Abstract
The majority of animals show external bilateral symmetry, precluding the observation of multiple internal left-right (L/R) asymmetries that are fundamental for organ packaging and function1,2. In vertebrates, left identity is mediated by the left-specific Nodal-Pitx2 axis that is repressed on the right-hand side by the epithelial-mesenchymal transition (EMT) inducer Snail13,4. Despite some existing evidence3,5, it remains unclear whether an equivalent instructive pathway provides right-hand specific information to the embryo. Here we show that in zebrafish, BMP mediates the L/R asymmetric activation of another EMT inducer, Prrx1a, in the lateral plate mesoderm (LPM) with higher levels on the right. Prrx1a drives L/R differential cell movements towards the midline leading to a leftward displacement of the cardiac posterior pole through an actomyosin-dependent mechanism. Downregulation of Prrx1a prevents heart looping and leads to mesocardia. Two parallel and mutually repressed pathways, respectively driven by Nodal and BMP on the left and right LPM, converge on the asymmetric activation of Pitx2 and Prrx1, two transcription factors that integrate left and right information to govern heart morphogenesis. This mechanism is conserved in the chicken embryo and, in the mouse, Snail1 fulfills the role played by Prrx1 in fish and chick. Thus, a differential L/R EMT produces asymmetric cell movements and forces, more prominent from the right, that drive heart laterality in vertebrates.
Keywords: Prrx1, EMT, left-right asymmetry, heart lateralization
Defects in L/R asymmetry arise in 1/10,000 humans6, and the associated morbidity and mortality usually imply congenital heart defects (CHDs)6,7. The EMT converts epithelial cells into migratory cells, and it is required in tissues and organs generated after profound cell movements, such as the mesoderm and the neural crest8. Thus, L/R asymmetries and EMT provoke severe congenital malformations or early embryonic lethality when deregulated1,9.
After an initial disruption of L/R symmetry in the vertebrate embryo, laterality is conferred to the organizer (node) and this information is transferred to the left LPM1. The left-specific programme in the LPM is driven by Nodal and its downstream target Pitx2. This pathway is conserved in deuterostomes, and in chick and mouse embryos it is repressed on the right-hand side by the EMT inducer Snail13,4. We have found that like Snail1 in amniotes, a prrx1 gene duplicate (prrx1a), is transiently expressed in the zebrafish embryo LPM with higher levels on the right (Fig. 1a). Prrx1, like Pitx2, is a paired-like homeobox transcription factor, and like Snail1, is an EMT inducer in embryos and cancer cells10.
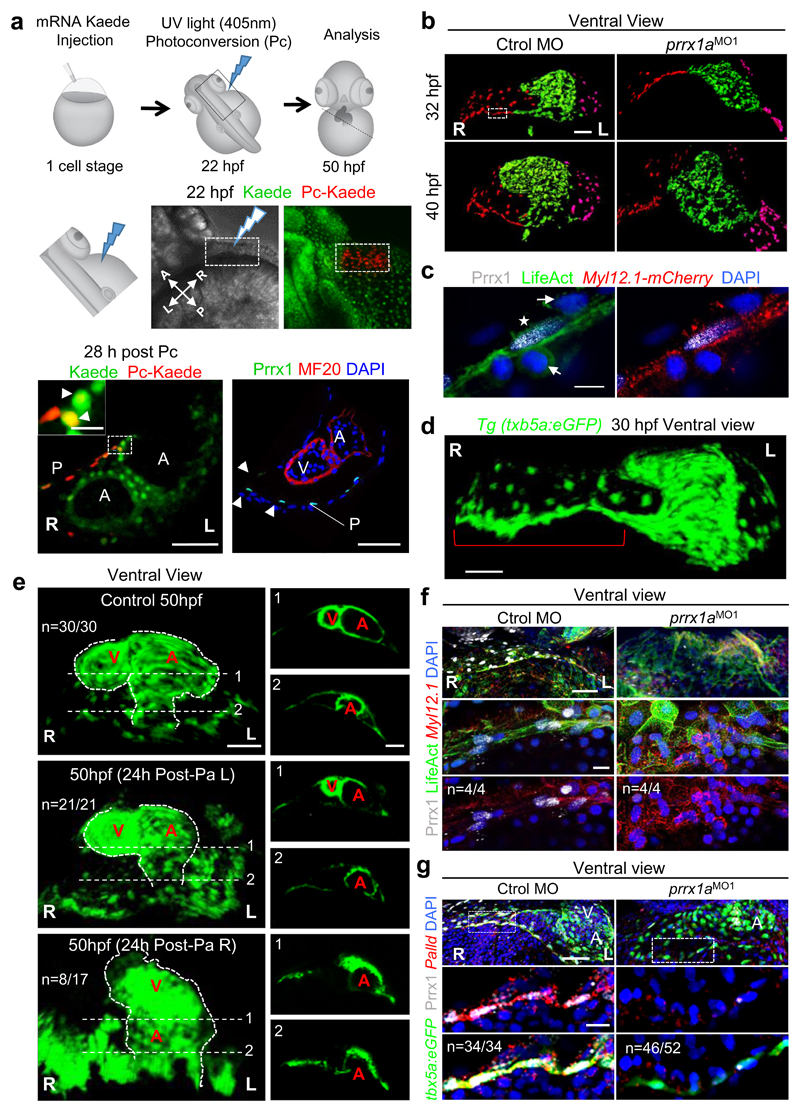
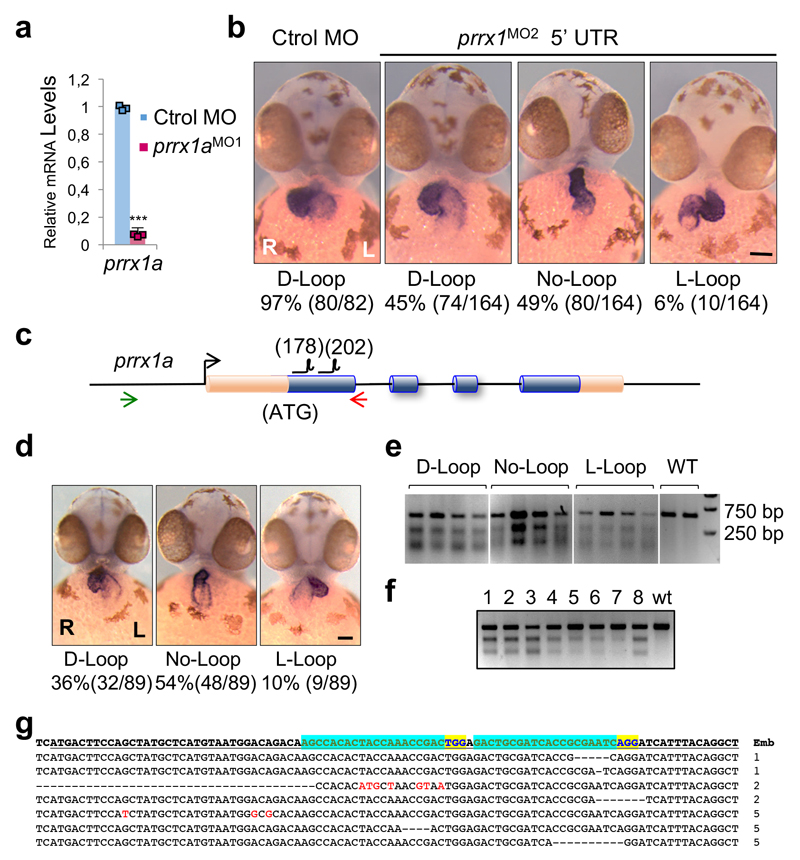
Figure 1. prrx1a L/R asymmetric expression.
(a) Whole mount in situ hybridization and transverse sections at the levels of the dotted lines. b) Heart situs in control and prrx1a morphant (prrx1aMO1) embryos analyzed with the cardiomyocyte marker myl7. (c) Quantification of heart situs. (d) Analysis of atrial (A) and ventricular (V) markers amhc and vmhc. (e) Confocal analysis of Prrx1 and Islet1 expression in transgenic zebrafish embryos Tg(tbx5a:eGFP). Nuclei stained with DAPI. Stars, areas of asymmetry. Scale bars, 50 µm except for high power images in (e), 10µm.
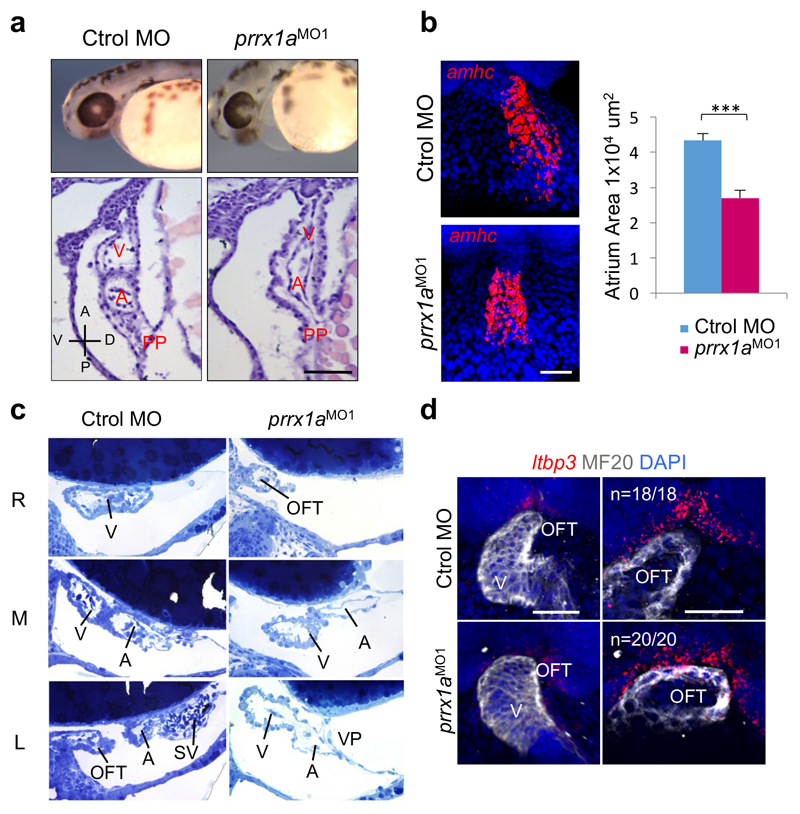
In vertebrates, the convergence of the left and right cardiogenic regions in the embryonic midline results in the formation of a linear primary heart tube (PHT). The subsequent bilateral addition of progenitor cells to the arterial (anterior) and venous (posterior) poles from the corresponding second heart fields (SHFs), respectively contribute to the elongation and growth of the heart at the outflow (OFT) and inflow (IFT) tracts 11. These additions occur at the time of heart looping, for which the underlying mechanisms remain poorly understood11 although defects in the posterior pole are responsible for numerous CHDs in humans. Considering Prrx1a L/R asymmetric expression and its described role in the induction of cell movements10, we examined whether prrx1a knockdown influenced heart situs. Both prrx1a morpholino oligonucleotide (prrx1aMO1 or prrx1aMO2)10 injections or CRISPR/Cas9-driven prrx1a mutations led to mesocardia, a straight heart that failed to undergo the normal dextral looping (Fig. 1b, c and Extended Data Fig. 1). Both atrial and ventricular chambers are specified in the morphants (Fig. 1d), but, in addition to defective looping, the heart presented,a smaller atrium and a defective posterior pole lacking a defined sinus venosus (SV) and the expression of its marker Islet112,13 (Fig. 1e and Extended Data Fig. 2a-c). However, the anterior pole was not overtly affected (Extended Data Fig. 2d).
As Tbx5a is a transcription factor expressed in the PHT and in the LPM cells that contribute to the pericardium and the posterior pole14,15, we generated a tbx5a reporter transgenic line tg(tbx5a:eGFP) that allowed us to see its coexpression with Prrx1a in the precursors of the posterior pole once the PHT had been formed (Extended Data Fig. 3) and to follow the movements and fate of this population of Prrx1a+ cells. Cell tracing experiments in this area using the photoconvertible (Pc) green-to-red Kaede potein indicated that Prrx1a+cells contributed to both the pericardium and the atrium, explaining the defective atrium in the prrx1a morphants (Fig. 2a). These atrial cells do not express Prrx1 any longer, indicating that prrx1a is downregulated after incorporation into the heart. As such, Prrx1 protein was never observed in the heart tube, positive for tbx5a (Fig. 2a and Extended Data Fig. 4a-d). Notably, other embryonic and cancer cells also downregulate EMT transcription factors, including Prrx1, when they lose their mesenchymal nature and undergo differentiation upon reaching their final destination8.
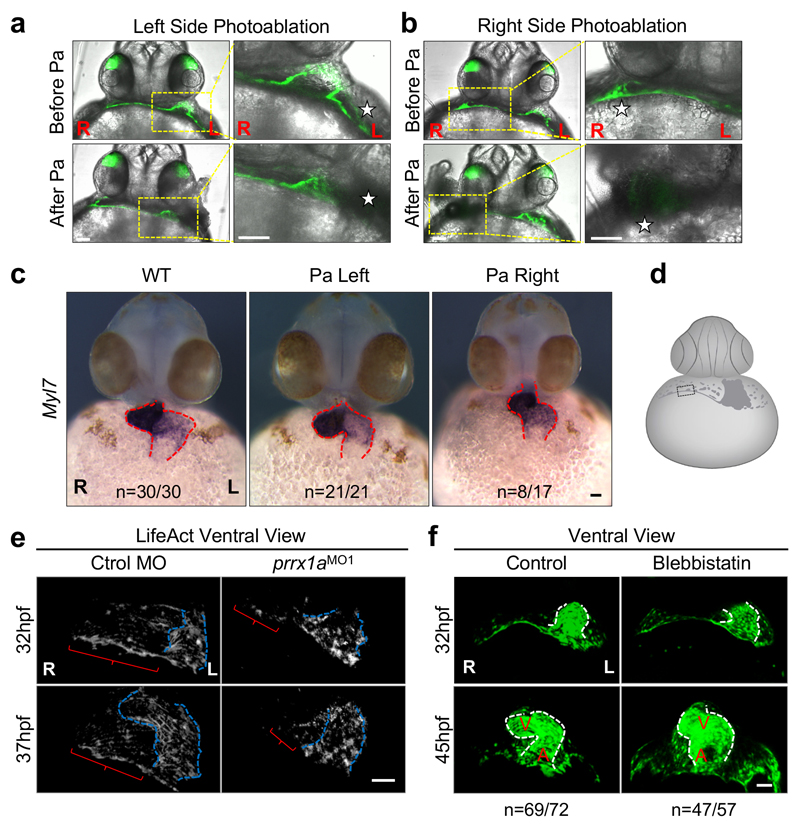
Figure 2. Asymmetric L/R Prrx1a expression drives heart looping in zebrafish.
(a) Cell lineage tracing, n=4. The heart tube is revealed by MF20 immunostaining. (b) Snapshots extracted from videos of control and prrx1a morphant Tg(tbx5a:eGFP) embryos. Red and magenta represent right and left cardiomyocyte precursors. (c) Prrx1 expression in a LifeAct injected Tg(actb2:myl12.1-mCherry) embryo to visualize F-actin and myosin II, respectively. Star, mesenchymal cell; arrows, epithelial cells. (d) Maximum intensity projection of a Tg(tbx5a:eGFP) embryo. Bracket highlights the “cable-like” structure. (e) Photoablation (Pa) in Tg(tbx5a:eGFP) embryos. Ventricles (V) and atria (A) are shown in transverse sections on the right. (f) High resolution images of the “cable-like” structure analyzed as in (c). (g) Palladin (Palld), Prrx1 and tbx5a expression in control and prrx1a morphant embryos. Scale bars 50 µm except for (c) and high power images in (a, f and g), 10 µm. P, pericardium.
We also followed the cell movements towards the posterior pole and found that they were defective in the morphants concomitantly with the failure of heart looping (Fig. 2b and Supplementary Videos 1 and 2). We detected actomyosin stress fibers in Prrx1+ cells, which also showed elongated cell shape and nuclei. These phenotypic traits, when associated with cell movements, are characteristic of mesenchymal cells undergoing EMT (Fig. 2c located in area boxed in Fig. 2b), which were absent in morphant embryos. In turn, morphants showed numerous tbx5a+ positive cells that failed to contribute to the posterior pole and the atrium, remaining dispersed throughout the pericardium (Extended Data Fig. 5a), again explaining their defective posterior pole (Fig. 1e and Extended Data Fig. 2a-c). Altogether, this indicates that Prrx1a might direct Tbx5a+ cardiac progenitors to the posterior pole in a bilaterally asymmetric manner, with a greater contribution from the right.
Directed cell migration has already been implicated in heart jogging, an initial leftward displacement of the PHT driven by Nodal in zebrafish16,17. While we found randomization of heart jogging in prrx1a morphants (Extended Data Fig. 5b, c), the majority of embryos developed mesocardia regardless of the direction of jogging (Supplementary Videos 3 and 4), indicating that the Prrx1-mediated driving force for heart looping occurs at later stages, compatible with looping being independent of both jogging and the Nodal pathway16,18.
The asymmetric contribution of right and left cells may generate asymmetric forces and greater tension on the right-hand side, inducing a maintained leftward displacement of the posterior pole that would promote the dextral torsion of the heart19–21, the two steps in looping morphogenesis. Compatible with this, the tbx5a reporter revealed cells that appeared to form a structure reminiscent of a cable (Fig. 2d). To assess the contribution of asymmetric tension to heart looping, we laser ablated a small population of tbx5a+/prrx1a+ LPM cells adjacent to the PHT on either side after jogging and before looping. Heart lateralization was only affected after ablating cells in the right LPM (Fig. 2e and Extended Data Fig. 6a-c). In vivo visualization of actin stress fibers and actin/myosin double labelling confirmed the formation of a cable during looping, a structure that was absent in prrx1a morphants (Fig. 2f, Extended Data Fig. 6d, e and Supplementary Videos 5 to 8).
We next found that Palladin, an actin-binding protein that promotes actin bundling in migratory and invasive cells22,23 was co-expressed with Prrx1 and tbx5a in the cable-like structure, and downregulated in the prrx1a morphants (Fig. 2g), linking Prrx1 with the cytoskeleton. This actomyosin cable is reminiscent of those that control forces during embryonic development24,25. Its function during looping is compatible with the mesocardia found when zebrafish PHT explants18 or embryos were exposed to the myosin inhibitor blebbistatin (Extended Data Fig. 6f and Supplementary Videos 9 and 10) and when chick embryos were treated with cytochalasin to inhibit actin polymerization26, suggesting that this mechanism may be conserved in the chick.
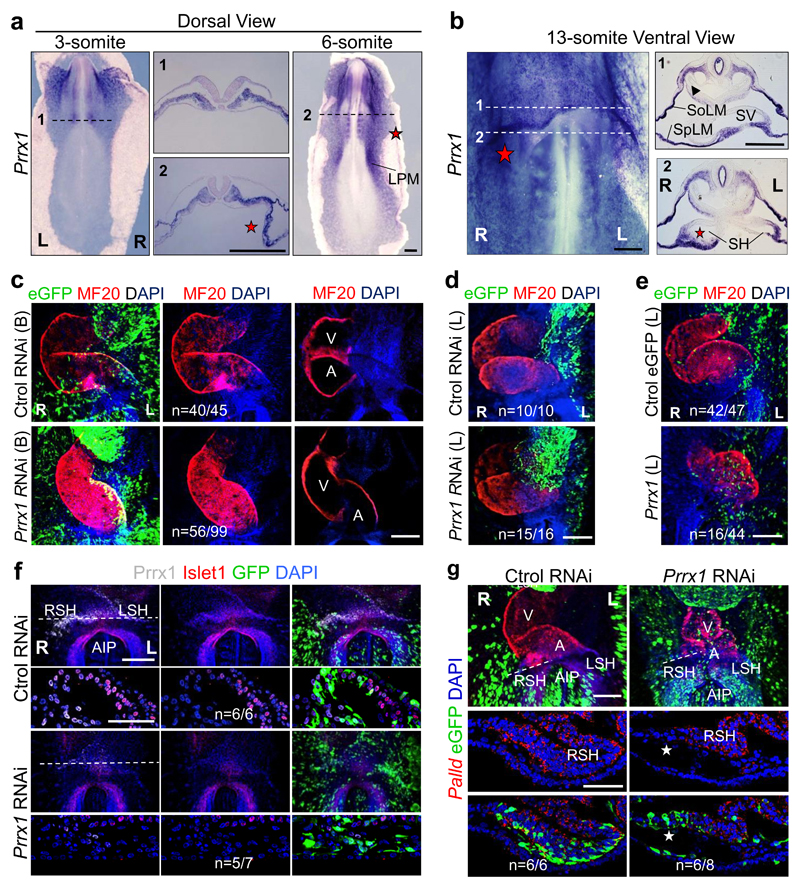
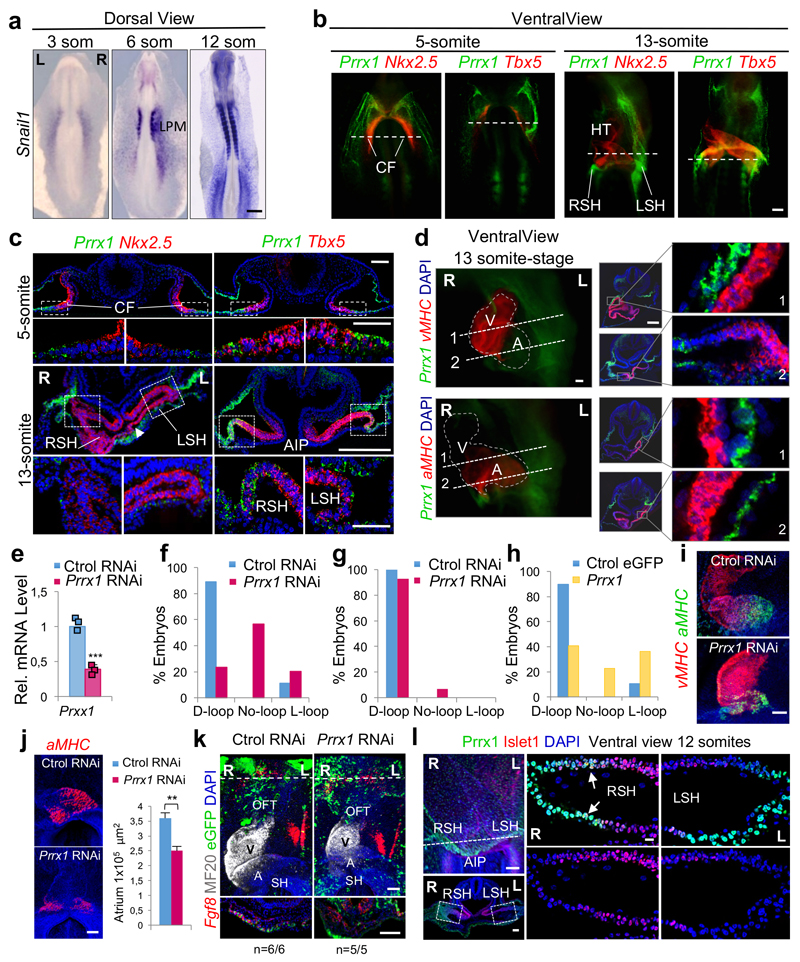
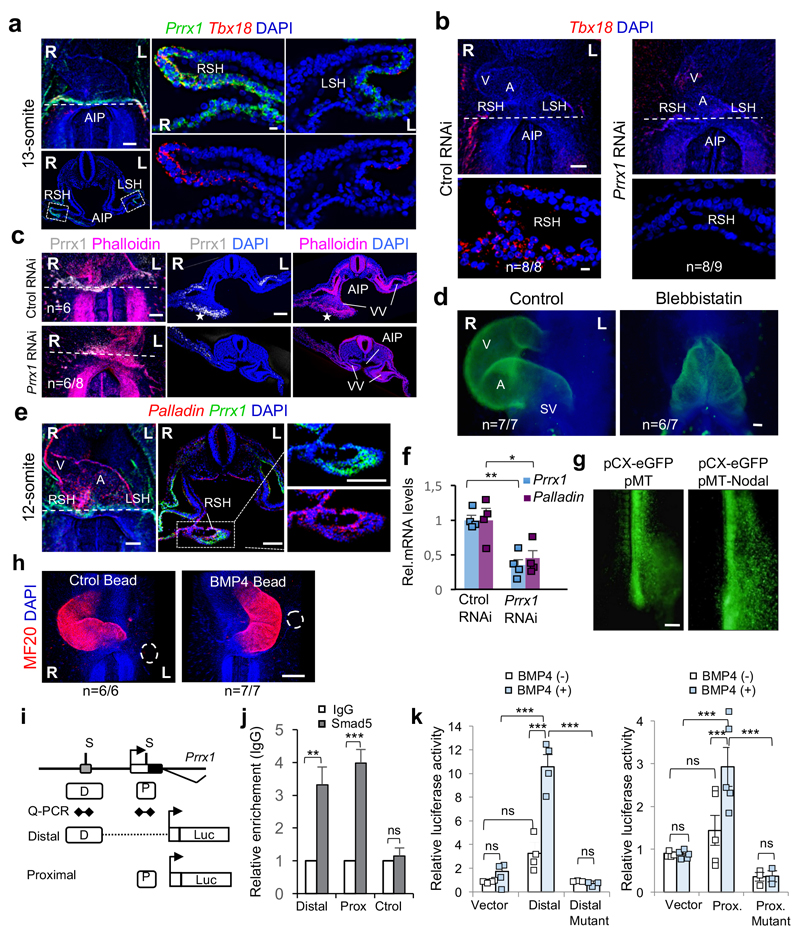
A transient asymmetric distribution of Prrx1 was evident in the chick LPM, with higher levels on the right-hand side (Fig. 3a) at stages in which Snail1 is also asymmetrically expressed (extended Data Fig. 7a and Ref. 27). Prrx1 was also asymmetrically expressed in the sinus venosus horns (SH, Fig. 3b). As in the fish, Prrx1 is not expressed in the cardiac folds (CF) or PHT, but it is expressed by the population of Tbx5+ cells lateral and posterior to the cardiac posterior pole (Extended Data Fig. 7b-d). Bilateral downregulation of Prrx1 in the chick embryo induced mesocardia in the majority of embryos (Fig. 3c and Extended Data Fig. 7e, f). However, Prrx1 unilateral downregulation on the left half did not affect heart looping (Fig. 3d and Extended Data Fig. 7g), whereas overexpression could induce reverse looping (Fig. 3e and Extended Data Fig. 7h), further evidence that Prrx1 L/R asymmetric expression drives heart laterality.
Figure 3. Prrx1 also drives heart looping in the chick embryo.
(a-b) Prrx1 expression in whole embryos and transverse sections. (c) Bilateral (B) electroporation with an eGFP reporter construct plus either RNAi against Prrx1 or control RNAi. (d-e) Unilateral electroporation (left, L) with control or Prrx1 RNAi vector (d) or with a vector containing the Prrx1-coding sequence (e). (f) Immunofluorescence of Islet1 and Prrx1 in control and Prrx1 RNAi electroporated embryos. (g) Palladin expression in similarly treated embryos. Stars, areas of asymmetry. Nuclei were stained with DAPI (blue). Dotted lines indicate the level of the sections shown. Scale bars, all 250 μm except for the high power images in (f) and (g), which correspond to 50 μm. AIP; anterior intestinal portal; L or R SH; left or right sinus venosus horns; SoLM or SpLM, somatic or splanchnic lateral mesoderm.
As in the zebrafish embryo, deficient Prrx1 expression did not prevent the specification of atrial and ventricular fates, the size of the atrium was reduced and did not seem to affect the anterior pole (see OFT) (Extended Data Fig. 7i-k). Again, Islet1 colocalized with Prrx1 in the posterior pole and its expression was downregulated in embryos electroporated with Prrx1 RNAi (Fig. 3f and Extended Data Fig. 7l). This population of sinus venosus progenitors expressing Tbx18 (Extended Data Fig. 8a), contributes to the posterior pole together with the SHF, and has been referred to as posterior SHF20,28 or tertiary heart field29, because it is located more laterally than the classical Nkx2.5+ SHF. Interestingly, Tbx18 is repressed when Prrx1 is downregulated (Extended Data Fig. 8b). Actin fibers were observed in a cable-like structure of Prrx1+ cells at the wall of the right vitelline vein (VV) and right sinus venosus horns in control embryos (Extended Data Fig. 8c), compatible with the displacement of the posterior pole to the left side. The loss of Phalloidin staining and L/R morphological asymmetry after Prrx1 silencing (Extended Data Fig. 8c) explains the mesocardia observed. The role of actomyosin is confirmed by the mesocardia obtained after incubation with blebbistatin (Extended Data Fig. 8d). As in the fish, Prrx1 downregulation also decreased Palladin expression, which was coexpressed with Prrx1 in a L/R asymmetric manner in control embryos (Fig. 3g and Extended Data Fig. 8e, f).
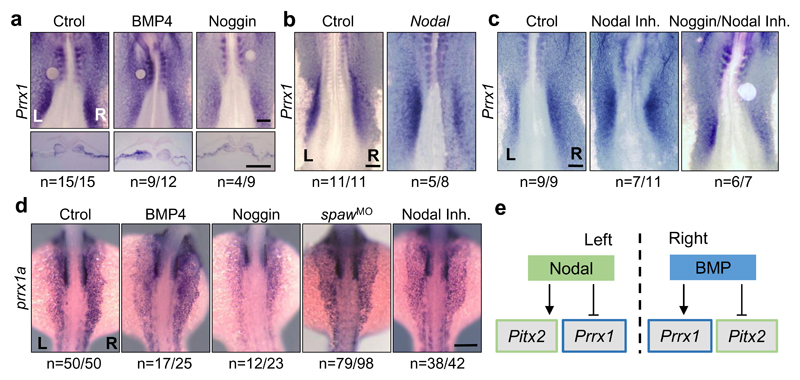
We next investigated the signals that induce Prrx1 expression and how Prrx1 is integrated into the pathways involved in L/R asymmetry. BMP induces Prrx1 and Snail1 expression in the chick embryo10, and the latter represses Pitx2 transcription on the right-hand side1,3,4. Administration of BMP4 on the left or its inhibitor Noggin on the right LPM, respectively induced or repressed Prrx1 expression (Fig. 4a). Ectopic Nodal expression on the right downregulated Prrx1 expression (Fig. 4b and Extended Data Fig. 8g), and incubation with a Nodal inhibitor increased Prrx1 levels (Fig. 4c), as previously shown for Snail130. Noggin can still decrease Prrx1 levels in the presence of Nodal inhibitor (Fig. 4c), suggesting that the BMP pathway controls Prrx1 expression directly. As such, BMP4 beads on the left side reversed both Prrx1 L/R asymmetric expression and heart looping (Fig. 4a and Extended Data Fig. 8h). The Prrx1 gene contains two consensus-binding sites (P and D) for the BMP downstream effectors Smad 1/5/8 (Extended Data Fig. 8i), and Smad5 binds to both (Extended Data Fig. 8j), compatible with its proposed role in heart looping in the mouse31. The activity of Prrx1 promoter increases upon BMP4 administration only when these binding sites are intact (Extended Data Fig. 8k). Thus, BMP signalling directly activates Prrx1 gene transcription.
Figure 4. Two parallel and mutually exclusive pathways integrate left and right information.
(a) Prrx1 expression in chick embryos treated with BSA-(Ctrol), BMP4- or Noggin-soaked beads.(b) Embryos electroporated on the right with a vector containing the Nodal coding sequence. (c) Embryos treated with Nodal inhibitor alone or in combination with a Noggin- soaked bead. (d) prrx1a expression in control zebrafish embryos and after treatment with BMP4 or Noggin, or after Nodal inhibition (inhibitor or spawMO microinjection). (e) The parallel Nodal and BMP pathways during heart looping. Scale bars 250 μm.
Incubation of whole zebrafish embryos with BMP4 or Noggin, respectively enhanced or diminished prrx1a expression, and Nodal inhibition abolished prrx1a L/R asymmetry by de-repressing Prrx1a on the left-hand side of the embryo (Fig. 4d). Hence, BMP-mediated Prrx1 expression is stronger on the right-hand side to drive normal heart looping, as this right pathway is downregulated on the left-hand side by Nodal both in fish and chick embryos. Thus, two parallel and mutually repressed pathways driven by two TGF-β superfamily members (Nodal and BMP), activate paired-like homeobox transcription factors, Pitx2 and Prrx1 respectively, integrating left-right information (Fig. 4e).
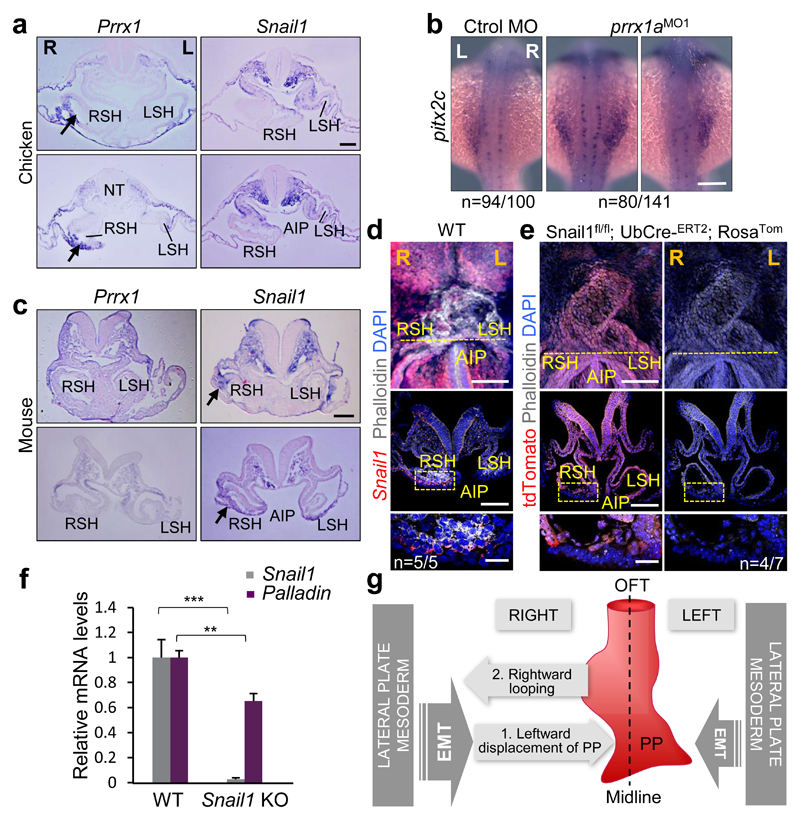
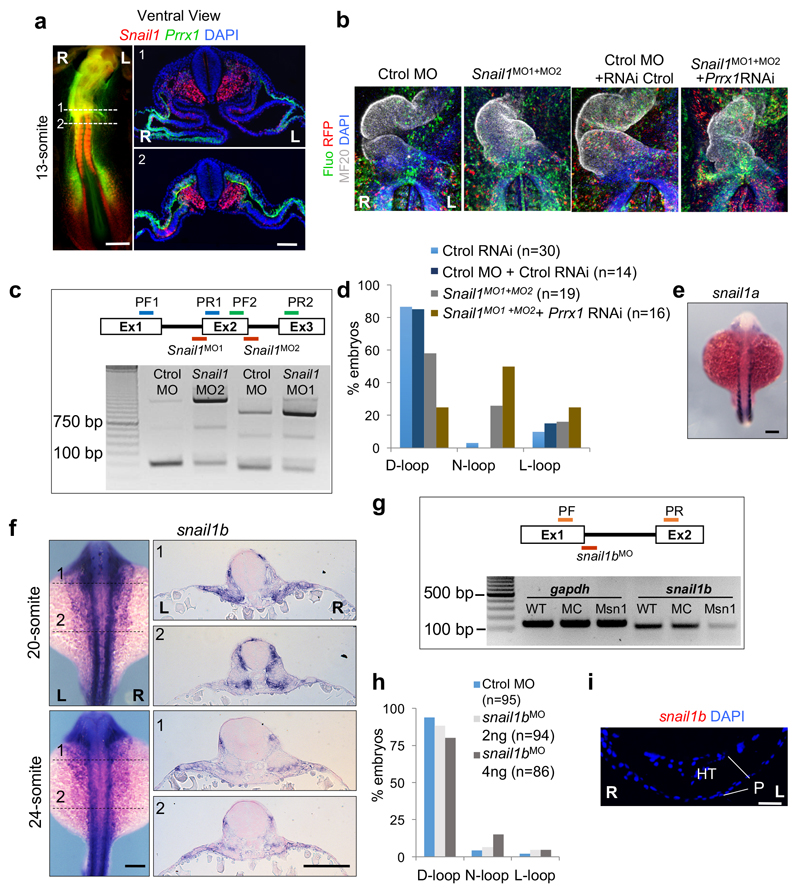
As Snail1 has also been implicated in heart laterality1,3, we examined Prrx1 and Snail1 in the chick and both are expressed in the SH with higher levels on the right, covering mutually exclusive territories in lateral mesoderm (Fig. 5a and Extended Data Figs. 9a and 10a). Snail1 downregulation with Snail1MO reproduced the effects described3, provoking 26,4% mesocardia and 15,7% L-loop (Extended Data Fig. 9 b-d). Simultaneous downregulation of Snail1 and Prrx1 induced mesocardia (50%) with a distribution similar to that found after Prrx1 downregulation (Extended Data Fig. 7f and Fig. 9b-d). Hence, Snail1 contributes to heart positioning in the chick acting independently from Prrx1, which induces mesocardia independently from Snail1 function.
Figure 5. A differential L/R EMT drives heart positioning in vertebrates.
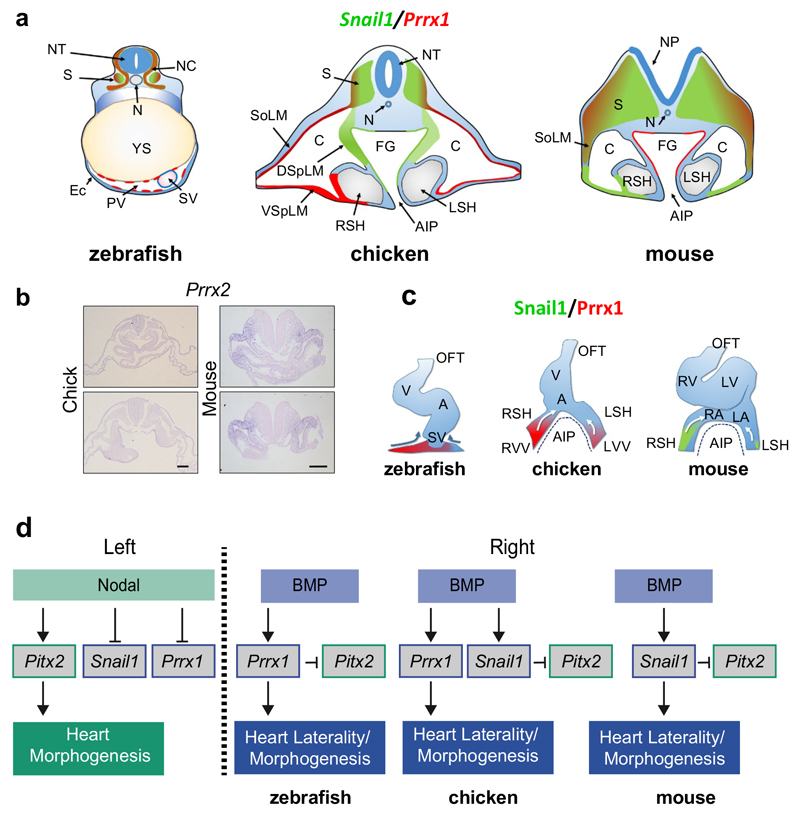
(a) Prrx1 and Snail1 expression in chick embryos (9-10 somite stage) at two levels of the posterior pole of the heart. (b) pitx2c expression in control and prrx1a morphant zebrafish embryos. (c) Similar analysis to that shown in (a) in mouse embryos (8.5 dpc). (d) Whole mount and transverse sections of 8.5 dpc mouse embryos showing Phalloidin staining and Snail1 expression. (e) Snail1 mutant cells visualized as tdTomato+ (red) cells and actin fibers (Phalloidin). (f) Palladin expression upon Snail1 depletion (KO). Data represent mean ± s.e.m. ** p<0.01, *** p<0.001 by Student t-test (n=6 per condition). (g) Proposed model for heart looping in vertebrates. Scale bars 100 µm except for the high powers in (d) and (e) 25 µm.
In the fish embryo, neither snail1a nor snail1b, result of the teleost whole genome duplication32, showed asymmetric L/R expression (Extended Data Figs. 9e, f and 10a), being therefore unlikely that they regulate heart positioning. This was confirmed in morphant embryos (Extended Data Fig. 9g-i). This lack of L/R asymmetry also suggests that, unlike in chick and mouse3,4, snail1 may not repress pitx2c on the right-hand side of the fish. However, prrx1a is expressed in the equivalent territories and we found that pitx2c was derepressed on the right hand side of the prrx1a morphant embryos (Fig. 5b).
Prrx1 or the Prrx1/Prrx2 compound mutant mice do not show any overt heart looping defect33,34, compatible with their absence in the territories relevant to heart looping (Fig. 5c and Extended Data Fig. 10a, b). Interestingly, the asymmetric expression of Prrx1 in the SH in chick embryos is replaced by that of Snail1 in the mouse (arrows in Fig. 5a and c, and Extended Data Fig. 10a), where actin stress fibers concentrate (Fig. 5d), explaining the heart laterality defects in Snail1 mutant mice4. We next depleted Snail1 after early gastrulation stages (see Methods) and found that mutant cells (red) do not accumulate actin fibers (Fig. 5e) and Palladin transcription was downregulated (Fig. 5f), confirming that the cellular and molecular pathway observed downstream of Prrx1 in the fish and the chick, operates in the mouse downstream of Snail1.
We provide here evidence of an asymmetric L/R EMT process that drives differential cell movements towards the midline to induce the leftward displacement of the posterior pole of the heart and subsequent dextral looping in vertebrates. The EMT is triggered by different transcription factors in distinct groups, Prrx1a in the zebrafish, Prrx1 and Snail1 in the chick and Snail1 in the mouse. The interchange in some expression sites of Prrx1 and Snail1 between chick and mouse is reminiscent of that observed for Snail1 and Snail2 in other migratory embryonic territories35, indicating that the use of different EMT inducers for similar processes in different species is not unique to heart laterality, and that an evolutionarily conserved role for EMT in the control of heart positioning operates from fish to mice (Fig. 5g and Extended Data Fig. 10c). In the mouse, Snail1 not only represses left-handed information on the right3,4, but it also instructs the right-handed pathway leading to heart looping. Similarly, Prrx1a fulfills both roles in the zebrafish (Extended Data Fig. 10d). In summary, while a well-described left-hand side Nodal-Pitx2 pathway promotes and stabilizes left identify1, a BMP pathway operates prominently on the right not only to repress the Nodal-Pitx2 pathway, but to provide instructive information that drives heart looping and morphogenesis.
While the Nodal-Pitx2 axis is not conserved in worms and flies, the BMP-Prrx1 pathway activates actomyosin, and myosin acts as a L/R determinant in Drosophila36, suggesting a conserved role in laterality during bilaterian evolution. Whether the unilateral leftward cell movement that breaks bilateral symmetry in the early chick embryo is also driven by a similar mechanism awaits further investigation37. Heart looping is essential for both atrio-ventricular and heart-vasculature concordance7, and hence, for proper organ function. Defining the events underlying this process may shed light on the evolution of the heart from the simple straight invertebrate tube acting as a peristaltic pump to a suction pump in zebrafish and a rhythmically beating structure in amniotes. These data should also help to better understand the CHDs derived from disrupted heart laterality in humans.
Methods
Embryos
Zebrafish strains AB wild-type, Tg(actb2:myl12.1-mCherry) to visualize non-cardiac myosin II38, and Tg(tbx5aBAC:eGFP) (see below) were maintained at 28 ºC under standard conditions, and the embryos were staged as described previously39. Fertilized hen eggs were purchased from Granja Gilbert (Tarragona, Spain) and incubated in a humidified incubator at 37.5 ºC. Chick embryos were staged according to Hamburger and Hamilton40 (HH) or somite number. Normal mouse embryos were obtained from natural matings of C57BL/6J mice. The age of mouse embryos was determined as days postcoitum (dpc), and by somite number. Snai1tm1.1Stjw; Tg(UBC-cre/ERT2)1Ejb; Gt(ROSA)26Sortm14(CAG-tdTomato)Hze [Snail1flox/flox41 Ubiquitin-driven cre/ERT242; tdTomato cre reporter43] embryos were obtained from pregnant females that were gavage fed 250 µl of 30 mg/ml Tamoxifen dissolved in 5% Ethanol in corn oil (Sigma) at 6.5 dpc. Embryos were obtained at 8.5 dpc. This combined genotype allows circumventing the early lethality caused by the loss of function of Snail1 before gastrulation44. UBC-cre/ERT2 provides a temporal control of CRE activity, the presence of Tamoxifen at 6.5 dpc allows CRE activity after gastrulation in all the cells of the embryo. Tamoxifen induced Cre activity was monitored by the presence of tdTomato fluorescence. An additional Snail1 conditional mutant model in which deletion was driven by the Meox-Cre line (Snail1 deleted in embryonic tissues from around 8dpc) has been used previously4. In both conditional models, Snail1 is deleted after early gastrulation and early L/R determination, and coinciding with the time when Snail1 is transiently expressed in a L/R asymmetric manner in the LPM45. Both models show heart laterality defects and our data in provide the cellular and molecular mechanism that explains this phenotype.
All animal studies were performed without randomization to form the experimental groups. Sample size was not predefined for experiments involving embryos. For ChIP experiments 5 biological replicates where done, Luciferase experiments were done on primary fibroblast using 4 biological replicates. No blinding was used in the experimental design; however, results were independently scored by two people. No particular selection of sex of the animals was performed since in all cases the embryos were younger than the sex determination stage. All the experimental animals used here were at embryonic stages.
All animal procedures were conducted in strict compliance with the European Community Council Directive (2010/63/EU) and Spanish legislation. The protocols were approved by the CSIC Ethical Committee and the Animal Welfare Committee at the Institute of Neurosciences.
Generation of the Tg(tbx5aBAC:eGFP) transgenic line
The bacterial artificial chromosome (BAC) containing the zebrafish tbx5a gene (CH73-99A14) was obtained from the BACPAC Resources Center. A GFP-polyA cassette was introduced into the zftbx5a-containing BAC by homologous recombination46. Briefly, a 588 bp 5’ homology arm encompassing part of the zftbx5a first intron and the 5’UTR of the second exon, as well as a 479 bp 3’ arm starting at the ATG of the tbx5a gene present in its second exon, were cloned into the PL451 vector (kindly provided by Dr Neal G. Copeland). The resulting targeting construct was electroporated into EL250 cells containing the CH73-99A14 BAC in order to enable homologous recombination and incorporation of the GFP-polyA cassette into the endogenous tbx5a ATG contained in the BAC. Homologous recombination was confirmed by sequencing the PCR fragments obtained with primers external to the targeting construct. The primer sequences used for both construction of the targeting construct and to confirm homologous recombination are available upon request. The NucleoBond BAC 100 Kit (MACHEREY-NAGEL) was used to obtain BAC DNA, and 1 nl of the 70 ng/μl BAC DNA was injected into the blastodisc of fertilized eggs, allowing the embryos to develop thereafter. On reaching sexual maturity, F0 fish were individually crossed to wild-type fish and EGFP+ F1 embryos were raised to adulthood to create the stable Tg(tbx5a:eGFP) transgenic line used herein. The analysis of the Tg(tbx5a:eGFP) reporter was carried out taking advantage of the native expression of GFP in all the time-lapse experiments. When the reporter was combined with immunofluorescence (i. e. Fig 2g and Extended Data Figs. 3 and 4), eGFP detection was performed with an anti-GFP antibody to ensure a correct detection of the reporter expression.
Morpholinos and mRNA Injections
All antisense morpholino oligonucleotides were obtained from Gene Tools LLC and used as described elsewhere47. To interfere with endogenous expression, one-cell zebrafish embryos were injected with 4 ng of a previously published prrx1aMO1 splice site blocking morpholino or with 10 ng of the 5’ UTR prrx1aMO2 morpholino, (both with proven efficiency and specificity10), with 5 ng of a spaw morpholino18, with 2 or 4 ng of a snail1bMO splice site blocking morpholino. Sequences in Supplementary table II. The dose for snail1bMO was chosen after testing its efficiency in preventing splicing. After injecting different amounts of snail1bMO, RNA was extracted from 48hpf embryos, and RT-PCR was performed with oligonucleotides in exons 1 and 2 (sequences in supplementary table I) that yield a fragment of 100 bp (Extended Data Fig. 9g). The standard control morpholino from Gene Tools was used in control injections. Capped nlsKaede48 and LifeAct-GFP/LifeAct-RFP49 mRNA were synthesized with the SP6 mMessage mMachine kit (Ambion), and 75 and 100 pg was injected into one-cell embryos, respectively.
Electroporation of siRNAs, morpholinos and vectors
Chick embryo explants (stage HH3-4) were placed ventral side up on filter paper rings and electroporated with selected plasmids, siRNAs or morpholinos as described previously50. Unilateral and bilateral electroporations were performed as follows: For unilateral electroporation embryos were injected with plasmid and/or siRNA into the left or right half sides and the positive platinum electrode was placed parallel to the primitive streak onto the samples. To electroporate the whole population of cardiomyocyte precursors (bilateral electroporation), the positive platinum electrode was placed transverse to the primitive streak after the sample injection. Electroporations were performed bilaterally except for the experiments shown in Fig. 3d, e, Fig. 4b, and Extended Data Fig 8g that were unilateral, left (L) or right (R). Gain of function experiments were performed by unilateral electroporation (Fig. 3e). pMT-cNodal51, a generous gift from Claudio Stern (UCL, UK) was used at 4 µg/µl (Fig. 4b). The previously described pCX-Prrx1 vector (1µg/µl)10 or the pMT or pCX empty vectors (for control embryos) were combined with pCX-eGFP plasmid (0.5 µg/µl) to follow electroporation. Loss of function experiments were performed by unilateral or bilateral electroporation as indicated. Two siRNA for Prrx1 were designed (See Supplementary table II for sequences). A mix of these siRNAs (2µM each), plus the pCX-eGFP plasmid (0.5 µg/µl) and 6% sucrose was injected. Control embryos were electroporated with a scrambled siRNA. To interfere with the Snail1 expression two splice site blocking fluorescein labelled morpholinos were designed (Snail1MO1 and Snail1MO2 Sequences in Supplementary table II) and a mix of these two morpholinos (1mM each) electroporated alone or in combination with the two siRNA for Prrx1 and pCX-RFP as a control for electroporation. The standard control fluorescein labelled morpholino from Gene Tools was used as a control. Snail1 MOs efficiency in preventing splicing was tested by RT-PCR as described above. Oligonucleotides in exons 1 and 2 for Snail1MO1 that yield a fragment of 255 bp, and in exon 2 and 3 for Snail1MO2 that yield fragments 288 bp were used. The unspliced fragments are 950 and 1244 respectively.
CRISPR-Cas9
Cas9-NLS-6xHis (pET-28b-Cas9-His; Addgene # 4732752, was expressed in E. coli, purified on a Ni-column and stored at -20 ºC in 300mM NaCl, 10 mM Tris-HCl, 0.1 mM EDTA, 1 mM DTT and 50% Glycerol pH 7.4 at 25 ºC. For sgRNA production, oligos were obtained from Sigma and cloned in Bsa I digested DR274 (Addgene #42250)53. The primer sequences are listed in Supplementary table II. The sgRNA was synthesized using MEGAshortscript™ T7 (Ambion) and purified with the MEGAclear™ kit (Ambion). For RNP formation and injection into one-cell zebrafish embryos, Cas9 (800 ng/µl) was incubated for 5 minutes at 37 ºC with equimolar amounts of in vitro transcribed gRNAs in the presence of 150 mM KCl. RNPs were injected into one-cell embryos. To perform T7 endonuclease test, DNA extracted from 24 hpf embryos was used to amplify a fragment containing the target region of prrx1a. After PCR, the amplicon was denatured and allowed to anneal. Digestion was performed with T7 Endonuclease I (New England Biolabs) according to the manufacturer’s instructions. Heart situs and presence of mutations was determined in 89 embryos. The tail of each embryo was used to individually extract DNA and perform a T7 endonuclease test. The rest of the embryo was fixed and used for in situ hybridization with myl7 to determine heart situs. Ratio of mesocardia in crispant was 54% (n=48/89) that is very similar to that of the prrx1MO morphants 49% (n=80/164) (Extended Data Fig. 1d-f). Analysis of the CRiSPR- induced mutations was performed by amplification of the exon1 region and subsequent cloning and sequencing of the products. We detected mutations in all the embryos, the nature of the mutations was diverse and in all cases consistent with NHEJ repair of the resulting cuts produced around the location of the guides.
Pharmacological treatments
Zebrafish embryos were treated from the 8 to 22 somite-stage with 0.02 mg/ml of human recombinant BMP4 protein (R&D Systems), Noggin (0.03 mg/ml; R&D Systems) or the Nodal inhibitor SB431542 (100 µM; Sigma). Embryos were also treated from 29 to 50 hpf with Blebbistatin (25µM; Sigma), while the control embryos received 0.1% DMSO alone. Before treatments the embryos were dechorionated to improve drug accessibility. Chick embryos were grown in easy culture as described elsewhere54 and exposed to 1 ml of SB431542 (100 µM) from the 1 to 10 somite-stage or to 1ml of Blebbistatin (50 µM) from the 1 to 14 somite stage. After fixation, the embryos were processed for in situ hybridization. BMP4 (0.2 mg/ml) or Noggin (1 mg/ml) was loaded onto heparin acrylic beads by soaking the beads for 3h at room temperature. The beads were then placed into one side of the LPM at stage HH7 and the embryos were harvested 7 h later. Beads soaked in PBS were used as controls.
Whole-Mount In Situ Hybridization
Whole-mount in situ hybridization using digoxigenin-labeled probes was performed on zebrafish, chick and mouse embryos55. Fluorescent in situ hybridizations were performed as described56. Probes for zfprrx1a, zfcrestin zfitbp3, zfsnail1a, zfsnail1b, zfpitx2c, cPrrx111and cPrrx2 were described previously. Templates for zfmyl7, zfamhc, zfvmhc, caMHC, cvMHC, cTbx5, cTbx18, cNkx2.5, cPalld, cPrrx2, mPrrx1 and mPrrx2 were amplified by PCR from their respective cDNAs and subcloned into the pGEMT-easy vector (Promega) or pCDNA3 (Thermo Fisher) with the primers indicated in Supplementary Table II and used to generate probes. Some embryos were embedded in paraffin, gelatin or low melting temperature agarose and sectioned at 7, 40 or 150 μm respectively. For detail about probes see Supplementary Table III.
Anti-Prrx1 antibody generation
To develop the rabbit polyclonal anti-Prrx1 antiserum, the N-terminus of axolotl (Ambystoma mexicanum) Prrx1 (amino acids 1-101) was cloned as a Glutathione S-Transferase (GST) and a Maltose Binding Protein (MBP) fusion protein. The GST-AxPrrx1-N and MBP-AxPrrx1-N fusion proteins were expressed in E. coli (BL21-DE3), and they were purified using Glutathione Sepharose 4B (GE) and amylose resin (New England Biolab), respectively. GST-AxPrrx1-N was injected into the rabbit to generate a polyclonal antiserum, which was affinity purified against MBP-AxPrrx1-N immobilized on a NHS-activated sepharose 4 Fast Flow column (GE). The antiserum was dialyzed and concentrated with Amicon Ultra-15 10K MWCO (Millipore) prior to use.
Immunofluorescence
Whole mount embryos were fixed with 4% PFA overnight at 4 °C. After fixation, the embryos were dehydrated and rehydrated in graduated PBT (PBS + 0.1 % Tween 20):methanol series. Antigen retrieval was performed by maintaining the embryos for 15 min at 70 ºC in 150 mM Tris-HCl buffer (pH 9) and the embryos were permeabilised with cold acetone for 20 min at -20ºC. After washing with PBT, embryos were blocked with 10 % FBS for 3 hours at room temperature and incubated with the primary antibodies overnight at 4 °C. After washing for several hours in PBT, the embryos were incubated with the secondary antibodies overnight at 4 °C. Some embryos were embedded in gelatin or low melting temperature agarose and sectioned at 150 μm. For immunofluorescence on sections, embryos were embedded in paraffin and sectioned at 7 μm. Immunostaining was performed by standard procedures. Antigen retrieval was performed on the paraffin sections by incubation for 20 min at 100 ºC in Sodium Citrate Buffer (10 mM Sodium Citrate, 0.05% Tween 20, pH 6.0). Sections were incubated with the primary antibodies overnight at 4ºC and then incubated with the secondary antibodies for 1 hour at RT. Nuclei were counterstained with DAPI (4',6-Diamidine-2'-phenylindole dihydrochloride) by incubation of the samples in 1 µg/ml DAPI in PBT. Embryos were incubated overnight at 4 ºC and sections for 1 h at room temperature. Samples were washed extensively in PBT before mounting.
For antibody details, see Supplementary Table I and III.
Detection of actin and stress fibers
Detection of stress fibers and actin was performed using either TRITC or FITC conjugated Phalloidin. Fixed embryos were treated with a 1/50 dilution of labelled Phalloidin in PBT at 4 ºC overnight. After extensive washing in PBT, embryos or sections were directly analyzed in the confocal microscope. For life imaging of actin polymerization, GFP or RFP conjugated LifeAct49 was used. Zebrafish one cell embryos were injected with mRNA encoding a GFP or RFP conjugated LifeAct and embryos were followed or fixed at the desired time. LifeAct was detected using an antibody against the conjugated fluorescent protein (GFP or RFP) in all cases in which the samples were previously fixed.
Semithin sections
Zebrafish embryos were fixed overnight at 4 ºC with 4% paraformaldehyde/2.5% glutaraldehyde in 0.1 M phosphate buffer (PB). After washing with PB, the embryos were post-fixed for 1 hour in the dark with 1% osmium tetroxide in PB at room temperature. After washing with PB, the embryos were dehydrated at 4 ºC through an acetone series (30%, 50%, 70%, 90%) followed by two washes in 100% acetone at room temperature. The embryos were embedded in Spurr's resin (EMS) following a progressive series of acetone:Spurr combinations: 1:1 overnight, 1:3 for eight hours, and pure resin overnight. On the next day, the embryos were transferred to fresh pure resin in flat molds, oriented and the blocks were polymerized at 70 ºC for 72 hours. Serial sections (500 nm) were obtained using a Histo diamond knife (Diatome), collecting every fifth section. The sections were placed on a deionized water drop on a glass slide using a steel loop, dried immediately at 90 ºC, stained with 30 μL of toluidine blue for 1 minute at the same temperature, washed with deionized water and dried for storage.
Microscopy and Image analysis
Different laser scanning confocal microscopes were used for image acquisition, including Leica TCS SP2, Leica TCS SP2 MP (Leica Microsystems GmbH) and Olympus FV1200 (Olympus). Whole-mount in situ images were taken using a Leica M125 dissecting scope and sections photographed using a Leica DMR microscope under Nomarski optics and equipped with a Leica DFC 7000T digital camera. In situ and immunofluorescence images were collected on an Olympus FV1200 microscope with a 20x objective (UPLSAPO 0.75NA dry) a 40x objective (UPLFLN 1.30 oil) and a 60x objective (PLAPON 1.40 oil). Different fluorophores were excited with the indicated laser lines as follows: DAPI at 405 nm, GFP, FITC and Alexa Fluor 488 at 488 nm, Cy3, RFP and Alexa Fluor 568 at 559 nm and Alexa Fluor 647 at 635 nm. DAPI was collected at 440-470 nm, GFP, FITC and Alexa Fluor 488 at 500-550 nm, Cy3, RFP and Alexa Fluor 568 at 570-620 nm and Alexa Fluor 647 at 650-700 nm. Time-lapse, Kaede photoconversion and laser ablation images were collected on a Leica TCS SP2 with a 20x objective (HC PL APO 0.7NA dry). Zebrafish actomyosin cable experiments were analysed using a confocal microscope Leica TCS SP8 with HyVolution with a 20x objective (HC PL APO CS2 0.75NA IMM) to improve the lateral resolution down to 140nm and to 300nm in the axial direction. This technique combines special acquisition settings (pinhole size 0.6 AU, hybrid detectors, step size between sections of 0.15 microns) and Huygens deconvolution in four different channels sequentially.
Raw data were analysed using Imaris software (Bitplane AG), to create 3D and 4D reconstructions of different views and for surface rendering. Single optical sectioning was used whenever possible, otherwise maximum intensity projection was used to generate images that contained enough information to be analyzed. Time-lapse movies were generated as Quick Time (.mov) format.
Quantification of the asymmetrical expression of Prrx1a
To identify Prrx1 positive cells in the LPM we took advantage of the Tg(tbx5a:eGFP) zebrafish line and the antibody against Prrx1. 22-somite transgenic embryos were fixed and subjected to double immunofluorescence against Prrx1 and GFP. Images were collected at a dorsal view and the images were analyzed using the Imaris software (Bitplane AG). Each individual cell was identified as single or double positive for the two markers. We developed a mask using ImageJ software (https://imagej.nih.gov/ij/).for all the double positive cells (Prrx1+, GFP+). The resulting nuclei were labelled with a yellow line in Extended Data Fig. 3b. These cells were counted and a heat map of its levels is shown in the Extended Data Fig. 3c. Both the number of cells and the levels of Prrx1 protein are higher on the right hand side.
Time-Lapse imaging
For whole embryo imaging, embryos were dechorionated, anesthetized with 0.02% tricaine (MS222; Sigma-Aldrich) and mounted in 3% methylcellulose on a glass-bottom 35 mm culture dish (MatTek) containing an imprinted pattern of 1% agarose to provide support. Confocal imaging of the Tg(tbx5a:eGFP) embryos was performed using an Spectral Confocal Microscope (Leica TCS SP2), acquiring z-stacks every 10 min (step-size of 5 µm up to 300 µm depth). During imaging, embryos were maintained in an incubation chamber heated to 28.5 °C.
Kaede Photoconversion
For cell tracing experiments embryos were injected with 75 pg of in vitro transcribed mRNA encoding the photoconvertible green-to-red fluorescent protein Kaede into one-cell stage. Embryos at 22 hpf were mounted as above for live imaging. Photoconversion of Kaede49 was performed unilaterally at the defined region of interest (ROI) illuminating with a diode laser 405 nm (Radius 405nm, 50 mW, Coherent Inc. Santa Clara. CA. USA) at 11x optical zoom and 80% laser power during 10 s. Z-stacks of green and red florescence were acquired immediately before and after photoconversion. To avoid further exposure to UV light, embryos were incubated in the dark until 50 hpf when they were imaged again.
Laser ablation
The Tg(tbx5a:eGFP) embryos were mounted at 26 hpf as described above for time-lapse imaging. Laser ablation was performed unilaterally with a two-photon laser (Femtosecond Mai Tai HP Ti: sapphire mode-locked laser system, Spectra Physics, Mountain View, CA, USA) integrated on a spectral confocal microscope (Leica TCS SP2 MP). Irradiation over the selected region of eGFP positive cells was performed using an optical zoom of 12x, laser wavelength of 800 nm and maximum laser power. Before and after laser ablation, z-stacks were taken and maximum projections were created in order to evaluate the damage of the targeted cells. Embryos were incubated at 28 °C and imaged again at 50 hpf to determine the post-ablation phenotype. Embryos were fixed in 4 % PFA and some of them were used to assess heart looping by in situ hybridization with the cardiac myosin light chain marker myl7.
Total RNA Extraction, cDNAs Isolation and RT-qPCR Analysis
Using the Illustra RNAspin Mini isolation kit (GE Healthcare 25-0500-70), total mRNA was extracted from pools of 10 zebrafish embryos at 48 hpf, from pools of 5 chick embryos at HH11 stage or from mouse embryos at stage 8.5 dpc. Reverse transcription of total RNA was performed using Maxima First Strand cDNA Synthesis Kit for RT-qPCR (Thermo Scientific), according to the manufacturer’s guidelines. RT-qPCRs were performed in a Step One Plus machine (Applied Biosystems) using Fast SYBR Green Mastermix (Applied Biosystems). RNA expression levels were calculated using the comparative Ct method normalized to the internal control genes indicated below. The final results were expressed as the relative RNA levels, calculated with the 2-(ΔΔCt) method. Data are represented as the mean±s.e.m. of at least three independent experiments with each point analysed in triplicate. The primers used are listed in Supplementary Table II.
Chromatin immunoprecipitation (ChIP) assay
A pool of approximately 80 chick embryos at stage HH10 were hand dissected and the region between the atrioventricular channel and the 3rd to 4th somite was used for Chromatin purification. Tissue was crosslinked with 1% formaldehyde in PBS for 10 min at room temperature and subsequently quenched with 0.125 M glycine for 15 min. Embryos were washed in cool PBS and resuspended in 300 µl of lysis buffer (1% SDS, 10mM EDTA, 50mM Tris-HCl pH8 and Protease inhibitor cocktail), followed by 15 min incubation on ice. Lysates were sonicated three times for 15 minutes in a Bioruptor (Diagenode) (H, 30 s on / 30 s off on ice). Sonicated lysates were diluted to 5 ml with dilution solution (0.01% SDS, 1% Tx100, 2mM EDTA, 20mM Tris-HCl pH 8, 150 mM NaCl, protease inhibitor cocktail), and used for antibody incubation. Antibodies (listed in the Supplementary table I) were added to the chromatin dilutions and incubated overnight with continuous rotation at 4°C. The chromatin-antibody mixes were added to overnight blocked Dynabeads-Protein A beads (Thermo Fisher) (Blocking solution is 0.05% BSA, 2 mg/ml salmon sperm DNA in dilution solution), and incubated for 4 hours with continuous rotation at 4°C. The beads were washed in four consecutive steps with wash buffers (WB) 1 to 4 using a magnetic concentrator (WB1: 0.1% SDS, 1% Triton X-100, 2mM Tris-HCl pH8 and 150 mM NaCl; WB2: 0.1% SDS, 1% Triton X-100, 2mM Tris-HCl pH8 and 500 mM NaCl; WB3: 1mM EDTA, 10mM Tris-HCl pH8, 1% Igepal (NP40), 1% sodium deoxycholate and 0.25M LiCl; WB4: 10mM Tris-HCl pH8 and 1mM EDTA). 10% Chelex was added to the beads, followed by vortexing and incubating at 95°C for 10 min. Proteinase K treatment (40 µg/ml in WB4) was performed by incubating at 55°C for 30 min with agitation followed by a 95°C incubation for 10 min with agitation. Samples were centrifuged at 13000 rpm for 2 min and the supernatant, which contains DNA, was used for qPCR assays with the primers indicated in Supplementary Table II.
Promoter activity assays
Distal and Proximal Smad containing regions were cloned from chick genomic DNA using the indicated primers. PCR fragments were cloned in the pGL3-Promoter vector (Promega). Mutagenesis was performed using the QuickChange Kit (Agilent) with the primers indicated in Supplementary Table II.
A pool of 5 chick embryos were collected after dissected at stage HH 35 to obtain embryonic fibroblast. Head, internal organs and bones were removed and the rest was minced with sterile scissors and digested with trypsin for 5 minutes at 37 C. Cells were pelleted and resuspended in DMEM supplemented with 5% FCS and 1% of penicillin/streptomycin. Fibroblasts were purified by their differential attachment to plates for 1 hour and posterior washing out of the other cell types. 24 hours later, 4×105 cells/well were seeded and transfected with 20 ng of the pRL-TK control vector plus 100ng of either the Prrx1 promoter fragments cloned in pGL3-promoter (Promega) or empty vector to be used in luciferase assays. Treatment with BMP4 (10ng/ml) was performed 24 h after transfection and lasted for 24 h. Cell lysates were harvested and Firefly and Renilla luciferase luminescence assays were performed using a Dual Luciferase Assay (Promega) as described by the manufacturer.
Statistical analysis
Statistical analysis was performed using the GraphPad Prism software package. Results were expressed as mean s.e.m. (standard error of mean). Differences among groups were tested by using two-tailed pair or unpaired Student’s t-test or one-way ANOVA test, as indicated in figure legends. Differences were considered statistically significant when *p < 0.05, **p < 0.01, ***p < 0.001.
Anatomical nomenclature
We have used herein the term “posterior pole” to refer to the cardiac structures that are located at the caudal end of the developing heart in all the animal models. This term is equivalent to “venous pole” or “inflow tract” used by other authors. In the posterior pole, cells migrating from the posterior secondary heart field incorporate and contribute to part of the atrium and to all the sinus venosus (SV), the most posterior cardiac chamber. This process occurs in fish, chick and mouse. The main difference between the anatomical structures developing at the posterior pole of these models is the lack of sinus venosus horns in zebrafish embryos. In both chick and mouse, the sinus venosus comprises a central common chamber expanded at both sides by lateral horns (RSH and LSH) that are posteriorly continuous with the vitelline veins (VV). The myocardial/smooth muscle transition marks the limit between the sinus horns and the VV. The anterior or arterial pole of the zebrafish heart has been referred as outflow tract (OFT), since this is the usual term in the literature. We have used the term anterior intestinal portal (AIP) for the anterior invagination of the endoderm that gives rise to the foregut. Sinus horns and vitelline veins are successively located at the lateral margins of the AIP. The lateral plate mesoderm splits into somatic and splanchnic lateral mesodermal layers during the formation of the coelomic cavity. The somatic lateral mesoderm (SoLM) is in contact with the ectodermal cell layer. We have used the term dorsal splanchnic lateral mesoderm (DSpLM) to refer the mesodermal layer close to the invaginated foregut endoderm, and ventral splanchic lateral mesoderm (VSpLM) for the mesoderm in contact with the non-invaginated endoderm lateral to the AIP. This mesoderm contributes to the walls of the vitelline veins and the sinus venosus horns. A full list of abbreviations of anatomical structures is described in Supplementary Table IV.
Extended Data
Extended Data Figure 1. prrx1a downregulation induces heart looping defects.
(a) Downregulation of prrx1a transcripts upon prrx1aMO1 microinjection. Data represent mean ± s.e.m. *** p<0.001 by Student´s t-test (n= 3 biological replicates). (b) Microinjection of a prrx1aMO2 morpholino located in the 5´UTR also impairs normal heart looping at 50 hpf. Heart looping assessed by in situ hybridization with a myl7 probe in embryos microinjected with control or prrx1aMO2 morpholino. The frequency of the phenotypes is indicated below the images. (c) Scheme of the prrx1a locus indicating its exon organization. The coding sequence is depicted in blue and the location of the guide RNAs (gRNAs) is indicated. The location of primers used for screening is indicated by the two coloured arrows. (d) Representative images of “crispants” obtained by injection of the indicated gRNAs and Cas9. The frequency of the phenotypes is indicated below the images. (e) Representative T7 endonuclease assays of 4 embryos per phenotype indicated in (d) and two wild type embryos (WT) used as negative controls. (f) T7 Endonuclease assay of 8 crispants and one WT embryo after injection of the indicated gRNAs. (g) The PCR products obtained from embryos 1, 2 and 5 in (f) were cloned and sequenced. Representative sequences of those embryos (embryo number indicated on the right), with the mutations indicated in red and the deletions with hyphens. The location of the gRNAs is indicated in green and the corresponding adjacent PAM in blue. Scale bars, 100 µm
Extended Data Figure 2. Cardiac phenotype of prrx1a morphant embryos.
(a) Bright-field images in whole mount and Hematoxylin/eosin staining in sagittal sections of control and prrx1a morphant embryos (n=5 embryos per condition). (b) Snapshots of surface rendering images of representative control and prrx1a morphant embryos hybridized with the atrial marker amhc. The graph represents the area of amhc expression (µm2; n=7 per condition). Scale bar 100 µm. Data represent mean ± s.e.m. ***p< 0.001. Student´s t-test. (c) Parasagittal semithin sections of control and morphant embryos (48hpf) obtained at the right (R), middle (M) and left (L) levels. The ventricle (V) is on the right and the sinus venosus (SV) is well developed in the control. However, the morphant has the ventricle aligned with the atrium (A), and lacks a SV in the cardiac posterior pole (PP). Outflow tract, OFT. (d) Ventral view of a maximum intensity projection and z-plane sections of confocal images (right) through the OFT showing the expression of the anterior SHF marker latent TGF-β binding protein-3 (ltbp3), in control and prrx1a morphant zebrafish embryos. The cardiomyocytes are visualized by MF20 immunofluorescence. Scale bars 50 µm.
Extended Data Figure 3. Prrx1a expression in the early embryo.
(a) Maximum intensity projection of confocal images showing Prrx1 expression in Tg(tbx5a:eGFP) embryos with DAPI staining of cell nuclei (blue). Prrx1 and tbx5a colocalize in the LPM (left image). The asymmetric L/R expression of Prrx1 in the right LPM is better seen in the single channel view (middle image) and the transverse sections at the level of the white dotted line (right panel). tbx5a is absent from the neural crest (NC) and the posterior LPM (PLPM). (b) Prrx1/tbx5 colocalization (yellow) identifies the Prrx1+ cardiac precursor population once the PHT has formed. Number of double-positive cells on the left and right halves of the LPM. Data represent mean ± s.e.m. ** p<0.01 by Student´s t-test (n=6). (c) Heat map of Prrx1 expression levels in the LPM cells identified in (b). Color code is shown on the right. Intensity was measured in individual cells of the selected regions in the left and right LPM and an average intensity value was obtained per side. The plot shows the average value obtained in the right relative to that in the left LPM. Data represent mean ± s.e.m * p<0.05 by Student´s paired t-test (n=6). (d) Dorsal view of zebrafish embryos to visualize the expression of prrx1a and the pan-neural crest marker crestin. Colocalization of prrx1a and crestin identifies the prrx1a+ neural crest cell population. tbx5 is excluded from the neural crest. NT, neural tube.
Extended Data Fig. 4. Prrx1 and Tbx5 co-localization in developing heart tissues.
(a-c) Ventral views or sagittal sections showing more Prrx1+/tbx5a+ cells in the visceral pericardium (P) in contact with the heart tube (HT) on the right side (arrowheads in high power panels in a and c). Prrx1 is absent from the HT. Boxes in (c) indicate the areas highlighted on the right. (d) Sagittal sections of a 48 hpf zebrafish embryo showing Prrx1 expression compared with the cardiomyocyte marker MF20, or the atrial marker S46. Scale bars, 50 µm. A, atrium; E, eye; PV, pericardial vesicle; SV, sinus venosus; V, ventricle.
Extended Data Figure 5. Cell movements during heart development in the zebrafish embryo.
(a) Snapshots from time lapse analysis of Tg(tbx5:eGFP) control and prrx1 morphant embryos at heart looping stages. (b) Randomization of cardiac jogging in prrx1a morphants. Representative images of control (Ctrol MO) and prrx1a morphant (prrx1aMO1) embryos hybridized with a myosin light-chain 7 (myl7) probe to assess heart tube jogging direction. Images are dorsal views with anterior to the top. Scale bar 100 μm. (c) Quantification of heart-jogging direction. A, atrium; PHT, primary heart tube; SV, sinus venosus; V, ventricle.
Extended Data Figure 6. Photoablation of mesodermal cells in the LPM.
(a, b) Photoablation was performed at 26-27hpf on the left (a) or the right side (b). Ventral views with merged bright field and z-plane confocal images, before and after unilateral photoablation (Pa) of tbx5:eGFP+ cells. The photoablated areas are located in the most anterior region of the LPM adjacent to the limit of the extraembryonic tissue. Higher power images of the photoablated areas in the right columns correspond to the boxed areas. (c) Analysis of heart situs at 50 hpf. Heart visualized after myl7 in situ hybridization in WT and left- or right-photoablated (Pa) embryos. Mesocardia only develops in embryos photoablated on the right-hand side. (d) Cartoon of a ventral view of a zebrafish embryo at 30-32hpf with a boxed area that corresponds to that in the high power confocal images shown in Figure 2f and g. (e) Snapshots of actomyosin fibers, generated by the merged images extracted from a time lapse confocal microscopy in live control (left panels) and prrx1a morphant embryos (right panels) of the Tg(tbx5a:eGFP) reporter line injected with LifeAct (n=7 embryos per condition). (f) Snapshots from time lapse videos of Tg(tbx5a:eGFP) embryos treated with Blebbistatin or with the vehicle alone. Scale bars, 50 μm.
Extended Data Figure 7. Prrx1 expression in chick embryos.
(a) Whole mount in situ hybridization in chick embryos showing the transient asymmetric expression of Snail1 in the LPM. (b) Expression analysis by double fluorescence in situ hybridization of Prrx1 and Nkx2.5 or Tbx5 in whole mount embryos. (c) Transverse sections showing Prrx1 and Nkx2.5 or Tbx5 expression at the levels indicated by dotted lines in (b). Boxes indicate the position of the corresponding higher power images. The arrowhead indicates the center of the posterior pole displaced to the left. (d) Dual fluorescent in situ hybridization with Prrx1 and the atrial (aMHC) or ventricular (vMHC) myosins. High power images of transverse sections as indicated on the left showing the absence of myocardial Prrx1 expression. (e) Downregulation of Prrx1 transcription upon RNAi electroporation. Data represent mean ± s.e.m. *** p<0.001 by Student´s t-test, (n= 3 biological replicates). (f-h) Quantification of heart situs from experiments shown in Fig. 3c-d, respectively. (i) aMHC and vMHC in control embryos and in those electroporated with Prrx1 RNAi. (j) Morphometric analysis of the atrium in chick embryos electroporated with Ctrol RNAi or Prrx1 RNAi. The graph represents the area of aMHC expression (µm2; 7 embryos per condition). Data represent mean ± s.e.m. **p < 0.01, Student t-test. (k) Expression of Fgf8, required for OFT formation, is not altered by Prrx1 downregulation. (l) Prrx1 and Islet1 coexpression in the lateral right sinus venosus horn (RSH) (arrows). Scale bars 100 μm except for (a), 500 μm, and high power images in (l), 10 μm. A, atrium; AIP; anterior intestinal portal; CF, cardiac folds; LPM, lateral plate mesoderm; L or R SH; left or right sinus horns; OFT, outflow tract; SH, sinus venosus horns; V, ventricle.
Extended Data Figure 8. Prrx1 is essential in the posterior pole of the heart.
(a) Double fluorescent in situ hybridization for Tbx18 and Prrx1 in chick embryos. (b) Tbx18 downregulation in embryos electroporated with control or Prrx1 RNAi. (c) Maximum intensity projection of dual fluorescent staining for Prrx1 and Phalloidin (a mycotoxin that stains actin stress fibers). Stars show area of asymmetry and colocalization. (d) Heart looping assessed by MF20 immunofluorescence in embryos treated with vehicle or Blebbistatin. (e) Maximum intensity projection showing the cells coexpressing Palladin and Prrx1. (f) Palladin is decreased upon Prrx1 downregulation. Data represent mean ± s.e.m. of 4 pools (5 embryos each) of control and Prrx1 knockdown embryos. * p<0.05, ** p<0.01 by Student t-test. (g) Visualization of electroporated areas in chick embryos shown in Fig. 4b. Embryos were co-electroporated on the right side with an eGFP reporter construct plus and empty vector or one containing the Nodal coding sequence. (h) Heart situs in embryos containing BSA- or BMP4-loaded beads. (i) Diagram of Prrx1 promoter. Proximal (P) and distal (D) binding sites for Smad1/5/8 and constructs used in (j and k). (j) Chromatin immunoprecipitation with antibodies against Smad5 or IgG (negative control). Data represent mean ± s.e.m. ** p<0.01, *** p<0.001, ns (not significant) by Student t-test (n=5 biological replicates). (k) Luciferase activity of Prrx1 promoter. Both distal (D) and proximal (P) Smad5 binding sites are functional. Fragments containing these sites can activate the promoter in primary chick fibroblasts upon BMP4 treatment. Mutations in those sites prevent the response to BMP4. Data represent mean ± s.e.m. *** p<0.001, ns (not significant) by one way ANOVA (n=4 biological replicates). Dotted lines show the level of the sections. Scale bars 100 μm except for the high power images shown in (a), (b) and (e), which correspond to 10 μm. A, atrium; AIP; anterior intestinal portal; CF, cardiac folds; LPM, lateral plate mesoderm; L or R SH; left or right sinus horns; OFT, outflow tract; SV, sinus venosus; V, ventricle; VV, vitelline veins.
Extended Data Figure 9. Snail and Prrx in heart looping in vertebrate embryos.
(a) Prrx1 and Snail1 expression in whole mount and transverse sections in chick embryos. (b) Maximum intensity projection of confocal images showing heart situs by triple immunofluorescence in embryos electroporated with control fluorescein-labelled morpholinos, with a mix of two fluorescein-morpholinos against Snail1 or a combination of Snail1 morpholinos plus Prrx1 RNAi. An RFP construct was used as a reporter of the electroporated cells. (c) Schematic drawing and knockdown efficiency assay of the Snail1 splice fluo-morpholinos. (d) Quantitative analysis of heart situs. (e) snail1a is not expressed in the LPM in zebrafish embryos. (f) snail1b expression in whole mounted and transverse sections at the levels of the dotted lines. (g) Schematic drawing and knockdown efficiency assay of the snail1b splicing site morpholino (snail1bMO). (h) Quantification of heart situs in control and snail1b morphants. (i) snail1b is not expressed in the pericardium (P) in the zebrafish embryo. Scale Bars 100 μm except for the whole mount in (a) and section shown in (i), which correspond to 500 and 50 μm, respectively.
Extended Data Figure 10. Model for heart looping in vertebrates.
(a) Cartoon depicting the comparative expression of Prrx1 and Snail1 in zebrafish, chick and mouse embryos. (b) Transverse sections showing the lack of Prrx2 expression in the SH of chick or mouse embryos at HH9-10 and 8.5dpc stages, respectively. Scale bar 100 μm. (c) Schematic representation of the proposed model for heart looping in vertebrates in relation to the expression of Prrx1 and Snail1. (d) A prominent right-handed pathway drives differential L/R EMT and heart looping in vertebrates. In addition to the left-specific Nodal-Pitx2 pathway that confers left identity and represses the right-handed pathway, BMP signalling activates EMT in the LPM in a L/R asymmetric manner to drive heart laterality and the repression of leftward information on the right through the repression of Pitx2. This conserved cellular mechanism is implemented through the activation of different EMT inducers in different vertebrates. A, atrium; AIP, anterior intestinal portal; C, coelom; DSpLM, dorsal splanchnic lateral mesoderm; Ec, ectoderm; FG, foregut; LA, left atrium; LSH, left sinus horn; LV, left ventricle; LVV, left vitelline vein; NC, neural crest; N, notochord; NP, neural plate; NT, neural tube; OFT, outflow tract; PV, pericardial vesicle; RA, right atrium; RSH, right sinus horn; RV, right ventricle; RVV, right vitelline vein; SoLM, somatic lateral mesoderm; S, somite; SV, sinus venosus; V, ventricle; VSpLM, ventral splanchnic lateral mesoderm; YS, yolk sac.
Supplementary Material
Acknowledgements
We thank members of Angela Nieto’s laboratory for continuous and helpful discussions, Berta Alsina (Universitat Pompeu Fabra) for the myl12.1-mCherry transgenic line (originally from C.P. Heisenberg´s lab22), Luis Miguel Gutiérrez (IN) for the LifeAct construct, Joana Expósito (IN) for her technical advice with confocal microscopy, Juan Félix López Téllez (BIONAND) for the semithin histological sections and Stuart Ingham (IN) for his help with graphics. This work was supported by the Spanish Ministry of Economy and Competitiveness (BFU2008-01042; BFU2014-53128-R co-financed by the European Regional Development Fund, ERDF), Generalitat Valenciana (PROMETEOII/2013/002) and European Research Council (ERC AdG 322694) to M.A.N, and BFU2014-52299-P, Instituto de Salud Carlos III-TERCEL network (RD12/0019-0022) and Junta de Andalucía (P11-CTS-07564) to R.M.C. M.A.N. acknowledges financial support from the A.E.I., through the ‘Severo Ochoa’ Programme for Centres of Excellence in R&D (SEV-2013-0317) to Instituto de Neurociencias (IN).
Footnotes
Author Contributions
O.H.O. performed the majority of the experiments, analyzed the data and contributed to writing. H.C. performed histological and expression studies. C.M. generated the tbx5a zebrafish transgenic line, P.M. generated the anti-Prrx1 antibody in E.T.´s laboratory, and J.G. designed and performed the CRISPR-mediated prrx1a mutagenesis and the Prrx1 promoter analysis. R.M.-C. carried out the morphological analysis and helped interpret the data. M.A.N. conceived the project, interpreted the data and wrote the manuscript.
The authors have no competing financial interests to declare.
Author Information
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
- 1.Raya A, Izpisúa Belmonte JC. Left-right asymmetry in the vertebrate embryo: from early information to higher-level integration. Nat Rev Genet. 2006;7:283–293. doi: 10.1038/nrg1830. [DOI] [PubMed] [Google Scholar]
- 2.López-Gracia ML, Ros MA. Left-right asymmetry in vertebrate development. Adv Anat Embryol Cell Biol. 2007;188:1–121. [PubMed] [Google Scholar]
- 3.Patel K, Isaac A, Cooke J. Nodal signalling and the roles of the transcription factors SnR and Pitx2 in vertebrate left-right asymmetry. Curr Biol. 1999;9:609–612. doi: 10.1016/s0960-9822(99)80267-x. [DOI] [PubMed] [Google Scholar]
- 4.Murray SA, Gridley T. Snail family genes are required for left-right asymmetry determination, but not neural crest formation, in mice. Proc Natl Acad Sci USA. 2006;103:10300–10304. doi: 10.1073/pnas.0602234103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schlueter J, Brand T. A right-sided pathway involving FGF8/Snai1 controls asymmetric development of the proepicardium in the chick embryo. Proc Natl Acad Sci USA. 2009;106:7485–7490. doi: 10.1073/pnas.0811944106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin AE, et al. Laterality defects in the national birth defects prevention study (1998-2007): birth prevalence and descriptive epidemiology. Am J Med Genet A. 2014;164A:2581–2591. doi: 10.1002/ajmg.a.36695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ramsdell AF. Left-right asymmetry and congenital cardiac defects: getting to the heart of the matter in vertebrate left-right axis determination. Dev Biol. 2005;288:1–20. doi: 10.1016/j.ydbio.2005.07.038. [DOI] [PubMed] [Google Scholar]
- 8.Nieto MA, Huang RY, Jackson RA, Thiery JP. EMT: 2016. Cell. 2016;166:21–45. doi: 10.1016/j.cell.2016.06.028. [DOI] [PubMed] [Google Scholar]
- 9.Acloque H, Adams MS, Fishwick K, Bronner-Fraser M, Nieto MA. Epithelial-mesenchymal transitions: the importance of changing cell state in development and disease. J Clin Invest. 2009;119:1438–1449. doi: 10.1172/JCI38019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ocaña OH, et al. Metastatic colonization requires the repression of the epithelial-mesenchymal transition inducer Prrx1. Cancer Cell. 2012;22:709–724. doi: 10.1016/j.ccr.2012.10.012. [DOI] [PubMed] [Google Scholar]
- 11.Kelly RG, Buckingham ME, Moorman AF. Heart fields and cardiac morphogenesis. Cold Spring Harb Perspect Med. 2014;4:10. doi: 10.1101/cshperspect.a015750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hami D, Grimes AC, Tsai HJ, Kirby ML. Zebrafish cardiac development requires a conserved secondary heart field. Development. 2011;138:2389–2398. doi: 10.1242/dev.061473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tessadori F, et al. Identification and functional characterization of cardiac pacemaker cells in zebrafish. PloS one. 2012;7:e47644. doi: 10.1371/journal.pone.0047644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ahn DG, Kourakis MJ, Rohde LA, Silver LM, Ho RK. T-box gene tbx5 is essential for formation of the pectoral limb bud. Nature. 2002;417:754–758. doi: 10.1038/nature00814. [DOI] [PubMed] [Google Scholar]
- 15.Xie L, et al. Tbx5-hedgehog molecular networks are essential in the second heart field for atrial septation. Dev Cell. 2012;23:280–291. doi: 10.1016/j.devcel.2012.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bakkers J, Verhoeven MC, Abdelilah-Seyfried S. Shaping the zebrafish heart: from left-right axis specification to epithelial tissue morphogenesis. Dev Biol. 2009;330:213–220. doi: 10.1016/j.ydbio.2009.04.011. [DOI] [PubMed] [Google Scholar]
- 17.Veerkamp J, et al. Unilateral dampening of Bmp activity by nodal generates cardiac left-right asymmetry. Dev Cell. 2013;24:660–667. doi: 10.1016/j.devcel.2013.01.026. [DOI] [PubMed] [Google Scholar]
- 18.Noël ES, et al. Nodal-independent and tissue-intrinsic mechanism controls heart-looping chirality. Nat Commun. 2013;4:2754. doi: 10.1038/ncomms3754. [DOI] [PubMed] [Google Scholar]
- 19.Taber LA, Voronov DA, Ramasubramanian A. The role of mechanical forces in the torsional component of cardiac looping. Ann NY Acad Sci. 2010;1188:103–110. doi: 10.1111/j.1749-6632.2009.05089.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Domínguez JN, Meilhac SM, Bland YS, Buckingham ME, Brown NA. Asymmetric fate of the posterior part of the second heart field results in unexpected left/right contributions to both poles of the heart. Circ Res. 2012;111:1323–1335. doi: 10.1161/CIRCRESAHA.112.271247. [DOI] [PubMed] [Google Scholar]
- 21.Bayraktar M, Männer J. Cardiac looping may be driven by compressive loads resulting from unequal growth of the heart and pericardial cavity. Observations on a physical simulation model. Front Physiol. 2014;5:112. doi: 10.3389/fphys.2014.00112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gurung R, et al. Actin polymerization is stimulated by actin cross-linking protein palladin. Biochem J. 2016;473:383–396. doi: 10.1042/BJ20151050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Najm P, El-Sibai M. Palladin regulation of the actin structures needed for cancer invasion. Cell Adh Migr. 2014;8:29–35. doi: 10.4161/cam.28024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Behrndt M, et al. Forces driving epithelial spreading in zebrafish gastrulation. Science. 2012;338:257–260. doi: 10.1126/science.1224143. [DOI] [PubMed] [Google Scholar]
- 25.Naganathan SR, Middelkoop TC, Fürthauer S, Grill SW. Actomyosin-driven left-right asymmetry: from molecular torques to chiral self organization. Curr Opin Cell Biol. 2016;38:24–30. doi: 10.1016/j.ceb.2016.01.004. [DOI] [PubMed] [Google Scholar]
- 26.Itasaki N, Nakamura H, Sumida H, Yasuda M. Actin bundles on the right side in the caudal part of the heart tube play a role in dextro-looping in the embryonic chick heart. Anat- Embryol- (Berl) 1991;183:29–39. doi: 10.1007/BF00185832. [DOI] [PubMed] [Google Scholar]
- 27.Morales AV, et al. Snail genes at the crossroads of symmetric and asymmetric processes in the developing mesoderm. EMBO Rep. 2007;8:104–109. doi: 10.1038/sj.embor.7400867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rana MS, et al. Tbx1 coordinates addition of posterior second heart field progenitor cells to the arterial and venous poles of the heart. Circulation research. 2014;115:790–799. doi: 10.1161/CIRCRESAHA.115.305020. [DOI] [PubMed] [Google Scholar]
- 29.Bressan M, Liu G, Mikawa T. Early mesodermal cues assign avian cardiac pacemaker fate potential in a tertiary heart field. Science. 2013;340:744–748. doi: 10.1126/science.1232877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pagán-Westphal SM, Tabin CJ. The transfer of left-right positional information during chick embryogenesis. Cell. 1998;93:25–35. doi: 10.1016/s0092-8674(00)81143-5. [DOI] [PubMed] [Google Scholar]
- 31.Chang H, Zwijsen A, Vogel H, Huylebroeck D, Matzuk MM. Smad5 is essential for left-right asymmetry in mice. Dev Biol. 2000;219:71–78. doi: 10.1006/dbio.1999.9594. [DOI] [PubMed] [Google Scholar]
- 32.Barrallo-Gimeno A, Nieto MA. Evolutionary history of the Snail/Scratch superfamily. Trends Genet. 2009;25:248–252. doi: 10.1016/j.tig.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 33.Martin JF, Bradley A, Olson EN. The paired-like homeo box gene MHox is required for early events of skeletogenesis in multiple lineages. Genes & Development. 1995;9:1237–1249. doi: 10.1101/gad.9.10.1237. [DOI] [PubMed] [Google Scholar]
- 34.Bergwerff M, et al. Loss of function of the Prx1 and Prx2 homeobox genes alters architecture of the great elastic arteries and ductus arteriosus. Virchows Arch. 2000;436:12–19. doi: 10.1007/pl00008193. [DOI] [PubMed] [Google Scholar]
- 35.Locascio A, Manzanares M, Blanco MJ, Nieto MA. Modularity and reshuffling of Snail and Slug expression during vertebrate evolution. Proc Natl Acad Sci USA. 2002;99:16841–16846. doi: 10.1073/pnas.262525399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Spéder P, Adám G, Noselli S. Type ID unconventional myosin controls left-right asymmetry in Drosophila. Nature. 2006;440:803–807. doi: 10.1038/nature04623. [DOI] [PubMed] [Google Scholar]
- 37.Gros J, Feistel K, Viebahn C, Blum M, Tabin CJ. Cell movements at Hensen's node establish left/right asymmetric gene expression in the chick. Science. 2009;324:941–944. doi: 10.1126/science.1172478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maître JL, Berthoumieux H, Krens SF, Salbreux G, Jülicher F, Paluch E, Heisenberg CP. Adhesion functions in cell sorting by mechanically coupling the cortices of adhering cells. Science. 2012;338:253–6. doi: 10.1126/science.1225399. [DOI] [PubMed] [Google Scholar]
- 39.Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- 40.Hamburger V, Hamilton Hl. A series of normal stages in the development of the chick embryo. J Morphol. 1951;88:49–92. [PubMed] [Google Scholar]
- 41.Rowe RG, Li XY, Hu Y, Saunders TL, Virtanen I, Garcia de Herreros A, Becker KF, Ingvarsen S, Engelholm LH, Bommer GT, Fearon ER, et al. Mesenchymal cells reactivate Snail1 expression to drive three-dimensional invasion programs. J Cell Biol. 2009;184(3):399–408. doi: 10.1083/jcb.200810113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ruzankina Y, Pinzon-Guzman C, Asare A, Ong T, Pontano L, Cotsarelis G, Zediak VP, Velez M, Bhandoola A, Brown EJ. Deletion of the developmentally essential gene ATR in adult mice leads to age-related phenotypes and stem cell loss. Cell Stem Cell. 2007;1(1):113–26. doi: 10.1016/j.stem.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, Lein ES, et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci. 2010;13(1):133–40. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Carver EA, Jiang R, Lan Y, Oram KF, Gridley T. The mouse snail gene encodes a key regulator of the epithelial-mesenchymal transition. Mol Cell Biol. 2001;21:8184–8. doi: 10.1128/MCB.21.23.8184-8188.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sefton M, Sánchez S, Nieto MA. Conserved and divergent roles for members of the Snail family of transcription factors in the chick and mouse embryo. Development. 1998;125(16):3111–21. doi: 10.1242/dev.125.16.3111. [DOI] [PubMed] [Google Scholar]
- 46.Lee EC, Yu D, Martinez de Velasco J, Tessarollo L, Swing DA, Court DL, Jenkins NA, Copeland NG. A highly efficient Escherichia coli-based chromosome engineering system adapted for recombinogenic targeting and subcloning of BAC DNA. Genomics. 2001;73:56–65. doi: 10.1006/geno.2000.6451. [DOI] [PubMed] [Google Scholar]
- 47.Nasevicius A, Ekker SC. The zebrafish as a novel system for functional genomics and therapeutic development applications. Curr Opin Mol Ther. 2001;3:224–228. [PubMed] [Google Scholar]
- 48.Riedl J, Crevenna AH, Kessenbrock K, Yu JH, Neukirchen D, Bista M, Bradke F, Jenne D, Holak TA, Werb Z, Sixt M, et al. Lifeact: a versatile marker to visualize F-actin. Nat Methods. 2008;5(7):605–7. doi: 10.1038/nmeth.1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lombardo VA, Sporbert A, Abdelilah-Seyfried S. Cell tracking using photoconvertible proteins during zebrafish development. J Vis Exp. 2012;67 doi: 10.3791/4350. pii: 4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Acloque H, Ocaña OH, Abad D, Stern CD, Nieto MA. Snail2 and Zeb2 repress P-cadherin to define embryonic territories in the chick embryo. Development. 2017;144:645–652. doi: 10.1242/dev.142562. [DOI] [PubMed] [Google Scholar]
- 51.Bertocchini F, Stern CD. The hypoblast of the chick embryo positions the primitive streak by antagonizing nodal signaling. Dev Cell. 2002;3:735–744. doi: 10.1016/s1534-5807(02)00318-0. [DOI] [PubMed] [Google Scholar]
- 52.Gagnon JA, Valen E, Thyme SB, Huang P, Akhmetova L, Pauli A, Montague TG, Zimmerman S, Richter C, Schier AF. Efficient mutagenesis by Cas9 protein-mediated oligonucleotide insertion and largescale assessment of single-guide RNAs. PLoS One. 2014;9:e98186. doi: 10.1371/journal.pone.0098186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hwang WY, Fu Y, Reyon D, Maeder ML, Tsai SQ, Sander JD, Peterson RT, Yeh JR, Joung JK. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat Biotechnol. 2013;31:227–229. doi: 10.1038/nbt.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chapman SC, Collignon J, Schoenwolf GC, Lumsden A. Improved method for chick whole-embryo culture using a filter paper carrier. Dev Dyn. 2001;220:284–289. doi: 10.1002/1097-0177(20010301)220:3<284::AID-DVDY1102>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 55.Nieto MA, Patel K, Wilkinson DG. In situ hybridization analysis of chick embryos in whole mount and tissue sections. Methods Cell Biol. 1996;51:219–235. doi: 10.1016/s0091-679x(08)60630-5. [DOI] [PubMed] [Google Scholar]
- 56.Acloque H, Wilkinson DG, Nieto MA. In situ hybridization analysis of chick embryos in whole-mount and tissue sections. Methods Cell Biol. 2008;87:169–185. doi: 10.1016/S0091-679X(08)00209-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.