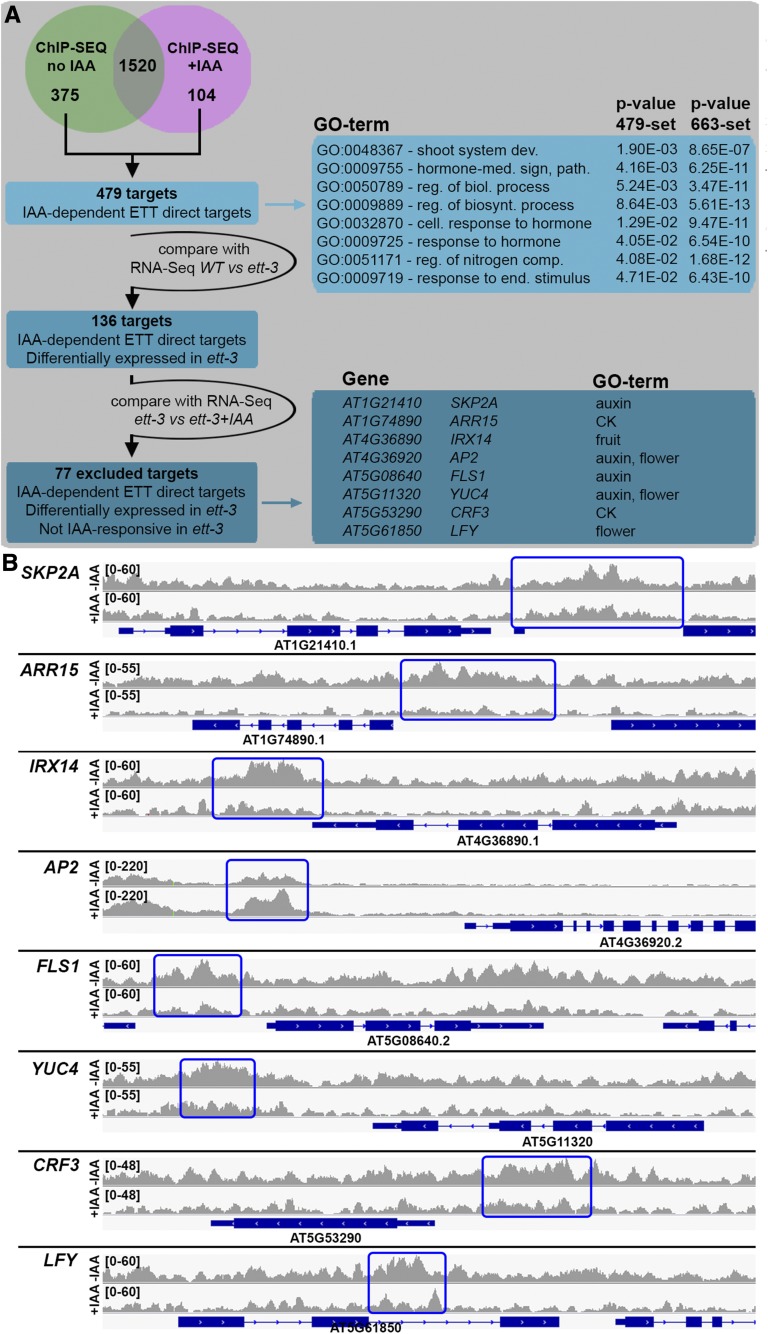
Figure 3.
ETT Regulates a Subset of Target Genes in an Auxin-Dependent Manner.
(A) Venn diagram of ChIP-seq ±IAA. Schematic representation of the strategy adopted to isolate ETT targets for which binding by ETT is IAA dependent and GO terms corresponding to the isolated set of targets.
(B) Representative raw ChIP-seq peaks (one replicate only) for SKP2A, ARR15, IRX14, AP2, FLS1, YUC4, CRF3, and LFY target genes. Blue rectangles mark the peak regions. Each gene is represented below with blue bars (exons) and lines (introns).
The overlap of ChIP-seq data sets from experiments conducted in the absence and presence of IAA revealed that 1520 loci (P < 0.01) are directly bound by ETT independently of IAA treatment (Figure 3A). Therefore, association of ETT with these genomic loci and regulation of their cognate genes by ETT appear to be auxin independent. These genes might constitute the core ETT targets in those tissues where auxin normally does not accumulate, such as the growing valves of the gynoecium.
Subtraction of these common 1520 targets from the initial 1895 of the ChIP-seq analyses in the absence of IAA revealed that 375 loci were specifically bound by ETT only under low auxin conditions (Figure 3A), thus suggesting that the increase in auxin levels negatively affected ETT association with these genomic loci. Similarly, 104 specific targets emerged from the subtraction of the 1520 common targets from the 1624 of the ChIP-seq analyses in presence of IAA (Figure 3A), thus suggesting that ETT recruitment to these genomic loci is stimulated by auxin. In summary, a total of 479 loci are recognized and bound by ETT in an IAA-dependent manner: 375 associations are destabilized and 104 are stimulated by high IAA levels. A comparison of these 479 IAA-dependent ETT direct targets with the transcriptome profile of ett-3 versus the wild-type gene list highlighted an overlap of 136 genes (Figure 3A). This set represents genes whose binding by ETT is affected by IAA and whose expression is controlled by ETT.
Notably, the order of GO categories in terms of significance was different between the 136 ETT targets for which binding was affected by IAA and the 663 targets that emerged from the comparison between ChIP-seq and RNA-seq conducted in the absence of IAA treatment (Supplemental Data Set 1). Genes belonging to GO categories such as hormone-mediated signaling pathway, cellular response to hormone stimulus, and regulation of biological processes were higher up the list among the set of 136 IAA-dependent genes than they were for the 663-gene set (Supplemental Data Set 1; Figure 3A). IAA may therefore preferentially affect how ETT regulates targets involved in processes of organ patterning and tissue specification.
To identify which genes among the 136 IAA-dependent ETT direct targets require ETT to mediate their responsiveness to increased IAA levels, the 136-gene set was compared with the list of genes whose expression is affected by IAA in the ett-3 mutant background (ett-3 +IAA versus ett-3 transcriptome; Supplemental Data Set 2). Of the 136 genes, 59 were differentially expressed in IAA-treated ett-3 mutant inflorescences, suggesting that their responsiveness to auxin is independent on ETT. By contrast, the expression of the remaining 77 genes was unchanged (Table 2), thus characterizing these targets as bound and regulated by ETT in an IAA-sensitive manner (Figure 3A). In other words, the IAA-sensitive interaction of ETT with these genes affected their expression during inflorescence development. These 77 targets encode proteins of diverse classes, such as transcription factors and enzymes involved in lipid metabolism (TRANSLOCASE11), vesicle transport (PRENYLATED RAB ACCEPTOR1.F2), oxidative stress (CATALASE3, FLAVONOL SYNTHASE1), and posttranslational modification (PRESEQUENCE PROTEASE2).
Table 2. IAA-Dependent ETT Direct Targets.
| Gene | Name | In ett-3 | Gene | Name | In ett-3 |
|---|---|---|---|---|---|
| AT1G05420 | OFP12 | −1.9836 | AT3G16370 | GDSL esterase | 0.217299 |
| AT1G15080 | LPP2 | −0.348794 | AT3G20270 | LBR-2 | 0.381359 |
| AT1G15370 | SNARE-like | 0.197164 | AT3G28150 | AXY4L | 0.737246 |
| AT1G16520 | Unknown protein | −0.133696 | AT3G55960 | Dehalogenase-like | −0.159613 |
| AT1G20620 | CAT3 | −0.477492 | AT3G58570 | Triphosphate hydrol. | −0.254681 |
| AT1G20970 | Unknown protein | 0.208903 | AT3G61850 | DAG1 | 0.395935 |
| AT1G21410 | SKP2A | −0.216251 | AT4G16590 | CSLA01 | 0.280685 |
| AT1G32190 | Hydrolases | −0.377913 | AT4G17430 | O-fucosyltransfer. | 0.184038 |
| AT1G49630 | PREP2 | −0.534247 | AT4G17695 | KAN3 | 0.789874 |
| AT1G52410 | TSA1 | −0.292903 | AT4G18270 | TRANS11 | 0.153913 |
| AT1G53887 | Unknown protein | −0.60934 | AT4G18280 | Glycine-rich cell wall | −0.533175 |
| AT1G55190 | PRA1.F2 | −0.230444 | AT4G24090 | Unknown protein | −0.212325 |
| AT1G55260 | LTPG6 | 0.435487 | AT4G30260 | YIP4B | −0.330087 |
| AT1G64620 | DOF1.8 | 0.257194 | AT4G30800 | Nucleic acid binding | 0.132119 |
| AT1G65910 | NAC028 | 0.294149 | AT4G31620 | REM36 | 0.3657 |
| AT1G67420 | Exopeptidases | 0.186317 | AT4G31890 | ARM repeat | 0.314066 |
| AT1G68070 | Zinc finger C3HC4 | −0.275877 | AT4G36030 | ARO3 | −0.572796 |
| AT1G69420 | DHHC-type zinc finger | 0.328207 | AT4G36160 | NAC076 | 0.342157 |
| AT1G71870 | BIGE1A | −1.49108 | AT4G36890 | IRX14 | 0.371547 |
| AT1G74890 | ARR15 | 0.555819 | AT4G36920 | AP2 | 0.369832 |
| AT1G75690 | LQY1 | −0.210166 | AT5G02260 | EXPA9 | 0.518721 |
| AT1G75810 | Unknown protein | −0.800871 | AT5G07800 | Flavin binding monox. | 0.259713 |
| AT1G76170 | 2-Thiocytidine tRNA | 0.28612 | AT5G08250 | Cytochrome P450 superfamily | −0.455767 |
| AT1G76580 | Squamosa binding | 0.51678 | AT5G08640 | FLS1 | 0.364151 |
| AT1G79160 | Unknown protein | −0.247587 | AT5G11320 | YUC4 | 0.739941 |
| AT1G79360 | OCT2 | −0.300869 | AT5G15230 | GASA4 | 0.44449 |
| AT1G79420 | Unknown function | −0.685551 | AT5G25820 | Exostosin family | −0.461257 |
| AT1G80720 | Glycoprotein | 0.193639 | AT5G27220 | Frigida-like protein | 0.966952 |
| AT2G28080 | UDP-glycosyltransf. | 0.16851 | AT5G38510 | Serine protease | 0.233273 |
| AT2G30890 | Ferric reductase | 0.250341 | AT5G40040 | 60S acidic ribosomal | −0.315939 |
| AT2G32010 | CVL1 | 0.25333 | AT5G46690 | BHLH071 | 0.343774 |
| AT2G33855 | Unknown protein | −0.704788 | AT5G53290 | CRF3 | −0.491374 |
| AT2G39020 | NATA2 | −0.395927 | AT5G57130 | SMXL5 | 0.320535 |
| AT2G44900 | ARABIDILLO1 | 0.301096 | AT5G57550 | XTH25 | −1.06424 |
| AT2G46970 | PIL1 | 1.3596 | AT5G59350 | Unknown protein | 0.417153 |
| AT2G47485 | Unknown protein | −0.713701 | AT5G61850 | LFY | −0.765972 |
| AT3G03140 | Tudor/PWWP/MBT | 0.1616 | AT5G67170 | SEC-C motif- protein | 0.292576 |
| AT3G07450 | Bifunctional inhibitor | −1.90179 | AT5G67360 | SBT1.7 | −0.384351 |
| AT3G15220 | Protein kinase | 0.147785 |
Expression level indicated is the log2 value compared to the wild type.
GO analysis of enriched terms for this relatively short list of ETT-dependent and IAA-affected targets failed to identify common significantly enriched descriptive words. With the GO-enriched terms obtained for the 663 differentially expressed ETT direct targets in mind (Supplemental Data Set 1), we instead manually isolated a small subgroup of targets that contained at least one word among the “auxin,” “ethylene,” “jasmonic acid,” “cytokinin,” “flower,” and “fruit” categories. Following these criteria, a subset of eight genes was identified that encoded proteins controlling transcription (APETALA2 [AP2], LEAFY [LFY], and CYTOKININ RESPONSIVE FACTOR3 [CRF3]), hormone dynamics (ARABIDOPSIS RESPONSE REGULATOR15 [ARR15], YUC4 and CRF3), protein stability (F-box protein SKP2A), and synthesis of cell wall components (IRREGULAR XYLEM14 [IRX14] and FLS1) (Figure 3B; Supplemental Figure 2). Since these genes are known to function in the specification of floral organs (Kunst et al., 1989; Weigel et al., 1992; Rashotte et al., 2006; Zhao et al., 2010), it appears that IAA modulation of ETT activities is particularly important in spatio-temporal regulation of floral tissue identity and differentiation.
ETT Functions Both as Transcriptional Repressor and Activator
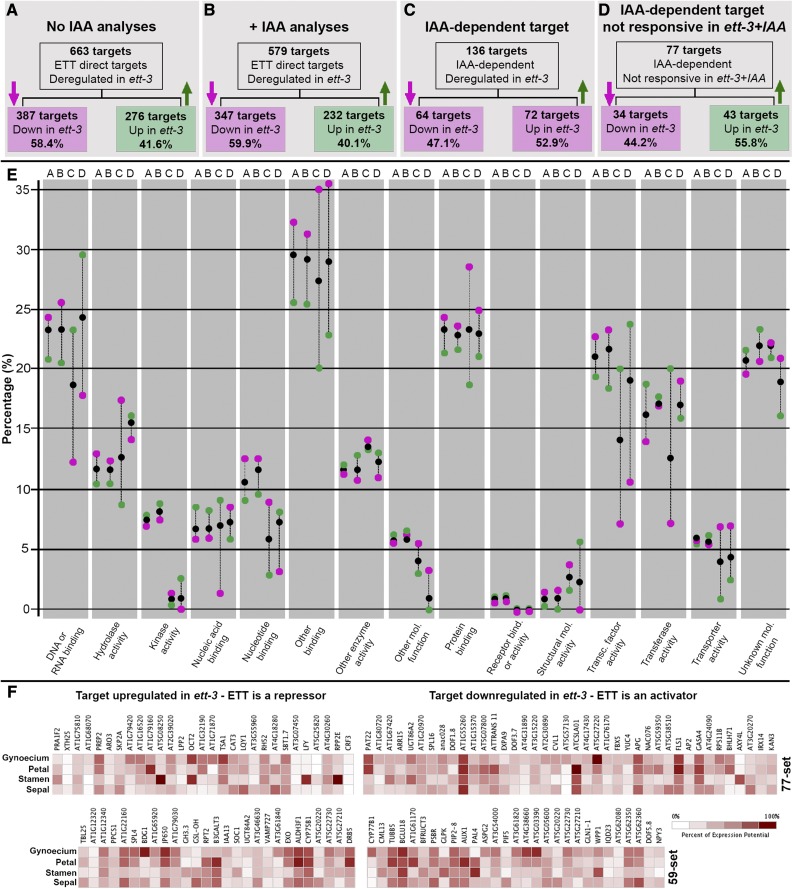
In 2007, Guilfoyle and Hagen (2007) proposed that ETT acts predominantly as a repressing ARF, considering the amino acid composition of its middle region. However, in our transcriptome analyses, ETT appeared to function both as a transcriptional repressor and an activator, as subgroups of downregulated and upregulated targets were represented in the sub-data sets. The proportion in percentage of downregulated-to-upregulated genes (down:up) in the 663 and 579 sets (Figure 1C), was 58.4/41.6 (ratio >1) and 59.9/40.1 (ratio >1), respectively (Figures 4A and 4B). Interestingly, the down:up ratio was inverted when looking at the IAA-dependent target data sets being 47.1/52.9% (ratio <1) and 44.2/55.8% (ratio <1), respectively, in the 136 and 77 sets (Figures 4C and 4D). Although speculative, these data may reflect a preference for ETT as a repressor/activator that is influenced by auxin levels.
Figure 4.
ETT Is Both a Transcriptional Repressor and Activator.
(A) to (D) Schematic representation of subdivision in downregulated (magenta arrow) and upregulated (green arrow) targets for each of the data sets emerging from the comparison of ChIP-seq and RNA-seq: 663 set (A), 579 set (B), 136 set (C), and 77 set (D).
(E) Schematic representation of GO term frequency associated with targets belonging to the upregulated and downregulated subgroups in the ett-3 mutant background. On the x axis are 15 GO term clusters of molecular function. On the y axis is the percentage of genes associated with the corresponding GO term cluster for each data set in (A) to (D). Black dots indicate the whole set, magenta dots indicate the subgroups of downregulated genes, and green dots indicate the subgroup of upregulated genes. A-B-C-D letters on the top of each GO cluster represent the data sets listed in (A) to (D).
(F) Heat map generated with Genevestigator showing expression of the 77-set and 59-set targets (which combined create the 136-set targets). Each set is divided into upregulated (left) and downregulated (right) targets. Color intensity reflects the expression level.
To understand if this difference in ETT transcriptional activity also reflected a preference of target category, the subset of downregulated and upregulated genes in each of the 663, 579, 136, and 77 sets (Figures 4A to 4D) was grouped according to 15 clusters of molecular function GO terms (The Arabidopsis Information Resource [TAIR]; www.arabidopsis.org) (x axis, Figure 4E) and the frequency of genes appearing in each cluster measured as percentage (y axis, Figure 4E). For GO clusters “Protein binding,” “Receptor binding/activity,” and “Transporter activity,” the down:up ratio was >1 across all four data sets, suggesting that ETT is predominantly an activator of genes belonging to these clusters and that its activity is only marginally influenced by an increase in auxin levels (Figure 4E). By contrast, particularly for GO clusters “DNA-RNA binding,” “Transcription factor activity,” and “Nucleic acid binding,” the down:up ratio was inverted from >1 of the 663 and 579 sets to <1 for the 136- and 77-IAA-dependent target data set (Figure 4E), suggesting that, following the increase in auxin levels, ETT preferentially acts as a repressor of genes encoding transcription-related factors.
A coexpression analysis of the 59- and 77-gene sets (composing the 136 targets previously described; Figure 3A) using floral organ-specific transcriptome data sets (gynoecium, stamen, petal, and sepal) publicly available at Genevestigator (genevestigator.com/gv/; experiment ID AT-00089, developmental baseline III from the Weigel Lab) confirmed the expression of the majority of the targets in floral tissues (Figure 4F). Although no particular trend could be observed comparing the expression of upregulated and downregulated targets in both the 59 and 77 sets (Figure 4F), genes that were upregulated in the ett-3 mutant (hence repressed by ETT in the wild type) showed more specific expression to one floral domain, particularly the stamen. It is therefore possible that ETT functions in excluding the expression of these targets from the gynoecium. Overall, the coexpression analysis supports an intricate relationship between ETT, auxin, and ETT-protein partners in guiding tissue differentiation and further suggests that organ identity is achieved by multiple layers of genetic regulation.
The IAA Sensitivity of ETT-TF Interactions Controls Target Gene Expression
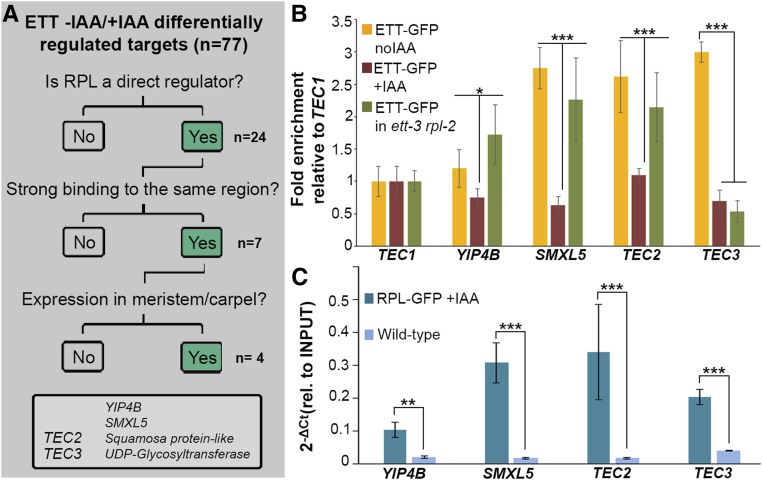
The reduction in ETT binding to a certain set of targets is in accordance with the model previously proposed, suggesting that IAA induces ETT to dissociate from its protein partner, thereby affecting its interaction with a specific genomic region (Simonini et al., 2016). ChIP-seq and transcriptome data sets for a number of ETT partners are already available in the public domain (Bencivenga et al., 2016; Santuari et al., 2016) and comparative studies are therefore possible to identify common targets and unravel modes of regulation. Here, we compared the list of 77 target genes bound and regulated by ETT in an IAA-dependent manner with the target gene data set of the ETT-interacting REPLUMLESS (RPL) protein (Bencivenga et al., 2016) (Figure 5A). The RPL gene encodes a homeobox transcription factor that was chosen because it, like ETT, has a prominent role in reproductive tissue development (Roeder et al., 2003). Out of a list of 24 targets shared between ETT and RPL, we identified seven where strong binding was detected in the same region (Figure 5A; Supplemental Data Set 1). Out of these seven, we decided to focus on four genes (Figure 5A), which according to the Bio Analytical Resource for Plant Biology (BAR; http://bar.utoronto.ca/) are expressed in meristematic and carpeloid tissues where both ETT and RPL are expressed (Sessions et al., 1997; Roeder et al., 2003; Bao et al., 2004; Bencivenga et al., 2016; Simonini et al., 2016).
Figure 5.
ETT Recruitment to the TEC3 Target Gene Locus Requires the Protein Partner RPL.
(A) Strategy adopted to search for targets shared by RPL and ETT that are differentially regulated ±IAA. Uncharacterized isolated genes are named TEC.
(B) ChIP analyses of pETT:ETT-GFP in untreated (yellow bars), IAA-treated (dark red bars), and ett-3 rpl-2 background (green bars) confirming dynamic changes in binding at the TEC2, TEC3, YIP4B, and SMXL5 loci after IAA treatment. Enrichment is represented as fold enrichment relative to the internal control TEC1 (set to 1).
(C) ChIP analyses of pRPL:RPL-GFP marker line and wild type as negative control.
Error bars indicate sd. Statistical analyses were conducted with one-way ANOVA (Supplemental File 1). *P value < 0.05; ***P value < 0.001.
The set of four ETT-RPL IAA-dependent common target genes included YPT/RAB GTPASE INTERACTING PROTEIN 4B (YIP4B) and SMAX1-LIKE5 (SMXL5), which are involved in secretion of cell wall polysaccharides and strigolactone signaling (Gendre et al., 2013; Wallner et al., 2017), respectively. In addition, the set included two undescribed genes: At1g76580, annotated as Squamosa binding-like protein, and At2g28080, annotated as UDP-glycosyltransferase. To further characterize the regulation of these genes by ETT and RPL, we included an IAA-independent internal control. To this end, we used the ETT target bHLH094 due to its consistent and reliable binding in both –IAA/+IAA conditions. For simplicity, all genes not previously characterized are referred to as TARGETS UNDER ETTIN CONTROL (TEC): TEC1/bHLH094, TEC2/At1g76580, and TEC3/At2g28080 (Figure 5A).
TEC2, TEC3, YIP4B, and SMLX5 targets all showed a significantly reduced signal in the ETT ChIP-seq data set upon IAA treatment (Supplemental Data Set 1), which was further confirmed by individual ChIP analyses using the pETT:ETT-GFP line (Figure 5B; Supplemental Figure 3A). The auxin-dependent dynamic binding to TEC2, TEC3, YIP4B, and SMLX5 was specific to ETT, since the same IAA treatment did not affect RPL recruitment at any of the targets tested (Figure 5B).
Subsequently, we questioned whether ETT was still capable of binding the TEC2, TEC3, YIP4B, and SMLX5 targets in the absence of RPL having established that these four targets are also bound by RPL (Figure 5C). For this purpose, the pETT:ETT-GFP construct was introduced into the ett-3 rpl-2 background. The resulting plants exhibited the rpl-2 phenotype and full complementation of the ett-3 defects by the pETT:ETT-GFP constructs, thus allowing assessment of the ETT-GFP binding in the rpl-2 mutant background.
While consistent enrichment could be detected for TEC2, YIP4B, and SMLX5, binding to the TEC3 region was significantly diminished in the ett-3 rpl-2 double mutant, to a similar level as in the ETT-GFP +IAA analysis (Figure 5B; Supplemental Figure 3B). These data suggest that RPL facilitates recruitment of ETT to the TEC3 locus and support a mechanism involving auxin-induced dynamic modulation of the ETT-partner dimerization status. Moreover, the ability of ETT to bind TEC2, YIP4B, and SMXL5 loci even in absence of RPL suggests the involvement of several ETT tissue-specific interactors and, therefore, the existence of multiple ETT-containing dimer/complexes, which might play role in different developmental stages of inflorescence tissue growth and differentiation.
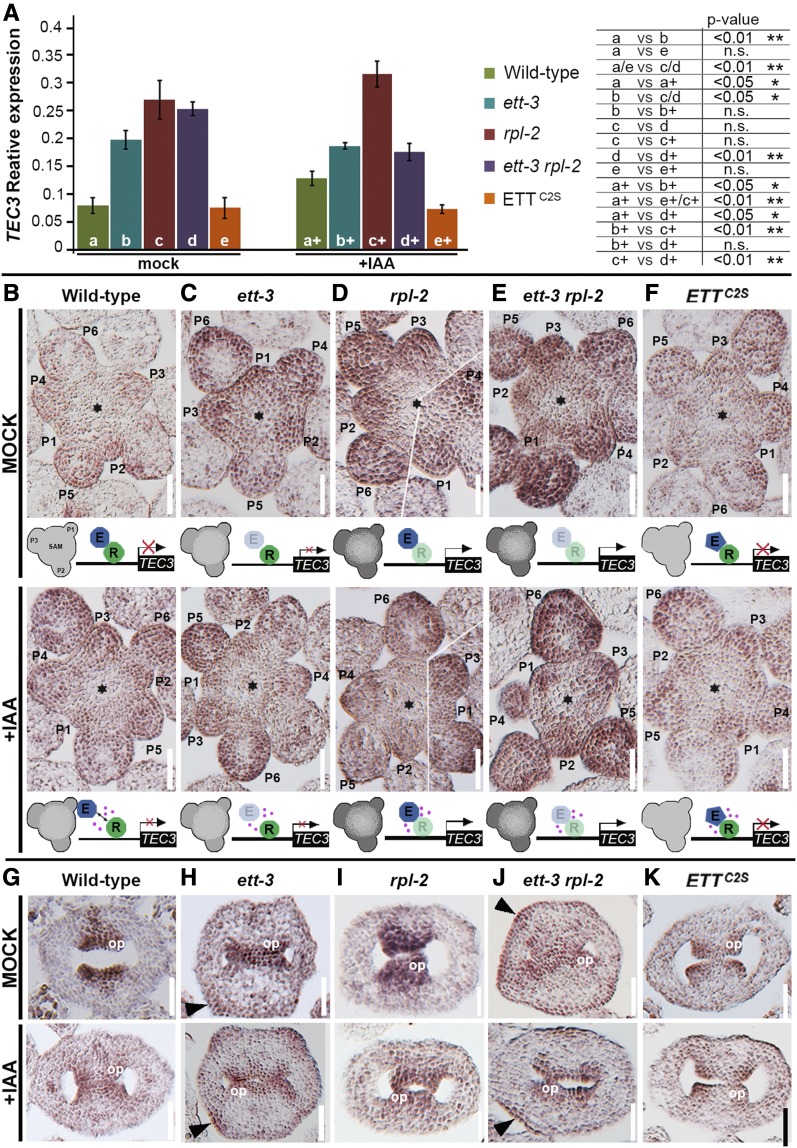
To test if the ETT-RPL interaction affects spatiotemporal control of TEC3 transcription, we used real-time PCR and in situ hybridization to compare TEC3 expression between mock- and IAA-treated wild type, ett-3, rpl-2, rpl-2 ett-3, and ETTC2S backgrounds (Figure 6). The ETTC2S line expresses an IAA-insensitive ETT variant, which is unable to mediate IAA-sensitive dissociation from its partners, in the ett-3 background (Simonini et al., 2016). TEC3 transcript was detected in inflorescence and floral meristems and in developing ovules (Figures 6B and 6G). In the ett-3 mutant, the TEC3 signal was more persistent in floral meristem tissues and in gynoecium epidermis, which is in agreement with a repressive effect of ETT on TEC3 expression (Figures 6A, 6C, and 6H). Furthermore, in agreement with RPL-dependent ETT recruitment to the TEC3 locus, comparable strong TEC3 expression was observed between rpl-2 and ett-3 rpl-2 (Figures 6A, 6D, and 6E), thus supporting a cooperative function for ETT and RPL in gene transcription. Finally, IAA treatment increased TEC3 expression in wild-type plants but not in the ETTC2S IAA-insensitive line (Figures 6A, 6B, and 6F), suggesting that TEC3 is IAA inducible and that IAA perception by ETT is necessary to achieve correct TEC3 transcriptional regulation. It is worth noting that the IAA-dependent combinatorial effect of ETT and RPL on TEC3 expression was not observed in developing ovules (Figures 6G to 6K), suggesting that a different regulatory route regulates TEC3 in this context.
Figure 6.
ETT Recruitment to TEC3 Locus by RPL Regulates TEC3 Expression.
(A) Expression analyses by real-time PCR of TEC3 in wild-type, ett-3, rpl-2, ett-3 rpl-2, and ETTC2S untreated (left, samples labeled a/e) and IAA-treated (right, samples labeled a+/e+) inflorescences and the comparison between the corresponding values (table). Error bars indicate sd. Statistical analyses were conducted with one-way ANOVA.
(B) to (F) In situ hybridization with a TEC3 antisense probe in wild-type, ett-3, rpl-2, ett-3 rpl-2, and ETTC2S untreated (upper row) and IAA-treated (lower row) shoot apical meristems. Stars indicate the center of the inflorescence meristems, and primordia (P1-6) are numbered from the youngest (1) to the oldest (6). Images in 3D result from the joining of two consecutive sections. A schematic representation of TEC3 expression is illustrated below each genotype. TEC3 signal intensity is shown with different shades of gray: light gray means low expression, and dark gray means strong expression. Blue circle with “E” represents ETT, and green circle with “R” represents RPL.
(G) to (K) TEC3 in situ hybridization in ovule primordia of wild-type, ett-3, rpl-2, ett-3 rpl-2, and ETTC2S untreated (upper row) and IAA-treated (lower row) inflorescences. Black arrowheads point to ectopic expression of TEC3 expanded to the epidermis of young ett-3 and ett-3 rpl-2 gynoecia.
pl, placenta; op, ovule primordia, ov, ovule; se, septum. Bars = 50 µm.
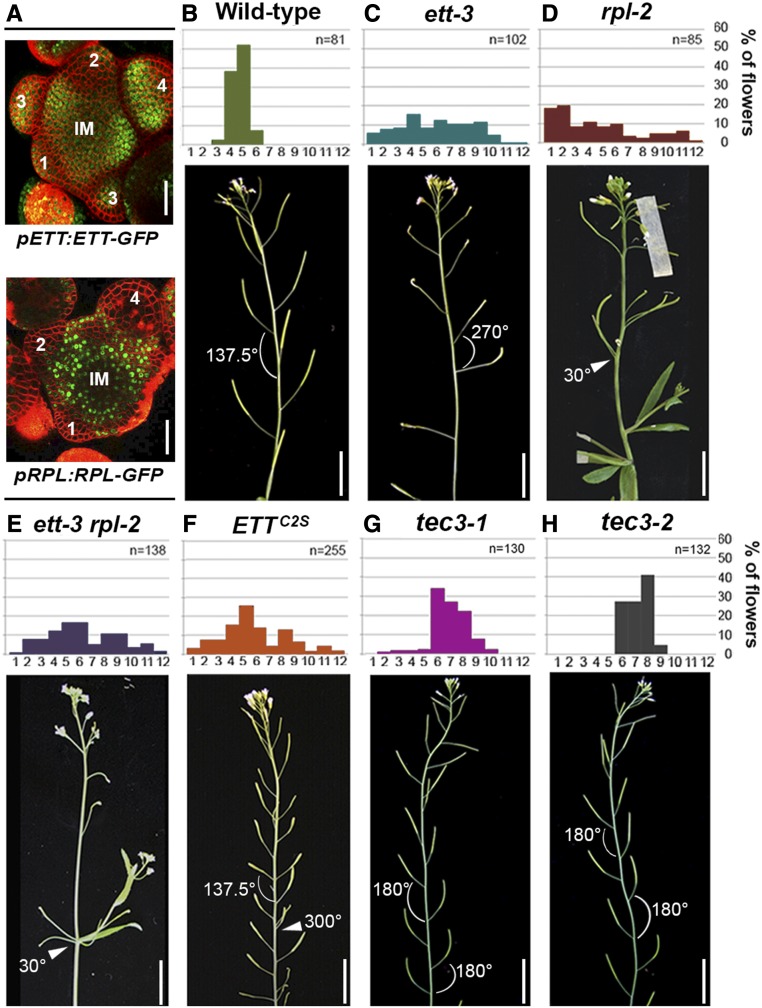
ETT expression in the meristem followed a phyllotactic pattern coinciding with the region where new floral primordia were emerging (Figure 7A) and overlapped with previously reported RPL expression in this region (Figure 7A; Bencivenga et al., 2016). Interestingly ett-3, rpl-2, ett-3 rpl-2, and ETTC2S mutant plants display dramatically altered phyllotaxis (Byrne et al., 2003) (Figure 7B to 7F). Moreover, in two independent TEC3 mutant alleles (named TEC3-1 and TEC3-2), the angle with which primordia emerged deviated from the canonical 137.5° exhibited by wild-type plants, with consecutive flowers appearing at angles between 150° and 210° (Figures 7G and 7H). These results suggest that ETT and RPL cooperate in an IAA-dependent manner to ensure correct positioning of floral primordium emergence and introduce a possible role for TEC3 in the maintenance of spiral phyllotaxis. Remarkably, TEC3 mutants did not exhibit reduced fertility, which may be due to a redundant role of TEC3 with other UDP-glycosyltransferases during ovule development.
Figure 7.
ETT, RPL, and TEC3 Regulate Phyllotactic Pattern in Arabidopsis.
(A) Expression pattern of pETT:ETT-GFP (upper panel) and pRPL:RPL-GFP (lower panel) in the shoot apical meristem.
(B) to (H) Analysis of angles at which successive flowers develop in wild-type, ett-3, rpl-2, ett-3 rpl-2, ETTC2S, tec3-1, and tec3-2 plants. Angle classes are indicated on the x axis subdivided into 12 categories. 1, Angles from 0 to 30; 2, angles from 31 to 60; 3, angles from 61 to 90; 4, angles from 91 to 120; 5, angles from 121 to 150; and so on until class 12, angles from 330 to 360. Below, phenotypes of stems of wild-type, ett-3, rpl-2, ett-3 rpl-2, ETTC2S, tec3-1, and tec3-2 plants.
Bars = 50 µm in (A) and 1 cm in (B) to (H).
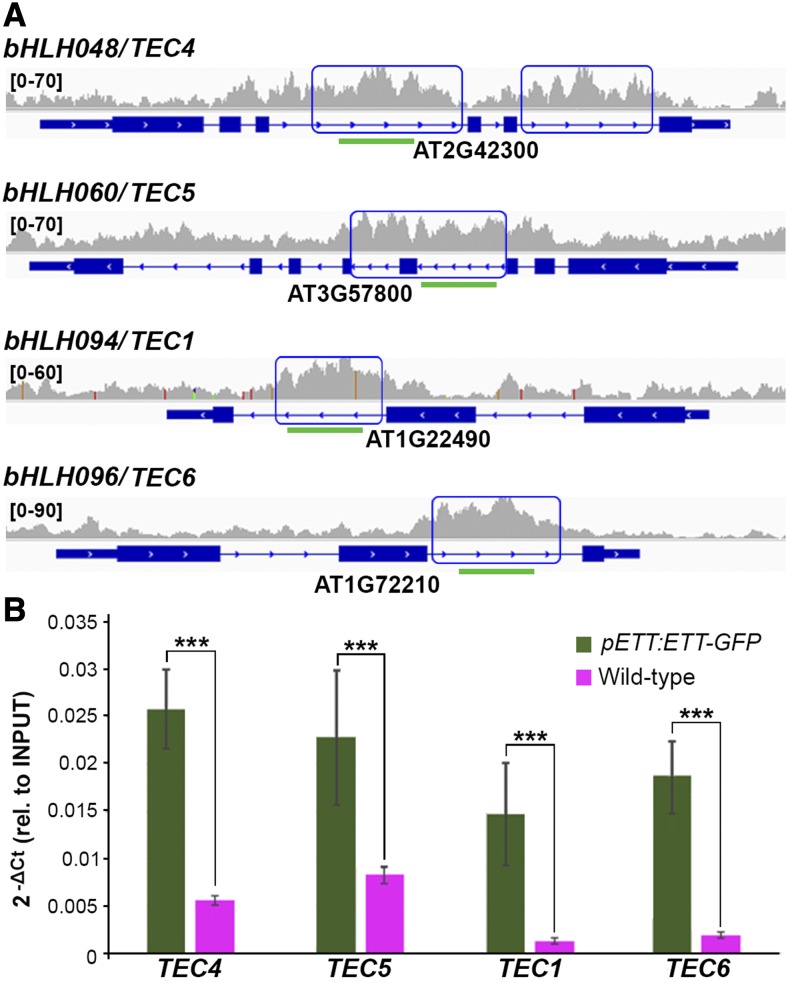
An ETT-Target Gene Encodes a bHLH Protein That Interacts with ETT in an IAA-Sensitive Manner
We have previously shown that ETT regulates gynoecium development in part via an IAA-sensitive interaction with the bHLH transcription factor INDEHISCENT (IND) (Simonini et al., 2016). The bHLH family was particularly highly represented in the ETT ChIP-seq list, with 21 bHLH proteins being direct targets of ETT (Supplemental Data Set 1). Out of these 21 targets, bHLH094 (TEC1 described above), bHLH048 (TEC4), bHLH060 (TEC5), and bHLH096 (TEC6) showed ETT binding primarily in intronic regions (Figure 8A; Supplemental Figure 4), and this was confirmed by independent ChIP analyses with the pETT:ETT-GFP line (Figure 8B). Whereas no information could be obtained for TEC6, results in the BAR database suggested that TEC1 and TEC4 are expressed in the shoot predominantly in meristem and gynoecium while TEC5 is strongly expressed in petals. Notably, these are all tissues where ETT is known to function (Sessions et al., 1997; Simonini et al., 2016).
Figure 8.
ETT Binds to Intronic Regions of a Set of bHLH Transcription Factors.
(A) Representative raw ChIP-seq peaks (one replicate only) for TEC1 and TEC4 to TEC6. Green lines represent the amplicons tested by real-time PCR.
(B) ChIP analyses with the pETT:ETT-GFP line confirming binding of ETT at the intronic regions of TEC1 and TEC4 to TEC6.
Error bars indicate sd. ***P value < 0.001.
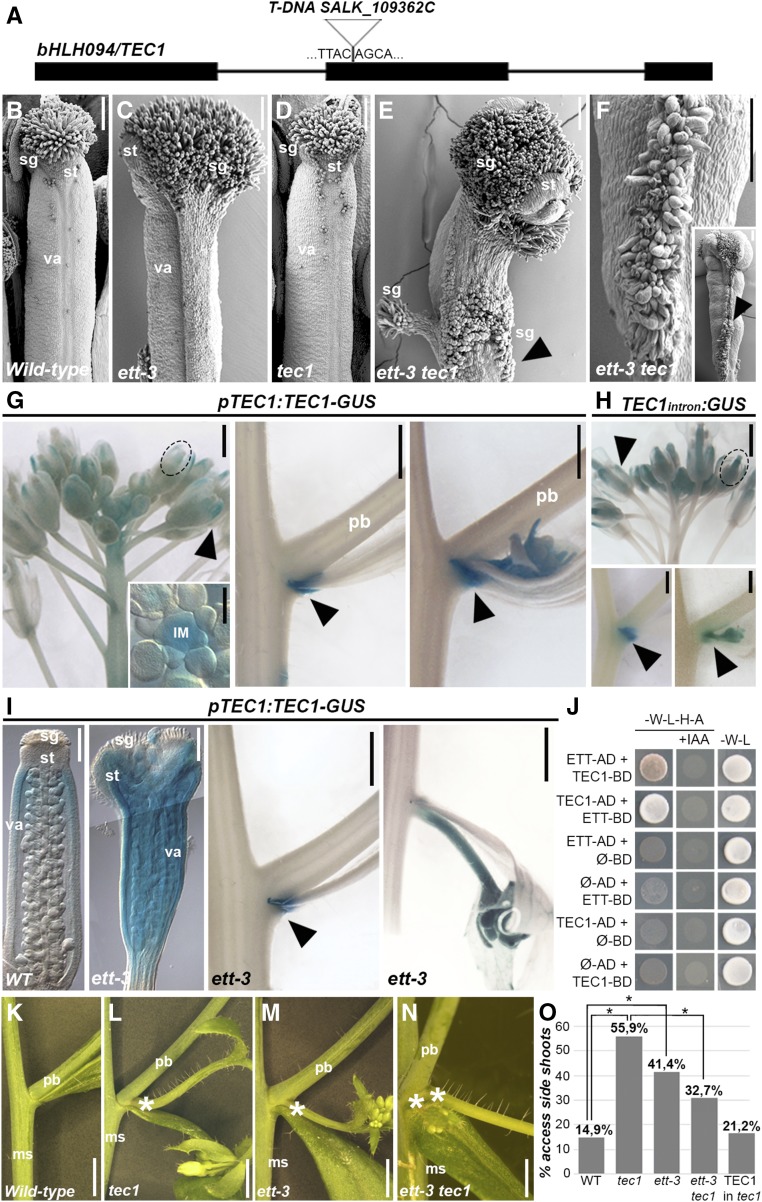
T-DNA insertional mutants were obtained for tec1 (Figure 9A), tec4, tec5, and tec6; however, none of these single mutants exhibited gynoecium or floral organ defects. Double mutant combinations between ett-3 and each of the tec1, tec4, tec5, and tec6 single mutants were obtained to uncover any genetic interactions. No obvious differences from the ett phenotype could be observed in the ett-3 tec4, ett-3 tec5, and ett-3 tec6. By contrast, ett-3 tec1 double mutant gynoecia developed conspicuous and widespread stigmatic tissue compared with the ett-3 and tec1 single mutants (Figures 9B to 9F), suggesting a synergistic role between ETT and TEC1 in carpel tissue patterning.
Figure 9.
TEC1 Is a bHLH Transcription Factor Involved in Auxin-Related Developmental Responses.
(A) Schematic representation of the position of T-DNA insertion in the TEC1 locus.
(B) to (F) Scanning electron microscopy images of wild-type (B), ett-3 (C), tec1 (D), and ett-3 tec1 double mutant ([E] and [F]) gynoecium with excessive production of stigmatic tissue (arrowhead).
(G) GUS staining of pTEC1:TEC1-GUS marker line in inflorescence, shoot apical meristem (inset), at the lower (abaxial) side, and throughout a developing primary branch (arrowhead).
(H) GUS staining of p35S(TEC1-2Intron):GUS in inflorescence and developing branch.
(I) Expression profile of pTEC1:TEC1-GUS in ett-3 mutant background in the gynoecium and at the lower (abaxial) side of a developing primary branch (arrowhead).
(J) Yeast-two-hybrid interaction assay between ETT and TEC1 and respective negative control with empty AD and BD vectors. Interactions are tested on selection media lacking Trp, Leu, His, and adenine (-W-L-H-A) and supplemented with 100 µM IAA.
(K) to (O) Emergence of extra accessory shoots (marked with an asterisk) in the wild type (K), tec1 (L), ett-3 (M), and ett-3 tec1 (N) and its frequency in percentage (O). Statistical analysis is conducted with one-way ANOVA. *P < 0.01
ms, main stem; pb, primary branch; va, valves; sg, stigma; st, style. Bars = 100 µm in (B) to (F), (G) (meristem), and (I) (gynoecium) and 5 mm in (G) and (I) (branches) and (K) to (N).
Analysis of a pTEC1:TEC1-GUS line revealed that TEC1 is expressed in sepals, in the apical part of the gynoecium, in the inflorescence meristem, and throughout the emerging primary branches including the lower base (Figure 9G). Remarkably, we found that DNA sequences located within the second intron of the TEC1 gene where ETT bound were sufficient to guide expression similar to that observed in the pTEC1:TEC1-GUS line (Figure 9H). Considering the observed TEC1 expression domains, ETT and TEC1 expression coincides in the gynoecium and at the primary branch point (Simonini et al., 2016).
In agreement with a genetic interaction between ETT and TEC1, expression of the pTEC1:TEC1-GUS construct was dramatically expanded in ett-3 gynoecia compared with the wild type (Figure 9I), albeit not at the branch points (Figure 9I). Interestingly, yeast two-hybrid assays revealed an IAA-sensitive protein-protein interaction between ETT and TEC1 (Figure 9J), thus suggesting that, in parallel to regulating its expression, ETT also cooperates with TEC1 in regulating common aspects of gynoecium development.
A T-DNA mutant line for tec1 did not show any obvious floral defects; however, in agreement with the expression domain of TEC1 at the internode between the main stem and the primary branches, tec1 homozygous mutants developed accessory side shoots with high frequency (>50%) at this site (Figures 9K, 9L, and 9O). This defect was also detectable, although less frequently, in the ett-3 mutant and was fully rescued in the pTEC1:TEC1 in tec1 complementation line (Figures 9M and 9O). Moreover, growth of accessory side shoots was partially rescued in the ett-3 tec1 double mutant, although twin accessory side shoots occasionally developed from the same internode (Figures 9N ad 9O). Growth inhibition of such structures is tightly linked to auxin regulation (Greb et al., 2003; Raman et al., 2008; Yang et al., 2012; Wang et al., 2014); therefore, given the ability of ETT and TEC1 to dimerize, ETT and TEC1 may cooperate to repress the growth of accessory side shoot structures in an auxin-related manner.
DISCUSSION
A major challenge for auxin biologists is to explain how a small molecule such as IAA can trigger the vast range of intricate, complex, and tissue-specific responses throughout plant development (Paciorek and Friml, 2006; Teale et al., 2006; Benjamins and Scheres, 2008; Vanneste and Friml, 2009; Rademacher et al., 2012; Enders and Strader, 2015; Hagen, 2015). We have recently shown that the atypical auxin response factor ETT responds to changes in cellular auxin levels in a different way from the canonical TIR1-dependent auxin-signaling pathway (Simonini et al., 2016), thus providing an additional route for auxin to mediate its diverse effects.
In this work, we present a genome-wide analysis to show that the direct effect of auxin on ETT function modulates the direct downstream transcriptome of ETT. The analysis described here is guided by our focus on the ETT-mediated IAA-sensitive pathway during gynoecium development, and target genes for analysis were selected accordingly. Our approach revealed that a significant proportion of genes directly regulated by ETT are involved in transcriptional regulation, hormone dynamics, and plant organ development, which is in agreement with previous reports on ETT gene function (Sessions et al., 1997; Simonini et al., 2016). The data analysis also uncovered that ETT controls a number of genes involved in the dynamics of different plant hormones and may therefore serve as a central node for hormonal crosstalk. First, genes that function at all levels of auxin homeostasis (biosynthesis, transport, and signaling) were among the direct ETT targets. Therefore, while ETT activity is affected by auxin, our genome-wide analysis also implicates ETT in regulating auxin dynamics itself. Interestingly, all auxin-signaling and -response targets are regulated by ETT in an IAA-independent way and are primarily repressed (Table 1). It is therefore possible that ETT provides information on polarity and identity by dampening auxin dynamics in certain tissues. Genes connected with ethylene and JA were also enriched in the set of direct ETT targets. Since ett mutants are severely affected in stamen and pollen morphology (Sessions et al., 1997), and considering the function of ethylene and JA in stamen development and pollination (Dobritzsch et al., 2015), it is plausible that aspects of the ett phenotype are caused by defects in ethylene and JA homeostasis.
ARF proteins have been shown to recognize so-called AuxREs (Ulmasov et al., 1997; Guilfoyle, 2015). These were initially characterized as TGTCTC hexamers, but it has recently been revealed that any form of TGTCNN could potentially function as an ARF binding site (Boer et al., 2014; Franco-Zorrilla et al., 2014). We performed a search for potential AuxREs among the 663 direct ETT targets identified in our study. All of them contained TGTCNN elements, but there was no significant enrichment of any particular form. This correlates with previous observations that ETT can bind TGTCAT, TGTCAC, and TGTCGG (Franco-Zorrilla et al., 2014; Simonini et al., 2016). The aim of this study was to identify ETT targets that are relevant to the ETT-mediated auxin-signaling mechanism; therefore, we did not make any further attempts to identify exact binding sites.
Recognition of otherwise weak binding sites by ETT may be assisted by interaction with specific partners, which might be predominantly TFs given the interaction preferences exhibited by ETT in yeast two-hybrid library screenings (Simonini et al., 2016) and in interactome analyses (Vernoux et al., 2011). In accordance with ETT lacking the PB1 domain, interaction of ETT with AUX/IAA repressors has never been observed (Simonini et al., 2016; Vernoux et al., 2011). Recently, the DBD of ARFs has been proposed to mediate ARF-ARF homodimerization and heterodimerization (Boer et al., 2014). Although there are presently no data showing ETT interacting with other ARFs through their DBD, it is possible that such interactions exist, thus allowing crosstalk between the canonical TIR1/AFB and the ETT-mediated auxin-signaling pathways in planta.
As a subset of the 77 genes identified as being directly bound by the ETT protein and regulated by ETT in an IAA-sensitive manner, seven genes overlapped with targets of the RPL homeobox protein, four of which have expression patterns that overlap with ETT and RPL. RPL was previously shown to interact with ETT in an IAA-sensitive manner (Simonini et al., 2016). Binding of ETT to three of these targets (TEC2, YIP4B, and SMLX5) did not require RPL, a finding which either reflects that ETT binding is assisted by another partner or that ETT recognizes a target sequence sufficiently strongly for independent binding. However, RPL was required to recruit ETT to the TEC3 gene and to properly regulate TEC3 expression; genetic analysis revealed the biological relevance of this regulation and its involvement in controlling phyllotactic patterning. The TEC3 gene encodes a UDP-glycosyltransferase. UDP-glycosyltransferases are enzymes capable of adding a glucose molecule to a set of molecular targets, including hormones. Indeed, the UDP-glycosyltransferases UGT84B1, UGT74E2, and UGT74D1 can all add sugar molecules to auxins, in particular, IAA and indole-3-butyric acid, and may thereby alter the activity, solubility, and cellular localization of auxin (Jackson et al., 2001; Tognetti et al., 2010; Jin et al., 2013). As phyllotaxis is tightly dependent on auxin accumulation and transport (Reinhardt et al., 2003; Jönsson et al., 2006; Smith et al., 2006), TEC3 may therefore contribute to the control of auxin homeostasis in the shoot apical meristem.
It is possible that ETT only weakly interacts with a target sequence in TEC3 and that RPL strengthens the interaction. Out of the initial 77 ETT-regulated IAA-dependent targets, only one exhibited RPL-dependent ETT binding. This observation might reflect the intricate network of ETT-containing dimers that can be formed and disrupted according to auxin levels in the cell. Since ETT is relatively widely expressed throughout floral tissues (Sessions et al., 1997; Simonini et al., 2016), the association of ETT with a specific DNA sequence might be influenced and guided by tissue-specific protein partners. Therefore, the multiple combinations of ETT-partner modules in different tissues at consecutive developmental stages will likely contribute to shaping the transcriptome.
An intriguing mechanism emerged from this study with the identification of a gene encoding a bHLH-type transcription factor, TEC1. ETT regulates the expression of TEC1 in an IAA-independent manner; however, analysis of single and double mutants for the ETT and TEC1 genes suggests that ETT and TEC1 synergistically cooperate in regulating gynoecium tissue identity and in repressing accessory side-shoot emergence. Indeed, direct protein-protein interactions in yeast were sensitive to IAA, similarly to what has been reported previously for other ETT-interacting TFs (Simonini et al., 2016), and further suggesting that ETT and TEC1 function via a feedback mechanism. The genetic relationship that coordinates ETT and TEC1 is intricate: In a simple scenario, one would expect that introgression of tec1 mutation in ett-3 mutant background would restore to some extent the defects in gynoecium development and in parallel would enhance the percentage of primary branches developing accessory side shoots. However, an ett-3 tec1 double mutant showed enhanced gynoecium defects compared with ett-3 single mutant and partial recovery of the percentage of branches developing accessory side shoot.
TEC1 is required for repressing accessory side-shoot development, as the single mutant already displays side shoot defects. However, other transcription factors are likely to be involved in this process as well. For instance, the bHLH096/TEC6 transcription factor described in this manuscript is the closest homolog of TEC1 in Arabidopsis. It is therefore possible that TEC1 both collaborates with and regulates the expression of other factors involved in these developmental aspects. Therefore, misregulated expression of TEC1 may have both direct and indirect consequences on tissue morphology, due to its role in regulating tissue identity genes while controlling the expression of synergistic and antagonistic transcription factors involved in these processes.
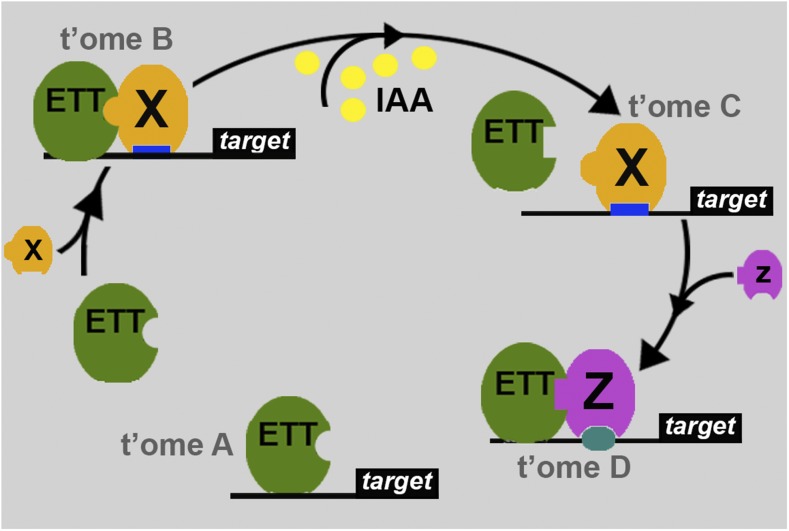
Together, this work demonstrates that auxin provides developmental cues that are translated by the auxin response factor ETT into multiple transcriptional responses during the development and patterning of plant organs (Figure 10). Specifically, our data show that certain combinations of ETT with protein partners regulate sets of targets involved in processes such as cell differentiation, tissue specification, and hormonal dynamics. Auxin in turn modulates the interaction of ETT with its partner(s), thereby affecting expression of its target genes (Figure 10).
Figure 10.
Proposed Model of Action for ETT and Auxin Effects on ETT Dimerization and Transcription.
ETT controls expression of target genes either alone (transcriptome A, t’ome A) or in combination with process-specific protein partners (“X”) (t’ome B). At increased auxin levels, dimerization of ETT with a set of partners is affected, thereby releasing ETT from a set of genomic loci, ultimately leading to a different transcriptional outcome (t’ome C). As auxin destabilizes ETT dimerization, it also promotes ETT association with a set of target sequences possibly through interaction of ETT with other partners (“Z”) (t’ome D).
In conclusion, the genome-wide approach presented here reveals how a noncanonical auxin-signaling mechanism employing just auxin and an ETT-partner module ensures perception, interpretation, and actuation of the auxin message to facilitate multiple levels of transcriptional control and developmental diversity. The TIR1-dependent canonical auxin-signaling pathway may be the dominant mechanism through which auxin affects plant growth and development. However, the work described here suggests that direct effects of auxin on protein factors, such as ETT-containing complexes, comprise an important part of auxin biology and therefore contribute to the vast number of biological processes that are affected by this simple molecule.
METHODS
Plant Materials and Growth Conditions
Plants were grown on soil in long-day conditions (16 h light/8 h dark, 90 µmol m−2 s−1 sodium lamp). The mutant line ett-3, pETT:ETT-GFP complementation line, and pETT:ETTC2S transgenic line (Simonini et al., 2016); the pRPL:RPL-GFP complementation line (Bencivenga et al., 2016); and the DR5:GFP, DII-VENUS, pPIN1:PIN1-GFP, pPIN3:PIN3-GFP, pASB1:GUS, and pTAA1:GFP-TAA1 lines (Benková et al., 2003; Brunoud et al., 2012; Zádníková et al., 2010; Stepanova et al., 2005, 2008) have been previously described. We obtained the following T-DNA insertional lines from the SALK collection (Alonso et al., 2003): AT1G22490 (bHLH094/TEC1), SALK_109362C; AT2G28080 (TEC3), SALK041660 (tec3-1) and SALK_139804 (tec3-2); AT2G42300 (bHLH048/TEC4), SALK_135489C and SALK_013047C; AT3G57800 (bHLH060/TEC5), SALK_134005C; and AT1G72210 (bHLH096/TEC6), SALK_060037C.
Scanning Electron Microscopy, GUS Assay, and Phyllotaxis Analyses
Scanning electron microscopy and GUS assay were performed as previously described (Moubayidin and Østergaard, 2014). Analyses of divergence angle were performed as previously reported (Pinon et al., 2013).
RNA-Seq and Data Analyses
Three samples of three inflorescences each were harvested on five consecutive days under the same conditions and RNA extracted immediately. Whole inflorescences were collected and mature flowers and siliques removed. This means the material used for the RNA-seq included meristem and young flowers up to and including stage 10 (Smyth et al., 1990). Samples were then combined to create a biological triplicate for sequencing. Auxin treatments were performed according to previous reports (Nagpal et al., 2005; Paponov et al., 2008): A few drops of auxin solution (10 µM IAA in 1% methanol) or mock solution (1% methanol) were applied on a whole inflorescence; after 30 min, samples were collected and immediately frozen. RNA was extracted and sequenced (50-bp single-end reads) using a HiSeq 2500 (Rapid-Run mode) as described by the manufacturer (Illumina).
RNA-seq reads obtained from the replicates were aligned against the TAIR10 reference sequence with TopHat version 2.0.10 (Trapnell et al., 2012) implementing Bowtie version 2.1.0.0 (Langmead and Salzberg, 2012). The Cuffdiff program from the Cufflinks suite version 2.2.1 (Trapnell et al., 2010) was then used to identify genes with significant expression differences between the various genotype-treatment combinations.
To test for overrepresented GO terms, the Gene Ontology Consortium-Panther website was used (http://www.geneontology.org/).
ChIP and Deep Sequencing
ChIP was performed in triplicate as previously described (Simonini et al., 2016; Bencivenga et al., 2016) using MultiMACS GFP isolation kits, pETT:ETT-GFP in ett-3 line, and wild-type plants (negative control). As with RNA-seq, the material used included meristem and young flowers up to and including stage 10 (Smyth et al., 1990). For ChIP-seq, seven Illumina TruSeq ChIP-seq libraries (three pETT:ETT-GFP replicates −/+ IAA and one wild-type control derived from the merge of three biological wild-type replicates) were produced as described (Bencivenga et al., 2016) and sequenced (50-bp single-end reads) using a HiSeq 2500 (Rapid-Run mode) as described by the manufacturer (Illumina).
IAA treatment was performed by spraying plants with a solution containing 100 µM IAA, 10 μM NPA, and 0.03% Silwet and collecting material 6 h after treatment.
Reads from the replicate treatments and the replicate controls were aligned against the TAIR10 Col-0 reference sequence with Bowtie2 (Langmead and Salzberg, 2012). MACS version 2.0.10 (Zhang et al., 2008; Feng et al., 2012) was used to identify enriched peaks over three comparisons of replicate treatments and controls. A Perl script was developed to identify peaks with P values <0.01 that were common to the majority (2/3) of comparisons and to then identify their closest cognate genes by programmatically interrogating the TAIR10 annotation, allowing peak positions to be located either in 5′ or 3′ regions or gene bodies.
To test for overrepresented GO terms, the Gene Ontology Consortium-Panther website was used (http://www.geneontology.org/). The ChIP-seq data of pRPL:RPL-GFP are available online (Bencivenga et al., 2016).
Cloning of TEC1 Locus and Intron
The genomic locus for bHLH094/TEC1 was amplified using gene-specific primers (Supplemental Table 1) with genomic DNA and cloned into the pPZP222 vector by the Gibson method. The fragment contained 5 kb of the promoter region and the coding region including introns. Fusion with the GUS reporter gene was achieved by the Gibson method via cloning of the GUS gene with the appropriate overhangs. The tNOS terminator was cloned downstream of the GUS gene. For complementation analyses, the same promoter-gene fragment was cloned into the pPZP222 vector by the Gibson method. A similar approach was used to clone the TEC1 second intron upstream the 35S minimal promoter (the sequence was included in the reverse primer for the cloning). Fusion with the GUS reporter gene was done by the Gibson method by cloning of the GUS gene with the appropriate overhangs, and the tNOS terminator was cloned downstream of the GUS gene.
Vectors were transformed into Agrobacterium tumefaciens GV3101 and plants were dipped following the floral-dip method (Clough and Bent, 1998).
Yeast Two-Hybrid Assay
The yeast two-hybrid assays were performed at 28°C in the yeast strain AH109 (Clontech) using the cotransformation technique (Egea-Cortines et al., 1999). Coding sequences were cloned into Gateway vector GAL4 system (pGADT7 and pGBKT7; Clontech) passing through pDONR207 (Life Technologies). Strength of interaction was tested on selective yeast synthetic dropout medium (YSD) lacking Leu (L), Trp (W), adenine (A), and His (H) supplemented with different concentrations of 3-aminotriazole (1, 2.5, and 5 mM of 3-aminotriazole). IAA was added at the final concentration of 100 μM in the cooling medium as previously described (Simonini et al., 2016).
In Situ Hybridization and Real-Time PCR
The TEC3 digoxygenin-labeled antisense RNA probe (Supplemental Table 1) was generated by in vitro transcription according to the instructions provided with the DIG RNA Labeling Kit (SP6/T7; Roche) using cDNA as template. Developing inflorescences were fixed and embedded in Paraplast Plus embedding medium, cut in 8-μm sections, and then hybridized as previously described (Dreni et al., 2007). Treatment of inflorescences has been performed as for the RNA-seq analyses previously described reports (Nagpal et al., 2005; Paponov et al., 2008). After treatment, the inflorescences were immediately fixed in FAA solution (3.7% formaldehyde, 50% ethanol, and 5% acetic acid).
Real-time PCR was performed using a Bio-Rad LightCycler LC480 and LightCycler Sybr green I Master Mix. Primers are listed in Supplemental Table 1.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: ETT, AT2G33860; TEC1/BHLH094, AT1G22490; YIP4B, AT4G30260; SMXL5, AT5G57130; TEC2, AT1G76580; TEC3, AT2G28080; TEC4/bHLH048, AT2G42300; TEC5/bHLH060, AT3G57800; TEC6/bHLH096, AT1G72210; ARR15, AT1G74890; SKP2A, AT1G21410; IRX14, AT4G36890; AP2, AT4G36920; FLS1, AT5G08640; YUC4, AT5G11320; CRF3, AT5G53290; and LFY, AT5G61850. ChIP-seq and RNA-seq raw data have been deposited in the EBI-ENA database under accession number PRJEB19862
Supplemental Data
Supplemental Figure 1. Raw ChIP-seq data for PIN1, PIN3, PIN7, AUX1, LAX1, ABCB19, and ASB1/WEI7.
Supplemental Figure 2. Raw ChIP-seq peaks of –IAA and +IAA analyses for SKP2A, ARR15, IRX14, AP2, FLS1, YUC4, CRF3, and LFY.
Supplemental Figure 3. ChIP analysis of TEC2-3, YIP4B, and SMXL5 with ETT.
Supplemental Figure 4. Raw ChIP-seq data for TEC1, TEC4, TEC5, and TEC6.
Supplemental Table 1. Primers used in this study.
Supplemental Data Set 1. ChIP-seq pETT:ETT-GFP ±IAA
Supplemental Data Set 2. RNA-seq wild type ±IAA versus ett-3 ±IAA.
Supplemental File 1. ANOVA tables.
Acknowledgments
We thank the Earlham Institute for ChIP-seq and RNA-seq, in particular Leah Clissold and Myriam Chapman for technical support, Robert Sablowski for technical help, and Teva Vernoux, Jose M. Alonso, and Jiri Friml for seeds. We are grateful for discussions, contributions, and helpful comments from Laila Moubayidin, Xinran Li, Yang Dong, Marie Brüser, Martin Kater, Andre Kuhn, Billy Tasker-Brown, Pauline Stephenson, and Nicola Stacey. This work was supported by grants from the BBSRC to L.Ø. (BB/M004112/1) and to the John Innes Centre (BB/J004588/1).
AUTHOR CONTRIBUTIONS
S.S., S.B., and L.Ø. conceived the experiments. S.S. performed the experiments and S.B helped with ChIP-seq protocol and analyses. M.T. did the bioinformatics analyses. S.S., S.B., and L.Ø. analyzed the data. S.S and L.Ø. wrote the manuscript and S.B. and M.T. commented on it. All authors read and approved the manuscript.
Glossary
- AuxREs
auxin response elements
- TFs
transcription factors
- ChIP-seq
chromatin immunoprecipitation coupled with deep sequencing
- GO
Gene Ontology
- JA
jasmonic acid
Footnotes
Articles can be viewed without a subscription.
References
- Alonso J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657. [DOI] [PubMed] [Google Scholar]
- Bao X., Franks R.G., Levin J.Z., Liu Z. (2004). Repression of AGAMOUS by BELLRINGER in floral and inflorescence meristems. Plant Cell 16: 1478–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bencivenga S., Serrano-Mislata A., Bush M., Fox S., Sablowski R. (2016). Control of oriented tissue growth through repression of organ boundary genes promotes stem morphogenesis. Dev. Cell 39: 198–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamins R., Scheres B. (2008). Auxin: the looping star in plant development. Annu. Rev. Plant Biol. 59: 443–465. [DOI] [PubMed] [Google Scholar]
- Benjamins R., Quint A., Weijers D., Hooykaas P., Offringa R. (2001). The PINOID protein kinase regulates organ development in Arabidopsis by enhancing polar auxin transport. Development 128: 4057–4067. [DOI] [PubMed] [Google Scholar]
- Benková E., Michniewicz M., Sauer M., Teichmann T., Seifertová D., Jürgens G., Friml J. (2003). Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115: 591–602. [DOI] [PubMed] [Google Scholar]
- Boer D.R., Freire-Rios A., van den Berg W.A., Saaki T., Manfield I.W., Kepinski S., López-Vidrieo I., Franco-Zorrilla J.M., de Vries S.C., Solano R., Weijers D., Coll M. (2014). Structural basis for DNA binding specificity by the auxin-dependent ARF transcription factors. Cell 156: 577–589. [DOI] [PubMed] [Google Scholar]
- Brunoud G., Wells D.M., Oliva M., Larrieu A., Mirabet V., Burrow A.H., Beeckman T., Kepinski S., Traas J., Bennett M.J., Vernoux T. (2012). A novel sensor to map auxin response and distribution at high spatio-temporal resolution. Nature 482: 103–106. [DOI] [PubMed] [Google Scholar]
- Byrne M.E., Groover A.T., Fontana J.R., Martienssen R.A. (2003). Phyllotactic pattern and stem cell fate are determined by the Arabidopsis homeobox gene BELLRINGER. Development 130: 3941–3950. [DOI] [PubMed] [Google Scholar]
- Calderón Villalobos L.I., et al. (2012). A combinatorial TIR1/AFB-Aux/IAA co-receptor system for differential sensing of auxin. Nat. Chem. Biol. 8: 477–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler J.W. (2009). Auxin as compère in plant hormone crosstalk. Planta 231: 1–12. [DOI] [PubMed] [Google Scholar]
- Chandler J.W. (2016). Auxin response factors. Plant Cell Environ. 39: 1014–1028. [DOI] [PubMed] [Google Scholar]
- Cheng Y., Dai X., Zhao Y. (2006). Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes Dev. 20: 1790–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Dobritzsch S., Weyhe M., Schubert R., Dindas J., Hause G., Kopka J., Hause B. (2015). Dissection of jasmonate functions in tomato stamen development by transcriptome and metabolome analyses. BMC Biol. 13: 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreni L., Jacchia S., Fornara F., Fornari M., Ouwerkerk P.B., An G., Colombo L., Kater M.M. (2007). The D-lineage MADS-box gene OsMADS13 controls ovule identity in rice. Plant J. 52: 690–699. [DOI] [PubMed] [Google Scholar]
- Egea-Cortines M., Saedler H., Sommer H. (1999). Ternary complex formation between the MADS-box proteins SQUAMOSA, DEFICIENS and GLOBOSA is involved in the control of floral architecture in Antirrhinum majus. EMBO J. 18: 5370–5379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enders T.A., Strader L.C. (2015). Auxin activity: Past, present, and future. Am. J. Bot. 102: 180–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J., Liu T., Qin B., Zhang Y., Liu X.S. (2012). Identifying ChIP-seq enrichment using MACS. Nat. Protoc. 7: 1728–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco-Zorrilla J.M., López-Vidriero I., Carrasco J.L., Godoy M., Vera P., Solano R. (2014). DNA-binding specificities of plant transcription factors and their potential to define target genes. Proc. Natl. Acad. Sci. USA 111: 2367–2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendre D., McFarlane H.E., Johnson E., Mouille G., Sjödin A., Oh J., Levesque-Tremblay G., Watanabe Y., Samuels L., Bhalerao R.P. (2013). Trans-Golgi network localized ECHIDNA/Ypt interacting protein complex is required for the secretion of cell wall polysaccharides in Arabidopsis. Plant Cell 25: 2633–2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greb T., Clarenz O., Schafer E., Muller D., Herrero R., Schmitz G., Theres K. (2003). Molecular analysis of the LATERAL SUPPRESSOR gene in Arabidopsis reveals a conserved control mechanism for axillary meristem formation. Genes Dev. 17: 1175–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilfoyle T.J. (2015). The PB1 domain in auxin response factor and Aux/IAA proteins: a versatile protein interaction module in the auxin response. Plant Cell 27: 33–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilfoyle T.J., Hagen G. (2007). Auxin response factors. Curr. Opin. Plant Biol. 10: 453–460. [DOI] [PubMed] [Google Scholar]
- Hagen G. (2015). Auxin signal transduction. Essays Biochem. 58: 1–12. [DOI] [PubMed] [Google Scholar]
- Jackson R.G., Lim E.K., Li Y., Kowalczyk M., Sandberg G., Hoggett J., Ashford D.A., Bowles D.J. (2001). Identification and biochemical characterization of an Arabidopsis indole-3-acetic acid glucosyltransferase. J. Biol. Chem. 276: 4350–4356. [DOI] [PubMed] [Google Scholar]
- Jin S.H., Ma X.M., Han P., Wang B., Sun Y.G., Zhang G.Z., Li Y.J., Hou B.K. (2013). UGT74D1 is a novel auxin glycosyltransferase from Arabidopsis thaliana. PLoS One 8: e61705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jönsson H., Heisler M.G., Shapiro B.E., Meyerowitz E.M., Mjolsness E. (2006). An auxin-driven polarized transport model for phyllotaxis. Proc. Natl. Acad. Sci. USA 103: 1633–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korasick D.A., Jez J.M., Strader L.C. (2015). Refining the nuclear auxin response pathway through structural biology. Curr. Opin. Plant Biol. 27: 22–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunst L., Klenz J.E., Martinez-Zapater J., Haughn G.W. (1989). AP2 gene determines the identity of perianth organs in flowers of Arabidopsis thaliana. Plant Cell 1: 1195–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Salzberg S.L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9: 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moubayidin L., Østergaard L. (2014). Dynamic control of auxin distribution imposes a bilateral-to-radial symmetry switch during gynoecium development. Curr. Biol. 24: 2743–2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moubayidin L., Østergaard L. (2017). Gynoecium formation: an intimate and complicated relationship. Curr. Opin. Genet. Dev. 45: 15–21. [DOI] [PubMed] [Google Scholar]
- Muday G.K., Rahman A., Binder B.M. (2012). Auxin and ethylene: collaborators or competitors? Trends Plant Sci. 17: 181–195. [DOI] [PubMed] [Google Scholar]
- Nagpal P., Ellis C.M., Weber H., Ploense S.E., Barkawi L.S., Guilfoyle T.J., Hagen G., Alonso J.M., Cohen J.D., Farmer E.E., Ecker J.R., Reed J.W. (2005). Auxin response factors ARF6 and ARF8 promote jasmonic acid production and flower maturation. Development 132: 4107–4118. [DOI] [PubMed] [Google Scholar]
- Nemhauser J.L., Feldman L.J., Zambryski P.C. (2000). Auxin and ETTIN in Arabidopsis gynoecium morphogenesis. Development 127: 3877–3888. [DOI] [PubMed] [Google Scholar]
- Paciorek T., Friml J. (2006). Auxin signaling. J. Cell Sci. 119: 1199–1202. [DOI] [PubMed] [Google Scholar]
- Paponov I.A., Paponov M., Teale W., Menges M., Chakrabortee S., Murray J.A., Palme K. (2008). Comprehensive transcriptome analysis of auxin responses in Arabidopsis. Mol. Plant 1: 321–337. [DOI] [PubMed] [Google Scholar]
- Pinon V., Prasad K., Grigg S.P., Sanchez-Perez G.F., Scheres B. (2013). Local auxin biosynthesis regulation by PLETHORA transcription factors controls phyllotaxis in Arabidopsis. Proc. Natl. Acad. Sci. USA 110: 1107–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rademacher E.H., Lokerse A.S., Schlereth A., Llavata-Peris C.I., Bayer M., Kientz M., Freire Rios A., Borst J.W., Lukowitz W., Jürgens G., Weijers D. (2012). Different auxin response machineries control distinct cell fates in the early plant embryo. Dev. Cell 22: 211–222. [DOI] [PubMed] [Google Scholar]
- Raman S., Greb T., Peaucelle A., Blein T., Laufs P., Theres K. (2008). Interplay of miR164, CUP-SHAPED COTYLEDON genes and LATERAL SUPPRESSOR controls axillary meristem formation in Arabidopsis thaliana. Plant J. 55: 65–76. [DOI] [PubMed] [Google Scholar]
- Rashotte A.M., Mason M.G., Hutchison C.E., Ferreira F.J., Schaller G.E., Kieber J.J. (2006). A subset of Arabidopsis AP2 transcription factors mediates cytokinin responses in concert with a two-component pathway. Proc. Natl. Acad. Sci. USA 103: 11081–11085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt D., Pesce E.R., Stieger P., Mandel T., Baltensperger K., Bennett M., Traas J., Friml J., Kuhlemeier C. (2003). Regulation of phyllotaxis by polar auxin transport. Nature 426: 255–260. [DOI] [PubMed] [Google Scholar]
- Roeder A.H., Ferrándiz C., Yanofsky M.F. (2003). The role of the REPLUMLESS homeodomain protein in patterning the Arabidopsis fruit. Curr. Biol. 13: 1630–1635. [DOI] [PubMed] [Google Scholar]
- Salehin M., Bagchi R., Estelle M. (2015). SCFTIR1/AFB-based auxin perception: mechanism and role in plant growth and development. Plant Cell 27: 9–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santuari L., et al. (2016). The PLETHORA gene regulatory network guides growth and cell differentiation in Arabidopsis roots. Plant Cell 28: 2937–2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessions A., Nemhauser J.L., McColl A., Roe J.L., Feldmann K.A., Zambryski P.C. (1997). ETTIN patterns the Arabidopsis floral meristem and reproductive organs. Development 124: 4481–4491. [DOI] [PubMed] [Google Scholar]
- Simonini S., Deb J., Moubayidin L., Stephenson P., Valluru M., Freire-Rios A., Sorefan K., Weijers D., Friml J., Østergaard L. (2016). A noncanonical auxin-sensing mechanism is required for organ morphogenesis in Arabidopsis. Genes Dev. 30: 2286–2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith R.S., Guyomarc’h S., Mandel T., Reinhardt D., Kuhlemeier C., Prusinkiewicz P. (2006). A plausible model of phyllotaxis. Proc. Natl. Acad. Sci. USA 103: 1301–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth D.R., Bowman J.L., Meyerowitz E.M. (1990). Early flower development in Arabidopsis. Plant Cell 2: 755–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanova A.N., Hoyt J.M., Hamilton A.A., Alonso J.M. (2005). A link between ethylene and auxin uncovered by the characterization of two root-specific ethylene-insensitive mutants in Arabidopsis. Plant Cell 17: 2230–2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanova A.N., Robertson-Hoyt J., Yun J., Benavente L.M., Xie D.Y., Dolezal K., Schlereth A., Jürgens G., Alonso J.M. (2008). TAA1-mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell 133: 177–191. [DOI] [PubMed] [Google Scholar]
- Teale W.D., Paponov I.A., Palme K. (2006). Auxin in action: signalling, transport and the control of plant growth and development. Nat. Rev. Mol. Cell Biol. 7: 847–859. [DOI] [PubMed] [Google Scholar]
- Tognetti V.B., et al. (2010). Perturbation of indole-3-butyric acid homeostasis by the UDP-glucosyltransferase UGT74E2 modulates Arabidopsis architecture and water stress tolerance. Plant Cell 22: 2660–2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D.R., Pimentel H., Salzberg S.L., Rinn J.L., Pachter L. (2012). Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7: 562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Williams B.A., Pertea G., Mortazavi A., Kwan G., van Baren M.J., Salzberg S.L., Wold B.J., Pachter L. (2010). Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28: 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov T., Hagen G., Guilfoyle T.J. (1997). ARF1, a transcription factor that binds to auxin response elements. Science 276: 1865–1868. [DOI] [PubMed] [Google Scholar]
- Vanneste S., Friml J. (2009). Auxin: a trigger for change in plant development. Cell 136: 1005–1016. [DOI] [PubMed] [Google Scholar]
- Vernoux T., et al. (2011). The auxin signalling network translates dynamic input into robust patterning at the shoot apex. Mol. Syst. Biol. 7: 508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallner E.S., et al. (2017). Strigolactone- and karrikin-independent SMXL proteins are central regulators of phloem formation. Curr. Biol. 27: 1241–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Kohlen W., Rossmann S., Vernoux T., Theres K. (2014). Auxin depletion from the leaf axil conditions competence for axillary meristem formation in Arabidopsis and tomato. Plant Cell 26: 2068–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel D., Alvarez J., Smyth D.R., Yanofsky M.F., Meyerowitz E.M. (1992). LEAFY controls floral meristem identity in Arabidopsis. Cell 69: 843–859. [DOI] [PubMed] [Google Scholar]
- Weijers D., Wagner D. (2016). Transcriptional responses to the auxin hormone. Annu. Rev. Plant Biol. 67: 539–574. [DOI] [PubMed] [Google Scholar]
- Yang F., Wang Q., Schmitz G., Müller D., Theres K. (2012). The bHLH protein ROX acts in concert with RAX1 and LAS to modulate axillary meristem formation in Arabidopsis. Plant J. 71: 61–70. [DOI] [PubMed] [Google Scholar]
- Zádníková P., et al. (2010). Role of PIN-mediated auxin efflux in apical hook development of Arabidopsis thaliana. Development 137: 607–617. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W., Liu X.S. (2008). Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9: R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z., Andersen S.U., Ljung K., Dolezal K., Miotk A., Schultheiss S.J., Lohmann J.U. (2010). Hormonal control of the shoot stem-cell niche. Nature 465: 1089–1092. [DOI] [PubMed] [Google Scholar]