Abstract.
We evaluated Tuberculosis-Spoligo-Rifampicin-Isoniazid Typing (TB-SPRINT), a microbead-based method for spoligotyping and detection of rifampicin and isoniazid resistance in Mycobacterium tuberculosis. For that, 67 M. tuberculosis complex strains were retrospectively selected. Membrane-based spoligotyping, restriction fragment length polymorphism, DNA sequencing/pyrosequencing of rpoB, katG, and inhA promoter, TB-SPRINT, and SNP typing were performed. Concordance between spoligotyping methods was 99.6% (2,785/2,795 spoligotype data points). For most of the discordant cases, the same lineage was assigned with both methods. Concordance between phenotypic drug susceptibility testing and TB-SPRINT for detecting rifampicin and isoniazid resistance was 98.4% (63/64) and 93.8% (60/64), respectively. Concordance between DNA sequencing/pyrosequencing and TB-SPRINT for detecting mutations in rpoB, katG, and inhA were 98.4% (60/61), 100% (64/64), and 96.9% (62/64), respectively. In conclusion, TB-SPRINT is a rapid and easy-to-perform assay for genotyping and detecting drug resistance in a single tube; therefore, it may be a useful tool to improve epidemiological surveillance.
During the last decades, emergence and spread of drug-resistant strains of Mycobacterium tuberculosis have posed challenges to tuberculosis (TB) control.1 Molecular genotyping methods have been used to detect transmission chains and outbreaks for local TB control, and to study the genetic diversity of the M. tuberculosis strains disseminated worldwide.2 A reference method for M. tuberculosis genotyping has been IS6110-restriction fragment length polymorphism (RFLP), which presents a high discriminatory power but is slow and laborious.3 Another widely used method is spoligotyping (spacer oligonucleotide typing), which is based on a polymerase chain reaction (PCR) amplification of the clustered regularly interspaced short palindromic repeats (CRISPR) locus and detection of the presence of different spacers between the repeats by reverse hybridization on membrane.4 Another strategy for TB control is the rapid detection of drug resistance to implement an adequate treatment.1 Because of the slow growth rate of M. tuberculosis, phenotypic drug susceptibility testing (DST) takes several weeks, and different molecular methods have been developed for rapid detection of mutations associated with drug resistance.5 The objective of this study was to evaluate a molecular method based on multiplex PCR and hybridization on microbeads for simultaneous spoligotyping and detection of mutations associated with rifampicin (RIF) and isoniazid (INH) resistance, termed Tuberculosis-Spoligo-Rifampicin-Isoniazid Typing (TB-SPRINT, Beamedex SAS, Orsay, France).6
A total of 67 M. tuberculosis complex strains isolated in Spain were retrospectively selected from the collection in Instituto de Investigación Sanitaria de Aragón, Zaragoza, Spain. The strains were selected including different lineages with diverse IS6110-RFLP patterns. DST was performed with VersaTREK Myco Susceptibility Kit (Trek Diagnostics, Cleveland, OH) or Bactec MGIT960 (Becton Dickinson, Sparks, MD). Critical concentrations used were 1 µg/mL for RIF, and 0.4 µg/mL and 0.1 µg/mL for INH.7 Genomic DNA was extracted from strains cultured on solid medium following the cetyltrimethylammonium bromide protocol. Conventional spoligotyping on membrane and IS6110-RFLP were performed as described previously.3,8 The spoligotyping method allowed analysis of 22 DNA samples in approximately 5 hours, whereas with IS6110-RFLP 16 samples could be analyzed in 3 days. DNA sequencing for detecting mutations in rpoB, and pyrosequencing for detecting mutations in katG codon 315 and inhA promoter were performed as previously described.9,10
Strains were blindly analyzed with the TB-SPRINT assay (Beamedex SAS, Orsay, France; www.beamedex.com).6 Briefly, the CRISPR region, rpoB, katG, and the promoter region of inhA were simultaneously amplified by PCR using dual-priming oligonucleotide primers. Subsequently, PCR product was hybridized to oligonucleotide-precoupled microbeads, and detection was performed either with the flow cytometry-based Luminex 200 system (Luminex Corp, Austin, TX) and XPONENT software for LX100/LX200 (version 3.1.871.0), or BioPlex200 (Biorad, Hercules, CA) running under Bio-Plex Manager 5.0. Raw TB-SPRINT results regarding relative fluorescence intensity values for each probe were interpreted as previously described.6,11 Numerical data were uploaded to BioNumerics version 6.1 software (Applied Maths, Sint-Martens-Latem, Belgium). The TB-SPRINT method allowed analysis of 96 DNA samples in approximately 2.5 hours. Spoligotyping patterns obtained with either membrane-spoligotyping or TB-SPRINT were compared with those in the International Spoligotyping Database (SITVITWEB) of the Pasteur Institute of Guadeloupe (http://www.pasteur-guadeloupe.fr:8081/SITVIT_ONLINE/), and Spoligotyping International Types (SITs) were assigned.
Strains were also subjected to typing of seven lineage-specific SNPs (SNP typing) with multiplex PCR, microbead-based hybridization, and detection with the Luminex 200 system as previously described.12
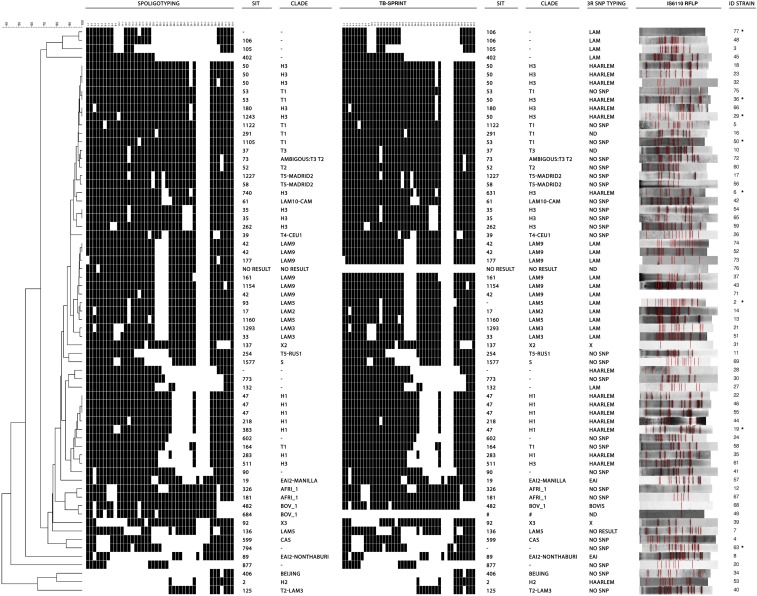
Genotyping results obtained for the 67 M. tuberculosis complex strains included in this study are shown in Figure 1. Among all the strains tested, TB-SPRINT result could not be obtained for one strain (strain 76) likely because of DNA degradation. For an additional strain (strain 49), TB-SPRINT spoligotyping result suggested the presence of two different populations, which was confirmed with analysis of mycobacterial interspersed repetitive units—variable number tandem repeats (MIRU-VNTR). Therefore, TB-SPRINT may be useful for detecting mixed infections, a relevant aspect in the management of TB patients. MIRU-VNTR typing has been also useful for this purpose, and has become another widespread used method for epidemiological studies.13
Figure 1.
Genotyping results obtained for the 67 Mycobacterium tuberculosis complex strains included in this study. From left to right are shown the dendrogram, spoligotypes, SIT, and clade obtained with membrane-based spoligotyping; spoligotypes, SIT, and clade obtained with TB-SPRINT; clade identified with SNP typing, IS6110- RFLP pattern, and strain ID. SIT and clades were identified according to SITVITWEB (http://www.pasteur-guadeloupe.fr:8081/SITVIT_ONLINE/index.jsp). Strains with any discordance between membrane-based spoligotyping and TB-SPRINT are marked with an asterisk. TB-SPRINT spoligotyping result for strain 49 (marked with #) suggested the presence of two different populations and it is not shown. TB-SPRINT = Tuberculosis-Spoligo-Rifampicin-Isoniazid typing; SNP typing = typing of lineage-specific single nucleotide polymorphisms; RFLP = restriction fragment length polymorphism; ND = not done. Dendrogram built with BioNumerics version 6.1.
According to TB-SPRINT results, distribution of families among the 65 strains with a result was as follows: H, N = 18 (27.7%); LAM, N = 13 (20.0%); T super-family (T), N = 13 (20.0%); AFRI, N = 2 (3.1%); EAI, N = 2 (3.1%); X, N = 2 (3.1%); Beijing, N = 1 (1.5%); Bovis (BOV-1), N = 1 (1.5%); Central Asian (CAS), N = 1 (1.5%); S, N = 1 (1.5%); for nine strains (13.8%) a SIT number could be assigned but the family to which they belonged was unknown; finally, the remaining two strains (3.1%) did not match any pattern of the SITVITWEB (Figure 1). Considering the 65 strains, the concordance between membrane-based spoligotyping and TB-SPRINT was 99.6% (2,785/2,795 spoligotype data points). For eight of the 65 strains, discordant results between membrane-based spoligotyping and TB-SPRINT were obtained for some spacers (Table 1).
Table 1.
Membrane-based spoligotyping, TB-SPRINT, and SNP typing results of the strains with discordant results between spoligotyping methods
| Strain | Spacer | Membrane-based spoligotyping | TB-SPRINT | SNP typing | ||||
|---|---|---|---|---|---|---|---|---|
| Spacer | SIT | Lineage | Spacer | SIT | Lineage | |||
| 2 | 37 | Present | 93 | LAM5 | Possibly absent* | Orphan | LAM5 | LAM |
| 6 | 23 | Absent | 740 | H3 | Present† | 631 | H3 | Haarlem |
| 19 | 9, 10 | Absent | 383 | H1 | Present† | 47 | H1 | Haarlem |
| 29 | 10 | Absent | 1,243 | H3 | Present† | 50 | H3 | Haarlem |
| 36 | 31 | Present | 53 | T1 | Absent‡ | 50 | H3 | Haarlem |
| 50 | 16 | Absent | 1,105 | T1 | Present† | 53 | T1 | NR§ |
| 63 | 22, 37 | Present | 794 | CAS1-Delhi | Possibly absent* | Orphan | Orphan | NR¶ |
| 77 | 16 | Absent | Orphan | Orphan | Present† | 106 | Orphan | LAM |
NR = no result; SIT = Spoligo-International type; SNP typing = typing assay based on detection of lineage-specific single nucleotide polymorphisms; TB-SPRINT = Tuberculosis-Spoligo-Rifampicin-Isoniazid typing.
Relative fluorescence intensity (RFI) values for the discordant spacers were near the cutoff.
RFI values for the discordant spacers were significantly high.
RFI values for the discordant spacers were significantly low.
No T1-specific SNP was included in this analysis.
No CAS-specific SNP was included in this analysis.
TB-SPRINT detected six clusters that were resolved by IS6110-RFLP (Figure 1). On the other hand, IS6110-RFLP detected only one cluster of two strains (strains 49 and 68) that harbored a single IS6110 copy, which was resolved by membrane-based spoligotyping (Figure 1). The higher discriminatory power of spoligotyping for strains with less than five IS6110 copies has been previously reported.14 It is of note that despite strain 49 was excluded from the TB-SPRINT analysis, the different microbead-based spoligotyping results allowed discriminating these strains.
Table 2 shows the drug susceptibility results for RIF and INH for the strains with any resistance or mutation detected by either phenotypic methods, DNA sequencing/pyrosequencing, or TB-SPRINT. TB-SPRINT showed good concordance with both phenotypic DST and DNA sequencing/pyrosequencing results (Table 3), although the number of resistant strains was low. In a previous work, Gomgnimbou and others obtained complete concordance with DNA sequencing for detecting resistance to both RIF and INH, and with phenotypic DST for RIF.6
Table 2.
Phenotypic and molecular drug susceptibility result for rifampicin and isoniazid for the strains with any resistance or mutation detected by either phenotypic drug susceptibility testing, DNA sequencing/pyrosequencing, or TB-SPRINT
| Strain | Phenotypic DST | DNA sequencing/pyrosequencing | TB-SPRINT | |||||
|---|---|---|---|---|---|---|---|---|
| RIF | INH | rpoB | katG | inhA | rpoB | katG | inhA | |
| 5 | S | S | wt | wt | C-15T | wt | wt | C-15T |
| 7 | S | S | wt | wt | wt | No 531 wt* | wt | wt |
| 16 | S | S | NR | wt | wt | wt | wt | No −15 wt† |
| 21 | S | R | wt | wt | C-15T | wt | wt | C-15T |
| 39 | S | R | wt | wt | wt | wt | wt | wt |
| 73 | R | R | S531L | wt | C-15T | S531L | wt | C-15T |
| 74 | R | R | H526D | wt | wt | H526D | wt | wt |
| 75 | R | R | H526Y | S315T | C-15T | No 516 wt* | S315T | wt |
| 76 | R | R | S531L | S315T | C-15T | NR‡ | NR‡ | NR‡ |
| 77 | R | R | S531L | wt | C-15T | S531L | wt | C-15T |
DST = drug susceptibility testing; INH = isoniazid; NR = no result; RIF = rifampicin; R = resistant; TB-SPRINT = Tuberculosis-Spoligo-Rifampicin-Isoniazid typing; S = susceptible; wt = wild-type.
The result of the specified probe was considered negative since the RFI value was low, and the strain was regarded as RIF resistant by TB-SPRINT.
The result of the specified probe was considered negative since the RFI value was low, and the strain was regarded as INH resistant by TB-SPRINT.
TB-SPRINT result could not be obtained for strain 76, likely because of DNA degradation.
Table 3.
Sensitivity, specificity, and concordance of TB-SPRINT for detecting drug resistance using phenotypic drug susceptibility testing or DNA sequencing/pyrosequencing as reference methods
| Sensitivity (no. detected/total no. [%], [95% CI]) | Specificity (no. detected/total no. [%] [95% CI]) | Concordance (no. detected/total no. [%]) | |
|---|---|---|---|
| RIF* | 4/4 (100) (39.6–100) | 59/60 (98.3) (89.9–99.9) | 63/64 (98.4) |
| INH* | 4/6 (66.7) (24.1–94.0) | 56/58 (96.6) (87.0–99.4) | 60/64 (93.8) |
| rpoB† | 4/4 (100) (39.6–100) | 56/57 (98.2) (89.4–99.9) | 60/61 (98.4) |
| katG† | 1/1 (100) (0.05–100) | 63/63 (100) (92.8–100) | 64/64 (100) |
| inhA† | 4/5 (80.0) (29.9–98.9) | 58/59 (98.3) (89.7–99.9) | 62/64 (96.9) |
CI = confidence interval; INH = isoniazid; RIF = rifampicin.
Phenotypic drug susceptibility testing was used as reference method for sensitivity, specificity, and concordance calculations.
DNA sequencing/pyrosequencing was used as reference methods for sensitivity, specificity, and concordance calculations.
Current technologies to diagnose, treat, and prevent dissemination of drug-resistant TB have limitations, and there is an increased need for more rapid, simple, sensitive, and affordable methods. In this study, we present additional evidence of the utility of TB-SPRINT, a microbead-based assay for simultaneous spoligotyping and detection of mutations in rpoB, katG, and inhA, associated with resistance to RIF and INH. Either TB-SPRINT or microbead-based spoligotyping has been successfully used in previous works.15–18 Compared with conventional membrane-based spoligotyping, this method improves the throughput and flexibility for first-line screening of potential epidemiological links. In addition, due to the multiplexing capacity, this method has the potential to simultaneously target a well-defined set of mutations associated with drug resistance, increasing the sensitivity of molecular resistance detection. Although molecular testing cannot replace phenotypic DST yet, it may be valuable as a complementary tool, especially to rule out the considered drug for treatment in case that drug resistance is detected.19 In the medium term, and subject to availability of a Luminex device, the combined approach of genotyping and detection of drug resistance may be attractive for low/middle-income countries with a high burden of drug-resistant TB where spoligotyping is routinely performed.6,15
In conclusion, spoligotyping results obtained with TB-SPRINT are in agreement with those obtained with conventional spoligotyping. In addition, TB-SPRINT is a more rapid and high-throughput assay that allows simultaneous detection of molecular resistance to RIF and INH in the same tube. Implementation of this method would be useful to improve epidemiological surveillance, and to obtain a preliminary drug susceptibility profile before phenotypic results are available, thus improving the management of TB patients and preventing further spread of drug-resistant M. tuberculosis strains.
Acknowledgments:
We thank all the laboratory members involved in this work in Universidad de Zaragoza and Hospital Universitario Miguel Servet. The authors thank also Oriol Martos, Nayanne Dantas, and Lizania Spinasse for their invaluable technical help.
Disclaimer: Partial results of this work have been previously presented in the 45th Union World Conference on Lung Health held in Barcelona (Spain) in 2014.
REFERENCES
- 1.Gunther G, 2014. Multidrug-resistant and extensively drug-resistant tuberculosis: a review of current concepts and future challenges. Clin Med (Lond) 14: 279–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wlodarska M, Johnston JC, Gardy JL, Tang P, 2015. A microbiological revolution meets an ancient disease: improving the management of tuberculosis with genomics. Clin Microbiol Rev 28: 523–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Embden JD, et al. , 1993. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J Clin Microbiol 31: 406–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kanduma E, McHugh TD, Gillespie SH, 2003. Molecular methods for Mycobacterium tuberculosis strain typing: a users guide. J Appl Microbiol 94: 781–791. [DOI] [PubMed] [Google Scholar]
- 5.Molina-Moya B, Latorre I, Lacoma A, Prat C, Domínguez J, 2014. Recent advances in tuberculosis diagnosis: IGRAs and molecular biology. Curr Treat Opt Infect Dis 6: 377e91. [Google Scholar]
- 6.Gomgnimbou MK, Hernandez-Neuta I, Panaiotov S, Bachiyska E, Palomino JC, Martin A, del Portillo P, Refregier G, Sola C, 2013. Tuberculosis-spoligo-rifampin-isoniazid typing: an all-in-one assay technique for surveillance and control of multidrug-resistant tuberculosis on Luminex devices. J Clin Microbiol 51: 3527–3534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Espasa M, Salvado M, Vicente E, Tudo G, Alcaide F, Coll P, Martin-Casabona N, Torra M, Fontanals D, Gonzalez-Martin J, 2012. Evaluation of the VersaTREK system compared to the Bactec MGIT 960 system for first-line drug susceptibility testing of Mycobacterium tuberculosis. J Clin Microbiol 50: 488–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kamerbeek J, et al. , 1997. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J Clin Microbiol 35: 907–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Portugal I, Maia S, Moniz-Pereira J, 1999. Discrimination of multidrug-resistant Mycobacterium tuberculosis IS6110 fingerprint subclusters by rpoB gene mutation analysis. J Clin Microbiol 37: 3022–3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garcia-Sierra N, Lacoma A, Prat C, Haba L, Maldonado J, Ruiz-Manzano J, Gavin P, Samper S, Ausina V, Dominguez J, 2011. Pyrosequencing for rapid molecular detection of rifampin and isoniazid resistance in Mycobacterium tuberculosis strains and clinical specimens. J Clin Microbiol 49: 3683–3686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gomgnimbou MK, Abadia E, Zhang J, Refregier G, Panaiotov S, Bachiyska E, Sola C, 2012. “Spoligoriftyping,” a dual-priming-oligonucleotide-based direct-hybridization assay for tuberculosis control with a multianalyte microbead-based hybridization system. J Clin Microbiol 50: 3172–3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abadia E, et al. , 2010. Resolving lineage assignation on Mycobacterium tuberculosis clinical isolates classified by spoligotyping with a new high-throughput 3R SNPs based method. Infect Genet Evol 10: 1066–1074. [DOI] [PubMed] [Google Scholar]
- 13.Perez-Lago L, Lirola MM, Navarro Y, Herranz M, Ruiz-Serrano MJ, Bouza E, Garcia-de-Viedma D, 2015. Co-infection with drug-susceptible and reactivated latent multidrug-resistant Mycobacterium tuberculosis. Emerg Infect Dis 21: 2098–2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Flores L, Jarlsberg LG, Kim EY, Osmond D, Grinsdale J, Kawamura M, Desmond E, Hopewell PC, Kato-Maeda M, 2010. Comparison of restriction fragment length polymorphism with the polymorphic guanine-cytosine-rich sequence and spoligotyping for differentiation of Mycobacterium tuberculosis isolates with five or fewer copies of IS6110. J Clin Microbiol 48: 575–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang J, Abadia E, Refregier G, Tafaj S, Boschiroli ML, Guillard B, Andremont A, Ruimy R, Sola C, 2010. Mycobacterium tuberculosis complex CRISPR genotyping: improving efficiency, throughput and discriminative power of ‘spoligotyping’ with new spacers and a microbead-based hybridization assay. J Med Microbiol 59: 285–294. [DOI] [PubMed] [Google Scholar]
- 16.de Freitas FA, et al. , 2014. Multidrug resistant Mycobacterium tuberculosis: a retrospective katG and rpoB mutation profile analysis in isolates from a reference center in Brazil. PLoS One 9: e104100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yasmin M, Gomgnimbou MK, Siddiqui RT, Refregier G, Sola C, 2014. Multi-drug resistant Mycobacterium tuberculosis complex genetic diversity and clues on recent transmission in Punjab, Pakistan. Infect Genet Evol 27C: 6–14. [DOI] [PubMed] [Google Scholar]
- 18.Dantas NG, et al. , 2015. Genetic diversity and molecular epidemiology of multidrug-resistant Mycobacterium tuberculosis in Minas Gerais State, Brazil. BMC Infect Dis 15: 306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dominguez J, et al. , 2016. Clinical implications of molecular drug resistance testing for Mycobacterium tuberculosis: a TBNET/RESIST-TB consensus statement. Int J Tuberc Lung Dis 20: 24–42. [DOI] [PubMed] [Google Scholar]