Abstract
Background
Translation initiation is the rate limiting step of protein synthesis and is highly regulated. Eukaryotic initiation factor 3C (EIF3C), an oncogene overexpressed in several human cancers, plays an important role in tumorigenesis and cell proliferation.
Material/Methods
Immunohistochemistry was used to determine the expression of EIF3C in breast cancer tissues from 42 patients. We investigated whether EIF3C silencing decreases breast cancer cell proliferation as assessed by colony formation assay, and whether EIF3C gene knockdown induces apoptosis as assessed by flow cytometry analysis. We utilized the stress and apoptosis signaling antibody array kit, while p-ERK1/2, p-Akt, p-Smad2, p-p38 MAPK, cleaved caspase-3, and cleaved caspase-7 were explored between EIF3C-siRNA and controls. Furthermore, the effects of EIF3C gene knockdown in mTOR pathway were analyzed by western blotting for different cell lines.
Results
In EIF3C-positive tumors, 32 out of 42 showed significantly higher frequencies of high grade group by immunoreactivity (p=0.0016). BrdU incorporation after four days of cell plating was significantly suppressed in MDA-MB-231 cells by EIF3C knockdown compared with controls, with average changes of 7.8-fold (p<0.01). Clone number was significantly suppressed in MDA-MB-231 cells by EIF3C knockdown compared with controls (p<0.05). Cell apoptosis was significantly increased in the EIF3C-siRNA group when compared with the cells that were transfected with scrambled siRNA (3.51±0.0842 versus 13.24±0.2307, p<0.01). The mTOR signaling pathway was involved in decreasing EIF3C translational efficiency.
Conclusions
Unveiling the mechanisms of EIF3 action in tumorigenesis may help identify attractive targets for cancer therapy.
MeSH Keywords: Eukaryotic Initiation Factor-3, Breast Neoplasms, Signal Transduction
Background
Breast cancer is one of the main causes of cancer-related deaths particularly among women from 35 to 75 years old [1]. Breast cancer mortality has declined in the past few decades, but the incidence of breast cancer remains at a high level in developing countries [2,3]. Current treatments include surgery, radiation, hormone therapy, chemotherapy, and targeted therapy. But the curative effects of patients with breast cancer remains poor, especially in the case of rapid progress, relapse of refractory disease and drug resistance. Through an understanding genome sequencing of breast cancer, we need to develop effective anti-cancer agents that target novel cancer-driven genes and induce apoptosis of cancer cells [4].
Protein synthesis is considered a critical step in gene expression while translation regulation is pivotal for metabolism, homeostasis, and cell physiology. Obviously, disordered protein synthesis makes for tumorigenesis and malignant progression. Abnormalities in protein synthesis result in transcriptional disorders of mRNAs, which were essential for cell survival, proliferation, and migration [5]. The formation of stabilized proteins plays a fundamental role in the functions of cell-cell interactions. These proteins help to stabilize the activation of the functional ribosome by the formation of the start codon and provide a mechanism for translation initiation [6]. The entire process is orchestrated by numerous individual proteins and three protein complexes commonly called eukaryotic initiation factors (EIFs) [7,8]. In the majority of cancers, improper translation of oncogenes, tumor suppressors, and EIFs are a critical process for cancer cell proliferation and apoptosis [9].
EIF3C (eukaryotic initiation factor 3C) is considered a housekeeping gene, and plays an important role in the cytoplasm for the assembly of the EIF3 complex [10,11]. Components of EIF3 complex are required for several steps in the initiation of protein synthesis [12,13]. In the past few years, it has been reported that EIF3C is essential for cell proliferation in various human tumors, such as glioma [14,15], hepatocellular carcinoma [16], testicular seminomas [17], meningioma [18], and colon cancer [19,20]. Downregulated EIF3C may result in switching off polysomes and inducing cell death due to translation depressed at the initiation stage. As a consequence, EIF3C may be a novel anti-cancer candidate in different malignancies.
Several randomized trials have shown that mammalian target of rapamycin (mTOR) inhibitors can promote patient outcomes with hormone receptor-positive or human epidermal growth factor receptor-2-positive [21]. Several studies showed that PI3K/AKT/mTOR signaling pathway was activated in TNBC patients, which conferred high aggressiveness and relapse risk [22]. The mTOR pathway is mainly considered an inhibitor of protein synthesis, which regulates downstream signaling transmission by combining the EIF’s cap with EIF4B. When cells are stimulated by hormones or mitogenic signals, mTOR binds to the EIF3 complex and phosphorylates S6K1. This process results in the dissociation of S6K1 from EIF3 and S6K1 activation [23,24].
In our study, EIF3C protein was detected in breast cancer tissues, which was related with high tumor grade status. The breast cancer cells with EIF3C knockdown were established for exploring its functions in tumorigenesis, proliferation, and apoptosis. The mTOR signaling pathway was involved in decreasing EIF3C translational efficiency. Our findings in this article provide new understandings of bio-function of EIF3C in breast cancer and recognize EIF3C as a potential prognosis or therapeutic target in clinic for breast cancer.
Material and Methods
Cell culture
Breast cancer cells, BT474 (invasive ductal carcinoma, luminal B subtype), and MDA-MB-231 (invasive ductal carcinoma, triple-negative subtype) were purchased from the National Platform of Experimental Cell Resources for Sci-Tech (Shanghai, China). Cells were cultured in D-MEM (Dulbecco’s modified Eagle’s medium) with 10% FBS (fetal bovine serum), 2 mM L-glutamine (Gibco, USA), penicillin (100 U/mL) and streptomycin (100 μg/mL) (Gen-View, USA). Cells were incubated at 37°C with humidified atmosphere of 5% CO2.
Patients and samples
The study included 42 patients who had undergone mastectomy or breast-conserving surgery for invasive ductal carcinoma of the breast at the People’s Liberation Army General Hospital between 2015 and 2017. Pathological stage was re-evaluated and determined using the present TNM classification as revised in the AJCC TNM Staging 8th Edition classification criteria. The study was approved by the Ethics Committee (Faculty of Medicine, People’s Liberation Army General Hospital).
Immunohistochemistry
The 4-um thick slices were prepared for incubating sequentially with primary antibody (EIF3C antibody, 1: 500, Catalog# PA5-62137, Invitrogen; Ki-67 antibody, 1: 500, Catalog# ab15580, Abcam), as previously described [25]. There is no criterion cutoff value for Ki-67 LI. Sara Bustreo et al. showed that Ki-67 LI cutoff value to distinguish luminal A from luminal B was 20% [26]. In our study, Ki-67 LI was more than 20% classified as a high group. The percentage of Ki-67 LI was calculated by comparing total number of tumor cells in the field (400×). For EIF3C expression, we use the same cutoff value in evaluation of immunohistochemistry.
Preparation of EIF3C siRNA lentivirus
A lentiviral system of siRNA was used to depress EIF3C expression, as previously described [15]. The targeting siRNA sequence was gtcactaaaggtctgttta designed in the website (www.invivogen.com/sirnawizard/), and homologous negative control siRNA was ttctccgaacgtgtgtgtacgt. Stem-loop DNA was inserted into the lentiviral hU6-MCS-CMV-EGFP vector (GeneChem Co., Ltd., Shanghai, China). Lentivector Expression Systems (GeneChem Co., Ltd., Shanghai, China) were used for EIF3C siRNA-expressing lentivirus. To evaluate the EIF3C knock-down efficiency in MDA-MB-231 and BT474 cells, gene level and protein were detected by RT-PCR and western blotting.
BrdU cell proliferation assay
MDA-MB-231 cells were infected by lentivirus of EIF3C-siRNA and scrambled siRNA respectively. Cell proliferation was determined by BrdU Cell Proliferation ELISA kit (Roche Applied Science, Switzerland). Seeded into 96-well plates, cells were cultured for an extra 1 to 4 days. Diluted BrdU reagents added into each well (10 μL/well). Labeling and detection of BrdU cell proliferation were determined according to the manufacturer’s procedure as previously described [14].
Colony formation assay
EIF3C-siRNA and scrambled siRNA transfected MDA-MB-231 cells were respectively cultured for five days and transferred to 6-well plates with a density of 500 cells per well. After a few weeks, the cells were fixed with 4% polyformaldehyde, and then stained with fresh prepared diluted Giemsa stain for 20 minutes. After rinsing with distilled water, colonies of 50 or more cells were detected by fluorescence microscopy.
Q-PCR
Total RNA was extracted from cells using TRIzol reagent and synthesized into cDNA by M-MLV reverse transcriptase. Quantitative real-time PCR was performed on BioRad Connect Real-Time PCR platform using SYBR Green Master Mix Kit. GAPDH was used as an internal reference. The primers were as following:
EIF3C (forward): 5′-CCATCCTCTGCCACATCTACC-3′,
EIF3C (reverse): 5′-CCACCTTCTCCTGCTCCTG-3′.
GAPDH (forward): 5′-TGACTTCAACAGCGACACCCA-3′,
GAPDH (reverse): 5′-CACCCTGTTGCTGTAGCCAAA-3′.
Western blotting
In order to estimate EIF3C expression status in MDA-MB-231 and BT474 cells western blotting analysis was performed. The proteins were probed by anti-EIF3C Antibody (1: 1,000; #HPA050112, Sigma), AKT Antibody (1: 1,000; #9272, Cell Signaling Technology), Phospho-Akt (Ser473) Antibody (1: 1,000; #9271, Cell Signaling Technology), ERK Antibody(1: 1000; #9102, Cell Signaling Technology), phospho-ERK Antibody (1: 1,000; #9101, Cell Signaling Technology), p38 MAP Kinase Antibody (1: 1,000; #9212, Cell Signaling Technology), phospho-p38 MAPK(Thr180/Tyr182) Antibody (1: 1,000;#9216S, Cell Signaling Technology),mTOR Antibody (1: 1,000; #2971, Cell Signaling Technology), Raptor Antibody (1: 1,000; #2280, Cell Signaling Technology) and detected by Electrochemiluminescence (ECL) kit (Amersham). GAPDH and β-actin were used as normalized controls (Santa Cruz Biotechnology, USA).
Apoptosis assay
We determined cell apoptosis via Annexin V-allophycocyanin (APC) staining assays. Briefly, EIF3C and scrambled siRNA infected MDA-MB-231 cells were collected after 96 hours of incubation, washed with PBS and resuspended in a final density of 1×106/mL in staining buffer. Mixed with 5 μL Annexin V-APC, 100 μL cell suspension was incubated at room temperature for 10–15 minutes. Cell apoptosis was analyzed by using FACS Calibur (Becton-Dickinson, USA).
Stress and apoptosis signaling array
According to the manufacturer’s instructions of PathScan Stress and Cell Apoptosis Signaling array kits (Cell Signaling Technology, Cell Signaling Technology), 20 candidates related with the pathways of stress and cell apoptosis signaling were determined, at the same time the phosphorylated proteins were also determined respectively.
Statistical analysis
All data are expressed as mean ±SEM values. Statistical data analysis was performed using SPSS v.17.0 (SPSS, Chicago, IL, USA) and GraphPad Prismv 5.0 with Student’s t-test, one-way or Fisher’s exact test. The significance level was set at p<0.05.
Results
Correlations of clinicopathological factors with EIF3C immunoreactivity and Ki-67 LI status
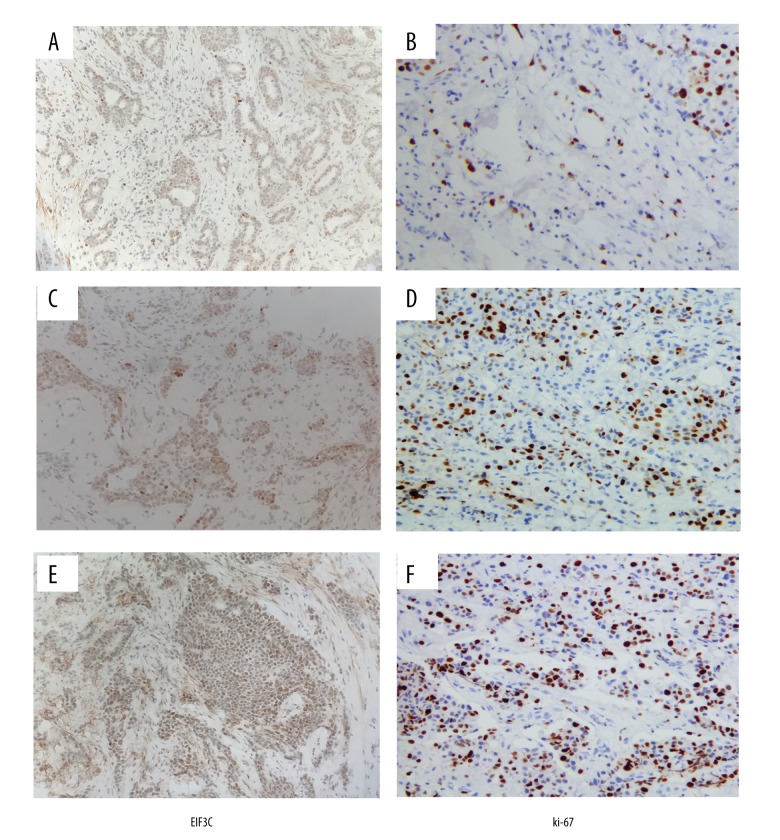
Representative images of immunostaining for EIF3C and Ki-67 examined in this study are shown in Figure 1. EIF3C positive substances with brownish-yellow particles were mainly located in the nucleus and cytoplasm of cancer cells. Ki-67 positive substances were mainly located in the nucleus. Among 42 breast tumors, there were 32 EIF3C-positive tumors (76.1%) and 17 high Ki-67 LI tumors (40.4%). The ratio of expression of EIF3C in nuclear grade 1 and 2 groups was lower than high nuclear grade 3 group (p=0.0016). The tumors with high Ki-67 LI showed significantly higher ratio of expression of high grade group (p=0.0017).There was no significant correlations between EIF3C positive and clinicopathological factors age, tumor size, nodal status, and pathological stages (Table 1).
Figure 1.
Expression of EIF3C and Ki-67 in breast cancer tissues. (A, C, E) EIF3C antigen expression in breast cancer tissues. Low expression in A, moderate expression in C and high expression in E. (B, D, F) Positive nuclear Ki-67 staining in breast cancer tissues. Low expression in B, moderate expression in D and high expression in F. (SP, magnification, 400×).
Table 1.
Correlation of clinicopathologic features and expression of EIF3C and ki-67 in patients with breast cancer.
| Clinicopathological factors | Expression of EIF3C | Expression of ki-67 | ||||||
|---|---|---|---|---|---|---|---|---|
| Total (n=42) | Negative (n=10) | Positive (n=32) | P-value | Total (n=42) | Low LI (n=25) | High LI (n=17) | P-value | |
| Age (years) | ||||||||
| ≤45 | 22 | 5 | 17 | 0.7037 | 22 | 13 | 9 | 0.6006 |
| >45 | 20 | 5 | 15 | 20 | 12 | 8 | ||
| Tumor size | ||||||||
| <5.0 cm | 36 | 8 | 28 | 0.4434 | 36 | 22 | 14 | 0.4661 |
| ≥5.0 cm | 6 | 2 | 4 | 6 | 3 | 3 | ||
| Lymph node metastasis | ||||||||
| (−) | 24 | 4 | 20 | 0.1869 | 24 | 14 | 10 | 0.5551 |
| (+) | 18 | 6 | 12 | 18 | 11 | 7 | ||
| Nuclear grade | ||||||||
| 1, 2 | 27 | 2 | 25 | 0.0016* | 27 | 21 | 6 | 0.0017* |
| 3 | 15 | 8 | 7 | 15 | 4 | 11 | ||
| Pathological stages | ||||||||
| I+II | 21 | 5 | 16 | 0.6407 | 21 | 12 | 9 | 0.5 |
| III+IV | 21 | 5 | 16 | 21 | 13 | 8 | ||
Fisher exact probability test shows the ratio of expression of EIF3C in nuclear grade 1, 2 is lower than high nuclear grade group (P<0.05). Ratio of expression of ki-67 in nuclear grade 1, 2 is lower than high nuclear grade group (P<0.05).
Lv-shRNA decreased EIF3C expression in breast cancer cells and cell proliferation was inhibited after EIF3C gene knockdown
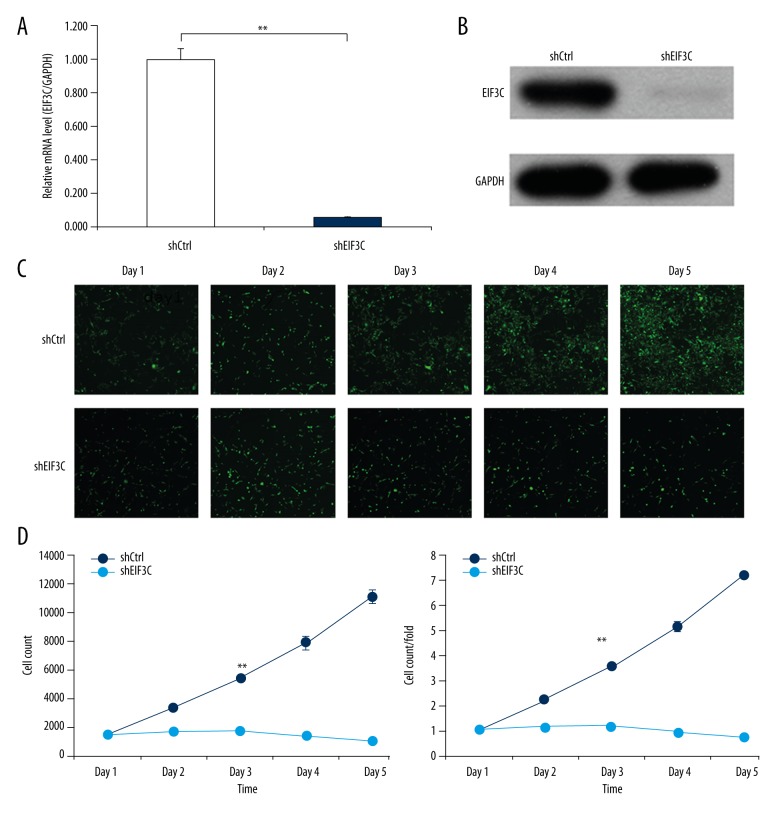
We knocked down EIF3C with lentivirus-mediated siRNA in the breast cancer cell (MDA-MB-231), while knockdown efficiency was confirmed by qPCR and western blotting. The EIF3C mRNA levels were significantly downregulated with Lv-shRNA (p<0.05, knockdown efficiency=94.2%) (Figure 2A). EIF3C protein was knocked down by Lv-shRNA which evaluated by western blotting (Figure 2B).
Figure 2.
Expression of EIF3C in breast cancer cell lines and EIF3C knockdown in MDA-MB-231 by lentivirus-based siRNA strategy. Knockdown of EIF3C expression inhibits proliferation of MDA-MB-231. (A) Efficient knock down of EIF3C at the mRNA level was achieved by lentivirus expressing EIF3C siRNA in MDA-MB-231 and normalized to GAPDH mRNA. Data shown are the mean ±SD of three independent reverse transcript quantitative PCR experiments (** p<0.01). (B) Western blot analysis showed efficient knock down of EIF3C at the protein level achieved by lentivirus expressing EIF3C siRNA in MDA-MB-231, and GAPDH was used as an internal reference. (C) Representative images of MDA-MB-231 cells infected with lentivirus expressing scrambled siRNA or EIF3C siRNA at different time points subsequent to lentivirus transfection. (D) Proliferation of MDA-MB-231 cells was monitored by Cellomics ArrayScan VTI. Proliferation data are shown as cell numbers (** p<0.01). (E) Proliferation data are shown as fold-changes of cell numbers. The results are expressed as the mean ± standard deviation of six separate experiments (** p<0.01).
To explore the role of EIF3C in the proliferation of MDA-MB-231 cells, the breast cell line MDA-MB-231 was infected with EIF3C or scrambled siRNA-expressing lentivirus. In the determination of BrdU, cell proliferation was evaluated by DNA synthesis, which was quantified by BrdU blending ratio (Figure 2C). Compared with the control group, the BrdU of EIF3C knockdown group was significantly inhibited after three days and the average was 7.8-folds p<0.05 (Figure 2D, 2E).
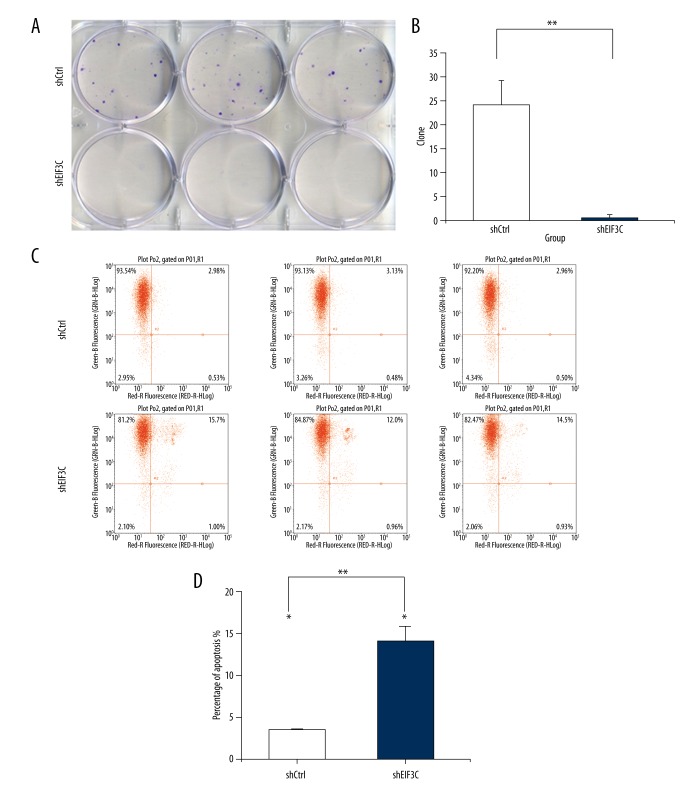
The EIF3C silencing decreased proliferation in colony formation assay and induces apoptosis in MDA-MB-231 cells
MDA-MB-231 cells could not form typical colonies in the plates. There are rare cells in the plates of EIF3C-siRNA (Figure 3A). Clone number was significantly decreased in EIF3C knockdown group compared with controls (Figure 3B) (p<0.05). With Annexin V staining, cell apoptosis was determined by FAC (Figure 3C). Cell apoptosis was significantly increased in the EIF3C-siRNA group compared with controls (3.51±0.0842 versus. 13.24±0.2307, p<0.01) (Figure 3D).
Figure 3.
Silencing of EIF3C using lentivirus-mediated EIF3C siRNA suppresses colony formation in MDA-MB-231. Downregulation of EIF3C results in cell cycle arrest and induces cell apoptosis in MDA-MB-231. The Scr-siRNA group consists of MDA-MB-231 cells infected with lentivirus expressing scrambled siRNA. EIF3C-siRNA represents MDA-MB-231 cells infected with lentivirus expressing EIF3C siRNA. (A) Representative images of colony formation of MDA-MB-231 cells in six-well plates. The Scr-siRNA group consists of the MDA-MB-231 cells infected with lentivirus expressing scrambled siRNA. The EIF3C-siRNA groups consist of the MDA-MB-231 cells infected with lentivirus expressing EIF3C siRNA. (B) Colony formation of MDA-MB-231 cells treated with EIF3C siRNA. The results are expressed as the mean ± standard deviation of three separate experiments (* p<0.01). (C) Cell apoptosis by Annexin-V staining was analyzed with FACS. (D) Cell apoptosis in cells treated with EIF3C siRNA. The graph expresses the mean ± standard deviation of cell percentage in apoptosis from three separate experiments (** p<0.01).
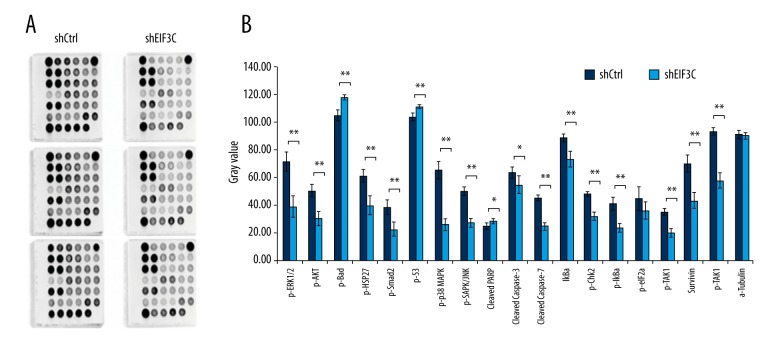
Stress and apoptosis-related gene expression patterns responding to EIF3C gene knockdown
The findings of the silent EIF3C enhanced apoptosis indicate that it may trigger certain signaling pathways directly. We hypothesized how EIF3C gene elicited cellular stress and apoptosis responses in breast cancer cells. We continued to explore the EIF3C related pattern in stress and apoptosis responses pathways. Stress and apoptosis signaling antibody array kit (Cell Signaling Technology) was utilized when 20 signaling molecules involved in the regulation of stress response and apoptosis. The expressed luminance differences were presented as histograms with statistical significance, which repeated three times (Figure 4A). Results showed in knockdown EIF3C proteins p-ERK1/2, p-Akt, p-Smad2, p-p38 MAPK, cleaved caspase-3, and cleaved caspase-7 were downregulated (Figure 4B). The results revealed the different patterns from the stress and apoptosis genes responding to silencing EIF3C in the breast cancer cell lines.
Figure 4.
Chemiluminescent array images of the PathScan Stress and Apoptosis Signaling array kit revealing various phosphorylated signaling nodes. Effects of shEIF3C on cell growth-related cell signaling in MDA-MB-231 cells. (A) Images were captured following brief exposure of the slide to standard chemiluminescent film between shCtrl and shEIF3C. (B) Array image pixel intensity ratio of phosphorylated signaling molecules. (** p<0.01, versus control).
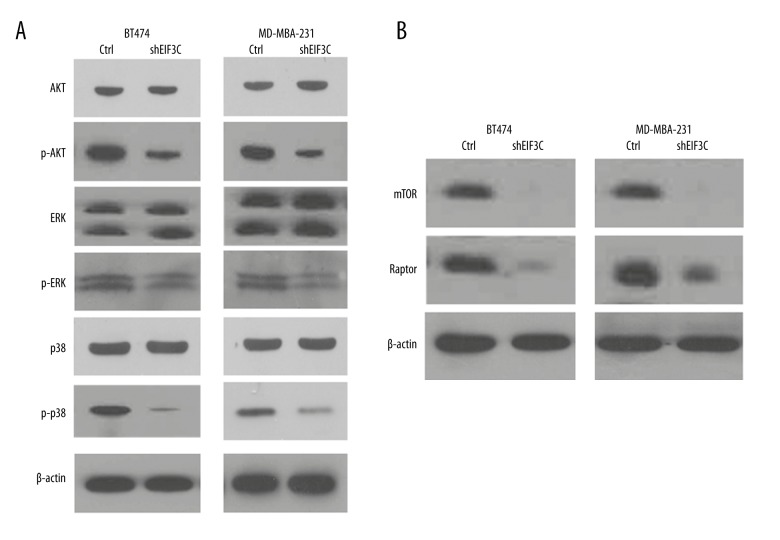
Involvement of mTOR in cell signaling pathway
We examined the molecular mechanisms by which inflammatory stress silencing EIF3C protein expression. The data showed that inflammatory stress suppresses the phosphorylation of AKT, ERK, and P38 were downregulated in BT474 and MDA-MB-231 cells (Figure 5A). We further examined the mTOR and raptor signaling pathway components which suggested that mTOR signaling pathway is involved in decreasing EIF3C translational efficiency (Figure 5B).
Figure 5.
Silencing EIF3C enhanced apoptosis in BT474 and MDA-MB-231 cells trigger related signaling pathways. (A) The AKT/ERK/P38 and their phosphorylated protein were determined between shCtrl and shEIF3C cells. (B) The mTOR signaling molecules related with EIF3C regulation were detected by western blotting.
Discussion
Breast cancer occurs in multistep process, which is related to the continuous and cumulative genetic changes caused by internal and ecological carcinogenic factors. Gene mutations of BRCA-1/2, p53, and PTEN are known to be the critical step in development of breast cancer. The proteomics, epigenetic modification, and functional proteomics in the tumorigenesis of breast cancer are not thoroughly understood. To explore protein functions, studies have looked at translation initiation focused on rate-limiting steps of protein synthesis which is highly regulated in cancer cells [27].
Mechanisms utilizing elements of protein translation, specifically in the rate-limiting step of initiation, offer potential methods to diagnose and treat cancer. Translational regulation is capable of efficiently altering specific protein levels in physiological stress conditions that are typical in breast cancer. Translation is mediated by EIFs, which have varying roles in regulating the rate of initiation [28,29]. The main mechanisms include EIF3 subunit interactions with mTOR and EIFs necessary activation through post-translational hypusination, important to the mediation of cell proliferation, apoptosis, and inflammatory response. Varying levels of EIFs in various cancer lines and stages, along with the mechanistic background, enforce the use of EIFs to regulate gene expression in cancer. During cellular stress inhibiting translation is expected to have negative/positive effects on the cells [10].
Several studies have evaluated the clinical significance of the Ki-67 LI among patients with breast cancer, and indicate high proliferation or histological grade [30,31]. For tumor size measurement, Jiang et al. reported that the anisotropy parameter of diffusion tensor imaging is an accurate method to determine its value [32]. In this study, we showed that EIF3C status may be similar to Ki-67 LI-high by clinico-pathologic features analysis. As is well known, Ki-67 LI alone predicts the prognosis factor for breast cancer patients. We continue to explore the functions of EIF3C by establishing stable knockdown EIF3C breast cell lines. We focus on the mTOR pathway which we screened with the stress and apoptosis-related gene expression assay.
We used quantitative phosphoproteomics to profile rapamycin-inhibition mediated phosphorylation changes in ribosomal proteins, which suggested that mTOR mediated the 80S ribosome formation and its binding to mRNA. It is known that mapped mTOR-regulated phosphorylation sites in eukaryotic translation initiation factors [28]. However, resistance of some cancers to mTOR-directed therapeutics has limited the success of mTOR inhibitors. Furthermore, everolimus is an oral mTOR inhibitor that is approved by the US Food and Drug Administration (FDA) in postmenopausal women with HR-positive breast cancer. Multiple trials have evaluated mTOR inhibitors in various settings in breast cancer [21,33]. The mTOR pathway, which promotes cell proliferation, presents an attractive target for cancer therapy since it is deregulated in a wide range of cancer types and a large proportion of cases of each type [33]. Beelen et al. reported that compensatory activation of the PI3K/AKT/mTOR pathway might indeed be a clinically relevant resistance mechanism resulting in acquired endocrine therapy resistance [34]. In the future, we will continue to explore the mechanism of mTOR inhibitors involved in translation initiation.
Conclusions
This study focused on the functions of EIF3C in the development of tumors. EIF3C-positive tumors showed significantly higher frequencies of the high grade group. The inhibitory effect of EIF3C knockdown on breast cancer growth was validated in colony formation assay. Furthermore, the underlying mechanisms, including the influence of EIF3C knockdown on tumor proliferation and apoptosis were explored through screening by stress and apoptosis signaling arrays. The mTOR signaling pathway has been shown to be involved in decreasing EIF3C translational efficiency. The further mechanisms remain to be explored. As potential drug targets promising cancer biomarkers and drug targets for tumor treatment will be explored. The results suggested that EIF3C may be a potential target for breast cancer treatment.
Footnotes
Conflicts of interest
None.
Source of support: The present study was supported by National Natural Science Foundation of China (grant nos. 81472183 and 81572875)
References
- 1.Fanale D, Amodeo V, Corsini LR, et al. Breast cancer genome-wide association studies: There is strength in numbers. Oncogene. 2012;31(17):2121–28. doi: 10.1038/onc.2011.408. [DOI] [PubMed] [Google Scholar]
- 2.Weir HK, Thompson TD, Soman A, et al. The past, present, and future of cancer incidence in the United States: 1975 through 2020. Cancer. 2015;121(11):1827–37. doi: 10.1002/cncr.29258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Krieger N, Bassett MT, Gomez SL. Breast and cervical cancer in 187 countries between 1980 and 2010. Lancet. 2012;379(9824):1391–92. doi: 10.1016/S0140-6736(12)60596-0. [DOI] [PubMed] [Google Scholar]
- 4.Wang Y, Waters J, Leung ML, et al. Clonal evolution in breast cancer revealed by single nucleus genome sequencing. Nature. 2014;512(7513):155–60. doi: 10.1038/nature13600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Truitt ML, Ruggero D. New frontiers in translational control of the cancer genome. Nat Rev Cancer. 2017;17(5):332. doi: 10.1038/nrc.2017.30. [DOI] [PubMed] [Google Scholar]
- 6.Aitken CE, Lorsch JR. A mechanistic overview of translation initiation in eukaryotes. Nat Struct Mol Biol. 2012;19(6):568–76. doi: 10.1038/nsmb.2303. [DOI] [PubMed] [Google Scholar]
- 7.Sonenberg N, Dever TE. Eukaryotic translation initiation factors and regulators. Curr Opin Struct Biol. 2003;13(1):56–63. doi: 10.1016/s0959-440x(03)00009-5. [DOI] [PubMed] [Google Scholar]
- 8.Jackson RJ, Hellen CU, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol. 2010;11(2):113–27. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Spilka R, Ernst C, Mehta AK, Haybaeck J. Eukaryotic translation initiation factors in cancer development and progression. Cancer Lett. 2013;340(1):9–21. doi: 10.1016/j.canlet.2013.06.019. [DOI] [PubMed] [Google Scholar]
- 10.Emmanuel R, Weinstein S, Landesman-Milo D, Peer D. eIF3c: A potential therapeutic target for cancer. Cancer Lett. 2013;336(1):158–66. doi: 10.1016/j.canlet.2013.04.026. [DOI] [PubMed] [Google Scholar]
- 11.Rezende AM, Assis LA, Nunes EC, et al. The translation initiation complex eIF3 in trypanosomatids and other pathogenic excavates – identification of conserved and divergent features based on orthologue analysis. BMC Genomics. 2014;15:1175. doi: 10.1186/1471-2164-15-1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Masutani M, Sonenberg N, Yokoyama S, Imataka H. Reconstitution reveals the functional core of mammalian eIF3. EMBO J. 2007;26(14):3373–83. doi: 10.1038/sj.emboj.7601765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee AS, Kranzusch PJ, Doudna JA, Cate JH. eIF3d is an mRNA cap-binding protein that is required for specialized translation initiation. Nature. 2016;536(7614):96–99. doi: 10.1038/nature18954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hao J, Wang Z, Wang Y, et al. Eukaryotic initiation factor 3C silencing inhibits cell proliferation and promotes apoptosis in human glioma. Oncol Rep. 2015;33(6):2954–62. doi: 10.3892/or.2015.3881. [DOI] [PubMed] [Google Scholar]
- 15.Hao J, Liang C, Jiao B. Eukaryotic translation initiation factor 3, subunit C is overexpressed and promotes cell proliferation in human glioma U-87 MG cells. Oncol Lett. 2015;9(6):2525–33. doi: 10.3892/ol.2015.3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li T, Li S, Chen D, et al. Transcriptomic analyses of RNA-binding proteins reveal eIF3c promotes cell proliferation in hepatocellular carcinoma. Cancer Sci. 2017;5(108):877–85. doi: 10.1111/cas.13209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rothe M, Ko Y, Albers P, Wernert N. Eukaryotic initiation factor 3 p110 mRNA is overexpressed in testicular seminomas. Am J Pathol. 2000;157(5):1597–604. doi: 10.1016/S0002-9440(10)64797-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Scoles DR, Yong WH, Qin Y, et al. Schwannomin inhibits tumorigenesis through direct interaction with the eukaryotic initiation factor subunit c (eIF3c) Hum Mol Genet. 2006;15(7):1059–70. doi: 10.1093/hmg/ddl021. [DOI] [PubMed] [Google Scholar]
- 19.Yu X, Zheng B, Chai R. Lentivirus-mediated knockdown of eukaryotic translation initiation factor 3 subunit D inhibits proliferation of HCT116 colon cancer cells. Biosci Rep. 2014;34(6):e00161. doi: 10.1042/BSR20140078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Z, Chen J, Sun J, et al. RNA interference-mediated silencing of eukaryotic translation initiation factor 3, subunit B (EIF3B) gene expression inhibits proliferation of colon cancer cells. World J Surg Oncol. 2012;10:119. doi: 10.1186/1477-7819-10-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vicier C, Dieci MV, Arnedos M, et al. Clinical development of mTOR inhibitors in breast cancer. Breast Cancer Res. 2014;16(1):203. doi: 10.1186/bcr3618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Massihnia D, Galvano A, Fanale D, et al. Triple negative breast cancer: Shedding light onto the role of pi3k/akt/mtor pathway. Oncotarget. 2016;7(37):60712–22. doi: 10.18632/oncotarget.10858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Merrick WC. eIF4F: A retrospective. J Biol Chem. 2015;290(40):24091–99. doi: 10.1074/jbc.R115.675280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu Y, Vertommen D, Rider MH, Lai YC. Mammalian target of rapamycin-independent S6K1 and 4E-BP1 phosphorylation during contraction in rat skeletal muscle. Cell Signal. 2013;25(9):1877–86. doi: 10.1016/j.cellsig.2013.05.005. [DOI] [PubMed] [Google Scholar]
- 25.Zhao W, Wang J, Zhu B, et al. IGFBP7 functions as a potential lymphangiogenesis inducer in non-small cell lung carcinoma. Oncol Rep. 2016;35(3):1483–92. doi: 10.3892/or.2015.4516. [DOI] [PubMed] [Google Scholar]
- 26.Bustreo S, Osella-Abate S, Cassoni P, et al. Optimal Ki67 cut-off for luminal breast cancer prognostic evaluation: A large case series study with a long-term follow-up. Breast Cancer Res Treat. 2016;157(2):363–71. doi: 10.1007/s10549-016-3817-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pola C, Formenti SC, Schneider RJ. Vitronectin-alphavbeta3 integrin engagement directs hypoxia-resistant mTOR activity and sustained protein synthesis linked to invasion by breast cancer cells. Cancer Res. 2013;73(14):4571–78. doi: 10.1158/0008-5472.CAN-13-0218. [DOI] [PubMed] [Google Scholar]
- 28.Lee AS, Kranzusch PJ, Cate JH. eIF3 targets cell-proliferation messenger RNAs for translational activation or repression. Nature. 2015;522(7554):111–14. doi: 10.1038/nature14267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sharma DK, Bressler K, Patel H, et al. Role of eukaryotic initiation factors during cellular stress and cancer progression. J Nucleic Acids. 2016;2016:8235121. doi: 10.1155/2016/8235121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Viale G, Giobbie-Hurder A, Regan MM, et al. Prognostic and predictive value of centrally reviewed Ki-67 labeling index in postmenopausal women with endocrine-responsive breast cancer: Results from Breast International Group Trial 1–98 comparing adjuvant tamoxifen with letrozole. J Clin Oncol. 2008;26(34):5569–75. doi: 10.1200/JCO.2008.17.0829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cheang MC, Chia SK, Voduc D, et al. Ki67 index, HER2 status, and prognosis of patients with luminal B breast cancer. J Natl Cancer Inst. 2009;101(10):736–50. doi: 10.1093/jnci/djp082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jiang R, Zeng X, Sun S, et al. Assessing detection, discrimination, and risk of breast cancer according to anisotropy parameters of diffusion tensor imaging. Med Sci Monit. 2016;22:1318–28. doi: 10.12659/MSM.895755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paplomata E, O’Regan R. The PI3K/AKT/mTOR pathway in breast cancer: Targets, trials and biomarkers. Ther Adv Med Oncol. 2014;6(4):154–66. doi: 10.1177/1758834014530023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Beelen K, Hoefnagel LD, Opdam M, et al. PI3K/AKT/mTOR pathway activation in primary and corresponding metastatic breast tumors after adjuvant endocrine therapy. Int J Cancer. 2014;135(5):1257–63. doi: 10.1002/ijc.28769. [DOI] [PMC free article] [PubMed] [Google Scholar]