Abstract
We evaluated usability of a previously developed genetically encoded molecular crowding sensor in various biological phenomena. Molecular crowding refers to intracellular regions that are occupied more by proteins and nucleotides than by water molecules and is thought to have a strong effect on protein function. To evaluate intracellular molecular crowding, usually the diffusion coefficient of a probe is used because it is related to mobility of the surrounding molecular crowding agents. Recently, genetically encoded molecular crowding sensors based on Förster resonance energy transfer were reported. In the present study, to evaluate the usability of a genetically encoded molecular crowding sensor, molecular crowding was monitored during several biological events. Changes in molecular crowding during stem cell differentiation, cell division, and focal adhesion development and difference in molecular crowding in filopodia locations were examined. The results show usefulness of the genetically encoded molecular crowding sensor for understanding the biological phenomena relating to molecular crowding.
Keywords: histone, chromatin packing, filopodia, focal adhesion, FRET
Significance.
Cells are densely packed with various macro-molecules with water molecules filling the gaps, a phenomenon known as molecular crowding. There has been an increasing interest for molecular crowding as it was shown to influence many biological reactions within cells. In this paper, molecular crowding was monitored during several biological events using a genetically encoded molecular crowding sensor we previously developed. Changes in molecular crowding during stem cell differentiation, cell division, and focal adhesion development and difference in molecular crowding among various filopodia locations were examined. Our results show the usefulness of the genetically encoded molecular crowding sensor for understanding the biological phenomena relating to molecular crowding.
Cells are densely packed with macromolecules such as proteins, lipids, and nucleic acids [1]. Protein concentration in cells is estimated to be 300–400 mg/mL, however, this high protein concentration is difficult to achieve in in vitro studies [2,3]. These crowded conditions influence many important characteristics of cell physiology, such as reaction rates, protein stability, or assembly/disassembly of supramolecular complex such as polymerization/de-polymerization of actin filament or microtubule. For example, molecularly crowded conditions slow down diffusion of molecules thus decreasing the reaction rate; however, the effect of confinement increases the reaction rate [4,5]. It has been reported that protein folding is more stable under molecularly crowded conditions than under dilute conditions [6,7]. Actin filament polymerization is known to be enhanced under crowded conditions because of the excluded-volume effect [8]. These examples show the importance of molecular crowding in biological phenomena. Furthermore, a recent study revealed that local molecular crowding, for example, at cell adhesion sites or on microtubules, influences behavior of the related proteins [9,10]. Thus, it is important to know the molecular crowding distribution within the cell to correctly interpret biological observations.
Because molecular crowding is directly related to viscosity, molecular crowding in the cell can be briefly estimated by the measurement of the diffusion constant of a probe [11,12]. There are other methods for estimation of molecular crowding in the cell. Silver nanoclusters synthesized in poly(methacrylic acid) are a good candidate for a molecular crowding sensor [13]. Digitally recorded interference microscopy with automatic phase shifting can estimate protein concentration in a live cell [14]. Raman microscopy is also capable of assessing protein concentration in the cell [15]. Recently, genetically encoded Förster resonance energy transfer (FRET)-based molecular crowding sensors were developed, which enable direct measurement of macromolecular crowding in live cells [16,17]. By means of these genetically encoded molecular crowding sensors, a change in molecular crowding during a cell volume change has been evaluated in these reports, but practical applications of the molecular crowding sensor have not been addressed.
To assess the possibility of a genetically encoded crowding sensor, here, we created various fusion proteins with our previously developed genetically encoded molecular crowding sensor GimRET (glycine inserted mutant FRET) [17], and assessed the change in molecular crowding under different conditions or at different locations. Changes in molecular crowding during stem cell differentiation, in the cell nucleus during cell division, during focal adhesion development, and at filopodia locations were evaluated in this study. The results show the ability of GimRET to reveal new biological insights into molecular crowding.
Materials and Methods
DNA constructs
GimRET was constructed as reported previously [17]. To generate transient expression vectors for H2B-GimRET and vinculin-GimRET, cDNAs of histone 2B (H2B) and vinculin were amplified by PCR with sense primers specific for the HindIII and NheI sites and reverse primers specific for the KpnI and NheI sites. The PCR products were ligated into a mammalian expression vector for GimRET between the HindIII and KpnI sites or into the NheI site and then transformed into Escherichia coli DH5α cells. To generate transient expression vectors for myosin-X-GimRET, cDNAs of GimRET were amplified by PCR using sense and reverse primers containing the Nhe I sites. The PCR products were ligated into a mammalian expression vector for myosin-X between the Nhe I/Nhe I sites and then transformed into DH5α. For creation of cells stably expressing the reporters, the GimRET fusion constructs were subcloned into the lentiviral expression vector pCDH-CMV-EF1-Puro (CD510B-1, System Biosciences). Oct3/4 reporter was generated by replacing RFP to mKate2 in Mouse Oct4 reporter plasmid pRedZeroTM –Oct4 Differentiation Reporter Virus (SR10045VA, System Biosciences). Lentiviral infection was carried out according to the manufacturer’s protocol.
Cell culture
Mouse embryonic stem (mES) cells (E14Tg2a) were maintained in high-glucose Dulbecco’s modified Eagle’s medium (DMEM) containing 10% of fetal bovine serum (FBS), 1% of a penicillin/streptomycin solution (Sigma-Aldrich), 1% of GlutaMAX-1 (Gibco), 1% of a solution of nonessential amino acids (Gibco), 1% of a solution of nucleosides (Millipore), 1% of a solution of sodium pyruvate (Sigma-Aldrich), 0.1% of 2-mercaptoethanol (Sigma-Aldrich), and 0.1% of leukemia inhibitory factor (LIF) (Nacalai USA Inc.). The mES cells were cultured in 0.1% gelatin-coated 10-cm dishes (Matsunami), and the medium was refreshed daily. To visualize the pluripotency of mES cells, Oct3/4 reporter was transfected to mES cells, i.e., undifferentiated cells show mKate2 fluorescence. Differentiation of mES cells was induced by removal of LIF from the medium. HeLa cells and COS7 cells were cultured in DMEM (Sigma-Aldrich) supplemented with 10% of FBS (Thermo Fisher Scientific) and 1% of penicillin/streptomycin (Sigma-Aldrich).
Microscopic observation
The microscopy setup consisted of an epifluorescence microscope (Ti-E, Nikon), an objective lens (40× CFI Plan Apo Lambda, 0.95 NA, Nikon), a relay optics box for dual-color imaging (GA03; G-Angstrom, Japan), and an electron multiplier-type CCD camera (EM-CCD, iXon DV887 or DU897; Andor Technology PLC, UK). A slit was placed at the imaging surface of the microscope in the relay optics. A dichroic mirror (FF458-Di02, Semrock) located just outside the slit split the optical pathway after the imaging surface into two pathways for cyan (CFP) and yellow (YFP1G) fluorescence. The two pathways converged on the acceptance surface of the EM-CCD camera side by side. Band-pass filters were set for each pathway (467–499 nm for CFP and 510–560 nm for YFP). Multiphoton fluorescence recovery after photobleaching (MP-FRAP) experiments were conducted as described elsewhere [17]. Image analyses were carried out in the ImageJ software (NIH, USA).
Results and Discussion
Molecular crowding in pluripotent cells
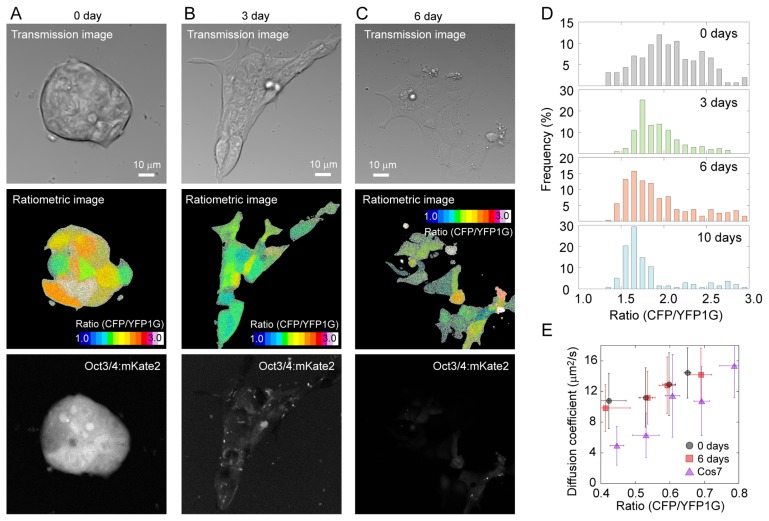
We first transfected GimRET into mES cells to test whether molecular crowding changes during cell differentiation because molecular crowding in the nucleus, e.g., chromatin packing, is thought to influence pluripotency [18]. GimRET-transfected mES cells in the presence of LIF and 3, 6, or 10 days after withdrawal of LIF were visualized using a multiphoton microscope. Intensity ratio of the CFP channel to YFP1G channel in each pixel to visually describe molecular crowding was calculated. Ratiometric images of the mES cells showed a color variation within a colony (Fig. 1A). As mES cells differentiated by withdrawal of LIF, the CFP/YFP1G ratio gradually decreased; however, the color variation was still observed (Fig. 1B & C). The peak value of the histogram of the ratio gradually decreased during the progression of differentiation, up to 10 days after removal of LIF, indicating less molecular crowding in the differentiated state (Fig. 1D). Nonetheless, we detected no differences in the diffusion coefficients between undifferentiated mES cells and differentiated mES cells according to MP-FRAP of GimRET (Fig. 1E), indicating that the mobility of proteins in the mES cells did not change with differentiation despite the lower molecular crowding. These results indicated that molecular crowding changes during cell differentiation, which cannot be detected by means of a diffusion coefficient of a fluorescent protein: a common method for estimation of molecular crowding in the cell.
Figure 1.
Live cell imaging of mES cells expressing GimRET. (A–C) Images of undifferentiated mES cells (A, with LIF), mES cells after 3 days of differentiation (B, 3 days after LIF removal), and mES cells after 6 days of differentiation (C, 6 days after LIF removal). mES cells were transfected with GimRET. Top panels, transmission images; middle panels, ratiometric images; bottom panels, Oct3/4 expression reporting mKate2 fluorescence images. The CFP/YFP1G ratio was calculated by dividing the data from the CFP channel (467–499 nm) by data from the YFP1G channel (510–560 nm) in each pixel. (D) Histogram of the intensity ratio of the GimRET during differentiation. (E) The relation between the ratio and the diffusion coefficient obtained by MP-FRAP of mES cells at 0 (circles) and 6 days (squares) after the removal of LIF, and that of COS7 cells (triangles). N=66, 52, and 41 cells, respectively. Error bars denote standard deviation.
Imaging of cell division with GimRET-fused H2B
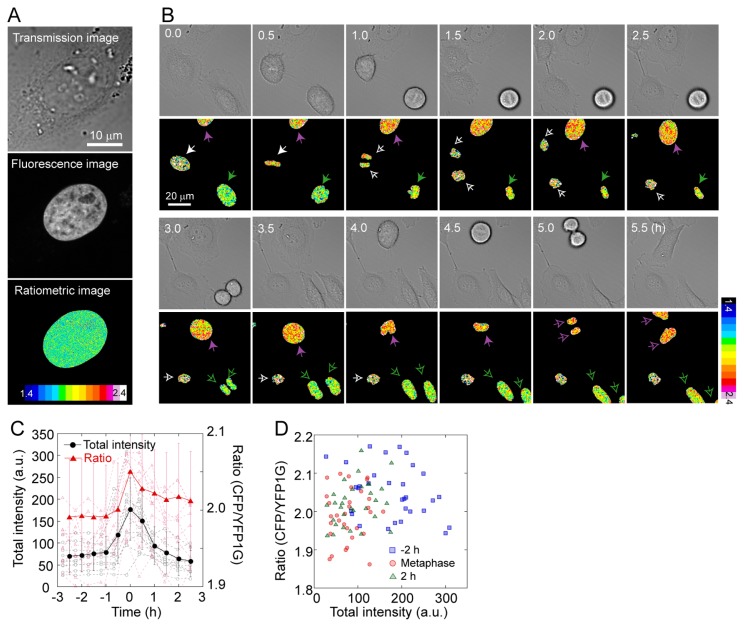
To demonstrate the applicability of GimRET to bioimaging, we fused GimRET to histone 2B (H2B-GimRET), which is one of the five main histone proteins controlling chromatin structure [19]. Figure 2A shows the intensity ratio of the whole cell, where H2B-GimRET was localized to chromatin. The change in the H2B-GimRET intensity ratio was quite different from that of GimRET: the intensity ratio transiently increased in the late G2 phase, which is when the nucleospindle is formed (Fig. 2B & C). To confirm that increase in the ratio is not due to the increase in the total fluorescence intensity, we calculated the relationship between the ratio and the total fluorescence intensity in different cells at different stages of cell division, which showed that the ratio and the total fluorescence intensity does not correlate (Fig. 2D). Because GimRET can sense protein and DNA crowding [17], we hypothesized that the condensation of DNA contributed to the increase in the H2B-GimRET intensity ratio. Rapid increase in the GimRET ratio during mitosis contradicts the finding that the unfused-GimRET ratio only show slow change during whole cell cycle [17], and indicates that when GimRET is fused to a functional protein, it can reveal a change in crowding in the local nanoenvironment. On the other hand, unfused GimRET can report the cytoplasmic environment where proteins can freely diffuse, which imply that observation of molecular crowding in fused and unfused GimRET at the same cellular location is important in the in depth understanding of the molecular environment at the site of interest.
Figure 2.
Live cell imaging with GimRET-fused H2B. (A) Transmission (top), total fluorescence (middle), and ratiometric images (bottom) of HeLa cells transfected with H2B-GimRET. (B) Time-lapse transmission (upper panel) and ratiometric images (lower panel) of HeLa cells transfected with H2B-GimRET. (C) Total fluorescence intensity (black), and the intensity ratio (red) during cell division. Error bars denote standard deviation. (D) The relation between total fluorescence intensity and the ratio 2 h before metaphase (blue), at metaphase (red), and 2 h after metaphase (green).
Molecular crowding at focal adhesion
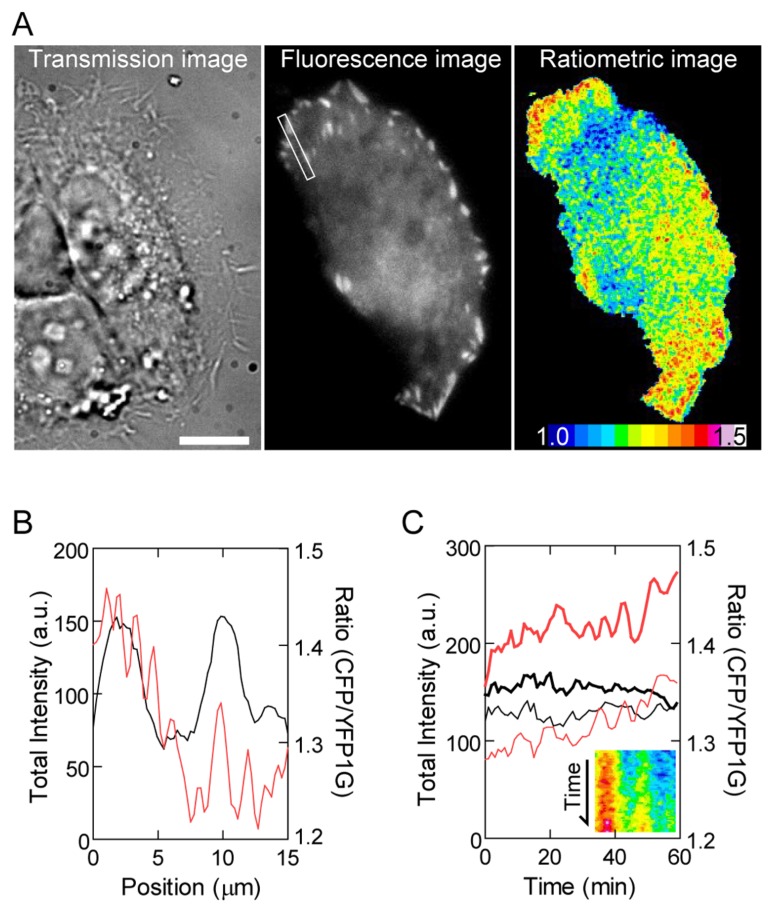
To monitor this molecular crowding, we chose vinculin as a focal adhesion marker because the residence time of vinculin molecules at focal adhesion is much longer than that of other focal adhesion proteins [20,21]. We examined HeLa cells transfected with GimRET-vinculin by Total Internal Reflection Fluorescence (TIRF) microscopy and calculated the CFP/YFP1G ratio (Fig. 3A). Focal adhesion sites in the cell periphery showed a relatively higher CFP/YFP1G ratio than did other cell regions including focal adhesion located inside the cell (Fig. 3A & B), indicating that focal adhesion proteins are densely packed within focal adhesion in the cell periphery. The higher CFP/YFP1G ratio at peripheral focal adhesion sites was stable for 1 h (Fig. 3C). Because focal adhesion at the base of the cell protrusion area tends to disassemble as new focal adhesion forms at the leading edge [22], low molecular crowding within the focal adhesion inside the cell seems to indicate the process of disassembly of focal adhesion. Furthermore, considering that the majority of tyrosine-phosphorylated focal adhesion proteins and catalytically activated Src kinases are located at peripheral focal adhesion sites [23,24] and because protein phosphorylation is key to signal transduction, the above results suggest that high molecular crowding within focal adhesion expectedly facilitates signal transduction at the interface between the extracellular matrix and cell.
Figure 3.
(A) Transmission (left), total fluorescence (center), and ratiometric images (right) of HeLa cells transfected with GimRET-vinculin. Scale bar, 10 μm. (B) Line profiles of total fluorescence intensity (black) and the ratio (red) along the rectangular region indicated in panel A. (C) Total fluorescence intensity (black) and the ratio (red) within focal adhesion in the cell periphery (bold curve) and inside the cell (solid curve) are plotted against time. The inset shows a kymograph of the ratio profile in the rectangular region indicated in panel A.
Molecular crowding in filopodia
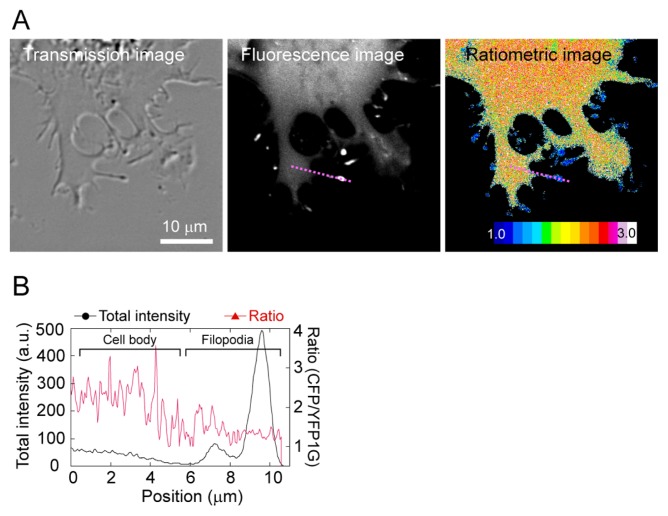
To demonstrate that GimRET can visualize local crowing conditions, we fused it to myosin-X, a vertebrate-specific unconventional myosin motor involved in filopodium production [25]. Filopodium is thin projection of cytoplasm and one may assume that biological characteristic should be similar to the cytoplasm. However, protein composition in filopodium is expected to differ from cytoplasm in cell body, where actin filament bundle and actin binding proteins are abundant [26]. Thus, it is of interest whether molecular crowding differ in filopodium and cytoplasm in cell body. Myosin-X widely distributed in the cytoplasm, with a proportion assembled at the tips of filopodia, and the differences in intensity ratio between the cell body and the filopodia tips were apparent (Fig. 4A). Interestingly, the whole filopodium showed a low degree of crowding (Fig. 4B). Since this difference was not observed in cells expressing GimRET only, the observation of GimRET-myosin-X indicated that the crowding condition around myosin-X was locally different between the cell body and the filopodia. Thus, GimRET-fused proteins enable monitoring of the temporal and spatial distribution of local molecular crowding in cells. While molecular crowding in the cell body was uniform [17], the differences between the filopodia and the cell body were observed by fusion of GimRET to myosin-X (Fig. 4). The combination of GimRET and GimRET-fused proteins allowed observation of differences in global and local intracellular crowding conditions.
Figure 4.
(A) Transmission (left), total fluorescence (middle), and ratiometric images (right) of COS7 cells transfected with myosin-X-GimRET. (B) One-dimensional cross-section of the total intensity (black) and the intensity ratio (red) for the purple dotted line shown in A.
Conclusion
Here, we showed the suitability of the GimRET molecular crowding sensor for elucidation of molecular crowding in live cells. Because GimRET is a genetically encoded probe, it can be easily attached to functional proteins, thereby enabling detection of molecular crowding near this protein. To deeply understand a biological function, it recently requires monitoring physiological and physical conditions at the site where the function occur. Since molecular crowding affects not only the mobility of molecules but also protein stability and kinetics, quantification of molecular crowding using GimRET fused to subcellular localization markers can be an in situ comprehensive parameter of biological functions. Thus, the genetically encoded molecular crowding sensor GimRET should advance the understanding of local nanoenvironments.
Acknowledgments
The authors thank Keiko Yoshizawa for technical assistance.
Footnotes
Conflict of Interests
The authors declare that there is no conflict of interests regarding the publication of this paper.
Author Contribution
H. M. performed imaging experiment, T. J. M. made the constructs, K. O. made cells expressing constructs, T. M. W. performed analysis and H.F. planned the project and wrote the manuscript.
References
- 1.Goodsell DS. The machinery of life. Copernicus Books; New York: 2009. [Google Scholar]
- 2.Fulton AB. How crowded is the cytoplasm? Cell. 1982;30:345–347. doi: 10.1016/0092-8674(82)90231-8. [DOI] [PubMed] [Google Scholar]
- 3.Zimmerman SB, Trach SO. Estimation of macromolecule concentrations and excluded volume effects for the cytoplasm of Escherichia coli. J Mol Biol. 1991;222:599–620. doi: 10.1016/0022-2836(91)90499-v. [DOI] [PubMed] [Google Scholar]
- 4.Ovadi J, Saks V. On the origin of intracellular compartmentation and organized metabolic systems. Mol Cell Biochem. 2004;256–257:5–12. doi: 10.1023/b:mcbi.0000009855.14648.2c. [DOI] [PubMed] [Google Scholar]
- 5.Kim JS, Yethiraj A. Effect of macromolecular crowding on reaction rates: a computational and theoretical study. Biophys J. 2009;96:1333–1340. doi: 10.1016/j.bpj.2008.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van den Berg B, Ellis RJ, Dobson CM. Effects of macromolecular crowding on protein folding and aggregation. EMBO J. 1999;18:6927–6933. doi: 10.1093/emboj/18.24.6927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tokuriki N, Kinjo M, Negi S, Hoshino M, Goto Y, Urabe I, et al. Protein folding by the effects of macromolecular crowding. Protein Sci. 2004;13:125–133. doi: 10.1110/ps.03288104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lindner RA, Ralston GB. Macromolecular crowding: effects on actin polymerisation. Biophys Chem. 1997;66:57–66. doi: 10.1016/s0301-4622(97)00011-2. [DOI] [PubMed] [Google Scholar]
- 9.Zeiger AS, Loe FC, Li R, Raghunath M, Van Vliet KJ. Macromolecular crowding directs extracellular matrix organization and mesenchymal stem cell behavior. PLoS ONE. 2012;7(e37904) doi: 10.1371/journal.pone.0037904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leduc C, Padberg-Gehle K, Varga V, Helbing D, Diez S, Howard J. Molecular crowding creates traffic jams of kinesin motors on microtubules. Proc Natl Acad Sci USA. 2012;109:6100–6105. doi: 10.1073/pnas.1107281109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Muller CB, Eckert T, Loman A, Enderlein J, Richtering W. Dual-focus fluorescence correlation spectroscopy: a robust tool for studying molecular crowding. Soft Matter. 2009;5:1358–1366. [Google Scholar]
- 12.Kalwarczyk T, Ziebacz N, Bielejewska A, Zaboklicka E, Koynov K, Szymanski J, et al. Comparative Analysis of Viscosity of Complex Liquids and Cytoplasm of Mammalian Cells at the Nanoscale. Nano Lett. 2011;11:2157–2163. doi: 10.1021/nl2008218. [DOI] [PubMed] [Google Scholar]
- 13.Wang KH, Chang CW. The spectral relaxation dynamics and the molecular crowding effect of silver nanoclusters synthesized in the polymer scaffold. Phys Chem Chem Phys. 2015;17:23140–23146. doi: 10.1039/c5cp03175c. [DOI] [PubMed] [Google Scholar]
- 14.Zicha D, Genot E, Dunn GA, Kramer IM. TGFbeta1 induces a cell-cycle-dependent increase in motility of epithelial cells. J Cell Sci. 1999;112:447–454. doi: 10.1242/jcs.112.4.447. [DOI] [PubMed] [Google Scholar]
- 15.Uzunbajakava N, Lenferink A, Kraan Y, Willekens B, Vrensen G, Greve J, et al. Nonresonant Raman imaging of protein distribution in single human cells. Biopolymers. 2003;72:1–9. doi: 10.1002/bip.10246. [DOI] [PubMed] [Google Scholar]
- 16.Boersma AJ, Zuhorn IS, Poolman B. A sensor for quantification of macromolecular crowding in living cells. Nat Methods. 2015;12:227–229. doi: 10.1038/nmeth.3257. [DOI] [PubMed] [Google Scholar]
- 17.Morikawa TJ, Fujita H, Kitamura A, Horio T, Yamamoto J, Kinjo M, et al. Dependence of fluorescent protein brightness on protein concentration in solution and enhancement of it. Sci Rep. 2016;6:22342. doi: 10.1038/srep22342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bancaud A, Huet S, Daigle N, Mozziconacci J, Beaudouin J, Ellenberg J. Molecular crowding affects diffusion and binding of nuclear proteins in heterochromatin and reveals the fractal organization of chromatin. EMBO J. 2009;28:3785–3798. doi: 10.1038/emboj.2009.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Campos EI, Reinberg D. Histones: annotating chromatin. Annu Rev Genet. 2009;43:559–599. doi: 10.1146/annurev.genet.032608.103928. [DOI] [PubMed] [Google Scholar]
- 20.Machiyama H, Hirata H, Loh XK, Kanchi MM, Fujita H, Tan SH, et al. Displacement of p130Cas from focal adhesions links actomyosin contraction to cell migration. J Cell Sci. 2014;127:3440–3450. doi: 10.1242/jcs.143438. [DOI] [PubMed] [Google Scholar]
- 21.Pasapera AM, Schneider IC, Rericha E, Schlaepfer DD, Waterman CM. Myosin II activity regulates vinculin recruitment to focal adhesions through FAK-mediated paxillin phosphorylation. J Cell Biol. 2010;188:877–890. doi: 10.1083/jcb.200906012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laukaitis CM, Webb DJ, Donais K, Horwitz AF. Differential dynamics of alpha 5 integrin, paxillin, and alpha-actinin during formation and disassembly of adhesions in migrating cells. J Cell Biol. 2001;153:1427–1440. doi: 10.1083/jcb.153.7.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kwong L, Wozniak MA, Collins AS, Wilson SD, Keely PJ. R-Ras promotes focal adhesion formation through focal adhesion kinase and p130(Cas) by a novel mechanism that differs from integrins. Mol Cell Biol. 2003;23:933–949. doi: 10.1128/MCB.23.3.933-949.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Machiyama H, Yamaguchi T, Sawada Y, Watanabe TM, Fujita H. SH3 domain of c-Src governs its dynamics at focal adhesions and the cell membrane. FEBS J. 2015;282:4034–4055. doi: 10.1111/febs.13404. [DOI] [PubMed] [Google Scholar]
- 25.Berg JS, Derfler BH, Pennisi CM, Corey DP, Cheney RE. Myosin-X, a novel myosin with pleckstrin homology domains, associates with regions of dynamic actin. J Cell Sci. 2000;113:3439–3451. doi: 10.1242/jcs.113.19.3439. [DOI] [PubMed] [Google Scholar]
- 26.Mattila PK, Lappalainen P. Filopodia: molecular architecture and cellular functions. Nat Rev Mol Cell Biol. 2008;9:446–454. doi: 10.1038/nrm2406. [DOI] [PubMed] [Google Scholar]