Abstract.
Contributions of species other than Plasmodium falciparum to human malaria in sub-Saharan Africa are uncertain. We collected blood from children aged 6 months to 10 years diagnosed with malaria by Giemsa-stained blood smears (176 subjects) or histidine rich protein-2-based rapid diagnostic tests (323 subjects) in 2016; 50 samples from each of 10 sites across Uganda were studied to identify infecting species. Of 499 available samples, 474 demonstrated plasmodial infection by polymerase chain reaction amplification of 18S ribosomal RNA genes, including P. falciparum in 472, Plasmodium malariae in 22, Plasmodium ovale in 15, and Plasmodium vivax in four; 435 were pure P. falciparum, two did not contain P. falciparum, and the remainder were mixed infections including P. falciparum. The prevalence of nonfalciparum species varied geographically. Stratifying based on recent history of indoor residual spraying (IRS) of insecticides, nonfalciparum infections were seen in 27/189 (14.8%) samples from sites that received and 13/285 (4.6%) samples from sites that did not receive IRS since 2010 (P = 0.0013). Overall, 39/474 (8.2%) samples from individuals diagnosed with malaria included nonfalciparum infections. Thus, a substantial proportion of episodes of malaria in Uganda include infections with plasmodial species other than P. falciparum.
INTRODUCTION
Four plasmodial species cause human malaria in sub-Saharan Africa, with the large majority of cases caused by Plasmodium falciparum.1 Plasmodium vivax, the other species commonly causing malaria around the world, is uncommon in most of Africa, presumably due to absence of the Duffy erythrocyte receptor in most African populations.2 The other human malaria species seen in Africa, Plasmodium malariae and Plasmodium ovale, are generally considered uncommon. However, data describing the prevalences of different plasmodial species in Africa are limited, some studies have shown high prevalences of nonfalciparum infections, and recent evidence suggests that P. vivax can infect Duffy-negative individuals.3 Data on prevalences of infecting plasmodial species are helpful, as different species require different therapeutic and control strategies.
Plasmodium falciparum is clearly the dominant human malaria pathogen in sub-Saharan Africa, and measures to treat and control malaria in this region focus on this species. Therefore, in Africa the most widely used rapid diagnostic tests (RDTs) for malaria detect histidine rich protein-2 (HRP2), which is present in P. falciparum, but not other human malaria parasites,4 and artemisinin-based combination therapies, which are targeted against drug-resistant P. falciparum infections, are recommended to treat all episodes of uncomplicated malaria.5
Plasmodium vivax has fairly high prevalence in some regions of Africa, including Ethiopia and Madagascar, but appears to be at low prevalence in most of sub-Saharan Africa.3 Reported prevalences of P. malariae and P. ovale infections have varied, with most data from west Africa.6 Microscopic surveys in the late twentieth century reported mean prevalences for P. malariae of 12–21% in Burkina Faso, Sierra Leone, Senegal, Ghana, and Cameroon.7–11 and for P. ovale of 1–14% in Sierra Leone, Senegal, and Ghana.8–10 A recent report from Benin showed seroprevalence of 68% for P. malariae and 57% for P. ovale.12 Seroprevalence for P. malariae and P. ovale was also high at three sites in Zimbabwe.13 A recent survey of children from two provinces of Zambia that used molecular diagnostics found that 88% of isolates with plasmodial infection had pure P. falciparum infections, 11% mixed infections, and 1% nonfalciparum monoinfections.14 In Kenya, P. malariae was identified by molecular analysis in 5% of asymptomatic and 3% of symptomatic malaria infections.15 In Uganda, molecular analysis of samples from a Kampala cohort for which genotyping did not identify P. falciparum suggested that 4.6% of plasmodial infections were due to P. malariae, 0.9% to P. ovale, and 0.5% to P. vivax.16 A study of children from eastern Uganda noted frequent mixed infections when malaria was diagnosed by molecular methods, with 41% of infections P. falciparum and P. malariae, 9% P. falciparum and P. ovale, and 8% all three of these species.17 To gain further insights into the species composition in symptomatic malaria, we surveyed the prevalence of different plasmodial species in samples from children presenting with malaria at multiple sites in Uganda.
MATERIALS AND METHODS
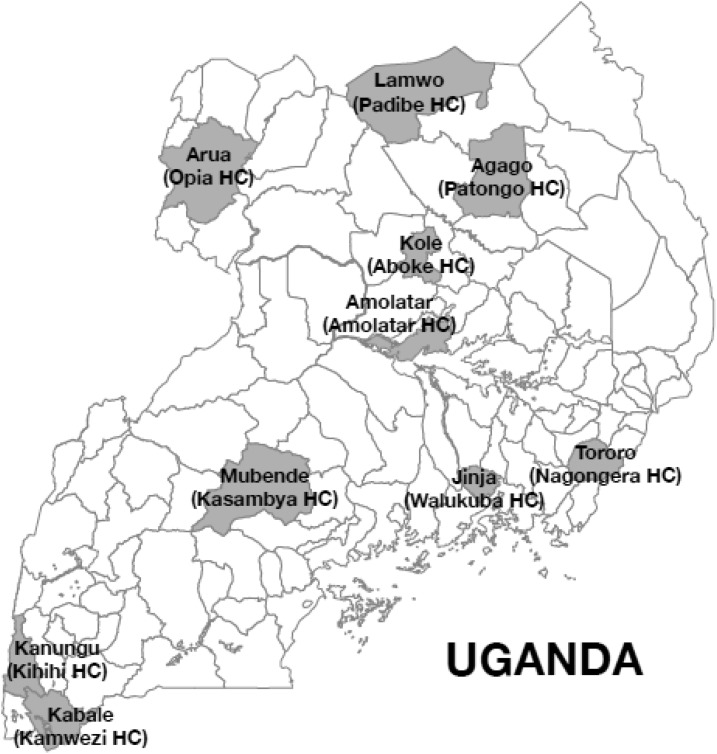
Ten sites around Uganda, all part of a regular surveillance program, were selected as representative of different regions of the country (Figure 1).18 At each site health-care personnel evaluated children 6 months to 10 years of age with clinical syndromes suggestive of malaria with either Giemsa stained blood smears or an HRP2-based RDT, following World Health Organization guidelines, and depending on local availability of these tests. For both microscopy and RDTs, participating laboratories underwent regular quality control training, but repeat readings of individual tests were not performed. Children diagnosed with malaria and their parents or guardians were approached for enrollment, and if consent was obtained, blood was collected as four blood spots dried on Whatman 3MM filter paper (Whatman GE). Filter paper samples were stored in zip lock bags with desiccant at room temperature and transported to our laboratory in Kampala for evaluation. This study was approved by the Makerere University Research and Ethics Committee, the Uganda National Council for Science and Technology, and the University of California, San Francisco Committee on Human Research.
Figure 1.
Study districts and health centers.
Genomic DNA was extracted from blood spots using Chelex 100.19 From each spot, 6-mm-diameter discs were cut using a clean hole punch and soaked overnight in 1 mL of phosphate-buffered saline (PBS) with 50 µL 10% saponin at 4°C. The solution was then aspirated, the discs were soaked in 1 mL PBS for 15–30 minutes at 4°C, and the solution was again aspirated. We then added 50 μL 20% Chelex 100 (BioRad Laboratories) and 100 μL DNAse free water, the mixture was heated for 10 minutes at 95°C, with vortexing every after 2 minutes, tubes were centrifuged for 5 minutes, supernatants were placed in fresh tubes, these were centrifuged for 10 minutes, and supernatants were again transferred to fresh tubes.
For identification of plasmodial DNA we performed nested PCR with primers specific for the 18S small subunit ribosomal RNA gene of all human plasmodial species and then species-specific primers, as previously described.20,21 PCR reactions were performed in 25 μL containing 1× standard Taq buffer, 200 µM deoxynucleoside triphosphates, 200 µM of each primer, 2 µL of template DNA (from Chelex extraction or the prior cycle of PCR), and 1 unit of Taq polymerase. PCR conditions were an initial denaturation at 94°C for 1 minute; followed by 35 cycles (30 cycles for the nested reaction) of 94°C for 1 minute, 58°C for 2 minutes, and 72°C for 5 minutes; followed by a final incubation at 72°C for 5 minutes. All reactions included negative controls (water) and positive controls, obtained from the Malaria Research and Reference Reagent Resource Center, BEI Resources. Primers for the first PCR reaction were rPLUf: 5′-TTAAATTTGTTGCAGTTAAAACG-3′ and rPLUr: 5′-CCTGTTGTTGCCTTAAACTTC-3′. Primers for the second (nested) reactions were rFALr: 5′-TTAAACTGGTTTGGGAAAACCAAATAT-3′ and rFALf: 5′-ACACAATGAACTCAATCATGACTACCC-3′ for P. falciparum; rMALr: 5′-AAAATTCCCATGCATAAAAAATTATACAAA-3′ and rMALf: 5′-ATAACATAGTTGTACGTTAAGAATAACCG-3′ for P. malariae; rOVAr: 5′-GGAAAAGGACACATTAATTGTATCCTA 3′ and rOVAf: 5′-ATCTCTTTTGCTATTTTTTAGTATTGG-3′ for P. ovale; and rVIVr: 5′-ACTTCCAAGCCGAAGCAAAGAAAGTCC-3′ and rVIVf: 5′-CGCTTCTAGCTTAATCCACATAACTGA-3′ for P. vivax.
PCR products were resolved by electrophoresis on 2% agarose gels, stained with ethidium bromide, and visualized by ultraviolet illumination. Sizes of amplicons were identified based on comparison with standard fragments of known size. Results were deemed valid when the negative control had no amplification product and positive controls had the expected band sizes. Samples with ambiguous or negative results were repeated with twice the volume of DNA template, and repeatedly negative results were reported as such. Positive nonfalciparum results were also repeated, and only consistently positive samples were reported. Statistical analyses used the Fisher’s exact and t tests. A P value < 0.05 was considered statistically significant.
RESULTS
Study population.
PCR assays were conducted for a total of 499 samples, each from a child diagnosed at a Ugandan health center with malaria in 2016. The mean age of study participants was 4.67 years. Malaria was diagnosed using available methodology; diagnoses were based on blood smears for 176 and RDTs for 323 children.
Plasmodial species identified.
After repeated PCR as described in Methods, no results were deemed ambiguous. PCR identified plasmodial infection in 474/499 (95.0%) samples. The 25 children who did not have the diagnosis confirmed by highly sensitive PCR methods included 5/176 (2.8%) children diagnosed by microscopy and 20/323 (6.2%) children diagnosed by RDT. Plasmodium falciparum was identified in 472 samples, P. malariae in 22, P. ovale in 15, and P. vivax in four (Table 1). Of all infections, 435 were pure P. falciparum, two did not contain P. falciparum (one pure P. malariae, one P. malariae/P. vivax), and the remainder were mixed infections including P. falciparum (Table 2). Nonfalciparum species varied geographically, with 0–13 samples demonstrating P. malariae, 0–10 demonstrating P. ovale, and 0–2 demonstrating P. vivax at different sites. The mean age of subjects was 4.64 years for those with pure P. falciparum infection and 4.96 years for those with pure or mixed nonfalciparum infections (P = 0.49). No pure nonfalciparum infections were identified in RDT-positive samples. Overall, 39/474 (8.2%) samples with molecular identification of plasmodial infection included nonfalciparum infections.
Table 1.
Plasmodium species identified at 10 sites in Uganda
| Site | IRS duration | N | Method of diagnosis | Plasmodium species identified (% of total) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| District | Health Center | Microscopy | RDT | Any | P. falciparum | P. malariae | P. ovale | P. vivax | ||
| Agago | Patongo | 2010–2014 | 50 | 1 | 49 | 49 | 49 (100) | 0 | 2 (4.1) | 0 |
| Amolatar | Amolatar | 2014– | 50 | 2 | 48 | 50 | 50 (100) | 13 (26) | 0 | 2 (4.0) |
| Arua | Opia | None | 50 | 0 | 50 | 42 | 42 (100) | 0 | 0 | 0 |
| Jinja | Walukuba | None | 50 | 35 | 15 | 49 | 47 (95.9) | 5 (10.2) | 1 (2.0) | 2 (4.1) |
| Kabale | Kamwezi | None | 50 | 50 | 0 | 49 | 49 (100) | 0 | 0 | 0 |
| Kanungu | Kihihi | None | 50 | 50 | 0 | 48 | 48 (100) | 1 (2.1) | 1 (2.1) | 0 |
| Kole | Aboke | 2010–2014 | 50 | 0 | 50 | 46 | 46 (100) | 0 | 9 (19.6) | 0 |
| Lamwo | Padibe | 2010–2014 | 49 | 0 | 49 | 44 | 44 (100) | 0 | 1 (2.3) | 0 |
| Mubende | Kasambya | None | 50 | 15 | 35 | 48 | 48 (100) | 3 (6.3) | 1 (2.1) | 0 |
| Tororo | Nagongera | 2014– | 50 | 23 | 27 | 49 | 49 (100) | 0 | 0 | 0 |
| Total | 499 | 176 | 323 | 474 | 472 (99.6) | 22 (4.6) | 15 (3.2) | 4 (0.8) | ||
IRS = indoor residual spraying; RDT = rapid diagnostic test.
Table 2.
Distribution of plasmodial infections
| Species | Number (% of total) |
|---|---|
| P. falciparum | 435 (91.8) |
| P. falciparum and P. malariae | 19 (4.0) |
| P. falciparum and P. ovale | 14 (3.0) |
| P. falciparum and P. vivax | 3 (0.6) |
| P. falciparum, P. malariae, and P. ovale | 1 (0.2) |
| P. malariae | 1 (0.2) |
| P. malariae and P. vivax | 1 (0.2) |
Potential impact of indoor residual spraying of insecticides on plasmodial species.
The prevalence of different plasmodial species might be impacted by malaria control activities. Indoor residual spraying (IRS) with carbamate or organophosphate insecticides was instituted at multiple sites in Uganda from 2010 to 2016. The use of IRS has been associated with remarkable decreases in the prevalence and incidence of malaria in some regions of the country.22 We stratified speciation results based on the use of IRS since 2010 at study sites. Five sites had IRS programs, either from 2010 to 2014 (three sites) or 2014–2016 (two sites; Table 1). Stratifying based on history of IRS at collection sites, mixed species and pure nonfalciparum infections were seen in 27/189 (14.8%) samples from IRS sites and 13/285 (4.6%) samples from non-IRS sites (P = 0.0013). The prevalence of nonfalciparum infections was not significantly different between sites where IRS ended in 2014 (12/149; 8.1%) and sites with ongoing IRS (15/100; 15.0%; P = 0.15), but sample sizes were small for this comparison, and prevalences differed markedly between sites.
DISCUSSION
We explored the presence of different species of malaria parasites in children diagnosed with malaria at health centers across Uganda. As expected, PCR verified P. falciparum infections in nearly all malaria cases. Only two of 499 infections (two of 176 infections diagnosed by microscopy) did not include P. falciparum. However, 8.2% of infections included nonfalciparum parasites. Thus, although P. falciparum was clearly the dominant species causing malaria in Ugandan children, other species were quite common when children presented with symptomatic malaria.
Of children diagnosed with malaria at Ugandan health centers, 5% did not have the diagnosis confirmed by highly sensitive PCR methods (3% diagnosed by microscopy and 6% diagnosed by RDT). These false positive results might be due to errors by clinic personnel or, for the RDT assay, persistence of P. falciparum antigen after an infection was cleared from the bloodstream.4 In addition, there is the possibility of false-negative PCR results on samples stored at room temperature before analysis.
We identified an association between recent use of IRS at a surveillance site and increased prevalence of nonfalciparum malaria infections. This result suggests that decreasing overall malaria prevalence may be accompanied by increasing prevalence of nonfalciparum infections. However, inferences based on this analysis should be cautious, as IRS was conducted over two different time frames, and discontinuation of IRS at some sites was soon followed by marked increases in prevalence.23 Thus, impacts on malaria prevalence likely varied between sites with different IRS schedules. Indeed, species prevalence varied greatly between sites, likely due to factors in addition to overall parasite prevalence. There are plausible explanations for increasing relative prevalence of nonfalciparum species with decreasing malaria transmission, but more studies are needed to determine whether IRS and other malaria control measures will in fact lead to changes in the relative prevalence of different plasmodial species.
Our study had an important limitation. Health centers used either of two different diagnostic methods for malaria, microscopy, which identifies all species, albeit with uncertain accuracy, and an HRP2-based RDT, which only detects P. falciparum. Thus, for about two-thirds of our samples pure nonfalciparum infections would not have been recognized. Cases with diagnosis based on microscopy offered data on the overall prevalence of nonfalciparum malaria, including monoinfections; 2/176 subjects had nonfalciparum malaria infections. Diagnosis based on RDT would be expected to miss nonfalciparum infections, and indeed none of these cases had pure nonfalciparum infections. Despite this limitation, our study offers a useful measure of the prevalence of nonfalciparum malaria in children diagnosed by standard methods at Ugandan health centers. Many infections included nonfalciparum parasites, with important ramifications for malaria control.
The identification of one species, P. vivax, in Ugandan children with malaria might be seen as surprising. It is well known that P. vivax typically requires the Duffy antigen for entry into erythrocytes and that most Africans do not express this receptor, explaining much lower prevalence of P. vivax in most of sub-Saharan Africa compared with that in other malarious regions.2,3 However, recent studies suggest transmission of P. vivax infections across Africa, due to the presence of a large minority of Duffy-positive hosts on the continent and/or infection in Duffy-negative individuals.3,24,25 Data on P. vivax infections in Uganda are limited. In a prior analysis of 1,464 samples from a cohort study in Kampala from 2004 to 2008 that identified nonfalciparum species only when initial genotyping did not identify P. falciparum, 0.5% of infections were due to P. vivax.16 Our current results add to these and additional data from multiple sources3 indicating that P. vivax is present at low levels in Uganda and that it contributes to occasional episodes of symptomatic malaria.
The identification of nonfalciparum infections in nearly 10% of children diagnosed with malaria at Ugandan health centers has important ramifications for malaria control. First, two species, P. vivax and P. ovale, optimally require therapy with primaquine to eliminate dormant liver hypnozoites, and thus provide definitive cure. Primaquine is little used in Africa due to logistical and safety challenges, in particular potential toxicity in those with glucose-6-phosphate dehydrogenase deficiency. Without primaquine therapy, relapses of P. vivax and P. ovale infections after therapy directed against asexual parasites may be seen. Second, standard artemisinin-based combination therapy may have lower efficacy against P. malariae infection than against P. falciparum infection, due to relatively long preerythrocytic and erythrocytic life cycles of P. malariae. This factor may explain anecdotal reports of recurrent episodes of malaria caused by P. malariae after treatment of P. falciparum.26 Third, it can be expected that each plasmodial species occupies different ecological niches. Prior reports noted relatively increased prevalence of P. malariae in savanna regions and of P. ovale in forested regions,10 and higher relative prevalence of P. malariae, compared with P. falciparum during the dry season.6 Ecological differences between sites might similarly explain differences in parasite prevalence in children presenting with malaria at different sites in Uganda. Attention to ecological differences may allow optimized focus on nonfalciparum species with particular treatment requirements. Finally, as we move toward malaria elimination, nonfalciparum parasites may be responsible for an increasing proportion of infections, due to the factors noted earlier, and as suggested by the association seen in our study between recent IRS and nonfalciparum malaria. Consistent with this possibility, a recent report studying areas of relatively low transmission intensity in southwestern Uganda noted a surprisingly high prevalence of 45% for P. malariae in samples from asymptomatic parasitemic children.27 Continued surveillance for the species causing malaria in regions undergoing interventions that decrease transmission intensity will be warranted.
Acknowledgments:
We thank the staffs of the 10 study health centers and Michelle Roh for assistance with preparation of the figure. This study (AI089674 and AI075045) and trainees (TW007375 and TW010132) were supported by funding from the National Institutes of Health.
REFERENCES
- 1.White NJ, Pukrittayakamee S, Hien TT, Faiz MA, Mokuolu OA, Dondorp AM, 2014. Malaria. Lancet 383: 723–735. [DOI] [PubMed] [Google Scholar]
- 2.Howes RE, et al. , 2011. The global distribution of the Duffy blood group. Nat Commun 2: 266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Howes RE, et al. , 2015. Plasmodium vivax transmission in Africa. PLoS Negl Trop Dis 9: e0004222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mouatcho JC, Goldring JP, 2013. Malaria rapid diagnostic tests: challenges and prospects. J Med Microbiol 62: 1491–1505. [DOI] [PubMed] [Google Scholar]
- 5.World Health Organization, 2015. Guidelines for the Treatment of Malaria. Geneva, Switzerland: WHO. [Google Scholar]
- 6.Mueller I, Zimmerman PA, Reeder JC, 2007. Plasmodium malariae and Plasmodium ovale–the “bashful” malaria parasites. Trends Parasitol 23: 278–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boudin C, Robert V, Verhave JP, Carnevale P, Ambroise-Thomas P, 1991. Plasmodium falciparum and P. malariae epidemiology in a West African village. Bull World Health Organ 69: 199–205. [PMC free article] [PubMed] [Google Scholar]
- 8.Barnish G, Maude GH, Bockarie MJ, Eggelte TA, Greenwood BM, 1993. The epidemiology of malaria in southern Sierra Leone. Parassitologia 35 (Suppl): 1–4. [PubMed] [Google Scholar]
- 9.Trape JF, et al. , 1994. The Dielmo project: a longitudinal study of natural malaria infection and the mechanisms of protective immunity in a community living in a holoendemic area of Senegal. Am J Trop Med Hyg 51: 123–137. [DOI] [PubMed] [Google Scholar]
- 10.Browne EN, Frimpong E, Sievertsen J, Hagen J, Hamelmann C, Dietz K, Horstmann RD, Burchard GD, 2000. Malariometric update for the rainforest and savanna of Ashanti region, Ghana. Ann Trop Med Parasitol 94: 15–22. [PubMed] [Google Scholar]
- 11.Bonnet S, Paul RE, Gouagna C, Safeukui I, Meunier JY, Gounoue R, Boudin C, 2002. Level and dynamics of malaria transmission and morbidity in an equatorial area of South Cameroon. Trop Med Int Health 7: 249–256. [DOI] [PubMed] [Google Scholar]
- 12.Doderer-Lang C, et al. , 2014. The ears of the African elephant: unexpected high seroprevalence of Plasmodium ovale and Plasmodium malariae in healthy populations in Western Africa. Malar J 13: 240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Amanfo SA, Mduluza T, Midzi N, Cavanagh DR, Mutapi F, 2016. Seroepidemiology of Plasmodium species infections in Zimbabwean population. Malar J 15: 267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sitali L, Chipeta J, Miller JM, Moonga HB, Kumar N, Moss WJ, Michelo C, 2015. Patterns of mixed Plasmodium species infections among children six years and under in selected malaria hyper-endemic communities of Zambia: population-based survey observations. BMC Infect Dis 15: 204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lo E, Nguyen K, Nguyen J, Hemming-Schroeder E, Xu J, Etemesi H, Githeko A, Yan G, 2017. Plasmodium malariae prevalence and csp gene diversity, Kenya, 2014 and 2015. Emerg Infect Dis 23: 601–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clark TD, Njama-Meya D, Nzarubara B, Maiteki-Sebuguzi C, Greenhouse B, Staedke SG, Kamya MR, Dorsey G, Rosenthal PJ, 2010. Incidence of malaria and efficacy of combination antimalarial therapies over 4 years in an urban cohort of Ugandan children. PLoS One 5: e11759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Betson M, Sousa-Figueiredo JC, Atuhaire A, Arinaitwe M, Adriko M, Mwesigwa G, Nabonge J, Kabatereine NB, Sutherland CJ, Stothard JR, 2014. Detection of persistent Plasmodium spp. infections in Ugandan children after artemether-lumefantrine treatment. Parasitology 141: 1880–1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yeka A, et al. , 2012. Malaria in Uganda: challenges to control on the long road to elimination: I. Epidemiology and current control efforts. Acta Trop 121: 184–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Plowe CV, Djimde A, Bouare M, Doumbo O, Wellems TE, 1995. Pyrimethamine and proguanil resistance-conferring mutations in Plasmodium falciparum dihydrofolate reductase: polymerase chain reaction methods for surveillance in Africa. Am J Trop Med Hyg 52: 565–568. [DOI] [PubMed] [Google Scholar]
- 20.Snounou G, Viriyakosol S, Jarra W, Thaithong S, Brown KN, 1993. Identification of the four human malaria parasite species in field samples by the polymerase chain reaction and detection of a high prevalence of mixed infections. Mol Biochem Parasitol 58: 283–292. [DOI] [PubMed] [Google Scholar]
- 21.Nsobya SL, Parikh S, Kironde F, Lubega G, Kamya MR, Rosenthal PJ, Dorsey G, 2004. Molecular evaluation of the natural history of asymptomatic parasitemia in Ugandan children. J Infect Dis 189: 2220–2226. [DOI] [PubMed] [Google Scholar]
- 22.Katureebe A, et al. , 2016. Measures of malaria burden after long-lasting insecticidal net distribution and indoor residual spraying at three sites in Uganda: a prospective observational study. PLoS Med 13: e1002167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Raouf S, et al. , 2017. Resurgence of malaria following discontinuation of indoor residual spraying of insecticide in a previously high transmission intensity area of Uganda. Clin Infect Dis 65: 453–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Poirier P, et al. , 2016. The hide and seek of Plasmodium vivax in West Africa: report from a large-scale study in Beninese asymptomatic subjects. Malar J 15: 570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rogier E, Moss DM, Chard AN, Trinies V, Doumbia S, Freeman MC, Lammie PJ, 2017. Evaluation of immunoglobulin G responses to Plasmodium falciparum and Plasmodium vivax in Malian school children using multiplex bead assay. Am J Trop Med Hyg 96: 312–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Calleri G, Balbiano R, Caramello P, 2013. Are artemisinin-based combination therapies effective against Plasmodium malariae? J Antimicrob Chemother 68: 1447–1448. [DOI] [PubMed] [Google Scholar]
- 27.Roh ME, Oyet C, Orikiriza P, Wade M, Kiwanuka GN, Mwanga-Amumpaire J, Parikh S, Boum Y, 2016. Asymptomatic Plasmodium infections in children in low malaria transmission setting, southwestern Uganda. Emerg Infect Dis 22: 1494–1498. [DOI] [PMC free article] [PubMed] [Google Scholar]