Abstract
Kinase-mediated signaling cascades regulate a number of different molecular mechanisms involved in cellular homeostasis, and are viewed as one of the most common intracellular processes that are robustly dysregulated in the pathophysiology of mood disorders such as depression. Newly emerged, rapid acting antidepressants are able to achieve therapeutic improvement, possibly in part, through stimulating activity of kinase-dependent signaling pathways. Thus, advancements in our understanding of how kinases may contribute to development and treatment of depression seem crucial. However, current investigations are limited to a single or small number of kinases and are unable to detect novel kinases. Here, we review fast developing kinome profiling approaches that allow identification of multiple kinases and kinase network connections simultaneously, analyze technical limitation and challenges, and discuss their future applications to mood disorders and antidepressant treatment.
Keywords: Fast-acting antidepressant, Major depressive disorders, Protein kinase, Kinase inhibitor, Kinome profiling
Pathophysiology of mood disorders and role of protein kinases
Major depressive disorder (MDD) is one of the most common and discomforting psychiatric illnesses that are characterized by high prevalence (approximately 6–7% of the US adult population) and pertinent socioeconomic burden (Simon, 2003; Kessler et al., 2005). Broad range of clinical symptoms associated with MDD, including feelings of sadness, helplessness, despair and anhedonia, suggests a complex pathophysiology likely due to alterations in molecular pathways and cellular function at multiple sites in the central nervous system (CNS) (Duric & Duman, 2013). Recent preclinical and clinical evidence supports this notion showing that chronic stress and depression can negatively affect the CNS and brain resulting in region-specific alterations in gene expression, protein activity, intracellular signaling, and neurotransmitter release. Prolonged stress-related dysregulation of these basic molecular mechanisms eventually can lead to cellular destabilization and loss of synaptic connectivity followed by overall reduction in cell function and survival (i.e., neurons and glia) that seem to underlie changes in behavioral phenotype. Several limbic brain areas within the mood-regulating neurocircuitry, primarily the hippocampus and prefrontal cortex (PFC), seem especially vulnerable to these negative effects of chronic stress (Duric et al., 2016). The hippocampus and PFC are involved in control of stress responses, hormonal homeostasis and cognitive function, and thus, significant alterations or loss of function in these brain regions may lead to development of behavioral deficits and mental illness, such as depression and anxiety. Indeed, examinations of depressed human brains and stressed rodent brains show functional deficits of the hippocampus and PFC thought to be associated with robust neuronal atrophy and altered morphology (e.g., decreased dendritic arborization and spine density), diminished production of new cells and overall reduction in tissue volume (Sheline et al., 1996; Cotter et al., 2001; Stockmeier et al., 2004; Videbech & Ravnkilde, 2004). Ultimately, the effects of chronic stress on function of individual neurons in specific regions, such as the hippocampus and PFC, can also negatively affect the connectivity and function of entire neural networks including both limbic and cortical brain areas (Duric & Duman, 2013).
To date, the exact molecular events and cellular remodeling mechanisms involved in the above mentioned stress-evoked brain alterations and development of depressive behavioral phenotype remain relatively unknown. The traditionally prevalent monoamine hypothesis links monoamine deficiencies with depression and other mood disorders (Hirschfeld, 2000), which has led to the development of currently available antidepressants. Going beyond the monoamine hypothesis, recent human and animal studies have related stress and antidepressant responses to alterations in activity of a number of different molecular modulators and intracellular pathways, such as the neurotrophic factor signaling [e.g., decreased levels of brain-derived neurotrophic factor (BDNF)], that are involved in regulation of neuronal growth, differentiation and survival (Duric et al., 2016). Furthermore, many of these cellular processes have been linked to changes in activity of specific protein kinases and related signaling cascades, primarily including the mitogen-activated protein kinase (MAPK), mammalian target of rapamycin (mTOR), and glycogen-synthase kinase-3 (GSK-3) pathways (Figure 1). Specific roles of these kinases in depression pathophysiology and antidepressant responses will be reviewed in detail in the following sections.
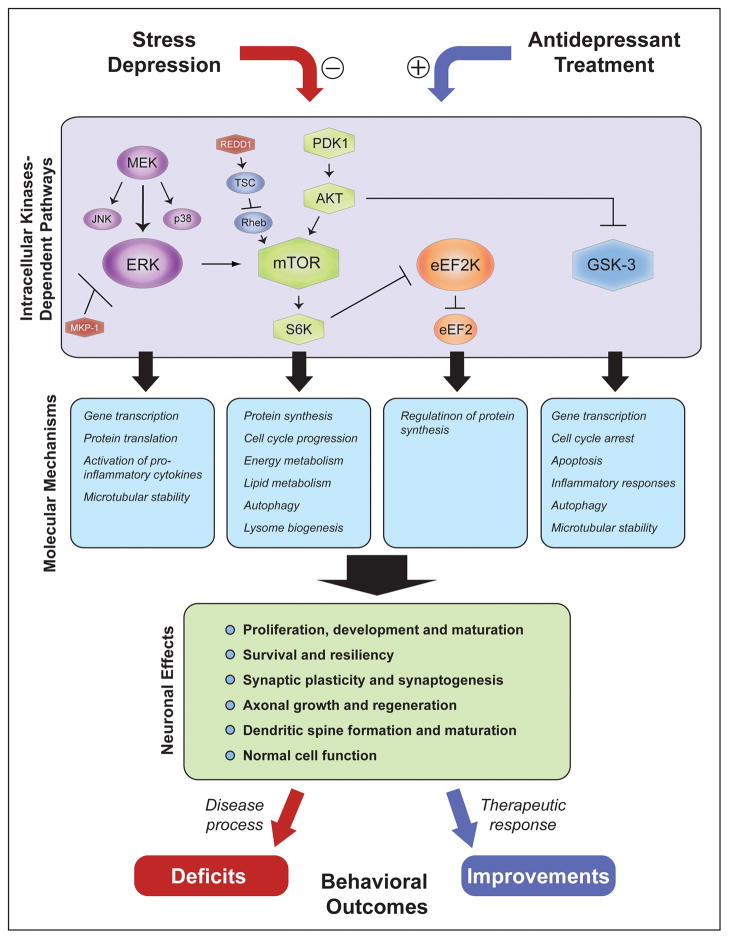
Figure 1. Kinase-regulated intracellular signaling cascades play important roles in cellular actions of chronic stress and antidepressant treatments.
Kinases pathways such as MAPK, mTOR, GSK-3 and eEF2K are involved in regulation of basic molecular mechanisms including gene transcription, protein synthesis, autophagy, metabolism and cell cycle that control numerous cellular functions critical for neuronal plasticity and survival. Stress-related negative regulation of these mechanisms can lead to neuronal dysfunction at specific brain regions within mood-controlling neurocircuitry resulting in behavioral deficits associated with depressive state. In contrast, antidepressant-related positive regulation of these kinase-dependent neural mechanisms produces improvements in behavioral outcomes associated with therapeutic response and remission.
Mitogen-activated protein kinase (MAPK)
Postmortem studies of depressed human brains have showed decreased activity and expression of several different MAPK proteins in the hippocampus, especially extracellular signal-regulated kinases 1/2 and 5 (i.e., ERK1/2 and ERK5, respectively) (Dwivedi et al., 2001; Dwivedi et al., 2006; Dwivedi et al., 2007). Moreover, animal studies further demonstrated that antidepressant treatments activate MAPK signaling, while pharmacological inhibition of these pathways diminishes effects of antidepressants and can result in development of depressive-like behaviors (Kodama et al., 2005; Duman et al., 2007). MAPK signaling is thought to promote neuronal plasticity, survival and neuroprotection (Grewal et al., 1999), and prolonged attenuation of this pathway may have severe adverse effects on neuronal function that could lead to depressive pathophysiology. Indeed, expression of MAPK phosphatase-1 (MKP-1), one of the main negative regulators of MAPK cascade, was found significantly increased in the hippocampus of depressed human brains, as well as animals exposed to chronic stress (Duric et al., 2010). Activation of hippocampal MKP-1 in non-stressed rodent hippocampus was further shown to be sufficient to evoke depressive-like behaviors, while deletion of MKP-1 (i.e., MKP-1 knockout mice) was found to increase resiliency to stress (Duric et al., 2010). Thus, sustained disruption or activation of MAPK signaling may represent one of the key cellular mechanisms contributing to development and treatment of depression, respectively.
Glycogen-synthase kinase-3 (GSK-3)
GSK-3 is a Ser/Thr kinase that plays an important role in control of numerous cellular processes and inflammatory responses. Generally, activation of intracellular GSK-3 promotes cell cycle arrest and apoptosis, while inhibition of GSK-3 function stimulates cell proliferation and resiliency (Kockeritz et al., 2006). Recently, the regulation of GSK-3 signaling has also been suggested to play an active role in antidepressant responses. Several preclinical studies have demonstrated that administration of both rapid-acting antidepressant agents, such as ketamine, as well as conventional antidepressants inhibits GSK-3 activity in rodent brains by serine phosphorylation (Beurel, 2014). Furthermore, reduction in GSK-3 function was shown to attenuate depressive-like behaviors in rodent models of depression, while genetic alterations causing expression of constitutively active GSK-3 result in resistance to antidepressant treatments (Beurel et al., 2011). In addition, inhibition of GSK-3 is also thought to promote increases in BDNF, which further implicates GSK-3 in neurotrophic factor-mediated antidepressant mechanisms (Zunszain et al., 2013).
Protein kinase pathways targeted by rapid acting antidepressant ketamine
Currently available antidepressants, whose therapeutic mechanisms of action are primarily based on the monoamine hypothesis of depression, are among the most prescribed drugs, including Monoamine Oxidase Inhibiters (MAOIs), Tricyclic Antidepressants (TCAs), and Selective Serotonin Reuptake Inhibitors (SSRIs). It has been increasingly appreciated that the mechanisms of their antidepressant effects depend on but may go well beyond their primary actions on monoamine transporters. It is proposed that signaling cascades induced by long-term antidepressant treatments lead to functional and structural remodeling of the brain circuitry (Hyman & Nestler, 1996). A number of different protein kinases have been suggested to play a role in this process, such as PKA, PKC and CaMKII (Popoli et al., 2000). However, those pharmacological treatments have significant therapeutic limitations including slow on-set, a high incidence of relapse, side effects, and treatment resistance, which is likely due to their specific targeting of monoamine systems. Accumulating evidence suggests that glutamatergic system is closely associated with stress and depression (Hercher et al., 2009). Postmortem analysis of depressive patients reveals downregulation of specific glutamate receptor subunits in brain regions that are involved in mood regulation, such as the prefrontal cortex (PFC) (Feyissa et al., 2009). In animal models, repetitive exposure to stress diminishes neuronal activity in the PFC, at least in part, through down-regulation of glutamatergic transmission (Yuen et al., 2009; Li et al., 2011). These results raise the possibility that glutamate receptors represent a therapeutic target for stress and depression.
More than a decade ago, a low dose administration of ketamine was shown to produce a rapid and lasting antidepressant response (Berman et al., 2000). Unlike traditional antidepressants that are primarily based on the monoaminergic systems, ketamine is believed to target glutamatergic transmission. Moreover, ketamine has a relatively acute clinical onset of therapeutic action (~ 2 hours) and is effective in a large number of otherwise treatment-resistant patients (Mathew et al., 2012). The proposed cellular mechanism of ketamine actions pertains to rapid enhancement of glutamate transmission and synaptogenesis (Li et al., 2010; Autry et al., 2011). While the exact mechanisms by which ketamine enhances glutamate transmission are unknown, it has been postulated that ketamine at low doses acts on NMDA receptors located on GABAergic interneurons, reducing inhibitory driving force in the network and leading to enhancement of overall activity (Seamans, 2008). It appears that the functional restoration induced by ketamine is associated with activation of signaling cascade within a few hours, which is followed by a second wave of increased expression in synaptic proteins within 24 hours (Li et al., 2010; Autry et al., 2011). Together, these molecular events lead to rapid synaptogenesis and reverse of neural atrophy (Li et al., 2010).
Mammalian target of rapamycin (mTOR)
Increased activation of mTOR signaling pathway was recently identified as a critical signaling mechanism that, in part, underlies fast antidepressant effects of ketamine (Li et al., 2010). Following enhancement of glutamatergic transmission and neuronal activity, through mechanisms yet to be identified, ketamine seems to rapidly activate PI3K-AKT and MEK-ERK signaling cascades within the PFC that leads to the postsynaptic activation of mTOR and a subsequent induction of synaptic proteins [e.g., glutamate-AMPA receptor-1 (GluR1), postsynaptic density protein-95 (PSD95), and synapsin I] and increase in neurotrophic factors signaling (i.e., activation of BDNF synthesis) (Autry et al., 2011). This rapid induction of mTOR, BDNF and synaptic proteins is thought to stimulate increases in production of new synapses (i.e, synaptogenesis) and overall synaptic function that are required for development of fast antidepressant responses (Li et al., 2010; Li et al., 2011).
Furthermore, in the hippocampus, another brain region that is also involved in stress and mood regulation, ketamine regulates a different set of molecular events. It deactivates eukaryotic elongation factor (eEF2) kinase, resulting in reduced eEF2 phosphorylation and subsequent enhancement of BDNF translation (Autry et al., 2011), which is also thought to be a key molecular event contributing to rapid antidepressant actions of ketamine.
Despite limitations of ketamine use as a long-term treatment, particularly its dissociative/psychotomimetic effects and abuse potential, ketamine represents a new line of antidepressant products that offer quick onset and long-lasting effects. The contribution of ketamine metabolites to its antidepressant effects has long been suspected in the field. A recent discovery from the Gould lab identified the first effective component of ketamine metabolites, (2R, 6R)-hydroxynorketamine (HNK) that is responsible for its antidepressant actions but with minimal side effects (Zanos et al., 2016). While those actions appear to be independent of NMDA receptor blockade, exact molecular mechanisms are still not clear. Like ketamine, HNK was shown to suppress eEF2 kinase activity in the hippocampus, but it failed to regulate mTOR pathway in either hippocampus or the prefrontal cortex (Zanos et al., 2016).
This latest finding further demonstrates the complexity of neural mechanisms involved in ketamine’s antidepressant actions, including the functional role of mTOR pathway. In general, recent progress in rapid acting antidepressants definitively reveals an incomplete understanding of their neural mechanisms of action. Furthermore, the lack of a consensus in molecular mechanisms among various studies highlights the necessity of unbiased screening for previously unknown molecular drug targets including specific kinases. In the following section we will review a selected technology to screen for kinases involved in etiology and treatment of depression that could lead towards discovery of novel treatment strategies for this and other psychiatric disorders.
Unbiased screening for kinase/signaling network
Kinases are emerging as important therapeutic targets in cancer and other diseases. There are more than 500 protein kinases in the human genome, and many of them can be mapped to disease loci (Manning et al., 2002). Traditional bioassays have identified important roles of a few individual kinases in the pathophysiology of a particular neurological disease. However, multiple kinases usually form complicated networks to regulate intracellular signaling. Thus, a comprehensive kinase profiling approach to investigate the potential role that each of the 500 kinases plays in neurological diseases would be advantageous. One of the direct ways to study kinases is to measure the catalytic activity of individual kinases. High-throughput kinase assay methods have recently been commercialized under the trademarks Kinase HotSpot (Reaction Biology Corporation) and KinaseProfiler™ (EMD Millipore). A recent study used this approach to analyze the selectivity of kinase inhibitors on 300 kinases (Anastassiadis et al., 2011). One drawback to this method is that in vitro kinase activity assays do not capture the highly regulated dynamic events, such as the spatial and temporal regulation of protein-protein interactions or compartmentalization, which occur in biological systems. This method is also limited by the fact that knowledge of substrates or consensus phosphorylation sites is needed, information which is not yet available for many understudied kinases.
Furthermore, technological breakthroughs in the early 2000’s allowed for the advent of protein and peptide arrays, which are currently used for kinome analysis (MacBeath & Schreiber, 2000). Kinase peptide and protein microarrays consist of glass or plastic slides spotted with substrate peptides or proteins, respectively. Cell or tissue lysate is incubated with these arrays and several different labeling techniques can be used to detect the phosphorylation of substrate peptides or proteins. There are many commercially available peptide and protein microarrays designed for kinome analysis. While these are relatively low cost and rapid methods to analyze the kinome, there are numerous limitations. Multiple kinases can often phosphorylate the same peptide or protein, making it difficult to distinguish between phosphorylating kinases. Additionally, as with other in vitro assays, important regulatory events that occur in intact cells are lost (Yamamoto et al., 2014). Another type of protein microarray used to study the kinome is the phospho-antibody microarray. These arrays are typically spotted with phospho-specific antibodies targeting known phosphorylation sites in kinases and cell signaling molecules. A major drawback of this method is that it is limited by the specificity and availability of phospho-antibodies.
Recent technical advances in quantitative mass spectrometry, protein enrichment, and informatics are facilitating exciting new approaches to studying the kinome (Graves et al., 2013; Xiao & Wang, 2016). Early efforts to profile kinase activities using mass spectrometry were based on large-scale phosphoproteomic analyses (Olsen et al., 2006). This approach involves enriching phosphopeptides from a biological sample with affinity chromatography and identifying the peptides and phosphorylated residues with mass spectrometry. The likely phosphorylating kinase of each protein then can be determined by matching the phosphorylation site and surrounding amino acids with consensus phosphorylation motifs that have been identified for individual kinases. While studies using this approach have been useful, the complexity of the proteome and the fact that closely related kinases can have similar or identical consensus phosphorylation motifs limit the usefulness of this technique in determining changes in the activity of specific kinases.
One of the challenges of analyzing kinases in cell or tissue lysates with mass spectrometry is that the proteome is complex and many kinases are expressed at low levels. Thus, new techniques for kinase enrichment have been instrumental in advancing the study of the kinome (Graves et al., 2013; Xiao & Wang, 2014). One such technique is the capture of kinases by immobilized kinase inhibitors. Knockaert et al. introduced this technique by covalently linking a single kinase inhibitor to agarose beads to identify intracellular targets of that inhibitor (Knockaert et al., 2000). The early studies utilizing this method were designed to investigate the specificity of kinase inhibitors (Godl et al., 2003; Fabian et al., 2005). This method has more recently been further developed and combined with phosphoproteomic approaches to analyze changes in the phosphorylation and expression of kinases. Importantly, the phosphorylation status of kinases can be used as readouts of enzyme activity (Wissing et al., 2007; Daub et al., 2008). An exciting extension of kinase affinity capture was used by Duncan et al. to selectivity enrich activated kinases (Duncan et al., 2012). Layers of sepharose beads conjugated to both specific and non-specific type I kinase inhibitors, which are ATP-competitive inhibitors that bind preferentially to active kinases at the ATP binding site, were placed in a single column and cell lysates were passed through. After extensive washing steps, the captured kinases were eluted and digested with trypsin. The peptides then were labeled with isobaric tags for relative and absolute quantitation (iTRAQ) and identified and quantitated with mass spectrometry (Figure 2). This approach was successfully used to identify kinome-wide changes in the activity and expression of kinases in breast cancer and leukemia in response to drug treatment. Importantly, this data will likely be useful in rationally designing combinational treatment regimens to help reduce the development of drug-resistant breast cancer and leukemia (Duncan et al., 2012; Cooper et al., 2013; Graves et al., 2013).
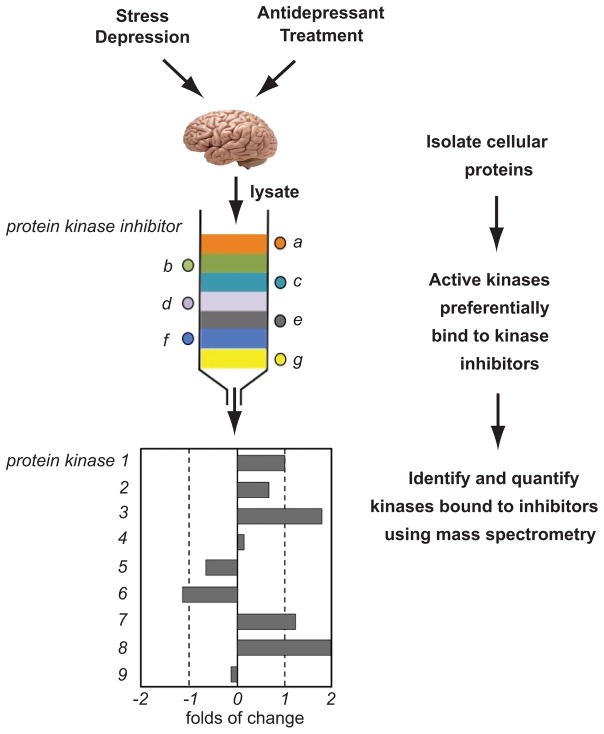
Figure 2. Proposed kinome profiling applications to major depressive disorders and antidepressant treatment.
As described in Duric et al., 2010, cellular extracts can be obtained from post-mortem brain tissue with MDD pathology or long-term antidepressant treatment. Protein fractions are added to a column of sepharose beads covalently linked to a number of different type I kinase inhibitors (adapted from Graves et al., 2013). The active kinases preferentially bind to the inhibitors and remain on the column. The kinases are then eluted from the columns and digested with trypsin. The tryptic peptides are labeled with isobaric tags for relative and absolute quantitation (iTRAQ) and identified and quantitated with mass spectrometry. If a kinase is activated by a specific treatment or pathology, more of that kinase will be bound to the column. Quantity of kinase bound from a treatment over control yields kinome activity profile.
Current (and proposed) applications of kinome profiling to neurological diseases
Kinase profiling approaches have increasingly become a prevalent methodology in cancer research and demonstrated a great potential in drug discovery. However, their application to neurological diseases and pharmaceutical treatments targeting mental illness is limited. Nonetheless, a few studies have tapped into this promising yet largely unexplored field. One of the efforts was to identify protein kinases and signaling pathways that are activated by Alzheimer’s disease (AD) pathology (Hoozemans et al., 2012). Microarray of peptides derived from known kinase substrates was used to profile kinome activity of postmortem brain tissue of AD patients. This approach revealed novel kinases and signaling pathways involved in AD pathology that otherwise would not be uncovered with conventional biochemical assays. Contrary to the above target (phosphorylation site)-based screening, Al-Ali et al conducted a phenotypic screening to identify drugs that have the potential to promote axonal growth (Al-Ali et al., 2013). Based on axonal morphology, the neuronal kinome activity of primary culture was probed with a diverse library of small molecule protein kinase inhibitors. Kinase inhibitors identified from such an in vitro functional screening hold the potential for pharmacological intervention of axonal degeneration in vivo.
Given the complexity of kinases involved in rapid acting antidepressant treatment and inconsistence among different studies, as reviewed above, it is likely there exist additional positive and negative regulators of the mTOR and the eEF2 pathways. It is also likely kinase network activated by ketamine treatment extends beyond these two pathways. Likewise, there are also likely unmet therapeutic opportunities in targeting kinases for reducing the morbidity and mortality of major depressive disorders. To uncover novel targets and profile aberrant expression and activity of the whole kinome, we propose the following direct applications of the kinome profiling approach. Specifically, protein kinases can be enriched from post-mortem brain tissue with MDD pathology by affinity purification with immobilized protein kinase inhibitors (Figure 2). This affinity chromatograph coupled with the mass spectrometry approach is expected to identify and simultaneously measure the activity of hundreds of kinases in a single sample. Similar approaches can also be applied to post-mortem brain tissue from patients who have received long-term antidepressant treatment. Additionally, animal models of depression and antidepressant treatments could offer an opportunity to execute kinome profiling analysis in a more controlled manner.
Conclusions and future directions
The ability to monitor kinome activity in an unbiased way has opened up a new realm of opportunities to link a specific pathology or pharmaceutical treatment with its underlying mechanisms at the proteomics level. We expect the application of kinome profiling to neurological disease conditions and their treatment to increase exponentially in the next decade. Equally important are the strategies to be implemented after potentially important kinases have been identified through the kinome screening. Finally, in addition to phosphorylation, other forms of posttranscriptional modification may also contribute to the pathophysiology of depression, and thus, may represent valid therapeutic targets for antidepressant actions that should be explored in a similar manner.
Acknowledgments
This review is written in honor of Dr. Barry Ganetzky’s scientific career, his contributions to the community of Drosophila genetics, and his dedication to his students and trainees. LY wishes to express the most sincere gratitude to Dr. Ganetzky for his support, encouragement, and sense of humor that goes well beyond her graduate studies.
We thank Dr. Graves for valuable comments on the manuscript. This work was supported by grants from NIH (MH108043 to LY), PhRMA Foundation (to VD), American Heart Association (to EW) and Iowa Osteopathic Education and Research (IOER) Funds (to EW, LY, and VD).
Abbreviations
- 4E-BP1
eukaryotic translation initiation factor 4E-binding protein 1
- AD
Alzheimer’s disease
- BDNF
brain-derived neurotrophic factor
- CaMKII
Calcium/calmodulin-dependent protein kinase II
- eEF2
eukaryotic elongation factor 2
- eEF2K
eEF2 kinase
- ERK
extracellular signal-regulated kinases
- GluR1
glutamate-AMPA receptor-1
- GSK-3
glycogen-synthase kinase-3
- HNK
hydroxynorketamine
- iTRAQ
isobaric tags for relative and absolute quantitation
- MAOI
monoamine oxidase inhibiter
- MAPK
mitogen-activated protein kinase
- MDD
major depressive disorder
- MKP-1
MAP kinase phosphatase-1
- mTOR
mammalian target of rapamycin
- NMDAR
N-methyl-D-aspartate receptor
- p70S6K
p70S6kinase
- PFC
prefrontal cortex
- PI3K
phosphoinositide 3-kinase
- PKA
protein kinase A
- PKB/AKT
protein kinase B
- PKC
protein kinase C
- PSD95
postsynaptic density protein-95
- S6
ribosomal protein S6
- TCA
tricyclic antidepressant
- SSRI
selective serotonin reuptake inhibitor
References
- Al-Ali H, Schurer SC, Lemmon VP, Bixby JL. Chemical interrogation of the neuronal kinome using a primary cell-based screening assay. ACS Chemical biology. 2013;8:1027–1036. doi: 10.1021/cb300584e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anastassiadis T, Deacon SW, Devarajan K, Ma H, Peterson JR. Comprehensive assay of kinase catalytic activity reveals features of kinase inhibitor selectivity. Nature biotechnology. 2011;29:1039–1045. doi: 10.1038/nbt.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Autry AE, Adachi M, Nosyreva E, Na ES, Los MF, Cheng PF, Kavalali ET, Monteggia LM. NMDA receptor blockade at rest triggers rapid behavioural antidepressant responses. Nature. 2011;475:91–95. doi: 10.1038/nature10130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman RM, Cappiello A, Anand A, Oren DA, Heninger GR, Charney DS, Krystal JH. Antidepressant effects of ketamine in depressed patients. Biological psychiatry. 2000;47:351–354. doi: 10.1016/s0006-3223(99)00230-9. [DOI] [PubMed] [Google Scholar]
- Beurel E. Regulation of inflammation and T cells by glycogen synthase kinase-3: links to mood disorders. Neuroimmunomodulation. 2014;21:140–144. doi: 10.1159/000356550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beurel E, Song L, Jope RS. Inhibition of glycogen synthase kinase-3 is necessary for the rapid antidepressant effect of ketamine in mice. Molecular psychiatry. 2011;16:1068–1070. [Google Scholar]
- Cooper MJ, Cox NJ, Zimmerman EI, Dewar BJ, Duncan JS, Whittle MC, Nguyen TA, Jones LS, Ghose Roy S, Smalley DM, Kuan PF, Richards KL, Christopherson RI, Jin J, Frye SV, Johnson GL, Baldwin AS, Graves LM. Application of multiplexed kinase inhibitor beads to study kinome adaptations in drug-resistant leukemia. PloS one. 2013;8:e66755. doi: 10.1371/journal.pone.0066755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotter D, Mackay D, Landau S, Kerwin R, Everall I. Reduced glial cell density and neuronal size in the anterior cingulate cortex in major depressive disorder. Archives of general psychiatry. 2001;58:545–553. doi: 10.1001/archpsyc.58.6.545. [DOI] [PubMed] [Google Scholar]
- Daub H, Olsen JV, Bairlein M, Gnad F, Oppermann FS, Korner R, Greff Z, Keri G, Stemmann O, Mann M. Kinase-selective enrichment enables quantitative phosphoproteomics of the kinome across the cell cycle. Molecular cell. 2008;31:438–448. doi: 10.1016/j.molcel.2008.07.007. [DOI] [PubMed] [Google Scholar]
- Duman CH, Schlesinger L, Kodama M, Russell DS, Duman RS. A role for MAP kinase signaling in behavioral models of depression and antidepressant treatment. Biological psychiatry. 2007;61:661–670. doi: 10.1016/j.biopsych.2006.05.047. [DOI] [PubMed] [Google Scholar]
- Duncan JS, Whittle MC, Nakamura K, Abell AN, Midland AA, Zawistowski JS, Johnson NL, Granger DA, Jordan NV, Darr DB, Usary J, Kuan PF, Smalley DM, Major B, He X, Hoadley KA, Zhou B, Sharpless NE, Perou CM, Kim WY, Gomez SM, Chen X, Jin J, Frye SV, Earp HS, Graves LM, Johnson GL. Dynamic reprogramming of the kinome in response to targeted MEK inhibition in triple-negative breast cancer. Cell. 2012;149:307–321. doi: 10.1016/j.cell.2012.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duric V, Banasr M, Licznerski P, Schmidt HD, Stockmeier CA, Simen AA, Newton SS, Duman RS. A negative regulator of MAP kinase causes depressive behavior. Nature medicine. 2010;16:1328–1332. doi: 10.1038/nm.2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duric V, Clayton S, Leong ML, Yuan LL. Comorbidity Factors and Brain Mechanisms Linking Chronic Stress and Systemic Illness. Neural plasticity. 2016;2016:5460732. doi: 10.1155/2016/5460732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duric V, Duman RS. Depression and treatment response: dynamic interplay of signaling pathways and altered neural processes. Cellular and molecular life science. 2013;70:39–53. doi: 10.1007/s00018-012-1020-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwivedi Y, Rizavi HS, Conley RR, Pandey GN. ERK MAP kinase signaling in post-mortem brain of suicide subjects: differential regulation of upstream Raf kinases Raf-1 and B-Raf. Molecular psychiatry. 2006;11:86–98. doi: 10.1038/sj.mp.4001744. [DOI] [PubMed] [Google Scholar]
- Dwivedi Y, Rizavi HS, Roberts RC, Conley RC, Tamminga CA, Pandey GN. Reduced activation and expression of ERK1/2 MAP kinase in the post-mortem brain of depressed suicide subjects. Journal of neurochemistry. 2001;77:916–928. doi: 10.1046/j.1471-4159.2001.00300.x. [DOI] [PubMed] [Google Scholar]
- Dwivedi Y, Rizavi HS, Teppen T, Sasaki N, Chen H, Zhang H, Roberts RC, Conley RR, Pandey GN. Aberrant extracellular signal-regulated kinase (ERK) 5 signaling in hippocampus of suicide subjects. Neuropsychopharmacology. 2007;32:2338–2350. doi: 10.1038/sj.npp.1301372. [DOI] [PubMed] [Google Scholar]
- Fabian MA, Biggs WH, 3rd, Treiber DK, Atteridge CE, Azimioara MD, Benedetti MG, Carter TA, Ciceri P, Edeen PT, Floyd M, Ford JM, Galvin M, Gerlach JL, Grotzfeld RM, Herrgard S, Insko DE, Insko MA, Lai AG, Lelias JM, Mehta SA, Milanov ZV, Velasco AM, Wodicka LM, Patel HK, Zarrinkar PP, Lockhart DJ. A small molecule-kinase interaction map for clinical kinase inhibitors. Nature biotechnology. 2005;23:329–336. doi: 10.1038/nbt1068. [DOI] [PubMed] [Google Scholar]
- Feyissa AM, Chandran A, Stockmeier CA, Karolewicz B. Reduced levels of NR2A and NR2B subunits of NMDA receptor and PSD-95 in the prefrontal cortex in major depression. Progress in neuropsychopharmacol and biological Psychiatry. 2009;33:70–75. doi: 10.1016/j.pnpbp.2008.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godl K, Wissing J, Kurtenbach A, Habenberger P, Blencke S, Gutbrod H, Salassidis K, Stein-Gerlach M, Missio A, Cotten M, Daub H. An efficient proteomics method to identify the cellular targets of protein kinase inhibitors. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:15434–15439. doi: 10.1073/pnas.2535024100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves LM, Duncan JS, Whittle MC, Johnson GL. The dynamic nature of the kinome. The Biochemical journal. 2013;450:1–8. doi: 10.1042/BJ20121456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal SS, York RD, Stork PJ. Extracellular-signal-regulated kinase signalling in neurons. Current opinion in neurobiology. 1999;9:544–553. doi: 10.1016/S0959-4388(99)00010-0. [DOI] [PubMed] [Google Scholar]
- Hercher C, Turecki G, Mechawar N. Through the looking glass: examining neuroanatomical evidence for cellular alterations in major depression. Journal of psychiatric research. 2009;43:947–961. doi: 10.1016/j.jpsychires.2009.01.006. [DOI] [PubMed] [Google Scholar]
- Hirschfeld RM. History and evolution of the monoamine hypothesis of depression. Journal of clinical psychiatry. 2000;61(Suppl 6):4–6. [PubMed] [Google Scholar]
- Hoozemans JJ, Hilhorst R, Ruijtenbeek R, Rozemuller AJ, van der Vies SM. Protein kinase activity profiling of postmortem human brain tissue. Neurodegenerative diseases. 2012;10:46–48. doi: 10.1159/000335914. [DOI] [PubMed] [Google Scholar]
- Hyman SE, Nestler EJ. Initiation and adaptation: a paradigm for understanding psychotropic drug action. American journal of psychiatry. 1996;153:151–162. doi: 10.1176/ajp.153.2.151. [DOI] [PubMed] [Google Scholar]
- Kessler RC, Chiu WT, Demler O, Merikangas KR, Walters EE. Prevalence, severity, and comorbidity of 12-month DSM-IV disorders in the National Comorbidity Survey Replication. Archives of general psychiatry. 2005;62:617–627. doi: 10.1001/archpsyc.62.6.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knockaert M, Gray N, Damiens E, Chang YT, Grellier P, Grant K, Fergusson D, Mottram J, Soete M, Dubremetz JF, Le Roch K, Doerig C, Schultz P, Meijer L. Intracellular targets of cyclin-dependent kinase inhibitors: identification by affinity chromatography using immobilised inhibitors. Chemistry & biology. 2000;7:411–422. doi: 10.1016/s1074-5521(00)00124-1. [DOI] [PubMed] [Google Scholar]
- Kockeritz L, Doble B, Patel S, Woodgett JR. Glycogen synthase kinase-3--an overview of an over-achieving protein kinase. Current drug targets. 2006;7:1377–1388. doi: 10.2174/1389450110607011377. [DOI] [PubMed] [Google Scholar]
- Kodama M, Russell DS, Duman RS. Electroconvulsive seizures increase the expression of MAP kinase phosphatases in limbic regions of rat brain. Neuropsychopharmacology. 2005;30:360–371. doi: 10.1038/sj.npp.1300588. [DOI] [PubMed] [Google Scholar]
- Li N, Lee B, Liu RJ, Banasr M, Dwyer JM, Iwata M, Li XY, Aghajanian G, Duman RS. mTOR-dependent synapse formation underlies the rapid antidepressant effects of NMDA antagonists. Science. 2010;329:959–964. doi: 10.1126/science.1190287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Liu RJ, Dwyer JM, Banasr M, Lee B, Son H, Li XY, Aghajanian G, Duman RS. Glutamate N-methyl-D-aspartate receptor antagonists rapidly reverse behavioral and synaptic deficits caused by chronic stress exposure. Biological psychiatry. 2011;69:754–761. doi: 10.1016/j.biopsych.2010.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacBeath G, Schreiber SL. Printing proteins as microarrays for high-throughput function determination. Science. 2000;289:1760–1763. doi: 10.1126/science.289.5485.1760. [DOI] [PubMed] [Google Scholar]
- Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- Mathew SJ, Shah A, Lapidus K, Clark C, Jarun N, Ostermeyer B, Murrough JW. Ketamine for treatment-resistant unipolar depression: current evidence. CNS Drugs. 2012;26:189–204. doi: 10.2165/11599770-000000000-00000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, Mortensen P, Mann M. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 2006;127:635–648. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- Popoli M, Brunello N, Perez J, Racagni G. Second messenger-regulated protein kinases in the brain: their functional role and the action of antidepressant drugs. Journal of neurochemtry. 2000;74:21–33. doi: 10.1046/j.1471-4159.2000.0740021.x. [DOI] [PubMed] [Google Scholar]
- Seamans J. Losing inhibition with ketamine. Nature chemical biology. 2008;4:91–93. doi: 10.1038/nchembio0208-91. [DOI] [PubMed] [Google Scholar]
- Sheline YI, Wang PW, Gado MH, Csernansky JG, Vannier MW. Hippocampal atrophy in recurrent major depression. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:3908–3913. doi: 10.1073/pnas.93.9.3908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon GE. Social and economic burden of mood disorders. Biological psychiatry. 2003;54:208–215. doi: 10.1016/s0006-3223(03)00420-7. [DOI] [PubMed] [Google Scholar]
- Stockmeier CA, Mahajan GJ, Konick LC, Overholser JC, Jurjus GJ, Meltzer HY, Uylings HB, Friedman L, Rajkowska G. Cellular changes in the postmortem hippocampus in major depression. Biological psychiatry. 2004;56:640–650. doi: 10.1016/j.biopsych.2004.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Videbech P, Ravnkilde B. Hippocampal volume and depression: a meta-analysis of MRI studies. The American journal of psychiatry. 2004;161:1957–1966. doi: 10.1176/appi.ajp.161.11.1957. [DOI] [PubMed] [Google Scholar]
- Wissing J, Jansch L, Nimtz M, Dieterich G, Hornberger R, Keri G, Wehland J, Daub H. Proteomics analysis of protein kinases by target class-selective prefractionation and tandem mass spectrometry. Molecular & cellular proteomics : MCP. 2007;6:537–547. doi: 10.1074/mcp.T600062-MCP200. [DOI] [PubMed] [Google Scholar]
- Xiao Y, Wang Y. Global discovery of protein kinases and other nucleotide-binding proteins by mass spectrometry. Mass spectrometry reviews. 2016;35:601–619. doi: 10.1002/mas.21447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto T, Mori T, Katayama Y. Microarray technologies for intracellular kinome analysis. Current medicinal chemistry. 2014;21:2542–2552. doi: 10.2174/0929867321666131212154153. [DOI] [PubMed] [Google Scholar]
- Yuen EY, Liu W, Karatsoreos IN, Feng J, McEwen BS, Yan Z. Acute stress enhances glutamatergic transmission in prefrontal cortex and facilitates working memory. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:14075–14079. doi: 10.1073/pnas.0906791106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanos P, Moaddel R, Morris PJ, Georgiou P, Fischell J, Elmer GI, Alkondon M, Yuan P, Pribut HJ, Singh NS, Dossou KS, Fang Y, Huang XP, Mayo CL, Wainer IW, Albuquerque EX, Thompson SM, Thomas CJ, Zarate CA, Jr, Gould TD. NMDAR inhibition-independent antidepressant actions of ketamine metabolites. Nature. 2016 doi: 10.1038/nature17998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zunszain PA, Horowitz MA, Cattaneo A, Lupi MM, Pariante CM. Ketamine: synaptogenesis, immunomodulation and glycogen synthase kinase-3 as underlying mechanisms of its antidepressant properties. Molecular psychiatry. 2013;18:1236–1241. doi: 10.1038/mp.2013.87. [DOI] [PMC free article] [PubMed] [Google Scholar]